Abstract
Mastomys are the most widespread African rodent and carriers of various diseases such as the plague or Lassa virus. In addition, mastomys have rapidly gained a large number of mammary glands. Here, we generated a genome, variome, and transcriptomes for Mastomys coucha. As mastomys diverged at similar times from mouse and rat, we demonstrate their utility as a comparative genomic tool for these commonly used animal models. Furthermore, we identified over 500 mastomys accelerated regions, often residing near important mammary developmental genes or within their exons leading to protein sequence changes. Functional characterization of a noncoding mastomys accelerated region, located in the HoxD locus, showed enhancer activity in mouse developing mammary glands. Combined, our results provide genomic resources for mastomys and highlight their potential both as a comparative genomic tool and for the identification of mammary gland number determining factors.
Keywords: Mastomys, Comparative genomics, mammary glands
Mastomys are the most widespread African rodent (Colangelo et al. 2013) (fig. 1B) and are members of the Praomyini tribe, the closest clade to Murini, which includes Mus musculus (Lecompte et al. 2008). Mastomys are of immense research interest as they are carriers of various diseases such as the plague and Lassa virus (Bonwitt et al. 2017), are a good animal model for gastric cancer (Nilsson et al. 1992), Papillomavirus (Helfrich et al. 2004) and Schistosoma (Lurie and De Meillon 1956), and are a model for chromosomal diversity and speciation. They are also notable in that females carry a well-developed prostate gland (Snell and Stewart 1965).
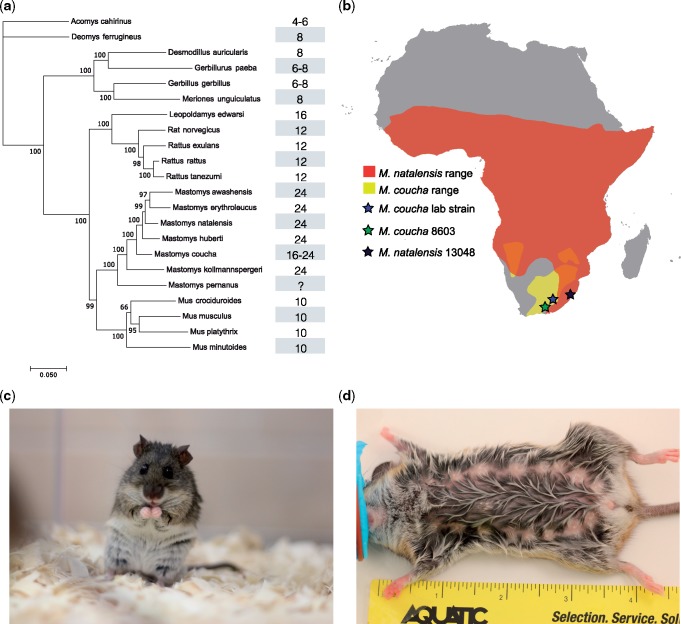
Fig. 1.
Mastomys coucha phylogeny, range, and morphology. (a) Rodent phylogeny showing the number of mammary glands for each species. (b) Mastomys range and sampling locations in Africa obtained from the International Union of Conservation red list of threatened species. The range of M. coucha is marked in yellow and Mastomys natalensis in red. (c) A picture of a M. coucha from our colony. (d) The ventral side of a M. coucha showing 16 mammary glands.
An intriguing trait found in the genus Mastomys is the presence of an unusually high number of mammary glands, anywhere between 16 and 24, which is a rapid gain of 6–14 glands in the 10.2 ± 0.6 My since their divergence with Mu. musculus and 11.3 ± 0.5 from Rattus (Lecompte et al. 2008) (fig. 1A). This is not correlated with their body size, which is on average 90 mm, compared with mouse and rat at 79 and 220 mm, respectively (Hayssen et al. 1993). Mammary glands form during early embryogenesis along the parallel lines on the ventral ectodermal surface. This line, also called the mammary line, is a region where Wnt family member 6 (Wnt6), Wnt10A, and Wnt10B (Chu 2004; Eblaghie et al. 2004; Veltmaat et al. 2004) and other signaling molecules interact to define the ectodermal placodes. Previous work on the five pairs of glands in the lab mouse found that knockouts of various developmental genes (Eda, Edar, Hoxc6, Hoxc8, Fgf10, Gli3, Tbx3, etc.) can lead to changes in the number of mammary glands (Veltmaat 2017). Enhancers that regulate these genes have also been associated with mammary gland development. For example, mammary expression of HoxD9 was found to be regulated by eutherian-specific changes in a nearby enhancer (Schep et al. 2016). Combined, this work suggests that each pair of mammary glands is the result of a distinct combination of signaling pathways. Despite having a partial gene and pathway list that is important in mammary development, the molecular components that determine mammary gland number remain largely unknown.
We established a colony of Mastomys coucha in our lab (fig. 1C). This colony was derived from a colony that was founded in the National Institute of Health and has been inbred since 1983 (Modlin et al. 1988). Analysis of individuals from the colony found that they have eight pairs of mammary glands: three pectoral, three abdominal, and two inguinal with occasional supernumerary mammary glands in the anterior region (fig. 1D). The average litter size we obtained from this colony was 3.2.
To generate a genome for M. coucha, we extracted DNA from the liver of an adult male and generated both short and mate-pair (2-, 5–6-, and 8–10-kb) Illumina sequencing libraries. The M. coucha genome, which we named mc1, was mapped to 26 pseudochromosomes. Mc1 is 2.48 Gb in size with 78× coverage and a contig and scaffold N50 of 29 kb and 3.6 Mb, respectively. The heterozygosity rate is 0.86 per kb, consistent with a highly inbred genome. The mitochondrial genome was assembled separately providing a 16.2-kb genome containing a complete set of mitochondrial genes. The genome was also annotated with transcriptomes from five tissues: adult testis, skin, spleen, embryonic day (E) 17.5 brain, and E11.5 developing mammary glands. These transcriptomes were de novo assembled and the mc1 assembly was assessed for completeness with BUSCO (Simão et al. 2015), finding it to contain 85% of the core vertebrate genes. Using RepeatMasker (Smit et al. 2015), we found that repeats make up 36.4% of the M. coucha genome.
As our colony was inbred, we also sequenced at low coverage (10×) a M. coucha collected from the wild to determine nucleotide variation. We found that the wild samples have a heterozygosity rate of 5.78 per kb, which is over five times higher than the lab strain, adding further support that the mastomys we selected for genome sequencing is inbred. We also sequenced at low-coverage (11×) Mastomys natalensis to determine genus level conservation. We found an even higher rate of heterozygosity, 13.41 per kb, compared with our M. coucha collected in the wild, suggesting that this species has a higher population size, as observed in a previous study (Sands et al. 2015).
As Mastomys diverged 10.2 ± 0.6 My from mouse and 11.3 ± 0.5 from rats (Lecompte et al. 2008) (fig. 1A), it occupies a strategic position in the rodent phylogenetic tree and hence can serve as a robust comparative genomic tool for these commonly used animal models (fig. 2A). Whole-genome alignment found that mc1 contains 90.1% and 85.5% of the mouse and rat genomes, respectively. The average length of synteny between mouse and mastomys is 3.8 Mb, comparable to the 2.5 Mb between mouse and rat (Bourque et al. 2004). We calculated the percent of mouse variation in the whole-genome alignment with all three species (mouse-mastomys-rats) and found that 84% of the mouse-rat divergence could be assigned to a branch of either the rat or the mouse lineage, providing a resource for rodent-specific evolution. Combined, our results show that mastomys provide a useful comparative genomics tool as a sister group to mouse and rat.
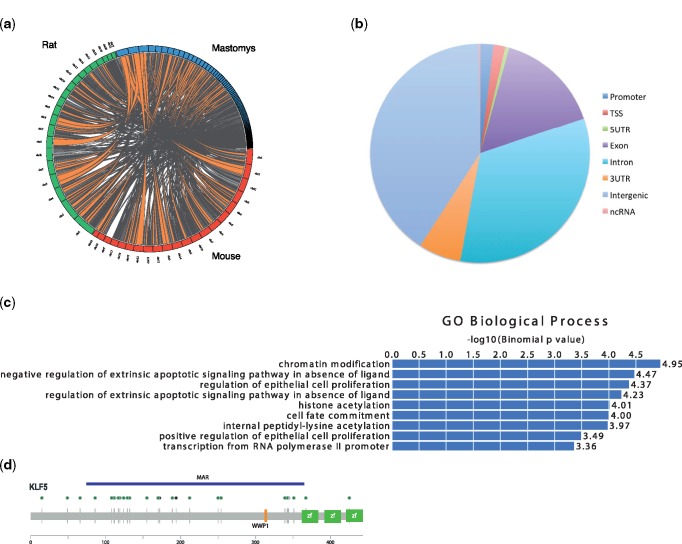
Fig. 2.
Comparative genomics. (a) Synteny comparisons between mastomys-mouse-rat. A circos plot showing chromosomal segments (in orange) that are conserved between mastomys (blue), mouse (red), and rat (green). (b) Pie chart showing the different genomic locations of MARs. (c) GREAT analysis for MARs showing their GO biological process. (d) Diagram showing the location of the coding MAR178 within the Klf5 protein. Green circles mark nonsynonymous protein changes and black circles mark amino acid insertions in mastomys. The orange line represents the ubiquitin binding domain (WWP1) and the green rectangles represent the zinc finger binding domains. The blue line shows the location of MAR178, which overlaps exon 2 of the Klf5 gene.
Accelerated regions represent a good starting point to search for genetic differences leading to lineage-specific traits and have been frequently found to function as enhancers in other species (Capra et al. 2013; Booker et al. 2016). We thus set out to identify sequences that are specifically accelerated in mastomys, as they could pose as potential drivers of mammary gland number increase in mastomys. A seven-way mammalian multiple genome alignment was created using the mastomys, mouse, rat, beaver, human, goat, and cat genomes. We used phastCons (Siepel 2005) to find highly conserved regions in the mammalian alignments (without including mc1). We then used phlyoP (Pollard et al. 2010) to identify accelerated evolution in Mastomys relative to a neutral model of evolution. Using a FDR cutoff of 0.05, we found 515 Mastomys accelerated regions (MARs), the majority (85%) of which are noncoding (fig. 2B). Using the Genomic Regions Enrichment of Annotations Tool (GREAT [McLean et al. 2010]), we found that MARs are adjacent to genes enriched for several gene ontology (GO) biological processes including “chromatin modification” and “regulation of epithelial cell proliferation” (fig. 2C).
We found that over 15% (N = 80) of our MARs were located in exons, with three of them located in exons of genes associated with mammary gland development (Baz2a, Flt4, and Klf5; supplementary table S1, Supplementary Material online). For instance, the MAR found in the Kruppel like factor 5 (Klf5), a gene that is known to promote epithelial growth in breast tissues (Chen et al. 2002), leads to 20 amino acid nonsynonymous changes compared with its mouse homolog (fig. 2D). Although these changes are not within the zinc finger binding domains, they are located in regions that determine protein–protein interactions.
As the majority of MARs are within noncoding sequences (85%), we set out to investigate the possibility of MARs functioning as gene regulatory elements for mammary gland development in Mastomys. To this end, we looked at possible co-occurrence of MARs and expressed genes within topologically associated domains (TADs) previously identified in mouse (Dixon et al. 2012). TADs are conserved genomic regions where DNA sequences interact more frequently with each other than outside and represent functional genomic units. We found 242 MARs residing in TADs containing mammary gland expressed genes which were derived from our RNA-seq data set from the E11.5 developing mammary glands (supplementary table S1, Supplementary Material online). Interestingly, two reside within a TAD with genes that have a known mammary gland phenotype when mutated in mice: sclerostin domain containing 1 (Sostdc1) and integrin subunit alpha 1 (Itga1) (Veltmaat 2017) (supplementary fig. S1, Supplementary Material online). Sostdc1 is of particular interest as it is known to modulate Bmp and Wnt signaling pathways and control the shape and size of mammary buds (Närhi et al. 2012).
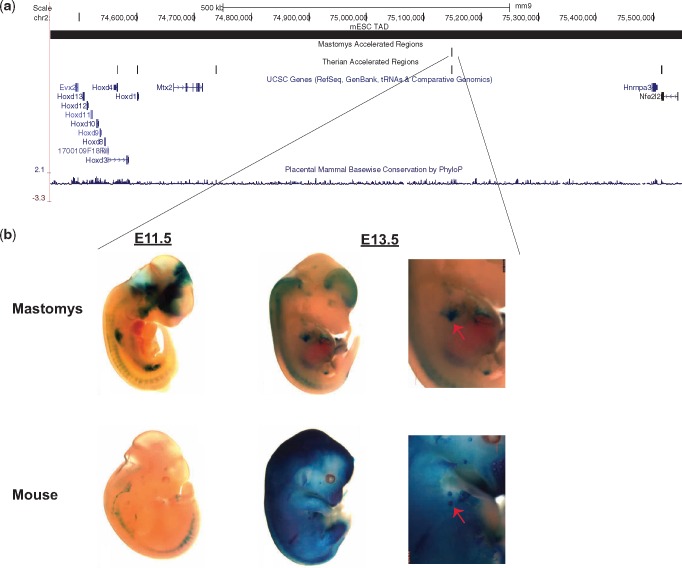
To specifically test whether MARs could function as enhancers within the developing mammary gland, we selected MAR456, which is located nearby the HoxD cluster (fig. 3A), for mouse transgenic enhancer assays. Interestingly, this MAR was also found to be a therian-specific accelerated region, sequences that were found to be accelerated in eutherians and are associated with the evolution of mammalian-specific traits (Holloway et al. 2016). We cloned both the M. coucha and mouse sequence into a mouse enhancer assay vector and tested it for E11.5 and E13.5 enhancer activity in transgenic mice. At E11.5, we observed an overall stronger enhancer expression for the mastomys sequence compared with mouse (fig. 3B and supplementary fig. S2, Supplementary Material online). However, we should note that this assay is not quantitative, as each embryo can have a different transgene copy number. We also noted that several of the embryos having the mastomys sequence showed enhancer activity in the limb and brachial arch, which was not observed in the embryos injected with the mouse sequence. At E13.5, a stage where the mammary glands are visible, we observed staining for both sequences in the developing mammary glands and did not see any inherent expression differences between mastomys and mouse (fig. 3B).
Fig. 3.
MAR456 enhancer expression. (a) UCSC Genome Browser snapshot showing the mouse HoxD genomic locus along with the mastomys and therian accelerated regions tracks. (b) Representative transgenic embryo showing MAR456 enhancer activity for either the mouse or the mastomys sequence at E11.5 and E13.5. Zoom in of the mammary gland expression (red arrow) for the E13.5 embryos are shown in the right panel.
In summary, using various genomic tools, we characterized the multimammate M. coucha. Mastomys are close relatives of mouse and rat and as such pose as a great comparative genomic tool for these commonly used animal models. Our mc1 genome showed extensive alignments and synteny blocks (average length 3.8 Mb) with these two genomes. In addition, it allowed assignment for 84% of the mouse-rat divergence to a branch of either the rat or mouse lineage, highlighting the comparative genomic advantage of this genome as a sister group to mouse and rat. Using low-coverage sequencing of M. coucha and M. natalensis collected from the wild, we provide additional information on nucleotide variation and diversity in these species. The generation of additional mastomys genomes could enhance the use of this species as a strong comparative genomics tool for the rodent clade in general and for the identification of sequences controlling mammary gland number.
Materials and Methods
Genome Assembly
DNA was extracted from the liver of a male M. coucha using the Qiagen AllPrep DNA/RNA Mini Kit. Modified versions of the 4-μg protocol of the Nextera Mate-Pair Sample Preparation kit (Illumina) were used to generate libraries with insert sizes of 2, 5–6, and 8–10 kb. Multiple reactions were pooled before size selection, with five reactions of 4.5 μg grouped for the 5–6- and 8–10-kb size range and five reactions of 2 μg to enrich the 2-kb size range. The smaller-insert libraries, 220 and 350 bp, were generated with the TruSeq DNA PCR-Free Library Preparation kit (Illumina), following the manufacturer’s instructions. DNA was sonicated with the Diagenode Bioruptor Plus, end repaired, A-tailed, and ligated with barcoded adaptors. All libraries were sequenced on the Illumina HiSeq 2500 platform with 2× 125-bp high output mode. The 220- and 350-bp paired reads were trimmed on either side using Trimgalore (Krueger 2015). The mate-pair read pairs were trimmed with Skewer(Jiang et al. 2014). All libraries were error corrected with lighter. Trimmed and corrected short-fragment libraries were used to determine the optimal kmer size of 59 using KmerGenie (Chikhi and Medvedev 2014). We used Meraculous (Chapman et al. 2011) to assemble the genome using the short-fragment libraries for contig creation and the mate-pairs for scaffolding. Ragout (Kolmogorov et al. 2016) was used for pseudochromosome mapping. The mitochondrial genome was assembled separately using the short insert library with NOVOPlasty (Dierckxsens et al. 2016). The assembly was submitted as whole-genome sequencing data under BioProject PRJNA406979. Heterozygosity was estimated with BWA (Li and Durbin 2010) and samtools (Li et al. 2009).
Genome Annotation
The M. coucha genome was annotated using the MAKER2 pipeline (Holt and Yandell 2011). Repeat masking was done using RepeatMasker (Smit et al. 2015) and the RepBase annotation set (Bao et al. 2015) to softmask 36% of the genome. Preliminary ab initio gene models were created using SNAP (Korf 2004), AUGUSTUS (Stanke and Morgenstern 2005), and GeneMark-ES (Besemer and Borodovsky 2005). EST data from transcriptomes were included from mapping embryonic brain and mammary gland, as well as adult testis, skin, and spleen. Reads from these libraries were mapped to the draft genome using HISAT2 (Kim et al. 2015) and gene models were created with StringTie (Pertea et al. 2015). The transcriptomes were also de novo assembled with Trinity (Grabherr et al. 2011) and all models with TMP > 1 were retained. De novo assembled contigs from M. natalensis blood previously published (Andersen et al. 2015) were also used. Mouse evidence from Ensemble 89 cDNA was mapped with BlastN and proteins were mapped using TBlastN (Camacho et al. 2009). Noncoding annotation of tRNAs was performed with tRNAscan (Lowe and Eddy 1997). All de novo and functional evidence was merged in MAKER2 to create gene models. Gene models with annotation edit distance <0.9 were annotated with blast matches to UniProt/Sprot (UniProt Consortium 2017). Ensemble transcript IDs were mapped with reciprocal best blast hits to the MAKER2 transcriptomes. Assessment of completeness was performed against the BUSCO3 eukaryotic gene set (Simão et al. 2015).
RNA Extraction, Sequencing, and Assembly
RNA from the embryonic brain tissues and mammary glands was collected from samples stored in RNAlater (Thermo Fisher). Adult tissues were collected from mastomys and processed immediately. All RNA was extracted using the Qiagen RNeasy micro kit. RNA libraries for the mouse and Mastomys embryonic mammary glands were made using the NuGen V2 RNA library kits and the other Mastomys libraries were created with the NuGen RNA-Seq Strand-Specific kits. For prostate, liver, and adult mammary tissues, RNA libraries were constructed with NEBNext Ultra RNA Library Prep Kit. All libraries were sequenced on a HiSeq 4000 2× 150 bp. De novo transcriptomes were created using each tissue following the Oyster River Protocol best practices (MacManes 2018). Briefly, reads were trimmed using Trimmomatic (Bolger et al. 2014) to remove adapters and no quality trimming was performed. After read correction with Rcorrector (Song and Florea 2015), multiple assemblies were performed using Trinity (Grabherr et al. 2011), Shannon (Kannan et al. 2016), and Spades (Chikhi and Medvedev 2014), and the assemblies were merged using OrthoFuser (MacManes 2018). Contigs were filtered by mapping reads back to the transcriptomes with Kallisto (Pimentel et al. 2016) and all contigs with TMP > 1 were annotated with dammit (Scott 2016).
Comparative Genomics
The mastomys genome was aligned to the Mus. musculus genome (mm10) and Rattus norvegicus (rn6) using Mercator (Dewey 2007). Pairwise genome alignments were made using LAST (Kiełbasa et al. 2011) between M. coucha and human (hg38), cat (felCat8), goat (Bickhart et al. 2017), mouse (mm10), rat (rn6), and beaver (Lok et al. 2017). These genomes were chosen due to their high quality and ability to represent specific clades. The pairwise MAF alignments were chained and netted into a mammalian multiple species alignment using ROAST (Blanchette et al. 2004). A neutral model of evolution was constructed using 4-fold degenerate sites with the Ensembl 89 annotation of mm10. The mammalian alignment was used to find regions of conservation with phastCons (Siepel 2005) ignoring mastomys. phastCons tree input models of neutral evolution and conservation were created using 4D sites mapped onto the alignment from Ensemble gene models and created with phyloFit (Siepel and Haussler 2004). Regions of mammalian conservation were examined in mastomys for accelerated evolution using phyloP and those with significant acceleration (FDR < 0.05) were used (Pollard et al. 2010). Annotation of MARs was performed relative to the mouse using regions with greater than 40% identity to the mouse mm10 assembly using liftOver. HOMER v4.9 (Heinz et al. 2010) was used to obtain genomic annotation classes with RefSeq and GO term enrichment was performed with GREAT 3.0 (McLean et al. 2010).
Population Diversity and History
In addition to the lab strain of M. coucha, low-coverage sequencing was performed on a wild specimen of M. coucha and of M. natalensis. Samples collected in South Africa were provided by the Durban Natural Science Museum and DNA from liver tissue was extracted using DNeasy Blood and tissue kit (Qiagen). Libraries were created by Novogene Co., Ltd using the NEB Next Ultra DNA Library Prep Kit and sequenced to 10× coverage on the Illumina HiSeq X platform with HiSeq X Ten Reagent Kit v2.5 for paired-end reads with 150 bp each (2× 150 bp). After mapping with BWA, single nucleotide polymorphisms (SNPs) were called using the GATK3.6 pipeline (Van der Auwera et al. 2013) following best practices for nonmodel organisms. Briefly, an initial SNP and indel set was called and hard filters were applied to obtain a preliminary variant set. This set was used to recalibrate read quality scores and variants were called again. The second variant set was again filtered with hard cutoffs to obtain a high-quality variant set of SNPs and indels for both M. coucha and M. natalensis.
Mouse Enhancer Assays
MAR456 was amplified using polymerase chain reaction (PCR) on mastomys or mouse genomic DNA using the following primers: mastomys forward, agatccaaatcacatgcagc; mastomys reverse, gctgcatgtgatttggatct; mouse forward, gaagtaggctggcacaagtaga; mouse reverse, acttgttgaaattttgctagttcttg. PCR products were cloned into the Hsp68-LacZ vector and sequence verified. All transgenic mice were generated by Cyagen Biosciences using standard procedures (Nagy et al. 2002) and harvested and stained for LacZ expression at either E11.5 or E13.5 as previously described (Pennacchio et al. 2006). Pictures were obtained using an M165FC stereo microscope.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Irvin Modlin and Mark Kidd (Yale University) for providing M. coucha from their colony, David Safronetz (Rocky Mountain Laboratories) for Mastomys advice, Jeff Wall (UCSF) for assistance with the genome assembly, Leigh Richards and the Durban Natural Science Museum for wild Mastomys samples, Nicola Illing and Mandy Mason (University of Cape Town) for extracting DNA from these samples, and Katie Pollard (Gladstone Institute and UCSF) for advice with comparative genomics. This work was supported in part by the National Human Genome Research Institute (NHGRI) grant number 1R01HG010333 (for N.A. and L.C.). L.C. is supported in part by the NIH Office of the Director, NIH/OD P51 OD011092 and the National Science Foundation (grant 1613856). N.A. is supported in part by the National Human Genome Research Institute 1UM1HG009408, National Institute of Mental Health grant numbers 1R01MH109907 and 1U01MH116438, National Institute of Child & Human Development 1P01HD084387, National Institute of Diabetes and Digestive and Kidney Diseases 1R01DK116738, and National Heart Lung and Blood Institute 1R01HL138424.
Author Contributions
A.H. and N.A. conceived the project and planned experiments. A.H. performed experiments. K.A.N. and L.C. generated genome sequencing libraries. A.H., W.L.E., and N.A. analyzed data and A.H. and N.A. wrote the manuscript.
References
- Andersen KG, Shapiro BJ, Matranga CB, Sealfon R, Lin AE, Moses LM, Folarin OA, Goba A, Odia I, Ehiane PE, et al. 2015. Clinical sequencing uncovers origins and evolution of Lassa virus. Cell 1624:738–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O.. 2015. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, Borodovsky M.. 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33(Web Server):W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickhart DM, Rosen BD, Koren S, Sayre BL, Hastie AR, Chan S, Lee J, Lam ET, Liachko I, Sullivan ST, et al. 2017. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet. 494:643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AFA, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 144:708–715., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 3015:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonwitt J, Sáez AM, Lamin J, Ansumana R, Dawson M, Buanie J, Lamin J, Sondufu D, Borchert M, Sahr F, et al. 2017. At home with Mastomys and Rattus: human–rodent interactions and potential for primary transmission of Lassa virus in domestic spaces. Am J Trop Med Hyg. 964:935–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker BM, Friedrich T, Mason MK, VanderMeer JE, Zhao J, Eckalbar WL, Logan M, Illing N, Pollard KS, Ahituv N.. 2016. Bat accelerated regions identify a bat forelimb specific enhancer in the HoxD locus. PLoS Genet. 123:e1005738.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Pevzner PA, Tesler G.. 2004. Reconstructing the genomic architecture of ancestral mammals: lessons from human, mouse, and rat genomes. Genome Res. 144:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, Erwin GD, McKinsey G, Rubenstein JL, Pollard KS.. 2013. Many human accelerated regions are developmental enhancers. Philos Trans R Soc Lond B Biol Sci. 3681632:20130025.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JA, Ho I, Sunkara S, Luo S, Schroth GP, Rokhsar DS.. 2011. Meraculous: de novo genome assembly with short paired-end reads. PLoS One 68:e23501.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Bhalala HV, Qiao H, Dong J-T.. 2002. A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene 2143:6567.. [DOI] [PubMed] [Google Scholar]
- Chikhi R, Medvedev P.. 2014. Informed and automated k-mer size selection for genome assembly. Bioinformatics 301:31–37. [DOI] [PubMed] [Google Scholar]
- Chu EY. 2004. Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development 13119:4819–4829. [DOI] [PubMed] [Google Scholar]
- Colangelo P, Verheyen E, Leirs H, Tatard C, Denys C, Dobigny G, Duplantier J-M, Brouat C, Granjon L, Lecompte E.. 2013. A mitochondrial phylogeographic scenario for the most widespread African rodent, Mastomys natalensis. Biol J Linn Soc. 1084:901. [Google Scholar]
- Dewey CN. 2007. Aligning multiple whole genomes with Mercator and MAVID. Methods Mol Biol. 395:221–236. [DOI] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 454:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518:331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eblaghie MC, Song S-J, Kim J-Y, Akita K, Tickle C, Jung H-S.. 2004. Interactions between FGF and Wnt signals and Tbx3 gene expression in mammary gland initiation in mouse embryos. J Anat. 2051:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 297:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayssen VD, Van Tienhoven A, Van Tienhoven A.. 1993. Asdell’s patterns of mammalian reproduction: a compendium of species-specific data. Ithaca, New York: Cornell University Press. [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 384:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich I, Chen M, Schmidt R, Fürstenberger G, Kopp-Schneider A, Trick D, Gröne H-J, zur Hausen H, Rösl F.. 2004. Increased incidence of squamous cell carcinomas in Mastomys natalensis papillomavirus E6 transgenic mice during two-stage skin carcinogenesis. J Virol. 789:4797–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway AK, Bruneau BG, Sukonnik T, Rubenstein JL, Pollard KS.. 2016. Accelerated evolution of enhancer hotspots in the mammal ancestor. Mol Biol Evol. 334:1008–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Lei R, Ding S-W, Zhu S.. 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15:182.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S, Hui J, Mazooji K, Pachter L, Tse D.. 2016. Shannon: an information-optimal de novo RNA-Seq assembler. bioRxiv: 039230. doi: https://doi.org/10.1101/039230.
- Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC.. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res. 213:487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 124:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Armstrong J, Raney BJ, Streeter I, Dunn M, Yang F, Odom D, Flicek P, Keane T, Thybert D.. 2016. Chromosome assembly of large and complex genomes using multiple references. Genome Res 28(11):1720–1732. [DOI] [PMC free article] [PubMed]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5:59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F. 2015. Trim Galore. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. Cambridge, UK: Babraham Institute.
- Lecompte E, Aplin K, Denys C, Catzeflis F, Chades M, Chevret P.. 2008. Phylogeny and biogeography of African Murinae based on mitochondrial and nuclear gene sequences, with a new tribal classification of the subfamily. BMC Evol Biol. 81:199.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 265:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup G.. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2516:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok S, Paton TA, Wang Z, Kaur G, Walker S, Yuen RKC, Sung WWL, Whitney J, Buchanan JA, Trost B, et al. 2017. De novo genome and transcriptome assembly of the Canadian beaver (Castor canadensis). G3 (Bethesda) 72:755–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR.. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 255:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie H, De Meillon B. 1956. Experimental bilharziasis in laboratory animals. III. A comparison of the pathogenicity of S. bovis, South African and Egyptian strains of S. mansoni and S. haematobium. S Afr Med J. 30:79–82. [PubMed]
- MacManes MD. 2018. The Oyster River Protocol: a multi assembler and Kmer approach for de novo transcriptome assembly. PeerJ. 6:e5428. [DOI] [PMC free article] [PubMed]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G.. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 285:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlin IM, Zucker KA, Zdon MJ, Sussman J, Adrian TE.. 1988. Characteristics of the spontaneous gastric endocrine tumor of Mastomys. J Surg Res. 443:205–215. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R.. 2002. Manipulating the mouse embryo: a laboratory manual. New York: Cold Spring Harbor. [Google Scholar]
- Närhi K, Tummers M, Ahtiainen L, Itoh N, Thesleff I, Mikkola ML.. 2012. Sostdc1 defines the size and number of skin appendage placodes. Dev Biol. 3642:149–161. [DOI] [PubMed] [Google Scholar]
- Nilsson O, Wängberg B, Johansson L, Modlin I, Ahlman H.. 1992. Praomys (Mastomys) natalensis: a model for gastric carcinoid formation. Yale J Biol Med. 656:741.. [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 4447118:499–502. [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL.. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 333:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel HJ, Bray N, Puente S, Melsted P, Pachter L.. 2017. Differential analysis of RNA-Seq incorporating quantification uncertainty. Nature Methods, 14(7):687–690. [DOI] [PubMed]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A.. 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 201:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands AF, Matthee S, Mfune JK, Matthee CA.. 2015. The influence of life history and climate driven diversification on the mtDNA phylogeographic structures of two southern African Mastomys species (Rodentia: Muridae: Murinae). Biol J Linn Soc. 1141:58–68. [Google Scholar]
- Schep R, Necsulea A, Rodríguez-Carballo E, Guerreiro I, Andrey G, Nguyen Huynh TH, Marcet V, Zákány J, Duboule D, Beccari L.. 2016. Control of Hoxd gene transcription in the mammary bud by hijacking a preexisting regulatory landscape. Proc Natl Acad Sci U S A. 11348:E7720–E7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C. 2016. dammit: an open and accessible de novo transcriptome annotator. Davis, CA: University of California Davis.
- Siepel A. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 158:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Haussler D.. 2004. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol Biol Evol. 213:468–488. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 3119:3210–3212. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P.. 2015. RepeatMasker Open-4.0. 2013–2015. Available from: http://repeatmasker.org.
- Snell KC, Stewart HL.. 1965. Adenocarcinoma and proliferative hyperplasia of the prostate gland in female Rattus (Mastomys) natalensis. J Natl Cancer Inst. 351:7–14. [DOI] [PubMed] [Google Scholar]
- Song L, Florea L.. 2015. Rcorrector: efficient and accurate error correction for Illumina RNA-seq reads. GigaScience 4:48.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B.. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33(Web Server issue):W465–W467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy‐Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J.. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltmaat JM. 2017. Prenatal mammary gland development in the mouse: research models and techniques for its study from past to present. Methods Mol Biol. 1501:21–76. [DOI] [PubMed] [Google Scholar]
- Veltmaat JM, Van Veelen W, Thiery JP, Bellusci S.. 2004. Identification of the mammary line in mouse by Wnt10b expression. Dev Dyn. 2292:349–356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.