Abstract
For disordered proteins, ligand binding can be a critical event that changes their structural dynamics. The ability to characterize such changes would facilitate the development of drugs designed to stabilize disordered proteins, whose mis-folding is important for a number of pathologies, including neurodegenerative diseases such as Parkinson’s and Alzheimer’s diseases. In this study, we used hydrogen/deuterium exchange, differential crosslinking, differential surface modification, and molecular dynamics (MD) simulations to characterize the structural changes in disordered proteins that result from ligand binding. We show here that both an ATP-independent protein chaperone, Spy L32P, and the FK506 binding domain of a prolyl isomerase, FKBP-25 F145A/I223P, are disordered, yet exhibit structures that are distinct from chemically denatured unfolded states in solution, and that they undergo transitions to a more structured state upon ligand binding. These systems may serve as models for the characterization of ligand-induced disorder-to-order transitions in proteins using structural proteomics approaches.
Keywords: Structural proteomics, mass spectrometry, discrete molecular dynamics simulations, protein-ligand interaction, hydrogen/deuterium exchange, crosslinking/mass spectrometry, surface modification, conformational change, quantitative protein structure, intrinsically disordered protein, protein folding
Introduction
Disordered proteins have been implicated in the pathogenesis of a variety of “protein misfolding” diseases [1]. Critical conformational changes may cause mis-folding events which result in the formation of cell-toxic protein aggregates. An attractive therapeutic concept for combating these diseases is to stabilize the native conformation of the proteins with small-molecule ligands and to shift the folding equilibrium toward the native monomeric state or to stabilize other soluble forms of the protein to prevent the accumulation of aggregation-prone conformations [2–5].
In a search for model systems to represent this type of scenario, we have returned to the study of the interaction of the bacterial chaperone Spy with its peptide ligand Im7 [6]. These proteins undergo an increase in secondary-structure content upon binding, which can be evidenced by circular dichroism [6, 7] and by an increase in protection of the peptide-bond amide protons in hydrogen/deuterium exchange experiments [6, 7]. Certain Spy mutants, particularly Spy L32P, have exhibited in-solution deuteration levels that are considerably higher than that of the wild type, as well as a higher shift in the amide protection upon client binding [6]. This might be evidence of a nearly complete loss of secondary structure in the mutant protein and a dramatic restructuring of the protein upon peptide ligand binding. In looking for small molecules which can induce order in the unfolded protein, we came across a “destabilization domain” system: Banaszynski et al. have shown that proteins fused with certain mutants of the FKBP domain -- F36V/L106P in particular -- are effectively degraded in cells when expressed in vivo, but can be reversibly protected from degradation by the presence of its small-molecule ligand, rapamycin [8]. Although, the system was not structurally characterized, the phenomenon can be explained by structurization of the protein upon ligand binding [9].
In our current work, we have studied both the Spy and the FKBP protein-ligand systems by structural proteomics and molecular-dynamics simulations. We have used hydrogen/deuterium exchange, differential crosslinking, and surface modification to characterize the conformational changes that occur upon both peptide binding (Im7 with Spy) and small molecule binding (rapamycin with FKBP) to the protein. We have shown that, in both cases, the proteins are considerably disordered but their structures are different from the unfolded structure obtained with 8M urea in solution, and both proteins undergo a dramatic increase in secondary structure content upon ligand binding. These systems, therefore, can serve as models for ligand-induced protein disorder-to-order transitions, and the structural proteomic techniques demonstrated here are shown to be effective tools for the characterization of the disorder-to-order transitions of these systems.
Materials and methods.
Proteins.
The N-terminally His-tagged FKBP domains (Uniprot Q00688, residues 107–224, residues 110–224 correspond to the 1–107 residues of the homologous domain of FKBP-12) of FKBP-25 wt and FKBP-25 F145A I223P mutant (mutation sites F145 I223 correspond to F36 L106 residues in FKBP-12) were expressed in Escherichia coli BL21 DE3 using pET-15 plasmid (Novagen) and were purified using Ni-NTA beads (Qiagen) as described previously [10]. Spy L32P protein and Im7–45 peptide were gifts from James Bardwell and were prepared as described previously [6].
Hydrogen-deuterium exchange.
FKBP-25 F145A I223P +/− Rapamycin.
Samples were dialyzed into 10 or 40 mM ammonium acetate and were incubated at a 1:1 ratio of protein:rapamycin for 1 hour in order to occupy the available binding sites. Samples were mixed with D2O online and were given 2.5 s to exchange, before mixing at a 1:1 ratio with a quenching buffer containing 0.4% formic acid (FA) and 20% acetonitrile (ACN), and being directly infused into a Bruker 12 T Apex-Qe hybrid Fourier transform mass spectrometer (Bruker Daltonics), equipped with an Apollo II electrospray source. Deuteration values were determined using HDX Match [11] by fitting the experimental isotopic envelopes of the intact protein to the theoretically predicted ones for 80% D2O content in solution.
Spy L32P+/−Im7.
Spy L32P and Im7 protein stock solutions (7.7 mM and 779 μM, respectively) in 40 mM HEPES, 150 mM NaCl, pH 7.5 were diluted with 10 mM ammonium acetate to 50 μM final Spy L32P and 100 μM Im7. Samples were prepared one day prior to HDX analysis and incubated overnight at 4°C to allow Spy L32P-Im7 complex formation equilibrium. HDX analysis was performed in the same way as described above for the FKBP-25 F145A I223P experiment.
Differential crosslinking.
FKBP-25 F145A I223P +/− Rapamycin.
FKBP-25 F145A I223P at a concentration of 13.3 μM in PBS, pH 7.4, 0.5 mM TCEP, with or without a molar excess of rapamycin at a concentration of 150 μM were crosslinked using a 0.25-mM final concentration of either the light or the heavy form of the 13C-isotopically-coded disuccinimidyladipate (DSA) crosslinker (Creative Molecules, Inc.). Crosslinking reagents were incubated for 15 minutes at 25°C and the reaction was quenched with 10 mM ammonium bicarbonate. A portion of each crosslinking reaction was checked by SDS-PAGE gel to determine the extent of intermolecular crosslinked products and to confirm the equivalent crosslinking efficiencies of the light and heavy forms of the crosslinking reagent. Samples were subsequently combined according to the scheme shown in supplementary table S2, S3 and digested with trypsin at a protein:enzyme ratio of 20:1 for 18 hours at 37°C. Samples were then desalted and acidified with FA for the analysis by mass spectrometry as described below.
Spy L32P +/− Im7 +/− Urea.
SpyL32P at concentration of 50 μM in 40 mM HEPES, 100 mM NaCl, pH 7.5 with or without a molar excess of Im7 ligand at a concentration of 100 μM and with or without 8M urea were crosslinked with a 0.1-mM final concentration of either the light or the heavy form of the 13C-isotopically-coded DSA crosslinker (Creative Molecules, Inc.). After crosslinking, the samples were treated in the same way as described above for the FKBP-25 F145A I223P experiment.
Crosslinking data were searched against a concatenated target-decoy (reverse) database using Kojak (v.1.5.1) [12]. The parameter files are included in the supplementary material. Inter-, intra-, and loop crosslink peptide spectral matches (PSMs) were validated with Percolator (v.3.0) [13]. Quantitation was performed on target PSMs from the Percolator output using XiQ (v.0.2) [14]. Only PSMs that met the following criteria were considered for further analysis: PPM Error = +/−2.5; label_count = 1; q-value = 0.05; forward-reverse fold-change delta ≤ 1. The difference in crosslinking yield between experimental conditions (+/−ligand or +/−urea) was expressed as “fold-change”. The fold-change for forward-label experiments (where the light isotopic form of the crosslinking reagent is used in the ligand(−) condition and heavy isotopic form of the crosslinking reagent used in the ligand(+) condition) was calculated as the binary logarithm of the observed crosslink H/L ratio (log2(H/L)) as reported by XiQ. For the reverse-label experiments (light and heavy isotopic forms of the reagents used in the reverse conditions: ligand(+) and ligand(−) respectively), this was calculated as the binary logarithm of the inverse of the observed crosslink H/L ratio (log2(1/(H/L)).
In order to compensate for systematic errors introduced by slight differences in the mixing ratios or reaction efficiencies of the light and heavy isotopic forms of the crosslinking reagent, the fold-changes were normalized to the median of all observed fold-changes between the forward- and reverse-label experiments. The fold-change for a particular crosslinked pair of residues was then calculated as the median of all its crosslinked peptides. Sample standard deviation for a crosslinked pair was calculated using all its crosslinked peptides.
The fold-change threshold used to classify an observed crosslinked pair of residues as “enriched” in a particular condition, or as exhibiting no change between conditions, was defined as the average sample standard deviation of the within-condition control experiments (e.g., the same ligand(−) or ligand(+) condition is prepared twice -- once with the light isotopic reagent and once with the heavy isotopic reagent -- and the labeled samples are subsequently combined as described above) (Supplementary Table S1). All quantified crosslinks included are in supplementary Microsoft Excel workbooks.
Differential surface modification.
FKBP-25 F145A I223P +/− Rapamycin.
Chemical surface modification with pyridine carboxylic acid N-hydroxysuccinimide ester (PCAS) was performed as previously described [15]. Briefly, proteins were prepared at a concentration of 50 μM in PBS, pH 7.4 (defined as the disordered state), or with a molar excess of ligand in PBS (folded state). Either the light or the heavy form of the isotopically-coded reagent (PCAS-H4 or PCAS-D4 (Creative Molecules)) was then added to give a concentration of 10 mM. Reaction mixtures were incubated for 30 minutes and then quenched with 50 mM ammonium bicarbonate. Samples were then mixed at a 1:1 ratio, combining folded (PCAS-H4) with unfolded (PCAS-D4) samples, as well as in reverse as a control. Samples were acidified with 150 mM acetic acid and then digested with pepsin at a 20:1 protein:enzyme ratio overnight at 37°C. After digestion, samples were prepared for mass spectrometry analysis using C18 zip-tips (Millipore) and eluted with 50% ACN/0.1% aqueous trifluoroacetic acid (TFA). Samples were analyzed by LC-MS/MS as described below.
SpyL32P+/−Im7 +/− Urea.
SpyL32P at concentration of 50 μM in 40 mM HEPES, 100 mM NaCl, pH 7.5 with or without a molar excess of Im7 ligand at a concentration of 100 μM, and with or without 8 M urea, were prepared in the same way as described above for the FKBP-25 F145A I223P experiment.
Surface modification data were searched using PEAKS 7 [16]. Quantitation results for the PCAS-modified residues were exported in .csv format, and further analysis was performed using Microsoft Excel. The difference in surface modification yield was calculated in the same manner as described above for qCL. All quantified PCAS-modified peptides have been included in a supplementary Microsoft Excel workbooks.
Molecular visualization.
To facilitate visualization of the crosslinking and surface modification results on the molecular level, the crystal structures of the wild-type Spy protein (PDB ID:3O39), and wild-type FKBP25 (1PBK) were supplemented with I-TASSER models to represent any residues not present in the crystallographic model. Specifically, residues including an N-terminal serine (which was introduced from the Sumo fusion constructs that were used for Spy L32P purification), A1–M28, and E125-E138 of Spy (PDB ID:3O39) and residues including an N-terminal His-tag (MRGSHHHHHHGLVPRGSM) and G107-P108 of FKBP25 (PDB ID:1PBK) were added. Quantified crosslinked residue pairs and surface modified residues were visualized on the supplemented models using PyMOL (v.1.3) [17]. The PyMOL session files used for the preparation of the figures have been included in the supplementary material.
LC-MS/MS analysis.
Mass spectrometric analysis was performed using a nano-HPLC system (Easy-nLC II, ThermoFisher Scientific), coupled to the ESI-source of an LTQ Orbitrap Velos (ThermoFisher Scientific), using conditions described in [18, 19]. Briefly, samples were injected onto a 100 μm ID, 360 μm OD trap column packed with Magic C18AQ (Bruker-Michrom, Auburn, CA), 100 Å, 5 μm pore size (prepared in-house) and desalted by washing with Solvent A (2% acetonitrile:98% water, both containing 0.1% FA. Peptides were separated with a 60-min gradient (0–60 min: 4–40% solvent B (90% ACN, 10% water, 0.1% FA), 60–62 min: 40–80% B, 62–70 min: 80% B), on a 75 μm ID, 360 μm OD analytical column packed with Magic C18AQ 100 Å, 5 μm pore size (prepared in-house), with IntegraFrit (New Objective Inc., Woburn, MA) and equilibrated with solvent A. MS data were acquired using a data dependent method utilizing dynamic exclusion, with an exclusion window of 10 ppm and exclusion duration of 60 seconds. MS and MS/MS events used 60000 and 30000 resolution FTMS scans, respectively, with a scan range of 400–2000 m/z in the MS. For MS/MS, the CID collision energy was set to 35%. The LC-MS/MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [20] partner repository with the dataset identifier PXD015275.
Molecular dynamics simulations.
To get structural characterization of the ligand-induced conformational change we performed all-atom MD simulations for three different systems: FKBP, double mutant FKBP F145A I223P and double mutant FKBP F145A I223P in presence of rapamycin ligand. A high-resolution X-ray structure (PDB ID: 1PBK) was used as initial structure for our simulations. Mutations and removal of rapamycin from the structure were done using PyMol 2.1 (Schrödinger, LLC) software. All starting structures were further optimized and all hydrogen were reconstructed using Maestro Protein and Ligand preparation tools (Schrödinger, LLC). MD simulations were conducted using GROMACS 2018 software [21, 22] using CHARMM36 force field [23]. The rapamycin was parametrized using the official CHARMM General Force Field server (CGenFF) [24, 25]. All systems were simulated at a constant pressure and temperature of 1 atm and 298 K. The systems were solvated in the TIP3P water model and neutralized by adding sodium and chloride ions. Particle mesh Ewald (PME) was used to calculate electrostatic interactions with a 10-Å cutoff for non-bonded interactions. All systems were equilibrated using NVT and NPT thermostats. The rapamycin molecule was constrained to the initial position with harmonic constraints during the equilibration process. After that, productive runs for 1μs for each system were conducted. Based on the analysis of the root-mean-square deviation (RMSD) of backbone Cα positions, the first 20 ns of each simulation was omitted prior to further analysis as the time required for the system to reach equilibrium. The structures and representative snapshots of the MD trajectories were inspected visually using PyMol 2.1 (Schrödinger, LLC). Representative structures for each protein were selected as the centroids of most populated clusters. The RMSD-based clustering analysis was done using a GROMACS clustering algorithm on the simulation trajectories [26]. To determine the clustering cutoff values for each system, we calculated the RMSD distributions along the corresponding trajectory, and the RMSD value corresponding to the maximum of the distribution were selected as the cutoff.
Real-mean-square-fluctuation (RMSF) values calculated along the trajectory were used as a measure of the conformational flexibility for each protein. To estimate the changes in the secondary structure, we analyzed the dynamics of the hydrogen bonds along the trajectory by monitoring the distances between backbone hydrogen bond-forming donor-acceptor atom pairs. We considered a hydrogen bond to be broken in given frame of the trajectory if the distance between N and O atoms exceeded the cutoff value of 3.2 Å. The total frequency of hydrogen bond disruption for each pair was calculated as a sum of the open states in all frames along the entire trajectory.
Results and discussion
Detection of ligand-induced disorder-to-order transitions with full-length protein hydrogen-deuterium exchange.
In top-down HDX analysis of proteins, deuterated full-length proteins are injected into mass spectrometer and fragmented by fast fragmentation techniques [27, 28]. In this experimental setup, measuring the mass of the intact deuterated protein prior to the fragmentation provides data on the total number of protected peptide-bond amide protons in the whole protein. This number can be used as a measure of the extent of secondary structure of the protein in solution. Indeed, using this technique, we previously obtained good correlation of the observed total number of protected peptide bond amide protons for a 2.5-s exchange time with the number of residues involved in secondary-structure elements for proteins with known three-dimensional structure [18]. Thus, for the wt Spy and wt FKBP domains, we previously obtained values of 32 and 70 protected residues [6, 18], respectively, which correlate well with the 35 and 72 protected residues found within secondary-structure motifs in the crystal structures of these proteins (PDB 3O39 and 1FKB). Fully unfolded proteins should be deuterated to their maximum extent, due to the absence of any main-chain amide hydrogen bonding and therefore the absence of secondary structure. When we tested Spy L32P and the FKBP-25 F145A I223P mutants, we found 3 and 13 protons protected in these proteins, respectively (Figure 1). Addition of the Im7–45 and rapamycin ligands to the corresponding proteins restored the protection values of 50 and 60, respectively, which were close to the values of 63 [6] and 72 (data not shown), respectively, which had been observed for the wild type protein-ligand complexes. Thus, using this technique, we have detected a transition from a disordered to a more folded protein states upon ligand binding.
Figure 1. Total HDX of intact proteins in free and ligand-bound states.
A. Spy L32P +/− Im7–45 analysis. B. FKBP25 F145A/I223P +/− rapamycin analysis. The number of protected protons, which reflects the total content of secondary structure, is very low in the ligand-free mutant proteins and dramatically increases upon ligand binding.
Characterization of ligand-induced disorder-to-order transitions with additional structural proteomics techniques.
To further characterize the observed structural transitions, we applied differential surface modification and differential crosslinking analyses [15] to the free and ligand-bound proteins. Isotopically-coded modification and crosslinking reagents were used for both approaches. For differential surface modification, the protein samples in the disordered (ligand-free) and folded (ligand-bound) states were modified with light and heavy isotopic forms of the reagent, respectively. Reactions were quenched, combined in a 1:1 ratio, digested with pepsin, and the resulting peptides were analyzed by LC-MS/MS. In this experimental design, surface-exposed residues will be equally modified in both folding states and result in doublets of peaks of equal intensity in the mass spectra. In contrast, buried residues will show a lower degree of modification in the folded state, resulting in doublets of peaks with unequal intensity ratios. As controls we performed the same reactions with light and heavy isotopic forms of the reagents swapped between states and with equimolar mixtures of light and heavy reagents. Those modifications which were detected as an unequal doublet of signals when an equimolar mixture of reagents was used, and whose signal ratios “flipped” when the isotopic forms of the reagents were swapped, were considered to be differentially modified.
The isotopically-coded reagent (PCAS-H4 or PCAS-D4) [15] was used for these differential surface modification studies. This reagent modifies Lys, Tyr, His, Ser, and Thr residues. Using this reagent, we detected a few residues that were differentially modified between the free and ligand-bound states of Spy-L32P, and most of the residues were equally modified in FKBP (Figure 2, S1, S2, S3). The distributions of the fold-changes in degree of modifications were similar for the ligand-free and ligand-bound states of both Spy L32P (Figure S1a) and FKBP (Figure S1b) proteins. These data were consistent with the hypothesis that overall structure of the proteins does not undergo a dramatic change upon ligand binding. These data were consistent with the assumption that the overall structure of the proteins does not undergo a dramatic change upon ligand binding.
Figure 2. Differential surface modification analysis of the proteins in free and ligand-bound states.
A. Experimental scheme of the analysis. Proteins in the two different states were reacted with light or heavy isotopic forms of the modification reagent. Samples were combined in a 1:1 ratio, enzymatically digested, and analyzed by LC-MS. Preferential modification in one of the condition is manifested by a higher intensity of the corresponding peak in a doublet of signals representing the modified peptide. B. Spy L32P +/− Im7–45 analysis. C. FKBP domain F145A/I223P +/− rapamycin analysis. Differentially modified functional groups in residues are shown as spheres with those residues preferentially modified in the presence of ligand shown in blue and those equally modified in the presence or absence of ligand shown in yellow. No residues were found to be preferentially modified in the absence of ligand, therefore there are no red labels in this figure. The data obtained are consistent with the hypothesis that no dramatic change in overall protein structure takes place upon ligand binding.
Similar to the differential surface modification experiment, for the differential quantitative crosslinking analysis, the protein samples in the disordered (ligand-free) and ordered (ligand-bound) states were modified with light and heavy isotopic forms of the crosslinking reagent. Reactions were quenched, combined, digested with pepsin, and the resulting peptides were analyzed by LC-MS/MS. In this experimental design, those residues which are pulled apart due to conformational change will be less extensively crosslinked and, in contrast, residues in close proximity will be crosslinked more efficiently, thus resulting in a doublet of peaks with unequal intensity ratios in the mass spectra. As before, we performed the same reactions with light and heavy isotopic forms of the reagents swapped between states, and with equimolar mixtures of light and heavy reagents for each state as controls. Those crosslinks, which were detected as doublets of signals with unequal intensities and as doublets in control reactions when an equimolar mixture of reagents was used --and whose signal ratios flipped appropriately when the isotopic forms of the reagents in the reactions were swapped -- were considered as differentially modified. We were able to detect several crosslinks that were preferentially formed in the ligand-bound states of Spy-L32P and FKBP (Figure 3, S4, S5, S6). Note that the wt Spy molecule exists as a dimer [6]. Four helices of each chain form a cradle-like shape with the putative ligand-binding site located inside the concave surface of the assembly, and with the N- and C-terminal sequences being disordered [6, 7]. We have determined that the crosslinks that preferentially formed upon ligand binding are mainly those that involve the disordered terminal sequences. This can be interpreted as these regions surrounding the ligand binding site becoming more compact upon formation of the protein-ligand complex, while preserving the overall shape of the molecule. The crosslinks in FKBP that preferentially form upon ligand binding are consistent with the concept that the portions of protein molecule surrounding the binding site are brought closer together as a result of the protein-ligand complex formation. Mutant Spy-L32P protein exhibited especially low HDX protection values, so it was intriguing to see if its ligand-free state was indeed fully unfolded or still somewhat different from the protein that had been fully unfolded with 8 M urea. To get an idea of the degree of unfolding/disorder in the ligand-free state of the mutant proteins, we performed similar differential surface modification and crosslinking analyses for the Spy L32P and compared it to the unfolded-with-8M-urea state (Figure 4, S1a, S2a, S3a, S4a, S5a). Interestingly, a higher degree of modification was observed in some of the residues in the unfolded-with-urea state, indicating that these residues were still somewhat more buried in the ligand-free state. A similar effect was also observed between the ligand-bound and unfolded with urea states.
Figure 3. Differential quantitative crosslinking analysis of the proteins in free and ligand-bound states.
A. Experimental scheme of the analysis. Proteins in the two different states were reacted with light or heavy isotopic forms of the crosslinking reagent. Samples were combined in a 1:1 ratio, enzymatically digested, and analyzed by LC-MS. Preferential modification in one of the condition is manifested by a higher intensity of the corresponding peak in a doublet of signals representing the crosslinked peptide. B. Spy L32P +/− Im7–45 analysis. C. FKBP domain F145A/I223P +/− rapamycin analysis. Differential crosslinks, predominantly formed in the ligand-bound states are shown as blue lines, those that decreased upon ligand binding are shown in red, and those that are unchanged are shown in yellow. The data obtained can be interpreted as indicating some compaction of the regions surrounding the ligand-binding site and are consistent with there being no dramatic change of overall protein structure upon ligand binding.
Figure 4. Differential surface modification and crosslinking analysis of Spy L32P protein in native (ligand-free and ligand-bound) and fully unfolded with 8 M urea states.
Color scheme is the same as in Figure 3. A. Spy L32P +/− 8M Urea differential surface modification analysis. B. Spy L32P + Im7–45 +/− 8M Urea differential surface modification analysis. C. Spy L32P +/− 8M Urea differential quantitative crosslinking analysis. D. Spy L32P + Im7–45 +/− 8M Urea differential quantitative crosslinking analysis. The data obtained are consistent with a presence of somewhat compact residual structure in the ligand-free native state of Spy L32P.
Molecular dynamics simulations of free and ligand-bound mutant proteins.
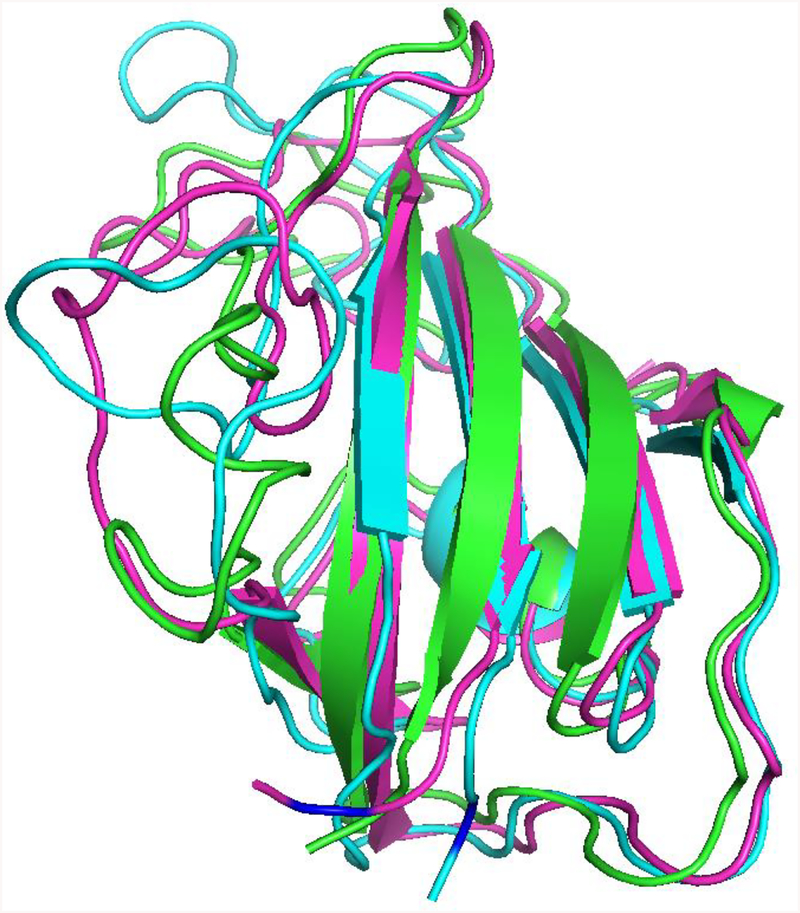
To further characterize the structural differences between the disordered and ordered states of the protein we performed molecular dynamics simulations for the wild type (wt), ligand-free double mutant (dm), and ligand-bound double mutant FKBP domains (we refrained from performing MD simulations on Spy-Im7 due to uncertainties in the dimerization status of the ligand-free Spy-L32P and the undefined location of the Im7 binding site). We did not find dramatic changes in overall structure between the ligand-free wild type and the double-mutant proteins for the time of the simulations. Some loss of secondary structure was observed in the region of the mutations -- especially at the I223P mutation site -- but this effect was not large enough to account for all of the loss of secondary structure observed with HDX. Similarly, ligand binding did not dramatically change the overall structure of the protein, although it did restore some of the secondary structure in the vicinity of the mutation sites, but, again, not to a sufficient extent to explain the gain of secondary structure observed with HDX (Figure 5). We have further compared main-chain fluctuations along the MD trajectory (i.e., the sequence of structures that are produced frame-by-frame during the simulation) between the free and ligand-bound structures. Here, we noticed dramatic differences between both the wt vs. dm, and the ligand-free dm vs. ligand-bound dm samples (Figure S7). This can be interpreted as the dm being much more conformationally flexible and “breathing” more than the wt, and becoming more stabilized and tighter upon ligand binding.
Figure 5. Molecular dynamics analysis of the rapamycin-induced FKBP-25 F145A/I223P double mutant disorder-to-order transition.
Wild type, ligand-free, and ligand-bound mutant proteins are shown in green, cyan, and magenta, respectively. The I223P mutation is indicated in blue. The overall protein structure is preserved between all three proteins. The β-sheet secondary structure in the region of the mutation is lost in the non-ligand-bound mutant protein and is recovered in the ligand-bound form of the mutant protein.
To relate the observed differences to the HDX experimental data, we calculated the frequencies of opening along the MD trajectory of the backbone-amide donor-acceptor pairs which constitute the hydrogen bonds measurable by HDX (Figure S8). We were able to obtain fairly good correlation of the calculated frequency values with the observed HDX data. Indeed, for 72 pairs (70 protected hydrogens in wt), 61 pairs exhibited higher frequencies of opening for ligand-free dm vs. wt, and 11 pairs stayed unchanged (13 protected hydrogens in the ligand-free dm). 44 pairs showed decreases in opening frequencies upon ligand binding (an increase in protection of 47 protons observed with HDX). NMR-derived hydrogen exchange protection factors have been proposed for use as experimental constraints in molecular dynamics simulations [29–31]. Here, we show that the simplified empirical approach of determining the opening frequencies of the backbone amide hydrogen-bond-forming donor-acceptor pairs in molecular dynamics simulations can be used to interpret mass spectrometry-derived HDX values, and provide an estimation of the secondary structure content in the proteins. Indeed, unstructured peptides can exchange in less than 1 s [32] and a single measurement of total exchange -- in the range of seconds used here -- would reflect the total content of hydrogen-bonded backbone amide protons, which can correspond to the presence of stable [18] and transient [19] secondary structure.
Nature of unfolded state of the ligand-free Spy L32P and FKBP-25 F145A I223P proteins.
Even in absence of secondary structure in the ligand-free proteins, it would be interesting to know the degree of unfolding/disorder of the native states of these proteins. Both structural proteomics and MD analyses suggest the presence of more or less compact conformations of the disordered mutant proteins in the ligand-free state. The existence of compact “molten globule”-like states for ligand-free Spy and FKBP mutant proteins agrees well with our recent findings that the intrinsically disordered proteins α-synuclein and tau can be represented by conformational ensembles of fairly compact globular nature [19, 33]. That a similar phenomenon was observed in this study could indicate a general pattern for neurodegenerative disease-related intrinsically-disordered proteins upon the binding of a small-molecule ligand, thus providing a conceptual basis for the development of native-structure-stabilizing therapeutic agents. In the light of these considerations, it would also be intriguing to know which specific structural differences between the disordered and folded states are critical for the activation of a cell’s protein degradation machinery and which would lead to protein elimination, as is the case for the FKBP-based destabilizing domain [8]. From the data presented here, it appears that it could be the presence of secondary structure in the protein that stabilizes the folded state. Following this logic, intrinsically disordered proteins in a cellular environment would have to possess similar features to escape degradation. We have recently detected transient secondary structure in the α-synuclein and tau conformational ensembles in solution, which may partially account for this type of protection from degradation [19, 33]. Another possibility could be complex formation with interacting proteins or with as-yet-to-be identified naturally occurring ligand(s) in the cellular matrix, leading to the structural stabilization similar to that described here.
Conclusions.
Spy L32P – Im7–45 and FKBP F145A/I223P – rapamycin were characterized by hydrogen/deuterium exchange, differential surface modification, differential quantitative crosslinking, and molecular dynamics simulations. Both systems exhibit a transition from a disordered state to a folded (ordered) state upon ligand binding, and can therefore serve as model systems for ligand-induced protein disorder-to-order transitions. The observed data are consistent with disordered proteins existing as compact globular states, characterized by a considerable increase in secondary-structure content upon ligand binding (i.e., a ligand-induced disorder-to-order transition). A combination of structural proteomics and molecular dynamics methods, applied to the characterization of this transition, could expedite the search for protein structure-stabilizing drugs as potential therapeutic agents for neurodegenerative mis-folding diseases. This analytical approach can be used to explore a range of basic structural questions in the study of intrinsically disordered proteins.
Supplementary Material
Significance.
In this study, we used hydrogen/deuterium exchange, differential crosslinking, differential surface modification, and molecular-dynamics simulations to characterize the structural changes in disordered proteins that result from ligand binding. The protein-ligand systems studied here (the ATP-independent protein chaperone, Spy L32P, and the FK506 binding domain of a prolyl isomerase, FKBP-25 F145A/I223P) may serve as models for understanding ligand-induced disorder-to-order transitions in proteins. Additionally, the structural proteomic techniques demonstrated here are shown to be effective tools for the characterization of disorder-to-order transitions and can be used to facilitate study of other systems in which this class of structural transition can be used for modulating major pathological features of disease, such as the abnormal protein aggregation that occurs with Parkinson’s disease and Alzheimer’s disease.
Acknowledgements.
The University of Victoria-Genome British Columbia Proteomics Centre is grateful to Genome Canada and Genome British Columbia for financial support through the (project Genomics Innovation Network (codes 204PRO for operations and 214PRO for technology development) and the Genomics Technology Platform (264PRO). CHB is also grateful for support from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Leading Edge Endowment Fund (University of Victoria), and for support from the Segal McGill Chair in Molecular Oncology at McGill University (Montreal, Quebec, Canada). CHB is also grateful for support from the Warren Y. Soper Charitable Trust and the Alvin Segal Family Foundation to the Jewish General Hospital (Montreal, Quebec, Canada). NVD also acknowledges support from NIH grants R01GM114015 and R01GM123247.
References.
- [1].Chiti F, Dobson CM, Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade, Annu Rev Biochem 86 (2017) 27–68. [DOI] [PubMed] [Google Scholar]
- [2].Shamsi TN, Athar T, Parveen R, Fatima S, A review on protein misfolding, aggregation and strategies to prevent related ailments, Int J Biol Macromol 105(Pt 1) (2017) 993–1000. [DOI] [PubMed] [Google Scholar]
- [3].Martinelli AHS, Lopes FC, John EBO, Carlini CR, Ligabue-Braun R, Modulation of Disordered Proteins with a Focus on Neurodegenerative Diseases and Other Pathologies, Int J Mol Sci 20(6) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Eisele YS, Monteiro C, Fearns C, Encalada SE, Wiseman RL, Powers ET, Kelly JW, Targeting protein aggregation for the treatment of degenerative diseases, Nat Rev Drug Discov 14(11) (2015) 759–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Heller GT, Aprile FA, Vendruscolo M, Methods of probing the interactions between small molecules and disordered proteins, Cell Mol Life Sci 74(17) (2017) 3225–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Quan S, Wang L, Petrotchenko EV, Makepeace KA, Horowitz S, Yang J, Zhang Y, Borchers CH, Bardwell JC, Super Spy variants implicate flexibility in chaperone action, Elife 3 (2014) e01584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Horowitz S, Salmon L, Koldewey P, Ahlstrom LS, Martin R, Quan S, Afonine PV, van den Bedem H, Wang L, Xu Q, Trievel RC, Brooks CL, Bardwell JC, Visualizing chaperone-assisted protein folding, Nat Struct Mol Biol 23(7) (2016) 691–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ, A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules, Cell 126(5) (2006) 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dagliyan O, Shirvanyants D, Karginov AV, Ding F, Fee L, Chandrasekaran SN, Freisinger CM, Smolen GA, Huttenlocher A, Hahn KM, Dokholyan NV, Rational design of a ligand-controlled protein conformational switch, Proc Natl Acad Sci U S A 110(17) (2013) 6800–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gudavicius G, Dilworth D, Serpa JJ, Sessler N, Petrotchenko EV, Borchers CH, Nelson CJ, The prolyl isomerase, FKBP25, interacts with RNA-engaged nucleolin and the pre-60S ribosomal subunit, RNA 20(7) (2014) 1014–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Petrotchenko EV, Borchers CH, HDX match software for the data analysis of top-down ECD-FTMS hydrogen/deuterium exchange experiments, J Am Soc Mass Spectrom 26(11) (2015) 1895–8. [DOI] [PubMed] [Google Scholar]
- [12].Hoopmann MR, Zelter A, Johnson RS, Riffle M, MacCoss MJ, Davis TN, Moritz RL, Kojak: efficient analysis of chemically cross-linked protein complexes, J Proteome Res 14(5) (2015) 2190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ, Semi-supervised learning for peptide identification from shotgun proteomics datasets, Nat. Methods 4(11) (2007) 923. [DOI] [PubMed] [Google Scholar]
- [14].Fischer L, Chen ZA, Rappsilber J, Quantitative cross-linking/mass spectrometry using isotope-labelled cross-linkers., J. Prot 88 (2013) 120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Serpa JJ, Patterson AP, Pan J, Han J, Wishart DS, Petrotchenko EV, Borchers CH, Using multiple structural proteomics approaches for the characterization of prion proteins, J Proteomics 81 (2013) 31–42. [DOI] [PubMed] [Google Scholar]
- [16].Ma B, Zhang K, Hendrie C, Liang C, Li M, Doherty‐Kirby A, Lajoie G, PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry, Rapid communications in mass spectrometry 17(20) (2003) 2337–2342. [DOI] [PubMed] [Google Scholar]
- [17].DeLano WL, Pymol: An open-source molecular graphics tool, CCP4 Newsletter on protein crystallography 40(1) (2002) 82–92. [Google Scholar]
- [18].Brodie NI, Popov KI, Petrotchenko EV, Dokholyan NV, Borchers CH, Solving protein structures using short-distance cross-linking constraints as a guide for discrete molecular dynamics simulations, Sci. Adv (2017) e1700479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brodie NI, Popov KI, Petrotchenko EV, Dokholyan NV, Borchers CH, Conformational ensemble of native α-synuclein in solution as determined by short-distance crosslinking constraint-guided discrete molecular dynamics simulations, PLoS Comput. Biol 15(3) (2019) e1006859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA, The PRIDE database and related tools and resources in 2019: improving support for quantification data, Nucleic Acids Res. 47(D1) (2019) D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hess B, Kutzner C, van der Spoel D, Lindahl E, GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation., Journal of Chemical Theory and Computation 4(3) (2008) 435–47. [DOI] [PubMed] [Google Scholar]
- [22].Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E, GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit., Bioinformatics 29(7) (2013) 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, Mackerell AD, Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles., Journal of Chemical Theory and Computation 8(9) (2012) 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD, CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields, J Comput Chem 31(4) (2010) 671–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yu W, He X, Vanommeslaeghe K, MacKerell AD, Extension of the CHARMM General Force Field to sulfonyl-containing compounds and its utility in biomolecular simulations, J Comput Chem 33(31) (2012) 2451–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Daura X, Gademann K, Jaun B, Seebach D, van Gunsteren WF, Mark AE, Peptide Folding: When Simulation Meets Experiment, Angewandte Chemie 38(1–2) (2004) 236–240. [Google Scholar]
- [27].Pan J, Han J, Borchers CH, Konermann L, Electron capture dissociation of electrosprayed protein ions for spatially resolved hydrogen exchange measurements, J. Am. Chem. Soc 130(35) (2008) 11574–5. [DOI] [PubMed] [Google Scholar]
- [28].Brodie NI, Huguet R, Zhang T, Viner R, Zabrouskov V, Pan J, Petrotchenko EV, Borchers CH, Top-down hydrogen-deuterium exchange analysis of protein structures using ultraviolet photodissociation (UVPD), Anal. Chem [Epub ahead of print] (2018). [DOI] [PubMed] [Google Scholar]
- [29].Vendruscolo M, Paci E, Dobson CM, Karplus M, Rare fluctuations of native proteins sampled by equilibrium hydrogen exchange, Journal of the American Chemical Society 125(51) (2003) 15686–15687. [DOI] [PubMed] [Google Scholar]
- [30].Best RB, Vendruscolo M, Structural interpretation of hydrogen exchange protection factors in proteins: characterization of the native state fluctuations of CI2, Structure 14(1) (2006) 97–106. [DOI] [PubMed] [Google Scholar]
- [31].Dixon RDS, Chen Y, Ding F, Khare SD, Prutzman KC, Schaller MD, Campbell SL, Dokholyan NV, New Insights into FAK Signaling and Localization Based on Detection of a FAT Domain Folding Intermediate, Structure 12(12) (2004) 2161–2171. [DOI] [PubMed] [Google Scholar]
- [32].Keppel TR, Weis DD, Analysis of disordered proteins using a simple apparatus for millisecond quench-flow H/D exchange, Anal Chem 85(10) (2013) 5161–8. [DOI] [PubMed] [Google Scholar]
- [33].Popov KI, Makepeace KA, Petrotchenko EV, Dokholyan NV, Borchers CH, Insight into the structure of the “unstructured” tau protein, Structure (2019), in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.