Abstract
Background:
The problem of variable selection for risk factor modeling is an ongoing challenge in statistical practice. Classical methods that select one subset of exploratory risk factors dominate the medical research field. However, this approach has been criticized for not taking into account the uncertainty of the model selection process itself. This limitation can be addressed by a Bayesian model averaging approach: instead of focusing on a single model and a few factors, Bayesian model averaging considers all the models with non-negligible probabilities to make inference.
Methods:
This paper reports on a simulation study designed to emulate a matched case-control study and compares classical versus Bayesian model averaging selection methods. We used Matthews’s correlation coefficient to measure the quality of binary classifications. Both classical and Bayesian model averaging were also applied and compared for the analysis of a matched case-control study of patients with methicillin-resistant Staphylococcus aureus infections after hospital discharge 2011–2013.
Results:
Bayesian model averaging outperformed the classical approach with much lower false positive rates and higher Matthew’s correlation scores. Bayesian model averaging also produced more reliable and robust effect estimates.
Conclusion:
Bayesian model averaging is a conceptually simple, unified approach that produces robust results. It can be used to replace controversial P-values for case-control study in medical research.
Keywords: Bayesian model averaging, Gibbs variable selection, matched case control, model selection, Zellner’s g-prior
INTRODUCTION
In early 2016, the American Statistical Association (ASA) Board issued a policy statement on the use of P-values and statistical significance that was unprecedented. According to the ASA board’s statement, scientific conclusions should not be based solely on whether or not P-values pass a threshold. The reason is that this practice “encourages the use of terminology such as significant/non-significant, and converts a probability into certainty” [1], which is contrary to the purpose of using statistics: to provide evidence incrementally for decision-making rather than make an immediate decision [2]. Bayesian probability is an alternative paradigm of statistical inference: while P-values quantify the probability of the data given the null hypothesis: P(D|H0), Bayesians calculate the probability of the hypothesis given the data: P(H1|D). Although far less used than P-values, Bayesian inference is more intuitive: it assigns a probability to a hypothesis based on how likely we think it to be true [2].
In this paper, we focus on model selection with a Bayesian approach. Model selection has posed significant challenges for many statisticians, numerous strategies have been developed [3–5] and yet no universal agreed-upon standard has emerged. In conventional model selection, a single model typically is selected based on P-values, and only those variables ‘selected’ by the model are considered. Also, because a single universally approved model selection strategy is unavailable, different approaches are used, which can result in different subsets of variables selected in a final model and, in turn, different results and conclusions. Bayesian model averaging (BMA) is a solution that closes an important methodological gap and obviates the need for complicated or sometimes confused modeling strategies. Instead of focusing on a single model and a few factors, BMA considers all the models with non-negligible probabilities and the posterior probabilities for all variables are summarized at the end.
A variable selection method is a way of selecting a particular set of independent variables for use in a regression model. Stepwise variable selection has been very popular for many years for its simplicity. However, stepwise selection applies methods intended for one test to many tests: as one author has stated, “the maximum F-to- enter statistic is not even remotely like an F-distribution”[6]; for a large enough data-set, all P-values would be ‘significant’ even for non-plausible variables. BMA could provide a way around these problems for better predictive power, effect estimation and hypothesis testing [7–10].
In recent years, investigators have applied BMA in numerous medical and epidemiological analytic studies. However, most of these investigations have used the Bayesian approximate computational approach [8, 9, 11], which is still model oriented and, according to O’Hara et al., is only feasible to use with a maximum of up to several dozens of candidate models [12]. Simulation is a more exact method for calculating model probabilities, and Markov chain Monte Carlo (MCMC) methods are the most common way to simulate from a posterior [13]. An MCMC algorithm was used to generate posterior distributions of parameters and marginal probabilities for selected models.
We applied this technique to simulated data as well as to data from a previously published medical research paper: a matched case control study for risk factors among patients with invasive Methicillin-Resistant Staphylococcus aureus (MRSA) infection after hospital discharge [14]. By comparing BMA with the classical approach, we aim to show the importance of accounting for model uncertainty, the need to replace P-values with probabilities and to evaluate the feasibility of applying MCMC-based BMA to general medical research practice.
METHODS
Classical approach
In this paper, we use the forward, stepwise and backward variable selection approaches implemented by SAS 9.4,
Bayesian Model Averaging Approach
The BMA approach uses fundamental Bayesian methods as follows. Given the data D, if f(m) is the prior probability of model m out of a set of competing models M, the posterior probability is given by
Where f(D|m) is the marginal likelihood calculated using f(D|m)= ∫f(D|βm, m)f(βm|m)dβm), f(D|βm) is the likelihood of model with parameter βm, and f(βm|m) is the prior of βm under model m [15]. The problem with this approach is that these integrals cannot be computed analytically in most case; the set of possible models M increases exponentially as the number of variables grows. For instance, the number of all possible models is equal to 230 = 1,073,741,824 with only p = 30 variables, and the authors have been involved with many studies in which many more variables have been considered. The calculation or approximation of f(D|m) for all m ∈M becomes infeasible. Therefore, MCMC methods which generate observations from the joint posterior distribution f(m, βm|D) of (m, βm) have become popular to estimate f(m|D) and f(βm|m, D) recently.
In details, for a generalized linear model, if an indicator vector is used to represent specific sets of variables that are included among the possible sets of variables such that (γi = 1) or not included (γi = 0) in the model, the linear predictor can be written as
Where Xi is the design matrix and βi the parameter vector of the full model (including all p available variables in the linear predictor) related to the ith term and η is the dependent variable. The model selection process partitions β into (βγ, β\γ) corresponding to those components of β that are included (γi = 1) or not included (γi = 0). Hence the vector βγ corresponds to the active parameters of the model (βm), while β\γ corresponds to the remaining parameters, which are not included in the model defined by γ. In most cases, the prior information is not available, so it is necessary to specify the prior distribution to allow the data to determine which variables are important. In this study we used the popular Zellner’s g-prior with Gibbs variable selection due to its simplicity and efficiency [10].
The g-prior for β is the multivariate normal distribution with μγ,β = 0, i.e.
The prior for the inclusion indicator is defined as γj ~ Bernoulli(0.5) with an inclusion probability of 50%. The elements of mean μγ,β for all j = 0,1,…,p is defined as following:
For the prior precision matrix T, each element Tj,k is equal to the elements of matrix c−2 δ−2XTX in the case where both variables Xj and Xk are included in the model. When at least one of them is not included in the model, then Tj,k = 0 for j ≠ k. Diagonal elements Tj,j denote the pseudo prior precision for γj = 0 that is when Xj is excluded from the model. Hence we set
with c2 = n (sample size) which means the prior has the equivalent weight of 1 observation, and δ2 ~ Inverse-Gamma(10−4, 10−4). The proposed and are estimated through maximum likelihood estimation of the full model with all variables included. The posterior distribution of β and γ are obtained through Gibbs sampling, which generates a sample from the distribution of each variable in turn, conditional on the current values of the other variables. The sequence of the samples constitutes a Markov chain, and the stationary distribution of that Markov chain is the sought-after joint distribution of P(β, γ). The details of this implementation can be found in the book by Ntzoufras[10] and the program was written in WinBugs [16] and R [10, 17, 18]. Finally, we monitored β and γ from the posterior distribution and summarized the results.
Simulation study
In accordance with the BMA approach we have outlined an illustrative example of its application using simulated data that resembles the matched case-control studies carried out in practice. We simulated data in a similar manner to what has been described by Viallefont et al. [19], in which the design of the simulated study was based on the literature review of the studies reported in the American Journal of Epidemiology in 1996 [19]. We modified simulations into a matched 1:2 case-control study for our study purpose. The number of variables initially under consideration was 32. Among these variables, the number actually associated with the health outcome, i.e., the typical dimension reported in the literature of a ‘final model’ found for a case control study, was chosen as 10 (nominal significant) [19]. For the 10 variables designed to be associated with the health outcome, 5 were correlated with each other, and 5 were independent of each other. The remaining 22 variables not related to the health outcome were correlated with each other, factors associated with the health outcome or were independent. The goal was to simulate the scenario where ‘explanatory’ variables are recorded, as in a typical epidemiological study. All variables were dichotomized for simplicity purpose, which is also common practice in logistic regression [19]. The model can easily be extended to include variables of any types. We generated a matrix having 50,000 rows by 33 columns to represent a population under study, with 32 columns representing variables and one column to represent the health outcome. The correlated variables were simulated with correlated probabilities in the range of [0.3–0.6]. The variables linked to outcome were simulated with absolute odds ratios in the interval [1.4, 3.5] and exposure rates in the range of [0.2, 0.6], while the odds ratio for remaining variables were set to 1. The outcome variable was simulated using the following equation:
with β0 was adjusted so as to yield the prevalence rate of approximately 1% to reflect the rare disease often found in epidemiological study, with βi (i = 23,…, 32) associated with Y as mentioned above. From the population 50,000, we randomly selected 200 cases and 400 control to form a 1:2 matched case-control study, and 10 such case-control data-sets were generated.
Analysis
The model selection results may or may not match the nominal status of the variables. In this setting:
True positive (TP): nominal significant variable correctly selected into model
False positive (FP): nominal non-significant variable incorrectly selected into model
True negative (TN): nominal non-significant variable correctly not selected into model
False negative (FN): nominal significant variable incorrectly not selected into model
The above four outcomes can be formulated in a 2 × 2 table called a confusion matrix. Based on the cell values of the confusion matrix, we calculated the false positive rate = FP/ (FP+TN), and false negative rate = FN / (TP+FN). To describe the confusion matrix of true and false positives and negatives by a single number, we used the Matthews correlation coefficient (MCC) [20], which is generally regarded as being one of the best such measures [21]:
The MCC is in essence a correlation coefficient between the observed and predicted binary classifications; it returns a value between −1 and +1. A coefficient of +1 represents a perfect prediction, 0 no better than random prediction and −1 indicates total disagreement between prediction and observation. For 10 data samples and 32 variables, we have 320 P-values to consider in the classical approach and 320 γ for BMA. The conventional rule of thumb for interpreting γ is that if it is less than 50%, there is no evidence for Xγ being a risk factor; if it is between 50% and 75% there is weak evidence for Xγ being a risk factor; if it is between 75% and 95% there is positive evidence, between 95% and 99% the evidence is strong, and beyond 99% the evidence is very strong [22]. Therefore we considered P-values significant at P < 0.05 for the classical approach and mean of γ > 75% for BMA, which is equivalent to P < 0.05 in classical approach [19]. In addition, for nominal significant variables, we also considered the ‘weak evidence’ with γ > 50% for BMA and P < 0.1 for classical approach. In this paper, all the P-values are two-sided.
MRSA matched case-control study
To demonstrate the application of BMA to epidemiological research, we re-analyzed a case-control study for invasive MRSA infections, for which the original data analysis was reported by Epstein et al [14]. A case was defined as MRSA cultured from a normally sterile body site in a patient discharged from a hospital within the prior 12 weeks. For each case patient, two controls were matched for hospital, month of hospital discharge, and age group. Potential risk factors present during the hospitalization and post-discharge period were collected. A total of 194 case patients and 388 matched controls were enrolled. The Centers for Disease Control and Prevention review boards approved the study. Because the study posed no greater than minimal risk to participants, a waiver of informed consent was granted to review medical records in both the hospitals and nursing homes. Verbal consent was obtained from all participants who were interviewed.
In the original published paper, the data set was analyzed as a matched set. The classical approach in the paper was based on Hosmer’s book ‘Applied logistic regression’ [3], and the details of modeling strategies can be seen in the original paper [14]. As a selection strategy, they first performed conditional logistic regression for all 32 variables independently. Conditional logistic regression with backward selection was then performed for the set of variables with P ≤ 0.25 from univariable regression, at the end, the variables with P > 0.25 were also entered to test whether they become significant after controlling for other confounders. The authors did not include any interaction terms, and did not focus on any specific factor of the 32. The definition of the variables can be seen in the original paper [14]. We reanalyzed the data using BMA with the same data definition from [14]. We also did not consider any interaction terms and used conditional logistic regression to analyze the data.
RESULTS
Table 1 summarizes the results of running logistic regression on the entire simulated population with all 32 variables included. Coefficients and P-values are shown for X23-X32 which are designed to be correlated with Y; P-values were not significant for X1-X22 as designed (not shown in the table). When running the full model with all 32 variables for 10 matched case-control subsets, we noticed that 22 out of 220 (10%) variables designed not to be significant (nominal non-significant) at the p < 0.05 level became significant, and only 75 out of 100 (75%) variables designed to be significant (nominal-significant) remained significant in the 10 subsets.
TABLE 1.
Design of the Simulations: Correlation and Strength of the Simulated Associations
| VARIABLES INDEPENDENT OF Y | |
|---|---|
| X1–10 | Independent of each other |
| X11–15 | Correlated with each other |
| X16–20 | Correlated with each other |
| X21 and X22 | Correlate with X23–27 |
| VARIABLES ASSOCIATED WITH Y | |||
|---|---|---|---|
| βi | OR | P | |
| Correlated with each other | |||
| X23 | 1.11 | 3.03 | <0.0001 |
| X24 | −0.79 | 0.45 | <0.0001 |
| X25 | 0.35 | 1.42 | <0.0001 |
| X26 | −0.97 | 0.38 | <0.0001 |
| X27 | −1.24 | 0.29 | <0.0001 |
| Independent of each other | |||
| X28 | −0.47 | 0.63 | <0.0001 |
| X29 | 0.89 | 2.44 | <0.0001 |
| X30 | −0.59 | 0.55 | <0.0001 |
| X31 | −0.54 | 0.58 | <0.0001 |
| X32 | 0.35 | 1.42 | <0.0001 |
Abbreviations: OR, odds ratio.
We analyzed the 10 matched case control data-sets using the classical approach and BMA. The total number of variable selected by BMA was 69 (TP+FP) with γ > 75% and 85 with additional nominal significant variables included at γ > 50%. The total number of variables selected by stepwise, forward and backward selection was 84, 86 and 91 for P < 0.05 and 96, 96 and 95 with nominal significant variables included at P < 0.1.
Table 2 shows that for P < 0.05 / γ> 75%, BMA has a much lower false positive rate of 2% compared to an average of 7% from classical approach. BMA has a slightly higher false negative rate of 35% compared to an average of 28% from classical approach. With weak evidence considered for X23-X32, BMA has lower false positive as well as false negative rate compared to the classical approach: 2% vs. 8% and 19% vs. 22%, respectively. If we use MCC as a single score to evaluate the overall performance for both approaches. BMA outperformed classical approaches with higher MCC of 0.71 vs. 0.69 (average) for strong evidence, and 0.83 vs. 0.71 for including nominal significant variables with weak evidence.
TABLE 2.
Classical Approach vs. Bayesian Model Averaging: True and False Positive and Negative rates and MCC
| METHOD | P<0.05/γ >75% | P<0.1/γ >50%C | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FP (%a) | TN | TP | FN (%b) | MCC | FP (%a) | TN | TP | FN (%b) | MCC | |
| forward | 14(6) | 206 | 72 | 28(28) | 0.69 | 18(8) | 202 | 78 | 22(22) | 0.71 |
| backward | 17(8) | 203 | 74 | 26(26) | 0.68 | 17(8) | 203 | 78 | 22(22) | 0.71 |
| stepwise | 13(6) | 207 | 71 | 29(29) | 0.69 | 18(8) | 202 | 78 | 22(22) | 0.71 |
| BMA | 4(2) | 216 | 65 | 35(35) | 0.71 | 4(2) | 216 | 81 | 19(19) | 0.83 |
Abbreviations: BMA, Bayesian model averaging; FN (false negative); FP (false positive); MCC (Matthews’s correlation coefficient); TP (true positive); TN (true negative).
False positive rate = FP/(FP+TN).
False negative rate = FN/(TP+FN).
P < 0.1 / γ > 50% for X23-X32 + P < 0.05 / γ > 75% for X1-X22
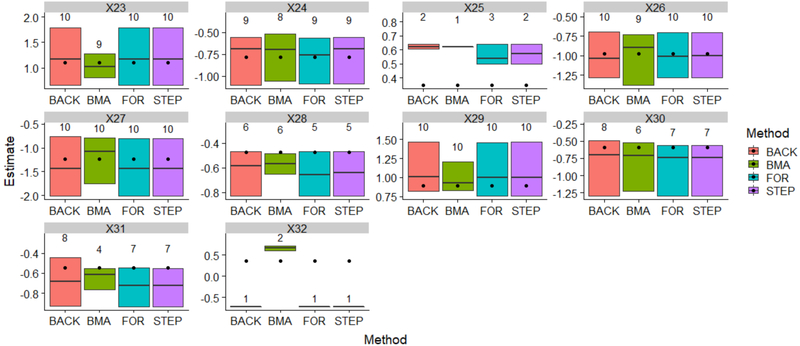
In Figure 1, the estimated effects were visualized by different methods for the variables designed to be associated with outcome (X23-X32). The bottom and top of the box represent the minimum and maximum of the effect estimates, respectively. The line inside the box represents the median. The number on the top of each box represents the variable selection frequency. The selection frequency is based on P < 0.05 / γ > 75%, the black dots are the “true” effects from the population. They have been repeated for each box for comparison purposes. As we can see, for all methods, the variables with larger effects tended to be selected more frequently (X23, X24, X26, X27, X29), and the ranges of the estimates cover the true effects; when the effects are small, the variables are selected less frequently, and the ranges of the boxes would not cover the true effects or barely cover them (X25, X28, X31, X32). Compared to the classical approaches, BMA provides tighter (X23, X27, X28, X29, X31) or comparable range of estimates around the true effects (X24, X26, X30). For X32, all classical method estimate this effect as negative, while BMA estimates it correctly as positive.
FIGURE 1. Estimated effects and variable selection frequency, BMA vs. classical approach.
Abbreviations: BMA: Bayesian model averaging; BACK, backward selection; For: forward selection; STEP: stepwise selection
Table 3 shows the results of BMA vs. classical approach for MRSA matched case-control dataset. As we can see, for γ > 75%, the 5 variables selected by BMA were all selected by classical approach; for γ > 50%, BMA would include all the variables selected by classical approach except admission diagnosis. In addition, BMA also included length of hospital, antibiotic exposure, dialysis and other wound during post-discharge period. The collection of model selected by BMA is 17,944 out of 232 = 4,294,967,296 possible models.
TABLE 3.
Risk Factors for Post-discharge Invasive MRSA, 2011–2013, Classical Approach vs. Bayesian Model Averaging
| CLASSICAL APPROACH | BMA | |||||
|---|---|---|---|---|---|---|
| Variable | OR | 95% CI | P | OR | 95% HDI | Mean(γ) |
| MRSA colonization | 7.7 | 3.60, 16.51 | <0.0001 | 5.6 | 2.83, 11.47 | 1 |
| Chronic wound during post-discharge period | 4.4 | 2.14, 9.09 | <0.0001 | 5.2 | 2.37, 11.97 | 1 |
| Male sex | 2.2 | 1.31, 3.63 | 0.0028 | 2.8 | 1.81, 4.61 | 1 |
| CVC at discharge | 2.2 | 1.13, 4.11 | 0.0191 | 2.7 | 1.23, 1.92 | 0.9 |
| Charlson comorbidity index >1 | 1.4 | 1.17, 1.55 | <0.0001 | 1.2 | 1.02, 1.35 | 0.8 |
| Length of hospital stay (>10 day vs. <=10 day) | 2.1 | 1.10, 4.05 | 0.7 | |||
| Other wound during post-discharge period | 2.4 | 1.12, 5.17 | 0.7 | |||
| Discharge to nursing home | 2.7 | 1.41, 4.99 | 0.0024 | 2.0 | 1.05, 3.80 | 0.6 |
| Antibiotic exposure during post-discharge period | 1.9 | 0.92, 4.02 | 0.6 | |||
| Dialysis during post-discharge period | 2.0 | 0.38, 9.09 | 0.6 | |||
| Discharge with non-CVC invasive device | 3.3 | 1.24, 7.39 | 0.0096 | 1.9 | 0.86, 4.16 | 0.5 |
| Admission Diagnosis | 1.8 | 1.05, 3.22 | 0.0405 | 1.0 | 0.41, 2.39 | 0.2 |
Abbreviations: BMA, Bayesian model averaging; CI, confidence interval; CVC, central venous catheter; HDI, highest density interval; MRSA, methicillin-resistant Staphylococcus aureus; OR, odds ratio; γ, inclusion indicator.
PERFORMANCE
The program was executed on a laptop with Intel® Core™ i5–5300 CPU @2.30GHz, 2.29GHz dual processors, 8.00GB installed memory (RAM) and 64-bit operating system. The convergence diagnostics were based on 1) Autocorrelation of Markov chain (ρlag < 30); 2) Gelman-Rubin diagnostic [27]; 3) Markov chain errors (< 1% of standard errors of model parameters for all β and γ) [24]. Using the MRSA research data as an example, convergence was observed with 150,000 iterations, for three chains of 50,000 iterations each. On average, each iteration required about 0.07 seconds, so 150,000 iterations took approximately 3 hours.
DISCUSSION
The results of our analysis underscores the possibility that popular model development strategies for matched-case control study can produce parameters that show strong associations with the outcome only by chance. Our simulation study showed that about 10% of parameters that were designed to have no association with the outcome in a population of 50,000 became significant in the randomly selected subset of 600 (false positive variables). The classical model strategy selected the false positive variables into a final model on an average of 7% of the time, while BMA selected them 2% of the time. BMA has slightly higher false negative rate of 35% compared to an average of 28% from classical approach. Alternatively, if we treat nominal significant variables as known risk factors supported by literature review and relax the selection standard for BMA to γ > 50%, the correspond false negative rate dropped to 19% and the false positive rate remained the same. There is no universal agree on standard on how to relax the P value for the classical approach, if we use P < 0.1 for the comparison purpose, the classical approach has both higher false positive rate and false negative rate compared to BMA (Table 2). We also introduced MCC, a single score to evaluate the overall performance of the binary classification, the results showed that BMA outperformed classical approach under both selection cut point of P < 0.05 / γ > 75% and additional P < 0.10 / γ > 50% for nominal significant variables. For variables designed to be associated with outcome, BMA produced better estimates with tighter ranges around the true values, and most importantly, BMA consistently quantified the effect with correct signs (+/−), while classical approaches reversed the sign of one effect from positive to negative (Figure 1). This demonstrates the potential shortcomings of depending on one model without taking into account the uncertainty associated with the selection of the model itself. Another shortcoming is that several different models may all seem reasonable given the data, in our simulation study, different subset of variables were selected by stepwise, forward and backward selection (Table 2, FP+TP), arbitrarily selecting a single model can lead to biased inference on the effect of interest.
We applied BMA to data from a published medical research study and compared the results to the original analysis. Because all the variables under consideration were based on a literature review or biological/clinical plausibility, we used γ > 50% as an indicator of association to outcome. As a result, BMA selected seven out of eight variables in the final model of classical approach. The only exception was the variable “admission diagnosis”, a categorical variable created by a data-driven process that collapsed 16 different diagnoses into two categories, each of which included very heterogeneous diagnoses. This variable was selected into final 8 parameter model by the classical approach, while the selection probability for this parameter was only 24% based on BMA. Furthermore, BMA detected evidence of association between the following four variables with the outcome: length of hospital stay, other wound, dialysis and total antibiotic exposure during post-discharge period. These variables have found to be risk factors for MRSA in other literature [25–28]. They were collected and tested but did not enter the final model with the P < 0.05 cut point. As a result, they were not treated as risk factors for informing strategies to prevent MRSA, although other literature provides evidence that they may be important factors.
We have proposed BMA as a comprehensive way to account for model uncertainty. Instead of relying on one model, the inference was carried out based on hundreds of thousands models and the results are more reliable and robust. Another advantage of BMA is that it provides a transparent interpretation through the variable’s posterior selection probability: the higher the probability, the stronger the association between the variable and the health outcome. In practice, this information can help to focus the limited resources on what matters the most. In contrast, classical P-values is hard to interpret, for example, the expression “fail to reject the null hypothesis” doesn’t mean to “accept the null hypothesis”, either, as a result, P-values are one of the most misunderstood and misinterpreted quantities in research [4].
In summary, BMA eliminates the need for complicated model selection and cross validation strategies that can lead to different results and conclusions. Our study demonstrates that BMA is a conceptually simple, unified approach that produces robust results. Bayesian way of dealing with the model uncertainty problem has been found to be the only way[14]. Compared to the classical approach, BMA can be both highly specific and sensitive if coupled with proper use of prior information. Another advantage is that results are easy to interpret. With advances in computer technology and computing power, a laptop alone can be used with ease for a typical medical research study. Computations involving hundreds of observations and dozens of variables can be completed within hours.
Bayesian inference has been controversial because it uses the prior distribution, which is subjectively determined by the user. However, prior can be totally non-informative, or equivalent to the weight of 1 observation in our study, which has little influence on the posterior estimates for the typical medical research with hundreds of thousands observations. A word of caution is that BMA is not a substitute for careful incorporation of available scientific knowledge or for careful data analysis. This together can lead to a set of possible confounders, or potential risk factors for further consideration with BMA.
References
- 1.Hurlbert SH and Lombardi CM Final collapse of the Neyman-Pearson decision theoretic framework and rise of the neoFisherian in Annales Zoologici Fennici. 2009. BioOne. [Google Scholar]
- 2.Vidgen B and Yasseri T.J.F.i.P., P-values: misunderstood and misused. Frontiers in Physics, 2016. 4: p. 6. [Google Scholar]
- 3.Hosmer D and Lemeshow S, Applied Logistic Regression. 2005: Wiley: New York, NJ. [Google Scholar]
- 4.Harrell F, Regression Modeling Strategies: with applications to linear models, logistic regression, and survival anlaysis. 2015: Springer: New York, NJ [Google Scholar]
- 5.Kleinbaum DG and Klein M, Logistic regression: A self-learning text (2nd ed.). 2002: Springer: New York, NJ. [Google Scholar]
- 6.Miller AJ, Selection of subsets of regression variables. Journal of the Royal Statistical Society: Series A, 1984. 147(3): p. 389–410. [Google Scholar]
- 7.Hoeting JA, et al. , Bayesian model averaging: a tutorial. 1999: p. 382–401.
- 8.Genell A, et al. , Model selection in medical research: a simulation study comparing Bayesian model averaging and stepwise regression. BMC Med Res Methodol, 2010. 10: p. 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, Zhang W, and Bakhai A, Comparison of Bayesian model averaging and stepwise methods for model selection in logistic regression. Stat Med, 2004. 23(22): p. 3451–67. [DOI] [PubMed] [Google Scholar]
- 10.Ntzoufras I, Bayesian modeling using WinBUGS. Wiley series in computational statistics. 2009, Hoboken, N.J.: Wiley; xxiii, 492 p. [Google Scholar]
- 11.R. AE, Bayesian Model Selection in Social Research, in Sociological Methodology. 1995. p. 111–163. [Google Scholar]
- 12.Hara RBO and Sillanpaa MJ, A Review of Bayesian Variable Selection Methods. Bayesian Anal, 2009. 4(2). [Google Scholar]
- 13.Gilks WR, Richardson S, and Spiegelhalter D, Markov chain Monte Carlo in practice. 1995: CRC press. [Google Scholar]
- 14.Epstein L, et al. , Risk Factors for Invasive Methicillin-Resistant Staphylococcus aureus Infection After Recent Discharge From an Acute-Care Hospitalization, 2011–2013. Clin Infect Dis, 2016. 62(1): p. 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dellaportas P, Forster JJ, and Ntzoufras I, On Bayesian model and variable selection using MCMC. Statistics and Computing, 2002. 12: p. 10. [Google Scholar]
- 16.Spiegelhalter D, et al. , WinBUGS user manual. 2003, version.
- 17.Team, R.C., R: A language and environment for statistical computing. 2013.
- 18.Sturtz S, Ligges U, and Gelman AE, R2WinBUGS: a package for running WinBUGS from R. 2005.
- 19.Viallefont V, Raftery AE, and Richardson S, Variable selection and Bayesian model averaging in case-control studies. Stat Med, 2001. 20(21): p. 3215–30. [DOI] [PubMed] [Google Scholar]
- 20.Matthews BW, Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochimica et Biophysica Acta (BBA)-Protein Structure, 1975. 405(2): p. 442–451. [DOI] [PubMed] [Google Scholar]
- 21.Powers DM, Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. 2011.
- 22.Kass AE and Raftery AE, Bayes factors. Joural of American Statistical Association, 1995. 90: p. 773–795. [Google Scholar]
- 23.Gelman A and Rubin DB, Inference from iterative simulation using multiple sequences. Statistical science, 1992: p. 457–472. [Google Scholar]
- 24.Ntzoufras I, Gibbs variable selection using BUGS. Journal of statistical software, 2002. 7: p. 1–19. [Google Scholar]
- 25.Nguyen DB, et al. , Invasive methicillin-resistant Staphylococcus aureus infections among patients on chronic dialysis in the United States, 2005–2011. Clin Infect Dis, 2013. 57(10): p. 1393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Safdar N and Maki DG, The commonality of risk factors for nosocomial colonization and infection with antimicrobial-resistant Staphylococcus aureus, enterococcus, gram-negative bacilli, Clostridium difficile, and Candida. Annals of Internal Medicine, 2002. 136(11): p. 834–844. [DOI] [PubMed] [Google Scholar]
- 27.Asensio A, et al. , Colonization and infection with methicillin-resistant Staphylococcus aureus: associated factors and eradication. Infection Control & Hospital Epidemiology, 1996. 17(1): p. 20–28. [DOI] [PubMed] [Google Scholar]
- 28.Big C and Malani PN, Staphylococcus aureus Bloodstream Infections in Older Adults: Clinical Outcomes and Risk Factors for InHospital Mortality. Journal of the American Geriatrics Society, 2010. 58(2): p. 300–305. [DOI] [PubMed] [Google Scholar]
- 29.Wasserstein RL and Lazar NA, The ASA’s statement on p-values: context, process, and purpose. The American Statistician, 2016: p. 00–00. [Google Scholar]