Abstract
Micronutrient deficiencies are common in undernourished societies yet remain inadequately assessed due to the complexity and costs of existing assays. A plasma proteomics-based approach holds promise in quantifying multiple nutrient:protein associations that reflect biological function and nutritional status. To validate this concept, in plasma samples of a cohort of 500 6- to 8-y-old Nepalese children, we estimated cross-sectional correlations between vitamins A (retinol), D (25-hydroxyvitamin D), and E (α-tocopherol), copper, and selenium, measured by conventional assays, and relative abundance of their major plasma-bound proteins, measured by quantitative proteomics using 8-plex iTRAQ mass tags. The prevalence of low-to-deficient status was 8.8% (<0.70 μmol/L) for retinol, 19.2% (<50 nmol/L) for 25-hydroxyvitamin D, 17.6% (<9.3 μmol/L) for α-tocopherol, 0% (<10 μmol/L) for copper, and 13.6% (<0.6 μmol/L) for selenium. We identified 4705 proteins, 982 in >50 children. Employing a linear mixed effects model, we observed the following correlations: retinol:retinol-binding protein 4 (r = 0.88), 25-hydroxyvitamin D:vitamin D-binding protein (r = 0.58), α-tocopherol:apolipoprotein C-III (r = 0.64), copper:ceruloplasmin (r = 0.65), and selenium:selenoprotein P isoform 1 (r = 0.79) (all P < 0.0001), passing a false discovery rate threshold of 1% (based on P value-derived q values). Individual proteins explained 34–77% (R2) of variation in their respective nutrient concentration. Adding second proteins to models raised R2 to 48–79%, demonstrating a potential to explain additional variation in nutrient concentration by this strategy. Plasma proteomics can identify and quantify protein biomarkers of micronutrient status in undernourished children. The maternal micronutrient supplementation trial, from which data were derived as a follow-up activity, was registered at clinicaltrials.gov as NCT00115271.
Introduction
Micronutrient deficiencies due to dietary inadequacy are widespread in the developing world, especially in rural South Asia (1–3), where they may contribute to risks of morbidity, mortality, poor growth, and impaired cognition (4–8), making their prevention a global public health goal. Yet their burden, referred to as “hidden hunger,” remains infrequently assessed in vulnerable populations. Obstacles that limit comprehensive and frequent assessment of multiple micronutrient status include technical difficulty, logistical challenges, and costs of performing multiple, nutrient-specific assays (9). Incomplete or outdated estimates of burden, stemming from infrequent assessment, have left national and global agencies poorly informed, unable to accurately and rapidly assess deficiencies, target and design effective interventions, and monitor changes in population micronutrient status. A few field methods are currently under development to concurrently assess status for a limited number of micronutrients of known health consequence, such as vitamin A and iron (10, 11). However, the breadth of nutritional need from dietary deficiencies and environmental stresses in poor settings is likely to span many essential nutrients, flagging a need for broader assessments and better informed prevention. In low-resource settings, meeting this public health need will require in the future more efficient, affordable, and comprehensive micronutrient status assays. Furthermore, because biochemical concentrations alone do not reflect nutrient function, a new assessment approach would ideally add value if it were to generate biomarkers linked to nutrient metabolism and function.
Quantitative proteomics, in which hundreds of plasma proteins can be identified and quantified in relative abundance in a single MS experiment using mass tags (12, 13), may offer a basis for discovering proteins and protein clusters that reflect nutrient functions and predict micronutrient status. Ultimately, such informative protein combinations could be simultaneously assessed using other high-throughput techniques, such as antibody chip screening. Using proteomics to estimate micronutrient deficiencies would rely on identifying plasma protein biomarkers that sufficiently covary, via binding or less directly through complex metabolic networks, with population nutrient distributions.
The application of proteomics to human nutrition has been widely proposed (14–17), but there have been, to our knowledge, no studies to date evaluating the correlation of plasma proteomic biomarkers determined by MS with population distributions of multiple plasma nutrient concentrations measured by conventional assays. This void may exist for several reasons: 1) lack of access to large plasma archives obtained from undernourished populations adequately characterized for multiple nutrient status; 2) need for substantial investment in state-of-art mass spectrometric, bioinformatic, and high through-put data analytic instrumentation; and 3) the required levels of effort to discover plasma protein biomarkers that covary with micronutrient status. A first step toward validating this approach would be to conduct a plasma micronutrient and proteomic assessment that quantifies strength of association between concentrations of nutrients and their cognate, bound proteins in circulation (nutrient:protein dyads). Observing strong associations would offer a biological proof of concept, strengthen confidence about nonclassical nutrient:protein associations that may appear, and encourage methodological development to quantify, analyze, and interpret proteomics data for potential public health application.
Using plasma biospecimens from a population cohort of Nepalese children, the present study explores the ability to combine plasma proteomics, bioinformatics, and a novel statistical modeling approach to reveal correlations between selected micronutrients and their cognate circulating proteins: specifically, retinol with its major transport protein, retinol binding protein 4 (RBP4)11 (18); 25-hydroxyvitamin D with vitamin D binding protein (VDBP), the major carrier protein for ergocalciferol (vitamin D2), cholecalciferol (vitamin D3), and 25-hydroxyvitamin D (19); α-tocopherol with apo C-III, one of the first apolipoproteins released with vitamin E from the liver (20); copper with ceruloplasmin (Cp), to which ∼95% of plasma copper is bound (21); and selenium with selenium protein P1 (SEPP1), the major hepatic-derived protein that transports Se to peripheral tissues (22). Beyond confirming expected correlations, we explored, for each nutrient, gains in explained variance achieved by adding a second plasma protein to each regression model based on statistical criteria. Further model building is currently limited by missing protein data, inherent to mass spectrometric analysis, for which extensive imputation is required to overcome. However, the analytic approach described here represents an initial step toward revealing protein combinations that may enable, in the future, plasma proteomic data to describe micronutrient status and predict levels of deficiency in populations.
Materials and Methods
We set out to quantify micronutrient concentrations and protein relative abundance in archived plasma samples obtained in 2006–2008 from 500 children, 6–8 y of age, living in the District of Sarlahi, Nepal. The area is located in the rural, southern plains of the country, where micronutrient deficiencies with preventable consequences have been documented in preschool-aged children (4, 5). The 500 children in this study comprised a random 50% subset of 1000 children in the same age range whose plasma multiple micronutrient and inflammation status was characterized by conventional biochemical tests (K. Schulze, P. Christian, L. Wu, M. Arguello, H. Cui, A. Nanayakkara-Bind, C. Stewart, S. Khatry, S. LeClerq, K. West, unpublished results). This assessment formed part of a nutrition, health, and cognitive follow-up study in 2006–2008 of a larger cohort of children (7, 23) whose mothers had participated in a randomized, antenatal micronutrient supplementation trial in 2000–2001 (6).
The field procedures for the follow-up study, which included histories of illness, anthropometry, blood pressure, urine collection, and phlebotomy, were previously described (23). Anthropometric status was summarized as Z-scores for weight-for-age, height-for-age, and BMI-for-age in relation to the WHO reference (24). Relevant to the current analysis, early morning blood samples were obtained by venipuncture following an overnight fast and transported light protected to a field laboratory on ice packs. Following centrifugation, plasma was analyzed for lipids, glycated hemoglobin, and glucose concentrations and three 1-mL aliquots stored and air-freighted under liquid nitrogen vapor to Johns Hopkins University where samples were stored at −80°C until analysis (23).
The original field trial was carried out among consenting mothers and was approved by the Nepal Health Research Council, Kathmandu, Nepal and the Institutional Review Board of the Johns Hopkins Bloomberg School of Public Health, Baltimore, MD. The follow-up study protocol was approved by Institutional Review Boards at the Institute of Medicine of Tribhuvan University, Kathmandu, Nepal and at Johns Hopkins University. Follow-up study procedures were carried out in children following parental consent.
Plasma micronutrient assays.
Laboratory assays were carried out to measure plasma concentrations of vitamins A, D, and E, copper, and selenium, among other nutrients. Plasma retinol and α-tocopherol were simultaneously measured by a conventional, reverse-phase HPLC method following protein precipitation and hexane extraction of the fat-soluble contents of the plasma. The assay was calibrated against Standard Reference Material 968e (National Institute of Standards and Technology). Chromatography was performed on an Alliance 2795 HPLC system (Waters) with autosampler and photodiode array detector (Waters 2475) and analyzed with Empower 2 software. The separation was achieved using a Supelcosil LC-18 25-cm × 4.6-mm, 5-μm column (Sigma-Aldrich).
A commercial competitive enzyme immunoassay (IDS) was used to measure 25-hydroxyvitamin D. According to the kit insert, the method had 100% reactivity with 25-hydroxyvitamin D3, 75% for 25-hydroxyvitamin D2, and 100% for 24, 25-dihydroxyvitamin D3.
Plasma copper and selenium were measured by graphite furnace atomic absorption spectroscopy (Perkin Elmer) with background correction using modifications of the manufacturer's recommended conditions. Assays were run against aqueous standards and accuracy was checked using commercial serum quality control materials with certified contents of copper and selenium (Seronorm Trace Elements Serum, Sero). Assay repeatability was established by running a pooled sample at regular intervals. Copper was diluted 1:15 in deionized water with 10 μL added by autosampler to 5 μL of a 10,000 mg/L magnesium nitrate matrix modifier (Perkin Elmer) prepared at a 0.1% v:v dilution in deionized water. Samples were read for 5 s during a 2000°C atomization step following an injection temperature of 80°C, 30 s of drying each at 110°C and 130°C, and 20 s of pyrolysis at 1200°C. Selenium was diluted 1:10 in an ascorbic acid solution prior to analysis and 10 μL was deposited by autosampler into the graphite tube with 5 μL of a 10,000 mg/L palladium nitrate matrix modifier prepared at a 12% v:v dilution and 3 μL of a 10,000 mg/L magnesium nitrate matrix modifier (Perkin Elmer) prepared at a 1.2% dilution in deionized water. Samples were read for 5 s during a 1900°C atomization step following drying steps at 110°C, 130°C, and 200°C, and 20 s of pyrolysis at 1050°C.
Plasma proteomics assays.
Plasma aliquots of 25 μL from each of the larger set of 1000 children in whom multiple micronutrient status assessment had been carried out were combined to create a “master plasma pool” (25). Plasma samples (40 μL) from each of the 500 participants randomly chosen for proteomics evaluation, plus 40 μL from each of the 72 aliquots of the master pool plasma bioarchive, were immuno-depleted of 85–90% of 6 high abundance proteins (albumin, IgG, IgA, transferrin, haptoglobin, and antitrypsin) using a Human-6 Multiple Affinity Removal System LC column (Agilent Technologies). Immuno-depleted samples (100 μg) were digested overnight with trypsin. Tryptic peptide samples from 7 individual samples plus a master pool were randomly labeled with iTRAQ 8-plex reagents (AB Sciex) according to manufacturer's instructions. The 7 samples and master pool were mixed and fractionated into 24 fractions by strong cation exchange chromatography. iTRAQ-labeled peptides in each strong cation exchange fraction were desalted and loaded directly on to a reverse-phase nanobore column and eluted using a 2–50% acetonitrile and 0.1% formic acid gradient for 110 min at 300 nL/min. Eluting peptides were sprayed through a 10-μm emitter tip into an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific) interfaced with a NanoAcquity ultra-HPLC (Waters). From each survey scan, up to 10 peptide masses (precursor ions) were individually isolated and fragmented. Precursors and the fragment ions were analyzed at 30,000 and 15,000 resolution, respectively. Isotopically resolved masses in mass spectrometric and MS/MS spectra were extracted with and without deconvolution using Thermo Scientific Xtract software and searched against the RefSeq 40 protein database using Mascot (Matrix Science) through Proteome Discoverer software (v1.3, Thermo Scientific) specifying Homo sapiens, trypsin as the enzyme allowing one missed cleavage, fixed cysteine methylthiolation and 8-plex-iTRAQ labeling of N-termini, and variable methionine oxidation and 8-plex-iTRAQ labeling of lysine and tyrosine. Peptide identifications from Mascot searches were filtered within the Proteome Discoverer to identify peptides with ≥95% confidence [i.e., false discovery rate (FDR) <5%].
Statistical analysis.
Protein relative abundances within each iTRAQ experiment were estimated using the medians of the log2-transformed and normalized reporter ion intensities derived from Proteome Discoverer v1.3, as described in detail elsewhere (25). We initially used a conventional approach to assess protein abundances by normalizing reporter ion intensities to those of a master pooled plasma sample included in every iTRAQ experiment. However, we ultimately employed linear mixed effects models (LME) to combine the proteomic data from different experiments and to assess the association of protein relative abundances with measured micronutrient concentrations. We used logarithmic transformations of plasma vitamin E and selenium data due to their skewed distributions. For each univariate nutrient-protein analysis, we fit a random intercept model via restricted maximum likelihood estimation, specifically:
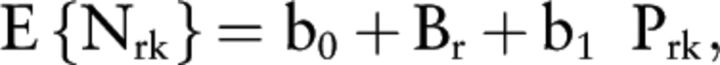 |
where Nrk denotes the observed (or logarithmic transformed) plasma concentrations for vitamins A, D, and copper (vitamin E and selenium) indexed by sample k in iTRAQ experiment r, and Prk are the respective protein relative abundance estimates. The variable b0 is the fixed effect for the intercept, Br denotes the random deviation from this fixed effect in experiment r, and the variable b1 denotes the slope of the nutrient:protein association. This approach allows for the determination of the strength of nutrient:protein associations via statistical inference for the slope variable b1 and to decompose the observed variability in the micronutrient concentrations into variability explained by protein abundances, differences between the samples in different iTRAQ experiments, and experimental error. For the mixed effect models, R2 was based on the observed nutrient concentrations and their respective best linear unbiased predictions from the MS data (26).
We summarize each nutrient:protein comparison by presenting a series of 3 figures that include a histogram of the serum nutrient concentrations, a scatterplot of the nutrient:protein association using the pooled plasma protein abundance, and a scatterplot of association using the LME-based protein abundance estimates, displayed as panels A, B, and C, respectively ( ). The R2 values show the proportion of variance explained by the fitted values of the nutrient:protein regression models. The P value reported in each panel B is derived from testing the hypothesis of no association between nutrient concentration and protein abundance, and the P value in each panel C is derived from testing the fixed effects slope of nutrient concentration on protein abundance in the LME model (b1). P values are not provided for correlations involving LME-based protein relative abundance values (i.e., nutrient:protein or protein:protein correlations), because within-experiment protein concentrations violate the assumption of independent observations required for hypothesis testing.
Finally, we extended the above mixed effects approach to a multivariate LME model for each nutrient, identifying the protein with the best explanatory power (i.e., maximizing the coefficient of determination, LME R2) for the nutrient with the original transport protein in the model, thus combining relative abundances of 2 proteins to explain variability in the micronutrients (25). All analyses were carried out using in-house developed open source software implemented in the statistical environment R (27).
Results
Nutritional profile of children.
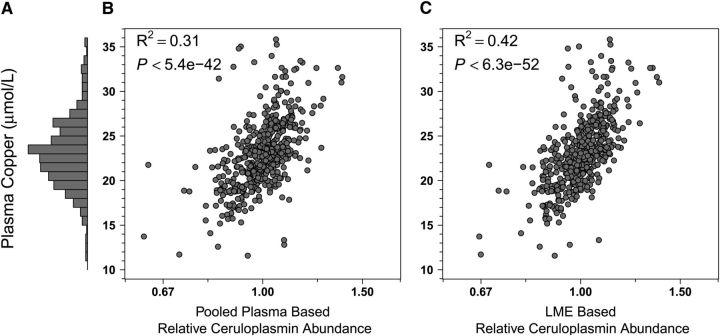
Study children (n = 500) were generally undernourished, reflected by low anthropometric Z-scores in relation to the WHO reference for children 5–19 y old (24). Children were, on average, underweight (weight-for-age Z-score = −1.98 ± 0.90), stunted (height-for-age Z-score = −1.77 ± 0.99), and mildly wasted (BMI-for-age Z-score = −1.20 ± 0.91). They were also marginal to deficient in status for most micronutrients, reflected by the plasma concentrations of retinol (1.04 ± 0.27 μmol/L) (28), 25-hydroxyvitamin D (65.9 ± 19.3 nmol/L) (29), and α-tocopherol (12.1 ± 3.2 μmol/L) (30). Copper status (23.2 ± 5.7 μmol/L) was within a normal range (31), whereas selenium status was marginal (0.86 ± 0.26 μmol/L) (32) (Figs. 1–5, panels A). The percentages of children classified as deficient were 8.8, 19.2, 17.6, 0, and 13.6% for the 5 nutrients, respectively.
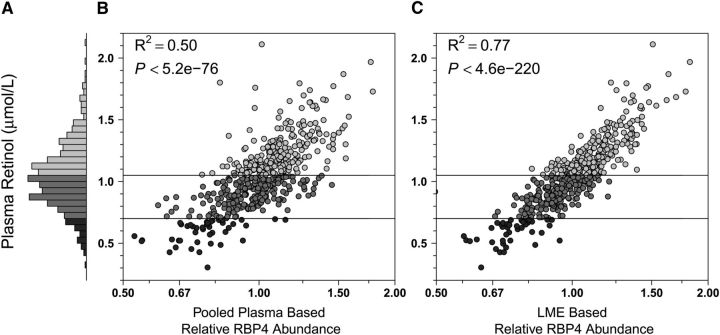
FIGURE 1.
Plasma retinol and RBP4 relative abundance distributions in Nepalese children 6–8 y of age (n = 500). (A) Histogram showing the frequency distribution of retinol concentrations: range = 0.30–2.11 μmol/L, 8.8% (n = 44) deficient (<0.70 μmol/L, dark gray), 45.6% (n = 228), marginal (0.70 to <1.05 μmol/L, medium gray), and 45.6% (n = 228) adequate (≥1.05 μmol/L, light gray) in status. (B) Plasma retinol by relative abundance of RBP4 by a traditional estimation method using a master plasma pool in one randomly assigned iTRAQ channel within each 8-plex experiment to normalize the protein distribution across iTRAQ runs. (C) Plasma retinol by relative abundance of RBP4 by an estimation method that relies on an LME model that combines abundance estimates from all 72 iTRAQ experiments (25). R2 values represent the proportion of variance in the nutrient explained by the fitted values of the nutrient-protein regression models. The P value in B is derived from testing the hypothesis of no association between the nutrient and protein abundance, whereas the P value in C is derived from testing the fixed effects slope for the protein abundance in the LME model. Shading of circles in B and C corresponds to bars. Horizontal lines indicate cutoffs for changes in micronutrient status. iTRAQ, isobaric tags for relative and absolute quantification; LME, linear mixed effects (model); RBP4, retinol binding protein isoform 4.
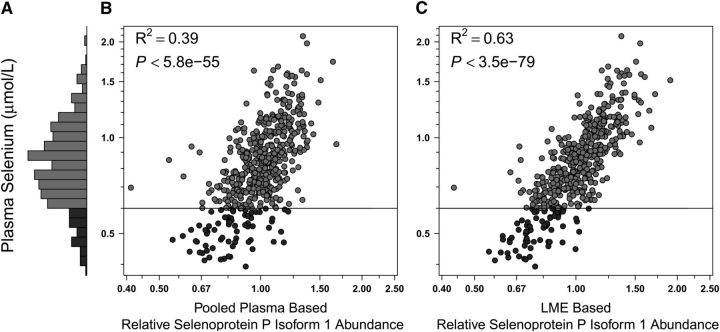
FIGURE 5.
Plasma selenium and SEPP1 relative abundance distributions in Nepalese children 6–8 y of age (n = 499). (A) Plasma selenium concentrations: range, 0.4–2.1 μmol/L; 13.6% (n = 68) deficient (<0.6 μmol/L, dark gray) and 86.4% (n = 431) adequate (≥0.6 μmol/L, medium gray) in status. (B,C) Plasma selenium by relative abundance of SEPP1 by traditional master plasma pool normalization and LME-adjusted methods, respectively (see Fig. 1 for details). LME, linear mixed effects (model); SEPP1, selenoprotein P isoform 1.
Proteomic profile of children.
Across seventy-three 8-channelled iTRAQ experiments, we identified 4705 nonredundant proteins at least one time, with high mass accuracy (<10 ppm) and a FDR of 5%. Of this number, the relative abundance of 982 proteins was quantified in >10% of all 500 child plasma samples (i.e., n > 50), of which 455 (46%) comprised extracellular, secretory, membrane, or lipoprotein-associated proteins (Supplemental Table 1). One hundred and forty-six (15%) of the listed plasma proteins were quantified in all 500 children.
Nutrient:protein dyad correlations.
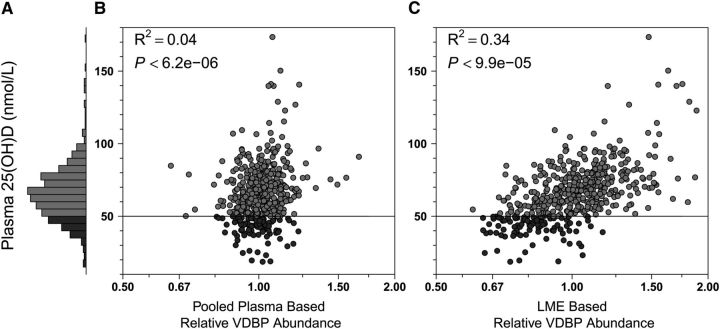
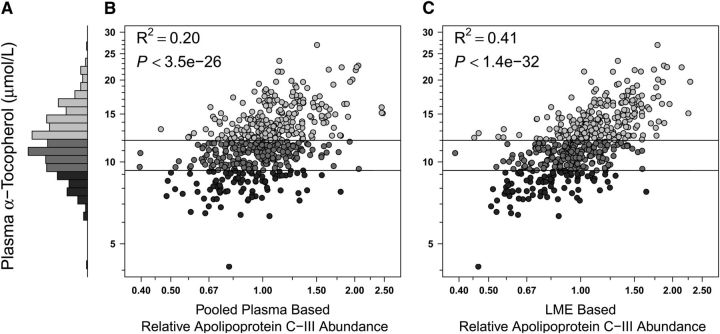
With respect to vitamin A, using the master pool sample as a reference for normalization among iTRAQ experiments, we observed a coefficient of determination (R2) of 0.50 (i.e., explaining 50% of variance in nutrient concentration) between plasma retinol concentration and relative abundance of RPB4 (Fig. 1B). Using a linear mixed effects model (LME) (25) for normalization, the R2 increased to 0.77 (Fig. 1C). This modeled approach also markedly increased explained variance in concentration for other nutrients. The correlation between plasma 25-hydroxyvitamin D concentrations and the relative abundance of VDBP was absent when using the pooled sample reference for normalization (Fig. 2B) but became evident under the LME model (Fig. 2C), increasing the explained variance in nutrient concentration from 4 to 34%. Marked improvements were also observed when assessing the relation of plasma α-tocopherol and apo C-III (increasing explained variance from 20 to 41%) (Fig. 3B,C), plasma copper and Cp (increasing explained variance from 31 to 42%) (Fig. 4B,C), and plasma selenium and SEPP1 (increasing explained variance from 39 to 63%) (Fig. 5B,C). All LME-based associations were observed at P values ranging from 9.9 × 10−5 to 4.6 × 10−220 and q values ranging from 2.1 × 10−29 to 5.9 × 10−217 for 4 of 5 comparisons, with one (the vitamin D dyad) having a FDR (q) of 0.026 (Table 1).
FIGURE 2.
Plasma 25-hydroxyvitamin D and VDBP relative abundance distributions in Nepalese children 6–8 y of age (n = 500). (A) Frequency distribution of 25-hydroxyvitamin D concentrations: range, 18.6–173.5 nmol/L, 19.2% (n = 96) deficient (<50 nmol/L, dark gray), and 80.8% (n = 404, medium gray) adequate (≥50 nmol/L) in status. (B,C) Plasma 25-hydroxyvitamin D by relative abundance of VDBP by traditional master plasma pool normalization and LME-adjusted methods, respectively (see Fig. 1 for details). LME, linear mixed effects (model); VDBP, vitamin D binding protein; 25(OH)D, 25-hydroxyvitamin D.
FIGURE 3.
Plasma α-tocopherol and Apo C-III relative abundance distributions in Nepalese children 6–8 y of age (n = 500). (A) Frequency distribution of α-tocopherol concentrations: range, 4.1–26.9 μmol/L, 17.6% (n = 88) deficient (<9.3 μmol/L, dark gray), 37.4% (n = 187) marginal (9.3 to <12 μmol/L, medium gray), and 45% (n = 225) adequate (≥12 μmol/L, light gray) in status. (B,C) Plasma α-tocopherol by relative abundance of Apo C-III by traditional master plasma pool normalization and LME-adjusted methods, respectively (see Fig. 1 for details). LME, linear mixed effects (model).
FIGURE 4.
Plasma copper and Cp relative abundance distributions in Nepalese children 6–8 y of age (n = 494). (A) Plasma copper concentrations: range, 11.6–35.8 μmol/L, 100% were adequate (>10 μmol/L, gray). Six implausible values (4 <5 μmol/L, and 1 each at 62.3 μmol/L and 100.5 μmol/L) were removed from this analysis. (B,C) Plasma copper by relative abundance of Cp by traditional master plasma pool normalization and LME-adjusted methods, respectively (see Fig. 1 for details). Cp, ceruloplasmin; LME, linear mixed effects (model).
TABLE 1.
Individual and combined estimates of association between plasma micronutrient concentrations derived by conventional assays and protein relative abundance derived by iTRAQ MS and linear mixed effects models in Nepalese children 6–8 y of age (n = 500)
| Micronutrient/candidate protein1 (accession no.) | Samples | Nutrient:protein association2 | ||||
|---|---|---|---|---|---|---|
| r | b1 | P | q | LME3R2 | ||
| n | % | |||||
| Retinol | ||||||
| RBP4 (gi55743122) | 500 | 0.88 | 0.83 | 4.6 × 10−220 | 5.9 × 10−217 | 79 |
| Complement C1r (gi66347875) | 500 | −0.49 | −0.33 | 5.6 × 10−05 | 1.2 × 10−03 | |
| 25-hydroxyvitamin D | ||||||
| VDBP (gi32483410) | 500 | 0.58 | 25.6 | 9.9 × 10−05 | 0.026 | 48 |
| Plexin-D1 (gi157694524) | 117 | 0.69 | 44.2 | 3.6 × 10−06 | 0.0056 | |
| α-Tocopherol | ||||||
| Apo C-III (gi4557323) | 500 | 0.64 | 36.6 | 1.4 × 10−32 | 2.1 × 10−29 | 65 |
| RGS8 (gi156416024) | 56 | −0.64 | −9.0 | 1.0 × 10−03 | 2.4 × 10−02 | |
| Copper | ||||||
| Cp (gi4557485) | 494 | 0.65 | 16.1 | 6.3 × 10−52 | 7.5 × 10−49 | 61 |
| CDC42BPKαA (gi30089960) | 143 | 0.70 | 14.4 | 4.0 × 10−22 | 9.5 × 10−20 | |
| Selenium | ||||||
| SEPP1 (gi62530391) | 499 | 0.79 | 106.9 | 3.5 × 10−79 | 5.7 × 10−76 | 64 |
| GPx-3 (gi6006001) | 499 | 0.60 | 30.3 | 7.7 × 10−06 | 4.2 × 10−03 | |
For each model, the first protein was chosen based on biological information and the second protein identified as a covariate that maximized the coefficient of determination in a multivariate LME model (LME R2) for each plasma nutrient (dependent variable). CDC42BPKαA, CDC42-binding protein kinase alpha-isoform A; Cp, ceruloplasmin; FDR, false discovery rate; GPx-3, glutathione peroxidase-3; iTRAQ, isobaric tags for relative and absolute quantification; LME, linear mixed effects (model); RBP4, retinol binding protein isoform 4; RGS8, regulator of G-protein signaling 8 (isoform 2); SEPP1, selenoprotein P isoform 1; VDBP, vitamin D binding protein.
Association between the nutrient and single-protein LME fitted values from the fixed effects hypothesis tests (26): r, the nutrient:protein correlation; b1, the slope of the nutrient-protein association, with b1 representing the change in nutrient concentration [for retinol, 25-hydroxyvitamin D, copper] or percent change (for log-transformed nutrients α-tocopherol and selenium) per 2-fold change in protein relative abundance; P value for the null hypothesis that b1 = 0; q values, FDRs.
Variance in nutrient distribution (R2) explained by fitted values from a 2-protein covariate linear mixed effects model (25).
Linear mixed effects model estimation.
The above analysis confirms that MS measurement of relative abundance can generate high correlations between expected plasma nutrient:protein dyads. There were also an additional 3–108 proteins from among the 982 quantified in >10% of all subjects (Supplemental Table 1) that substantially correlated (q < 0.05) (33) with plasma concentrations of each nutrient. Most proteins, however, were measured in fewer than 500 children (data not shown). This missingness, inherent in tandem MS-generated data, limits the ability to construct multivariable models without imputation. Still, to explore the predictive potential with the primary protein entered, we modeled one additional, substantially correlated protein from each nutrient-specific protein cluster that explained the most additional variability in nutrient concentration. With vitamin A, we obtained relative abundance estimates for complement C1r in all 500 samples, a protein that was negatively associated with plasma retinol (Table 1) but not correlated with RBP4 (r = 0.04), and thus potentially added information, independent of RBP4, about plasma retinol concentration. Including both RBP4 and complement C1r in a LME model explained 79% (vs. 77% with RPB4 alone) of the variability in the plasma retinol concentration (Fig. 1C).
Plexin-D1, a protein associated with plasma 25-hydroxyvitamin D (Table 1), was measured in only 117 of 500 samples. While also correlated with VDBP (r = 0.69), plexin-D1 still provided sufficient additional information about the plasma concentration of 25-hydroxyvitamin D to raise the explained variance in vitamin D from 34 to 48% in the LME model (Fig. 2C). For vitamin E, we measured relative abundance of the regulator of G-protein signaling 8 isoform 2 in 56 of 500 samples, a protein negatively correlated with α-tocopherol and weakly correlated with apo C-III (r = 0.12) (Table 1). Modeling both proteins, 65% of the variability in α-tocopherol concentration was explained compared with 41% achieved by apo C-III alone. For copper, CDC42-binding protein kinase alpha-isoform A (CDC42BPKαA) was observed in 143 samples. Strong in its marginal association with plasma copper and Cp (r = 0.69), CDC42BPKαA modeled with Cp increased the explained variation in plasma copper concentration from 42 to 61% (Table 1). Finally, the relative abundance of glutathione peroxidase-3 (GPx-3) observed in 499 samples was highly correlated with plasma selenium but weakly correlated with SEPP1 (r = 0.19) (Table 1). Modeled, these proteins together explained 64% of the variability in plasma selenium concentration, representing a small but substantial increase over 63% obtained with SEPP1 alone.
Discussion
This study offers credible evidence of correlation between plasma distributions of proteins, measured by quantitative proteomics, and micronutrient ligands, measured by conventional assays, in an undernourished Nepalese child population. The strength and expected directions of association observed between 3 vitamins (A, D, and E) and 2 minerals (copper and selenium) and their cognate plasma proteins, with explained variation reaching 34–77%, suggests that a nutrient-linked plasma proteome can be detected by MS. Establishing this proof of concept further suggests that comparably strong, but less well-understood nutrient:protein correlations, are likely to reflect metabolic networks with functional biomarkers that can also reflect plasma micronutrient concentrations. In this regard, we identified for each nutrient a second protein that, when entered into a linear mixed effects model (25), added important, independent information about plasma nutrient variability.
Our analysis revealed expected and novel nutrient:protein associations. With respect to vitamin A, we identified a strong correlation (r = 0.88) with RBP4, its cognate plasma protein. On release from hepatic stores, retinol circulates in an equimolar complex with RBP4 and a larger protein, transthyretin, which delivers vitamin A to peripheral tissues for cellular uptake (18). The observed correlation between plasma retinol and RPB4 was found to lie within an often-reported range of 0.62–0.93 (34), explaining about three-fourths of the variance in retinol concentration. The remaining, unexplained variation could in part reflect lack of specificity, because RBP4 also circulates as an apo-protein when lacking its ligand and may further participate in energy regulatory pathways apart from its association with vitamin A (35). In our statistical model, we found complement C1r, a protease involved in initiating the classical complement cascade (36) and negatively correlated with plasma vitamin A and RBP4, adding independent information and raising the explained variance in plasma retinol to nearly 80%, a level considered adequate for population prediction.
Vitamin D status was measured by an immunoassay method that captures total 25-hydroxyvitamin D, a conventional biomarker of vitamin D intake and photoproduction, and the major ligand for VDBP. Although strongly correlated with VDBP (r = 0.56), the relatively low observed variation in plasma vitamin 25-hydroxyvitamin D explained by VDBP (34%) may be because VDBP circulates in concentrations 100-fold >25-hydroxyvitamin D, binds to other vitamin D metabolites, and has many non-vitamin D-related functions such as actin scavenging and fatty acid binding (37). Our findings demonstrate a need to find other vitamin D-networked proteins to increase explained variance and strengthen the potential to predict vitamin D status. The glycoprotein plexin-D1 entered our model, raising explained variance to 48%. Interestingly, although it was observed in only 23% of samples, plexin-D1 exhibited a stronger correlation with 25-hydroxyvitamin D than did VDBP (Table 1). Plexin-D1 is a member of transmembrane surface receptors that transduce pleiotropic signals of semaphorins, widely involved in genesis and maintenance of neural, vascular, immune, and osteoid tissues (38–40). Metabolic linkages between plexin-D1 and vitamin D have not been established but are plausible given the roles of both plexins and vitamin D metabolites in skeletal (39–41), immune (39, 42, 43), angiogenic, and vascular (39, 44–46) development and homeostasis.
Vitamin E, a major lipid-soluble membrane and lipoprotein antioxidant protectant, has no specific plasma carrier protein. Rather, following absorption, different forms of vitamin E are released into circulation associated with chylomicrons, redistributed to other plasma lipoproteins and tissues, and delivered to the liver (47). Hepatic α-tocopherol reenters circulation initially associated with VLDL prior to being redistributed to other low- to intermediate-density lipoproteins (47) for transport to the periphery. Strong correlations were expected and found between plasma α-tocopherol and apolipoproteins, especially with apo C-III (r = 0.62), which is a principal component of VLDL (48), and explained 41% of the vitamin's variability in plasma. In our exploratory regression analysis, the regulator of G-protein signaling 8 (RGS8) protein, although evident in only 11% of specimens, was sufficiently strong in its positive, independent association with vitamin E to raise the explained variance to 65%. Although no direct link with vitamin E has been identified, RGS8 is a cytosolic protein that modulates neuronal G-protein signaling in myelinated, lipid-rich regions of the brain (49, 50), where α-tocopherol–dependent lipid redox homeostasis is likely critical for maintaining the stability, structure, and function of transduction proteins.
Analysis of the mineral:protein dyads revealed additional facets of a plasma nutriproteome. Copper is a transition metal ubiquitously involved in gene transcription, cellular respiration, and enzyme activation whose deficiency impairs neural and immune function (21). Its plasma concentration is considered to poorly reflect individual hepatic or total body copper nutriture (21). However, the distribution of the plasma copper concentration has been shown to respond to copper supplementation and may reflect population status (51). Although copper binds to numerous intracellular and extracellular proteins, up to 95% of its plasma content is bound to Cp, a largely hepatic-derived, acute-phase reactant and ferroxidase that regulates iron metabolism and homeostasis (52, 53). A strong association was expected and found (r = 0.65) between plasma copper concentration and relative abundance of Cp, explaining 42% of the mineral's variance. However, an unexpected protein, the Ras-subfamily member CDC42BPKαA, next entered the regression model, increasing the explained variance in plasma copper to 61% and reflecting predictive potential. Although a specific role for copper in the metabolism of CDC42BPKαA has not been elucidated, upstream copper influx across the cell plasma membrane is known to activate Ras and mitogen-activated protein kinase signaling within the cytoplasm of the cell (54), suggesting a metabolic basis for the existence and direction of the observed correlation.
SEPP1, a glycoprotein expressed and secreted largely from the liver, comprises the major circulatory protein that delivers selenium to tissues throughout the body (22). In humans, circulating SEPP1 has been shown to decrease in response to selenium deficiency (55) and respond to selenium supplementation (32). In animals, experimental deletion of the SEPP1 gene increases whole-body selenium excretion (56). Thus, a strong association, confirmed by an r = 0.79 and explained variance of 63%, was anticipated with this dyad. Residual, unexplained variation in plasma selenium may be reflecting varied strengths of its binding with SEPP1 isoforms and other plasma proteins, including albumin and glutathione peroxidases (57). GPx-3, a seleno-enzyme synthesized in the kidney that circulates in plasma (58), emerged as the second most informative protein following SEPP1, building a model that explained 64% of the variance in plasma selenium. GPx-3 is also considered a protein biomarker of selenium status (32, 56), possibly explaining the small increase in fit following its introduction into the model.
The quantitative proteomics and computational methods employed in this study were well suited for protein discovery and assessing linear associations between plasma nutrient concentrations and protein abundance. The observed correlations, markedly higher than those based on a conventional master pool approach to normalization, were obtained by utilizing LME models that incorporate nutrient status information into each correlation estimate (25). However, limitations remain to be solved before a proteomic approach can be applied to reliably predict multiple micronutrient status. For example, although 982 proteins were identified in >10% of subjects (Supplemental Table 1), missing data were common within iTRAQ runs such that only 146 proteins were observed in all 500 children. Missingness can be expected when assessing protein abundance by data-dependent tandem MS (59), a phenomenon that affects more proteins as the number of samples under evaluation increases. The resulting incomplete database for proteins of potential interest limited our ability to use multivariate analyses to explore nutrient status prediction, restricting present models to 2 protein covariates. Imputation (60) or likelihood-based methods (61) applied to missing proteomic data can be expected to markedly increase available proteins for estimation of nutrient status. These statistical techniques will be employed for more extensive, individual, nutrient-specific proteomic analyses in the future.
Notably, whereas nearly one-half (46%) of the 982 proteins presented in Supplemental Table 1 have been classified as extracellular, secretory, membrane, or lipoprotein associated, others, including the second proteins added to our current models, are not typically considered plasma proteins but are frequently observed in plasma proteomic studies. More than a decade ago, Anderson and Anderson (62) estimated that the plasma proteome consists of more than a half-million proteins in multitudes of isoforms and other variants, including proteins involved in transport, leakage, and cell turnover. Recently, Farrah et al. (13) constructed a high-confidence human plasma proteome reference set with estimated concentrations using raw MS data from several large-scale studies, reporting 1929 proteins identified with a 1% FDR threshold. Their list similarly contains transcriptional-regulating proteins, RNA-processing proteins, cell growth-related proteins, histone-related proteins, IL-related proteins, methyltransferases, nuclear pore complex proteins, and upstream element binding proteins, as was observed in the present study. The vast majority of these proteins are classically thought to be restricted to the intracellular compartments rather than secreted into plasma. The degree to which identified proteins may be due to normal homodynamics, tissue growth, and other developmental, disease, or sample collection processes is an important issue to explore. Notwithstanding, highly substantial, strong nutrient:protein correlations in either direction can be considered evidence of cellular processes that covary with micronutrient nutriture. Whether their presence is a reflection of cause, effect, or an indirect association does not detract from the protein being a potential marker of population micronutrient status.
We have reported in this study evidence of a strong correlation between plasma concentrations of micronutrients and their proteomics-derived, cognate plasma protein biomarkers. Although expected, we suggest that these validating associations may strengthen confidence in other, metabolically less direct and understood but precisely estimated plasma nutrient:protein pairs revealed by proteomics, illustrated by several second proteins added to our models. We expect that micronutrients lacking bound plasma proteins may have less recognizable, but nonetheless valid, correlated protein partners, which we are currently exploring. These findings from a large population sample of Nepalese children suggest that quantitative plasma proteomics may provide a new basis for identifying functional biomarkers that will eventually improve our ability to assess micronutrient status and deficiencies in populations.
Supplementary Material
Proteins Quantified by iTRAQ Mass Spectrometry in More than Ten Percent of Plasma Samples (n > 50) from 500 Children, 6–8 y of Age, Sarlahi, Nepal.
Acknowledgments
The Johns Hopkins Nutriproteomics Research Team, in addition to coauthors, includes Shahed Ahmed, Margia Arguello, Joshua Betz, Hongie Cui, Jaime Johnson, Subarna K. Khatry, Alain Labrique, Ashika Nanayakkara-Bind, and Fredrick Van Dyke. We thank C. Conover Talbot Jr for assistance with the HUGO gene annotation. R.N.C., I.R., K.S., P.C., J.D.Y., J.G., and K.P.W. designed research; R.N.C., I.R., K.S., P.C., and K.P.W. performed research; R.N.C., I.R., K.S., L.R.D., R.N.O., S.S., and T.N.B. contributed new reagents or analytic tools; I.R., S.H. and L.W. analyzed data; P.C., L.W., and K.P.W. conducted the original field study; R.N.C., I.R., K.S., J.G., and K.P.W. wrote the paper; and K.P.W. had primary responsibility for final content. All authors read and approved the final manuscript.
Abbreviations
- CDC42BPKαA
CDC42-binding protein kinase alpha-isoform A
- Cp
ceruloplasmin
- FDR
false discovery rate
- GPx-3
glutathione peroxidase-3
- iTRAQ
isobaric tags for relative and absolute quantification
- LME
linear mixed effects (model)
- RBP4
retinol binding protein isoform 4
- RGS8
regulator of G-protein signaling 8 (isoform 2)
- SEPP1
selenoprotein P isoform 1
- VDBP
vitamin D binding protein
Footnotes
Supported by the “Assessment of Micronutrient Status by Nutriproteomics” grant (GH 5241) from the Bill and Melinda Gates Foundation, Seattle, WA (plasma nutriproteomics study). The cohort study in Nepal fromwhich plasma sampleswere obtained was supported by the “Global Control of Micronutrient Deficiency” grant (GH 614) fromthe Bill andMelinda Gates Foundation. The original field trial in Nepal in 1999-2001 in which mothers of studied children were enrolled was supported by a “Micronutrients for Health” Cooperative Agreement (no. HRN-A-00-97-00015-00) between the Office of Health, Infectious Diseases and Nutrition, U.S. Agency for International Development,Washington, DC, and Center for Human Nutrition, Johns Hopkins Bloomberg School of Public Health, Baltimore,MD, and the Bill andMelinda Gates Foundation (GH 614). Additional assistance was received from the Sight and Life Research Institute, Baltimore, MD.
Literature Cited
- 1. Arlappa N, Laxmaiah A, Balakrishna N, Harikumar R, Kodavanti MR. Gal Reddy Ch, Saradkumar S, Ravindranath M, Brahmam GN. Micronutrient deficiency disorders among the rural children of West Bengal, India. Ann Hum Biol. 2011;38:281–9. [DOI] [PubMed] [Google Scholar]
- 2. Pasricha SR, Shet AS, Black JF, Sudarshan H, Prashanth NS, Biggs BA. Vitamin B-12, folate, iron, and vitamin A concentrations in rural Indian children are associated with continued breastfeeding, complementary diet, and maternal nutrition. Am J Clin Nutr. 2011;94:1358–70. [DOI] [PubMed] [Google Scholar]
- 3. Jiang T, Christian P, Khatry SK, Wu L, West KP Jr. Micronutrient deficiencies in early pregnancy are common, concurrent, and vary by season among rural Nepali pregnant women. J Nutr. 2005;135:1106–12. [DOI] [PubMed] [Google Scholar]
- 4. West KP Jr, Pokhrel RP, Katz J, LeClerq SC, Khatry SK, Shrestha SR, Pradhan EK, Tielsch JM, Pandey MR, Sommer A. Efficacy of vitamin A in reducing preschool child mortality in Nepal. Lancet. 1991;338:67–71. [DOI] [PubMed] [Google Scholar]
- 5. Tielsch JM, Khatry SK, Stoltzfus RJ, Katz J, LeClerq SC, Adhikari R, Mullany LC, Black R, Shresta S. Effect of daily zinc supplementation on child mortality in southern Nepal: a community-based, cluster randomised, placebo-controlled trial. Lancet. 2007;370:1230–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christian P, Khatry SK, Katz J, Pradhan EK, LeClerq SC, Shrestha SR, Adhikari RK, Sommer A, West KP Jr. Effects of alternative maternal micronutrient supplements on low birth weight in rural Nepal. A double-masked randomized community trial. BMJ. 2003;326:571–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Christian P, Murray-Kolb LE, Khatry SK, Katz J, Schaefer BA, Cole PM, LeClerq SC, Tielsch JM. Prenatal micronutrient supplementation and intellectual and motor function in early school-aged children in Nepal. JAMA. 2010;304:2716–23. [DOI] [PubMed] [Google Scholar]
- 8. Christian P, Stewart CP, LeClerq SC, Wu L, Katz J, West KP Jr, Khatry SK. Antenatal and postnatal iron supplementation and childhood mortality in rural Nepal: a prospective follow-up in a randomized, controlled community trial. Am J Epidemiol. 2009;170:1127–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Raiten DJ, Namasté S, Brabin B, Combs G Jr, L'Abbe MR, Wasantwisut E, Darnton-Hill I. Executive summary: biomarkers of nutrition for development: building a consensus. Am J Clin Nutr. 2011;94:S633–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Erhardt JG, Estes JE, Pfeiffer CM, Biesalski HK, Craft NE. Combined measurement of ferritin, soluble transferrin receptor, retinol binding protein, and C-reactive protein by an inexpensive, sensitive, and simple sandwich enzyme-linked immunosorbent assay technique. J Nutr. 2004;134:3127–32. [DOI] [PubMed] [Google Scholar]
- 11. Bechir M, Schelling E, Kraemer K, Schweigert F, Bonfoh B, Crump L, Tanner M, Zinsstag J. Retinol assessment among women and children in Sahelian mobile pastoralists. EcoHealth. 2012;9:113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pierce A, Unwin RD, Evans CA, Griffiths S, Carney L, Zhang L, Jaworska E, Lee C-F, Blinco D, Okoniewski MJ, et al. Eight-channel iTRAQ enables comparison of the activity of six leukemogenic tyrosine kinases. Mol Cell Proteomics. 2008;7:853–63. [DOI] [PubMed] [Google Scholar]
- 13. Farrah T, Deitsch EW, Omenn GS, Campbell DS, Sun Z, Bletz JA, Mallick P, Katz JE, Malmstrom J, Ossola R, et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol Cell Proteomics. 2011;10:M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barnes S, Kim H. Nutriproteomics: identifying the molecular targets of nutritive and non-nutritive components of the diet. J Biochem Mol Biol. 2004;37:59–74. [DOI] [PubMed] [Google Scholar]
- 15. Schweigert FJ. Nutritional proteomics: methods and concepts for research in nutritional science. Ann Nutr Metab. 2007;51:99–107. [DOI] [PubMed] [Google Scholar]
- 16. Zhang X, Yap Y, Wei D, Chen G, Chen F. Novel omics technologies in nutrition research. Biotechnol Adv. 2008;26:169–76. [DOI] [PubMed] [Google Scholar]
- 17. Sénéchal S, Kussmann M. Nutriproteomics: technologies and applications for identification and quantification of biomarkers and ingredients. Proc Nutr Soc. 2011;70:351–64. [DOI] [PubMed] [Google Scholar]
- 18. Quadro L, Hamburger L, Colantuoni V, Gottesman ME, Blaner WS. Understanding the physiological role of retinol-binding protein in vitamin A metabolism using transgenic and knockout mouse models. Mol Aspects Med. 2003;24:421–30. [DOI] [PubMed] [Google Scholar]
- 19. Christakos S, Ajibade DV, Dhawan P, Fechner AJ, Mady LJ. Vitamin D: metabolism. Endocrinol Metab Clin North Am. 2010;39:243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Borel P, Moussa M, Reboul E, Lyan B, Defoort C, Vincent-Baudry S, Maillot M, Gastaldi M, Darmon M, Portugal H, et al. Human fasting plasma concentrations of vitamin E and carotenoids, and their association with genetic variants in apo C–III, cholesteryl ester transfer protein, hepatic lipase, intestinal fatty acid binding protein and microsomal triacylglycerol transfer protein. Br J Nutr. 2009;101:680–7. [DOI] [PubMed] [Google Scholar]
- 21. Danzeisen R, Araya M, Harrison B, Keen C, Solioz M, Thiele D, McArdle HJ. How reliable and robust are current biomarkers for copper status? Br J Nutr. 2007;98:676–83. [DOI] [PubMed] [Google Scholar]
- 22. Burk RF, Hill KE. Selenoprotein P-expression, functions, and roles in mammals. Biochim Biophys Acta. 2009;1790:1441–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stewart CP, Christian P, Schulze KJ, LeClerq SC, West KP Jr, Khatry SK. Antenatal micronutrient supplementation reduces metabolic syndrome in 6- to 8-year-old children in rural Nepal. J Nutr. 2009;139:1575–81. [DOI] [PubMed] [Google Scholar]
- 24. WHO Growth reference data for 5–19 years. Geneva: WHO; 2007. [cited 2012 Oct 11]. Available from: http://www.who.int/growthref/en/.
- 25. Herbrich SM, Cole RN, West KP Jr, Schulze K, Yager JD, Groopman JD, Christian P, Wu L, O'Meally RN, May DH, et al. Statistical inference from multiple iTRAQ experiments without using common reference standards. J Proteome Res. 2013;12:594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson GK. That BLUP is a good thing: the estimation of random effects. Stat Sci. 1991;6:15–32. [Google Scholar]
- 27. The Comprehensive R Archive Network Available from: http://cran.r-project.org/.
- 28. West KP., Jr Extent of vitamin A deficiency among preschool children and women of reproductive age. J Nutr. 2002;132:S2857–66. [DOI] [PubMed] [Google Scholar]
- 29. Greer FR. 25-hydroxyitamin D: functional outcomes in infants and young children. Am J Clin Nutr. 2008;88:S529–33. [DOI] [PubMed] [Google Scholar]
- 30. Institute of Medicine Dietary reference intakes for vitamin C, vitamin E, selenium, and carotenoids. Washington: National Academy Press; 2000. p. 186–283. [PubMed] [Google Scholar]
- 31. Harvey LJ, Ashton K, Hooper L, Casgrain A, Fairweather-Tait SJ. Methods of assessment of copper status in humans: a systematic review. Am J Clin Nutr. 2009;89:S2009–24. [DOI] [PubMed] [Google Scholar]
- 32. Ashton K, Hooper L, Harvey LJ, Hurst R, Casgrain A, Fairweather-Tait SJ. Methods of assessment of selenium status in humans: a systematic review. Am J Clin Nutr. 2009;89:S2025–39. [DOI] [PubMed] [Google Scholar]
- 33. Storey JD. A direct approach to false discovery rates. JRSS-B. 2002;64:479–98. [Google Scholar]
- 34. de Pee S, Dary O. Biochemical indicators of vitamin A deficiency: serum retinol and serum retinol binding protein. J Nutr. 2002;132:S2895–901. [DOI] [PubMed] [Google Scholar]
- 35. Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, Kotani K, Quadro L, Kahn BB. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436:356–62. [DOI] [PubMed] [Google Scholar]
- 36. Mayilyan KR. Complement genetics, deficiencies, and disease associations. Protein Cell. 2012;3:487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Speeckaert M, Huang G, Delanghe JR, Taes YEC. Biological and clinical aspects of the vitamin D binding protein (Gc-globulin) and its polymorphism. Clin Chim Acta. 2006;372:33–42. [DOI] [PubMed] [Google Scholar]
- 38. Takamatsu H, Kumanogoh A. Diverse roles for semaphorin-plexin signaling in the immune system. Trends Immunol. 2012;33:127–35. [DOI] [PubMed] [Google Scholar]
- 39. Kang S, Kumanogoh A. Semaphorins in bone development, homeostasis, and disease. Semin Cell Dev Biol. 2013;24:163–71. [DOI] [PubMed] [Google Scholar]
- 40. Sutton AL, Zhang X, Dowd DR, Kharode YP, Komm BS, Macdonald PN. Semaphorin 3B is a 1,25-dihydroxyvitamin D3-induced gene in osteoclasts that promotes osteoclastogenesis and induces osteopenia in mice. Mol Endocrinol. 2008;22:1370–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fischer PR, Thacher TD, Pettifor JM. Pediatric vitamin D and calcium nutrition in developing countries. Rev Endocr Metab Disord. 2008;9:181–92. [DOI] [PubMed] [Google Scholar]
- 42. Holl EK, O'Connor BP, Holl TM, Roney KE, Zimmermann AG, Jha S, Kelsoe G, Ting JP. Plexin-D1 is a novel regulator of germinal centers and humoral immune responses. J Immunol. 2011;186:5603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peelen E, Knippenberg S, Muris AH, Thewissen M, Smolders J, Tervaert JW, Hupperts R, Damoiseaux J. Effects of vitamin D on the peripheral adaptive immune system: a review. Autoimmun Rev. 2011;10:733–43. [DOI] [PubMed] [Google Scholar]
- 44. Serini G, Valdembri D, Zanivan S, Morterra G, Burkhardt C, Caccavari F, Zammataro L, Primo L, Tamagnone L. Class 3 semaphorins control vascular morphogenesis by inhibiting integrin function. Nature. 2003;424:391–7. [DOI] [PubMed] [Google Scholar]
- 45. Gu C, Yoshida Y, Livet J, Reimert DV, Mann F, Merte J, Henderson CE, Jessell TM, Kolodkin AL, Ginty DD. Semaphorin 3E and plexin-D1 control vascular pattern independently of neuropilins. Science. 2005;307:265–8. [DOI] [PubMed] [Google Scholar]
- 46. Garcia LA, Ferrini MG, Norris KC, Artaza JN. 1,25(OH)2vitamin D3 enhances myogenic differentiation by modulating the expression of key angiogenic growth factors and angiogenic inhibitors in CC skeletal muscle cells. J Steroid Biochem Mol Biol. 2013;133:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morrissey PA, Kiely M. Vitamin E/physiology and health effects. In: Caballero B, Allen L, Prentice A eds. Encyclopedia of human nutrition. 2nd ed Amsterdam: Elsevier Academic Press; 2005. p. 389–98. [Google Scholar]
- 48. Mendivil CO, Zheng C, Furtado J, Lel J, Sacks FM. Metabolism of VLDL and LDL containing apolipoprotein C–III and not other small apolipoproteins-R2. Arterioscler Thromb Vasc Biol. 2010;30:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saitoh O, Yoshihiro K. Biochemical and electrophysiological analyses of RGS8 function. Methods Enzymol. 2004;390:129–48. [DOI] [PubMed] [Google Scholar]
- 50. Miyamoto-Matsubara M, Saitoh O, Maruyama K, Aizaki Y, Saito Y. Regulation of melanin-concentrating hormone receptor 1 signaling by RGS8 with the receptor third intracellular loop. Cell Signal. 2008;20:2084–94. [DOI] [PubMed] [Google Scholar]
- 51. Harvey LJ, McArdle HJ. Biomarkers of copper status: a brief update. Br J Nutr. 2008;99:S10–3. [DOI] [PubMed] [Google Scholar]
- 52. Hellman NE, Gitlin JD. Ceruloplasmin metabolism and function. Annu Rev Nutr. 2002;22:439–58. [DOI] [PubMed] [Google Scholar]
- 53. Meyer LA, Durley AP, Prohaska JR, Harris ZL. Copper transport and metabolism are normal in aceruloplasminemic mice. J Biol Chem. 2001;276:36857–61. [DOI] [PubMed] [Google Scholar]
- 54. Tsai C-Y, Finley JC, Ali SS, Patel HH, Howell SB. Copper influx transporter 1 is required for FGF, PDGF and EGF-induced MAPK signaling. Biochem Pharmacol. 2012;84:1007–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang JG, Hill KE, Burk RF. Dietary selenium intake controls rat plasma selenoprotein P concentration. J Nutr. 1989;119:1010–2. [DOI] [PubMed] [Google Scholar]
- 56. Burk RF, Hill KE, Motley AK, Austin LM, Norsworthy BK. Deletion of selenoprotein P upregulates urinary selenium excretion and depresses whole-body selenium content. Biochim Biophys Acta 2006;1760:1789–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Deagen JT, Butler JA, Zachara BA, Whanger PD. Determination of the distribution of selenium between glutathione peroxidase, selenoprotein P, and albumin in plasma. Anal Biochem. 1993;208:176–81. [DOI] [PubMed] [Google Scholar]
- 58. Burk RF, Olsen GE, Winfrey VP, Hill KE, Yin D. Glutathione peroxidase-3 produced by the kidney binds to population basement membranes in the gastrointestinal tract and other tissues. Am J Physiol Gastrointest Liver Physiol. 2011;301:G32–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang P, Tang H, Zhang H, Whiteaker J, Paulovich AG, McIntosh M. Normalization regarding non-random missing values in high-throughput mass spectrometry data. Pac Symp Biocomput. 2006;315–26. [PubMed] [Google Scholar]
- 60. Rubin DB. Multiple imputation after 18+ years (with discussion). JASA. 1996;91:473–89. [Google Scholar]
- 61. Little RJA, An H. Robust likelihood-based analysis of multivariate data with missing values. Statist Sinica. 2004;14:949–68. [Google Scholar]
- 62. Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteins Quantified by iTRAQ Mass Spectrometry in More than Ten Percent of Plasma Samples (n > 50) from 500 Children, 6–8 y of Age, Sarlahi, Nepal.