Abstract
Background: Nicotinamide riboside (NR) is a recently discovered NAD+ precursor vitamin with a unique biosynthetic pathway. Although the presence of NR in cow milk has been known for more than a decade, the concentration of NR with respect to the other NAD+ precursors was unknown.
Objective: We aimed to determine NAD+ precursor vitamin concentration in raw samples of milk from individual cows and from commercially available cow milk.
Methods: LC tandem mass spectrometry and isotope dilution technologies were used to quantify NAD+ precursor vitamin concentration and to measure NR stability in raw and commercial milk. Nuclear magnetic resonance (NMR) spectroscopy was used to test for NR binding to substances in milk.
Results: Cow milk typically contained ∼12 μmol NAD+ precursor vitamins/L, of which 60% was present as nicotinamide and 40% was present as NR. Nicotinic acid and other NAD+ metabolites were below the limits of detection. Milk from samples testing positive for Staphylococcus aureus contained lower concentrations of NR (Spearman ρ = −0.58, P = 0.014), and NR was degraded by S. aureus. Conventional milk contained more NR than milk sold as organic. Nonetheless, NR was stable in organic milk and exhibited an NMR spectrum consistent with association with a protein fraction in skim milk.
Conclusions: NR is a major NAD+ precursor vitamin in cow milk. Control of S. aureus may be important to preserve the NAD+ precursor vitamin concentration of milk.
Keywords: LC-MS, metabolomics, milk, nicotinamide adenine dinucleotide, pellagra-preventive factor
Introduction
One hundred years ago, pellagra was common in the rural American South. One of the early treatments for pellagra was consumption of a 1.5–2 pints of cow milk (1). In 1937, nicotinamide and nicotinic acid (NA)8 were identified as pellagra-preventive (PP) factors (2, 3), and tryptophan was subsequently discovered as a molecule with PP activity (4). Nicotinamide and NA, which are collectively termed niacin, contain a pyridine ring that can be salvaged to form NAD+ in 2 or 3 enzymatic steps, whereas tryptophan is the de novo precursor of NAD+, requiring 7 enzymatic steps (5). Largely because tryptophan can be incorporated into protein, oxidized as a fuel, and converted to many other metabolites such as serotonin, 50–60 mg tryptophan is considered the niacin equivalent of 1 mg nicotinamide or NA. In addition, much of the niacin equivalent in food is, in fact, NAD+ (6).
NAD+ and its phosphorylated and oxidized derivatives, NAD(P)+, NAD(H), and NAD(P)H, are essential hydride transfer cofactors in hundreds of oxidoreductase reactions and consumed substrates of several classes of enzymes with activities required for DNA repair, gene expression, regulation of energy metabolism, and calcium mobilization (7). NAD+ is one of the most abundant metabolites in the human body and is turned over at a rapid pace (8), requiring near-constant replenishment. Hence, although pellagra is described as niacin deficiency, at a cellular level, pellagra is a disease of NAD+ depletion as a result of diets deficient in NAD+ precursors.
It has long been known that the NAD+ precursors in milk include nicotinamide (9) and tryptophan (10). More recently, it has been discovered that milk also contains nicotinamide riboside (NR), another salvageable NAD+ precursor vitamin (11). Boosting NAD+ concentrations with NR extends life span in yeast (12) and has been shown to prevent and treat metabolic (13–16) and neurodegenerative (17, 18) conditions in mouse models. Although these studies suggest that dairy products as a source of NR could be beneficial to many aspects of human health, the amount of NR in milk has not been established. In this work, we determine the complete NAD+ precursor vitamin concentration in individual and pooled commercial samples of cow milk with the use of an LC tandem MS (LC-MS/MS)–based method (19). Our results show that milk from Staphylococcus aureus–positive samples contained lower concentrations of NR and nicotinamide, and that milk sold as organic milk contained lower concentrations of NR than conventionally sourced milk. Moreover, we show that NR is stable in milk and bound by substances in milk and that ~40% of the NAD+ precursor vitamin concentration of cow milk is present as NR.
Methods
Milk quality and herd health measurements.
Milk flows and representative samples were obtained from 19 conventionally raised cows with a Dairy Herd Improvement Association testing meter. Samples were dispensed into 2-oz snap-cap Dairy Herd Improvement Association vials containing liquid bronopol for analysis by Dairy Lab Services of fat, protein, lactose, other solids, milk urea nitrogen, and somatic cells (FOSS). Additional aseptic individual milk samples were obtained for bacterial analysis after teats were sterilized with 70% ethanol before collecting 3 mL milk from each teat into 12 × 75–mm culture tubes. All samples were frozen before further analysis. Blood agar culture plates were inoculated with sample and then incubated at 37°C and evaluated for bacterial growth at 24 and 48 h. Bacterial growth was characterized by morphology, and samples were subjected to confirmatory tests to identify genus and species.
Milk sample acquisition and preparations.
Nineteen milk samples from individual cows plus 8 skim milk samples (4 organic and 4 conventional) purchased in the Iowa City area were analyzed by LC-MS/MS. Two 50-μL aliquots were extracted from each milk sample. Each aliquot was dosed with either solution A [18.75 pmol [18O1]-NR, 18.75 pmol [18O1]-nicotinamide, 18.75 pmol [D3, 18O1]–1-methyl nicotinamide, and 150 pmol [D4]-NA] or solution B, a 13C-labeled yeast extract at 1:50 dilution. Aliquots were extracted with 0.5 mL of 1.5% formic acid at room temperature. Each aliquot was vortexed for 10 s and then sonicated for 10 min in a bath sonicator. Aliquots were then centrifuged at a speed of 16,100 × g for 10 min at room temperature. Extracts were transferred to fresh 1.5-mL centrifuge tubes and dried overnight via speed vacuum. Recovery was >90% for all metabolites of interest.
NMR spectroscopy.
NR 1H resonances were assigned with 1H/13C 2-dimensional heteronuclear multiple-quantum coherence and heteronuclear multiple-bond coherence experiments. NR binding to skim milk was analyzed with the use of water-ligand observed via gradient spectroscopy (WaterLOGSY) (20, 21). To analyze fractions of milk for NR-binding activity, 2 mL total milk was centrifuged for 1 h at 4°C at 16,100 × g. The supernatant was termed the soluble fraction, whereas the pellet resuspended in 2 mL 50 mmol/L sodium phosphate (pH 7) was termed the particulate fraction. NMR samples were prepared by adding 150 μL skim milk, skim milk soluble fraction, or resuspended skim milk particulate fraction to 352.2 μL buffer that contained 300 μL 50 mmol/L sodium phosphate (pH 7), 50 μL D2O, and 2.2 μL NR stock, giving a final NR concentration in the NMR samples of 0.3 mmol/L. For the WaterLOGSY experiment, a T2 relaxation filter of 100 ms was used just before data acquisition to suppress signals derived from macromolecules, and a water nuclear Overhauser effect mixing time of 1 s was used in the experiment. All NMR data were acquired with a Bruker Avance II 800-MHz NMR spectrometer equipped with a sensitive cryoprobe and recorded at 25°C. The 1H chemical shifts were referenced to 2,2-dimethyl-2-silapentane-5-sulfonate. NMR spectra were processed with the NMRPipe package software (22) and analyzed with NMRView (23).
NR stability assays.
[18O]-labeled and [13C1, D1]-NR were synthesized as described (24) (S Trammell, M Schmidt, B Weidemann, P Redpath, F Jaksch, R Dellinger, M Migaud, C Brenner, unpublished results, 2016). [18O]-NR was suspended in conventional and organic milk samples or in water at pH 5, 7, or 11 at a concentration of 10 μmol/L and allowed to sit at room temperature. In total, 20-μL aliquots were collected at 0, 0.5, 1, 2, 4, and 8 h and extracted as described above.
S. aureus growth experiments.
Strain RN3170 was a kind gift from Patrick Schlievert (University of Iowa) (25). Bacteria were streaked onto Todd-Hewitt (Becton & Dickinson) 2% agar plates, incubated overnight at 37°C, and then stored at 4°C. S. aureus was then inoculated into Todd-Hewitt media containing 50 mmol/L Bis-Tris (pH 6.7) and 10 μmol/L [13C1, D1]-NR at a starting OD600 nm of 0.1. Noninoculated medium was used as control for NR stability. All cultures were incubated at 37°C with constant shaking at 220 rpm, and 15-mL aliquots were collected at 0, 1, 2, 4, 6, and 8 h. OD600 nm and pH values were recorded at each time point. Aliquots were centrifuged at 2060 × g for 30 min at 4°C, at which time 1 mL culture medium was collected and snap-frozen in liquid nitrogen. The remainder of culture medium was aspirated and cell pellets were washed with 1 mL ice-cold PBS and centrifuged at 16,100 × g for 10 min at 4°C. PBS was aspirated and pellets were flash frozen. Then, 50 μL media was analyzed with LC-MS/MS as described below. Cells were extracted with buffered ethanol (3 parts ethanol to 1 part 10 mmol/L HEPES, pH 7.1) heated to 80°C for 3 min with constant shaking at 1050 rpm. Extracts were clarified by centrifugation (16,100 × g, 10 min, 4°C). Pellets were extracted again following the same procedure as above. Supernatants from both rounds of extraction were combined and dried via speed vacuum. Extracts were analyzed with LC-MS as described below.
LC-MS and LC-MS/MS.
Media samples were diluted 1:1 with double-distilled H2O. Standard solutions in double-distilled H2O were diluted 1:1 with noninoculated Todd-Hewitt media containing 50 mmol/L Bis-Tris and [13C1, D1]-NR, producing a standard curve with the final concentrations of 0, 0.1, 0.3, 0.5, 1, 3, 5, and 10 μmol/L. Quality control samples at a final concentration of 0.75 and 7.5 μmol/L were also prepared by diluting standard 1:1 with media. Then, 10 μL of media samples, quality controls, and standards was injected and quantified with a Waters TQD mass spectrometer with the use of the acid separation chromatographic conditions described previously (19). Media were quantified with raw peak areas and converted to micromoles per liter with background-subtracted standard curves.
For cow milk, standards (final concentrations of 0.08, 0.24, 0.8, 2.4, 8, 24, 80, and 120 μmol/L) and 2 quality control samples (final concentration of 2.5 and 25 μmol/L) were treated in the same manner as the samples and as described above. In total, 5 μL of samples, quality controls, and standards containing solution A or 10 μL of those containing solution B was loaded onto the column and quantified with a Waters TQD mass spectrometer according to the procedures previously described (19). Newly quantified metabolites in the acidic separation, N-methyl-nicotinamide, N-methyl-2-pyridone-5-carboxamide, and N-methyl-4-pyridone-5-carboxamide, were assayed with the following transitions: 1-methyl nicotinamide (m/z = 137 > 94), N-methyl-2-pyridone-5-carboxamide (m/z = 153 > 107), and N-methyl-4-pyridone-5-carboxamide (m/z = 153 > 136). Analyte peak areas were normalized to internal standard peak areas and converted to μmol/L with the standard curve. S. aureus cell pellets were suspended in 50 μL of 10 mmol/L ammonium acetate in LC-MS grade water. A260 nm values for each sample were measured with a Thermo Scientific 2000c Nanodrop spectrophotometer operated in nucleic acid mode. Samples at 0- and 1-h time points were diluted 1:1 with either solution A or solution B. Samples at all remaining time points were diluted to a final A260 nm value of 14 and then diluted 1:1 with solution A or B. All samples were analyzed according to the chromatography protocols previously described (19) and detected and quantified with a Waters Q-Tof Premier mass spectrometer operated in positive full-scan mode. The alkaline separation was altered by increasing the flow rate to 0.55 mL/min and shortening the runtime to 11.6 min. Separation was performed with the use of a modified gradient with initial equilibration at 3% B; a 0.9-min hold; a gradient to 50% B over 6.3 min, followed by a 1-min wash at 90% B; and a 3-min reequilibration at 3% B. When performing the alkaline separation, the scanning window was set to m/z = 120–800 with a scan rate of 0.1 and an interscan rate of 0.01. When performing the acid separation, the scanning window was set to m/z = 120–600 with a scan rate of 0.5 and an interscan delay of 0.05. In both cases, leucine enkephalin was infused and used for mass accuracy correction. Analyte peak areas were normalized to internal standard peak areas and converted to μmol/L with the standard curve. Nicotinamide concentrations were corrected for the contribution of [12C1]-nicotinamide and [13C1]-nicotinamide to the [18O1]-nicotinamide internal standard area counts. Enrichment for all metabolites was corrected for the natural abundance of the analyte, 13C abundance, and the purity of the doubly labeled NR (95/5% [13C1, D1]-NR/[13C1]-NR). The corrected concentrations of each analyte were converted to intracellular concentrations by calculating the total intracellular volume of S. aureus with an intracellular volume of 0.28 fL (26) and an assumption of 1 × 109 cells/mL per OD600 nm (27).
Statistical analysis.
Unless otherwise stated, all values are expressed as means ± SDs. Two-tailed, unpaired t tests were performed on all comparisons involving 2 groups. Outliers were identified with the robust regression and outlier removal method (28). Two-factor, repeated-measures ANOVA followed by Holm-Sidak's multiple-comparisons test was performed on experiments involving S. aureus. Media samples were compared with noninoculated medium within time points. Intracellular samples were compared with initial concentrations within condition. Spearman rank correlation coefficient was calculated for the concentration of nicotinamide and NR compared with the milk quality, herd health metrics, and breed. P < 0.05 was considered significant. Statistical analyses were performed with GraphPad Prism version 6.00 for Windows (GraphPad Software).
Results
NR is a major component of the NAD+ precursor vitamin concentration in cow milk.
The NAD+ metabolome of 19 individual cow milk samples was determined with LC-MS/MS and isotope dilution techniques. We define the NAD+ precursor vitamin concentration as the concentrations of salvageable NAD+ precursor (nicotinamide, NA, and NR) plus concentrations of the higher molecular weight species [nicotinic acid riboside, nicotinic acid mononucleotide, NAD+, nicotinic acid adenine dinucleotide, and NAD(P)+] from which a vitamin can be released by enzymatic or chemical decomposition. NAD(H) and NAD(P)H are oxidized in extraction, such that these metabolites, if present, would contribute to the peaks of NAD+ and NAD(P)+.
As shown in Table 1, in all 19 samples, nicotinamide and NR and no other NAD+ metabolite were quantifiable. Thus, neither NAD+ nor NA is a PP factor in milk. Excluding one unusual milk sample that contained 24 μmol nicotinamide/L and 27 μmol NR/L (Supplemental Table 1), milk samples contained 7.3 ± 1.5 μmol nicotinamide/L and 4.3 ± 2.6 μmol NR/L. To determine whether other parameters correlate with NAD+ precursor vitamin concentrations in the 18 remaining samples, breed was recorded and metrics of the health and milk quality of each cow were measured (Supplemental Table 2). As shown in Table 2, concentrations of NR positively correlated with concentrations of lactose (P = 0.013) and milk urea nitrogen (P = 0.018), whereas nicotinamide negatively correlated with somatic cell count (P = 0.029) and positively correlated with NR (P = 0.011). NR concentration negatively correlated with S. aureus infection (P = 0.014). Nicotinamide concentration also negatively correlated with S. aureus infection, but the correlation was not significant (P = 0.09). When we recalculated concentrations of NR and nicotinamide in the 12 samples without S. aureus infection or extremely high concentrations of NAD+ precursor vitamins, nicotinamide rose to 7.7 ± 1.2 μmol/L and NR rose to 5.1 ± 2.6 μmol/L. Thus, although it was clear from previous work that there is no NA in cow milk (9), there has been a substantial underreporting of NAD+ precursor vitamin on account of lack of an assay for NR.
TABLE 1.
Mean NAD+ metabolomes of 18 raw cow milk samples1
| Metabolome, μmol/L | Total (n = 18) | S. aureus negative (n = 12) | S. aureus positive (n = 6) |
|---|---|---|---|
| Nicotinamide | 7.3 ± 1.52 | 7.7 ± 1.2 | 6.4 ± 1.7 |
| NR | 4.3 ± 2.6 | 5.1 ± 2.6 | 2.7 ± 1.9 |
| NA | <1.0 | <1.0 | <1.0 |
| NMN | <0.4 | <0.4 | <0.4 |
| NAD+ | <0.08 | <0.08 | <0.08 |
| NAR | <0.04 | <0.04 | <0.04 |
| NAD(P)+ | <0.02 | <0.02 | <0.02 |
| NAAD | <0.01 | <0.01 | <0.01 |
NA, nicotinic acid; NAAD, nicotinic acid adenine dinucleotide; NAR, nicotinic acid riboside; NMN, nicotinamide mononucleotide; NR, nicotinamide riboside.
Mean ± SD (all such values).
TABLE 2.
Correlations of cow milk NR and nicotinamide concentrations with breed and indicators of milk quality1
| NR, μmol/L | Nicotinamide, μmol/L | |
|---|---|---|
| Breed | −0.28 | −0.21 |
| Fat, % | −0.14 | −0.39 |
| Protein, % | −0.21 | −0.13 |
| Lactose, % | 0.58* | 0.5 |
| Nonfat solids, % | −0.13 | −0.07 |
| Total solids, % | −0.08 | −0.3 |
| Somatic cell count, ×103 cells/mL | −0.38 | −0.52* |
| Urea nitrogen, mg/dL | 0.55* | 0.27 |
| S. aureus | −0.57* | 0.41 |
| NR, μmol/L | 0.58* |
Spearman correlation coefficients were determined between NAD+ precursor vitamins and milk quality and cow breed with data from Supplemental Table 1 (except cow 3) and Supplemental Table 2. *P < 0.05. NR, nicotinamide riboside.
S. aureus depletes NR and nicotinamide.
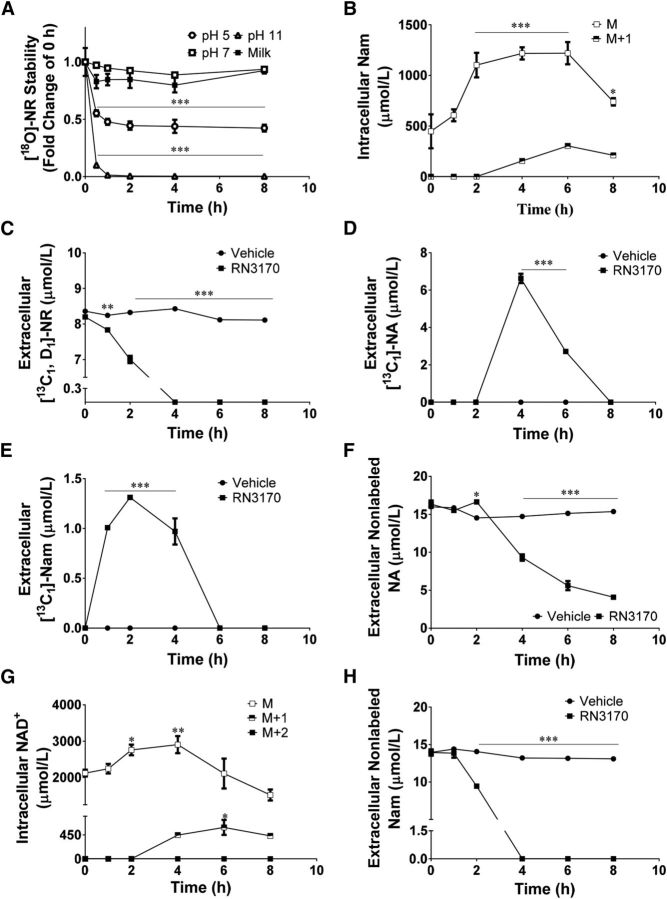
Because the presence of S. aureus was associated with lower concentrations of NR and nicotinamide, we tested whether S. aureus growth might directly alter the concentrations of these metabolites in rich media. Before testing stability in the presence of S. aureus, we investigated the stability of [18O]-NR in pasteurized cow milk or in water adjusted to pH values of 5.0, 7.0, and 11.0. As shown in Figure 1A, [18O]-NR was stable in pasteurized milk and in water at neutral pH but exhibited lesser stability at pH 5.0 and pH 11.0, with pH 11.0 producing nearly complete hydrolysis within 1 h. We measured the pH of cow milk in 4 store-bought milk brands and determined the pH to be 6.72 ± 0.01.
FIGURE 1.
NR is stable in milk and is degraded by S. aureus. (A) Stability of [18O]-NR in 4 store-bought brands of skim milk and in water at pH 5, 7, and 11 (n = 3) was assessed with LC-MS/MS. NR metabolism by S. aureus was determined with [13C1, D1]-NR and LC-MS (B–H). (B–F) Extracellular concentration of [13C1, D1]-NR, nonlabeled Nam, [13C1]-Nam, nonlabeled NA, and [13C1]-NA in Todd-Hewitt + 50 mmol/L Bis-Tris (pH 6.7) media containing 10 μmol/L [13C1, D1]-NR incubated at 37°C without or with S. aureus RN3170 inoculation (n = 3 for inoculated media). In each panel, the concentration of the metabolite is shown in the noninoculated media (n = 1). (G) Intracellular concentration of endogenous or 13C1 enriched Nam. (H) Intracellular concentration of endogenous 13C1-, or 13C1, D1–enriched NAD+ from extracts of S. aureus RN3170 cell pellets (n = 3/time point). For intracellular measurements, a Holm-Sidak multiple-comparisons post hoc test was performed to test for statistical significance compared with time point zero for each metabolite (B and G). For extracellular measurements, a Holm-Sidak multiple-comparisons test was performed to test for statistical significance compared with noninoculated medium within each time point (C–F and H). Data are represented as means ± SEMs. *P < 0.05, **P < 0.01, and ***P < 0.001. NA, nicotinic acid; Nam, nicotinamide; NR, nicotinamide riboside.
Bacteria might alter concentrations of NR found in milk by incorporating NR intact into NAD+ and/or by converting NR into nicotinamide or NA, either of which could be subsequently incorporated into NAD+. To distinguish between these possibilities, we synthesized a double-labeled NR containing a 13C in the nicotinamide moiety and a D1 in the ribose. Incorporation of NR into the bacterial NAD+ pool would be accompanied by a 2-Da mass shift (m/z 664 → 666), whereas breakdown of NR to nicotinamide or NA would be accompanied by appearance of 1-Da shifts in the peaks of these metabolites and a 1-Da mass shift in bacterial NAD+.
Three individual colonies of S. aureus strain RN3170 were cultured separately in Todd-Hewitt media supplemented with 10 μmol [13C1, D1]-NR/L and buffered at pH 6.7 with 50 mmol Bis-Tris/L. The inoculated media and a noninoculated medium control were incubated at 37°C with constant shaking over an 8-h period. Clarified media and cell pellets were collected and analyzed by LC-MS/MS and LC-MS. The pH of the clarified media was also recorded at each time point. The pH consistently remained between 6.5 and 6.7 until between the 6- and 8-h time points, at which time the pH rose to 7.8. As shown in Figure 1B, [13C1, D1]-NR was stable in noninoculated medium over the time course of the experiment. However, S. aureus inoculation significantly decreased the concentration of extracellular NR within 1 h and eliminated the presence of NR as an extracellular metabolite within 4 h. As shown in Figure 1C, singly labeled nicotinamide appeared in growth media within 1 h. As shown in Figure 1D–F, at 4 h, there was a simultaneous rise in singly labeled cellular NAD+, singly labeled cellular nicotinamide, and extracellular NA.
Todd-Hewitt media contain beef heart extract, nicotinamide, and NA (29). As shown in Figure 1G, H, nonlabeled nicotinamide was exhausted within 4 h, whereas the nonlabeled NA slowly declined. Thus, S. aureus principally uses NR as an extracellular source of nicotinamide. Consistent with nicotinamide deamination (30–32), S. aureus can also degrade NR and nicotinamide to NA. Because NR was eliminated by 4 h and the rise in pH occurred after 6 h, pH-mediated mechanisms cannot be responsible for S. aureus–mediated NR instability.
NR concentration as a function of organic certification.
Milk with organic certification requires avoidance of synthetic chemical inputs, irradiation, genetically modified seed, and adherence to certain standards of feed, housing, and breeding (33). Because one or more of these variables could affect NAD+ precursor vitamin concentrations in milk, we purchased 4 brands of conventional and 4 brands of organic milk and quantified the NAD+ metabolome. As observed in milk samples from individual cows, only nicotinamide and NR were above the limit of quantification (Table 3). In 3 of 4 conventional samples and 3 of 4 organic samples, the concentration of nicotinamide exceeded that of NR. Moreover, the concentration of nicotinamide was similar in conventional (5.2 ± 3.4 μmol/L) and organic (5.6 ± 2.5 μmol/L) milk. In the samples we obtained, NR tended to have a higher concentration in conventional (3.1 ± 1.6 μmol/L) compared with organic (1.9 ± 1.0 μmol/L) milk. We note that only one brand of store-bought milk had a combined NAD+ precursor vitamin concentration (8.9 μmol nicotinamide/L plus 5.4 μmol NR/L) that was higher than the mean of S. aureus–negative individual cow samples (7.7 μmol nicotinamide/L plus 5.1 μmol NR/L). NR concentration may be lower as a function of organic dairy practices, but sample sizes were too small for adequate assessment. Future work following NR concentration before and after processing in conventional and organic dairies will be necessary to assess NR concentration as a function of dairy practices.
TABLE 3.
NAD+ precursor concentrations in commercial cow milk1
| Organic | Conventional | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Brand A | Brand B | Brand C | Brand D | All | Brand A | Brand B | Brand C | Brand D | All | |
| Nicotinamide, μmol/L | 2.4 | 7.1 | 5.0 | 7.9 | 5.6 ± 2.5 | 5.6 | 0.67 | 8.9 | 5.4 | 5.2 ± 3.4 |
| NR, μmol/L | 3.1 | 0.84 | 2.2 | 1.4 | 1.9 ± 1.0 | 2.5 | 1.7 | 5.4 | 2.7 | 3.1 ± 1.6 |
Values are expressed as means ± SDs, n = 4. NR, nicotinamide riboside.
NR is a bound metabolite in cow milk.
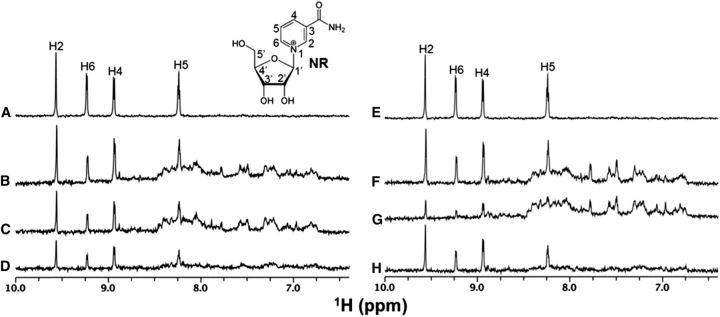
Although a higher level of S. aureus infection in organic milk production (32) could potentially account for lower concentrations of NR in organic milk, there was no change in nicotinamide and no appearance of NA that would be consistent with bacterial exposure. As shown in Figure 1, NR is more stable in milk than in water, suggesting that the metabolite might be complexed to a protective factor.
Organic dairies frequently ultrapasteurize milk at 135°C for 2 s, whereas most conventional dairies pasteurize at 72°C for 15 s (34). Although ultrapasteurization is employed to kill bacterial spores, it might damage a macromolecule responsible for the stabilization of NR. WaterLOGSY NMR measurements were used to detect and map protons in NR (Figure 2A, E) that are potentially bound by slower rotating macromolecules in milk (20, 21). As shown in Figure 2B and 2F, when NR was added to conventional or organic skim milk, 4 aromatic protons (H2, H4, H5, and H6) from the nicotinamide moiety of NR produced positive WaterLOGSY signals consistent with protein binding from both sources of milk. Interestingly, when conventional and organic milk were separated into soluble and resuspended particulate fractions, the conventional soluble fraction retained more NR-binding activity than did the organic soluble fraction (Figure 2C and 2G). Consistent with denaturation of an NR-binding protein by heat, the solubilized organic particulate fraction produced stronger NR WaterLOGSY signals than did the solubilized conventional particulate fraction (Figure 2D and 2H).
FIGURE 2.
NR binding to milk demonstrated by NMR. Organic and conventional skim milk was separated into soluble and resuspended particulate fractions. NR was added to the nonfractionated milk and to the fractions. NR binding was analyzed with NMR as described. (A, E) Normal 1-dimensional 1H NMR spectra of NR. (B–D, F–H) WaterLOGSY spectra of NR in the presence of milk. (A) NR alone. (B) NR + total conventional milk. (C) NR + conventional milk (soluble fraction). (D) NR + conventional milk (particulate fraction). (E) NR alone. (F) NR + total organic milk. (G) NR + organic milk (soluble fraction). (H) NR + organic milk (particulate fraction). The assigned 1H resonances of the nicotinamide ring aromatic protons are labeled. 1H, proton; NR, nicotinamide riboside; ppm, parts per million; WaterLOGSY, water-ligand observed via gradient spectroscopy.
Discussion
The data presented in this article show that ∼60% of the NAD+ precursor vitamin concentration of cow milk is present as nicotinamide, whereas ∼40% is present as NR. Although we occasionally detected cow milk samples with higher concentrations of NR than nicotinamide, we did not detect NAD+, NA, or any other NAD+ metabolite in any sample. In samples from individual cows, the presence of S. aureus, the most common cause of cattle mastitis (35, 36), was associated with lower concentrations of NR and nicotinamide. We also showed that S. aureus degraded NR into nicotinamide and NA and used the nicotinamide as a precursor of intracellular NAD+. These data are consistent with the ability of S. aureus to use either nicotinamide or NA for NAD+ synthesis (37) with nicotinamide utilization requiring deamination to NA (30–32).
Multiple aspects of cow nutrition are expected to contribute to concentrations of nicotinamide and NR in milk. In particular, feeding of herds with nicotinamide and NA (38, 39) is likely to produce cows that transmit higher concentrations of nicotinamide and NR into milk. In addition, it will be interesting to determine whether NR is actively transported by mammary glands (40).
Although PP factors in food include NAD+, NAD+ precursors, and tryptophan, high doses of NR appear to have some protective activities for metabolic and neurodegenerative conditions. The ability of milk to bind and preserve the integrity of NR makes dairy products potentially good sources of supplemented NR. Further research is needed to maximize NR concentration of conventional and organic milk and to identify the molecular basis of NR binding to milk.
Supplementary Material
NAD+ Precursor Concentrations in 19 Individual Milk Samples1
Acknowledgments
We thank Leo L Timms at Iowa State University for individual milk samples, Patrick M Schlievert for the S. aureus strain, and Fiorenza Ianzini for assistance in manuscript preparation. SAJT and CB designed the research, analyzed the results, and wrote the manuscript; SAJT performed the LC-MS with heavy standards synthesized by PR and MEM; and LY performed the NMR measurements. All authors read and approved the final manuscript.
Abbreviations
- NA
nicotinic acid
- NR
nicotinamide riboside
- PP
pellagra preventive
- WaterLOGSY
water-ligand observed via gradient spectroscopy
Footnotes
Supported by the Roy J. Carver Trust.
References
- 1. Goldberger J, Waring CH, Willets DG. The treatment and prevention of pellagra. Public Health Reports (1896–1970) 1914;29:2821–5. [Google Scholar]
- 2. Koehn CJ, Elvehjem CA. Further studies on the concentration of the antipellagra factor. J Biol Chem 1937;118:693–9. [Google Scholar]
- 3. Spies TD, Bean WB, Ashe WF. Recent advances in the treatment of pellagra and associated deficiencies. Ann Intern Med 1939;12:1830–44. [Google Scholar]
- 4. Krehl WA, Teply LJ, Sarma PS, Elvehjem CA. Growth-retarding effect of corn in nicotinic acid–low rations and its counteraction by tryptophane. Science 1945;101:489–90. [DOI] [PubMed] [Google Scholar]
- 5. Bogan KL, Brenner C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu Rev Nutr 2008;28:115–30. [DOI] [PubMed] [Google Scholar]
- 6. Oduho GW, Baker DH. Quantitative efficacy of niacin sources for chicks: nicotinic acid, nicotinamide, NAD and tryptophan. J Nutr 1993;123:2201–6. [DOI] [PubMed] [Google Scholar]
- 7. Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends Biochem Sci 2007;32:12–9. [DOI] [PubMed] [Google Scholar]
- 8. Mori V, Amici A, Mazzola F, Di Stefano M, Conforti L, Magni G, Ruggieri S, Raffaelli N, Orsomando G. Metabolic profiling of alternative NAD biosynthetic routes in mouse tissues. PLoS One 2014;9:e113939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krehl WA, de la Huerga J, Elvehjem CA, Hart EB. The distribution of niacinamide and niacin in natural materials. J Biol Chem 1946;166:53–7. [PubMed] [Google Scholar]
- 10. Sutton TS, Esh GC. The nutrition of the newborn dairy calf: I. Changes in the tryptophan content of the blood plasma following birth and the ingestion of colostrum. J Dairy Sci 1947;31:183–7. [Google Scholar]
- 11. Bieganowski P, Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 2004;117:495–502. [DOI] [PubMed] [Google Scholar]
- 12. Belenky P, Racette FG, Bogan KL, McClure JM, Smith JS, Brenner C. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 2007;129:473–84. [DOI] [PubMed] [Google Scholar]
- 13. Cantó C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, et al. . The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab 2012;15:838–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khan NA, Auranen M, Paetau I, Pirinen E, Euro L, Forsstrom S, Pasila L, Velagapudi V, Carroll CJ, Auwerx J, et al. . Effective treatment of mitochondrial myopathy by nicotinamide riboside, a vitamin B3. EMBO Mol Med 2014;6:721–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gariani K, Menzies KJ, Ryu D, Wegner CJ, Wang X, Ropelle ER, Moullan N, Zhang H, Perino A, Lemos V, et al. Eliciting the mitochondrial unfolded protein response by nicotinamide adenine dinucleotide repletion reverses fatty liver disease in mice. Hepatology 2016;63:1190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu W, Barrientos T, Mao L, Rockman Howard A, Sauve Anthony A, Andrews Nancy C. Lethal cardiomyopathy in mice lacking transferrin receptor in the heart. Cell Rep 2015;13:533–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gong B, Pan Y, Vempati P, Zhao W, Knable L, Ho L, Wang J, Sastre M, Ono K, Sauve AA, et al. . Nicotinamide riboside restores cognition through an upregulation of proliferator-activated receptor-gamma coactivator 1alpha regulated beta-secretase 1 degradation and mitochondrial gene expression in Alzheimer's mouse models. Neurobiol Aging 2013;34:1581–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brown KD, Maqsood S, Huang JY, Pan Y, Harkcom W, Li W, Sauve A, Verdin E, Jaffrey SR. Activation of SIRT3 by the NAD(+) precursor nicotinamide riboside protects from noise-induced hearing loss. Cell Metab 2014;20:1059–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trammell SA, Brenner C. Targeted, LCMS-based metabolomics for quantitative measurement of NAD+ metabolites. Comput Struct Biotechnol J 2013;4:e201301012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dalvit C, Fogliatto G, Stewart A, Veronesi M, Stockman B. WaterLOGSY as a method for primary NMR screening: practical aspects and range of applicability. J Biomol NMR 2001;21:349–59. [DOI] [PubMed] [Google Scholar]
- 21. Dalvit C, Pevarello P, Tatò M, Veronesi M, Vulpetti A, Sundström M. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J Biomol NMR 2000;18:65–8. [DOI] [PubMed] [Google Scholar]
- 22. Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 1995;6:277–93. [DOI] [PubMed] [Google Scholar]
- 23. Johnson BA, Blevins R. NMR View: a computer program for the visualization and analysis of NMR data. J Biomol NMR 1994;4:603–14. [DOI] [PubMed] [Google Scholar]
- 24. Yang T, Chan NY, Sauve AA. Syntheses of nicotinamide riboside and derivatives: effective agents for increasing nicotinamide adenine dinucleotide concentrations in mammalian cells. J Med Chem 2007;50:6458–61. [DOI] [PubMed] [Google Scholar]
- 25. Thurmond MC, Tyler JW, Luiz DM, Holmberg CA, Picanso JP. The effect of pre-enrichment on recovery of Streptococcus agalactiae, Staphylococcus aureus and mycoplasma from bovine milk. Epidemiol Infect 1989;103:465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maass S, Sievers S, Zuhlke D, Kuzinski J, Sappa PK, Muntel J, Hessling B, Bernhardt J, Sietmann R, Volker U, et al. . Efficient, global-scale quantification of absolute protein amounts by integration of targeted mass spectrometry and two-dimensional gel-based proteomics. Anal Chem 2011;83:2677–84. [DOI] [PubMed] [Google Scholar]
- 27. Rasigade JP, Moulay A, Lhoste Y, Tristan A, Bes M, Vandenesch F, Etienne J, Lina G, Laurent F, Dumitrescu O. Impact of sub-inhibitory antibiotics on fibronectin-mediated host cell adhesion and invasion by Staphylococcus aureus. BMC Microbiol 2011;11:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Motulsky HJ, Brown RE. Detecting outliers when fitting data with nonlinear regression—a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics 2006;7:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Todd EW, Hewitt LF. A new culture medium for the production of antigenic streptococcal hæmolysin. J Pathol Bacteriol 1932;35:973–4. [Google Scholar]
- 30. Bieganowski P, Brenner C. The reported human NADsyn2 is ammonia-dependent NAD synthetase from a pseudomonad. J Biol Chem 2003;278:33056–9. [DOI] [PubMed] [Google Scholar]
- 31. Gazzaniga F, Stebbins R, Chang SZ, McPeek MA, Brenner C. Microbial NAD metabolism: lessons from comparative genomics. Microbiol Mol Biol Rev 2009;73:529–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sorci L, Blaby IK, Rodionova IA, De Ingeniis J, Tkachenko S, de Crecy-Lagard V, Osterman AL. Quinolinate salvage and insights for targeting NAD biosynthesis in group A streptococci. J Bacteriol 2013;195:726–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. USDA National Organic Program [Internet]. [cited 2016 Jan 3]. Available from: http://www.ams.usda.gov/about-ams/programs-offices/national-organic-program.
- 34. Cappozzo JC, Koutchma T, Barnes G. Chemical characterization of milk after treatment with thermal (HTST and UHT) and nonthermal (turbulent flow ultraviolet) processing technologies. J Dairy Sci 2015;98:5068–79. [DOI] [PubMed] [Google Scholar]
- 35. Ruegg P. Management of mastitis on organic and conventional dairy farms. J Anim Sci 2009;87(Suppl):43–55. [DOI] [PubMed] [Google Scholar]
- 36. Cicconi-Hogan KM, Gamroth M, Richert R, Ruegg PL, Stiglbauer KE, Schukken YH. Associations of risk factors with somatic cell count in bulk tank milk on organic and conventional dairy farms in the United States. J Dairy Sci 2013;96:3689–702. [DOI] [PubMed] [Google Scholar]
- 37. Knight BC. The nutrition of Staphylococcus aureus: the activities of nicotinamide, aneurin (vitamin B(1)) and related compounds. Biochem J 1937;31:966–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jaster EH, Ward NE. Supplemental nicotinic acid or nicotinamide for lactating dairy cows. J Dairy Sci 1990;73:2880–7. [DOI] [PubMed] [Google Scholar]
- 39. Cervantes A, Smith TR, Young JW. Effects of nicotinamide on milk composition and production in dairy cows fed supplemental fat. J Dairy Sci 1996;79:105–13. [DOI] [PubMed] [Google Scholar]
- 40. Shennan DB, Peaker M. Transport of milk constituents by the mammary gland. Physiol Rev 2000;80:925–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NAD+ Precursor Concentrations in 19 Individual Milk Samples1