Staphylococcus aureus is responsible for numerous community outbreaks and is one of the most frequent causes of nosocomial infections with significant morbidity and mortality. While the function of lytic transglycosylases (LTs) in relation to cell division, biofilm formation, and antibiotic resistance has been determined for several bacteria, their role in S. aureus remains largely unknown. The only known LTs in S. aureus are immunodominant staphylococcal antigen A (IsaA) and Staphylococcus epidermidis D protein (SceD).
KEYWORDS: MRSA, Staphylococcus aureus, antibiotic resistance, β-lactams, biofilms, peptidoglycan, transglycosylases
ABSTRACT
Staphylococcus aureus is responsible for numerous community outbreaks and is one of the most frequent causes of nosocomial infections with significant morbidity and mortality. While the function of lytic transglycosylases (LTs) in relation to cell division, biofilm formation, and antibiotic resistance has been determined for several bacteria, their role in S. aureus remains largely unknown. The only known LTs in S. aureus are immunodominant staphylococcal antigen A (IsaA) and Staphylococcus epidermidis D protein (SceD). Our study demonstrates that, in strains of methicillin-resistant S. aureus (MRSA), IsaA and SceD contribute differently to biofilm formation and β-lactam resistance. Deletion of isaA, but not sceD, led to decreased biofilm formation. Additionally, in isaA-deleted strains, β-lactam resistance was significantly decreased compared with that of wild-type strains. Plasmid-based expression of mecA, a major determinant of β-lactam resistance in MRSA, in an isaA-deleted strain did not restore β-lactam resistance, demonstrating that the β-lactam susceptibility phenotype is exhibited by the isaA mutant regardless of the production level of PBP2a. Overall, our results suggest that IsaA is a potential therapeutic target for MRSA infections.
INTRODUCTION
Staphylococcus aureus is an opportunistic pathogen responsible for infections ranging from skin infections to life-threatening diseases (1, 2). Since the 1960s, the emergence of antibiotic-resistant strains, such as methicillin-resistant S. aureus (MRSA), has contributed to its spread, and S. aureus is now the most common cause of nosocomial infections with significant morbidity and mortality (3). Currently, the emergence of new strains with increased virulence and colonization capacity is responsible for an unprecedented number of community outbreaks in the United States, resulting in the number of deaths caused by S. aureus infection to be comparable to those caused by HIV, tuberculosis, and viral hepatitis combined (4, 5). The pathogenicity of S. aureus relies on a wide range of virulence factors, including proteins conferring resistance to host antimicrobial peptides (6), secreted toxins (1, 2, 4), and factors enhancing its ability to form matrix-encased biofilms on tissues and medical devices (2, 7). S. aureus biofilms have been shown to be resistant to killing by antibiotics at concentrations 10 to 1,000 times greater than the concentration required to kill planktonic cells, leading to the failure of conventional antibiotic therapy and recurrence of infections (3, 8).
It has been reported that bacterial lytic transglycosylases (LTs) are involved in various cell functions and phenotypes. LTs cleave the β-1,4-glycosidic bonds between N-acetylmuramic and N-acetylglucosamine residues of peptidoglycan (9) and affect peptidoglycan turnover and septum cleavage during cell division (10, 11). Additionally, in some Gram-negative bacteria, LTs are involved in virulence mechanisms during infection (11). The peptidoglycan fragments produced by LTs are recycled in the cytoplasm for peptidoglycan biosynthesis (12–14). During infection, released peptidoglycan fragments may also be recognized by host cells, leading to general symptoms, such as fever, lack of appetite, or sleepiness, and inducing inflammation via the production of cytokines (11, 15). This mechanism contributes to virulence in Gram-negative bacteria, such as Bordetella pertussis and Neisseria gonorrhoeae (11). LTs are known to affect biofilm formation in both Gram-positive and Gram-negative bacteria (16–18). Moreover, deletion of LTs can affect β-lactamase induction (19) and the extent of β-lactam resistance in some Gram-negative bacteria (18, 20, 21).
The only known S. aureus LTs are immunodominant staphylococcal antigen A (IsaA) and Staphylococcus epidermidis D protein (SceD), both of which have been shown to possess cell wall hydrolytic activity (22). IsaA was identified as a major antigen expressed during sepsis or bacteremia caused by S. aureus (23, 24). IsaA is both excreted into the extracellular environment and bound to the cell wall, where it localizes mainly to the septal region of dividing cells (25). Deletion of isaA results in increased expression of SceD (22). SceD plays an essential role in nasal colonization in cotton rats, and its inactivation results in impaired cell separation (22). Increased SceD levels in the cell wall fraction of vancomycin-intermediate S. aureus strains can partly explain the altered peptidoglycan structure of these strains (26).
Considering the functions LTs reportedly play in the cell cycle, resistance to antibiotics, and virulence in other bacteria, S. aureus LTs are likely to have major clinical significance. However, their roles in S. aureus have been the subject of only a few investigations and remain largely unknown. The present study was conducted to characterize the roles of LTs in MRSA by investigating their effects on biofilm formation and antibiotic susceptibility.
RESULTS
IsaA is involved in biofilm formation.
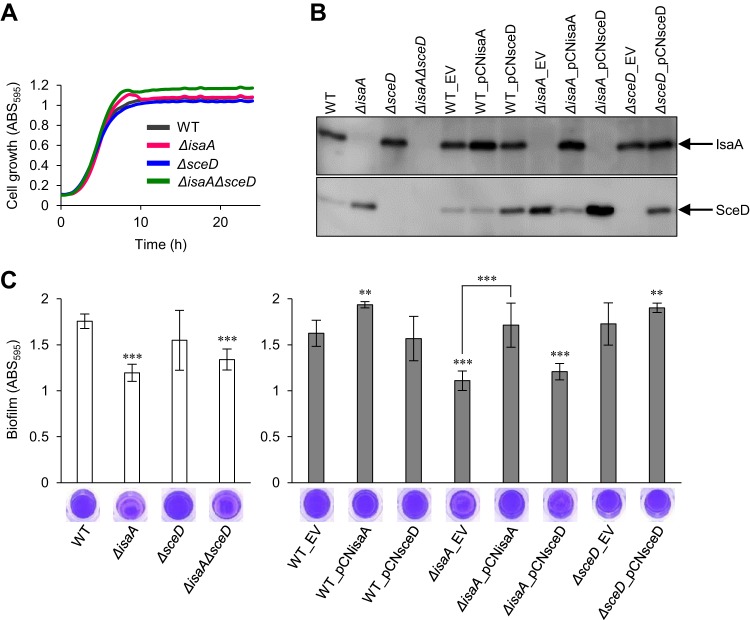
LT gene mutations have been reported to affect biofilm formation in Lactococcus lactis (16), Salmonella enterica (17), and Pseudomonas aeruginosa (18). Here, we investigated the biofilm-forming capacity of wild-type (WT) and LT mutants of S. aureus MR23 (27), a MRSA clinical isolate that produces a large amount of biofilm, as well as the effects of overexpression and complementation of LT genes using a plasmid-based expression system shown in Table 1. The cell growth of WT and LT mutants of MR23 was almost identical, except for the final cell density being slightly higher in the ΔisaAsceD mutant than in the WT (Fig. 1A). Note that there was no difference in CFUs between WT and all LT mutants in stationary phase (ca. 2 × 109 CFU/ml). We analyzed the levels of IsaA and SceD in S. aureus culture supernatants using Western blotting (Fig. 1B). In agreement with previous findings (22), deletion of isaA resulted in an increased level of SceD compared with that of the WT. In the opposite case, no IsaA overproduction was detected in the ΔsceD mutant. We then confirmed the effect of complementation with plasmids harboring each LT gene on the production of LTs. Notably, the SceD-overproduction phenotype in the ΔisaA mutant (WT_EV versus ΔisaA_EV) was abolished by plasmid-based expression of isaA (ΔisaA_EV versus ΔisaA_pCNisaA). IsaA and SceD protein production in the supernatant fractions was almost identical in whole-cell fractions that contained cell wall proteins and cytoplasmic proteins (see Fig. S1 in the supplemental material).
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Genotype or characteristic | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5a | F-, Φ80d lacZΔM15, Δ(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17(rK− mK+), phoA, supE44, λ-, thi-1, gyrA96, relA1 | Toyobo |
| BL21(DE3) | F-, ompT, hsdSB(rB- mB-), gal(λcI857, ind1, Sam7, nin5, lacUV5-T7gene1), dcm(DE3) | Promega |
| DC10B | E. coli K12, Δdcm mutant | 49, 50 |
| S. aureus | ||
| MR23 | Clinical isolate of MRSA (ST380) | 27 |
| MR23ΔisaA | isaA was deleted from MR23 | This study |
| MR23ΔsceD | sceD was deleted from MR23 | This study |
| MR23ΔisaAΔsceD | isaA and sceD were deleted from MR23 | This study |
| MR23ΔmecA | mecA was deleted from MR23 | This study |
| MR10 | Clinical isolate of MRSA (ST5) | 27 |
| MR10ΔisaA | isaA was deleted from MR10 | This study |
| MR10ΔsceD | sceD was deleted from MR10 | This study |
| MR10ΔisaAΔsceD | isaA and sceD were deleted from MR10 | This study |
| USA300 | Clinical isolate of MRSA (ST8) | ATCC BAA1717 |
| USA300ΔisaA | isaA was deleted from USA300 | This study |
| USA300ΔsceD | sceD was deleted from USA300 | This study |
| USA300ΔisaAΔsceD | isaA and sceD were deleted from USA300 | This study |
| Plasmids | ||
| pET29a | E. coli expression vector; KmR | Novagen |
| pETsceD | sceD cloned in pET29a | This study |
| pKOR1 | An E. coli/S. aureus shuttle vector plasmid for knockout of S. aureus genes by allelic exchange; ApR, CmR | 48 |
| pCN51 | An E. coli/S. aureus shuttle vector plasmid; ApR, EmR | 54 |
| pCN51Cm | An E. coli/S. aureus shuttle vector plasmid; ApR, CmR | This study |
| pCNisaA | isaA cloned in pCN51Cm | This study |
| pCNsceD | sceD cloned in pCN51Cm | This study |
| pCNmecA | mecA cloned in pCN51Cm | This study |
KmR, kanamycin resistance; ApR, ampicillin resistance; EmR, erythromycin resistance; CmR, chloramphenicol resistance; ST, sequence type.
FIG 1.
Effect of LT gene deletion on cell growth and biofilm formation of S. aureus MR23. (A) Cell growth of wild-type (WT) and LT mutants. The average of three independent cultures is shown. (B) Western blot showing levels of IsaA and SceD in culture supernatants of WT and LT mutants. (C) Biofilm formation of WT and LT mutants without (white bars) or with (gray bars) plasmid. Biofilms formed on the bottom of the 96-well plate were stained with crystal violet; subsequently, absorbance at 595 nm was measured. Representative images of stained biofilms are shown below. The values are the means of six independent cultures. The error bars indicate the standard error of the mean. Differences between WT and mutants were compared using Student’s t test. **P < 0.01, ***P < 0.001. EV, pCN51Cm (empty vector); pCNisaA, pCN51Cm containing isaA; pCNsceD, pCN51Cm containing sceD.
Deletion of isaA significantly decreased biofilm formation but sceD deletion did not (Fig. 1C). The biofilm-forming capacity of the ΔisaAΔsceD mutant was nearly identical to that of the ΔisaA mutant. Plasmid-based expression of isaA in the WT strain enhanced biofilm formation. We also confirmed that plasmid-based expression of isaA complemented the decreased biofilm phenotype of the ΔisaA mutant (ΔisaA_EV versus ΔisaA_pCNisaA). In contrast, plasmid-based expression of sceD did not affect biofilm formation in either the WT or ΔisaA mutant. These results demonstrate that, of the two LTs, only IsaA significantly affects biofilm formation in this strain.
Deletion of isaA induces the β-lactam susceptibility phenotype in MRSA.
In other bacterial species, deletion of LTs has been shown to affect β-lactam resistance (18, 20, 21); however, no such information is available with regard to S. aureus LTs. We determined the MICs of antibiotics against WT and LT mutants of MR23 in Mueller-Hinton II (MH) broth (Table 2). Four-fold or greater differences in MIC values were regarded as significant. No significant increase in MIC was observed for any of the LT mutants. Notably, deletion of isaA decreased the MICs of 5/7 β-lactams (oxacillin, cefazolin, flomoxef, cefoxitin, and imipenem), and deletion of sceD decreased the MICs of 2/7 β-lactams (ampicillin and cefoxitin). In the assay performed using brain heart infusion broth supplemented with 1% glucose (BHIG), deletion of isaA decreased the MICs of 4/7 β-lactams (oxacillin, ampicillin, flomoxef, and cefoxitin), whereas deletion of sceD showed no effect (see Table S1 in the supplemental material). Plasmid-based complementation of isaA significantly increased the MICs of β-lactams against the ΔisaA mutant (ΔisaA_EV versus ΔisaA_pCNisaA). In contrast, plasmid-based expression of sceD in the ΔisaA mutant did not lead to such an effect (Table 2). These results indicate that S. aureus LTs contribute to β-lactam resistance and that IsaA is more important in this role than SceD.
TABLE 2.
MICs of antibiotics for WT and LT mutants of S. aureus MR23 cultured in Mueller-Hinton brotha
| Antibiotic | MIC (μg/ml) of: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | ΔisaA | ΔsceD | ΔisaAΔsceD | WT_EV | WT_pCNisaA | WT_pCNsceD | ΔisaA_EV | ΔisaA_pCNisaA | ΔisaA_pCNsceD | ΔsceD_EV | ΔsceD_pCNsceD | |
| β-lactam | ||||||||||||
| Oxacillin | 64 | 1 | 32 | 1 | 64 | 64 | 32 | 2 | 16 | 2 | 64 | 64 |
| Ampicillin | 4 | 2 | 0.5 | 0.5 | 8 | 8 | 8 | 4 | 8 | 4 | 0.5 | 8 |
| Cefazolin | 8 | 1 | 8 | 1 | 16 | >16 | >16 | 1 | 16 | 2 | 1 | >16 |
| Cefmetazole | 8 | 4 | 8 | 2 | 8 | >32 | >32 | 4 | 16 | 4 | 8 | 32 |
| Flomoxef | 8 | 2 | 4 | 1 | 8 | >16 | >16 | 1 | 8 | 2 | 4 | >16 |
| Cefoxitin | >16 | 8 | 8 | 8 | >16 | >16 | >16 | 8 | >16 | 16 | 8 | >16 |
| Imipenem | 8 | <0.25 | 8 | <0.25 | 8 | >8 | >8 | <0.25 | 8 | <0.25 | 8 | 8 |
| Glycopeptide | ||||||||||||
| Vancomycin | 1 | 2 | 1 | 2 | 2 | 4 | 4 | 2 | 4 | 2 | 1 | 2 |
| Teicoplanin | 4 | 4 | 4 | 2 | 1 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Oxazolidinone | ||||||||||||
| Linezolid | 8 | 8 | 8 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Lincomycin | ||||||||||||
| Clindamycin | 0.5 | 0.5 | 0.12 | 0.25 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.12 | 0.5 |
WT, wild type; ΔisaA, isaA-deleted strain; ΔsceD, sceD-deleted strain; ΔisaAΔsceD, isaA and sceD-deleted strain; EV, pCN51Cm (empty vector); pCNisaA, pCN51Cm containing isaA; pCNsceD, pCN51Cm containing sceD.
Correlation between PBP2a production and β-lactam resistance in isaA mutants.
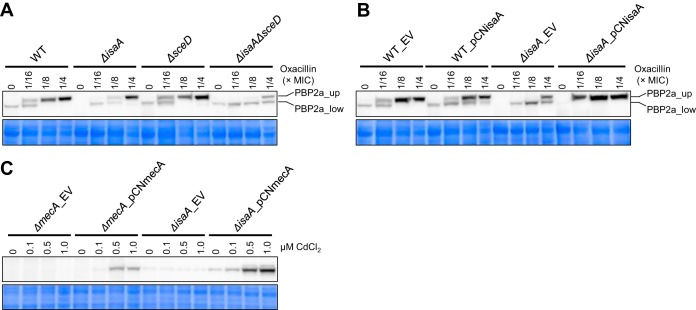
As β-lactam resistance of MRSA is conferred mainly by mecA-encoded PBP2a (28), we investigated the impact of isaA and sceD deletions on PBP2a production in the presence or absence of oxacillin as an inducer of PBP2a (29). The oxacillin concentrations used were 1/16, 1/8, and 1/4 of the MIC for each strain. Western blotting of WT using an anti-PBP2a antibody revealed two bands of different molecular size (Fig. 2A). Both bands were derived from PBP2a, as they were not detected in the ΔmecA mutant in the presence and absence of oxacillin (see Fig. S2 in the supplemental material). The cause of the two bands with different molecular weights corresponding to PBP2a peptides is presently unknown. In WT, expression of the upper PBP2a band (PBP2a_up) was oxacillin dependent. Only a weak lower PBP2a band (PBP2a_low) was detected in the absence of oxacillin. A similar phenomenon was observed in the ΔsceD strain. Instead, induction of PBP2a_up was decreased in the ΔisaA strain and dramatically decreased in the ΔisaAΔsceD strain compared with the WT (Fig. 2A). Importantly, the lower production of PBP2a_up in the ΔisaA strain was complemented by plasmid-based expression of isaA (ΔisaA_EV versus ΔisaA_pCNisaA) (Fig. 2B).
FIG 2.
Effect of LT gene deletion on PBP2a production in S. aureus MR23. (A and B) Western blot showing levels of PBP2a in wild-type (WT) and LT mutants in the presence (A) and absence (B) of plasmids. (C) Western blot showing levels of PBP2a in mecA and isaA mutants. Concentrations of oxacillin and CdCl2 used for induction are indicated above the panels. As loading controls, CBB-stained gels of the same area are shown below the panels. EV, pCN51Cm (empty vector); pCNisaA, pCN51Cm containing isaA; pCNmecA, pCN51Cm containing mecA.
From these results, we hypothesized that the expression of isaA is directly or indirectly involved in the production of PBP2a, thereby contributing to the β-lactam resistance of MR23. To establish a cause-effect relationship between the β-lactam susceptibility phenotype and decreased PBP2a production level, pCNmecA, a plasmid containing mecA cloned under the control of a cadmium-inducible promoter in pCN51Cm plasmid, was introduced in ΔmecA and ΔisaA mutants. In the mutants harboring pCNmecA, PBP2a production was induced depending on the concentration of cadmium added to the culture medium (Fig. 2C). Then, the effects of inducible PBP2a production on β-lactam resistance of the mutants were investigated. The MIC of oxacillin against ΔmecA_pCNmecA was dramatically increased depending on cadmium concentration (Table 3), confirming that PBP2a produced by the cadmium-inducible system was functional. On the other hand, contrary to our hypothesis, PBP2a induction had no effect on the MIC of oxacillin against ΔisaA_pCNmecA. Accordingly, these results indicated that the β-lactam susceptibility phenotype can be exhibited regardless of the production level of PBP2a in the ΔisaA mutant.
TABLE 3.
MICs of oxacillin for mecA and isaA mutants of S. aureus MR23 cultured in Mueller-Hinton brotha
| CdCl2 concn (μM) | MIC (μg/ml) of: |
|||
|---|---|---|---|---|
| ΔmecA_EV | ΔmecA_pCNmecA | ΔisaA_EV | ΔisaA_pCNmecA | |
| 0 | 0.5 | 1 | 2 | 2 |
| 0.1 | 0.5 | 32 | 2 | 2 |
| 0.5 | 0.5 | 128 | 2 | 2 |
| 1.0 | 0.5 | 128 | 2 | 4 |
ΔmecA, mecA-deleted strain; ΔisaA, isaA-deleted strain; EV, pCN51Cm (empty vector); pCNmecA, pCN51Cm containing mecA.
Effect of LT gene deletion in MRSA clones belonging to different sequence types.
Here, we showed the effect of LT gene deletion in S. aureus MR23. MR23 belongs to multilocus sequence type 380 (ST380) (27). ST380 has been reported to be involved in clonal complex 8 (CC8) (30). CC8 includes S. aureus USA300, which is one of the most common and well-characterized community-acquired MRSA (CA-MRSA) clones belonging to ST8 (31). To investigate whether deletion of LT genes caused a similar effect in health care-associated MRSA (HA-MRSA), S. aureus MR10 belonging to ST5 (27) was used. ST5 is a widely present HA-MRSA clone (32). Additionally, S. aureus USA300 was used to verify the generality of the phenotypes induced by LT mutations.
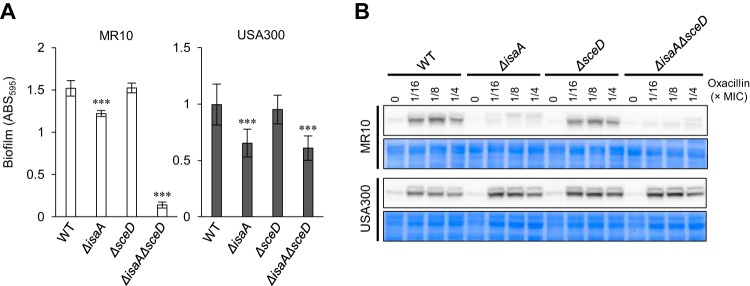
Deletion of LT genes did not significantly affect cell growth of MR10 and USA300 (see Fig. S3 in the supplemental material), echoing the case of MR23 (Fig. 1A). Deletion of isaA decreased the biofilm forming capacity of MR10 and USA300, whereas deletion of sceD did not (Fig. 3A). In USA300, the biofilm-forming capacity of the ΔisaAΔsceD strain was almost identical to that of the ΔisaA strain, as was the case with MR23. Interestingly, double deletion of isaA and sceD drastically decreased the biofilm-forming capacity of MR10, suggesting that SceD has a complementary role in biofilm formation in this strain.
FIG 3.
Effect of LT gene deletion in S. aureus MR10 and USA300. (A) Biofilm formation of wild-type (WT) and LT mutant. Biofilms formed on the bottom of the 96-well plate were stained with crystal violet; subsequently, absorbance at 595 nm was measured. The values are the means of six independent cultures. The error bars indicate the standard error of the mean. Differences between WT and mutants were compared using Student’s t test. ***P < 0.001. (B) Western blot showing levels of PBP2a in WT and LT mutants. Concentrations of oxacillin used for induction are indicated above the panels. As loading controls, CBB-stained gels of the same area are shown below the panels.
Deletion of isaA or both isaA and sceD resulted in a drastic drop of oxacillin MIC compared with that of the WT in both MR10 and USA300 but deletion of sceD did not (Table 4). Next, PBP2a production in MR10 and USA300 and their LT mutants was analyzed. In MR10, PBP2a production was induced by oxacillin in the WT and ΔsceD mutants but hardly induced in ΔisaA and ΔisaAΔsceD mutants (Fig. 3B). On the other hand, in USA300, PBP2a production was similarly induced in WT and LT mutants. These results suggest that the β-lactam susceptibility phenotype in ΔisaA and ΔisaAΔsceD mutants was induced in USA300 regardless of oxacillin-inducible PBP2a production, as was the case with MR23ΔisaA overproducing plasmid-derived PBP2a.
TABLE 4.
MICs of oxacillin for WT and LT mutants of S. aureus MR10 and S. aureus USA300 cultured in Mueller-Hinton brotha
| S. aureus strain | MIC (μg/ml) of: |
|||
|---|---|---|---|---|
| WT | ΔisaA | ΔsceD | ΔisaAΔsceD | |
| MR10 | 512 | 8 | 512 | 2 |
| USA300 | 256 | 1 | 256 | 1 |
WT, wild type; ΔisaA, isaA-deleted strain; ΔsceD, sceD-deleted strain; ΔisaAΔsceD, isaA and sceD-deleted strain.
DISCUSSION
The roles of S. aureus IsaA and SceD in cellular physiology, cell wall dynamics, and host-pathogen interaction, as well as subcellular localization of IsaA, have been already described (22, 25). We show here that the two S. aureus LTs IsaA and SceD contributed differently to biofilm formation; deletion of isaA significantly decreased biofilm formation, whereas deletion of sceD did not. A similar phenomenon has been described in Salmonella enterica serovar Typhimurium, where only 2/7 LTs, namely, MltE and MltC, were shown to be involved in the regulation of biofilm formation (17). As a double deletion of isaA and sceD drastically decreased biofilm-forming capacity compared with a single deletion of isaA in MR10 but not in MR23 and USA300, the complementary role of SceD in biofilm formation varies by S. aureus strain.
Deletion of isaA resensitized the three tested MRSA strains to β-lactams (Table 2; Table S1; Table 4). To the best of our knowledge, this is the first report to show that the loss of an LT gene alters antibiotic resistance in S. aureus. MRSA β-lactam resistance is acquired mainly through the production of PBP2a, which is encoded by mecA (28). Therefore, we first hypothesized that PBP2a production was decreased in isaA-deleted strains, thereby resulting in a β-lactam-sensitive phenotype. In MR23 and MR10, oxacillin-dependent production of PBP2a was remarkably reduced in isaA-deleted strains, suggesting that IsaA is involved in the oxacillin-dependent production of PBP2 in these strains. Induction of PBP2a is regulated by a three-component system encoded by mecR1-mecI-mecR2 regulatory genes consisting of a transcriptional repressor, MecI; a membrane-bound sensor inducer, MecR1; and an anti-repressor, MecR2; in MRSA strains with functional regulatory loci (33, 34). It has been reported that homologous blaR1-blaI regulatory genes, which are required for inducible production of β-lactamase encoded by blaZ, also regulate the production of PBP2a (35). Inactivation of MecI/BlaI through direct interaction with an intracellular peptidoglycan fragment induced by β-lactam has been proposed to constitute another mechanism of induction (36). Peptidoglycan is continuously synthesized, degraded, and recycled during cell growth (37). In Gram-negative bacteria, the peptidoglycan fragment released during peptidoglycan turnover is efficiently transported into the cytoplasm and recycled (12, 38). The presence of β-lactam antibiotics leads to inactivation of PBPs and, consequently, to the cytoplasmic accumulation of peptidoglycan fragments, which can act as a signal or coactivator for β-lactamase induction (39, 40). In mutants lacking LT genes, the inducibility of β-lactamase was greatly diminished (19). Gram-positive bacteria, including S. aureus, have been recently reported to recycle the peptidoglycan component N-acetylmuramic acid during growth (41). Therefore, it may be possible that the peptidoglycan fragments produced by IsaA are taken up by the cell via an unknown transport mechanism and recycled. In the presence of β-lactam antibiotics, the activity of IsaA may result in intracellular accumulation of a specific peptidoglycan fragment, ultimately leading to induction of mecA expression in MRSA through inhibition of MecI/BlaI.
The hypothetical mechanism mentioned above may partially explain the reduced β-lactam resistance in MR23 and MR10; however, other data suggested that the loss of resistance in the isaA-deleted strains cannot be explained solely by the production level of PBP2a. In USA300, although the β-lactam resistance level of isaA-deleted strains was dramatically decreased, PBP2a production was induced by oxacillin in the same manner as in WT. Moreover, plasmid-based expression of mecA in the ΔisaA strain of MR23 failed to restore β-lactam resistance regardless of successful PBP2a production. Taken together, in isaA-deletion mutants, it is considered that the β-lactam susceptibility phenotype is induced regardless of the production level of PBP2a. In previous work by Memmi et al., PBP4 was demonstrated as a key element in β-lactam resistance in CA-MRSA (42). In that report, it was also suggested that constitutive expression of PBP2a did not contribute to oxacillin resistance in the absence of PBP4. Considering the similarity of their data to our data here in terms of deletion of the cell wall-related gene that induces the β-lactam susceptibility phenotype regardless of PBP2a production, these phenomena might be explained by a common molecular mechanism. Further investigation is needed to elucidate the molecular basis of the loss of β-lactam resistance in the mutants lacking isaA.
IsaA has been shown to be immunogenic in both humans and mice (43), and a specific antibody response against IsaA has been reported in human sepsis and a murine skin infection model (23, 44). Additionally, it has been reported that monoclonal antibodies targeting IsaA are effective in the treatment of staphylococcal infections (45–47), suggesting it is a promising candidate for antibody-based immunotherapy during S. aureus infection. The present study suggests that IsaA is a promising target not only for immunotherapy but also for attenuating biofilm formation and β-lactam resistance, factors which make the treatment of S. aureus infections difficult. A better understanding of the role of IsaA in biofilm formation and β-lactam resistance may lead to the development of novel therapeutic strategies for S. aureus infections.
MATERIALS AND METHODS
Bacterial strains and mutants.
Bacterial strains and plasmids used in this study are listed in Table 1. Bacterial strains were grown overnight in BHI broth (Becton, Dickinson, Franklin Lakes, NJ) unless otherwise mentioned. All mutants were constructed from S. aureus MR23 and MR10, which are clinical MRSA isolates from Jikei University Hospital (27), and S. aureus USA300 (ATCC BAA1717). Genomic DNA extracted from S. aureus cells was used as a template for PCR. Allelic replacements of the entire sequences of isaA, sceD, and mecA were achieved according to the procedure reported by Bae et al. (48). Briefly, approximately 500-bp upstream and downstream sequences of the relevant gene were amplified by PCR, using KOD-plus v.2 DNA polymerase (Toyobo, Tokyo, Japan) and the primers listed in Table S2 in the supplemental material. The resulting PCR fragments were connected by overlap extension PCR. The generated PCR product was cloned into pKOR1 using the Gateway BP Clonase II enzyme mix (Life Technologies, Palo Alto, CA). The resulting plasmids were amplified in Escherichia coli DC10B (49, 50) and then introduced into S. aureus by electroporation. After allelic replacement, the generated mutant strains were verified by gene sequencing confirming that the entire open reading frame was accurately deleted.
Antibiotics and antibodies.
Ampicillin and gentamicin were obtained from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). All other antibiotics were purchased from Sigma-Aldrich (St. Louis, MO). Antibiotic stock solutions were prepared in sterile distilled water or sterile recommended solvent (51) and then diluted in fresh broth as described below. Anti-IsaA polyclonal antibody against a synthesized peptide fragment of IsaA was raised in rabbits as previously reported (52). Anti-IsaA antibody was purified from rabbit serum with a MAb Trap kit (GE Healthcare, Little Chalfont, UK) according to the manufacturer’s instructions. For overexpression and purification of the SceD protein, sceD was amplified from the genome of S. aureus MR23 by PCR using the primers listed in Table S2. The amplified PCR fragment was cloned into the pET29a vector (Novagen, Madison, WI) using an In-Fusion HD cloning kit (TaKaRa Bio, Otsu, Japan) according to the manufacturer’s protocol. The resulting plasmid pET_sceD was amplified in E. coli DH5α and subsequently introduced into E. coli BL21(DE3) by electroporation. E. coli BL21(DE3) harboring pET_sceD was cultured in Luria-Bertani broth containing 50 μg/ml kanamycin at 37°C. When the optical density at 660 nm (OD600) reached 0.5, the culture was cooled and kept at 15°C for 30 min. Then, isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 1 mM to induce expression. Purification of SceD was performed according to the protocol for the purification of S. aureus enolase described previously (53). Anti-SceD polyclonal antibody was raised in rabbits using purified SceD as an antigen and was purified in the same way as anti-IsaA antibodies. Mouse anti-PBP2a antibody (product code, 130-10096-200) was purchased from Raybiotech, Inc. (Norcross, GA). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG or HRP-conjugated goat anti-mouse IgG (Bio-Rad, Hercules, CA) was used as the secondary antibody.
Plasmid-based expression of genes.
For plasmid-based expression of LT genes and mecA in S. aureus MR23, an expression vector was constructed by replacing the erythromycin resistance gene of pCN51 (54) with a chloramphenicol resistance gene amplified from pKOR1 (48). PCR was performed using KOD-plus v.2 DNA polymerase and the primers listed in Table S2. The two fragments were fused using an In-Fusion HD cloning kit according to the manufacturer’s protocol, thereby producing pCN51Cm. For expression of isaA and sceD, the fragments from the respective native promoter sequences to the stop codon of each gene were cloned. For expression of mecA, the fragments from the ribosomal binding site to the stop codon of the mecA gene were cloned. Each fragment was amplified using KOD-plus v.2 DNA polymerase and the primers listed in Table S2. The PCR products were subsequently cloned into pCN51Cm, which had been predigested with BamHI and EcoRI using an In-Fusion HD cloning kit. The resulting plasmids were amplified in E. coli DC10B and then introduced into S. aureus MR23. The generated plasmids were verified by DNA sequencing.
Bacterial growth and biofilm assays.
Overnight cultures of S. aureus strains were diluted 500-fold in BHIG, and 5 μg/ml of chloramphenicol (final concentration) was added to BHIG for strains harboring plasmid pCN51Cm and its derivatives. Subsequently, aliquots (200 μl) of this suspension were transferred to the wells of a 96-well microtiter plate (Corning Inc., Corning, NY) and incubated for 24 h at 37°C under static conditions. Bacterial growth was determined by measuring absorbance at 595 nm using a microplate spectrophotometer (Infinite F200 Pro; Tecan, Mannedorf, Switzerland). For biofilm assays, after removing the supernatant, the biofilms were washed twice with 200 μl of phosphate-buffered saline (PBS) to remove loosely adherent cells, dried, stained with 100 μl of 0.05% crystal violet for 1 min, and subsequently washed twice with PBS. Biofilm formation was assessed by measuring absorbance at 595 nm using a microplate spectrophotometer (Tecan). As the biofilms can form on the bottom wells in a patchy manner, multiple reads (4 × 4) per well function of the Infinite F200 Pro were performed to obtain the average value of the well. The background was not subtracted because the background value was exceedingly low in the dynamic range of this assay. The average value obtained from six wells represented biofilm formation.
Western blotting.
IsaA and SceD in culture supernatants and whole-cell fractions were detected by Western blotting. S. aureus strains were grown in BHIG for 24 h and then harvested by centrifugation at 5,000 × g for 10 min. The supernatants (culture supernatant fraction) were stored at –30°C until use. The harvested cells were treated with 25 μg/ml of lysostaphin (Wako Pure Chemical Industries, Ltd.) in PBS for 30 min at 37°C and subsequently disrupted by sonication. After centrifugation at 10,000 × g for 10 min, the supernatants (whole-cell fractions) were stored at –30°C until use. Culture supernatants (5 μl) or whole-cell fractions (5 μg of protein) were subjected to SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane (ATTO Corp., Tokyo, Japan). The membrane was treated for 1 h at room temperature with a blocking buffer composed of 5% Easy Blocker (GeneTex, Irvine, CA) in Tris-buffered saline with 0.05% Tween 20. The membrane was probed with a primary antibody (30 min) followed by a secondary antibody (30 min), both of which were diluted 1:10,000 in Can Get Signal solution (Toyobo). Signals were detected using enhanced chemiluminescence (ECL) prime (GE Healthcare) on an Image Quant LAS 4000 instrument (GE Healthcare).
PBP2a was detected as described previously (50). S. aureus cells were cultured in BHIG in the presence or absence of oxacillin (below its MIC) for 24 h. For expression of mecA cloned under the control of the cadmium-inducible promoter, CdCl2 (final concentration of 0.1, 0.5, or 1.0 μM) was added to BHIG. Cells were harvested by centrifugation, treated with lysostaphin, and disrupted by sonication. To avoid degradation during sample preparation, cOmplete Mini EDTA-free protease inhibitor cocktail (Roche, Indianapolis, IN) was added to the buffer. After centrifugation, the supernatants (mixture of cytoplasm and membrane fraction) were collected, and 5 μg of proteins was first run on SDS-PAGE and then electroblotted onto a nitrocellulose membrane. To detect PBP2a, primary (mouse anti-PBP2a, catalog number 130-10096-200; Raybiotech, Inc.) and secondary (HRP-conjugated anti-mouse IgG; Bio-Rad) antibodies were used. Coomassie brilliant blue (CBB)-stained gels were used as loading controls.
Antibiotic susceptibility testing.
Overnight cultures of S. aureus were diluted in BHIG or MH broth (Becton, Dickinson, Franklin Lakes, NJ) to prepare inocula of 5 × 105 CFU/ml. Five micrograms per milliliter of chloramphenicol (final concentration) was added to the medium for strains harboring plasmids pCN51Cm and its derivatives. MICs were determined by the broth microdilution method following the EUCAST guidelines (http://www.eucast.org/) using a commercially available MIC plate (Dry Plate Eiken; Eiken Chemical Co., Tokyo, Japan) or a laboratory-made plate. Three independent replicates were analyzed. Plates were incubated for 24 h at 37°C, with the exception of oxacillin, whose MIC was determined at 35°C. The MIC was defined as the lowest concentration that prevented the appearance of visible growth.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by JSPS KAKENHI grant numbers 26860294 and 19K07546, The Jikei University Research Fund, and Takeda Science Foundation. The authors have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01277-19.
REFERENCES
- 1.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szweda P, Schielmann M, Kotlowski R, Gorczyca G, Zalewska M, Milewski S. 2012. Peptidoglycan hydrolases-potential weapons against Staphylococcus aureus. Appl Microbiol Biotechnol 96:1157–1174. doi: 10.1007/s00253-012-4484-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto M. 2012. MRSA virulence and spread. Cell Microbiol 14:1513–1521. doi: 10.1111/j.1462-5822.2012.01832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Hal SJ, Jensen SO, Vaska VL, Espedido BA, Paterson DL, Gosbell IB. 2012. Predictors of mortality in Staphylococcus aureus bacteremia. Clin Microbiol Rev 25:362–386. doi: 10.1128/CMR.05022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peschel A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol 10:179–186. doi: 10.1016/s0966-842x(02)02333-8. [DOI] [PubMed] [Google Scholar]
- 7.Otto M. 2008. Staphylococcal biofilms. Curr Top Microbiol Immunol 322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37:1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holtje JV, Mirelman D, Sharon N, Schwarz U. 1975. Novel type of murein transglycosylase in Escherichia coli. J Bacteriol 124:1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheurwater E, Reid CW, Clarke AJ. 2008. Lytic transglycosylases: bacterial space-making autolysins. Int J Biochem Cell Biol 40:586–591. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Dik DA, Marous DR, Fisher JF, Mobashery S. 2017. Lytic transglycosylases: concinnity in concision of the bacterial cell wall. Crit Rev Biochem Mol Biol 52:503–542. doi: 10.1080/10409238.2017.1337705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodell EW. 1985. Recycling of murein by Escherichia coli. J Bacteriol 163:305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodell EW, Higgins CF. 1987. Uptake of cell wall peptides by Salmonella Typhimurium and Escherichia coli. J Bacteriol 169:3861–3865. doi: 10.1128/jb.169.8.3861-3865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park JT. 1993. Turnover and recycling of the murein sacculus in oligopeptide permease-negative strains of Escherichia coli: indirect evidence for an alternative permease system and for a monolayered sacculus. J Bacteriol 175:7–11. doi: 10.1128/jb.175.1.7-11.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cloud-Hansen KA, Peterson SB, Stabb EV, Goldman WE, McFall-Ngai MJ, Handelsman J. 2006. Breaching the great wall: peptidoglycan and microbial interactions. Nat Rev Microbiol 4:710–716. doi: 10.1038/nrmicro1486. [DOI] [PubMed] [Google Scholar]
- 16.Mercier C, Durrieu C, Briandet R, Domakova E, Tremblay J, Buist G, Kulakauskas S. 2002. Positive role of peptidoglycan breaks in lactococcal biofilm formation. Mol Microbiol 46:235–243. doi: 10.1046/j.1365-2958.2002.03160.x. [DOI] [PubMed] [Google Scholar]
- 17.Monteiro C, Fang X, Ahmad I, Gomelsky M, Romling U. 2011. Regulation of biofilm components in Salmonella enterica serovar Typhimurium by lytic transglycosylases involved in cell wall turnover. J Bacteriol 193:6443–6451. doi: 10.1128/JB.00425-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamers RP, Nguyen UT, Nguyen Y, Buensuceso RN, Burrows LL. 2015. Loss of membrane-bound lytic transglycosylases increases outer membrane permeability and β-lactam sensitivity in Pseudomonas aeruginosa. Microbiologyopen 4:879–895. doi: 10.1002/mbo3.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraft AR, Prabhu J, Ursinus A, Holtje JV. 1999. Interference with murein turnover has no effect on growth but reduces β-lactamase induction in Escherichia coli. J Bacteriol 181:7192–7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng X, Gillespie B, Lin J. 2015. Important role of a putative lytic transglycosylase Cj0843c in β-lactam resistance in Campylobacter jejuni. Front Microbiol 6:1292. doi: 10.3389/fmicb.2015.01292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavallari JF, Lamers RP, Scheurwater EM, Matos AL, Burrows LL. 2013. Changes to its peptidoglycan-remodeling enzyme repertoire modulate β-lactam resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:3078–3084. doi: 10.1128/AAC.00268-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stapleton MR, Horsburgh MJ, Hayhurst EJ, Wright L, Jonsson IM, Tarkowski A, Kokai-Kun JF, Mond JJ, Foster SJ. 2007. Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J Bacteriol 189:7316–7325. doi: 10.1128/JB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenz U, Ohlsen K, Karch H, Hecker M, Thiede A, Hacker J. 2000. Human antibody response during sepsis against targets expressed by methicillin resistant Staphylococcus aureus. FEMS Immunol Med Microbiol 29:145–153. doi: 10.1111/j.1574-695X.2000.tb01517.x. [DOI] [PubMed] [Google Scholar]
- 24.Clarke SR, Brummell KJ, Horsburgh MJ, McDowell PW, Mohamad SA, Stapleton MR, Acevedo J, Read RC, Day NP, Peacock SJ, Mond JJ, Kokai-Kun JF, Foster SJ. 2006. Identification of in vivo-expressed antigens of Staphylococcus aureus and their use in vaccinations for protection against nasal carriage. J Infect Dis 193:1098–1108. doi: 10.1086/501471. [DOI] [PubMed] [Google Scholar]
- 25.Sakata N, Terakubo S, Mukai T. 2005. Subcellular location of the soluble lytic transglycosylase homologue in Staphylococcus aureus. Curr Microbiol 50:47–51. doi: 10.1007/s00284-004-4381-9. [DOI] [PubMed] [Google Scholar]
- 26.Pieper R, Gatlin-Bunai CL, Mongodin EF, Parmar PP, Huang ST, Clark DJ, Fleischmann RD, Gill SR, Peterson SN. 2006. Comparative proteomic analysis of Staphylococcus aureus strains with differences in resistance to the cell wall-targeting antibiotic vancomycin. Proteomics 6:4246–4258. doi: 10.1002/pmic.200500764. [DOI] [PubMed] [Google Scholar]
- 27.Sugimoto S, Sato F, Miyakawa R, Chiba A, Onodera S, Hori S, Mizunoe Y. 2018. Broad impact of extracellular DNA on biofilm formation by clinically isolated Methicillin-resistant and -sensitive strains of Staphylococcus aureus. Sci Rep 8:2254. doi: 10.1038/s41598-018-20485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stapleton PD, Taylor PW. 2002. Methicillin resistance in Staphylococcus aureus: mechanisms and modulation. Sci Prog 85:57–72. doi: 10.3184/003685002783238870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyce JM, Medeiros AA, Papa EF, O'Gara CJ. 1990. Induction of β-lactamase and methicillin resistance in unusual strains of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother 25:73–81. doi: 10.1093/jac/25.1.73. [DOI] [PubMed] [Google Scholar]
- 30.Sato T, Usui M, Konishi N, Kai A, Matsui H, Hanaki H, Tamura Y. 2017. Closely related methicillin-resistant Staphylococcus aureus isolates from retail meat, cows with mastitis, and humans in Japan. PLoS One 12:e0187319. doi: 10.1371/journal.pone.0187319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 32.Stefani S, Chung DR, Lindsay JA, Friedrich AW, Kearns AM, Westh H, Mackenzie FM. 2012. Meticillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int J Antimicrob Agents 39:273–282. doi: 10.1016/j.ijantimicag.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 33.Arêde P, Oliveira DC. 2013. Proteolysis of mecA repressor is essential for expression of methicillin resistance by Staphylococcus aureus. Antimicrob Agents Chemother 57:2001–2002. doi: 10.1128/AAC.02510-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arêde P, Milheirico C, de Lencastre H, Oliveira DC. 2012. The anti-repressor MecR2 promotes the proteolysis of the mecA repressor and enables optimal expression of β-lactam resistance in MRSA. PLoS Pathog 8:e1002816. doi: 10.1371/journal.ppat.1002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hackbarth CJ, Chambers HF. 1993. blaI and blaR1 regulate β-lactamase and PBP 2a production in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 37:1144–1149. doi: 10.1128/aac.37.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amoroso A, Boudet J, Berzigotti S, Duval V, Teller N, Mengin-Lecreulx D, Luxen A, Simorre JP, Joris B. 2012. A peptidoglycan fragment triggers β-lactam resistance in Bacillus licheniformis. PLoS Pathog 8:e1002571. doi: 10.1371/journal.ppat.1002571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson JW, Fisher JF, Mobashery S. 2013. Bacterial cell-wall recycling. Ann N Y Acad Sci 1277:54–75. doi: 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park JT, Uehara T. 2008. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol Mol Biol Rev 72:211–227. doi: 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobs C, Huang LJ, Bartowsky E, Normark S, Park JT. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J 13:4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobs C, Frere JM, Normark S. 1997. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in gram-negative bacteria. Cell 88:823–832. doi: 10.1016/s0092-8674(00)81928-5. [DOI] [PubMed] [Google Scholar]
- 41.Borisova M, Gaupp R, Duckworth A, Schneider A, Dalugge D, Muhleck M, Deubel D, Unsleber S, Yu W, Muth G, Bischoff M, Gotz F, Mayer C. 2016. Peptidoglycan recycling in gram-positive bacteria is crucial for survival in stationary phase. mBio 7:e00923-16. doi: 10.1128/mBio.00923-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Memmi G, Filipe SR, Pinho MG, Fu Z, Cheung A. 2008. Staphylococcus aureus PBP4 is essential for β-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob Agents Chemother 52:3955–3966. doi: 10.1128/AAC.00049-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van den Berg S, Koedijk DG, Back JW, Neef J, Dreisbach A, van Dijl JM, Bakker-Woudenberg IA, Buist G. 2015. Active immunization with an octa-valent Staphylococcus aureus antigen mixture in models of S. aureus bacteremia and skin infection in mice. PLoS One 10:e0116847. doi: 10.1371/journal.pone.0116847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Berg S, de Vogel CP, van Belkum A, Bakker-Woudenberg IA. 2015. Mild Staphylococcus aureus skin infection improves the course of subsequent endogenous S. aureus bacteremia in mice. PLoS One 10:e0129150. doi: 10.1371/journal.pone.0129150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorenz U, Lorenz B, Schmitter T, Streker K, Erck C, Wehland J, Nickel J, Zimmermann B, Ohlsen K. 2011. Functional antibodies targeting IsaA of Staphylococcus aureus augment host immune response and open new perspectives for antibacterial therapy. Antimicrob Agents Chemother 55:165–173. doi: 10.1128/AAC.01144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oesterreich B, Lorenz B, Schmitter T, Kontermann R, Zenn M, Zimmermann B, Haake M, Lorenz U, Ohlsen K. 2014. Characterization of the biological anti-staphylococcal functionality of hUK-66 IgG1, a humanized monoclonal antibody as substantial component for an immunotherapeutic approach. Hum Vaccin Immunother 10:926–937. doi: 10.4161/hv.27692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Berg S, Bonarius HP, van Kessel KP, Elsinga GS, Kooi N, Westra H, Bosma T, van der Kooi-Pol MM, Koedijk DG, Groen H, van Dijl JM, Buist G, Bakker-Woudenberg IA. 2015. A human monoclonal antibody targeting the conserved staphylococcal antigen IsaA protects mice against Staphylococcus aureus bacteremia. Int J Med Microbiol 305:55–64. doi: 10.1016/j.ijmm.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 48.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 49.Jones MJ, Donegan NP, Mikheyeva IV, Cheung AL. 2015. Improving transformation of Staphylococcus aureus belonging to the CC1, CC5 and CC8 clonal complexes. PLoS One 10:e0119487. doi: 10.1371/journal.pone.0119487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3:e00277-11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.European Committee for Antimicrobial Susceptibility Testing of the European Society of Clinical Microbiology and Infectious Diseases. 2000. EUCAST definitive document E.DEF 3.1, June 2000: determination of minimum inhibitory concentrations (MICs) of antibacterial agents by agar dilution. Clin Microbiol Infect 6:509–515. doi: 10.1046/j.1469-0691.2000.00142.x. [DOI] [PubMed] [Google Scholar]
- 52.Payne DE, Martin NR, Parzych KR, Rickard AH, Underwood A, Boles BR. 2013. Tannic acid inhibits Staphylococcus aureus surface colonization in an IsaA-dependent manner. Infect Immun 81:496–504. doi: 10.1128/IAI.00877-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshii Y, Okuda K, Yamada S, Nagakura M, Sugimoto S, Nagano T, Okabe T, Kojima H, Iwamoto T, Kuwano K, Mizunoe Y. 2017. Norgestimate inhibits staphylococcal biofilm formation and resensitizes methicillin-resistant Staphylococcus aureus to β-lactam antibiotics. NPJ Biofilms Microbiomes 3:18. doi: 10.1038/s41522-017-0026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl Environ Microbiol 70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.