A novel KPC variant, KPC-41, was identified in a Klebsiella pneumoniae clinical isolate from Switzerland. This β-lactamase possessed a 3-amino-acid insertion (Pro-Asn-Lys) located between amino acids 269 and 270 compared to the KPC-3 amino acid sequence.
KEYWORDS: KPC, ceftazidime-avibactam, Klebsiella pneumoniae
ABSTRACT
A novel KPC variant, KPC-41, was identified in a Klebsiella pneumoniae clinical isolate from Switzerland. This β-lactamase possessed a 3-amino-acid insertion (Pro-Asn-Lys) located between amino acids 269 and 270 compared to the KPC-3 amino acid sequence. Cloning and expression of the blaKPC-41 gene in Escherichia coli, followed by determination of MIC values and kinetic parameters, showed that KPC-41, compared to those of KPC-3, has an increased affinity to ceftazidime and a decreased sensitivity to avibactam, leading to resistance to ceftazidime-avibactam once produced in K. pneumoniae. Furthermore, KPC-41 exhibited a drastic decrease of its carbapenemase activity. This report highlights that a diversity of KPC variants conferring resistance to ceftazidime-avibactam already circulate in Europe.
INTRODUCTION
As part of the antibiotic resistance crisis, carbapenemase-producing Enterobacterales (CPE) are among the main concerns. Indeed, apart from their resistance to almost all β-lactams, including carbapenems, which is observed most of the time, they are frequently coresistant to other antibiotic families (1). The main carbapenemases identified worldwide are the serine carbapenemases of the KPC and OXA-48 types, and the metallo-β-lactamases (MBLs) of the NDM, VIM, and IMP types (1). Development of novel drugs is mostly driven by the spread of those determinants. Among the recently developed drugs, combinations of β-lactams and β-lactamases inhibitors, such as ceftazidime-avibactam (CZA) and ceftolozane-tazobactam, are very successful alternatives (2). Although serine β-lactamases are inhibited by avibactam (AVI), this combination is not active against MBLs (2). Despite a still limited use of CZA at a worldwide scale, KPC variants resistant to AVI have been already reported (3–9). Such resistance is mostly driven by amino acid substitutions in the sequence of the KPC carbapenemase that might be selected after a CZA treatment (22). Variants conferring CZA resistance are derivatives of either KPC-2 or KPC-3 enzymes. Substitutions in the omega loop (amino acid positions 164 to 179) have been often associated with concomitant enhanced affinity toward ceftazidime and prevention of binding to AVI (7–10).
Here we describe β-lactamase KPC-41, a variant of KPC-3 conferring resistance to CZA. This novel KPC variant possesses a 3-amino-acid insertion in its protein sequence compared to KPC-3 and has been identified from a Klebsiella pneumoniae clinical isolate.
RESULTS AND DISCUSSION
K. pneumoniae strains FF and UM were recovered from rectal swabs of a 72-year-old man with pancreatic cancer, hospitalized at the University Hospital Center of Lausanne, Switzerland. Actually, this patient was living and previously hospitalized in Sicily before his transfer with a known history of a ciprofloxacin-containing treatment for cholangitis. At his admission in Switzerland, he screened positive for a KPC-3-positive K. pneumoniae strain (isolate FF). After pancreatic surgery performed in Switzerland, he was treated with piperacillin-tazobactam. Following a peritonitis episode, colistin (loading dose of 3 g, followed by 1.5 g/12 h) for 28 days and CZA (2/0.5 g for 5 days followed by 1/0.25g/8h for 19 days) were administered (Fig. 1). Then systematic rectal screenings identified another K. pneumoniae strain (isolate UM) (see below). Finally, this patient developed a cholangitis episode as a result of a biliary stent occlusion. He was treated successfully with colistin (one loading dose of 3 g followed by 3 g/day) for 10 days and meropenem (2 g/8 h) for 12 days (Fig. 1).
FIG 1.
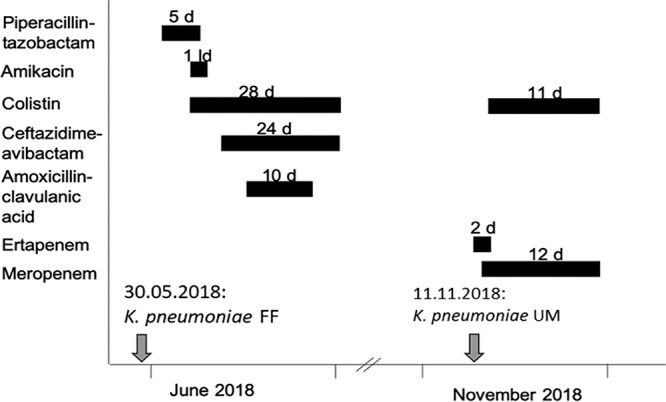
Timetable of antimicrobial treatments and isolation of the K. pneumoniae strains FF and UM. Black bars represent antimicrobial treatments, with numbers representing treatment length in days (d) or the loading dose (ld). Gray arrows indicate the dates of isolation of K. pneumoniae isolates FF and UM.
K. pneumoniae isolate UM was resistant to expanded-spectrum cephalosporins, ertapenem, CZA, and aztreonam and remained susceptible to imipenem and meropenem (Table 1). This isolate remained susceptible to colistin according to the negative result of the rapid polymyxin test (data not shown). Carbapenemase activity was assessed according to the results of the rapid Carba NP test, which was positive for both K. pneumoniae FF and K. pneumoniae UM isolates. Hence, common procedures to screen for carbapenem-resistant isolates and to detect any carbapenemase activity allowed the identification of K. pneumoniae UM.
TABLE 1.
MICs of β-lactams for K. pneumoniae clinical isolates, E. coli TOP10 transformants, and E. coli TOP10 recipient strains
| β-Lactama | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
|
K. pneumoniae |
E. coli |
||||
|
UM (KPC-41) |
FF (KPC-3) |
TOP10 (pTOPO-KPC-41) |
TOP10 (pTOPO-KPC-3) |
TOP10 | |
| Amoxicillin | >128 | >128 | >128 | >128 | 8 |
| Amoxicillin + CLA | >128 | >128 | >128 | >128 | 8 |
| Ticarcillin | >128 | >128 | >128 | >128 | 4 |
| Ticarcillin + CLA | >128 | >128 | >128 | >128 | 4 |
| Piperacillin | >128 | >128 | 128 | >128 | 2 |
| Piperacillin + TZB | >128 | >128 | 128 | >128 | 2 |
| Cephalothin | >128 | >128 | >128 | >128 | 16 |
| Cefotaxime | 32 | >128 | 16 | >128 | <0.125 |
| Ceftazidime | 1,024 | 1,024 | 2,048 | 2,048 | 0.25 |
| Ceftazidime + AVI | >128 | 4 | 128 | 2 | 0.25 |
| Ceftaroline | >256 | >256 | >256 | >256 | 0.25 |
| Cefepime | 16 | 128 | 16 | 128 | <0.125 |
| Ceftolozane + TZB | >256 | 64 | >256 | >256 | 0.06 |
| Cefoxitin | 32 | 32 | 8 | 128 | 8 |
| Aztreonam | >128 | >128 | 64 | 64 | <0.125 |
| Imipenem | 4 | 8 | 2 | 32 | 0.25 |
| Meropenem | 1 | 8 | 0.5 | 16 | 0.5 |
| Ertapenem | 4 | 16 | 0.5 | 32 | <0.12 |
Clavulanic acid (CLA) was added at a fixed of 2 μg/ml, tazobactam (TZB) at 4 μg/ml, and avibactam at 4 μg/ml.
Analysis of the whole-genome sequences of both isolates ruled out any clonal relationship. Isolate FF belonged to sequence type 307 (ST307), while isolate UM belonged to ST395. The antibiotic resistome of K. pneumoniae isolate UM included genes encoding β-lactamases TEM-1, SHV, and CMY.
Immediately after the isolation of isolate UM, it was found to be positive for blaKPC by PCR. Sequencing of the amplicon revealed a KPC variant possessing a 3-amino-acid insertion (Ambler 269-ProAsnLys-270) within a KPC-3 sequence, leading to a novel variant named KPC-41.
In order to evaluate the differences in term of hydrolysis spectra between KPC-41 and KPC-3, the corresponding genes were cloned and expressed in Escherichia coli TOP10. Once produced in this E. coli background, KPC-3 conferred resistance to all β-lactams, including ceftazidime, but it remained susceptible to CZA. On the other hand, KPC-41 conferred resistance to ceftazidime, reduced susceptibility to expanded-spectrum cephalosporins such as cefotaxime and cefepime, and yielded only a slight reduction in the susceptibility to carbapenems (Table 1). Noticeably, this recombinant E. coli strain producing KPC-41 was resistant to CZA (Table 1).
Some KPC variants conferring CZA resistance have been previously reported, exhibiting a single amino acid substitution often occurring in the omega loop, in particular at position Ambler 179 (8). In contrast, the amino acid insertion identified in KPC-41 was distantly located from the omega loop. Nevertheless, amino acid substitutions distantly located from the omega loop but leading to resistance to CZA have been also reported among KPC variants, such as KPC-8 (Val240Gly and His274Tyr) (6) and KPC-23 (Val240Ala and His274Tyr) (11).
The blaKPC-41-positive K. pneumoniae UM belonged to ST395, although the previously reported KPC producers exhibiting CZA resistance belonged to ST258 (6), ST307 (3), and ST1519 (11). It is noteworthy that an outbreak caused by a KPC-3-producing K. pneumoniae ST395 isolate (susceptibility to CZA not reported) has been reported in a neonatal intensive care unit in Palermo, Italy, and in our study the patient carrying K. pneumoniae UM also originated from Sicily (12).
Mating-out experiments performed with K. pneumoniae UM and using E. coli as a recipient (23) were successful and confirmed the plasmid location of the blaKPC-41 gene. Analysis of the corresponding plasmid revealed it was ca. 70 kb and belonged to the IncFII-type incompatibility group.
Purification of both KPC-3 and KPC-41 enzymes was performed, reaching a purity estimated to be 90%, with a single dominant band on the SDS-polyacrylamide gel for each protein (data not shown). Kinetic data showed that KPC-41 has a lower hydrolysis activity of cefotaxime, aztreonam, and imipenem than that of KPC-3 (Table 2). Similar decreased hydrolytic efficiencies for carbapenems of some KPC variants have previously been reported, such as for the Asp179Tyr KPC-2 mutant (10). In contrast, MIC values of aztreonam for KPC-41 and KPC-3 remained similar, as previously observed for the Thr243Ala KPC-3 variant (13).
TABLE 2.
Kinetic parameters of purified β-lactamases KPC-41 and KPC-3a
| β-Lactam | KPC-41 |
KPC-3 |
||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (mM−1·s−1) | kcat (s−1) | Km (μM) | kcat/Km (mM−1·s−1) | |
| Benzylpenicillin | 5.07 | 36 | 0.15 | 5.6 | 33 | 0.2 |
| Cephalothin | 4.75 | 102 | 0.03 | 47 | 113.5 | 0.4 |
| Cefotaxime | 1 | 116.7 | 0.008 | 34.9 | 532.8 | 0.065 |
| Ceftazidime | ND | ND | ND | >3.3 | >700 | >4.7 E−3 |
| Aztreonam | 1.9 | 190.16 | 0.01 | 5 | 194.8 | 0.03 |
| Imipenem | 0.17 | 43.8 | 0.004 | 4.7 | 71.5 | 0.07 |
| Meropenem | ND | ND | ND | 0.47 | 18.5 | 0.03 |
| Ertapenem | ND | ND | ND | 0.58 | 37 | 0.02 |
kcat, turnover; Km, Michaelis constant (affinity); kcat/Km, specificity constant (hydrolysis). ND, not determinable due to a low initial rate of hydrolysis.
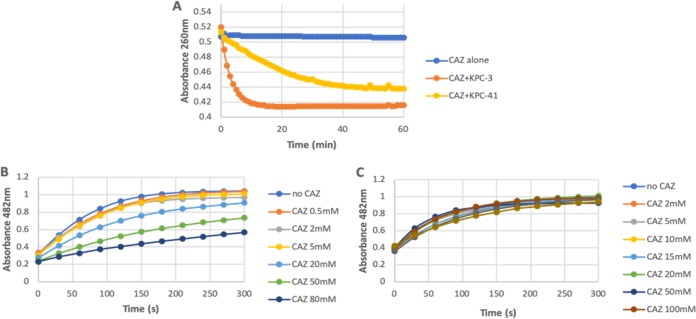
Kinetic analyses measuring the hydrolysis of ceftazidime were conducted for KPC-3 and KPC-41 enzymes. Although significant hydrolysis was detected with KPC-3, no hydrolysis could be detected with purified KPC-41 under normal conditions (measurement made over 5 min); however, by performing the assay for 1 h, ceftazidime hydrolysis was detected with KPC-41, but the hydrolysis rate was much lower than that of KPC-3 (Fig. 2A).
FIG 2.
Analysis of ceftazidime hydrolysis. (A) KPC-41 and KPC-3 (1 μM enzyme) hydrolysis of 25 μM ceftazidime (CAZ) at room temperature. (B and C) Competitive inhibition curves determined with 50 μM nitrocefin and increasing concentrations of CAZ with 0.1 μM KPC-41(B) and 0.1 μM KPC-3 (C) at room temperature. For panels B and C, nitrocefin absorbance was measured.
Considering that KPC-41 conferred significant resistance to ceftazidime once produced by the recombinant E. coli clone, it was therefore speculated that KPC-41 could strongly bind to ceftazidime but without efficient hydrolysis. Therefore, the affinities of KPC-3 and KPC-41 for ceftazidime were measured using various concentrations of ceftazidime, to inhibit the hydrolysis of a reporter substrate (nitrocefin). At the same ceftazidime concentrations, a higher level of inhibition of nitrocefin was observed in the presence of KPC-41 than in the presence of KPC-3 (Fig. 2B and C), and the Ki of ceftazidime was found to be 3-fold lower for KPC-41 than for KPC-3. Decreased ceftazidime hydrolysis and increased ceftazidime affinity have been also observed for the KPC-2 Asp179Asn variant conferring resistance to CZA (7).
Comparative inhibitory activities of clavulanic acid, tazobactam, and AVI were determined for KPC-41 and KPC-3 (Table 3). The inhibitory activity of AVI toward KPC-41 was much lower than toward KPC-3, while conversely, those of tazobactam and clavulanic acid were similar toward KPC-41 and KPC-3. An increased inhibitory activity of the β-lactamase inhibitor clavulanic acid, whose structure is basically very different from that of AVI but is more related to that of tazobactam, has been also reported for the Asp179Tyr mutant of KPC-2 (10).
TABLE 3.
Inhibitory concentrations and kinetic inhibition parameters of β-lactamase inhibitors against KPC-41 and KPC-3a
| Inhibitor | IC50 (μM) |
Ki (μM) |
||
|---|---|---|---|---|
| KPC-41 | KPC-3 | KPC-41 | KPC-3 | |
| Clavulanic acid | 15 | 20 | 10 | 20 |
| Tazobactam | 20 | 50 | 3 | 10 |
| Avibactam | 6 | 1 | 4 | 1 |
IC50 represents the concentration of a drug that is required for 50% inhibition of the enzymatic activity. Ki corresponds to a relative koff/kon to the inhibitor for the enzyme.
Together, these results indicate that the 269-ProAsnLys-270 insertion observed in the KPC-41 sequence was responsible for reduced hydrolysis of cephalothin, cefotaxime, and imipenem, associated with a higher affinity toward ceftazidime and with a reduced sensitivity to AVI. It is noteworthy that KPC-3 differs from KPC-2 by one amino acid substitution at position 273 that confers already a ca. 30-fold increase of hydrolytic efficiency towards ceftazidime (14).
Conclusion.
We report here a multidrug-resistant K. pneumoniae isolate producing KPC-41, a novel KPC variant conferring resistance to CZA. This is the first report of a K. pneumoniae isolate exhibiting such a resistance pattern in Switzerland. The patient had been treated with CZA. Several reports of KPC variants conferring CZA resistance originate from Italy, and the ST395 background of the KPC-41-producing strain has been shown to be widespread in some Italian hospitals. Therefore, it might be speculated that this Italian patient, hospitalized in Sicily, had been colonized there by K. pneumoniae producing KPC-41 and that the further treatment with CZA given in Switzerland likely contributed to its selection.
From a therapeutic point of view, full or partial recovery of susceptibility to carbapenems observed in such a KPC variant may be considered good news. In the present case, a therapeutic combination of meropenem and colistin was successful. Use of meropenem may be justified by the in vitro data of reversion of KPC-3 mutations leading to CZA resistance after serial passages of cultures with meropenem (15). We may also consider double-carbapenem therapy (ertapenem and meropenem, for example) as suggested for treating carbapenemase producers in Enterobacterales (16). Finally, the report underlines that such an isolate can be recovered by routine screening of carbapenem-resistant strains (at least by using the SuperCarba CHROMagar medium), since the isolate remained resistant to ertapenem, and subsequently by identification of the carbapenemase activity by using a biochemical test such as the rapid Carba NP test.
MATERIALS AND METHODS
Clinical strains and genome analyses.
The National Reference Center for Emerging Antibiotic Resistance (NARA; Fribourg, Switzerland) received K. pneumoniae isolates FF and UM. The genetic similarity of the K. pneumoniae isolates was evaluated by whole-genome sequencing using both the Illumina and Nanopore technologies. DNA was extracted with the Wizard Genomic DNA purification kit (Promega; A1120) and libraries were prepared with FC-131-1096 NexteraVRXT. Divergency between the two strains was evaluated by mapping (bwa 0.7.17) reads of K. pneumoniae UM on the assembled (Spades 3.11.1) genome of K. pneumoniae FF. Variant calling was performed with GATK’s HaplotypeCaller. Only genotype calls with ≥10 reads or supported by ≥75% of the observations were retained.
Conjugation experiments.
Mating-out assays were performed using K. pneumoniae UM as the donor and E. coli J53 (azide resistant) as the recipient. Selection was made using Trypticase-soy plates supplemented with amoxicillin (100 μg/ml) and sodium azide (100 μg/ml). Typing of plasmids was performed as described previously (17).
Cloning and sequencing.
The blaKPC-3 and blaKPC-41 genes were amplified from DNA of the K. pneumoniae FF and UM isolates using primers KPC-3-all-Fw (5′-TATATGAATTCAAGGGCGGCTGAAGGAATAC-3′) and KPC-3-all-Rev (5′-ATATAGAATTCCGCCATCGTCAGTGCTCTAC-3′). PCR products were cloned into pCR-BluntII-Topo (Invitrogen, Thermo Fisher). Recombinant plasmids were further transformed into E. coli strain TOP10. Sequencing of the amplicons and recombinant plasmids was performed by the company Microsynth (Balgach, Switzerland).
Carbapenemase activity and antimicrobial susceptibility testing.
Carbapenemase activity was detected by using the rapid Carba NP test (18). MIC determinations were performed in triplicate by the broth microdilution method in Mueller-Hinton broth (Bio-Rad, Marnes-La-Coquette, France), with the exception of the combinations CZA and ceftolozane-tazobactam, for which MICs were determined by Etest (AB bioMérieux, Solna, Sweden). Results were interpreted according to EUCAST breakpoints (19). The CZA resistance breakpoint is defined as >8 μg/ml for Enterobacterales. MICs of β-lactams were determined alone or in combination with a fixed concentration of clavulanic acid (2 μg/ml), tazobactam (4 μg/ml), or AVI (4 μg/ml). The antimicrobial agents were obtained from standard laboratory powders and were used immediately after their solubilization. The agents and their sources were as follows: amoxicillin, piperacillin, cefepime, cephalothin, ceftazidime, and clavulanic acid, Sigma (Saint-Quentin Falavier, France); ticarcillin and cefoxitin, ROTH (Arlesheim, Switzerland); benzylpenicillin and tazobactam, Abcam (Cambridge, UK); cefotaxime, aztreonam, and ertapenem, Acros Organic (Geel, Belgium); imipenem, Carbosynth (Berkshire, UK), meropenem, Combi-Blocks (San Diego, CA); AVI, MedChem Express (Luzern, Switzerland); and ceftolozane, ACS DOBFAR (Tribiano, Italy). Susceptibility to colistin was evaluated by using the rapid polymyxin NP test (20).
Screening of carbapenem-resistant Gram negatives was performed by using mSuperCARBA plates (CHROMagar, Paris, France).
β-Lactamase purification and relative molecular mass determination.
Cultures of E. coli TOP10 harboring plasmid pTOPO-KPC-3 and pTOPO-KPC-41 were grown overnight at 37°C in 2 liters of Luria broth with kanamycin (50 μg/ml). The bacterial suspensions were pelleted, resuspended in 20 ml of 50 mM morpholine ethanesulfonic acid (MES) buffer (pH 5.5), disrupted by sonication, and centrifuged at 11,000 × g for 1 h at 4°C. The enzyme extract was loaded onto a preequilibrated HiTrap Q HP column (GE Healthcare) with MES buffer. The resulting enzyme extract was recovered in the flowthrough and was then eluted with a linear NaCl gradient (0 to 1 M). The fractions showing the highest β-lactamase activity were pooled and dialyzed against 100 mM phosphate buffer (pH 7.0), prior to a 10-fold concentration with a Vivaspin 20 (GE Healthcare). The purified β-lactamase extract was immediately used for enzymatic determinations. To assess the relative purity of the extracts and to determine the molecular weight of the KPC-3 and KPC-41 β-lactamases, purified enzymes were subjected to SDS-PAGE analysis. Enzyme extracts and marker proteins were boiled for 10 min in a 1% SDS–3% β-mercaptoethanol solution before being separated by electrophoresis (100 V for 1 h) at room temperature. In parallel, molecular weights were determined in silico using the Compute pI/Mw tool on the Expasy server (21).
Kinetic measurements.
Purified β-lactamases were used for kinetic measurements (Km and kcat), performed at 30°C in 100 mM sodium phosphate (pH 7.0). A Genesys 10S UV-visible (UV-Vis) spectrophotometer (Thermo Scientific) was used to determine the initial rates of hydrolysis. The following wavelengths and absorption coefficients (Λε) were used: for benzylpenicillin, 232 nm and −1,100 M−1 cm−1; for cephalothin, 62 nm and –7,960 M−1 cm−1; for ceftazidime, 260 nm and –8,660 M−1 cm−1; for cefotaxime, 265 nm and –6,260 M−1 cm−1; for imipenem, 297 nm and –9,210 M−1 cm−1; for meropenem, 298 nm and –10,940 M−1 cm−1; and for aztreonam, 318 nm and –640 M−1 cm−1.
Determination of enzyme parameters (Km and kcat) was unsuccessful for ceftazidime because of the low level of hydrolysis. For this reason, to analyze ceftazidime hydrolysis, the antibiotic was incubated with 1 μM KPC-3 or KPC-41 enzyme and hydrolysis was measured for 1 h, as reported previously (9). To compare the affinities of ceftazidime for KPC-3 and for KPC-41, we performed competitive inhibition of 50 μM nitrocefin using 0.1 μM KPC-3 or 0.1 μM KPC-41 and various concentration of ceftazidime, as reported previously (7).
The 50% inhibitory concentrations (IC50) were determined for clavulanic acid, tazobactam, and AVI for each enzyme. Various concentrations of these inhibitors were preincubated with the purified enzyme for 5 min at 30°C to determine the concentrations that reduced the hydrolysis rate of 100 μM cephalothin by 50%. The results are expressed in micromolar units. The total protein content was measured by Bradford assay.
The Ki value was determined by direct competition assays using 100 μM cephalothin. Inverse initial steady-state velocities (1/V0) were plotted against the inhibitor concentration ([I]) to obtain a straight line. The plots were linear and provided y-intercept and slope values used for Ki determination. Ki was determined by dividing the value for the y-intercept by the slope of the line and then corrected by taking into account the cephalothin affinity by the following equation: Ki (corrected) = Ki (observed)/(1 + [S]/Km). Here, “[S]” is the concentration of cephalothin (100 μM) used in the assay and Km is the Michaelis constant determined for cephalothin (102 μM for KPC-41 and 113.5 μM for KPC-3). IC50 and Ki values were determined in triplicate.
Accession number(s).
Genome sequences were deposited at EMBL/EBI under EBI project PRJEB33694. The sequence of KPC-41 was deposited in the NCBI database under GenBank accession number MK497255.
ACKNOWLEDGMENTS
This work was financed by the University of Fribourg, Switzerland, and by the Swiss National Science Foundation (projects FNS-31003A_163432).
We thank Trestan Pillonel for his contribution in the bioinformatic analysis.
REFERENCES
- 1.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemases producers in Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Duin D, Bonomo RA. 2016. Ceftazidime/avibactam and ceftolozane/tazobactam: second-generation β-lactam/β-lactamase inhibitor combinations. Clin Infect Dis 63:234–241. doi: 10.1093/cid/ciw243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Humphries RM, Yang S, Hemarajata P, Ward KW, Hindler JA, Miller SA, Gregson A. 2015. First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 59:6605–6607. doi: 10.1128/AAC.01165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shields RK, Chen L, Cheng S, Chavda KD, Press EG. 2017. Emergence of ceftazidime-avibactam mutations during treatment. Antimicrob Agents Chemother 61:e02097-16. doi: 10.1128/AAC.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemarajata P, Humphries RM. 7 February 2019. Ceftazidime/avibactam resistance associated with L169P mutation in the omega loop of KPC-2. J Antimicrob Chemother doi: 10.1093/jac/dkz026. [DOI] [PubMed] [Google Scholar]
- 6.Gregory CJ, Llata E, Stine N, Gould C, Santiago LM, Vazquez GJ, Robledo IE, Srinivasan A, Goering RV, Tomashek KM. 2010. Outbreak of carbapenem‐resistant Klebsiella pneumoniae in Puerto Rico associated with a novel carbapenemase variant. Infect Control Hosp Epidemiol 31:476–484. doi: 10.1086/651670. [DOI] [PubMed] [Google Scholar]
- 7.Barnes MD, Winkler ML, Taracila MA, Page MG, Desarbre E, Kreiswirth BN, Shields RK, Nguyen M-H, Clancy C, Spellberg B, Papp-Wallace KM, Bonomo RA. 2017. Klebsiella pneumoniae carbapenemase-2 (KPC-2), substitutions at Ambler position Asp179, and resistance to ceftazidime-avibactam: unique antibiotic-resistant phenotypes emerge from β-lactamase protein engineering. mBio 8:e00528-17. doi: 10.1128/mBio.00528-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livermore DM, Warner M, Jamrozy D, Mushtaq S, Nichols WW, Mustafa N, Woodford N. 2015. In vitro selection of ceftazidime-avibactam resistance in Enterobacteriaceae with KPC-3 carbapenemase. Antimicrob Agents Chemother 59:5324–5330. doi: 10.1128/AAC.00678-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winkler ML, Papp-Wallace KM, Bonomo RA. 2015. Activity of ceftazidime/avibactam against isogenic strains of Escherichia coli containing KPC and SHV β-lactamases with single amino acid substitutions in the Ω-loop. J Antimicrob Chemother 70:2279–2286. doi: 10.1093/jac/dkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Compain F, Arthur M. 2017. Impaired inhibition by avibactam and resistance to the ceftazidime-avibactam combination due to the D179Y substitution in the KPC-2 β-lactamase. Antimicrob Agents Chemother 61:e00451-17. doi: 10.1128/AAC.00451-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galani I, Antoniadou A, Karaiskos I, Kontopoulou K, Giamarellou H, Souli M. 2019. Genomic characterization of a KPC-23-producing Klebsiella pneumoniae ST258 clinical isolate resistant to ceftazidime-avibactam. Clin Microbiol Infect 25:763.e5–763.e8. doi: 10.1016/j.cmi.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Maida CM, Bonura C, Geraci DM, Graziano G, Carattoli A, Rizzo A, Torregrossa MV, Vecchio D, Giuffrè M. 2018. Outbreak of ST395 KPC-producing Klebsiella pneumoniae in a neonatal intensive care unit in Palermo. Infect Control Hosp Epidemiol 39:496–498. doi: 10.1017/ice.2017.267. [DOI] [PubMed] [Google Scholar]
- 13.Shields RK, Nguyen MH, Press EG, Chen L, Kreiswirth BN, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance and restoration of carbapenem susceptibility in Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: a case report and review of literature. Open Forum Infect Dis 4:ofx101. doi: 10.1093/ofid/ofx101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alba J, Ishii Y, Thomson K, Moland ES, Yamaguchi K. 2005. Kinetics study of KPC-3, a plasmid-encoded class A carbapenem-hydrolyzing β-lactamase. Antimicrob Agents Chemother 49:4760–4762. doi: 10.1128/AAC.49.11.4760-4762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shields RK, Nguyen MH, Press EG, Chen L, Kreiswirth BN, Clancy CJ. 2017. In vitro selection of meropenem resistance among ceftazidime-avibactam-resistant, meropenem-susceptible Klebsiella pneumoniae isolates with variant KPC-3 carbapenemases. Antimicrob Agents Chemother 61:e00079-17. doi: 10.1128/AAC.00079-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bulik CC, Nicolau DP. 2011. Double-carbapenem therapy for carbapenemase-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 55:3002–3004. doi: 10.1128/AAC.01420-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 18.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.EUCAST. 2019. Breakpoint tables for interpretation of MICs and zone diameters. Version 9 http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9_Breakpoint_Tables.pdf.
- 20.Nordmann P, Jayol A, Poirel L. 2016. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg Infect Dis 22:1038–1043. doi: 10.3201/eid2206.151840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server In Walker JM. (ed), The proteomics protocols handbook. Humana Press, Totowa, NJ. [Google Scholar]
- 22.Gaibani P, Campoli C, Lewis RE, Volpe SL, Scaltriti E, Giannella M, Pongolini S, Berlingeri A, Cristini F, Bartoletti M, Tedeschi S, Ambretti S. 2018. In vivo evolution of resistant subpopulations of KPC-producing Klebsiella pneumoniae during ceftazidime/avibactam treatment. J Antimicrob Chemother 73:1525–1529. doi: 10.1093/jac/dky082. [DOI] [PubMed] [Google Scholar]
- 23.Kieffer N, Nordmann P, Aires-de-Sousa M, Poirel L. 2016. High prevalence of carbapenemase-producing Enterobacteriaceae among hospitalized children in Luanda, Angola. Antimicrob Agents Chemother 60:6189–6192. doi: 10.1128/AAC.01201-16. [DOI] [PMC free article] [PubMed] [Google Scholar]