Pretomanid (PA-824) is a nitroimidazole in clinical testing for the treatment of tuberculosis. A population pharmacodynamic model for pretomanid was developed using a Bayesian analysis of efficacy data from two early bactericidal activity (EBA) studies, PA-824-CL-007 and PA-824-CL-010, conducted in Cape Town, South Africa.
KEYWORDS: pretomanid, PA-824, tuberculosis, pharmacokinetics, pharmacodynamics, mathematical modeling
ABSTRACT
Pretomanid (PA-824) is a nitroimidazole in clinical testing for the treatment of tuberculosis. A population pharmacodynamic model for pretomanid was developed using a Bayesian analysis of efficacy data from two early bactericidal activity (EBA) studies, PA-824-CL-007 and PA-824-CL-010, conducted in Cape Town, South Africa. The two studies included 122 adult male and female participants with newly diagnosed pulmonary tuberculosis who received once-daily oral pretomanid doses of either 50, 100, 150, 200, 600, 1,000, or 1,200 mg for 14 days. The structural model described capacity-limited growth and saturable drug-induced bacterial killing with separate rate equations for sputum solid culture CFU counts and liquid culture time to positivity (TTP) that were linked through a time constant. The posterior population geometric means and interindividual variability coefficients of variation were, respectively, 0.152 ± 0.013 log10 CFU/ml sputum/day and 54% ± 6% for the maximum kill rate constant, 770 ± 140 ng/ml and 48% ± 17% for the pretomanid half-maximum effect plasma concentration, and 20.4 ± 1.0 h and 20.8% ± 0.1% for the time constant of proportionality between the CFU and TTP rate equations. Model simulations showed once-daily pretomanid at 100, 200, and 300 mg attained 58, 73, and 80%, respectively, of an expected maximum 14-day EBA of 0.136 log10 CFU/ml sputum/day. These results establish a pretomanid exposure-efficacy relationship with dual outcomes for CFU counts and TTP and with potential applications to dose optimization of pretomanid-containing regimens.
INTRODUCTION
There are several novel regimens in clinical testing for the treatment of tuberculosis (TB) that include the investigational new drug pretomanid (PA-824) (1). Pretomanid is a bicyclic nitroimidazole with activity against replicating and nonreplicating Mycobacterium tuberculosis (2, 3), including hypoxia-adapted (2, 4) and acid-growth-arrested (5, 6) phenotypes. Preclinical testing of pretomanid showed bactericidal activity similar to that of isoniazid during the initial phase of treatment in mouse TB infection models (2, 4, 7), established a time-dependent pattern of mycobacterial killing (8), and found no indication of cross-resistance with commonly employed TB drugs (2, 4). Early-phase clinical testing of orally administered pretomanid with dosages ranging across 50 to 1,500 mg/day in healthy adults (9–12) and 50 to 1,200 mg/day in adults with newly diagnosed pulmonary TB (13, 14) showed that pretomanid was safe and exhibited bactericidal activity at all tested doses, and 100 to 200 mg/day was optimal for further clinical testing in combination regimens (14, 15). Pretomanid at 200 mg once daily is the administered dosage in current phase 3 trials for regimens that include pretomanid with bedaquiline, moxifloxacin, and pyrazinamide (16), with bedaquiline and linezolid (17–19), and with bedaquiline, moxifloxacin or clofazimine, and linezolid (19). Consideration of further dose optimization for the component drugs in such regimens may be aided by the application of mathematical modeling and simulation (20) to identify dose combinations that maximize the efficacy and safety of the regimen as a whole. The objective here is to develop a pharmacodynamic (PD) model for pretomanid in the context of an early bactericidal activity (EBA) study as a step toward mathematical model-based dose optimization (21) of pretomanid-containing regimens.
Current EBA studies are typically conducted as randomized phase 2a clinical trials that assess the safety, tolerability, drug exposure, and efficacy of one or more drugs up to the first 14 days of therapy in a small number of adult participants with sputum smear-positive pulmonary TB (22). EBA is the outcome measure of efficacy, determined as the daily rate of change of viable M. tuberculosis bacilli in serial sputum collections and calculated for various predefined time intervals during treatment as the decrease of solid culture log10 CFU counts per milliliter of sputum per day or as the increase in liquid culture time to positivity (TTP) per day (23). There were two EBA studies for pretomanid, PA-824-CL-007 (CL-007) (13) and PA-824-CL-010 (CL-010) (14), and both were conducted by the same principal investigators with similar study populations in Cape Town, South Africa. Combined, there were 63 male and 59 female adult participants (median age, 27 years; age range, 18 to 56 years) who received once-daily oral pretomanid doses of either 50, 100, 150, 200, 600, 1,000, or 1,200 mg for 14 days. Intensive pharmacokinetic (PK) sampling of each participant provided detailed plasma drug concentration-time profiles, and serial overnight sputum collections provided the corresponding time course of mycobacterial load determined from parallel CFU and TTP measurements. The results included informative pretomanid dose-exposure and dose-efficacy relationships that were based on noncompartmental analysis of the plasma drug concentrations and EBA calculations using the observed CFU and TTP data from each treatment group. While results from CL-007 and CL-010 regarding the safety and tolerability of pretomanid were also reported in their respective publications (13, 14), the focus here is the exposure-efficacy relationship.
The present study describes a PD model for pretomanid in adult patients with pulmonary TB disease. A structural model for the initial time course of sputum mycobacterial load was developed by combining a standard model for antimicrobial drug effect (24) with previously reported PK equations for pretomanid concentrations in plasma (25) and with a functional relationship between CFU counts and TTP that was based on a previously described correlation between their corresponding EBA values (23). Population- and individual-level parameter estimates were obtained using Bayesian hierarchical modeling (26) with the individual participant data from CL-007 and CL-010. The Bayesian analysis included informative prior population distributions and Markov chain Monte Carlo (MCMC) sampling to estimate the posterior distribution. Model simulations of population- and individual-level sputum CFU and TTP profiles were conducted to assess agreement between model output and observation and to compare model predictions of EBA for once-daily and twice-daily pretomanid dosing.
RESULTS
Structural model.
The PD model equations were developed to represent sputum CFU and TTP profiles for pretomanid monotherapy under EBA study conditions. EBA time-kill profiles (22, 27–31) are characterized by baseline bacillary loads (∼105 to 107 CFU/ml) that remain approximately stable over the study duration in the absence of drug and by one or more log-linear phases with different rates of decline that depend on the effectiveness of the drug regimen against possibly different mycobacterial subpopulations (27). For pretomanid, the bactericidal activity requires bioactivation by an M. tuberculosis deazaflavin-dependent nitroreductase (Ddn), with subsequent production of several reactive nitrogen species and with resulting effects that include inhibition of cell wall synthesis and respiratory poisoning (32, 33). Parallel TTP and CFU measurements on serial sputum collections show an inverse relationship, with decreasing CFU counts corresponding to increasing TTP (23). Additionally, previous comparisons between EBA values using log10 CFU/ml and TTP (23, 34, 35) suggested a proportional relationship between their corresponding instantaneous time rates of change.
Using a minimal set of biologically interpretable functions and parameters to account for the net effect of resource and immune system constraints on mycobacterial population growth in the lungs, and for a single target drug-receptor interaction for the enzymatic activation of pretomanid, the PD model was defined with separate rate equations for CFU/ml sputum (B) and TTP (T) as
where B = B(t) and T = T(t) are functions of elapsed time (t ≥ 0) since the start of sputum collection, and where B(0) = B(0) and T(0) = T(0) denote the initial conditions. The first term in the equation for B describes a capacity-limited increase in sputum bacilli parameterized by a mycobacterial accumulation rate constant (λ) and carrying capacity (K), and the second term describes a saturable drug-induced decrease of bacilli that depends on drug exposure (C), a drug kill rate constant (kd), and a drug exposure half-maximum effect (C50). Drug exposure was defined as C = C(t) = f·Cp(t)/MIC, where f is the free drug fraction, MIC is the minimum inhibitory concentration, and Cp is the concentration of pretomanid in plasma. Pretomanid PK equations for the calculation of Cp were described previously (25) and require the specification of a dosage regimen and parameter values for oral bioavailability, absorption, volume of distribution, and clearance, which, together with values for f and MIC, were considered here as given model inputs and covariates. The equation for T was derived from the equation for B through an equality between the time rates of change for log(B) and T, with the liquid culture time constant (τL) as a conversion factor. An additional relation between the in vitro mycobacterial doubling time (τdouble) and the patient-level liquid culture time constant, τL = τdouble/log(2), was determined from an assumption of log-phase bacterial growth in the liquid culture system.
Individual participant data.
Estimates for the PD model parameters were based on the individual participant data from CL-007 and CL-010, which were combined as a single data set based on the similarities of the two study designs, experimental methods, and participant characteristics (13, 14). There were 122 total participants (61 in CL-007 and 61 in CL-010) in the pretomanid treatment groups and 16 total participants (8 in CL-007 and 8 in CL-010) in the standard treatment (RHZE [rifampin, isoniazid, pyrazinamide, and ethambutol]) control groups. The PD model analysis excluded the same 4 participants from the pretomanid treatment groups that were excluded in the previously described PK analysis (25), and the numbers of observed CFU, TTP, and MIC values from the remaining 118 included participants are summarized in Table 1. There were 11 total sputum collection time points, including two that were pretreatment. While none of the included participants had completely missing CFU and TTP values, one participant in the CL-010 50-mg group, one in the CL-010 200-mg group, and one in the CL-007 600-mg group had completely missing CFU counts. There were two MIC measurements (one pretreatment and one on day 14) for each individual, with 22 of 27 missing values being from 11 participants who were missing both measurements. All but five of the observed MICs were recorded as <0.1 μg/ml, with the other five recorded as >0.4 μg/ml, and with four of the latter being day 14 measurements. The single participant with a pretreatment MIC of >0.4 μg/ml had a day 14 value of <0.1 μg/ml. As the MIC measurements were not available throughout treatment, and as no participant with recorded MICs had both values different from <0.1 μg/ml, a single MIC value equal to 0.1 μg/ml was set as a covariate for each individual, with the implication of pretomanid-susceptible TB for all of the included study participants. PK covariates consisted of individual point values for total body weight, oral absorption, distribution, and clearance, which together with a free drug fraction of f = 0.1 were determined previously (25). The values for f and MIC in the PD model equations served as scale factors for the pretomanid plasma exposure. The pretreatment CFU and TTP measurements from the RHZE control groups were included only to specify prior population distributions for the PD model initial conditions.
TABLE 1.
CL-007 and CL-010 observed efficacy dataa
| Groupb | No. M/F | Total no. (missing) of values by: |
||
|---|---|---|---|---|
| CFU | TTP | MIC | ||
| CL-010 | ||||
| Pretomanid | ||||
| 50 mg | 6/6 | 132 (19) | 132 (8) | 24 (1) |
| 100 mg | 8/7 | 165 (2) | 165 (4) | 30 (0) |
| 150 mg | 7/8 | 165 (4) | 165 (4) | 30 (0) |
| 200 mg | 7/9 | 176 (19) | 176 (8) | 32 (0) |
| RHZEc | 6/2 | 16 (0) | 16 (0) | − |
| CL-007 | ||||
| Pretomanid | ||||
| 200 mg | 7/7 | 154 (14) | 154 (17) | 28 (6) |
| 600 mg | 9/6 | 165 (20) | 165 (13) | 30 (6) |
| 1,000 mg | 9/7 | 176 (18) | 176 (23) | 32 (6) |
| 1,200 mg | 9/6 | 165 (8) | 165 (20) | 30 (8) |
| RHZEc | 4/4 | 16 (0) | 16 (0) | − |
Shown are numbers of male (M) and female (F) participants and total and missing CFU, TTP, and MIC values from each treatment group that were included for the PD model analysis.
Shown are the pretomanid treatment groups and RHZE (rifampin, isonazid, pyrazinamide, and ethambutol) standard treatment groups.
Pretreatment CFU and TTP measurements only. −, not included.
Prior distributions.
The independent prior distributions that were assigned to the population means and variances of each PD model parameter, θ = (B0, λ, K, kd, C50, T0, τL, and to the residual errors, σ = (σB, σB, are summarized in Table 2. Typical values for population parameters were defined using the geometric mean (GM) for central tendency, and the geometric standard deviation (GSD) or the coefficient of variation (CV) for dispersion. Each component of the population mean vector (μ) was assigned a normal (N) distribution, μ = log(GM[θ]) ∼ N(Mμ, Sμ2), with values for the mean (Mμ) and variance (Sμ2) that were based on estimates of a typical value for the corresponding PD model parameter and the uncertainty in that typical value, respectively. These distributions were truncated to limit sampling of implausible values but with ranges that were large enough to account for measurement uncertainty and interindividual variability. The population variances (ω2) were assigned half-normal (HN) distributions, ω2 = log(CV2 + 1) ∼ HN(Sω), with mean values set to typical value estimates of interindividual variability (36). The log uniform residual error distributions were specified with lower bounds that accounted for known measurement error (23) and upper bounds for additional unknown error and model misspecification.
TABLE 2.
Prior PD model population distribution parametersa
| Parameter | μ |
Sω for ω2 | [a, b] for σ | ||
|---|---|---|---|---|---|
| Mμ | Sμ2 | Range | |||
| B0 (CFU/ml) | 13.9 | 0.13 | [6.91, 20.7] | 5.3 | − |
| λ (1/h) | −6.91 | 0.15 | [−8.11, −5.81] | − | − |
| K (CFU/ml) | 18.4 | 0.6 | [16.1, 20.7] | − | − |
| kd (1/h) | −3.91 | 0.039 | [−5.81, −2.12] | 0.2 | − |
| C50 | −0.105 | 0.039 | [−2.30, 1.79] | 0.2 | − |
| T0 (h) | 4.66 | 0.0035 | [3.18, 5.99] | 0.14 | − |
| τL (h) | 3.56 | 0.01 | [2.08, 5.01] | 0.2 | − |
| σB | − | − | − | − | [1.1, 10] |
| σT | − | − | − | − | [1.1, 3] |
Values are the normal distribution mean (Mμ), variance (Sμ2), and sampling range for each component of the population mean vector (μ), the half-normal distribution SD (Sω) for each component of the population variance (ω2), and the log uniform distribution range [a, b] for the residual error distributions (σ). −, not included or not applicable.
The prior distributions for the initial conditions, B0 and T0, were based on bootstrap mean and bootstrap standard deviation (SD) distributions that were generated by resampling the observed pretreatment CFU and TTP values from the standard treatment control groups. The distributions for the carrying capacity, K, and rate constant, λ, were based on observed pretreatment sputum CFU values, as well as EBA values from untreated control groups that were reported in several early studies of adult patients with pulmonary TB (28, 30, 37–39), and were specified with measurement uncertainty and interindividual variability combined. The distributions for the maximum pretomanid mycobacterial kill rate, kd, and for the half-maximum effect exposure, C50, were based on preclinical mouse studies that included pretomanid time-kill and dose fractionation results (4, 8). The prior distribution parameters for the liquid culture time constant, τL, were based on a standard typical value (40) and an observed in vitro distribution for M. tuberculosis generation times (41).
Posterior distribution.
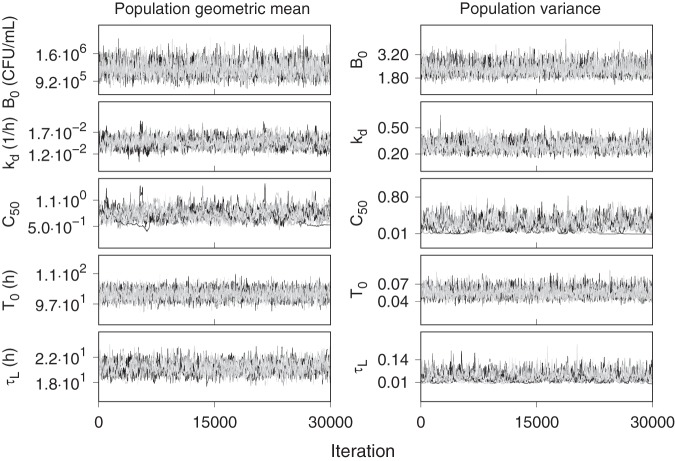
The prior distributions for the unmeasured PD model parameters were updated using Bayes’ rule, with a lognormal data likelihood, to a posterior distribution conditioned on the observed CFU and TTP profiles from the CL-007 and CL-010 pretomanid treatment group participants. The missing data mechanisms were assumed to be ignorable (42), and all CFU and TTP values were included for analysis without deletion of incomplete cases or missing data imputation and without prior identification or exclusion of outliers. The individual PK covariate data sets were fully specified, with no missing values. A 10,000-iteration sample of the posterior was obtained as the aggregate of 10 independent MCMC simulations, where each simulation consisted of 300,000 iterations with retention of every 30th iteration of the last 30,000. The distributions for λ and K were not updated in the posterior sampling but were treated as measured distributions that were randomly sampled with each of the MCMC iterations as individual covariate components. Figure 1 shows overlapping trace plots of the retained 1,000 values from each of the 10 MCMC sampling chains for each population parameter. Each set of chains fluctuated within well-defined ranges with stable patterns, which provided a basis for pooling the individual chains into a final aggregate posterior sample. Adequate convergence and precision of the posterior were found with the Gelman-Rubin statistic (R̂) less than 1.1, and an effective sample size (neff) greater than 500, respectively, for every sampled population- and individual-level parameter.
FIG 1.
Overlapping trace plots of the posterior population geometric means and population variances for the retained samples of the last 30,000 iterations from 10 independent 300,000-iteration MCMC simulations.
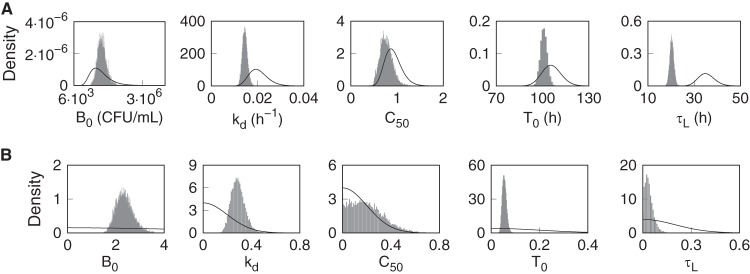
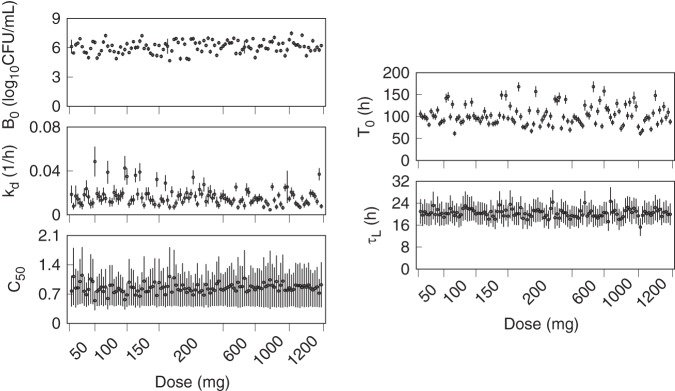
Figure 2 shows the prior population density functions and (normalized) histograms of the marginal population posterior distributions. The posterior mean distributions were unimodal and symmetric, with shifted peaks and smaller ranges than the priors, indicating informative data that reduced the prior uncertainty for each parameter. The posterior variance distributions were similarly well defined, with C50 being the least affected by the data. The mean and SD for each of the participant marginal posterior distributions were calculated and plotted versus the administered dose in Fig. 3. The distribution of the individual parameter means across the different pretomanid treatment groups did not indicate any strong dependence on the dose, but higher values for kd and lower values for C50 are evident for some individuals in the lower-dose groups. There was higher variation in the individual C50 mean values at the lower doses than for those above 200 mg, indicating a higher sensitivity of this parameter to the observed data from the lower-dose groups.
FIG 2.
Prior and posterior probability densities for the PD model parameters. (A) Prior (solid lines) and posterior (histograms) probability densities of the population geometric means. (B) Prior (solid lines) and posterior (histograms) probability densities of the population variances.
FIG 3.
Means (points) and SDs (error bars) of the individual marginal posterior parameter distributions and the administered dose for each pretomanid treatment group participant.
Summary statistics for the posterior marginal population distributions are shown in Table 3, and those for the individual-level marginal posterior distribution means, including for male and female participants separately, are shown in Table 4. Table 5 shows Pearson correlation coefficients between the individual PK covariate and posterior PD parameter means from the 118 individual participants. The highest correlations were among the drug effect parameters kd and C50 and the accumulation rate constant λ, as well as between the two initial conditions, T0 and B0. The negative correlation between the initial condition parameters was consistent with their relationship as inverse measures of the same mycobacterial cell density. The correlations among the accumulation rate and drug effect parameters reflect limiting cases for the rate equations, with a high average drug concentration and low cell density limit proportional to λ − kd and a low average drug concentration and high cell density limit proportional to kd⋅C̅/C50, where C̅ denotes the average concentration. As C̅ is proportional to dose, these latter correlations indicate the need for a wide range of tested doses to accurately estimate the drug effect parameters. Also, there were no significant correlations between the PK and PD parameter sets that would indicate redundant variables, which supported the separate development of the PK and PD model components.
TABLE 3.
Posterior population distributiona represented by summaries of the population geometric means, exp(μ), and CVs
| Parameter | exp(μ) |
CVb
|
||
|---|---|---|---|---|
| Mean (SD) | Median [2.5th, 97.5th]c | Mean (SD) | Median [2.5th, 97.5th]c | |
| B0 (CFU/ml) | 1.23 × 106 (1.71 × 105) | 1.22 × 106 [9.21 × 105, 1.60 × 106] | 3.1 (0.61) | 3.1 [2.2, 4.7] |
| kd (1/h) | 0.0146 (0.0012) | 0.0145 [0.0125, 0.0170] | 0.58 (0.07) | 0.58 [0.046, 0.072] |
| C50 | 0.77 (0.14) | 0.76 [0.53, 1.1] | 0.48 (0.17) | 0.46 [0.08, 0.82] |
| T0 (h) | 101 (2.21) | 101 [97.0, 106] | 0.24 (0.02) | 0.24 [0.21, 0.28] |
| τL (h) | 20.4 (1.02) | 20.3 [18.6, 22.2] | 0.21 (0.07) | 0.20 [0.08, 0.33] |
Sample size equal to 10,000.
CV = , where ω2 = population variance.
Values in brackets are the 2.5th and 97.5th percentiles.
TABLE 4.
Summaries of marginal posterior individual parameter means (SD) for the PD model parameters and comparison of values between male and female participants
| Parameter | Mean (SD) no. |
P valuea | ||
|---|---|---|---|---|
| Total (n = 118) | Male (n = 62) | Female (n = 56) | ||
| B0 (log10 CFU/ml) | 6.09 (0.62) | 6.12 (0.59) | 6.06 (0.66) | 0.59 |
| kd (1/h) | 0.017 (0.008) | 0.018 (0.010) | 0.015 (0.006) | 0.023 |
| C50 | 0.85 (0.11) | 0.82 (0.12) | 0.87 (0.10) | 0.013 |
| T0 (h) | 103.0 (23.3) | 103.0 (22.0) | 104.0 (24.8) | 0.81 |
| τL (h) | 20.4 (1.5) | 20.6 (1.4) | 20.2 (1.6) | 0.16 |
Comparison between males and females by Welch’s t test.
TABLE 5.
Correlation coefficientsa between the individual PKb covariates and PD marginal posterior parameter means
| Parameter | BW | ED50 | τF | ka | VC | CLC | Log10B0 | λ | K | kd | C50 | T0 | τL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BW | 1.00 | −0.08 | 0.00 | 0.35 | 0.10 | −0.08 | 0.01 | −0.07 | 0.01 | 0.12 | −0.11 | 0.03 | 0.13 |
| ED50 | 1.00 | −0.04 | −0.14 | −0.54 | −0.37 | −0.02 | 0.22 | 0.25 | −0.15 | 0.18 | −0.02 | −0.03 | |
| τF | 1.00 | 0.04 | 0.21 | 0.25 | −0.06 | 0.07 | 0.06 | −0.02 | 0.04 | 0.17 | −0.10 | ||
| ka | 1.00 | 0.34 | −0.26 | 0.02 | 0.10 | −0.03 | −0.17 | 0.17 | −0.08 | 0.11 | |||
| VC | 1.00 | −0.28 | 0.03 | 0.01 | −0.15 | −0.20 | 0.14 | −0.01 | 0.08 | ||||
| CLC | 1.00 | −0.09 | −0.24 | −0.05 | 0.36 | −0.38 | 0.16 | −0.05 | |||||
| Log10B0 | 1.00 | 0.16 | −0.01 | −0.01 | 0.05 | −0.65 | −0.44 | ||||||
| λ | 1.00 | 0.35 | −0.57 | 0.77 | 0.01 | −0.27 | |||||||
| K | 1.00 | −0.08 | 0.18 | −0.03 | −0.10 | ||||||||
| kd | 1.00 | −0.87 | −0.04 | 0.05 | |||||||||
| C50 | 1.00 | 0.04 | −0.16 | ||||||||||
| T0 | 1.00 | 0.26 | |||||||||||
| τL | 1.00 |
Pearson correlation calculated from 118 individual parameter sets.
PK parameters are from Lyons (25): BW, body weight; ED50, bioavailability oral dose at half-maximum effect; τF, oral bioavailability time constant; ka, oral absorption rate constant; VC, volume of distribution allometric constant; CLC, clearance allometric constant.
The rate equations and parameter values were based on CFU counts and the corresponding natural logarithm rate of change. For comparison with the standard log10 CFU scale, the population GM of the maximum pretomanid kill rate, kd = 0.0146 ± 0.0012/h ≈ 0.152 ± 0.013 log10 CFU/ml/day. A point estimate for the SD/mean calculated for log-transformed values of the posterior population distribution for B0 is obtained as . The population GM for the liquid culture time constant as an in vitro M. tuberculosis minimum doubling time is τdouble = log(2) · τL = 14.1 ± 0.7 h. Also, the half-maximum effect exposure expressed as a plasma concentration (Cp50) = MIC · C50/f = 770 ± 140 ng/ml.
Model simulations.
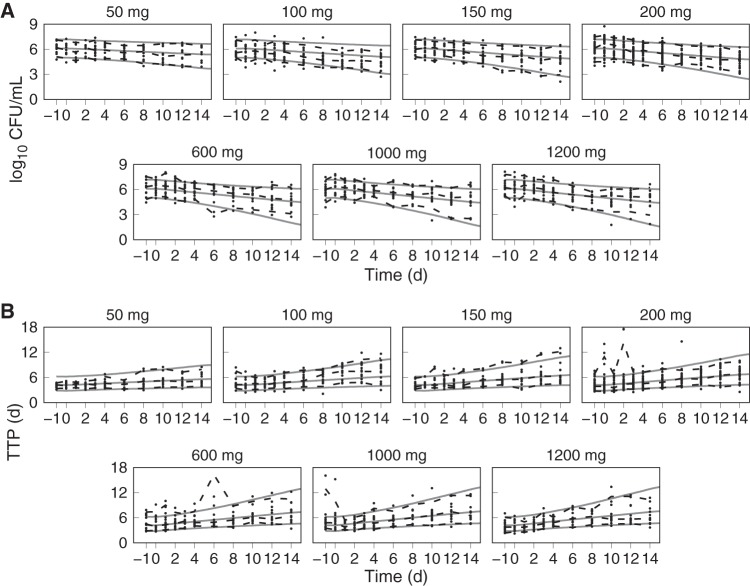
Monte Carlo (MC) simulations of sputum CFU and TTP profiles from pretreatment through the 14 consecutive days of pretomanid monotherapy, together with the observed individual participant data, are shown in Fig. 4. The simulations and data include the two pretreatment sputum collections, with the elapsed time in days shown relative to the first administered dose on day 0 and where the last administered dose was on day 13 and the last sputum collection on day 14. The population distributions of observed and model predicted data points are summarized in each plot with solid (prediction) and dashed (observed) lines that represent the 5th, median, and 95th percentiles. The 90% confidence intervals for uncertainty in the predicted percentiles (calculated using a binomial proportion with 10,000-MC-iteration sample sizes) corresponded to errors of less than 1%. The corresponding MC plasma PK simulation plots with the same percentile summaries were shown previously (25).
FIG 4.
PD model simulation and observed values for log10 CFU/ml (A) and time to positivity (TTP) (B) for the 50-, 100-, 150-, 200-, 600-, 1,000-, and 1,200-mg pretomanid dose groups versus time (days) since the start of treatment. The dashed lines represent the 5th, 50th, and 95th percentiles of the observed values (points). The solid lines represent the 5th, 50th, and 95th percentiles of 10,000 simulated individual log10 CFU/ml and TTP profiles using the joint posterior population distribution.
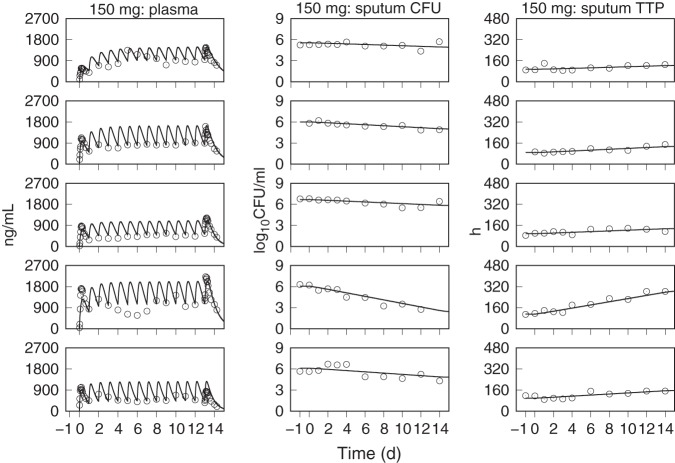
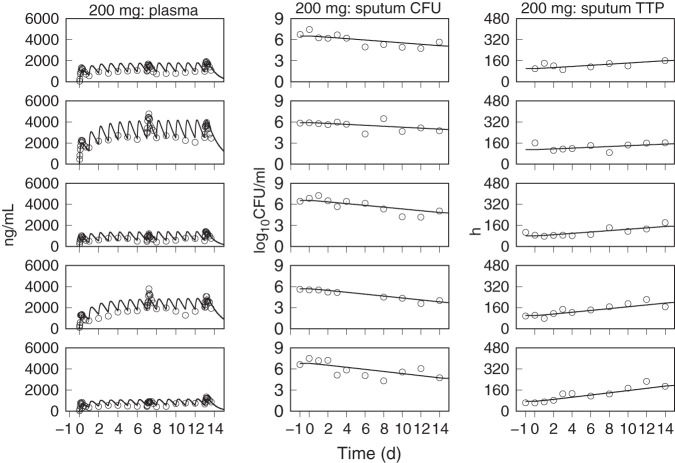
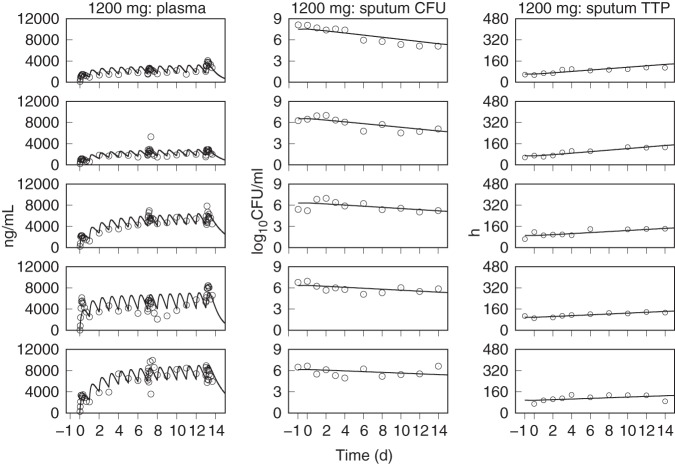
The predicted population distributions for CFU counts and TTP were used to calculate each of the primary and secondary efficacy outcomes for all of the pretomanid treatment arms, which consisted of the EBA measurements from day 0 to day 2 (EBA0−2), day 2 to day 14 (EBA2−14), and day 0 to day 14 (EBA0−14). The observed and model predicted distributions of EBA using CFU and TTP data are shown in Table 6 and Table 7, where the observed values are those that were reported in the published results (13, 14) and where the number of significant figures for the predicted values were chosen to match those for the observed values. These results show good agreement between the distributions of model predicted and observed values across the treatment duration and administered doses. The tabulated values also show good agreement between observed and predicted values, but with higher variability in the observed EBA0−2 values than predicted. For the 1,000- and 1,200-mg doses, there was some overprediction of the mean EBA values but adequate agreement between the observed and predicted ranges and between the observed and predicted median values, as shown in the population CFU and TTP versus time plots. Figures 5 to 11, respectively, show examples of individual participant simulations and observed values for the first 5 participants in the CL-010 50-, 100-, and 150-mg treatment groups and the CL-007 200-, 600-, 1,000-, and 1,200-mg treatment groups. The simulations for the 50-mg dose show examples of the model predictions for participants with missing data, including one participant with completely missing CFU data. These individual plots illustrate the extent of agreement between observed reduction in sputum mycobacterial load and the predicted log-linear CFU and linear TTP profiles.
TABLE 6.
Observed and PD model-predicted EBA as fall in log10 CFU/ml sputum/day for CL-010 and CL-007
| Groupa | Mean (SD) value forb
: |
|||||
|---|---|---|---|---|---|---|
| EBA0–2 |
EBA2–14 |
EBA0–14 |
||||
| Obs [n] | Pred | Obs [n] | Pred | Obs [n] | Pred | |
| CL-010 | ||||||
| 50 mg | 0.093 (0.211) [14] | 0.042 (0.037) | 0.059 (0.060) [12] | 0.057 (0.046) | 0.063 (0.058) [12] | 0.055 (0.045) |
| 100 mg | 0.111 (0.332) [15] | 0.064 (0.050) | 0.088 (0.085) [15] | 0.081 (0.059) | 0.091 (0.073) [15] | 0.079 (0.057) |
| 150 mg | −0.009 (0.290) [15] | 0.076 (0.056) | 0.096 (0.098) [14] | 0.094 (0.065) | 0.078 (0.074) [14] | 0.091 (0.063) |
| 200 mg | 0.160 (0.255) [14] | 0.084 (0.061) | 0.104 (0.083) [14] | 0.100 (0.068) | 0.112 (0.070) [14] | 0.099 (0.067) |
| CL-007 | ||||||
| 200 mg | 0.109 (0.487) [15] | 0.084 (0.061) | 0.106 (0.063) [12] | 0.100 (0.068) | 0.106 (0.049) [12] | 0.099 (0.067) |
| 600 mg | 0.096 (0.226) [13] | 0.110 (0.073) | 0.113 (0.079) [12] | 0.120 (0.077) | 0.107 (0.053) [14] | 0.120 (0.076) |
| 1,000 mg | 0.025 (0.340) [15] | 0.110 (0.077) | 0.095 (0.062) [14] | 0.130 (0.079) | 0.091 (0.083) [15] | 0.130 (0.078) |
| 1,200 mg | −0.035 (0.420) [15] | 0.120 (0.078) | 0.113 (0.099) [11] | 0.130 (0.080) | 0.088 (0.084) [11] | 0.130 (0.079) |
Pretomanid treatment group.
The values shown represent the fall in log10 CFU/ml sputum/day during treatment days 0 to 2 (EBA0–2), 2 to 14 (EBA2–14), and 0 to 14 (EBA0–14) in the CL-007 and CL-010 studies. EBA, early bactericidal activity; Obs, observed value; [n], number of participants for observed data; Pred, predicted value (sample size equal to 10,000). Observed values for the CL-007 and CL-010 studies are from references 13 and 14, respectively, by Diacon et al.
TABLE 7.
Observed and PD model-predicted EBA as rise in TTP for CL-010 and CL-007
| Groupa | Mean (SD) value forb
: |
|||||
|---|---|---|---|---|---|---|
| EBA0–2 |
EBA2–14 |
EBA0–14 |
||||
| Obs [n] | Pred | Obs [n] | Pred | Obs [n] | Pred | |
| CL-010 | ||||||
| 50 mg | 1.483 (8.153) [15] | 1.992 (1.833) | 2.958 (2.652) [13] | 2.728 (2.340) | 2.621 (2.534) [13] | 2.623 (2.265) |
| 100 mg | −1.345 (8.586) [14] | 3.037 (2.477) | 5.744 (3.973) [15] | 3.875 (3.015) | 4.969 (3.644) [14] | 3.756 (2.935) |
| 150 mg | 4.867 (12.755) [15] | 3.634 (2.832) | 4.594 (5.035) [15] | 4.489 (3.363) | 4.633 (3.687) [15] | 4.367 (3.284) |
| 200 mg | 3.096 (8.202) [13] | 4.024 (3.062) | 5.391 (3.608) [13] | 4.875 (3.580) | 4.640 (3.447) [16] | 4.753 (3.503) |
| CL-007 | ||||||
| 200 mg | 1.115 (15.256) [13] | 4.024 (3.062) | 3.833 (2.954) [11] | 4.875 (3.580) | 3.818 (2.327) [12] | 4.753 (3.503) |
| 600 mg | 5.788 (12.173) [13] | 5.126 (3.710) | 5.09 (2.768) [13] | 5.903 (4.160) | 4.776 (2.879) [14] | 5.792 (4.093) |
| 1,000 mg | 2.795 (9.230) [11] | 5.429 (3.890) | 4.069 (1.916) [12] | 6.169 (4.313) | 4.865 (3.461) [13] | 6.063 (4.250) |
| 1,200 mg | 1.400 (7.659) [15] | 5.511 (3.939) | 4.868 (3.224) [12] | 6.240 (4.354) | 4.440 (2.169) [12] | 6.136 (4.292) |
Pretomanid treatment group.
The values shown represent the rise in time to positivity (TTP) in hours per day during treatment days 0 to 2 (EBA0–2), 2 to 14 (EBA2–14), and 0 to 14 (EBA0–14) for the CL-010 and CL-007 studies. Obs, observed value; [n], number of participants for observed data; Pred, predicted value (sample size equal to 10,000). Observed values for the CL-007 and CL-010 studies are from references 13 and 14, respectively, by Diacon et al.
FIG 5.
Observed (points) and PD model simulations (lines) of the individual pretomanid plasma concentration, CFU, and TTP profiles for the first five participants in the 50-mg dose group of the CL-010 study.
FIG 6.
Observed (points) and PD model simulations (lines) of the individual pretomanid plasma concentration, CFU, and TTP profiles for the first five participants in the 100-mg dose group of the CL-010 study.
FIG 7.
Observed (points) and PD model simulations (lines) of the individual pretomanid plasma concentration, CFU, and TTP profiles for the first five participants in the 150-mg dose group of the CL-010 study.
FIG 8.
Observed (points) and PD model simulations (lines) of the individual pretomanid plasma concentration, CFU, and TTP profiles for the first five participants in the 200-mg dose group of the CL-007 study.
FIG 9.
Observed (points) and PD model simulations (lines) of the individual pretomanid plasma concentration, CFU, and TTP profiles for the first five participants in the 600-mg dose group of the CL-007 study.
FIG 10.
Observed (points) and PD model simulations (lines) of the individual pretomanid plasma concentration, CFU, and TTP profiles for the first five participants in the 1,000-mg dose group of the CL-007 study.
FIG 11.
Observed (points) and PD model simulations (lines) of the individual pretomanid plasma concentration, CFU, and TTP profiles for the first five participants in the 1,200-mg dose group of the CL-007 study.
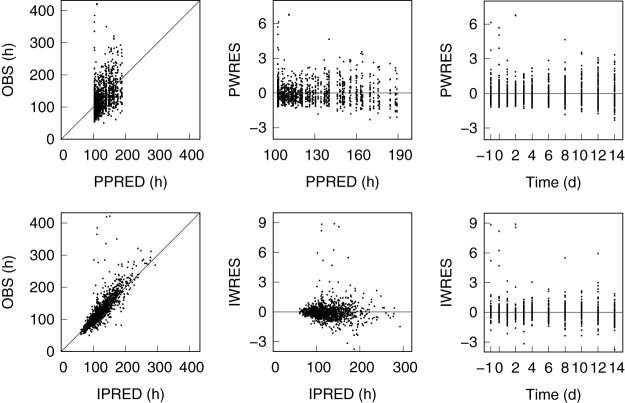
Goodness-of-fit plots (43) for observed versus predicted log10 CFU/ml and TTP values and standardized residuals are shown in Fig. 12 and 13, with corresponding summary statistics given in Table 9. For the CFU results, the residuals were approximately symmetric and uniform across time, with 3.7% and 0.17% of the population predicted residuals and 5.6% and 1.0% of the individual predicted residuals outside the ranges of ±2 and ±3, respectively. The results for TTP showed similarly symmetric and uniform residuals but with an unbalanced set of outliers that were biased toward larger than expected TTP values and can be seen in aggregate as points outside the 95th percentile lines in the population plots of Fig. 4. Inspection of the population residuals with absolute values greater than 2.58 (expected for 1% of the residuals) identified 21 points (1.7% of the total) from 15 individual participants across all but the 50-mg dose group. Each of these outliers resulted from significant differences in one or two measurements from otherwise smooth individual participant TTP profiles. There were no complete individual profiles that were outside an expected range, and there was no significant correlation between the 21 observed TTP outliers and the corresponding log10 CFU/ml values (Pearson coefficient, r = 0.46).
FIG 12.
Goodness-of-fit plots for CFU. Shown are the observed (OBS) versus individual predicted (IPRED) and population predicted mean (PPRED) log10 CFU/ml sputum values and the corresponding weighted residuals of the individual predictions (IWRES) and population mean predictions (PWRES). The diagonal lines in the observed versus predicted plots are lines of perfect fit, and the horizontal lines in the residual plots are lines of zero residual error.
FIG 13.
Goodness-of-fit plots for TTP. Shown are the observed (OBS) versus individual predicted (IPRED) and population predicted mean (PPRED) TTP values and the corresponding weighted residuals of the individual predictions (IWRES) and population mean predictions (PWRES). The diagonal lines in the observed versus predicted plots are lines of perfect fit, and the horizontal lines in the residual plots are lines of zero residual error.
Model simulations of CFU and TTP profiles for twice-daily dosing with 50, 100, 150, 200, 600, 1,000, and 1,200 mg were conducted to compare model predictions of EBA for once-daily and twice-daily dosing. Figure 14 shows the population distribution mean values from the EBA distributions that were calculated from the MC simulation output of CFU and TTP values for once-daily and twice-daily dosing, together with a least-squares fit to a hyperbolic dose-effect model (with effect being the EBA), EBA = E = Emax · D/(D50 + D), where Emax is the maximum effect, D is the administered dose, and D50 is the half-maximum effect dose. The curves for both dosing frequencies are similar up to a 200-mg total dose, with twice-daily administration showing slightly higher efficacy with further dose increases. The parameter estimates and standard errors for the best-fit curves are shown in Table 8. The EBA values calculated using either TTP or log10 CFU were ordered as EBA0−2 < EBA0−14 < EBA2−14 for all of the once- or twice-daily dosages. The ranking is consistent with the pretomanid plasma exposure (peak concentration, or 24-h area under the concentration-time curve) at the beginning of dose administration being nearly half that achieved as steady state is approached approximately 4 days after the start of treatment (25). The EBA values calculated with either TTP or CFU data are comparable, with the half-maximum effect doses being nearly equal for TTP and CFU calculated values. Based on the dose-effect model parameter values for the primary outcome measure, EBA0−14 (log10 CFU), the once-daily 100-, 200-, and 300-mg dosages result in 58, 73, and 80%, respectively, of the expected maximum EBA0−14 = 0.136 log10 CFU/ml/day, and the 200-mg once-daily dose yields approximately the same 0.1 log10 CFU/ml/day reduction as 100 mg twice daily.
FIG 14.

PD model simulations (points) of mean EBA values for total daily dose calculated from log10 CFU/ml (A) and TTP (B) for the 50-, 100-, 150-, 200-, 600-, 1,000-, and 1,200-mg doses administered once daily (squares) or twice daily (triangles) together with least-squares fits (solid and dashed lines) to a hyperbolic dose-response curve.
TABLE 8.
Dose-response model parameter values for EBA calculated from MC simulation mean CFU and TTP profiles
| EBA parametera | Mean (SE) value forb
: |
|||||
|---|---|---|---|---|---|---|
| EBA0–2 |
EBA2–14 |
EBA0–14 |
||||
| Emax | D50 | Emax | D50 | Emax | D50 | |
| CFU count | ||||||
| q.d. | 0.125 (0.0003) log10 CFU/ml/day | 98 (0.9) mg | 0.137 (0.0001) log10 CFU/ml/day | 70 (0.4) mg | 0.136 (0.0001) log10 CFU/ml/day | 73 (0.4) mg |
| b.i.d. | 0.136 (0.0001) log10 CFU/ml/day | 61 (0.2) mg | 0.148 (0.002) log10 CFU/ml/day | 37 (0.2) mg | 0.146 (0.0001) log10 CFU/ml/day | 40 (0.3) mg |
| TTP | ||||||
| q.d. | 5.96 (0.02) h/day | 97 (1.0) mg | 6.61 (0.004) h/day | 71 (0.2) mg | 6.51 (0.005) h/day | 74 (0.3) mg |
| b.i.d. | 6.48 (0.008) h/day | 62 (0.3) mg | 7.16 (0.007) h/day | 38 (0.2) mg | 7.06 (0.009) h/day | 41 (0.3) mg |
q.d., once daily; b.i.d., twice daily.
The values shown represent the parameter estimate (standard error [SE]) for the maximum effect (Emax) and half-maximum effect dose (D50) for treatment days 0 to 2 (EBA0–2), 2 to 14 (EBA2–14), and 0 to 14 (EBA0–14). EBA was calculated with CFU counts as log10 CFU/ml/day or with TTP as hours/day. The sample size was 10,000.
DISCUSSION
While PK/PD modeling and simulation can aid the development of novel drug regimens with quantitative analyses of clinical trial outcomes and designs (20), such mathematical and computational tools are not yet established for pretomanid-containing regimens that are currently in clinical testing (16–19). The central results of this study are the CFU and TTP rate equations for sputum mycobacterial load as functions of pretomanid plasma concentrations. Combined with the previously reported PK model of pretomanid (25), these results extend the empirical dose-exposure and dose-efficacy relationships established by Diacon et al. (13, 14) to a precisely defined dose-exposure-efficacy relationship and place into context the current 200-mg once-daily dosage as significantly more efficacious than 100 mg once daily, but with a smaller difference between the 80% maximum effect obtained with 300 mg once daily. These results also provide a novel approach to analysis of EBA studies in general with a full integration of plasma drug concentration-time profiles and serial sputum CFU and TTP measurements for every study participant and for the study population as a whole. The approach includes translation of preclinical results to inform prior distributions for human outcomes and provides a formal relationship between CFU and TTP kinetics with a biologically interpretable parameter that can potentially be used to rank order different drug regimens.
Mathematical modeling of antimicrobial time-kill profiles with logistic growth and Hill-type drug effect terms is based on resource-dependent bacterial cell division and mass action kinetics for drug-receptor interactions (44, 45). However, the interpretation remains approximate and descriptive and depends on the scale of experimental measurements used to determine the model parameter values. The logistic function was used here for in vivo kinetics of sputum cell density, and separately for in vitro kinetics of mycobacterial growth in a liquid culture system. As sputum collections are interval samples, the growth rate constant for the in vivo case represented changes in the overnight accumulation of bacilli in secreted mucus (analogous to the cumulative amount of drug excreted by the kidneys in a urine sample) rather than mycobacterial generation times within the lung and with prior distributions that were determined from observed sputum EBA values in separate studies with untreated controls. For the in vitro case, the growth rate constant represented the conventional intrinsic growth rate, with prior values set from known M. tuberculosis generation times. The in vivo carrying capacity represented a long-term potential increase in mycobacterial load in the absence of treatment, with sputum CFU values from older studies reflecting more advanced disease. Additionally, an analysis of time trends in EBA studies by De Jager et al. (31) noted a decrease in pretreatment CFU counts between early studies grouped from 1992 to 2001 and later studies grouped from 2007 to 2015 (or an increase in CFU counts looking back in time) with differences attributed to several factors, which include possibly earlier detection of pulmonary TB disease in the more recent studies. For the in vitro case, the carrying capacity was an unmeasured batch culture limit that was ignored in the approximation of log-phase growth, with the assumption being detection of bacilli before the stationary-phase limit is reached. The choice of a Hill function for the killing effect of pretomanid was motivated by its required enzymatic activation and represented the simplest nontrivial characterization of a drug-receptor interaction. While a linear approximation is often used in the absence of data required for parameter identification, the tested pretomanid dose range in the combined CL-007 and CL-010 studies was adequate to inform the maximum and half-maximum effects. These latter parameters for drug kill rate represented a composite effect of several molecular-scale transport and biochemical reaction processes for the drug-target interactions within the bacilli, including the effects of fluctuating and possibly asynchronous concentration gradients of pretomanid across plasma, lung tissues, and TB lesion microenvironnments (46, 47). As CL-007 and CL-010 were the first clinical studies to provide efficacy data for pretomanid, prior distributions for the drug effect parameters were specified from nonclinical data. Starting from an observed similarity between 14-day mouse and human EBAs for standard regimen drugs (48), mouse time-kill results that included CFU sampling time points in the first days of treatment were considered most informative for prior distributions. While both chronic and acute mouse TB infection model data were available (4, 7, 8, 49), only the acute mouse studies included a dose range and sampling schedule from which a maximum kill rate constant and half-maximum effect concentration could be identified. However, dose-ranging efficacy studies with several different mouse models may provide a more informative basis for mouse-to-human translation (50). In particular, conventional BALB/c and C57BL/6 mouse TB infection models exhibit uniform cellular lung lesions with mostly intracellular bacilli, while the chronic C3HeB/FeJ model develops several lesion types that include caseous necrotic lesions with bacilli in both intracellular and extracellular microenvironments (51, 52). The latter can provide for experimental measurements of a more complex lesion drug penetration kinetics and killing effect on multiple mycobacterial phenotypes that may better reflect human disease (46, 47).
The experimental assessment of sputum mycobacterial load in CL-007 and CL-010 was conducted using parallel CFU and TTP measurements. A comparison between EBA values that were calculated using log10 CFU counts and TTP by Diacon et al. (23) provided an initial motivation for a formal definition of equality between the corresponding instantaneous rates of change. This relation, together with a constant of proportionality (the liquid culture time constant, τL), provided for TTP and CFU to be treated as dual measures of efficacy that could be used simultaneously for PD model parameter estimation. Further interpretation of this relation was found from an analysis of a logistic growth model applied to a generic TTP-based liquid culture system, with the patient-level time constant equal to the reciprocal of the in vitro intrinsic growth rate. This suggests a dependence of the time constant on the M. tuberculosis strain or on the state of cultured bacilli in the collected sputum sample under the various drug treatments. In this context, the unimodal posterior population mean distribution and small interindividual variability for τL (Fig. 2 and Table 3) indicate a high degree of mycobacterial homogeneity in the pretomanid treatment group sputum samples. While τL was defined using instantaneous rates of change for CFU counts and TTP, an approximate value may be obtained as τL ≈ EBA(TTP)/EBA(log CFU) = EBA(TTP)/[log(10)·EBA(log10 CFU)]. Using the average of the mean values for EBA(log10 CFU) and EBA(TTP) given in the published tabulated results from the CL-007 (13) and CL-010 (14) standard treatment (RHZE) control groups, yields τL(RHZE) ≈ 30 h for each of the EBA study periods. With the corresponding τL(PA-824) ≈ 20 h, the ranking, τL(RHZE) > τL(PA-824) illustrates a possible use for τL as an index-based categorization of different drug treatments in a particular study population or of the same drug treatments in different study populations or in different TTP-based liquid culture systems.
The method used to combine TTP and CFU measurements through an equality between their time rates of change differs from previously described approaches to modeling M. tuberculosis liquid culture TTP that were based on regression (53, 54) or time-to-event analysis (55, 56). While the regression analyses specified a relationship between observed CFU and TTP values, they were based on free parameters without a kinetic description in terms of explicit PK and PD effects. However, once established, such regression curves could act as a conversion formula for new experiments and provide for TTP to be used as a single-outcome measure of efficacy to infer the corresponding CFU kinetics. The time-to-event methods were similarly applied to TTP as a single-outcome measure of efficacy, but with separate mathematical models for an unobserved sputum mycobacterial load and for in vitro mycobacterial growth in the liquid culture system. The unknown model parameters were estimated through a model fit using observed and predicted probabilities for a positive liquid culture measurement. In contrast, the PD model framework developed here was tailored to the parallel CFU and TTP measurements in CL-007 and CL-010. While the Bayesian hierarchical modeling provided for partial or missing values, the optimal use of this PD model framework includes paired baseline CFU and TTP measurements to specify the initial conditions as parameters in the TTP kinetic equation.
The killing effect of pretomanid against M. tuberculosis throughout the 2-week duration of the CL-007 and CL-010 studies was described by a monophasic decline in mycobacterial sputum cell density. The model-predicted and observed data plots of CFU and TTP versus time for each treatment group (Fig. 4), together with the individual participant profiles (Fig. 5 and 11) and the residual analysis (Fig. 12 and 13 and Table 9), showed good agreement between model prediction and observation, without significant systematic deviations that indicated a clear need to account for additional structural elements. In particular, there were no significant errors that would suggest evidence of additional log-linear phases for drug-susceptible mycobacterial killing, drug-resistant mutant growth, or delayed onset of drug effect. Additional effects that could be considered include deviations from unity of the Hill coefficient and a time dependence between sputum CFU counts and TTP found in the regression analysis of high-dose rifampin and standard regimen treatment data by Bowness et al. (53). The random pattern of the TTP outliers across dose groups and individual profiles, but biased toward a slow-growing subpopulation, suggested measurement uncertainty from differences in solid and liquid culture growth environments and methods of decontamination (23, 57). While multiple phenotypic subpopulations with possibly different pretomanid susceptibilities may be evident with a longer duration of treatment, the monophasic killing kinetics are consistent with the observed bactericidal activity of pretomanid against multiple M. tuberculosis phenotypes (2, 4–6), and biphasic killing kinetics were seen as expected in the observed CFU and TTP profiles of the RHZE control groups that were shown in the published CL-007 and CL-010 results (13, 14). This monophasic pattern of killing for pretomanid is consistent with the monophasic activity described for delamanid (58), which is also a nitroimidazole TB drug.
TABLE 9.
| Parametera | Mean (SD) | Median [5th, 95th]b |
|---|---|---|
| CFU count (log10 CFU/ml) (n = 1,194) | ||
| OBS | 5.57 (1.02) | 5.57 [3.90, 7.19] |
| PPRED | 5.57 (0.50) | 5.73 [4.58, 6.09] |
| IPRED | 5.67 (0.83) | 5.74 [4.19, 6.88] |
| PWRES | 0.0 (1.0) | 0.065 [−1.66, 1.58] |
| IWRES | 0.0 (1.0) | 0.038 [−1.78, 1.46] |
| TTP (n = 1,201) | ||
| OBS (h) | 129.7 (50.0) | 117.6 [75.6, 218.4] |
| PPRED (h) | 128.8 (24.1) | 120.4 [103.3, 175.6] |
| IPRED (h) | 125.7 (35.6) | 118.5 [80.9, 190.0] |
| PWRES | 0.0 (1.0) | −0.208 [−1.10, 1.64] |
| IWRES | 0.0 (1.0) | −0.084 [−0.99, 1.06] |
n, sample size; OBS, observed; PPRED, population predicted; IPRED, individual predicted; PWRES population predicted weighted residual; IWRES, individual predicted weighted residual.
Values in brackets are the 5th and 95th percentiles.
Summary statistics of the individual mean values of the marginal posterior parameter distributions (Table 4) showed a small but statistically significant difference between male and female values for the mean kill rate constant and mean half-maximum effect concentration, with an apparently higher efficacy of pretomanid in males than females. However, the scope of analysis and the included data for model development did not support conclusions on the source of this possible drug effect difference. There were no significant correlations (Table 5) between the individual PK and PD parameters, including body weight, that would provide a drug exposure explanation, and there were no strong correlations between baseline mycobacterial loads and the drug-dependent PD parameters that might suggest a disease-related correlation. Further understanding of covariate effects on these PD model parameters could be aided by better accounting for variation in the drug effect parameters based on more detailed MIC measurements with a larger range of tested concentrations, together with time course measurements that coincide with the CFU and TTP profiles. While concomitant medications, age, HIV status, and lung cavitation may also be important sources of variation in drug effect, they were not included as model elements in this study.
While the CL-007 and CL-010 studies were not powered to test the differences in efficacy outcomes between treatment arms (14), such differences were clearly defined in the calculated distributions shown for CFU counts in Table 6 and for TTP in Table 7. A key result of the CL-007 and CL-010 studies was the identification of 100 to 200 mg pretomanid daily as a proposed clinical dose for treatment of pulmonary TB (14). The simulated population EBA distributions with the dose-effect model fit to the population means shown in Fig. 14 and Table 8 provide additional detail and context for this dosage range, including an accounting of the efficacy within the experimentally untested 200- to 600-mg range. For example, from the EBA0−14 values for once-daily dosing, there is a 27% increase in EBA if 100-mg/day dosing is increased to 200 mg/day, while increasing from 200 mg/day to 300 mg/day yields a 10% increase in EBA. For dosages higher than 200 mg once daily, consideration of the decreasing rate of gain in efficacy against a possibly dose-proportional increase in the incidence of adverse events (13) supports the current 200-mg once-daily dosage (16–19).
Limitations on the interpretation and use of EBA study data are primarily those related to predictions of long-term outcomes for efficacy based on serial sputum measurements and the short-term duration of treatment (59, 60). The same limitations are inherent to the PD model developed here. While the PD model equations and parameter distributions provide for the simulation of CFU and TTP outcomes for any desired pretomanid dosage, the continuation of monophasic killing kinetics beyond 2 weeks and the EBA simulations of twice-daily dosing should be considered model-generated hypotheses. In addition to the lack of accounting for possible phenotypic variation, the model does not account for the growth of pretomanid-resistant mutants (32) that could significantly alter the killing kinetics profile with continued monotherapy. Similarly, comparison of observed and model-predicted pretomanid killing kinetics in patients from a more highly variable target population may indicate the need for modification of both the PK and PD structural models. While the mycobacterial sputum accumulation rate constant and cell density carrying capacity are study population-dependent parameters, untreated control measurements that could further inform their prior specification were not available with the CL-007 and CL-010 data, and these parameters were included as measured covariates with values sampled from fixed probability distributions. Although these fixed parameter distributions provide for standardized comparison of model-predicted EBA with other studies that measure EBA against a fixed zero baseline value (61), there is a loss of accounting for interindividual variability for these parameters that instead becomes part of the variability in the estimated drug effect parameters. The proportionality between log CFU and TTP rates of change is considered here only for the EBA study populations on which the relationship was based. As a conjectured continuum limit restatement of the observed correlation between EBA values (23), the same limits on interpretation and extrapolation from those data apply to the mathematical relationship. Also, while there was good agreement between model simulation and observation for the CL-007 and CL-010 data, a nonlinear relationship between mean log CFU and mean TTP, with higher-than-expected values for large CFU values, was noted by Diacon et al. (23) and would not be accounted for using the simple assumption of logistic growth for the liquid culture system. This suggests a more detailed mathematical model of mycobacterial growth in the liquid culture system—for example, by including a lag time (55) or multiple mycobacterial subpopulations and phenotypic states (62, 63), as well as possible differences in phenotypes that are detected under the different growth conditions of the liquid and solid culture systems.
In the context of a series of clinical studies that are used to evaluate and characterize various aspects of the dose-exposure-response relationships, the PD model developed here provided an additional quantitative assessment of the short-duration CL-007 and CL-010 EBA data and established a baseline or limiting-case model for subsequent intermediate and long-term clinical studies of pretomanid-containing regimens. A remaining task from the CL-007 and CL-010 EBA data is to include adverse event modeling (64) for more precise dose optimization of pretomanid that could apply to individual TB patients and clinical trial treatment groups. The method of relating the CFU and TTP rate equations through a time constant may be applied to PD modeling of other preclinical and clinical studies of TB drugs that include these two measurement types.
MATERIALS AND METHODS
The natural logarithm of a positive number x was denoted “log x,” and the base 10 logarithm was denoted “log10x.” EBA was calculated as the decrease in log10 CFU/ml sputum or increase in TTP from day x to day y with the following equations (23):
and
Structural model.
The PD model equations were developed as a biologically plausible and parsimonious representation of EBA sputum mycobacterial load measurements (22, 29, 65), with a specific application to pretomanid. The modeling approach was based on visual comparison between general large-scale features of experimental sputum CFU time-kill curves and the solutions to standard antimicrobial drug effect models for one or more bacterial subpopulations (24, 66–68). A logistic function for capacity-limited bacterial population growth and Hill functions for drug-receptor-mediated killing were chosen for a base model of the CFU kinetics. A dual kinetic model for parallel sputum TTP measurements was constructed from the CFU kinetics equation as a change of variables based on previous comparisons between CFU and TTP bactericidal activity measurements (23). A previously developed pretomanid PK model (25) was used for drug concentration input to the PD model equations. The PK and PD models were integrated as a single set of kinetic equations, and the model output provided a simultaneous relation between the EBA experimental data for plasma drug concentrations and sputum CFU and TTP measurements in the form that they were collected. While conventional PD modeling also includes dose or area under the concentration-time curve (AUC) as measures of drug exposure (45), the importance of the concentration-time curve shape for antibacterial response (69, 70) motivated the use of the fluctuating pretomanid plasma concentrations for the drug-bacillus interactions.
Liquid culture time constant.
A relationship between sputum CFU and TTP kinetics was previously described in terms of a correlation between EBA(log10 CFU) and EBA(TTP) (23), which was formalized here as an equality between the instantaneous time rates of change
with τL as a liquid culture time constant of proportionality. Additional interpretation of τL was obtained through consideration of sputum sample bacillary growth in a generic TTP-based liquid culture system as follows. Let N = N(tL) denote the number of bacilli in the liquid culture system at an elapsed time (tL ≥ 0) since the start of incubation. For logistic growth, N(tL) = KL/[1 + (KL/N0 −1) exp(−λL · tL)], where N(tL = 0) = N0 is the inoculum size, and λL and KL are the intrinsic growth rate constant and carrying capacity, respectively. Denoting the number of bacilli at the time of positivity by NTTP = N(tL) = TTP, and rewriting the growth equation in terms of tL = TTP gave
With the liquid culture inoculum obtained from a serial sputum collection at an elapsed time (t ≥ 0) since the first collection and with N0 = N0(t) and TTP = T(t)
where the approximate expression results from inoculum size much less than the carrying capacity, which is equivalent to log-phase growth. Denoting the time dependence of sputum CFU/ml as B(t), and setting N0(t) ∝ B(t) gave the relation between T and B as
where τL = 1/λL. The total bacterial population, B, was also written as the sum of a number (p) of subpopulations with cell densities, Bi, i = 1, …, p, that satisfied rate equations of the form dBi/dt = fi(B1, …, Bp), Bi(0) = Bi0, where fi are functions that describe the growth and drug effect for each subpopulation. This relation between rates, together with the solution B(t) = B0 · exp[−(T − T0)/τL], where B(t = 0) = B0 and T(t = 0) = T0 are the initial conditions, was used to rewrite the kinetic equation for B in terms of T.
Statistical model.
The PD model parameters were treated as random variables and estimated using a hierarchical statistical model with population- and individual-level probability distributions (26, 42) that were specified as:
with the normal (N), lognormal (LN), half-normal (HN), uniform (U), and log-uniform (LU) probability distributions specified using conventional parameterizations (71). The values yijm represent the individual-level observed data from i = 1, …, I study participants at j = 1, …, J sampling time points, tij, for each of m = 1, …, M measurement types. The corresponding model-predicted values for the data are given by the solutions to the PD model equations, gm (tij, θi, Di, φi), where θi = (θ1, …, θL)i is a vector, indexed by l = 1, …, L, of unknown PD model parameters for each individual, Di is the dosage, and φi is a vector of covariates. The individual-level parameters θi are sampled from population-level distributions for the mean, μ = (μ1, …, μL), and the variance, ω2 = (ω12, …, ωL2). The residual error distributions σ = (σ1, …, σM) for each measurement type are specified with a lower bound a and upper bound b. The parameters (Mμ, Sμ2) and Sω are hyperparameters that represent the PD model parameter typical values and expected values for interindividual variability, respectively.
The following definitions and formulas were used for specification of the probability distributions and for calculation of associated statistical quantities. For a random variable X, X ∼ LN(μ, σ) ⇔ log X ∼ N(μ, σ), where N(μ, σ) denotes the normal distribution with mean, μ, and standard deviation (SD), σ. The mean of X (Mean[X]) = exp(μ + σ2/2), the variance (Var[X]) = exp(2 μ + σ2) [exp(σ 2) − 1], and the coefficient of variation (CV[X]) = = SD(X)/Mean(X). The geometric mean of X (GM[X]) = exp(μ) = exp(Mean[log X]), and the geometric standard deviation (GSD[X]) = exp(σ) = exp(SD[log X]). For a positive number n, the normal distribution range, μ ± n·σ, corresponds to the lognormal distribution range, exp(μ ± n·σ) = GM ×/GSDn, where “×/” denotes “multiplied or divided by” and where GSD acts as a dimensionless multiplicative factor (72). The HN distribution was specified by an SD with Mean(HN[SD]) = . For the LU distribution, with lower bound a and upper bound b, Y ∼ LU[a, b] ⇔ log Y ∼ U[log(a), log(b)].
CL-007 and CL-010 participant data.
Anonymized individual participant data from two published pretomanid EBA studies, PA-824-CL-007 (CL-007 [ClinicalTrials.gov identifier NCT00567840]) (13) and PA-824-CL-010 (CL-010 [ClinicalTrials.gov identifier NCT00944021]) (14) were obtained from the Global Alliance for TB Drug Development (TB Alliance [http://www.tballiance.org/]) through the Platform Aggregation of Clinical TB Studies data platform (TB-PACTS [https://c-path.org/programs/tb-pacts]). Both studies were randomized phase 2 clinical trials to evaluate the plasma PK, EBA, and safety of 14 consecutive days of pretomanid monotherapy with a range of doses that were orally administered following an overnight fast. The enrollment criteria for both studies included male or female, age 18 to 64 years, body weight 40 to 90 kg, and newly diagnosed sputum smear-positive uncomplicated pulmonary TB. Both studies were conducted by the same principal investigators (A. Diacon and R. Dawson) at study sites (Task Applied Science, Intercare Hospital, Bellville; Tiervlei Trials Center-Karl Bremer Hospital, Bellville; and University of Cape Town Lung Institute) located in Cape Town, South Africa.
CL-007 enrolled 69 adult male and female participants, with 61 randomly assigned to one of four pretomanid treatment groups (200, 600, 1,000, or 1,200 mg once daily for 14 days) and 8 randomly assigned to a standard treatment (RHZE [rifampin, isoniazid, pyrazinamide, and ethambutol]) positive-control group. Similarly, CL-010 enrolled 69 adult male and female participants, with 61 randomly assigned to one of four pretomanid treatment groups (50, 100, 150, or 200 mg once daily for 14 days) and 8 randomly assigned to an RHZE standard treatment positive-control group. In both CL-007 and CL-010, sputum mycobacterial load was assessed from 16-h overnight sputum collections on each of the 2 days immediately preceding the start of treatment (days −2 and −1), and on treatment days 1, 2, 3, 4, 6, 8, 10, 12, and 14. During treatment, the individual sputum collections were obtained immediately preceding dose administration. From each sputum sample, a value for CFU/ml was obtained as the mean of a maximum of 4 CFU counts from solid culture 7H11 agar plates after 3 to 4 weeks of incubation, and a TTP value was obtained as the mean of a maximum of 2 liquid culture TTP measurements from a Bactec mycobacterial growth indicator tube (MGIT) 960 system (Becton, Dickinson). The MIC of pretomanid was measured from pretreatment and day 14 sputum collections, using the agar proportion method with a concentration range of 0.1 to 3.2 μg/ml. Additional details of the CL-007 and CL-010 study designs, participant characteristics and demographics, pretomanid PK measurements and analysis, processing of sputum samples, EBA assessments, and adverse event details are provided in their respective publications (13, 14).
Prior distributions.
Informative prior distributions for the PD model parameters were based on available data for (i) baseline CFU and TTP measurements from the standard treatment control groups of CL-007 and CL-010 and from summary results from several separate EBA studies (14, 31), (ii) the baseline CFU measurements and minimum EBA values from untreated (nil) groups in several early studies of adult patients with pulmonary TB (28, 30, 37–39), (iii) preclinical measures of pretomanid efficacy and potency (4, 8), (iv) in vitro measures of M. tuberculosis doubling times (40, 41), and (v) an analysis of CFU and TTP measurement errors in current EBA studies (23). These data were only used to specify the prior distributions and were not included for any further parameter estimation.
Estimates for population typical values and variances for the PD model initial conditions were obtained from bootstrap mean and bootstrap SD distributions, respectively, which were generated from the pretreatment sputum CFU and TTP measurements from the combined 16 participants of the CL-007 and CL-010 standard treatment control groups. Each of the 32 CFU and 32 TTP values was log transformed and resampled with replacement, with the sample mean and sample SD calculated for each of 10,000 iterations. Denoting the resulting bootstrap distributions for the means as MCFU and MTTP, and those for the SDs as SCFU and STTP, the typical values for CFU and TTP measurements were, respectively, GM(MCFU) = exp[Mean(MCFU)] = 1.094 × 106 CFU/ml, and GM(TTP) = exp[Mean(MTTP)] = 105.6 h, with the corresponding uncertainties GSD(MCFU) = exp[SD(MCFU)] = 1.43 and GSD(MTTP) = exp[SD(MTTP)] = 1.061. The mean values for the SD distributions were used to determine population variances as [Mean(SCFU)]2 = 4.263 and [Mean(STTP)]2 = 0.1089, which were set as the expected values for the half-normal population variance distributions. The sampling ranges for CFU and TTP were set as 103 to 109 CFU/ml and 24 to 400 h, respectively, to include the observed pretreatment CFU and TTP values that were reported in the analyses of several EBA studies in addition to CL-007 and CL-010 (14, 31).
A maximum sputum mycobacterial cell density was based on a distribution of observed pretreatment CFU/ml sputum values from an early EBA study (39). The upper 13th percentile (107 to 109) CFU/ml sputum, was chosen to represent a range of maximum values and to include the upper values for sputum CFU/ml counts reported in the older studies with adult pulmonary TB patients by Yeager et al. (37) and Hobby et al. (38). The typical value was set as the GM of this range, equal to 108 CFU/ml sputum, and the GSD, which here included both measurement uncertainty and interindividual variability, was set with GSD3 = 10, which provided for approximately 99.7% of the distribution within ±3 SDs of the mean. A maximum rate of change of sputum mycobacterial load was similarly set based on a range of EBA values, −0.0033 to −0.033 log10 CFU/ml sputum/day (3.1 × 10−4 to 3.1 × 10−3 CFU/ml/h), chosen to include the minimum EBA values from the nil (untreated) groups in the early studies by Donald et al. (30) and Jindani et al. (28).
A maximum pretomanid mycobacterial kill rate equal to 0.2 log10 CFU/day/lung was obtained as an average of previously reported pretomanid kill rates for M. tuberculosis Erdman in a rapid C57BL/6 gamma interferon gene-disrupted (GKO) mouse model (4) and for M. tuberculosis H37Rv in an acute infection BALB/c mouse model (8). An approximation for tissue volume of a TB-infected mouse lung of ≈1 ml was used to equate CFU per mouse lung to CFU/ml sputum. Also from the acute BALB/c model results (8), the best correlate of activity was found to be percentage of time that pretomanid unbound plasma concentrations were above the MIC (T>MIC), with 41.08% T>MIC reported as the half-maximum effect value. Based on previous PK model simulations (25), this T>MIC value was attained at steady state in the CL-007 and CL-010 study participants with an approximate 50% probability at the 100-mg daily dose. Using the 100-mg dose as a prior estimate for a half-maximum effect dose, a prior value for a half-maximum effect pretomanid plasma concentration was set as the corresponding model-predicted steady-state concentration: Css = AUC24/24 h = 0.9 μg/ml. A conservative estimate for measurement error in the mouse-based parameters was assigned with CV = 20%, and for interindividual variability of the study participants, CV = 40% was assigned as a possibly large variability and in accordance with the observed variability in the pretomanid AUC values (25).
A typical value and variance components for τL were based on M. tuberculosis doubling times (τdouble) through the relation τL = τdouble/log(2). An approximate 24-h generation time for M. tuberculosis noted by Cole et al. (40) was used for the prior typical value. CV = 10% for measurement uncertainty and CV = 40% for interindividual variability were based on MGIT system doubling time results (a confidence interval for measurement error and a total range for variability) reported by von Groll et al. (41).
An analysis of error in TTP and log10 CFU/ml measurements conducted as part of several EBA studies by Diacon et al. (23) indicated that approximately 30% of the total variation, measured by a total SD, was due to intraindividual variation and technical variation from laboratory measurements. This was accounted for in the log uniform residual error distributions with an approximate 10% CV measurement error in the lower bounds of the log uniform residual error distributions, based on comparison with the CL-007 standard group baseline mean and SD measurements for TTP and log10 CFU/ml (13).
Posterior distribution.
The PD model parameters were estimated as a joint posterior probability distribution conditioned on the drug efficacy-time measurements, dosage, individual covariates, and prior distributions, using Bayes’ rule (42)
where P(θ, μ, ω2, σ|y, t, D, φ, Mμ, Sμ2, Sω) is the posterior, P(y|θ, φ, D, t, σ) is the likelihood, P (θ|μ, ω2) is the population model, and P(μ, ω2|Mμ, Sμ2, Sω) P(σ) is the prior distribution. The parameters in each term refer to the same quantities as described above for the statistical model, with θ denoting the individual-level parameters to be estimated, φ the covariates, μ the population means, ω2 the population variances, σ the measurement SDs, y the observed data, D the dosage, t the measurement times, and (Mμ, Sμ2) and Sω the population mean and population variance hyperparameters, respectively. The joint posterior distribution was estimated using Markov chain Monte Carlo (MCMC) simulation with Metropolis-within-Gibbs sampling (73). Multiple independent sampling chains were generated, and convergence, mixing, and precision were assessed using trace plots, density plots, the Gelman-Rubin scale reduction factor (R̂), and the effective sample size (neff) (42, 74).
Model simulations.
The PD model differential equations were solved numerically to simulate individual and population CFU and TTP profiles for specified pretomanid dosage regimens. Individual simulations for each pretomanid treatment group participant were performed using the individual PK covariate values and the individual marginal posterior PD parameter distribution means. Population CFU/ml and TTP distributions were obtained using Monte Carlo (MC) simulation of the PD model equations with randomly generated individual PK and PD parameter sets. Each MC simulation consisted of 10,000 iterations, with each iteration providing the time course of CFU/ml sputum and TTP for the specified dosage regimen and a randomly generated individual PK and PD model parameter set. Each body weight was a random sample (with replacement) from the complete set of observed daily body weights from the combined CL-007 and CL-010 studies, and individual values for the PK parameters were sampled from the pretomanid PK model population distributions, with the lognormal distributions truncated to include approximately 95% of the distribution values. The individual PD model parameters were generated as samples from the individual parameter vector θ ∼ LN(μ, ω2), where μ and ω2 were sampled from the joint population distribution. EBA values were calculated from the model output for both CFU/ml and TTP using the same equations as used for the corresponding observed values.
Comparisons between observed and model predicted values for CFU and TTP measurements were conducted using plots of model predictions for individual participants and pretomanid treatment groups overlaid with observed data, together with corresponding summary statistics for model outputs. Standard goodness-of-fit plots of observed versus predicted values for the population means and individual participants were also generated, together with least-squares regression and weighted residual plots (calculated as standardized residuals) (43, 75).
Software.
The software environment was based on the Linux operating system (version 3.16.0-4-amd64; Debian distribution [https://www.debian.org]). The SQLite database engine (version 3.8.7.1; SQLite Development Team [https://www.sqlite.org]) was used for CL-007 and CL-010 database queries. The GNU MCSim Modeling and Simulation Suite (76) (version 5.6.5 [http://www.gnu.org/software/mcsim]) was used for the Bayesian data analysis, including numerical evaluation of the differential equations. gcc (version 4.9.2 [https://gcc.gnu.org]) was used for compiling the MCSim model files (C-programs). The R statistical software (version 3.3.3; R Development Team [https://www.R-project.org]) with the CODA package (version 0.18 [https://cran.r-project.org/web/packages/coda]) was used for statistical analysis of the experimental and model simulation data, and gnuplot (version 5.0 [http://www.gnuplot.info]) was used for generation of the data and function plots.
ACKNOWLEDGMENTS
I thank Stephen Murray (Global Alliance for TB Drug Development, TB Alliance), Khisi Mdluli (TB Alliance), Anna Upton (TB Alliance), Carl Mendel (TB Alliance), Bob Stafford (Critical Path to TB Drug Regimens [CPTR]), Debra Hanna (CPTR), and Klaus Romero (CPTR) for providing and facilitating the acquisition of the clinical data. I thank Anne Lenaerts (Colorado State University [CSU]), Gregory Robertson (CSU), Mercedes Gonzalez-Juerroro (CSU), and Jerry Nedelman (TB Alliance) for discussions on pretomanid and experimental methods for TB. I also thank Andreas Diacon (TASK Applied Science and Stellenbosh University, South Africa) for discussions on EBA studies and for suggestions and comments on the manuscript.
The National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), HHS, provided funding to M. A. Lyons under grant no. R01AI125454.
REFERENCES
- 1.Murray S, Mendel C, Spigelman M. 2016. TB Alliance regimen development for multidrug-resistant tuberculosis. Int J Tuberc Lung Dis 20:38–41. doi: 10.5588/ijtld.16.0069. [DOI] [PubMed] [Google Scholar]
- 2.Stover CK, Warrener P, VanDevanter DR, Sherman DR, Arain TM, Langhorne MH, Anderson SW, Towell JA, Yuan Y, McMurray DN, Kreiswirth BN, Barry CE, Baker WR. 2000. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405:962–966. doi: 10.1038/35016103. [DOI] [PubMed] [Google Scholar]
- 3.Singh R, Manjunatha U, Boshoff HI, Ha YH, Niyomrattanakit P, Ledwidge R, Dowd CS, Lee IY, Kim P, Zhang L, Kang S, Keller TH, Jiricek J, Barry CE. 2008. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science 322:1392–1395. doi: 10.1126/science.1164571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenaerts AJ, Gruppo V, Marietta KS, Johnson CM, Driscoll DK, Tompkins NM, Rose JD, Reynolds RC, Orme IM. 2005. Preclinical testing of the nitroimidazopyran PA-824 for activity against Mycobacterium tuberculosis in a series of in vitro and in vivo models. Antimicrob Agents Chemother 49:2294–2301. doi: 10.1128/AAC.49.6.2294-2301.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iacobino A, Piccaro G, Giannoni F, Mustazzolu A, Fattorini L. 2017. Fighting tuberculosis by drugs targeting nonreplicating Mycobacterium tuberculosis bacilli. Int J Mycobacteriol 6:213–221. doi: 10.4103/ijmy.ijmy_85_17. [DOI] [PubMed] [Google Scholar]
- 6.Baker JJ, Abramovitch RB. 2018. Genetic and metabolic regulation of Mycobacterium tuberculosis acid growth arrest. Sci Rep 8:4168. doi: 10.1038/s41598-018-22343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyagi S, Nuermberger E, Yoshimatsu T, Williams K, Rosenthal I, Lounis N, Bishai W, Grosset J. 2005. Bactericidal activity of the nitroimidazopyran PA-824 in a murine model of tuberculosis. Antimicrob Agents Chemother 49:2289–2293. doi: 10.1128/AAC.49.6.2289-2293.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmad Z, Peloquin CA, Singh RP, Derendorf H, Tyagi S, Ginsberg A, Grosset JH, Nuermberger EL. 2011. PA-824 exhibits time-dependent activity in a murine model of tuberculosis. Antimicrob Agents Chemother 55:239–245. doi: 10.1128/AAC.00849-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginsberg AM, Laurenzi MW, Rouse DJ, Whitney KD, Spigelman MK. 2009. Safety, tolerability, and pharmacokinetics of PA-824 in healthy subjects. Antimicrob Agents Chemother 53:3720–3725. doi: 10.1128/AAC.00106-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginsberg AM, Laurenzi MW, Rouse DJ, Whitney KD, Spigelman MK. 2009. Assessment of the effects of the nitroimidazo-oxazine PA-824 on renal function in healthy subjects. Antimicrob Agents Chemother 53:3726–3733. doi: 10.1128/AAC.00112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter H, Ginsberg A, Egizi E, Erondu N, Whitney K, Pauli E, Everitt D. 2013. Effect of a high-calorie, high-fat meal on the bioavailability and pharmacokinetics of PA-824 in healthy adult subjects. Antimicrob Agents Chemother 57:5516–5520. doi: 10.1128/AAC.00798-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dooley KE, Luetkemeyer AF, Park JG, Allen R, Cramer Y, Murray S, Sutherland D, Aweeka F, Koletar SL, Marzan F, Bao J, Savic R, Haas DW, Barr L, Egizi E, Hovind L, Janik J, Lizak P, Mendel C, Size T, Leonard M, Nicotera F, Dwyer J, Smith A, Clark J, Harber H, Clark J, Wiggins I, Weiss A. 2014. Phase I safety, pharmacokinetics, and pharmacogenetics study of the antituberculosis drug PA-824 with concomitant lopinavir-ritonavir, efavirenz, or rifampin. Antimicrob Agents Chemother 58:5245–5252. doi: 10.1128/AAC.03332-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diacon AH, Dawson R, Hanekom M, Narunsky K, Maritz SJ, Venter A, Donald PR, van Niekerk C, Whitney K, Rouse DJ, Laurenzi MW, Ginsberg AM, Spigelman MK. 2010. Early bactericidal activity and pharmacokinetics of PA-824 in smear-positive tuberculosis patients. Antimicrob Agents Chemother 54:3402–3407. doi: 10.1128/AAC.01354-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diacon AH, Dawson R, Du Bois J, Narunsky K, Venter A, Donald PR, van Niekerk C, Erondu N, Ginsberg AM, Becker P, Spigelman MK. 2012. Phase II dose-ranging trial of the early bactericidal activity of PA-824. Antimicrob Agents Chemother 56:3027–3031. doi: 10.1128/AAC.06125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawson R, Diacon A. 2013. PA-824, moxifloxacin and pyrazinamide combination therapy for tuberculosis. Expert Opin Invest Drugs 22:927–932. doi: 10.1517/13543784.2013.801958. [DOI] [PubMed] [Google Scholar]
- 16.Global Alliance for TB Drug Development. 2017. Trial to evaluate the efficacy, safety and tolerability of BPaMZ in drug-sensitive (DS-TB) adult patients and drug-resistant (DR-TB) adult patients. ClinicalTrials.gov. http://clinicaltrials.gov/ct2/show/NCT03338621.
- 17.Global Alliance for TB Drug Development. 2015. A phase 3 study assessing the safety and efficacy of bedaquiline plus PA-824 plus linezolid in subjects with drug resistant pulmonary tuberculosis. ClinicalTrials.gov. http://clinicaltrials.gov/ct2/show/NCT02333799.
- 18.Global Alliance for TB Drug Development. 2017. Safety and efficacy of various doses and treatment durations of linezolid plus bedaquiline and pretomanid in participants with pulmonary TB, XDR-TB, pre-XDR-TB or non-responsive/intolerant MDR-TB (ZeNix). ClinicalTrials.gov. http://clinicaltrials.gov/ct2/show/NCT03086486.
- 19.Medecins Sans Frontieres, Netherlands. 2015. Pragmatic Clinical Trial for a More Effective Concise and Less Toxic MDR-TB Treatment Regimen(s) (TB-PRACTECAL). ClinicalTrials.gov. http://clinicaltrials.gov/ct2/show/NCT02589782.
- 20.Ette E, Williams P. 2007. Pharmacometrics: the science of quantitative pharmacology. Wiley, Hoboken, NJ. [Google Scholar]
- 21.Lyons MA. 2014. Computational pharmacology of rifampin in mice: an application to dose optimization with conflicting objectives in tuberculosis treatment. J Pharmacokinet Pharmacodyn 41:613–623. doi: 10.1007/s10928-014-9380-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diacon AH, Donald PR. 2014. The early bactericidal activity of antituberculosis drugs. Expert Rev Anti Infect Ther 12:223–237. doi: 10.1586/14787210.2014.870884. [DOI] [PubMed] [Google Scholar]
- 23.Diacon AH, Maritz JS, Venter A, van Helden PD, Dawson R, Donald PR. 2012. Time to liquid culture positivity can substitute for colony counting on agar plates in early bactericidal activity studies of antituberculosis agents. Clin Microbiol Infect 18:711–717. doi: 10.1111/j.1469-0691.2011.03626.x. [DOI] [PubMed] [Google Scholar]
- 24.Czock D, Keller F. 2007. Mechanism-based pharmacokinetic-pharmacodynamic modeling of antimicrobial drug effects. J Pharmacokinet Pharmacodyn 34:727–751. doi: 10.1007/s10928-007-9069-x. [DOI] [PubMed] [Google Scholar]
- 25.Lyons MA. 2018. Modeling and simulation of pretomanid pharmacokinetics in pulmonary tuberculosis patients. Antimicrob Agents Chemother 62:e02359-17. doi: 10.1128/AAC.02359-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernillon P, Bois FY. 2000. Statistical issues in toxicokinetic modeling: a Bayesian perspective. Environ Health Perspect 108(Suppl 5):883–893. doi: 10.1289/ehp.00108s5883. [DOI] [PubMed] [Google Scholar]
- 27.Mitchison DA. 1979. Basic mechanisms of chemotherapy. Chest 76:771–781. doi: 10.1378/chest.76.6_supplement.771. [DOI] [PubMed] [Google Scholar]
- 28.Jindani A, Aber VR, Edwards EA, Mitchison DA. 1980. The early bactericidal activity of drugs in patients with pulmonary tuberculosis. Am Rev Respir Dis 121:939–949. [DOI] [PubMed] [Google Scholar]
- 29.Hafner R, Cohn JA, Wright DJ, Dunlap NE, Egorin MJ, Enama ME, Muth K, Peloquin CA, Mor N, Heifets LB. 1997. Early bactericidal activity of isoniazid in pulmonary tuberculosis. Optimization of methodology. The DATRI 008 Study Group. Am J Respir Crit Care Med 156:918–923. doi: 10.1164/ajrccm.156.3.9612016. [DOI] [PubMed] [Google Scholar]
- 30.Donald PR, Sirgel FA, Botha FJ, Seifart HI, Parkin DP, Vandenplas ML, Van de Wal BW, Maritz JS, Mitchison DA. 1997. The early bactericidal activity of isoniazid related to its dose size in pulmonary tuberculosis. Am J Respir Crit Care Med 156:895–900. doi: 10.1164/ajrccm.156.3.9609132. [DOI] [PubMed] [Google Scholar]
- 31.De Jager V, van der Merwe L, Venter A, Donald PR, Diacon AH. 2017. Time trends in sputum mycobacterial load and two-day bactericidal activity of isoniazid-containing antituberculosis therapies. Antimicrob Agents Chemother 61:e02088-16. doi: 10.1128/AAC.02088-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manjunatha UH, Boshoff H, Dowd CS, Zhang L, Albert TJ, Norton JE, Daniels L, Dick T, Pang SS, Barry CE. 2006. Identification of a nitroimidazooxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 103:431–436. doi: 10.1073/pnas.0508392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manjunatha U, Boshoff HI, Barry CE. 2009. The mechanism of action of PA-824: novel insights from transcriptional profiling. Commun Integr Biol 2:215–218. doi: 10.4161/cib.2.3.7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kayigire XA, Friedrich SO, Venter A, Dawson R, Gillespie SH, Boeree MJ, Heinrich N, Hoelscher M, Diacon AH, Pan-African Consortium for the Evaluation of Anti-tuberculosis Antibiotics. 2013. Direct comparison of Xpert MTB/RIF assay with liquid and solid mycobacterial culture for quantification of early bactericidal activity. J Clin Microbiol 51:1894–1898. doi: 10.1128/JCM.03290-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bark CM, Gitta P, Ogwang S, Nsereko M, Thiel BA, Boom WH, Eisenach KD, Joloba ML, Johnson JL. 2013. Comparison of time to positive and colony counting in an early bactericidal activity study of anti-tuberculosis treatment. Int J Tuber Lung Dis 17:1448–1451. doi: 10.5588/ijtld.13.0063. [DOI] [PubMed] [Google Scholar]
- 36.Gelman A. 2006. Prior distributions for variance parameters in hierarchical models. Bayesian Anal 1:515–533. doi: 10.1214/06-BA117A. [DOI] [Google Scholar]
- 37.Yeager H, Lacy J, Smith LR, LeMaistre CA. 1967. Quantitative studies of mycobacterial populations in sputum and saliva. Am Rev Respir Dis 95:998–1004. [DOI] [PubMed] [Google Scholar]
- 38.Hobby GL, Holman AP, Iseman MD, Jones JM. 1973. Enumeration of tubercle bacilli in sputum of patients with pulmonary tuberculosis. Antimicrob Agents Chemother 4:94–104. doi: 10.1128/aac.4.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen BW, Mitchison DA. 1992. Counts of viable tubercle bacilli in sputum related to smear and culture gradings. Med Lab Sci 49:94–98. [PubMed] [Google Scholar]
- 40.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 41.von Groll A, Martin A, Portaels F, da Silva PE, Palomino JC. 2010. Growth kinetics of Mycobacterium tuberculosis measured by quantitative resazurin reduction assay: a tool for fitness studies. Braz J Microbiol 41:300–303. doi: 10.1590/S1517-83822010000200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gelman A, Carlin JB, Stern HS, Rubin DB. 2003. Bayesian data analysis, 2nd ed. Chapman and Hall/CRC, Boca Raton, FL. [Google Scholar]
- 43.Nguyen THT, Mouksassi M-S, Holford N, Al-Huniti N, Freedman I, Hooker AC, John J, Karlsson MO, Mould DR, Pérez Ruixo JJ, Plan EL, Savic R, van Hasselt JGC, Weber B, Zhou C, Comets E, Mentré F. 2017. Model evaluation of continuous data pharmacometric models: metrics and graphics. CPT Pharmacometrics Syst Pharmacol 6:87–109. doi: 10.1002/psp4.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gabrielsson J, Weiner D. 2007. Pharmacokinetic and pharmacodynamic data analysis: concepts and applications, 4th ed Swedish Pharmaceutical Press, Stockholm, Sweden. [Google Scholar]
- 45.Vinks A, Derendorf H, Mouton J. 2013. Fundamentals of antimicrobial pharmacokinetics and pharmacodynamics. Springer, New York, NY. [Google Scholar]
- 46.Dartois V. 2014. The path of anti-tuberculosis drugs: from blood to lesions to mycobacterial cells. Nat Rev Microbiol 12:159–167. doi: 10.1038/nrmicro3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lenaerts A, Barry CE, Dartois V. 2015. Heterogeneity in tuberculosis pathology, microenvironments and therapeutic responses. Immunol Rev 264:288–307. doi: 10.1111/imr.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grosset J, Almeida D, Converse PJ, Tyagi S, Li SY, Ammerman NC, Pym AS, Wallengren K, Hafner R, Lalloo U, Swindells S, Bishai WR. 2012. Modeling early bactericidal activity in murine tuberculosis provides insights into the activity of isoniazid and pyrazinamide. Proc Natl Acad Sci U S A 109:15001–15005. doi: 10.1073/pnas.1203636109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lakshminarayana SB, Boshoff HI, Cherian J, Ravindran S, Goh A, Jiricek J, Nanjundappa M, Nayyar A, Gurumurthy M, Singh R, Dick T, Blasco F, Barry CE, Ho PC, Manjunatha UH. 2014. Pharmacokinetics-pharmacodynamics analysis of bicyclic 4-nitroimidazole analogs in a murine model of tuberculosis. PLoS One 9:e105222. doi: 10.1371/journal.pone.0105222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartelink IH, Zhang N, Keizer RJ, Strydom N, Converse PJ, Dooley KE, Nuermberger EL, Savic RM. 2017. New paradigm for translational modeling to predict long-term tuberculosis treatment response. Clin Transl Sci 10:366–379. doi: 10.1111/cts.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Irwin SM, Driver E, Lyon E, Schrupp C, Ryan G, Gonzalez-Juarrero M, Basaraba RJ, Nuermberger EL, Lenaerts AJ. 2015. Presence of multiple lesion types with vastly different microenvironments in C3HeB/FeJ mice following aerosol infection with Mycobacterium tuberculosis. Dis Model Mech 8:591–602. doi: 10.1242/dmm.019570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Irwin SM, Prideaux B, Lyon ER, Zimmerman MD, Brooks EJ, Schrupp CA, Chen C, Reichlen MJ, Asay BC, Voskuil MI, Nuermberger EL, Andries K, Lyons MA, Dartois V, Lenaerts AJ. 2016. Bedaquiline and pyrazinamide treatment responses are affected by pulmonary lesion heterogeneity in Mycobacterium tuberculosis infected C3HeB/FeJ mice. ACS Infect Dis 2:251–267. doi: 10.1021/acsinfecdis.5b00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bowness R, Boeree MJ, Aarnoutse R, Dawson R, Diacon A, Mangu C, Heinrich N, Ntinginya NE, Kohlenberg A, Mtafya B, Phillips PP, Rachow A, Plemper van Balen G, Gillespie SH. 2015. The relationship between Mycobacterium tuberculosis MGIT time to positivity and CFU in sputum samples demonstrates changing bacterial phenotypes potentially reflecting the impact of chemotherapy on critical sub-populations. J Antimicrob Chemother 70:448–455. doi: 10.1093/jac/dku415. [DOI] [PubMed] [Google Scholar]
- 54.Magombedze G, Pasipanodya JG, Srivastava S, Deshpande D, Visser ME, Chigutsa E, McIlleron H, Gumbo T. 2018. Transformation morphisms and time-to-extinction analysis that map therapy duration from preclinical models to patients with tuberculosis: translating from apples to oranges. Clin Infect Dis 67:S349–S358. doi: 10.1093/cid/ciy623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chigutsa E, Patel K, Denti P, Visser M, Maartens G, Kirkpatrick CM, McIlleron H, Karlsson MO. 2013. A time-to-event pharmacodynamic model describing treatment response in patients with pulmonary tuberculosis using days to positivity in automated liquid mycobacterial culture. Antimicrob Agents Chemother 57:789–795. doi: 10.1128/AAC.01876-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Svensson EM, Karlsson MO. 2017. Modelling of mycobacterial load reveals bedaquiline’s exposure-response relationship in patients with drug-resistant TB. J Antimicrob Chemother 72:3398–3405. doi: 10.1093/jac/dkx317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dhillon J, Fourie PB, Mitchison DA. 2014. Persister populations of Mycobacterium tuberculosis in sputum that grow in liquid but not on solid culture media. J Antimicrob Chemother 69:437–440. doi: 10.1093/jac/dkt357. [DOI] [PubMed] [Google Scholar]
- 58.Diacon AH, Dawson R, Hanekom M, Narunsky K, Venter A, Hittel N, Geiter LJ, Wells CD, Paccaly AJ, Donald PR. 2011. Early bactericidal activity of delamanid (OPC-67683) in smear-positive pulmonary tuberculosis patients. Int J Tuber Lung Dis 15:949–954. doi: 10.5588/ijtld.10.0616. [DOI] [PubMed] [Google Scholar]
- 59.Wallis RS, Nacy C. 2013. Early bactericidal activity of new drug regimens for tuberculosis. Lancet 381:111–112. doi: 10.1016/S0140-6736(13)60041-0. [DOI] [PubMed] [Google Scholar]
- 60.Diacon AH, Donald PR, Mendel CM. 2013. Early bactericidal activity of new drug regimens for tuberculosis—authors’ reply. Lancet 381:112–113. doi: 10.1016/S0140-6736(13)60042-2. [DOI] [PubMed] [Google Scholar]
- 61.Sirgel F, Venter A, Mitchison D. 2001. Sources of variation in studies of the early bactericidal activity of antituberculosis drugs. J Antimicrob Chemother 47:177–182. doi: 10.1093/jac/47.2.177. [DOI] [PubMed] [Google Scholar]
- 62.Davies GR. 2010. Early clinical development of anti-tuberculosis drugs: science, statistics and sterilizing activity. Tuberculosis (Edinb) 90:171–176. doi: 10.1016/j.tube.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 63.Svensson RJ, Simonsson U. 2016. Application of the multistate tuberculosis pharmacometric model in patients with rifampicin-treated pulmonary tuberculosis. CPT Pharmacometrics Syst Pharmacol 5:264–273. doi: 10.1002/psp4.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Vries Schultink AH, Suleiman AA, Schellens JH, Beijnen JH, Huitema AD. 2016. Pharmacodynamic modeling of adverse effects of anti-cancer drug treatment. Eur J Clin Pharmacol 72:645–653. doi: 10.1007/s00228-016-2030-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jindani A, Dore CJ, Mitchison DA. 2003. Bactericidal and sterilizing activities of antituberculosis drugs during the first 14 days. Am J Respir Crit Care Med 167:1348–1354. doi: 10.1164/rccm.200210-1125OC. [DOI] [PubMed] [Google Scholar]
- 66.Mouton JW, Vinks AA, Punt NC. 1997. Pharmacokinetic-pharmacodynamic modeling of activity of ceftazidime during continuous and intermittent infusion. Antimicrob Agents Chemother 41:733–738. doi: 10.1128/AAC.41.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nielsen EI, Friberg LE. 2013. Pharmacokinetic-pharmacodynamic modeling of antibacterial drugs. Pharmacol Rev 65:1053–1090. doi: 10.1124/pr.111.005769. [DOI] [PubMed] [Google Scholar]
- 68.Jacobs M, Gregoire N, Couet W, Bulitta JB. 2016. Distinguishing antimicrobial models with different resistance mechanisms via population pharmacodynamic modeling. PLoS Comput Biol 12:e1004782. doi: 10.1371/journal.pcbi.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Craig WA. 1998. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin Infect Dis 26:1–10. doi: 10.1086/516284. [DOI] [PubMed] [Google Scholar]
- 70.de Steenwinkel JE, de Knegt GJ, ten Kate MT, van Belkum A, Verbrugh HA, Kremer K, van Soolingen D, Bakker-Woudenberg IA. 2010. Time-kill kinetics of anti-tuberculosis drugs, and emergence of resistance, in relation to metabolic activity of Mycobacterium tuberculosis. J Antimicrob Chemother 65:2582–2589. doi: 10.1093/jac/dkq374. [DOI] [PubMed] [Google Scholar]
- 71.Armitage P, Colton T (ed). 2005. Encyclopedia of biostatistics. Wiley, Hoboken, NJ. [Google Scholar]
- 72.Limpert E, Stahel WA. 2011. Problems with using the normal distribution—and ways to improve quality and efficiency of data analysis. PLoS One 6:e21403. doi: 10.1371/journal.pone.0021403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gilks WR, Best N, Tan K. 1995. Adaptive rejection Metropolis sampling within Gibbs sampling. Appl Stat 44 :455–472. doi: 10.2307/2986138. [DOI] [Google Scholar]
- 74.Plummer M, Best N, Cowles K, Vines K. 2006. CODA: convergence diagnosis and output analysis for MCMC. R News 6:7–11. [Google Scholar]
- 75.Rosner B. 2011. Fundamentals of biostatistics, 7th ed Cengage Learning, Boston, MA. [Google Scholar]
- 76.Bois FY. 2009. GNU MCSim: Bayesian statistical inference for SBML-coded systems biology models. Bioinformatics 25:1453–1454. doi: 10.1093/bioinformatics/btp162. [DOI] [PubMed] [Google Scholar]