Four sequence type 664 (ST664) (serotype O:5) strains of Pseudomonas aeruginosa that were highly resistant to antibiotics, including ceftolozane-tazobactam and ceftazidime-avibactam, but were susceptible to colistin were found to harbor the gene encoding the rare class C β-lactamase PAC-1 on a chromosomally located Tn1721-like transposon. The blaPAC-1 gene was associated with the 16S rRNA methylase determinant rmtF2, which confers pan-aminoglycoside resistance.
KEYWORDS: Pseudomonas aeruginosa, β-lactamase, PAC-1, antibiotics, mechanisms of resistance
ABSTRACT
Four sequence type 664 (ST664) (serotype O:5) strains of Pseudomonas aeruginosa that were highly resistant to antibiotics, including ceftolozane-tazobactam and ceftazidime-avibactam, but were susceptible to colistin were found to harbor the gene encoding the rare class C β-lactamase PAC-1 on a chromosomally located Tn1721-like transposon. The blaPAC-1 gene was associated with the 16S rRNA methylase determinant rmtF2, which confers pan-aminoglycoside resistance. These genotypically related strains were isolated in repatriated patients from Mauritius and Afghanistan and were close to a lineage reported in Nepal, Pakistan, and India.
TEXT
In countries such as France, where the prevalence of extended-spectrum β-lactamases and carbapenemases in Pseudomonas aeruginosa is still low, resistance of the pathogen to the cephalosporins ceftazidime and cefepime mostly results from mutations upregulating the production of the intrinsic cephalosporinase AmpC. While transferable class C β-lactamases have spread significantly in Enterobacterales strains over the past decades (1, 2), their occurrence in P. aeruginosa has not been reported in the literature.
The present work reports on the characterization of four multidrug-resistant strains of P. aeruginosa that harbor a class C enzyme, named PAC-1, that has been poorly characterized to date. These bacterial strains were isolated between January 2017 and April 2019, from three male patients repatriated from Mauritius (Indian Ocean) and one repatriated from Afghanistan after a bombing attack. One strain was present in a rectal swab and was considered nonpathogenic. The second strain was collected from multiple body sites, including blood, and was responsible for the patient’s death. The third strain was involved in a diabetic foot infection. Finally, the fourth isolate was recovered from the urine of a patient suffering from renal colic.
The drug susceptibility (MICs) of the four isolates was determined by using customized Sensititre plates (Thermo Fisher Scientific), and results were interpreted according to the 2019 EUCAST breakpoints (3). As shown in Table 1, these strains were resistant to all of the antipseudomonal antibiotics tested except colistin and, for two of them, imipenem. They met the criteria of extensively drug-resistant P. aeruginosa (4). Their levels of resistance to ceftazidime (with or without 4 μg/ml avibactam), ceftolozane with 4 μg/ml tazobactam, cefepime, amikacin, and tobramycin were very high (MICs of >128 μg/ml). Addition of 2,000 μg/ml of the AmpC inhibitor cloxacillin to Mueller-Hinton agar (Bio-Rad, Marnes-la-Coquette, France) failed to restore the susceptibility of the bacteria to penicillins and cephalosporins, as assessed by the disk diffusion method. Intriguingly, screening of class A and class B β-lactamases by double-disk synergy tests also yielded negative results, despite the use of various disk combinations (imipenem-EDTA, ceftazidime-EDTA, ceftazidime-clavulanate, and cefepime-clavulanate). PCR sequencing experiments targeting genes coding for various class A (CTX-M, GES, PER, SHV, TEM, and VEB types), class B (IMP, NDM, and VIM types), and class D (OXA-1, OXA-2, and OXA-10 variants) β-lactamases potentially produced by clinical P. aeruginosa strains were performed and revealed only the presence of the blaOXA-10 gene in the four strains. Because production of penicillinase OXA-10 could not account for the resistance profiles of studied bacteria (5), the whole DNA content of two strains (17.4313 and 17.4319) was extracted and sequenced by hybrid de novo assembly of paired-end reads (2 by 75 reads), using the NextSeq Illumina technology (100× coverage). The resultant reads were assembled with CLC Genomics Workbench 10.0.1 software (Qiagen Aahrus A/S) and then analyzed by using the CARD Resistance Gene Identifier 4.2.2 (https://card.mcmaster.ca/analyze/rgi). The two strains were determined to contain rmtF2, the gene that codes for 16S rRNA methylase RmtF2, and aacA4-22, the determinant of aminoglycoside modification. Previously identified in P. aeruginosa isolates from India and Nepal (6, 7), RmtF2 confers high levels of resistance to all aminoglycosides (8). A third gene, exhibiting 60% sequence homology to that of β-lactamase SRT-2, was finally identified by BLAST comparisons (https://blast.ncbi.nlm.nih.gov/Blast.cgi) as blaPAC-1, which is indicated in the GenBank database as encoding a putative class C β-lactamase (GenBank accession number NG_052627.1). Since no transconjugants could be obtained, despite several attempts (data not shown), we cloned the blaPAC-1 gene in the broad-host-range vector pUCP24 (with gentamicin resistance) and transformed reference strain PAO1 with the resultant plasmid, pUCP24blaPAC-1. The resistance phenotypes of PAO1, PAO1(pUCP24::blaPAC-1), and the AmpC-overproducing mutant PAO1ΔdacB (9) are presented in Table 1. Compared with PAO1ΔdacB, PAO1(pUCP24::blaPAC-1) exhibited much higher levels of resistance to all of the cephalosporins tested, including ceftolozane-tazobactam and ceftazidime-avibactam (MICs of >128 μg/ml), while its levels of resistance to ticarcillin, piperacillin/tazobactam, and aztreonam were similar or lower. In contrast to AmpC, PAC-1 was poorly antagonized by cloxacillin even at the high concentration of 2,000 μg/ml (cephalosporin MICs of ≥32 μg/ml) (Table 1). Since the MICs of carbapenems were unaffected by the expression of blaPAC-1 in strain PAO1, we examined the sequence of the gene that codes for the carbapenem-specific porin OprD in the imipenem-resistant strains 17.4319 and 19.6465 (MICs of 16 μg/ml); we found a disruptive mutation (T209 deletion) in the oprD gene of 17.4319 and a premature stop codon (G831A, W277*) in that of 19.6465. Analysis of the nalB, nalC, and nalD genes, whose alteration is known to upregulate the efflux operon mexAB-oprM, did not provide an explanation for the 4- to 16-fold higher levels of resistance of all strains to meropenem, compared with imipenem (i.e., only meropenem is a substrate of the efflux pump MexAB-OprM) (10). This MIC difference between the two carbapenems is possibly due to OXA-10, because this β-lactamase was demonstrated to have stronger enzymatic activity with meropenem than with imipenem (11). The sequenced strains 17.4313 and 17.4319 both exhibited the canonical mutations T83I and S87L in the quinolone-resistance-determining regions of DNA gyrase subunit A and topoisomerase IV subunit C, respectively, accounting for the high levels of resistance of these bacteria to ciprofloxacin (MICs of 16 μg/ml).
TABLE 1.
Characteristics of strains producing class C β-lactamase PAC-1
| Strain | ST | MIC (μg/ml)a
|
Acquired β-lactamases | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TIC | TZPb | CAZ | FEP | CTb | CZAb | ATM | IPM | MEM | AMK | TOB | CIP | FOS | CST | |||
| 17.4313 | ST664 | >512 | 128 | >128 | >128 | >128 | >128 | 64 | 1 | 16 | >128 | >128 | 16 | 256 | 1 | PAC-1, OXA-10 |
| 17.4319 | ST664 | >512 | 256 | >128 | >128 | >128 | >128 | 64 | 16 | 64 | >128 | >128 | 16 | 256 | 1 | PAC-1, OXA-10 |
| 19.6465 | ST664 | >512 | 128 | >128 | >128 | >128 | >128 | 64 | 16 | 64 | >128 | >128 | >16 | 128 | 1 | PAC-1, OXA-10 |
| 19.6625 | ST664 | >512 | 128 | >128 | >128 | >128 | >128 | 128 | 2 | 32 | >128 | >128 | >16 | 128 | 1 | PAC-1, OXA-10 |
| PAO1 | 16 | 4 | 1 | 1 | ≤0.5 | 1 | 4 | 1 | ≤0.5 | 4 | 0.5 | ≤0.12 | 1 | |||
| PAO1ΔdacBc | 64 | 32 | 16 (1) | 8 (1) | 1 (0.5) | 2 (0.5) | 8 | 1 | ≤0.5 | ≤2 | 1 | ≤0.12 | 1 | |||
| PAO1(pUCP24) | 16 | 4 | 2 | 2 | ≤0.5 | 1 | 4 | 1 | ≤0.5 | ≤2 | 1 | ≤0.12 | 1 | |||
| PAO1(pUCP24blaPAC-1) | 32 | 16 | >128 (64) | >128 (32) | >128 (64) | >128 (64) | 8 | 1 | ≤0.5 | ≤2 | 1 | ≤0.12 | 1 | |||
MICs were determined by using customized Sensititre plates (Thermo Fisher Scientific). MIC values in parentheses were determined in the presence of 2,000 μg/ml of cloxacillin. TIC, ticarcillin; TZP, piperacillin-tazobactam; CAZ, ceftazidime; FEP, cefepime; CT, ceftolozane-tazobactam; CZA, ceftazidime-avibactam; ATM, aztreonam; IPM, imipenem; MEM, meropenem; AMK, amikacin; TOB, tobramycin; CIP, ciprofloxacin; FOS, fosfomycin; CST, colistin.
With a fixed concentration of 4 μg/ml tazobactam or avibactam.
PAO1ΔdacB was a gift from Antonio Oliver.
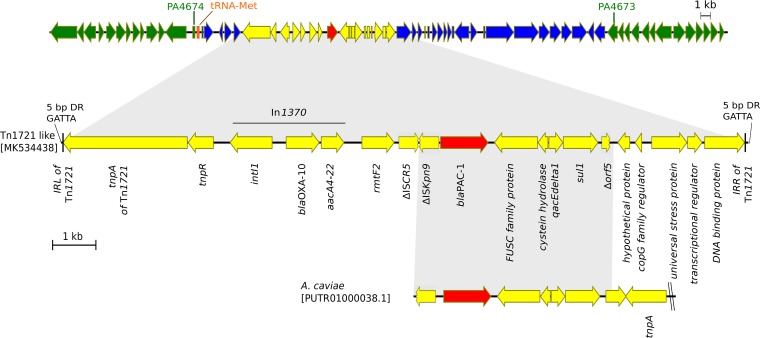
The enzyme PAC-1 is composed of 381 amino acids. This protein shares 65% sequence identity with the class C β-lactamase encoded by Cronobacter turicensis (formerly Enterobacter sakazakii) and 47% with the AmpC enzyme from P. aeruginosa strain PAO1 (PDC-1). As depicted in Fig. 1, the blaPAC-1 gene was localized on a complex Tn1721-like transposon, a partial sequence of which was previously established from a Nepalese strain named IOMTU487 (GenBank accession number LC224309.1) (6). In our strains, blaPAC-1 was found to be embedded in a 4,539-bp sequence containing a truncated insertion sequence (IS) element, ISKpn9, that was previously identified in an Aeromonas caviae strain (GenBank accession number PUTR01000038.1).
FIG 1.
Schematic representation of the genetic environment of blaPAC-1. The blaPAC-1 gene (red) is located on a Tn1721-like transposon (yellow). The genetic environment of the transposon (indicated in blue; 4,549 bp on the left and 22,312 bp on the right) is 99% homologous to the genome of strain S04-90 (GenBank accession number CP011369.1); open reading frames indicated in green are homologous to a genomic region present in reference strain PAO1 (GenBank accession number AE004091.2).
Analysis of the genetic environment of the Tn1721-like transposon indicated that this element was located on an ∼27-kb DNA sequence showing 99% homology with the genome of a P. aeruginosa strain deposited in the NCBI database (strain S04-90; GenBank accession number CP011369.1). This long fragment was inserted between genes PA4673 and PA4674, according to the genome annotation of strain PAO1, next to a tRNA-Met-encoding gene. Attempts to identify an integrative and conjugative element (ICE) bearing the Tn1721-like transposon, by using the ICEBerg website (http://db-mml.sjtu.edu.cn/ICEberg) or manual characterization of conserved features of ICEs (e.g., genes predicted to encode conjugative relaxases, type IV coupling proteins, and ATPases of type IV secretion systems), were unsuccessful (12, 13).
A whole-genome multilocus sequence typing analysis of 6,349 genetic loci with Bionumerics software package 7.6.3 revealed a close clonal relationship of the PAC-1-producing isolates with strain PA_041 from Pakistan (GenBank accession number NZ_RHSY00000000.1) (98% allelic identity), strain PA_038 from Pakistan (GenBank accession number NZ_RHSZ00000000.1) (97.7%), and strain VRFPA06 from India (92%) (14). Like the Nepalese strain IOMTU487, all eight isolates belonged to sequence type 664 (ST664) (serotype O:5).
In conclusion, PAC-1 is a cephalosporinase acquired by closely related strains of P. aeruginosa belonging to epidemic clone ST664 (15). Coinheritance of the blaPAC-1, blaOXA-10, and rmtF2 genes is sufficient to provide P. aeruginosa with a high degree of resistance to major antipseudomonal penicillins, cephalosporins, and aminoglycosides. PAC-1 thus should be added to the growing list of β-lactamases produced by multidrug-resistant P. aeruginosa strains, especially those occurring in the Indian subcontinent.
Accession number(s).
Genomic DNA sequences of the P. aeruginosa strains have been submitted to the GenBank database under accession numbers SISN00000000 (strain 17.4313) and SISM00000000 (strain 17.4319). The transposon sequence was deposited in the GenBank nucleotide database under accession number MK534438.
ACKNOWLEDGMENTS
The Centre National de Référence de la Résistance aux Antibiotiques is funded by the French Ministry of Health through the Santé Publique France agency.
We have no conflicts of interest to declare.
REFERENCES
- 1.Philippon A, Arlet G, Jacoby GA. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob Agents Chemother 46:1–11. doi: 10.1128/aac.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meini S, Tascini C, Cei M, Sozio E, Rossolini GM. 2019. AmpC β-lactamase-producing Enterobacterales: what a clinician should know. Infection 47:363–375. doi: 10.1007/s15010-019-01291-9. [DOI] [PubMed] [Google Scholar]
- 3.European Committee for Antimicrobial Susceptibility Testing. 2019. Breakpoint tables for interpretation of MICs and zone diameters, version 9.0. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf.
- 4.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 5.Poirel L, Naas T, Nordmann P. 2010. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob Agents Chemother 54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tada T, Shimada K, Satou K, Hirano T, Pokhrel BM, Sherchand JB, Kirikae T. 2017. Pseudomonas aeruginosa clinical isolates in Nepal coproducing metallo-β-lactamases and 16S rRNA methyltransferases. Antimicrob Agents Chemother 61:e00694-17a. doi: 10.1128/AAC.00694-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahman M, Prasad KN, Pathak A, Pati BK, Singh A, Ovejero CM, Ahmad S, Gonzalez-Zorn B. 2015. RmtC and RmtF 16S rRNA methyltransferase in NDM-1-producing Pseudomonas aeruginosa. Emerg Infect Dis 21:2059–2062. doi: 10.3201/eid2111.150271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galimand M, Courvalin P, Lambert T. 2012. RmtF, a new member of the aminoglycoside resistance 16S rRNA N7 G1405 methyltransferase family. Antimicrob Agents Chemother 56:3960–3962. doi: 10.1128/AAC.00660-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moya B, Dötsch A, Juan C, Blázquez J, Zamorano L, Haussler S, Oliver A. 2009. β-Lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog 5:e1000353. doi: 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:3322–3327. doi: 10.1128/aac.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antunes NT, Lamoureaux TL, Toth M, Stewart NK, Frase H, Vakulenko SB. 2014. Class D β-lactamases: are they all carbapenemases? Antimicrob Agents Chemother 58:2119–2125. doi: 10.1128/AAC.02522-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delavat F, Miyazaki R, Carraro N, Pradervand N, van der Meer JR. 2017. The hidden life of integrative and conjugative elements. FEMS Microbiol Rev 41:512–537. doi: 10.1093/femsre/fux008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murugan N, Malathi J, Umashankar V, Madhavan H. 2014. Comparative genomic analysis of multidrug-resistant Pseudomonas aeruginosa clinical isolates VRFPA06 and VRFPA08 with VRFPA07. Genome Announc 2:e00140-14. doi: 10.1128/genomeA.00140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pragasam A, Veeraraghavan B, Anandan S, Narasiman V, Sistla S, Kapil A, Mathur P, Ray P, Wattal C, Bhattacharya S, Deotale V, Subramani K, Peter J, Hariharan T, Ramya I, Iniyan S, Walia K, Ohri V. 2018. Dominance of international high-risk clones in carbapenemase-producing Pseudomonas aeruginosa: multicentric molecular epidemiology report from India. Indian J Med Microbiol 36:344. doi: 10.4103/ijmm.IJMM_18_294. [DOI] [PubMed] [Google Scholar]