Multidrug efflux pumps play an important role in antibiotic resistance in bacteria. In Pseudomonas aeruginosa, the MexXY pump provides intrinsic resistance to many antimicrobials, including aminoglycosides. The expression of the mexXY operon is negatively regulated by the MexZ repressor. This repression is alleviated in response to antibiotic-induced ribosome stress, which results in increased synthesis of the antirepressor ArmZ, interacting with MexZ.
Keywords: multidrug efflux pump, gene expression regulation, antirepressor, Pseudomonas aeruginosa, TetR transcriptional regulator, aminoglycoside resistance
ABSTRACT
Multidrug efflux pumps play an important role in antibiotic resistance in bacteria. In Pseudomonas aeruginosa, the MexXY pump provides intrinsic resistance to many antimicrobials, including aminoglycosides. The expression of the mexXY operon is negatively regulated by the MexZ repressor. This repression is alleviated in response to antibiotic-induced ribosome stress, which results in increased synthesis of the antirepressor ArmZ, interacting with MexZ. The molecular mechanism of MexZ inactivation by ArmZ is not known. Here, we show that the N-terminal part of MexZ, encompassing the DNA-binding domain, is required for the interaction with ArmZ. Using bacterial two-hybrid system-based mutant screening and pulldown analyses, we identified substitutions in MexZ that diminished (R3S, K6E, and R13H) or completely impaired (K53E) the interaction with ArmZ without blocking MexZ activity as a transcriptional repressor. The introduction of the corresponding mexZ missense mutations into the P. aeruginosa PAO1161 chromosome impaired (mexZK6E and mexZR13H) or blocked (mexZK53E) tetracycline-mediated induction of mexY expression. Concomitantly, the PAO1161 mexZK53E strain was more susceptible to aminoglycosides. The identified residues are highly conserved in MexZ-like transcriptional regulators found in bacterial genomes encoding both MexX/MexY/MexZ and ArmZ/PA5470 orthologs, suggesting that a similar mechanism may contribute to the induction of efflux-mediated resistance in other bacterial species. Overall, our data shed light on the molecular mechanism of ArmZ-mediated induction of intrinsic antimicrobial resistance in P. aeruginosa.
INTRODUCTION
Pseudomonas aeruginosa, a versatile Gram-negative bacterium, is the most detrimental infectious agent for cystic fibrosis (CF) patients (1, 2). Almost 80% of CF patients develop chronic pulmonary tract infections (3), which result in a progressive loss of lung function and are the primary cause of mortality (2, 4). Treatment of both initial and recurrent chronic infections involves the use of aminoglycosides (e.g., tobramycin and amikacin), fluoroquinolones (e.g., ciprofloxacin), or broad-spectrum β-lactams (e.g., imipenem) (5, 6). Remarkably, P. aeruginosa possesses intrinsic resistance to many antimicrobials due to low outer membrane permeability and the presence of multidrug efflux transporters (7, 8). The multidrug efflux pumps of the resistance-nodulation-division (RND) family are major determinants of efflux-based antimicrobial resistance in P. aeruginosa (7, 9). Twelve RND efflux systems with different substrate specificities have been characterized so far in P. aeruginosa (10). Of these, the inducible MexXY system was extensively studied due to its frequent overproduction in clinical isolates of P. aeruginosa from CF and non-CF patients and its implication in resistance to a broad spectrum of antimicrobials, especially the highly hydrophilic aminoglycosides (11–22).
This system is composed of a membrane fusion protein, MexX; a proton motive force-driven antiporter, MexY; and one of the outer membrane proteins, OprM (23), OprA in strain PA7 (24), or OpmB (25). The MexXY system accommodates aminoglycosides (e.g., tobramycin), fluoroquinolones (e.g., ciprofloxacin), macrolides (e.g., erythromycin), tetracyclines, and cephalosporins (15, 26). The expression of the MexXY-encoding operon is negatively controlled by MexZ, a TetR family transcriptional regulator encoded by a gene oriented divergently from mexXY (9). MexZ acts as a dimer binding to a 20-bp palindromic sequence in the mexXY-mexZ intergenic region (27, 28). The crystal structure of MexZ shows that it represents a classical TetR fold with nine α helices, with the N-terminal α1-α3 helices forming the DNA-binding domain (DBD) and α8-α9 helices forming the dimer interface (29). MexZ-mediated repression is alleviated in response to ribosome dysfunction following exposure to aminoglycosides, macrolides, and tetracyclines (30–33); oxidative stress (34); or decreased synthesis of ribosomal proteins (35). However, the antimicrobials inducing mexXY expression do not form a complex with MexZ to modulate its DNA-binding activity (27, 28). The increase in mexXY expression was shown to depend on the product of the armZ (PA5471) gene, encoding a large, 41-kDa protein with similarities to the RtcB family of RNA-splicing ligases (28, 36). This gene is expressed in an operon with PA5470, encoding a peptide-releasing factor (36, 37). The expression of the armZ-PA5470 operon is increased in response to subinhibitory concentrations of ribosome-targeting drugs (36). Induction involves a transcription attenuation mechanism (38, 39). Under drug-free conditions, only PA5471.1, preceding the armZ coding region and encoding a short leader peptide, is transcribed and translated. Drug-induced ribosome stalling on PA5471.1 leads to alternate mRNA folding, which allows transcription progression and consequently ArmZ translation (38). A direct ArmZ-MexZ interaction was previously shown using yeast two-hybrid and LexA-based bacterial two-hybrid systems (28, 40). A recent study showed that amino acid residues P68, G76, R216, R221, G231, and G252 of ArmZ are involved in the interaction with MexZ (40). ArmZ binding to MexZ is thought to modulate its repressor activity; however, the exact mechanism is not known (28, 40).
Here, using a series of MexZ truncations and a double-screening method to identify mexZ mutations which diminished MexZ-ArmZ interactions but had no major effect on MexZ repressor activity, we identified a putative ArmZ-binding region in MexZ. The data indicate that several positively charged amino acids in the N-terminal domain of MexZ are critical for the interaction with ArmZ and for drug-induced mexXY expression in P. aeruginosa. Moreover, our bioinformatics analyses suggested that similarly regulated efflux pumps may be present in other bacterial species.
RESULTS
Interactions of ArmZ with truncated forms of MexZ.
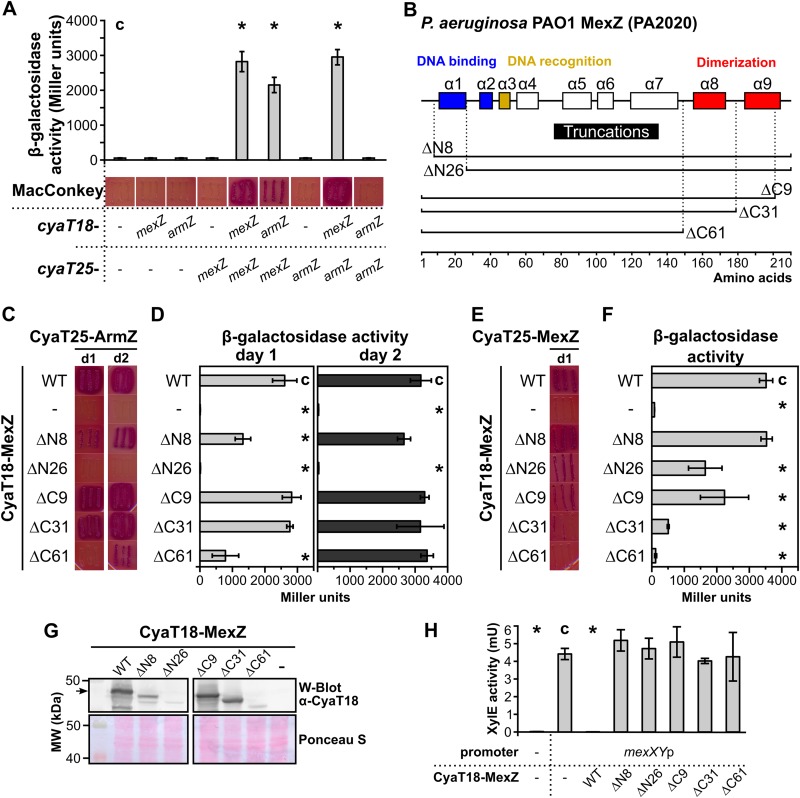
The part of MexZ involved in the interaction with ArmZ was defined by the use of a bacterial two-hybrid system, based on the reconstitution of Bordetella pertussis adenylate cyclase (CyaA) (BACTH) (41). Escherichia coli BTH101 (Δcya) cells were transformed with pairs of complementary BACTH vectors containing cyaT18 and cyaT25 fragments fused with mexZ or armZ. Transformants were selected on MacConkey plates with 1% maltose. Red color of colonies of transformants and high β-galactosidase activity in the corresponding cell extracts, indicating CyaA reconstitution, were observed only for transformants containing plasmids with cyaT18-mexZ plus cyaT25-mexZ, cyaT18-armZ plus cyaT25-mexZ, and cyaT18-mexZ plus cyaT25-armZ but not for remaining combinations, confirming that MexZ can self-interact and that MexZ and ArmZ interact with each other (Fig. 1A). These data indicated that the BACTH system could be used to study molecular details of the MexZ-ArmZ interaction.
FIG 1.
The N terminus of MexZ is involved in interactions with ArmZ. (A) MexZ-ArmZ interactions analyzed by using the BACTH system (41). E. coli BTH101 Δcya transformants carrying vectors expressing cyaT18 and cyaT25 fusions and grown on MacConkey agar were restreaked onto the same medium, photographed after 24 h, and used to inoculate cultures to perform β-galactosidase activity assays. (B) Schematic overview of the MexZ alpha-helical structure (29) and truncation alleles used in this study. (C and D) BACTH analysis of the interactions between truncated MexZ forms and ArmZ. Restreaked transformants were photographed after 1 day (d1) or 2 days (d2) (C) and used for β-galactosidase activity measurements (D). (E and F) Ability of truncated MexZ derivatives to interact with full-length MexZ, analyzed as described above for panels C and D. (G) CyaT18-fused protein levels in transformants from panel C analyzed by Western blotting (W-Blot) using extracts from exponentially growing cultures and anti-CyaT18 antibodies. A Ponceau S-stained membrane is shown as a loading control. The arrow indicates CyaT18-MexZ. MW, molecular weight. (H) Repression of mexXYp-xylE transcriptional fusion by CyaT18-MexZ and its truncated derivatives. XylE activity was assayed in extracts from E. coli DH5α cells containing pPT01 (promoterless xylE) or pKAB10.1 (mexXYp-xylE) and the pLKB4 vector or its derivatives with the indicated MexZ variants. Data represent mean values from at least three cultures ± standard deviations (SD). *, P < 0.05 (as determined by two-sided Student’s t test) relative to the control (indicated with “c”).
To identify the part of MexZ involved in interactions with ArmZ, we designed a series of truncated MexZ variants (Fig. 1B), based on the crystal structure of MexZ (29). mexZ alleles coding for truncated proteins deprived of short unstructured regions or adjacent alpha helices, from either the N terminus or the C terminus, were PCR amplified and cloned into BACTH vectors. Since MexZ-ArmZ interactions were observed only when the Cya fragment was linked to the N terminus of MexZ (data not shown), all shortened mexZ alleles were cloned into pLKB4, facilitating N-terminal translational fusion with CyaT18, and the constructed plasmids were introduced into E. coli BTH101 carrying pKAB25.2 (cyaT25-armZ) or pKAB25.1 (cyaT25-mexZ). Analysis of transformants grown on MacConkey plates and β-galactosidase activity measurements showed that the lack of the N-terminal 8 amino acids (aa) diminished the interaction with ArmZ, whereas the lack of the N-terminal 26 amino acids completely abrogated the MexZ-ArmZ interaction (Fig. 1C and D). Importantly, MexZ ΔN26 showed an interaction with full-length MexZ, proving that the hybrid protein was at least partially functional in the BACTH system (Fig. 1E and F). Concomitantly, the lack of C-terminal alpha helices (ΔC9, ΔC31, and even ΔC61) comprising the MexZ dimerization domain did not block MexZ-ArmZ heterodimerization completely (Fig. 1C and D), while it significantly reduced the self-association properties of MexZ (Fig. 1E and F). All truncated CyaT18-MexZ variants were expressed in the corresponding E. coli BTH101(pKAB25.2) cells, although a decreased level relative to that of full-length CyaT18-MexZ was observed for some of them (Fig. 1G). Remarkably, the reduced protein level not only might be caused by a deleterious effect of the truncation on protein stability but also might be a consequence of the impaired interaction with ArmZ, as we observed that the presence of CyaA reconstitution in E. coli BTH101 triggered by the interaction between MexZ and ArmZ stabilized CyaT18-containing fusion proteins (see Fig. S1 in the supplemental material). To check if the truncations affected the ability of MexZ to repress transcription from the mexXY promoter (mexXYp), we expressed the CyaT18-MexZ proteins in E. coli containing promoter-probe plasmid pKAB10.1 with the mexX-mexZ intergenic region cloned upstream of a promoterless xylE cassette (mexXYp-xylE). Measurement of XylE activity in extracts of cells producing full-length CyaT18-MexZ showed a strong repression of mexXYp-xylE (Fig. 1H). No XylE activity decrease was observed when CyaT18 fusions with truncated MexZ variants were expressed (Fig. 1H), indicating the significance of residues at both the N terminus (DBD) and the C terminus (dimerization domain) for the repressor activity of MexZ.
Identification of MexZ residues important for MexZ-ArmZ interaction.
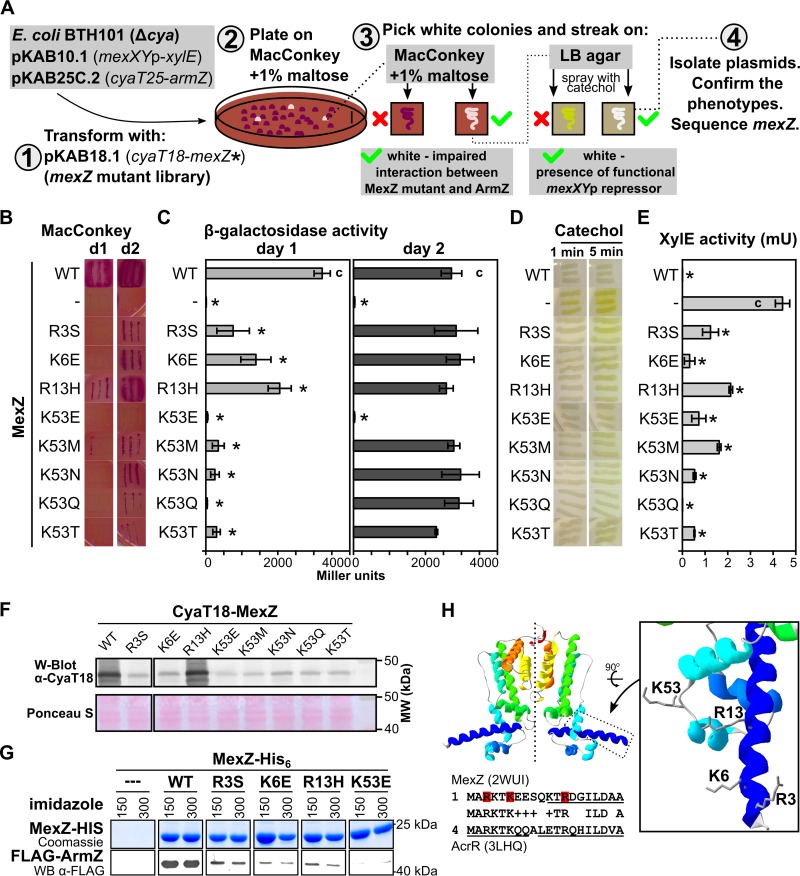
The truncation analysis pointed out that the N-terminal MexZ region involved in interactions with ArmZ was also crucial for MexZ DNA-binding activity. To search for amino acid substitutions in MexZ that abolish MexZ-ArmZ interactions without interfering with MexZ DNA-binding activity, we developed a BACTH-based screening procedure (Fig. 2A). E. coli BTH101 cells carrying pKAB10.1 (mexXYp-xylE) and pKAB25C.2 (cyaT25-armZ) were transformed with a mexZ mutant library in pKAB18.1 (cyaT18-mexZ*). The library was obtained by in vitro hydroxylamine mutagenesis of the PCR-amplified mexZ gene, followed by high-efficiency cloning of mutated mexZ into the BACTH vector pLKB4. Transformants were plated onto MacConkey medium with 1% maltose and grown for 1 to 2 days, and white colonies (comprising ∼0.4% of all colonies) were restreaked onto MacConkey and L agar plates (Fig. 2A). The white color of streaked bacteria on indicator plates could be caused by an effect of the mexZ mutation on the interaction of the corresponding protein product with ArmZ, but it could also be caused by the lack of a mexZ insert, the presence of a premature stop codon, or a mutation that drastically destabilizes the protein. Hence, the ability of CyaT18-MexZ* to repress mexXYp was tested by spraying the streaks onto L agar plates with a catechol solution to assess XylE activity. The white streaks on both media, MacConkey and L agar plates sprayed with catechol, indicated the production of a MexZ variant that has repressor activity but is defective in the interaction with ArmZ.
FIG 2.
Positively charged amino acids in the N-terminal part of MexZ are engaged in interaction with ArmZ. (A) Overview of the strategy used for identification of MexZ mutants with disturbed interactions with ArmZ but that retained the ability to repress mexXYp. (B) BACTH analysis of the interactions between MexZ single-substitution variants and ArmZ. Transformants carrying pKAB10.1, pKAB25C.2, and plasmids expressing cyaT18-mexZ (WT or the indicated mutants) were restreaked onto selective MacConkey plates and grown for 1 day (d1) or 2 days (d2). (C) β-galactosidase activity in cultures of transformants from panel B. (D) Ability of MexZ mutants to repress mexXYp-xylE. Transformants (as in panels B and C) were grown on L agar, sprayed with 10 mM catechol, and photographed after 1 and 5 min. White color indicates repression of mexXYp-xylE by MexZ. (E) XylE activity assayed in extracts from exponentially growing strains as described above for panel B. β-Galactosidase and XylE activity data represent mean values from at least three assays ± SD. *, P < 0.05 (as determined by two-sided Student’s t test) relative to controls (indicated with “c”). (F) Assessment of CyaT18-MexZ levels by Western blotting in cultures used for XylE activity assays. Anti-CyaT18 antibodies were used, and a Ponceau S-stained membrane is shown as a loading control. (G) Pulldown analysis of WT and mutant MexZ interactions with FLAG-ArmZ. A total of 150 μg of purified MexZ-His6 (WT or mutants) and 1 mg of proteins from an extract of E. coli cells overproducing FLAG-ArmZ were applied to Ni-NTA columns, followed by washing and elution using an imidazole gradient. Proteins were separated by SDS-PAGE, followed by Coomassie staining or Western blot analysis using anti-FLAG antibodies. For clarity, the images were cropped and the contrast was enhanced (original images are included in Fig. S2 in the supplemental material). (H) Amino acids participating in the MexZ-ArmZ interaction visualized on the refined MexZ structural model based on data reported previously (29). At the bottom is an alignment of the N-terminal part of MexZ and S. Typhimurium AcrR (42). Residues identified in this study as being involved in interactions with ArmZ are indicated in red. Alpha helices detected in crystal structures are underlined.
Overall, 28 such clones were isolated. Sequencing of mexZ in these plasmids showed nucleotide changes introducing the R3S (2 clones), K6E (2 clones), R13H (2 clones), K53E (8 clones), K53M (4 clones), K53N (8 clones), K53Q (1 clone), and K53T (1 clone) amino acid substitutions into MexZ. The mutations impaired the MexZ interaction with ArmZ; however, only the K53E mutation completely abrogated the interaction (Fig. 2B and C). Concomitantly, all mutants retained the ability to repress mexXYp, as assessed by a plate assay (Fig. 2D) and measurements of XylE activity in extracts from exponentially growing cultures (Fig. 2E). The level of repression varied significantly between MexZ variants despite similar levels of expression of fusion proteins (Fig. 2F). The strongest repression, comparable to that of wild-type (WT) MexZ, was observed for MexZK53Q. Altogether, BACTH screening led to the identification of MexZ amino acids important for the interaction with its antirepressor, pointing out the significance of K53. Since only the K53E mutation completely abrogated the MexZ-ArmZ interaction, this variant was chosen for further analysis from the set of substitutions at this position.
To check the impact of R3S, K6E, R13H, and K53E substitutions in MexZ on the interaction with ArmZ in vitro, MexZ-His6 (WT and mutants) was purified and used as the bait in a pulldown assay. Equal amounts of MexZ-His6 (WT and mutants) were mixed with extracts from E. coli cells overproducing FLAG-ArmZ and loaded onto Ni-nitrilotriacetic acid (NTA) columns, followed by washing and elution of bound proteins with increasing concentrations of imidazole. Eluates were separated on polyacrylamide gels, which were stained with Coomassie brilliant blue or used for Western blot analysis. MexZ-His6 eluted at imidazole concentrations of 0.15 M and 0.3 M (Fig. S2A). Concomitantly, these fractions contained large amounts of FLAG-ArmZ in the case of WT MexZ-His6, observed even after Coomassie staining (Fig. S2A) or in Western blot analysis with anti-FLAG antibodies (Fig. 2G and Fig. S2B), whereas no FLAG-ArmZ was bound to a control column containing the purified His6 tag (Fig. 2G and Fig. S2). The use of R3S, K6E, and R13H variants of MexZ-His6 resulted in a significantly reduced amount of FLAG-ArmZ bound, whereas in the case of MexZK53E-His6, FLAG-ArmZ was hardly detectable (Fig. 2G). Overall, these results are consistent with data from BACTH analyses and indicate that R3S, K6E, and R13H substitutions in MexZ disturb the interactions with ArmZ, whereas the K53E amino acid change completely abolishes them.
The N-terminal 8 residues encompassing R3 and K6, important for MexZ-ArmZ interactions, were missing in the solved structure of MexZ (29). Inspection of the structure of the N-terminal part of AcrR from Salmonella enterica serovar Typhimurium (42), the sequence of which is highly similar to that of the N-terminal part of MexZ (Fig. 2H, bottom), showed that the amino acids form two alpha helices with the N terminus bent toward other helices forming the DNA-binding domain (42). To visualize residues on a structural model of MexZ, we used I-TASSER (43) to refine the structure of the N-terminal part of MexZ. Prediction showed that the N terminus of MexZ forms a long alpha helix (Fig. 2H). In this model, R3, K6, R13, and K53 residues were localized in proximity to each other (Fig. 2H). This suggests that the identified amino acids may form an interface for interaction with ArmZ.
Impact of ArmZ on DNA binding by WT MexZ and its R3S, K6E, R13H, and K53E variants.
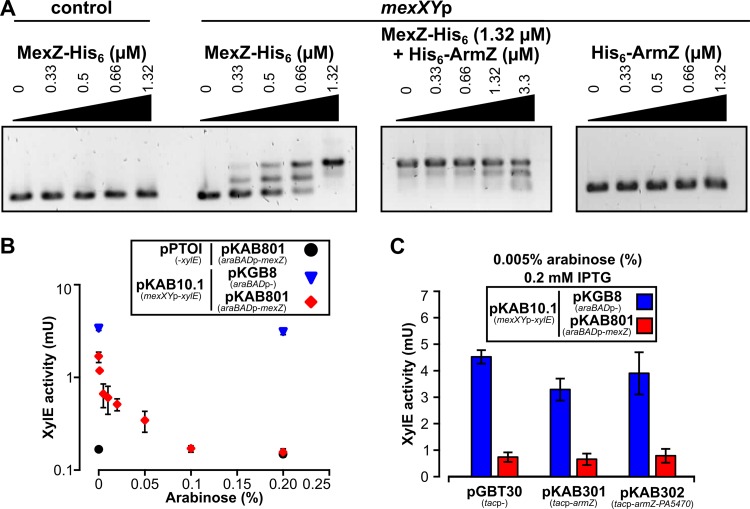
The impact of ArmZ on MexZ binding to DNA in vitro was analyzed using an electrophoretic mobility shift assay (EMSA) with purified MexZ-His6 and the mexXY-mexZ intergenic region (mexXYp) as a probe, with or without His6-ArmZ. MexZ-His6 bound to mexXYp DNA but not to a similar-length control fragment (Fig. 3A). mexXYp was not bound by His6-ArmZ. Strikingly, the addition of His6-ArmZ to MexZ-His6 prior to mixing it with mexXYp only partly interfered with MexZ-DNA binding under the conditions tested (Fig. 3A). Concomitantly, to test the impact of ArmZ on MexZ-mediated repression of mexXYp in vivo, we first used the E. coli DH5α(pKAB10.1 [mexXYp-xylE]/pKAB801 [araBADp-mexZ]) strain and determined that induction with 0.005% arabinose resulted in an amount of MexZ that was not capable of full mexXYp repression (Fig. 3B). However, even such moderate MexZ repression of mexXYp was not relieved by the overproduction of ArmZ. Similarly, the overproduction of ArmZ together with PA5470, a putative peptide-releasing factor (36, 37) encoded in the same operon, had no effect on MexZ-mediated repression (Fig. 3C). This strongly suggested that other factors present in P. aeruginosa, but not in the E. coli strain used, might participate in the control of the influence of ArmZ on MexZ binding to DNA.
FIG 3.
Impact of ArmZ on MexZ binding to mexXYp in vitro and in vivo in the heterologous host E. coli. (A) EMSA of MexZ-His6 and His6-ArmZ binding to a 184-bp mexXYp DNA fragment. DNA at a concentration of 0.1 μM was incubated with the indicated concentrations of MexZ-His6, His6-ArmZ, or both proteins; separated on a 1.5% agarose gel; stained with ethidium bromide; and photographed. A 204-bp pUC19 fragment (amplified using primers 46 and 7) was used to verify the specificity of MexZ binding. (B) MexZ repression of mexXYp in a heterologous host. XylE activity measurements were performed with cell extracts from cultures of E. coli DH5α strains containing the indicated plasmids grown in medium with different concentrations of arabinose, to overproduce MexZ (pKAB801 araBADp-mexZ). Data are presented on a semilogarithmic plot. (C) Effect of ArmZ or ArmZ/PA5470 overproduction on MexZ repression of mexXYp in E. coli DH5α. XylE activity was assayed in extracts of E. coli DH5α transformants grown in the presence of two inducers, IPTG and arabinose. Data represent mean activities from at least three cultures ± SD.
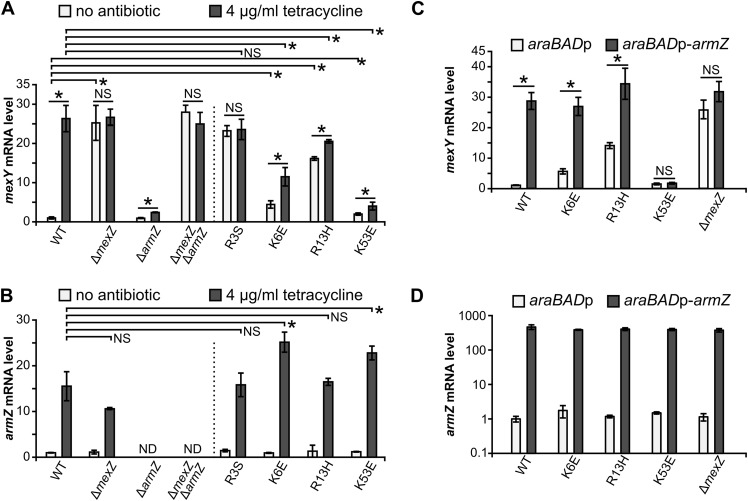
To analyze the impact of R3S, K6E, R13H, and K53E amino acid substitutions in MexZ on the ArmZ-mediated increase of mexXY expression in P. aeruginosa, we constructed a series of PAO1161 mutants containing the corresponding missense mutations in mexZ as well as control strains with inactivated mexZ (ΔmexZ) or armZ (ΔarmZ) and the ΔmexZ ΔarmZ double-deletion strain. WT and mutant strains were grown in medium with or without a subinhibitory concentration of tetracycline, known to induce mexXY expression (33), and mexY as well as armZ transcript levels were analyzed using reverse transcription-quantitative PCR (RT-qPCR) (Fig. 4A and B). The addition of tetracycline resulted in increased armZ mRNA production in the WT and all mexZ missense mutants, with a slightly higher level of armZ expression observed in strains producing MexZK6E and MexZK53E than in the WT strain (Fig. 4B). As expected, tetracycline induced mexY expression in WT PAO1161, whereas only a minor increase was observed in the ΔarmZ mutant, suggesting that the bulk of the induction was ArmZ dependent (Fig. 4A). The lack of MexZ (ΔmexZ) or the production of MexZR3S led to high mexY expression levels independently of the presence of tetracycline (Fig. 4A). In the absence of an inducer, a significant increase in the mexY transcript level was observed in strains expressing MexZK6E and MexZR13H but not to the extent observed in ΔmexZ or mexZR3S strains, suggesting ongoing partial repression of the mexXY promoter in these two mutants (Fig. 4A). However, these variants were impaired in the ArmZ-dependent response to tetracycline, since the observed increase of the mexY mRNA level was significantly lower than that in the WT strain. The strongest repression of mexXYp in the absence and presence of the inducer was observed in cells expressing MexZK53E, confirming a minor effect of this amino acid substitution on the DNA-binding ability of MexZ but a very strong effect on the interaction with ArmZ under native conditions. Altogether, these data indicate that mutations that block the MexZ-ArmZ interaction may hamper both MexZ repression and tetracycline-mediated induction of mexY expression.
FIG 4.
Disturbing the MexZ interaction with ArmZ decreases the extent of ArmZ-mediated induction of mexXY expression in P. aeruginosa. (A and B) RT-qPCR analysis of the expression of mexY (A) and armZ (B) by the indicated PAO1161 strains grown in L broth with or without 4 μg ml−1 tetracycline. (C and D) RT-qPCR analysis of the expression of mexY (C) and armZ (D) in the WT PAO1161 and mexZ mutant strains containing either araBADp (control) or an araBADp-armZ cassette inserted at a neutral site. Cells were grown in L broth containing 0.15% arabinose to induce expression from the araBADp promoter. Data in panel D are presented on a logarithmic scale. All RT-qPCR data represent means ± SD from at least three biological replicates. Expression levels were normalized to the value for the nadB gene and are presented relative to value for the PAO1161 WT strain not exposed to tetracycline (A and B) or the PAO1161 araBADp strain (C and D). *, P < 0.05 (as determined by two-sided Student’s t test assuming equal variance); NS, not significant (P value of >0.05). ND, not detected.
To check if excess ArmZ restores the defect in the MexZ response, the empty araC-araBADp expression cassette or the araC-araBADp-armZ cassette, allowing arabinose-induced overexpression of armZ, was introduced into the genomes of PAO1161 and its derivatives carrying mutant alleles of mexZ. Strains were grown in arabinose-containing medium, and RNA was isolated for RT-qPCR analysis (Fig. 4C and D). The extents of armZ overexpression were similar in all tested strains (Fig. 4D). The abundance of ArmZ triggered an increase in the mexY level in cells producing WT MexZ, to the level observed for the control ΔmexZ mutant strain (Fig. 4C). In agreement with the observed partial inhibition of the MexZ-ArmZ interaction by the K6E or R13H substitution in MexZ (Fig. 2), mexY expression was responsive to armZ overexpression in strains producing MexZK6E or MexZR13H (Fig. 4C). Strikingly, this was not observed in cells expressing MexZK53E (Fig. 4C), and akin to the ΔarmZ strain, the mutant producing MexZK53E was more susceptible to aminoglycosides and tetracycline than WT strain PAO1161 (Table 1), confirming the critical role of K53 for MexZ interactions with ArmZ. Overall, these data indicate that the mutations that disturb the MexZ-ArmZ interaction negatively affect ArmZ-mediated upregulation of genes encoding the MexXY multidrug efflux system, thus increasing the susceptibility of the cells to antimicrobials extruded by this pump.
TABLE 1.
Impact of mexZ and armZ mutations on antimicrobial resistance of P. aeruginosa PAO1161a
| Modified allele(s) of strain PAO1161 Rifr | MIC (μg/ml) |
|||
|---|---|---|---|---|
| Amikacin | Gentamicin | Tobramycin | Tetracycline | |
| None (WT) | 4 | 2 | 1 | 64 |
| ΔmexZ | 8 | 4 | 1 | 64 |
| ΔarmZ | 1 | 0.5 | 0.25 | 16 |
| ΔmexZ ΔarmZ | 8 | 4 | 1 | 64 |
| mexZR3S | 8 | 4 | 1 | 64 |
| mexZK6E | 4 | 2 | 0.5 | 64 |
| mexZR13H | 8 | 4 | 1 | 64 |
| mexZK53E | 2 | 1 | 0.25 | 32 |
Data represent MICs of the indicated antibiotics assayed with a microdilution method in cation-adjusted Mueller-Hinton medium. MIC analysis was performed twice, with identical results.
Identification of MexX/MexY/MexZ orthologs possibly regulated by ArmZ-like proteins.
To identify other bacterial multidrug efflux systems with expression potentially regulated by large ArmZ-like antirepressors alleviating the downregulation exerted by local TetR family transcription factors, we used MultiGeneBlast (44) to search for clusters of genes encoding proteins with similarity to P. aeruginosa MexX/MexY/MexZ in 1,748 representative and reference bacterial genomes included in the RefSeq database (release 91) (45). Genes encoding all three components in proximity to each other were identified in 141 strains (Data Set S1). Since ArmZ shows sequence similarity to RtcB family ligases, ubiquitous in prokaryotic genomes, the identification of ArmZ orthologs was based on the fact that in P. aeruginosa, armZ is transcriptionally coupled with PA5470 (36), a gene encoding a peptide chain release factor (37). A MultiGeneBlast search using P. aeruginosa ArmZ/PA5470 showed 61 genomes with operons containing putative orthologs (Data Set S2). Concomitantly, genomes of 27 strains contained genes encoding MexX/MexY/MexZ and ArmZ/PA5471 orthologs (Data Set S3). These were classified mostly as Enterobacteriales; however, one other Pseudomonas species (P. protegens CHA0) and seven Betaproteobacteria species were also identified (Fig. 5A and Data Set S3). Among these, Burkholderia ubonensis MSMB22 and Burkholderia cepacia ATCC 25416 contained multiple clusters encoding MexXYZ orthologs (Data Set S3). Phylogenetic analysis of MexZ orthologs showed that P. aeruginosa MexZ grouped with proteins identified in B. cepacia, B. ubonensis, or Achromobacter xylosoxidans (Fig. 5A). The alignment of amino acid sequences of MexZ orthologs showed a very high level of conservation of residues forming the N-terminal DNA-binding domain, including those corresponding to R3, K6, and R13 of P. aeruginosa MexZ, likely forming the MexZ-ArmZ interaction surface (Fig. 5B). Concomitantly, K53, critical for the MexZ-ArmZ interaction in P. aeruginosa, was replaced with serine only in the case of the MexZ ortholog from P. protegens and with alanine in the case of B. cepacia and B. ubonensis in one of the MexZ orthologs encoded by their genomes but not in the others (Fig. 5B). A substitution of arginine, which was unlikely to affect the interaction, was observed in the case of the B. cepacia protein under GenBank accession no. ALK16488.1 (Fig. 5B). A similar comparison of amino acid sequences of MexZ-like regulators from strains lacking ArmZ/PA5470 orthologs also demonstrated a high level of conservation of amino acid residues corresponding to R3, K6, and R13, whereas a lower level of conservation was observed for K53 (Fig. 5B and Fig. S4). These data show that a lysine is preferentially found at the position corresponding to K53 of P. aeruginosa MexZ in MexZ-like regulators in strains whose genomes also encode ArmZ/PA5470 orthologs. Taken together, our results suggest a widespread role of ArmZ-like proteins in the regulation of multidrug efflux pump expression in various bacterial species, including clinically important pathogens.
FIG 5.
Residues involved in the interaction with the ArmZ-like antirepressor are conserved in MexZ orthologs from various bacteria. Genes encoding clustered orthologs of MexZ/MexX/MexY or ArmZ/PA5470 were identified in bacterial genomes using MultiGeneBlast (44). Only 27 genomes containing two sets of MexZ/MexX/MexY and ArmZ/PA5470 orthologs were selected for the analysis (see Data Set S3 in the supplemental material). (A) Phylogenetic tree of the identified MexZ orthologs constructed using COBALT (67) by the use of the neighbor-joining method. C. serinivorans, Comamonas serinivorans; D. acidovorans, Delftia acidovorans; C. testosteroni, Comamonas testosteroni; R. solanacearum, Ralstonia solanacearum; N. elongata, Neisseria elongata; P. sneebia, Providencia sneebia; P. stuartii, Providencia stuartii; X. bovienii, Xenorhabdus bovienii; X. hominickii, Xenorhabdus hominickii; M. morganii, Morganella morganii; P. fontium, Pragia fontium; S. fonticola, Serratia fonticola; P. carotovorum, Pectobacterium carotovorum; K. cowanii, Kosakonia cowanii; S. flexneri, Shigella flexneri; S. dysenteriae, Shigella dysenteriae. (B) Sequence alignment of the N-terminal parts of MexZ orthologs (corresponding to amino acids 1 to 56 of P. aeruginosa MexZ). Sequences are ordered as in the tree, amino acids identical to those in P. aeruginosa MexZ are highlighted in gray, and residues corresponding to P. aeruginosa MexZ residues participating in the interaction with its antirepressor are indicated in red. The consensus sequence (amino acids occurring in more than 60% of proteins) and bars representing conservation are presented for MexZ orthologs identified in genomes with and without identified ArmZ/PA5470 orthologs. Full alignments are included in Fig. S3 and S4, respectively.
DISCUSSION
The expression of genes encoding the P. aeruginosa MexXY multidrug efflux system is negatively controlled by MexZ binding to the palindromic sequence in mexXYp (27, 28). The MexZ-mediated repression is alleviated by antirepressor ArmZ, interacting with MexZ (28, 36, 40). The inhibitory effect of ArmZ on DNA binding by MexZ was previously demonstrated using an EMSA with an oligonucleotide corresponding to the MexZ-binding site, cell extracts from tetracycline-treated PAO1 cells, or recombinant ArmZ (28). The EMSA performed here with MexZ-His6 showed only a minor disruption of MexZ-His6 binding to DNA by His6-ArmZ (Fig. 3A). The discrepancy between the results of in vitro experiments may be caused by the ratio of MexZ to ArmZ used; nevertheless, the minor effect observed in our study compromised the EMSA as a method for in vitro analysis of the MexZ-ArmZ interplay. Interestingly, the overproduction of ArmZ in E. coli expressing MexZ had no effect on its ability to repress mexXYp-xylE, whereas excess ArmZ readily increased the level of the mexY transcript in P. aeruginosa (Fig. 4C). These data suggest that another factor present in P. aeruginosa, but not in the E. coli strains used, may be responsible for the fine-tuning of ArmZ activity, but as we have shown, it is not a product of PA5470, the other cistron in the armZ-PA5470 operon (Fig. 3C). ArmZ shows amino acid similarity with RtcB RNA-splicing ligases involved in tRNA repair (46), suggesting that ArmZ may potentially bind small RNAs; however, the significance of this observation, if any, is unknown.
The mechanism by which ArmZ interferes with MexZ binding to DNA has been highly speculative (29, 40, 47). The truncation analysis indicated that the removal of the C-terminal α8 and α9 helices of MexZ, involved in dimerization, does not block its interaction with ArmZ, suggesting that MexZ self-association is not necessary for the interaction with ArmZ. While TetR repressors are thought to bind to DNA as dimers (48) and analytical gel filtration showed that MexZ forms dimers in solution (29), we cannot rule out that a MexZ monomer-dimer equilibrium exists in the cells and that binding of ArmZ to MexZ disrupts MexZ dimer formation.
Analysis of short N-terminal-deletion derivatives of MexZ demonstrated an impairment of both functions, DNA binding and interactions with the antirepressor (Fig. 1C, D, and H). To dissect these two activities, a search for mutations that did not abolish repressor function but that impaired MexZ-ArmZ interactions was undertaken (Fig. 2A). The double-screening procedure led to the identification of four positively charged amino acids in the N terminus of MexZ, R3, K6, R13, and K53 (Fig. 2B to E). Notably, K53 was crucial for the interaction with ArmZ, since several substitution mutants at this position were isolated. Interestingly, various amino acids at this position modulated DNA binding and interactions with ArmZ to different extents, with the K53E substitution showing a minor impact on repressor activity while eliminating sensitivity to the antirepressor. Residues R3, K6, and R13 are located in the putative α1 helix of the MexZ DBD (Fig. 2H), whereas K53 is located in the loop between the DNA recognition helix α3 and the α4 helix. Since in the crystal structure, α4 extensively interacts with α1-α3 (29), our data indicate that the ArmZ-binding domain overlaps the DNA-binding domain. Mutations in R3, K6, R13, and K53 participated to various extents not only in interactions with ArmZ but also in DNA binding; therefore, we speculate that ArmZ interferes with MexZ by blocking its DNA-binding domain.
The only other described P. aeruginosa antirepressor, ArmR (53 aa), inactivates the MarR family transcriptional regulator MexR by inducing a conformational change incompatible with DNA binding (49). Recently, several examples of proteins that mimic the DNA substrates of various proteins to block their DNA binding have been described (50, 51), including the CarS (111 aa) antirepressor from Myxococcus xanthus (52, 53), the antirepressor AbbA (68 aa) of Bacillus subtilis (54), and DMP19 (166 aa) of Neisseria meningitidis (55). The interaction between DMP19 and NHTF, a xenobiotic response element (XRE) family transcription factor, has been suggested to be driven by specific charge-charge interactions (55). Previously, two positively charged residues, R216 and R221, of ArmZ were also indicated to be part of the ArmZ-MexZ interaction domain, and substitution of these amino acids in ArmZ had the strongest negative effect on the spectinomycin-induced increase of mexXY expression (40). However, since positively charged residues were also identified in our study, together, these mutational analyses do not directly suggest that there are any residues in MexZ and ArmZ that could form charge-charge interactions. The examples listed above indicate proteins relatively smaller than ArmZ (41 kDa). Nevertheless, larger antirepressors may employ the same mechanisms as the ones indicated above, e.g., the antiactivator RapF (380 aa) from B. subtilis (56). Overall, these data suggest that inactivation of MexZ by ArmZ may involve changes in the DBD of MexZ; however, the exact mechanism requires further studies.
The search for efflux systems in other species whose expression could be regulated through an interaction between a TetR family transcriptional regulator and a large ArmZ-like antirepressor showed several candidates (see Data Set S3 in the supplemental material). These include efflux systems in pathogenic A. xylosoxidans (57, 58), members of the B. cepacia complex (59), and several enterobacteria, e.g., Proteus mirabilis (60). We are aware that the search is by no means exhaustive; however, it clearly shows that regulation of genes that control the expression of multidrug efflux pumps by large ArmZ-like antirepressors could be a common way of inducing efflux-mediated resistance to antimicrobials in bacteria.
Conclusions.
ArmZ interacts with MexZ to control the expression of genes encoding the MexXY efflux pump, which confers multidrug resistance to P. aeruginosa. The positively charged residues responsible for MexZ-ArmZ interactions are located at the N terminus of MexZ, suggesting an overlap between DNA- and ArmZ-binding domains. Similar cross talk may contribute to the induction of efflux-mediated antimicrobial resistance in other bacterial species. Further structural studies should shed light on the exact molecular mechanism of MexZ inactivation by ArmZ.
MATERIALS AND METHODS
Strains, growth conditions, and genetic manipulations.
Strains used and constructed in this work are listed in Table 2. All P. aeruginosa strains used in this study are derivatives of PAO1161 [leu(r− m−)] (restriction-modification defective, leucine-auxotrophic strain; provided by B. M. Holloway, Monash University, Clayton, Victoria, Australia). Escherichia coli strain DH5α was used for plasmid manipulations. Plasmids used and constructed in this study are listed in Tables S1 and S2, respectively, in the supplemental material. Oligonucleotides used in this work are listed in Table S3. All new plasmid constructs were verified by sequencing (Laboratory of DNA Sequencing and Oligonucleotide Synthesis, Institute of Biochemistry and Biophysics, Polish Academy of Sciences [IBB PAS]). Standard DNA manipulations were performed as described previously (61). Gibson assembly reactions were performed by the use of OverLap assembly mix (A&A Biotechnology, Poland).
TABLE 2.
Bacterial strains used and constructed in this study
| Strain | Genotype or description | Source and/or reference(s) |
|---|---|---|
| Escherichia coli | ||
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (λDE3) | Novagen Inc. |
| BTH101 | F− cya-99 araD139 galE15 galK16 rpsL1 (Smr) hsdR2 mcrA1 mcrB1 | Euromedex; 41 |
| DH5α | F− (ϕ80dlacZΔM15) recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U196 | 71 |
| S17-1 | thi pro ΔhsdR hsdM+ recA Tpr Smr RP4-Tc::Mu-Km::Tn7 | 72 |
| Pseudomonas aeruginosa | ||
| PAO1161 Rifr | leu (r− m−) Rifr | 63, 73 |
| PAO1161 Rifr ΔmexZ | Allele exchange in the PAO1161 Rifr genome by use of pKAB611 | This study |
| PAO1161 Rifr ΔarmZ | Allele exchange in the PAO1161 Rifr genome by use of pKAB612 | This study |
| PAO1161 Rifr ΔmexZ ΔarmZ | Allele exchange in the PAO1161 Rifr ΔmexZ genome by use of pKAB612 | This study |
| PAO1161 Rifr mexZR3S | Allele exchange in the PAO1161 Rifr genome by use of pKAB613 | This study |
| PAO1161 Rifr mexZK6E | Allele exchange in the PAO1161 Rifr genome by use of pKAB614 | This study |
| PAO1161 Rifr mexZR13H | Allele exchange in the PAO1161 Rifr genome by use of pKAB615 | This study |
| PAO1161 Rifr mexZK53E | Allele exchange in the PAO1161 Rifr genome by use of pKAB616 | This study |
| PAO1161 Rifr araBADp | araC-araBADp-rrnBt cassette inserted into the PA5412-PA5413 intergenic region of PAO1161 Rifr by use of pKAB601 | 65 |
| PAO1161 Rifr ΔmexZ araBADp | araC-araBADp-rrnBt cassette inserted into the PA5412-PA5413 intergenic region of PAO1161 Rifr ΔmexZ by use of pKAB601 | This study |
| PAO1161 Rifr mexZK6E araBADp | araC-araBADp-rrnBt cassette inserted into the PA5412-PA5413 intergenic region of PAO1161 Rifr mexZK6E by use of pKAB601 | This study |
| PAO1161 Rifr mexZR13H araBADp | araC-araBADp-rrnBt cassette inserted into the PA5412-PA5413 intergenic region of PAO1161 Rifr mexZR13H by use of pKAB601 | This study |
| PAO1161 Rifr mexZK53E araBADp | araC-araBADp-rrnBt cassette inserted into the PA5412-PA5413 intergenic region of PAO1161 Rifr mexZK53E by use of pKAB601 | This study |
| PAO1161 Rifr araBADp-armZ | araC-araBADp-armZ-rrnBt cassette inserted into the PA5412-PA5413 intergenic region of PAO1161 Rifr by use of pKAB610 | This study |
| PAO1161 Rifr ΔmexZ araBADp-armZ | araC-araBADp-armZ-rrnBt cassette inserted into the PA5412-PA5413 intergenic region of PAO1161 Rifr ΔmexZ by use of pKAB610 | This study |
| PAO1161 Rifr mexZK6E araBADp-armZ | araC-araBADp-armZ-rrnBt cassette inserted into the PA5412-PA5413 intergenic region of PAO1161 Rifr mexZK6E by use of pKAB610 | This study |
| PAO1161 Rifr mexZR13H araBADp-armZ | araC-araBADp-armZ-rrnBt cassette inserted into the PA5412-PA5413 intergenic region of PAO1161 Rifr mexZR13H by use of pKAB610 | This study |
| PAO1161 Rifr mexZK53E araBADp-armZ | araC-araBADp-armZ-rrnBt cassette inserted into the PA5412-PA5413 intergenic region of PAO1161 Rifr mexZK53E by use of pKAB610 | This study |
Mutated alleles to be introduced into the PAO1161 genome by homologous recombination were cloned into suicide vector pAKE600 (62). The E. coli S17-1 strain was transformed with pAKE600 derivatives, and transformants were used as donors for conjugation with PAO1161 Rifr. The allele exchange method was performed as described previously (63). The presence of the mexZ or armZ deletions as well as insertions in the PA5412-PA5413 intergenic region was verified by PCR, whereas the presence of mexZ missense mutations was verified by restriction analysis of PCR products (Table S2).
Bacteria were grown in L broth or on L agar (L broth with 1.5% [wt/vol] agar) at 37°C or 28°C. MacConkey agar (BD Difco) supplemented with 1% maltose was used in bacterial adenylate cyclase two-hybrid (BACTH) assays. Cation-adjusted Mueller-Hinton II broth (BBL) was used in antimicrobial susceptibility assays. Plasmids in E. coli strains were maintained by the addition of 150 μg ml−1 benzylpenicillin sodium salt in liquid medium (300 μg ml−1 for solid medium), 50 μg ml−1 kanamycin, or 10 μg ml−1 chloramphenicol. P. aeruginosa transconjugants were selected by the addition of 300 μg ml−1 carbenicillin and 300 μg ml−1 rifampin.
Analysis of protein-protein interactions using a two-hybrid system.
Protein-protein interactions were analyzed using the BACTH system (41). CyaT18 and CyaT25 fragments were fused to the N termini of the proteins of interest. Pairs of plasmids encoding the hybrid genes cyaT18-X and cyaT25-Y were transformed into E. coli BTH101 Δcya. The interaction between the proteins leading to the reconstitution of CyaA activity, triggering cAMP synthesis and the transcription of genes involved in the catabolism of carbohydrates (lactose or maltose), was identified as red color of colonies on MacConkey agar and by an increase of the β-galactosidase activity in the cell extracts. Bacteria were plated onto MacConkey medium with 1% maltose supplemented with appropriate antibiotics and grown for 1 day at 28°C. Three random clones for each type of transformant were streaked onto fresh MacConkey plates, incubated for 24 h or 48 h at 28°C, and photographed. The colonies were used to inoculate cultures in liquid L broth (grown for 20 h at 28°C) for β-galactosidase activity measurements as described previously (64).
Screening of mexZ mutants in the BACTH system.
To obtain the library of mexZ alleles in the pLKB4 vector (ΔcyaT18), the PCR-amplified open reading frame (ORF) was mutagenized in vitro and cloned into pLKB4. This strategy prevented the accumulation of random mutations in the plasmid backbone, including lacp-cyaT18. The PCR product encompassing mexZ, amplified from pKAB18.1 by the use of primers 44 and 45, was gel purified, and 2 μg of DNA in 1 ml of a solution containing 0.1 M potassium phosphate buffer (pH 6.0) and 1 mM EDTA was mixed with 0.8 ml of a solution containing 2 M hydroxylamine and 0.4 M NaOH and incubated for 90 min at 70°C, followed by purification with a Clean-up kit (A&A Biotechnology, Poland). The mutagenized PCR product, digested with EcoRI and SacI, was ligated with pLKB4 digested with same enzymes and treated with FastAP thermosensitive alkaline phosphatase (Thermo). The ligation mixtures were digested with BamHI to remove traces of the empty vector and used for the transformation of electrocompetent E. coli DH5α cells. Approximately 100,000 clones, with more than 99% containing the insert, were pooled, and the plasmid DNA was isolated to form the pKAB18.1 (cyaT18-mexZ*) library.
A search for MexZ variants impaired in interactions with ArmZ but capable of mexXYp repression was performed by transformation of the pKAB18.1 library into E. coli BTH101 Δcya cells carrying pKAB10.1 (mexXYp-xylE) and pKAB25C.2 (cyaT25-armZ) (Fig. 2A). Transformants were selected on MacConkey agar with 1% maltose supplemented with penicillin, kanamycin, and chloramphenicol at 28°C for 24 to 48 h. White colonies were streaked onto a fresh MacConkey agar plate and onto an L agar plate. After overnight growth, the L agar plate was sprayed with 10 mM catechol to assess the ability of the MexZ mutant to repress mexXYp-xylE. Overall, 120,000 transformants were screened, resulting in 28 clones producing proteins with a disturbed interaction with ArmZ but with the ability to repress mexXYp-xylE. Sequencing of mexZ in these clones showed point mutations resulting in 8 single-amino-acid substitutions (plasmids pKAB18.8 to pKAB18.15) (Table S2).
Protein purification.
E. coli BL21(DE3) cells with pET28 derivatives encoding MexZ-His6 (WT and mutants) or His6-ArmZ were grown to an optical density at 600 nm (OD600) of 0.6, induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and grown with shaking at 37°C for an additional 3 h. The cells were harvested by centrifugation and sonicated in lysis, elution, wash (LEW) buffer (50 mM NaPi [pH 8.0], 300 mM NaCl). His6-tagged proteins were captured using affinity chromatography on Macherey-Nagel Protino Ni-TED 1000 columns, followed by washing (10 ml of LEW buffer) and elution using LEW buffer with 300 mM imidazole. The fractions were dialyzed overnight against LEW buffer with 10% glycerol, and proteins were stored at −80°C. Protein yield and purity were estimated by SDS-PAGE using the Pharmacia Phast gel system.
Pulldown assay.
Purified MexZ-His6 (150 μg) was mixed with 1 mg of proteins from a cell extract obtained by sonication of E. coli BL21(pKAB28.6) cells overproducing FLAG-ArmZ in LEW buffer with 10% glycerol. The mixtures were incubated at 22°C for 10 min and applied to Protino Ni-TED 150 columns (Macherey-Nagel) equilibrated with LEW buffer. The columns were washed with 5 ml of LEW buffer, and protein complexes were eluted by applying LEW buffer with increasing concentrations of imidazole. Samples of E. coli BL21 cells with the empty vector pKAB28.7, treated as described above for the protein purification protocol, were used as a negative control. Eluates were separated on 10% polyacrylamide gels and stained with Coomassie brilliant blue or analyzed by Western blotting by the use of mouse monoclonal anti-FLAG antibodies (catalog no. MA1-91878; Thermo).
Electrophoretic mobility shift assay.
Binding of MexZ-His6 and His6-ArmZ to DNA was analyzed by an electrophoretic mobility shift assay (EMSA). Probes were prepared by gel purification of PCR products obtained using pKAB10.1 as a template and primers 1 and 2 (mexXYp, 184 bp) or using pUC19 as a template and primers 46 and 7 (control, 204 bp). The recombinant protein(s) at the indicated concentrations was added to 0.1 μM probe in 20 μl of binding buffer (10 mM Tris-HCl [pH 8.5], 10 mM MgCl2, 100 mM KCl, 0.1 mg ml−1 bovine serum albumin [BSA]) and incubated at 22°C for 15 min, and the mixtures were separated by electrophoresis on a 1.5% agarose gel in 0.5× TBE buffer (45 mM Tris-HCl [pH 8.0], 45 mM boric acid, 1 mM EDTA) at 4°C. Gels were placed in 1 μg ml−1 ethidium bromide in water for 10 min, followed by destaining in water (10 min or as long as required) before visualization of the bands using UV light. In competition experiments, MexZ-His6 and His6-ArmZ were mixed and equilibrated for 15 min in binding buffer before the addition of the probe.
RT-qPCR analysis.
RNA isolation was performed with the Total RNA minikit (A&A Biotechnology, Poland) using cells from 2 ml of cultures at an OD600 of 0.5. RT-qPCR was performed essentially as described previously (65). Briefly, cDNA was synthesized with a TranScriba cDNA synthesis kit (A&A Biotechnology, Poland) using random-hexamer primers. qPCRs with Hot FIREPol EvaGreen qPCR mix plus (Solis Biodyne) were run in a Roche LightCycler 480 apparatus and analyzed by the relative expression method using Roche LightCycler 480 software (v1.5.1.62). Three technical replicates were used for each gene-primer combination. Oligonucleotides are listed in Table S3. nadB (PA0761) was used as a reference gene.
Bioinformatics analyses.
The MexZ structure, including the N-terminal part missing from the previously solved structure (PDB accession no. 2WUI) (29), was modeled using I-TASSER (43). Swiss PDB Viewer v4.1 (http://www.expasy.org/spdbv/) was used for application of symmetry and visualization of the residues on the model (66). Identification of genes encoding clustered orthologs of MexZ/MexX/MexY or ArmZ/PA5470 was performed using MultiGeneBlast (44). The database used for searches encompassed 1,748 representative and reference bacterial genomes included in RefSeq release 91 (45). The minimal sequence coverage of BLAST hits was set to 25%, and the minimal percent identity was set to 30%. The maximum distance between genes was limited to 20 kb. Protein sequence alignment and phylogenetic tree construction by the use of the neighbor-joining method were conducted using COBALT (https://www.ncbi.nlm.nih.gov/tools/cobalt/) (67). The alignment was visualized using SnapGene viewer (SnapGene software; GSL Biotech).
Other assays.
The activity of catechol 2,3-dioxygenase (XylE) in cell extracts was determined spectrophotometrically as described previously (68) and normalized to the protein content in cell extracts determined by using the Bradford assay (69). One mU is defined as the activity of enzyme resulting in the formation of 1 nmol of 2-hydroxymuconic semialdehyde per min. Western blot analysis of CyaT18-fused proteins was performed by the use of mouse anti-CyaA antibodies (3D1, catalog no. SC-13582; Santa Cruz Biotechnology). For Western blot analysis, cell pellets were resuspended in loading buffer (50 mM Tris-HCl [pH 8.0], 0.1 M dithiothreitol [DTT], 2% SDS, 0.1% bromophenol blue, 10% glycerol) and boiled for 10 min. MICs were evaluated by using a microdilution method in microtiter plates with cation-adjusted Mueller-Hinton broth (70). MICs for the control strain ATCC 27853 were 2 μg ml−1 for amikacin, 1 μg ml−1 for gentamicin, 0.5 μg ml−1 for tobramycin, and 16 μg ml−1 for tetracycline and were in the ranges indicated for this reference strain by the CLSI.
Supplementary Material
ACKNOWLEDGMENTS
This work was financed by Polish National Science Centre grant no. 2013/11/B/NZ2/02555 awarded to G.J.-B. M.M. has been supported by Polish National Science Centre grant no. 2015/18/E/NZ2/00675.
A.K. designed the project, conducted most of the experiments, performed data analysis, and wrote the manuscript. B.Z. performed the MIC analysis. M.M. performed protein purification and EMSAs. A.A.B. and G.J.-B. contributed to acquisition and analysis of the data at various stages of the project. G.J.-B. and A.A.B. revised the manuscript. All authors approved the final version of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01199-19.
REFERENCES
- 1.Oliver A, Cantón R, Campo P, Baquero F, Blázquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1253. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 2.Emerson J, Rosenfeld M, McNamara S, Ramsey B, Gibson RL. 2002. Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr Pulmonol 34:91–100. doi: 10.1002/ppul.10127. [DOI] [PubMed] [Google Scholar]
- 3.European Cystic Fibrosis Society. 2018. ECFS patient registry: annual data report, 2016 data, version 1.2018. European Cystic Fibrosis Society, Karup, Denmark: https://www.ecfs.eu/sites/default/files/general-content-images/working-groups/ecfs-patient-registry/ECFSPR_Report2016_06062018.pdf. [Google Scholar]
- 4.Li Z, Kosorok MR, Farrell PM, Laxova A, West SEH, Green CG, Collins J, Rock MJ, Splaingard ML. 2005. Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA 293:581–588. doi: 10.1001/jama.293.5.581. [DOI] [PubMed] [Google Scholar]
- 5.Høiby N, Frederiksen B, Pressler T. 2005. Eradication of early Pseudomonas aeruginosa infection. J Cyst Fibros 4(Suppl 2):49–54. doi: 10.1016/j.jcf.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 6.Langan KM, Kotsimbos T, Peleg AY. 2015. Managing Pseudomonas aeruginosa respiratory infections in cystic fibrosis. Curr Opin Infect Dis 28:547–556. doi: 10.1097/QCO.0000000000000217. [DOI] [PubMed] [Google Scholar]
- 7.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breidenstein EBM, de la Fuente-Núñez C, Hancock REW. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Aires JR, Köhler T, Nikaido H, Plésiat P. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 43:2624–2628. doi: 10.1128/AAC.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X-Z, Plésiat P. 2016. Antimicrobial drug efflux pumps in Pseudomonas aeruginosa, p 359–400. In Li X-Z, Elkins CA, Zgurskaya HI (ed), Efflux-mediated antimicrobial resistance in bacteria: mechanisms, regulation, and clinical implications. Adis, Cham, Switzerland. [Google Scholar]
- 11.Westbrock-Wadman S, Sherman DR, Hickey MJ, Coulter SN, Zhu YQ, Warrener P, Nguyen LY, Shawar RM, Folger KR, Stover CK. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob Agents Chemother 43:2975–2983. doi: 10.1128/AAC.43.12.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frimodt-Møller J, Rossi E, Haagensen JAJ, Falcone M, Molin S, Johansen HK. 2018. Mutations causing low level antibiotic resistance ensure bacterial survival in antibiotic-treated hosts. Sci Rep 8:12512. doi: 10.1038/s41598-018-30972-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hocquet D, Vogne C, El Garch F, Vejux A, Gotoh N, Lee A, Lomovskaya O, Plésiat P. 2003. MexXY-OprM efflux pump is necessary for adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 47:1371–1375. doi: 10.1128/aac.47.4.1371-1375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hocquet D, Roussel-Delvallez M, Cavallo J-D, Plésiat P. 2007. MexAB-OprM- and MexXY-overproducing mutants are very prevalent among clinical strains of Pseudomonas aeruginosa with reduced susceptibility to ticarcillin. Antimicrob Agents Chemother 51:1582–1583. doi: 10.1128/AAC.01334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morita Y, Tomida J, Kawamura Y. 2012. MexXY multidrug efflux system of Pseudomonas aeruginosa. Front Microbiol 3:408. doi: 10.3389/fmicb.2012.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prickett MH, Hauser AR, McColley SA, Cullina J, Potter E, Powers C, Jain M. 2017. Aminoglycoside resistance of Pseudomonas aeruginosa in cystic fibrosis results from convergent evolution in the mexZ gene. Thorax 72:40–47. doi: 10.1136/thoraxjnl-2015-208027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riou M, Avrain L, Carbonnelle S, El Garch F, Pirnay J-P, De Vos D, Plésiat P, Tulkens PM, Van Bambeke F. 2016. Increase of efflux-mediated resistance in Pseudomonas aeruginosa during antibiotic treatment in patients suffering from nosocomial pneumonia. Int J Antimicrob Agents 47:77–83. doi: 10.1016/j.ijantimicag.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Sobel ML, McKay GA, Poole K. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 47:3202–3207. doi: 10.1128/aac.47.10.3202-3207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogne C, Aires JR, Bailly C, Hocquet D, Plésiat P. 2004. Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob Agents Chemother 48:1676–1680. doi: 10.1128/aac.48.5.1676-1680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guénard S, Muller C, Monlezun L, Benas P, Broutin I, Jeannot K, Plésiat P. 2014. Multiple mutations lead to MexXY-OprM-dependent aminoglycoside resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother 58:221–228. doi: 10.1128/AAC.01252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiser TH, Obritsch MD, Jung R, MacLaren R, Fish DN. 2010. Efflux pump contribution to multidrug resistance in clinical isolates of Pseudomonas aeruginosa. Pharmacotherapy 30:632–638. doi: 10.1592/phco.30.7.632. [DOI] [PubMed] [Google Scholar]
- 22.López-Causapé C, Rubio R, Cabot G, Oliver A. 2018. Evolution of the Pseudomonas aeruginosa aminoglycoside mutational resistome in vitro and in the cystic fibrosis setting. Antimicrob Agents Chemother 62:e02583-17. doi: 10.1128/AAC.02583-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother 43:415–417. doi: 10.1128/AAC.43.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morita Y, Tomida J, Kawamura Y. 2012. Primary mechanisms mediating aminoglycoside resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7. Microbiology 158:1071–1083. doi: 10.1099/mic.0.054320-0. [DOI] [PubMed] [Google Scholar]
- 25.Murata T, Gotoh N, Nishino T. 2002. Characterization of outer membrane efflux proteins OpmE, OpmD and OpmB of Pseudomonas aeruginosa: molecular cloning and development of specific antisera. FEMS Microbiol Lett 217:57–63. doi: 10.1111/j.1574-6968.2002.tb11456.x. [DOI] [PubMed] [Google Scholar]
- 26.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:3322–3327. doi: 10.1128/aac.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuo Y, Eda S, Gotoh N, Yoshihara E, Nakae T. 2004. MexZ-mediated regulation of mexXY multidrug efflux pump expression in Pseudomonas aeruginosa by binding on the mexZ-mexX intergenic DNA. FEMS Microbiol Lett 238:23–28. doi: 10.1111/j.1574-6968.2004.tb09732.x. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto M, Ueda A, Kudo M, Matsuo Y, Fukushima J, Nakae T, Kaneko T, Ishigatsubo Y. 2009. Role of MexZ and PA5471 in transcriptional regulation of mexXY in Pseudomonas aeruginosa. Microbiology 155:3312–3321. doi: 10.1099/mic.0.028993-0. [DOI] [PubMed] [Google Scholar]
- 29.Alguel Y, Lu D, Quade N, Sauter S, Zhang X. 2010. Crystal structure of MexZ, a key repressor responsible for antibiotic resistance in Pseudomonas aeruginosa. J Struct Biol 172:305–310. doi: 10.1016/j.jsb.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 30.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:2242–2246. doi: 10.1128/aac.44.9.2242-2246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dean CR, Visalli MA, Projan SJ, Sum P-E, Bradford PA. 2003. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob Agents Chemother 47:972–978. doi: 10.1128/aac.47.3.972-978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caughlan RE, Sriram S, Daigle DM, Woods AL, Buco J, Peterson RL, Dzink-Fox J, Walker S, Dean CR. 2009. Fmt bypass in Pseudomonas aeruginosa causes induction of MexXY efflux pump expression. Antimicrob Agents Chemother 53:5015–5021. doi: 10.1128/AAC.00253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeannot K, Sobel ML, Garch FE, Poole K, Plésiat P. 2005. Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J Bacteriol 187:5341–5346. doi: 10.1128/JB.187.15.5341-5346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraud S, Poole K. 2011. Oxidative stress induction of the MexXY multidrug efflux genes and promotion of aminoglycoside resistance development in Pseudomonas aeruginosa. Antimicrob Agents Chemother 55:1068–1074. doi: 10.1128/AAC.01495-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau CH-F, Fraud S, Jones M, Peterson SN, Poole K. 2012. Reduced expression of the rplU-rpmA ribosomal protein operon in mexXY-expressing pan-aminoglycoside-resistant mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother 56:5171–5179. doi: 10.1128/AAC.00846-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morita Y, Sobel ML, Poole K. 2006. Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product. J Bacteriol 188:1847–1855. doi: 10.1128/JB.188.5.1847-1855.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi J, Liu Y, Zhang Y, Jin Y, Bai F, Cheng Z, Jin S, Wu W. 2018. PA5470 counteracts antimicrobial effect of azithromycin by releasing stalled ribosome in Pseudomonas aeruginosa. Antimicrob Agents Chemother 62:e01867-17. doi: 10.1128/AAC.01867-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita Y, Gilmour C, Metcalf D, Poole K. 2009. Translational control of the antibiotic inducibility of the PA5471 gene required for mexXY multidrug efflux gene expression in Pseudomonas aeruginosa. J Bacteriol 191:4966–4975. doi: 10.1128/JB.00073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia X, Ling B-D, Li X-Z. 2016. Influence of regulatory RNAs on antimicrobial resistance and efflux mechanisms, p 625–650. In Li X-Z, Elkins CA, Zgurskaya HI (ed), Efflux-mediated antimicrobial resistance in bacteria: mechanisms, regulation, and clinical implications. Adis, Cham, Switzerland. [Google Scholar]
- 40.Hay T, Fraud S, Lau CH-F, Gilmour C, Poole K. 2013. Antibiotic inducibility of the mexXY multidrug efflux operon of Pseudomonas aeruginosa: involvement of the MexZ anti-repressor ArmZ. PLoS One 8:e56858. doi: 10.1371/journal.pone.0056858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osipiuk J, Mulligan R, Papazisi L, Anderson WF, Joachimiak A, Center for Structural Genomics of Infectious Diseases. 2010. X-ray crystal structure from “DNA-binding transcriptional repressor AcrR from Salmonella Typhimurium.” PDB http://www.rcsb.org/pdb/explore/explore.do?structureId=3LHQ (accession no. 3LHQ).
- 43.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER suite: protein structure and function prediction. Nat Methods 12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medema MH, Takano E, Breitling R. 2013. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol Biol Evol 30:1218–1223. doi: 10.1093/molbev/mst025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka N, Shuman S. 2011. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem 286:7727–7731. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Housseini B Issa K, Phan G, Broutin I. 2018. Functional mechanism of the efflux pumps transcription regulators from Pseudomonas aeruginosa based on 3D structures. Front Mol Biosci 5:57. doi: 10.3389/fmolb.2018.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuthbertson L, Nodwell JR. 2013. The TetR family of regulators. Microbiol Mol Biol Rev 77:440–475. doi: 10.1128/MMBR.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilke MS, Heller M, Creagh AL, Haynes CA, McIntosh LP, Poole K, Strynadka NCJ. 2008. The crystal structure of MexR from Pseudomonas aeruginosa in complex with its antirepressor ArmR. Proc Natl Acad Sci U S A 105:14832–14837. doi: 10.1073/pnas.0805489105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H-C, Chou C-C, Hsu K-C, Lee C-H, Wang AH-J. 2019. New paradigm of functional regulation by DNA mimic proteins: recent updates. IUBMB Life 71:539–548. doi: 10.1002/iub.1992. [DOI] [PubMed] [Google Scholar]
- 51.Wang H-C, Ho C-H, Hsu K-C, Yang J-M, Wang AH-J. 2014. DNA mimic proteins: functions, structures, and bioinformatic analysis. Biochemistry 53:2865–2874. doi: 10.1021/bi5002689. [DOI] [PubMed] [Google Scholar]
- 52.León E, Navarro-Avilés G, Santiveri CM, Flores-Flores C, Rico M, González C, Murillo FJ, Elías-Arnanz M, Jiménez MA, Padmanabhan S. 2010. A bacterial antirepressor with SH3 domain topology mimics operator DNA in sequestering the repressor DNA recognition helix. Nucleic Acids Res 38:5226–5241. doi: 10.1093/nar/gkq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitworth DE, Hodgson DA. 2001. Light-induced carotenogenesis in Myxococcus xanthus: evidence that CarS acts as an anti-repressor of CarA. Mol Microbiol 42:809–819. doi: 10.1046/j.1365-2958.2001.02679.x. [DOI] [PubMed] [Google Scholar]
- 54.Tucker AT, Bobay BG, Banse AV, Olson AL, Soderblom EJ, Moseley MA, Thompson RJ, Varney KM, Losick R, Cavanagh J. 2014. A DNA mimic: the structure and mechanism of action for the anti-repressor protein AbbA. J Mol Biol 426:1911–1924. doi: 10.1016/j.jmb.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H-C, Ko T-P, Wu M-L, Ku S-C, Wu H-J, Wang AH-J. 2012. Neisseria conserved protein DMP19 is a DNA mimic protein that prevents DNA binding to a hypothetical nitrogen-response transcription factor. Nucleic Acids Res 40:5718–5730. doi: 10.1093/nar/gks177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baker MD, Neiditch MB. 2011. Structural basis of response regulator inhibition by a bacterial anti-activator protein. PLoS Biol 9:e1001226. doi: 10.1371/journal.pbio.1001226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bador J, Amoureux L, Blanc E, Neuwirth C. 2013. Innate aminoglycoside resistance of Achromobacter xylosoxidans is due to AxyXY-OprZ, an RND-type multidrug efflux pump. Antimicrob Agents Chemother 57:603–605. doi: 10.1128/AAC.01243-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adewoye L, Topp E, Li X-Z. 2016. Antimicrobial drug efflux genes and pumps in bacteria of animal and environmental origin, p 561–593. In Li X-Z, Elkins CA, Zgurskaya HI (ed), Efflux-mediated antimicrobial resistance in bacteria: mechanisms, regulation, and clinical implications. Adis, Cham, Switzerland. [Google Scholar]
- 59.Scoffone VC, Coenye T, Riccardi G, Buroni S. 2016. Antimicrobial drug efflux pumps in Burkholderia, p 417–438. In Li X-Z, Elkins CA, Zgurskaya HI (ed), Efflux-mediated antimicrobial resistance in bacteria: mechanisms, regulation, and clinical implications. Adis, Cham, Switzerland. [Google Scholar]
- 60.Visalli MA, Murphy E, Projan SJ, Bradford PA. 2003. AcrAB multidrug efflux pump is associated with reduced levels of susceptibility to tigecycline (GAR-936) in Proteus mirabilis. Antimicrob Agents Chemother 47:665–669. doi: 10.1128/aac.47.2.665-669.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 62.El-Sayed AK, Hothersall J, Thomas CM. 2001. Quorum-sensing-dependent regulation of biosynthesis of the polyketide antibiotic mupirocin in Pseudomonas fluorescens NCIMB 10586. Microbiology 147:2127–2139. doi: 10.1099/00221287-147-8-2127. [DOI] [PubMed] [Google Scholar]
- 63.Lasocki K, Bartosik AA, Mierzejewska J, Thomas CM, Jagura-Burdzy G. 2007. Deletion of the parA (soj) homologue in Pseudomonas aeruginosa causes ParB instability and affects growth rate, chromosome segregation, and motility. J Bacteriol 189:5762–5772. doi: 10.1128/JB.00371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 65.Kawalek A, Glabski K, Bartosik AA, Fogtman A, Jagura-Burdzy G. 2017. Increased ParB level affects expression of stress response, adaptation and virulence operons and potentiates repression of promoters adjacent to the high affinity binding sites parS3 and parS4 in Pseudomonas aeruginosa. PLoS One 12:e0181726. doi: 10.1371/journal.pone.0181726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 67.Papadopoulos JS, Agarwala R. 2007. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 68.Zukowski MM, Gaffney DF, Speck D, Kauffmann M, Findeli A, Wisecup A, Lecocq JP. 1983. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc Natl Acad Sci U S A 80:1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 70.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing. Supplement M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 71.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 72.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 73.Kawalek A, Kotecka K, Modrzejewska M, Jagura-Burdzy G, Bartosik AA. 2019. Genome sequence of Pseudomonas aeruginosa PAO1161, a PAO1 derivative with the ICEPae1161 integrative and conjugative element. bioRxiv doi: 10.1101/494302. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.