Lethal mutagenesis is an antiviral approach that consists of extinguishing a virus by an excess of mutations acquired during replication in the presence of a mutagenic agent, often a nucleotide analogue. One of its advantages is its broad-spectrum nature, which renders the strategy potentially effective against emergent RNA viral infections. Here we describe the synergistic lethal mutagenesis of hepatitis C virus (HCV) by a combination of favipiravir (T-705) and ribavirin.
KEYWORDS: favipiravir, ribavirin, viral quasispecies, antiviral therapy
ABSTRACT
Lethal mutagenesis is an antiviral approach that consists of extinguishing a virus by an excess of mutations acquired during replication in the presence of a mutagenic agent, often a nucleotide analogue. One of its advantages is its broad-spectrum nature, which renders the strategy potentially effective against emergent RNA viral infections. Here we describe the synergistic lethal mutagenesis of hepatitis C virus (HCV) by a combination of favipiravir (T-705) and ribavirin. Synergy has been documented over a broad range of analogue concentrations using the Chou-Talalay method implemented in CompuSyn graphics software, with the average dose reduction index (DRI) being above 1 (68.02 ± 101.6 for favipiravir and 5.83 ± 6.07 for ribavirin) and the average combination indices (CI) being below 1 (0.52 ± 0.28). Furthermore, analogue concentrations that individually did not extinguish high-fitness HCV in 10 serial infections extinguished high-fitness HCV in 1 to 2 passages when used in combination. Although both analogues displayed a preference for G → A and C → U transitions, deep sequencing analysis of mutant spectra indicated a different preference of the two analogues for the mutation sites, thus unveiling a new possible synergy mechanism in lethal mutagenesis. The prospects for synergy among mutagenic nucleotides as a strategy to confront emerging viral infections are discussed.
INTRODUCTION
Lethal mutagenesis is an antiviral approach consisting of the achievement of viral extinction by an excess of mutations, an outcome supported by theoretical and experimental studies (1–10). Cell culture and in vivo infection experiments have documented the extinction of RNA viruses by base and nucleoside analogues (converted intracellularly into their active nucleotides), notably, favipiravir (T-705; 6-fluoro-3-hydroxy-2-pyrazinecarboxamide), favipiravir derivatives, and ribavirin (1-β-d-ribofuranosyl-1-H-1,2,4-triazole-3-carboxamide). Both purine analogues have been licensed for the treatment of some human viral infections, and they can act as lethal mutagens for some RNA viruses (reviewed in reference 10).
We are interested in exploring broad-spectrum antiviral treatments based on lethal mutagenesis using hepatitis C virus (HCV) replication in human hepatoma cells as a model system. HCV infections have an important public health impact, and the virus is a representative of the Flaviviridae family of human pathogens. Despite 95% sustained viral response rates with direct-acting antiviral agents (DAAs) against HCV, there is a trend toward the increased circulation of DAA-resistant, natural occurring HCV variants (11–13). Such a circulation is unfolding in parallel with continuing genotype and subtype HCV diversification (14). In addition, recent evidence suggests epigenetic-mediated hepatic pathological sequels once the virus is eliminated by DAAs, including hepatocellular carcinoma recurrence (15–19). If treatment escape mutants become epidemiologically dominant and the observations of pathological sequels following DAA-mediated virus clearance are corroborated, new treatments for HCV will be needed.
Ribavirin, used in combination with pegylated interferon alpha (IFN-α), was the standard anti-HCV therapy a decade ago, and ribavirin is still included in some DAA formulations (20). There is genetic and clinical evidence that lethal mutagenesis may be part of the anti-HCV mechanism of ribavirin (21–24). Regarding favipiravir and derivatives, Furuta and colleagues documented potent inhibitory activity against RNA viruses, notably, influenza virus (25–29). Picornaviruses, alphaviruses, flaviviruses, rhabdoviruses, orthomyxoviruses, paramyxoviruses, arenaviruses, hantaviruses, and bunyaviruses are inhibited by members of this pyrazinecarboxamide family of molecules (27, 30–48), thus rendering these as drug candidates to confront emerging viral infections (49, 50).
The participation of lethal mutagenesis in the antiviral activity of favipiravir and derivatives has been suggested for some virus-host systems by the increase of the mutant spectrum complexity when the virus was on its way toward extinction (51–60). A few studies have examined synergistic effects between nucleotide analogues or between an analogue and a standard, nonmutagenic inhibitor. Smee and colleagues demonstrated synergism between favipiravir and oseltamivir against influenza virus infections in mice (43), thus expanding the value of favipiravir as an antiviral agent (50). Favipiravir and ribavirin exerted a synergistic activity against Rift Valley fever virus and viral hemorrhagic fever viruses in animal models (46, 61, 62). Synergism between favipiravir and ribavirin may result from their independent mechanisms of activity (10, 63–66), and a role of lethal mutagenesis in the reinforcement of their effectiveness has not been established.
Our previous work documented the participation of lethal mutagenesis in the antiviral activity of favipiravir (53) and ribavirin (24) when present individually during HCV replication in human hepatoma cells. Here we show that favipiravir and ribavirin exert a synergistic activity against HCV in human hepatoma cells, including the extinction of high-fitness virus which is resistant to the analogues administered individually. Interestingly, despite the two analogues evoking a similar bias in favor of G → A and C → U transitions during lethal mutagenesis of HCV (24, 53), deep sequencing showed that the preferred mutation sites of the two analogues are not identical, therefore revealing a new potential synergism mechanism among mutagenic nucleotides.
RESULTS
Synergism of favipiravir and ribavirin against hepatitis C virus.
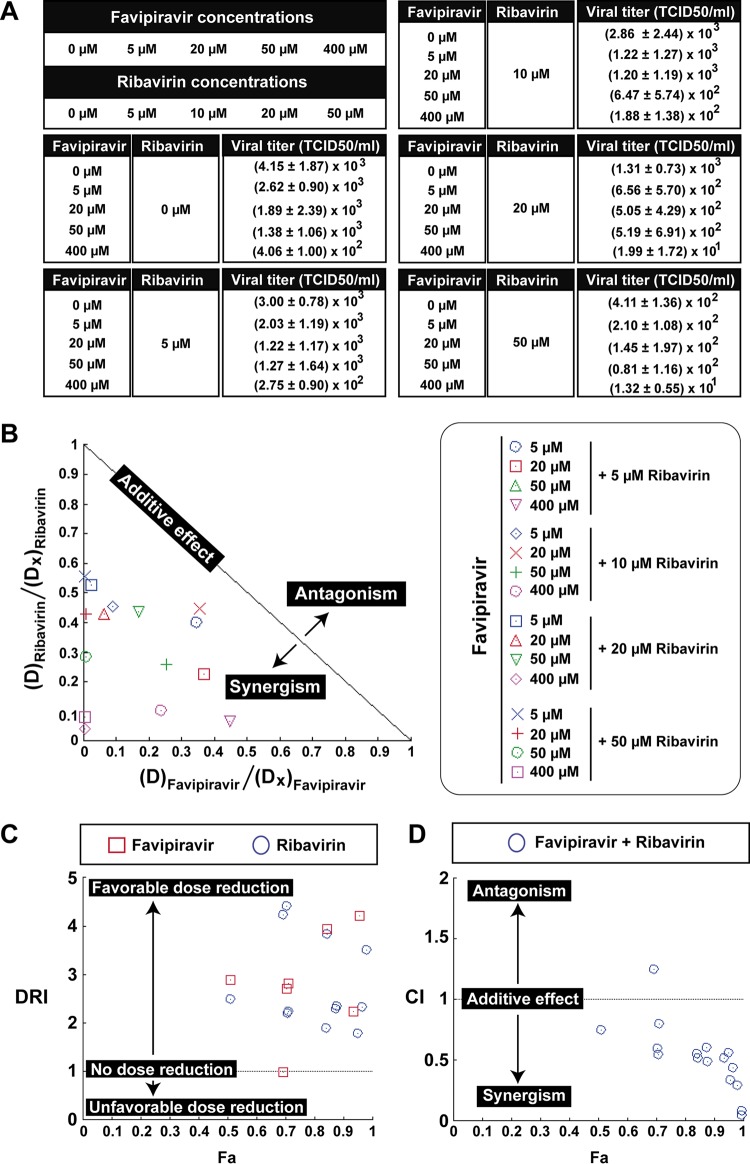
The inhibition of HCV infectious progeny production in single infections of Huh-7.5 cells was measured using a concentration range of 0 to 400 μM favipiravir (the maximum concentration is 0.46-fold the 50% cytotoxic concentration [CC50] value and 54.0-fold the 50% inhibitory concentration [IC50] value [53]) and 0 to 50 μM ribavirin (the maximum concentration is 0.46-fold the CC50 value and 5.9-fold the IC50 value [24]). The virus tested was the parental, low-fitness population of HCV at passage 0 (HCV p0) (67), derived from transcription of plasmid Jc1FLAG2(p7-nsGluc2A) (genotype 2a) (68). The analogues were present either individually or in combination during infection, and infectious progeny production was analyzed using CompuSyn software (69–71). The results (Fig. 1) indicated synergism, according to the normalized isobologram (Fig. 1B); a favorable dose reduction, based on an average dose reduction index (DRI) above 1 (68.02 ± 101.6 for favipiravir and 5.83 ± 6.07 for ribavirin, which are the average DRIs of 16 different concentration combinations of the two drugs; Fig. 1C and Table 1); and an average combination index (CI) below 1 (0.52 ± 0.28, which is the average CI of 16 different drug concentration combinations; Fig. 1D and Table 1). The values of all parameters are diagnostic of synergism between favipiravir and ribavirin acting on HCV p0.
FIG 1.
Synergistic activity of favipiravir and ribavirin against hepatitis C virus. (A) Infectious progeny production of HCV p0 upon infection of Huh-7.5 reporter cells. Values and standard deviations (from triplicate determinations) are given. (B) Dose-normalized isobologram for the nonconstant ratio combinations. The combination data points that fall on the hypotenuse indicate an additive effect, those that fall on the lower left indicate synergism, and those that fall on the upper right indicate antagonism. The program excluded the representation of the point corresponding to 50 μM favipiravir and 5 μM ribavirin. D, dose; Dx, dose of a single drug that exhibits a given percent inhibition. (C) Dose reduction index (DRI) for each drug for a given effect (Fa). DRI of >1, 1, and <1 indicate a favorable dose reduction, no dose reduction, and a negative dose reduction, respectively. Values larger than 5 are not represented in the plot. (D) Combination index (CI) values as a function of the effect levels (Fa), where CI values of <1, 1, and >1 indicate synergism, an additive effect, and antagonism, respectively. Panels B to D were obtained by analysis with CompuSyn software. The procedures are detailed in Materials and Methods.
TABLE 1.
Parameters obtained with the software CompuSyn to describe the synergistic effect of favipiravir and ribavirina
| Ribavirin + favipiravir doseb | Effect (Fa)c | Dosed
(μM) |
DRIe
|
CIf | ||
|---|---|---|---|---|---|---|
| Ribavirin | Favipiravir | Ribavirin | Favipiravir | |||
| 5 + 5 | 0.5101 | 12.4563 | 14.4863 | 2.49126 | 2.89727 | 0.74656 |
| 5 + 20 | 0.7059 | 22.0253 | 54.2846 | 4.40506 | 2.71423 | 0.59544 |
| 5 + 50 | 0.6935 | 21.1558 | 49.4461 | 4.23115 | 0.98892 | 1.24755 |
| 5 + 400 | 0.9338 | 73.7638 | 893.946 | 14.7528 | 2.23486 | 0.51524 |
| 10 + 5 | 0.7065 | 22.0688 | 54.5335 | 2.20688 | 10.9067 | 0.54481 |
| 10 + 20 | 0.7109 | 22.3922 | 56.4033 | 2.23922 | 2.82017 | 0.80117 |
| 10 + 50 | 0.8441 | 38.3742 | 196.573 | 3.83742 | 3.93147 | 0.51495 |
| 10 + 400 | 0.9547 | 97.0170 | 1687.07 | 9.70170 | 4.21768 | 0.34017 |
| 20 + 5 | 0.8421 | 37.9802 | 191.928 | 1.89901 | 38.3856 | 0.55264 |
| 20 + 20 | 0.8785 | 46.7487 | 310.618 | 2.33743 | 15.5309 | 0.49221 |
| 20 + 50 | 0.8751 | 45.7551 | 295.531 | 2.28776 | 5.91061 | 0.60630 |
| 20 + 400 | 0.9952 | 461.821 | 62,762.4 | 23.0911 | 156.906 | 0.04968 |
| 50 + 5 | 0.9494 | 89.6197 | 1,403.78 | 1.79239 | 280.757 | 0.56147 |
| 50 + 20 | 0.9650 | 116.543 | 2,580.56 | 2.33086 | 129.028 | 0.43678 |
| 50 + 50 | 0.9804 | 174.998 | 6,620.73 | 3.49996 | 132.415 | 0.29327 |
| 50 + 400 | 0.9968 | 609.716 | 119,493.0 | 12.1943 | 298.733 | 0.08535 |
The viral populations analyzed are described in Fig. 3.
Dose (concentration) of drug (in micromolar).
Fa, the fraction of the population that is affected by the dose.
The dose of a single drug that exhibits a given percent inhibition.
DRI, dose reduction index, calculated by the equation of Chou: DRI = Dx/D, where Dx is the dose of a single drug that exhibits a given percent inhibition, and D is the dose (69).
CI, combination index, which is calculated with the formula CI = D1/Dx1 + D2/Dx2, where D1 and D2 are the doses of ribavirin and favipiravir, respectively, and Dx1 and Dx2 are the doses of ribavirin and favipiravir that exhibit a given percent inhibition, respectively.
Effective extinction of high-fitness hepatitis C virus by favipiravir-ribavirin combinations.
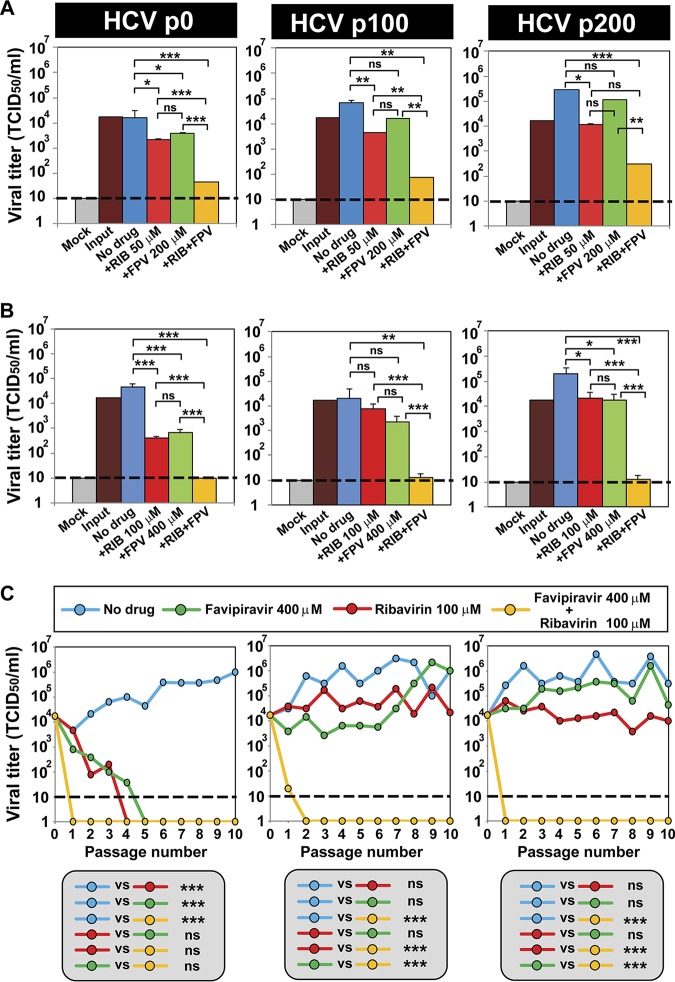
Two hundred serial passages of HCV p0 in Huh-7.5 cells resulted in population HCV p200, which displayed a 2- to 3-fold increase in replicative fitness, as calculated from progeny production in single and serial infections, as well as from growth competition experiments (72, 73). The high-fitness intermediate-passage HCV p100 and HCV p200 displayed a lower sensitivity to the anti-HCV agents than their parental virus, HCV p0, including to favipiravir and ribavirin (72, 74, 75), thus providing HCV populations for a stringent evaluation of synergistic activities. The infectious progeny production upon single infections of Huh-7.5 cells by HCV p0, HCV p100, and HCV p200 was 10- to 100-fold lower with favipiravir-ribavirin combinations than with the individual analogues (Fig. 2A and B). In serial infections in the presence of the drugs, HCV p100 and HCV p200 displayed sustained resistance to favipiravir (at a concentration 54.0-fold its IC50 value for HCV p0) and ribavirin (at a concentration 11.9-fold its IC50 value for HCV p0); in contrast, the analogue combination extinguished all HCV populations in one to two passages independently of their fitness (Fig. 2C). To ascertain that the decrease in viral replication correlates with the extinction of the HCV p0, HCV p100, and HCV p200 populations, we performed three blind passages in the absence of any drug starting at passage 10 for HCV p0 (with favipiravir, ribavirin, and the combination) and at passage 10 for HCV p100 and HCV p200 (with the combination). In all cases, at blind passage 3, no infectivity and no extracellular or intracellular viral RNA (using a highly sensitive reverse transcription-PCR [RT-PCR] protocol) was detected (data not shown). Thus, favipiravir-ribavirin combinations are effective in extinguishing low- and high-fitness HCV populations.
FIG 2.
Extinction of high-fitness hepatitis C virus by combinations of favipiravir and ribavirin. (A, B) Viral titers obtained after a single infection of HCV p0, HCV p100, or HCV p200 (the virus is indicated at the top) in the absence or presence of the drug concentrations given on the abscissae (Mock, mock infected; RIB, ribavirin; FPV, favipiravir). In each panel, +RIB and +FPV mean that the concentrations used in the combination were the same concentrations of the drugs used individually. Titrations were carried out in triplicate, and the statistical significance of the differences was determined by ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. (C) The response of HCV p0, HCV p100, and HCV p200 subjected to 10 serial infections in the absence or the presence of the drug concentrations indicated in the key. The statistical significance of the differences was determined by two-way ANOVA. ***, P < 0.001; ns, not significant. The origins of the viruses and further experimental details are described in Materials and Methods.
Mutation site preferences.
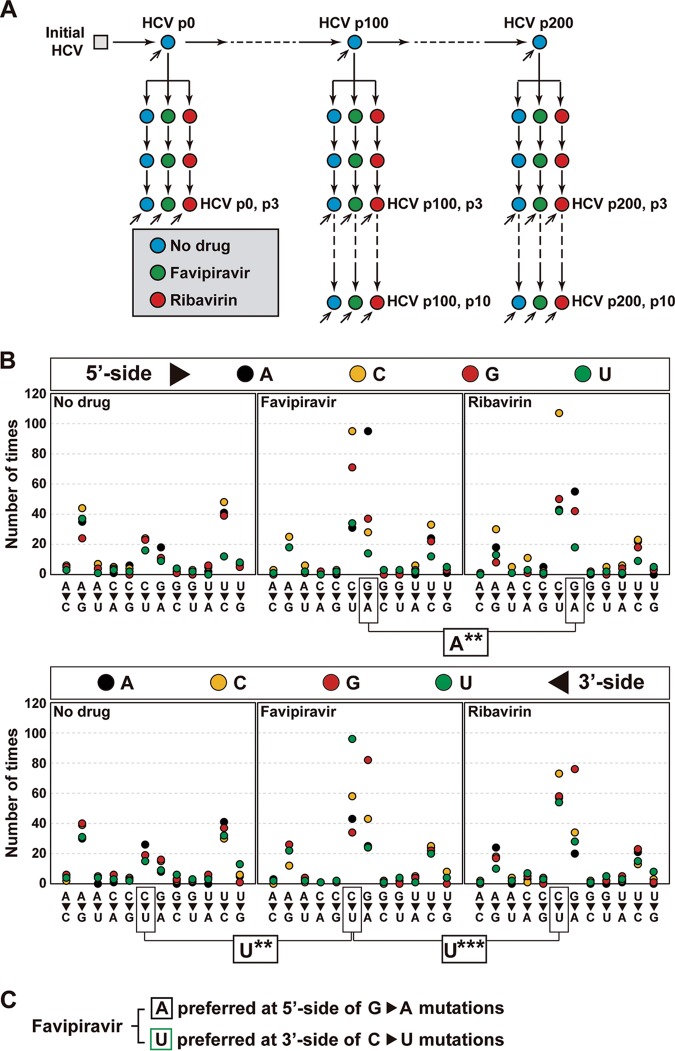
NS5B RNA from several HCV p0, HCV p100, and HCV p200 populations passaged in the absence or presence of favipiravir or ribavirin was analyzed by Illumina MiSeq deep sequencing, and for each mutant spectrum, the nucleotide types present at the 5′ and 3′ end sides of the mutation sites were compared using as a reference the consensus sequence of the corresponding population. Read cleaning and data processing were as previously described (76, 77). The incidence-based context at the 5′ side and the 3′ side of each mutated position was subjected to two statistical evaluations. The data are based on eight HCV populations passaged in the absence of drug, five populations passaged in the presence of favipiravir, and five populations passaged in the presence of ribavirin (Fig. 3A). For each sample (population), the different haplotypes were aligned without considering the haplotype abundance or the number of haplotypes in which a given mutation was present. Then, using all alignments for populations passaged under the same conditions (either in the absence of drug or in the presence of favipiravir or ribavirin), the distribution of nucleotides adjacent to each mutation site was determined.
FIG 3.
Influence of the immediate neighbors of each mutated site on mutant frequency. (A) Scheme of HCV passages in the absence (No drug) and the presence of favipiravir or ribavirin. Inclined arrows indicate the viral population analyzed by MiSeq Illumina deep sequencing of three amplicons of the NS5B-coding region. (B) Effect of the nucleotide type present at the 5′ and 3′ sides of the mutated residue. The mutation types are given on the abscissa, and the number of times that a given neighbor is present is given on the ordinate (the color code for the nucleotide is in the box at the top). Statistical significances (determined by the proportion test) are given below each panel group, with boxes linking the relevant comparisons. **, P < 0.01; ***, P < 0.001. Deep sequencing, data processing, and statistical procedures are described in Materials and Methods. (C) Summary of statistically significant preferred neighbor nucleotides for mutation sites.
Fisher’s test was used to test the null hypothesis of the independence of the presence of drug on the residues that flanked each mutation site (see Table S1 posted at http://babia.cbm.uam.es/~lab121/SupplMatGallego2). Regarding the nucleotide distribution at the 5′ side of any mutation type, no significant difference was observed in the comparison between the absence of drug and the presence of either favipiravir (P = 0.384) or ribavirin (P = 0.105). The corresponding P values by Fisher’s test for the nucleotide frequencies at the 3′ side of any mutation site were 0.391 and 0.516. No significant difference was noted either for the 5′- and 3′-side position in a direct comparison between samples passaged in the presence of favipiravir and ribavirin (P = 0.0712 and 0.137, respectively) (see Table S1 posted at http://babia.cbm.uam.es/~lab121/SupplMatGallego2).
When only transition mutations were considered, some significant differences were found. Specifically, the nucleotide type distribution at the 5′ side of the G → A transitions evoked by favipiravir differed from that evoked by ribavirin (P = 0.00362) (see Table S2 posted at http://babia.cbm.uam.es/~lab121/SupplMatGallego2). A difference was also quantified for the nucleotide distributions at the 3′ side of the C → U transitions generated by the two analogues (P = 7.96 × 10−5) (see Table S2 posted at http://babia.cbm.uam.es/~lab121/SupplMatGallego2) and also in the comparison between populations passaged in the absence of drug and the presence of favipiravir (P = 5.72 × 10−4) (see Table S3 posted at http://babia.cbm.uam.es/~lab121/SupplMatGallego2). A neighbor residue bias was not observed for any other transition type or any transversion (see Tables S2, S3, and S4 posted at http://babia.cbm.uam.es/~lab121/SupplMatGallego2), although the overall frequency of transitions was 6.24-fold higher than that of transversions, weakening the detection of possible differences in the distribution of transversion mutations.
Once the differences in the residues adjacent to mutation sites had been identified, the responsible nucleotide types were determined using the proportion test, with P value correction being performed using Bonferroni’s test (Fig. 3 and Table 2). For the comparison between favipiravir and ribavirin treatment, the proportion test indicated that A and C are preferential at the 5′ side of the G → A transitions in the presence of favipiravir, with only the preference for A reaching significance after P value correction. Likewise, G and U were observed to be dominant at the 3′ side of the C → U transitions evoked by favipiravir, with only the preference for U reaching significance after P value correction. In the comparison between populations passaged in the absence of any drug and the presence of favipiravir, U was significantly dominant at the 3′ side of the C → U transitions. Thus, the results (Fig. 3 and Table 2) indicate that favipiravir and ribavirin do not display an identical choice of mutation sites in the HCV NS5B-coding region, and such a difference may contribute to their synergism.
TABLE 2.
Statistical analysis of the difference between the nucleotides present on the 5′ or 3′ side of the indicated mutation type in HCV populations passaged in the absence and presence of favipiravir and ribavirina
| Mutation type | Side of immediate neighbors | Nucleotideb | Comparison | No.c |
P value |
Significanced | |
|---|---|---|---|---|---|---|---|
| Proportion test | Bonferroni correction | ||||||
| G → A | 5′ | A | Favipiravir | 95 | 0.00054 | 0.00216 | ** |
| Ribavirin | 55 | ||||||
| C | Favipiravir | 28 | 0.02530 | 0.10100 | NS | ||
| Ribavirin | 42 | ||||||
| G | Favipiravir | 37 | 0.29800 | 1.00000 | NS | ||
| Ribavirin | 42 | ||||||
| U | Favipiravir | 14 | 0.38700 | 1.00000 | NS | ||
| Ribavirin | 18 | ||||||
| C → U | 3′ | A | Favipiravir | 43 | 0.229000 | 0.917 | NS |
| Ribavirin | 57 | ||||||
| C | Favipiravir | 58 | 0.260000 | 1.00000 | NS | ||
| Ribavirin | 73 | ||||||
| G | Favipiravir | 34 | 0.015400 | 0.0614 | NS | ||
| Ribavirin | 58 | ||||||
| U | Favipiravir | 96 | 0.000011 | 0.0000439 | *** | ||
| Ribavirin | 54 | ||||||
| C → U | 3′ | A | Favipiravir | 43 | 0.013100 | 0.05250 | NS |
| No drug | 26 | ||||||
| C | Favipiravir | 58 | 0.971000 | 1.00000 | NS | ||
| No drug | 19 | ||||||
| G | Favipiravir | 34 | 0.083900 | 0.33500 | NS | ||
| No drug | 19 | ||||||
| U | Favipiravir | 96 | 0.000509 | 0.00203 | ** | ||
| No drug | 15 | ||||||
The viral populations analyzed are described in Fig. 3.
The nucleotide which is the 5′ or 3′ neighbor (indicated in the second column) of the mutation type given in the first column.
Number of times that the nucleotide is a neighbor to a mutation, without considering the haplotype abundance or the number of haplotypes in which a given mutation was present.
The statistical significance of the differences is given as follows: NS, not significant; **, P < 0.01; ***, P < 0.001.
DISCUSSION
Synergism permits a decrease in drug dosage and side effects while enhancing the therapeutic effects, thereby reducing the probability of selection of drug-resistant mutants (69). The search for synergistic antiviral combinations is particularly important for highly variable viruses whose adaptability is guided by quasispecies dynamics (78). Synergism is favored when the relevant drugs are directed to independent viral or cellular targets or act by different mechanisms on the same target (69). In the case of favipiravir and ribavirin, synergism may be prompted by two relevant differences that distinguish the two drugs: (i) the multiple and nonidentical antiviral mechanisms displayed by the two analogues and (ii) their different preferences for some mutation sites, as revealed in the present study. Concerning the first difference, favipiravir may act as a mutagenic agent and viral RNA chain terminator (63, 65); ribavirin may exert immunomodulatory activities and cause the depletion of intracellular GTP, the inhibition of mRNA cap formation, or the inhibition of viral polymerases, in addition to lethal mutagenesis (reviewed in references 64, 79, and 80). Concerning the second difference, the preference for different mutation sites, revealed by deep sequencing, even if operative for only a subset of preferred mutation types, should confer an advantage when the two mutagens act conjointly relative to the equivalent mutagenic activity relying on only one of the compounds. We have no evidence that the preferred mutation sites correspond to hot spots. The possibility that additional differences in mutational preferences might be revealed with larger sample sizes of the genome populations under comparison cannot be excluded. Additional spectrum analyses are necessary to further quantify mutation repertoire differences. Given the multiple mechanistic differences between the two analogues, it is not possible to evaluate the contribution of differences in mutation site preferences to the synergistic action.
Our study has benefited from the availability of monophyletic (descendant from the same initial genome) HCV populations that differ in fitness and the prior evidence that fitness is a determinant of drug resistance in HCV (reviewed in reference 10). As a significant comparison, combined doses of favipiravir and ribavirin at levels 54-fold and 11.9-fold their IC50 values, respectively, extinguished HCV p0 in one passage and HCV p100 in two passages, while sofosbuvir used at a concentration 60-fold its IC50 value required two passages to extinguish HCV p0 and six passages to extinguish HCV p100 under the same experimental conditions (compare the data in Fig. 2 with those in reference 75). Since nucleotide analogues often differ in mutation preferences (10), synergisms among this class of compounds are expected.
Synergistic interactions among drugs are particularly important when the objective is suppression of pathogen replication to prevent the selection of treatment-resistant escape mutants. This is an objective for any pathogenic entity, be they genetically variable and heterogeneous DNA and RNA viruses, protozoa, or cancer cells (71, 81–83). A previous case of mutation type-driven antiviral reinforcement involved APOBEC3G (A3G; a human deaminase naturally expressed in cells) and 5-azacytidine (5-AZC). A3G is mutagenic for HIV-1 and preferentially induces G → A mutations in plus-strand DNA through C deaminations in the minus-strand DNA (84); in turn, 5-AZC is also mutagenic for HIV-1 but has a preference for G → C transversions (85). Exposure of replicating HIV-1 to A3G and 5-AZC increased the frequency of G → A mutations relative to that with exposure to A3G alone, and this enhancement was accompanied by an even stronger reduction in the number of G → C transversions induced by 5-AZC alone (86). In this case, the two mutagenic activities potentiated the antiviral activity of each other by a range of 3- to 6-fold over the concentration range tested.
The potential of synergistic lethal mutagenesis is reinforced by several proof-of-principle experiments and clinical assays that have established the feasibility of the lethal mutagenesis approach to treat viral infections in vivo (52, 57, 87–89). Synergistic lethal mutagenesis offers the prospect of the broad-spectrum treatment of infections caused by newly arising RNA viral pathogens and a rescue treatment for established viral diseases when the circulation of inhibitor-resistant mutants acquires epidemiological relevance.
MATERIALS AND METHODS
Cells, viruses, and infections.
Huh-7.5 cells and Huh-7.5 reporter cells were grown in Dulbecco’s modification of Eagle’s medium (DMEM) at 37°C in 5% CO2 as previously described (74, 90, 91). Huh-7.5 reporter cells were used for all infections in the absence and the presence of drugs, while Huh-7.5 cells were used for titration of infectivity. Titration of HCV infectivity was performed by applying serial viral dilutions of the sample to be tested on Huh-7.5 cells that had been seeded 16 h earlier on 96-well plates at 6,400 cells/well. At 3 days postinfection, the monolayers were washed with phosphate-buffered saline (PBS), fixed with cold methanol, and stained using anti-NS5A monoclonal antibody 9E10 (92). Virus titers (expressed as the 50% tissue culture infective dose [TCID50] per milliliter) were calculated as previously described (67, 74).
The viruses used were HCV p0, a preparation derived by transcription from plasmid Jc1FLAG2(p7-nsGluc2A) (genotype 2a) (68) and then expanded into a working stock as previously described (67). HCV p100 and HCV p200 are the populations that resulted from subjecting HCV p0 to 100 and 200 serial passages in Huh-7.5 reporter cells, respectively (67, 73). Controls involving mock-infected cells and cells infected with replication-defective mutant HCV GNN [GNNFLAG2(p7-nsGluc2A)] (68) were included as previously described (67, 74).
For infections in the presence of favipiravir, ribavirin, or their combinations, the drugs were prepared and used as detailed previously (74). In brief, filter-sterilized stocks of favipiravir (20 mM in water; Atomax Chemicals Co. Ltd.) and of ribavirin (100 mM in PBS; Sigma) were stored at −70°C and diluted in DMEM prior to use to reach the desired concentration. Huh-7.5 reporter cells (4 × 105) were pretreated with the drugs (or DMEM without drug) for 16 h prior to infection, and then they were infected at a multiplicity of infection (MOI) of 0.03 TCID50/cell with a virus adsorption time of 5 h. The infection was continued in the absence or the presence of the drugs for 72 to 96 h. Serial passages in the absence or the presence of the drugs were performed in parallel by infecting 4 × 105 Huh-7.5 reporter cells with the virus contained in 0.5 ml of cell culture supernatant from the previous infection. This yielded a range of MOI of from 4.6 × 10−5 to 6 TCID50/cell, and the value in each infection can be calculated from the data given for each experiment. HCV was considered extinct when no infectivity or material amplifiable by RT-PCR could be detected in the cell culture or upon blind passages in HuH-7.5 reporter cells in the absence of any drug (53).
RNA extraction, cDNA amplification, and deep sequencing.
Total extracellular or intracellular viral RNA was extracted from infected or mock-infected cells using a QIAamp viral RNA kit and a Qiagen RNeasy kit (Qiagen, Valencia, CA, USA), respectively, according to the manufacturer’s instructions. RT-PCR amplification of HCV RNA for deep sequencing was performed using an AccuScript kit (Agilent Technologies) and primers specific for the NS5B-coding region (see Table S5 posted at http://babia.cbm.uam.es/~lab121/SupplMatGallego2). The amplified DNA products were analyzed by agarose gel electrophoresis with a Gene Ruler 1-kb Plus DNA ladder (Thermo Scientific) as a molar mass standard. For Illumina deep sequencing, PCR products were purified (QIAquick gel extraction kit; Qiagen), quantified (Qubit double-stranded DNA assay kit), and analyzed for quality (BioAnalyzer DNA 1000 LabChip) as previously described (77). The three amplicons used for the deep sequencing analyses covered the following NS5B genomic regions: A1, residues 7626 to 7962; A2, residues 7941 to 8257; and A3, residues 8229 to 8653. Controls without template RNA were included in parallel to ascertain the absence of contamination by template nucleic acids.
fastq data treatment.
The fastq files obtained from MiSeq deep sequencing were subjected to a data analysis pipeline (77, 93, 94) that was adapted to the Illumina MiSeq platform in a paired-end 2 × 300-bp mode. It involved the following main steps: (i) quality control evaluation, performed by inspecting the profiles of per site quality, read length, and general instrument parameters of quality; (ii) in paired-end experiments, determination of the overlap paired reads obtained with the FLASH tool (95), with a minimum of 20 bp of overlap with a maximum of 10% mismatches; (iii) determination of the quality profiles of the FLASH reads; (iv) demultiplexing of the reads by identifying the oligonucleotides within windows of expected positions in the sequenced reads; (v) haplotype alignment in each fasta file to the wild-type reference sequence or the master sequence in the file (the most abundant haplotype) and quality filter, with exclusion from the analysis of haplotypes not covering the full amplicon or with two indeterminations, three gaps, or differences of more than 30% with respect to the reference sequence; and (vi) the intersection of haplotypes in both strands with a minimum abundance of 0.1%, excluding haplotypes unique to one strand. The minimum coverage was 40,000 reads per amplicon, with the median coverage being 139,200 reads (interquartile range, 71,480 to 210,600 reads). The procedures for read cleaning and to determine reliable mutant detection (set at 0.2%) and the origin of the pipeline components were previously described (77, 96).
Computational and statistical analyses.
Synergism between favipiravir and ribavirin was tested using CompuSyn software (97, 98). To determine the statistical significance of differences in infectivity levels, one-way and two-way analyses of variance (ANOVA) were carried out using Prism (version 6) software (GraphPad). Fisher’s test was applied to detect differences in neighbor site-related mutational preferences in mutant spectra. The proportion test was used to identify the nucleotide residues responsible for the differences in mutational preferences. The Bonferroni correction was implemented for multiple determinations.
ACKNOWLEDGMENTS
C.P. is supported by the Miguel Servet program of the Instituto de Salud Carlos III (grant CP14/00121), cofinanced by the European Regional Development Fund (ERDF). The Centro de Investigación en Red de Enfermedades Hepáticas y Digestivas (CIBERehd) is funded by the Instituto de Salud Carlos III. The work in Madrid, Spain, was supported by grant SAF2014-52400-R from the Ministerio de Economía y Competitividad, grants SAF2017-87846-R, BFU2017-91384-EXP, and PI18/00210 from the Ministerio de Ciencia, Innovación y Universidades, grant S2013/ABI-2906 from PLATESA from the Comunidad de Madrid/FEDER, and grant P2018/BAA-4370 from PLATESA2 from the Comunidad de Madrid/FEDER. The work in Barcelona, Spain, was also funded by the Instituto de Salud Carlos III; by grant PI16/00337, cofinanced by the European Regional Development Fund (ERDF); and by the Centro para el Desarrollo Tecnológico Industrial (CDTI), Spanish Ministry of Economics and Competitiveness (MINECO) (grant IDI-20151125). Institutional grants from the Fundación Ramón Areces and Banco Santander to CBMSO are also acknowledged.
REFERENCES
- 1.Eigen M, Schuster P. 1979. The hypercycle. A principle of natural self-organization. Springer, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 2.Lynch M, Gabriel W. 1990. Mutation load and the survival of small populations. Evolution 44:1725–1737. doi: 10.1111/j.1558-5646.1990.tb05244.x. [DOI] [PubMed] [Google Scholar]
- 3.Holland JJ. 1990. Defective viral genomes, p 151–165. In Fields BM, Knipe DM, Chanock RM, Hirsch MS, Melnick JL, Monath TP, Roizman B (ed), Fields virology, 2nd ed Raven Press, New York, NY. [Google Scholar]
- 4.Loeb LA, Essigmann JM, Kazazi F, Zhang J, Rose KD, Mullins JI. 1999. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc Natl Acad Sci U S A 96:1492–1497. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crotty S, Maag D, Arnold JJ, Zhong W, Lau JYN, Hong Z, Andino R, Cameron CE. 2000. The broad-spectrum antiviral ribonucleotide, ribavirin, is an RNA virus mutagen. Nat Med 6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 6.Graci JD, Cameron CE. 2008. Therapeutically targeting RNA viruses via lethal mutagenesis. Future Virol 3:553–566. doi: 10.2217/17460794.3.6.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dapp MJ, Patterson SE, Mansky LM. 2013. Back to the future: revisiting HIV-1 lethal mutagenesis. Trends Microbiol 21:56–62. doi: 10.1016/j.tim.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuster P. 2016. Quasispecies on fitness landscapes. Curr Top Microbiol Immunol 392:61–120. doi: 10.1007/82_2015_469. [DOI] [PubMed] [Google Scholar]
- 9.Tejero H, Montero F, Nuno JC. 2016. Theories of lethal mutagenesis: from error catastrophe to lethal defection. Curr Top Microbiol Immunol 392:161–179. doi: 10.1007/82_2015_463. [DOI] [PubMed] [Google Scholar]
- 10.Perales C, Gallego I, De Avila AI, Soria ME, Gregori J, Quer J, Domingo E. 2019. The increasing impact of lethal mutagenesis of viruses. Future Med Chem 11:1645–1657. doi: 10.4155/fmc-2018-0457. [DOI] [PubMed] [Google Scholar]
- 11.Peres-da-Silva A, Brandão-Mello CE, Lampe E. 2017. Prevalence of sofosbuvir resistance-associated variants in Brazilian and worldwide NS5B sequences of genotype-1 HCV. Antivir Ther 22:447–451. doi: 10.3851/IMP3131. [DOI] [PubMed] [Google Scholar]
- 12.Huang W, Wang M, Gong Q, Yu D, Chen P, Lin J, Han Y, Su Y, Qu L, Zhang X. 2019. Comparison of naturally occurring resistance-associated substitutions between 2008 and 2016 in Chinese patients with chronic hepatitis C virus infection. Microb Drug Resist 25:944–950. doi: 10.1089/mdr.2018.0360. [DOI] [PubMed] [Google Scholar]
- 13.Echeverria N, Betancour G, Gambaro F, Hernandez N, Lopez P, Chiodi D, Sanchez A, Boschi S, Fajardo A, Sonora M, Moratorio G, Cristina J, Moreno P. 2016. Naturally occurring NS3 resistance-associated variants in hepatitis C virus genotype 1: their relevance for developing countries. Virus Res 223:140–146. doi: 10.1016/j.virusres.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Hedskog C, Parhy B, Chang S, Zeuzem S, Moreno C, Shafran SD, Borgia SM, Asselah T, Alric L, Abergel A, Chen JJ, Collier J, Kapoor D, Hyland RH, Simmonds P, Mo H, Svarovskaia ES. 2019. Identification of 19 novel hepatitis C virus subtypes—further expanding HCV classification. Open Forum Infect Dis 6:ofz076. doi: 10.1093/ofid/ofz076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Ambrosio R, Della Corte C, Colombo M. 2015. Hepatocellular carcinoma in patients with a sustained response to anti-hepatitis C therapy. Int J Mol Sci 16:19698–19712. doi: 10.3390/ijms160819698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conti F, Buonfiglioli F, Scuteri A, Crespi C, Bolondi L, Caraceni P, Foschi FG, Lenzi M, Mazzella G, Verucchi G, Andreone P, Brillanti S. 2016. Early occurrence and recurrence of hepatocellular carcinoma in HCV-related cirrhosis treated with direct-acting antivirals. J Hepatol 65:727–733. doi: 10.1016/j.jhep.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 17.Perez S, Kaspi A, Domovitz T, Davidovich A, Lavi-Itzkovitz A, Meirson T, Alison Holmes J, Dai CY, Huang CF, Chung RT, Nimer A, El-Osta A, Yaari G, Stemmer SM, Yu ML, Haviv I, Gal-Tanamy M. 2019. Hepatitis C virus leaves an epigenetic signature post cure of infection by direct-acting antivirals. PLoS Genet 15:e1008181. doi: 10.1371/journal.pgen.1008181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamdane N, Juhling F, Crouchet E, El Saghire H, Thumann C, Oudot MA, Bandiera S, Saviano A, Ponsolles C, Roca Suarez AA, Li S, Fujiwara N, Ono A, Davidson I, Bardeesy N, Schmidl C, Bock C, Schuster C, Lupberger J, Habersetzer F, Doffoel M, Piardi T, Sommacale D, Imamura M, Uchida T, Ohdan H, Aikata H, Chayama K, Boldanova T, Pessaux P, Fuchs BC, Hoshida Y, Zeisel MB, Duong FHT, Baumert TF. 2019. HCV-induced epigenetic changes associated with liver cancer risk persist after sustained virologic response. Gastroenterology 156:2313–2329.e7. doi: 10.1053/j.gastro.2019.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reig M, Marino Z, Perello C, Inarrairaegui M, Ribeiro A, Lens S, Diaz A, Vilana R, Darnell A, Varela M, Sangro B, Calleja JL, Forns X, Bruix J. 2016. Unexpected high rate of early tumor recurrence in patients with HCV-related HCC undergoing interferon-free therapy. J Hepatol 65:719–726. doi: 10.1016/j.jhep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 20.European Association for the Study of the Liver. 2018. EASL recommendations on treatment of hepatitis C 2018. J Hepatol 69:461–511. doi: 10.1016/j.jhep.2018.03.026. [DOI] [PubMed] [Google Scholar]
- 21.Asahina Y, Izumi N, Enomoto N, Uchihara M, Kurosaki M, Onuki Y, Nishimura Y, Ueda K, Tsuchiya K, Nakanishi H, Kitamura T, Miyake S. 2005. Mutagenic effects of ribavirin and response to interferon/ribavirin combination therapy in chronic hepatitis C. J Hepatol 43:623–629. doi: 10.1016/j.jhep.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 22.Cuevas JM, González-Candelas F, Moya A, Sanjuán R. 2009. Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo. J Virol 83:5760–5764. doi: 10.1128/JVI.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dietz J, Schelhorn SE, Fitting D, Mihm U, Susser S, Welker MW, Fuller C, Daumer M, Teuber G, Wedemeyer H, Berg T, Lengauer T, Zeuzem S, Herrmann E, Sarrazin C. 2013. Deep sequencing reveals mutagenic effects of ribavirin during monotherapy of hepatitis C virus genotype 1-infected patients. J Virol 87:6172–6181. doi: 10.1128/JVI.02778-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ortega-Prieto AM, Sheldon J, Grande-Perez A, Tejero H, Gregori J, Quer J, Esteban JI, Domingo E, Perales C. 2013. Extinction of hepatitis C virus by ribavirin in hepatoma cells involves lethal mutagenesis. PLoS One 8:e71039. doi: 10.1371/journal.pone.0071039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furuta Y, Takahashi K, Fukuda Y, Kuno M, Kamiyama T, Kozaki K, Nomura N, Egawa H, Minami S, Watanabe Y, Narita H, Shiraki K. 2002. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob Agents Chemother 46:977–981. doi: 10.1128/aac.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuta Y, Takahashi K, Kuno-Maekawa M, Sangawa H, Uehara S, Kozaki K, Nomura N, Egawa H, Shiraki K. 2005. Mechanism of action of T-705 against influenza virus. Antimicrob Agents Chemother 49:981–986. doi: 10.1128/AAC.49.3.981-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furuta Y, Takahashi K, Shiraki K, Sakamoto K, Smee DF, Barnard DL, Gowen BB, Julander JG, Morrey JD. 2009. T-705 (favipiravir) and related compounds: novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res 82:95–102. doi: 10.1016/j.antiviral.2009.02.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furuta Y, Komeno T, Nakamura T. 2017. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B Phys Biol Sci 93:449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakamoto K, Ohashi S, Yamazoe R, Takahashi K, Furuta Y. 2006. The inhibition of FMD virus excretion from the infected pigs by an antiviral agent, T-1105. Appendix 64. Session of the Research Group of the Standing Technical Committee of the EU FMD Food and Agriculture Organization (FAO), Rome, Italy. [Google Scholar]
- 30.Furuta Y, Takahashi K, Maekawa M, Maegawa H, Egawa H, Terashima N. 2004. T-1106, a novel pyrazine nucleoside, hepatitis C virus polymerase inhibitor, abstr F-487, p 199–200. Abstr 44th Intersci Conf Antimicrob Agents Chemother. American Society for Microbiology, Washington, DC. [Google Scholar]
- 31.Gowen BB, Wong MH, Jung KH, Sanders AB, Mendenhall M, Bailey KW, Furuta Y, Sidwell RW. 2007. In vitro and in vivo activities of T-705 against arenavirus and bunyavirus infections. Antimicrob Agents Chemother 51:3168–3176. doi: 10.1128/AAC.00356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrey JD, Taro BS, Siddharthan V, Wang H, Smee DF, Christensen AJ, Furuta Y. 2008. Efficacy of orally administered T-705 pyrazine analog on lethal West Nile virus infection in rodents. Antiviral Res 80:377–379. doi: 10.1016/j.antiviral.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smee DF, Hurst BL, Egawa H, Takahashi K, Kadota T, Furuta Y. 2009. Intracellular metabolism of favipiravir (T-705) in uninfected and influenza A (H5N1) virus-infected cells. J Antimicrob Chemother 64:741–746. doi: 10.1093/jac/dkp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Julander JG, Shafer K, Smee DF, Morrey JD, Furuta Y. 2009. Activity of T-705 in a hamster model of yellow fever virus infection in comparison with that of a chemically related compound, T-1106. Antimicrob Agents Chemother 53:202–209. doi: 10.1128/AAC.01074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gowen BB, Wong MH, Jung KH, Smee DF, Morrey JD, Furuta Y. 2010. Efficacy of favipiravir (T-705) and T-1106 pyrazine derivatives in phlebovirus disease models. Antiviral Res 86:121–127. doi: 10.1016/j.antiviral.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gowen BB, Smee DF, Wong MH, Hall JO, Jung KH, Bailey KW, Stevens JR, Furuta Y, Morrey JD. 2008. Treatment of late stage disease in a model of arenaviral hemorrhagic fever: T-705 efficacy and reduced toxicity suggests an alternative to ribavirin. PLoS One 3:e3725. doi: 10.1371/journal.pone.0003725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiso M, Takahashi K, Sakai-Tagawa Y, Shinya K, Sakabe S, Le QM, Ozawa M, Furuta Y, Kawaoka Y. 2010. T-705 (favipiravir) activity against lethal H5N1 influenza A viruses. Proc Natl Acad Sci U S A 107:882–887. doi: 10.1073/pnas.0909603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendenhall M, Russell A, Smee DF, Hall JO, Skirpstunas R, Furuta Y, Gowen BB. 2011. Effective oral favipiravir (T-705) therapy initiated after the onset of clinical disease in a model of arenavirus hemorrhagic fever. PLoS Negl Trop Dis 5:e1342. doi: 10.1371/journal.pntd.0001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendenhall M, Russell A, Juelich T, Messina EL, Smee DF, Freiberg AN, Holbrook MR, Furuta Y, de la Torre JC, Nunberg JH, Gowen BB. 2011. T-705 (favipiravir) inhibition of arenavirus replication in cell culture. Antimicrob Agents Chemother 55:782–787. doi: 10.1128/AAC.01219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gowen BB, Juelich TL, Sefing EJ, Brasel T, Smith JK, Zhang L, Tigabu B, Hill TE, Yun T, Pietzsch C, Furuta Y, Freiberg AN. 2013. Favipiravir (T-705) inhibits Junin virus infection and reduces mortality in a guinea pig model of Argentine hemorrhagic fever. PLoS Negl Trop Dis 7:e2614. doi: 10.1371/journal.pntd.0002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Safronetz D, Falzarano D, Scott DP, Furuta Y, Feldmann H, Gowen BB. 2013. Antiviral efficacy of favipiravir against two prominent etiological agents of hantavirus pulmonary syndrome. Antimicrob Agents Chemother 57:4673–4680. doi: 10.1128/AAC.00886-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Safronetz D, Rosenke K, Westover JB, Martellaro C, Okumura A, Furuta Y, Geisbert J, Saturday G, Komeno T, Geisbert TW, Feldmann H, Gowen BB. 2015. The broad-spectrum antiviral favipiravir protects guinea pigs from lethal Lassa virus infection post-disease onset. Sci Rep 5:14775. doi: 10.1038/srep14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smee DF, Tarbet EB, Furuta Y, Morrey JD, Barnard DL. 2013. Synergistic combinations of favipiravir and oseltamivir against wild-type pandemic and oseltamivir-resistant influenza A virus infections in mice. Future Virol 8:1085–1094. doi: 10.2217/fvl.13.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smee DF, Hurst BL, Wong MH, Bailey KW, Tarbet EB, Morrey JD, Furuta Y. 2010. Effects of the combination of favipiravir (T-705) and oseltamivir on influenza A virus infections in mice. Antimicrob Agents Chemother 54:126–133. doi: 10.1128/AAC.00933-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caroline AL, Powell DS, Bethel LM, Oury TD, Reed DS, Hartman AL. 2014. Broad spectrum antiviral activity of favipiravir (T-705): protection from highly lethal inhalational Rift Valley fever. PLoS Negl Trop Dis 8:e2790. doi: 10.1371/journal.pntd.0002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scharton D, Bailey KW, Vest Z, Westover JB, Kumaki Y, Van Wettere A, Furuta Y, Gowen BB. 2014. Favipiravir (T-705) protects against peracute Rift Valley fever virus infection and reduces delayed-onset neurologic disease observed with ribavirin treatment. Antiviral Res 104:84–92. doi: 10.1016/j.antiviral.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamada K, Noguchi K, Komeno T, Furuta Y, Nishizono A. 2016. Efficacy of favipiravir (T-705) in rabies postexposure prophylaxis. J Infect Dis 213:1253–1261. doi: 10.1093/infdis/jiv586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim JA, Seong RK, Kumar M, Shin OS. 2018. Favipiravir and ribavirin inhibit replication of Asian and African strains of Zika virus in different cell models. Viruses 10:E72. doi: 10.3390/v10020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delang L, Abdelnabi R, Neyts J. 2018. Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antiviral Res 153:85–94. doi: 10.1016/j.antiviral.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 50.de la Torre JC. 2018. Extending the antiviral value of favipiravir. J Infect Dis 218:509–511. doi: 10.1093/infdis/jiy153. [DOI] [PubMed] [Google Scholar]
- 51.Baranovich T, Wong SS, Armstrong J, Marjuki H, Webby RJ, Webster RG, Govorkova EA. 2013. T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J Virol 87:3741–3751. doi: 10.1128/JVI.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arias A, Thorne L, Goodfellow I. 2014. Favipiravir elicits antiviral mutagenesis during virus replication in vivo. Elife 3:e03679. doi: 10.7554/eLife.03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Avila AI, Gallego I, Soria ME, Gregori J, Quer J, Esteban JI, Rice CM, Domingo E, Perales C. 2016. Lethal mutagenesis of hepatitis C virus induced by favipiravir. PLoS One 11:e0164691. doi: 10.1371/journal.pone.0164691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Avila AI, Moreno E, Perales C, Domingo E. 2017. Favipiravir can evoke lethal mutagenesis and extinction of foot-and-mouth disease virus. Virus Res 233:105–112. doi: 10.1016/j.virusres.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 55.Escribano-Romero E, Jimenez de Oya N, Domingo E, Saiz JC. 2017. Extinction of West Nile virus by favipiravir through lethal mutagenesis. Antimicrob Agents Chemother 61:e01400-17. doi: 10.1128/AAC.01400-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiu L, Patterson SE, Bonnac LF, Geraghty RJ. 2018. Nucleobases and corresponding nucleosides display potent antiviral activities against dengue virus possibly through viral lethal mutagenesis. PLoS Negl Trop Dis 12:e0006421. doi: 10.1371/journal.pntd.0006421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guedj J, Piorkowski G, Jacquot F, Madelain V, Nguyen THT, Rodallec A, Gunther S, Carbonnelle C, Mentre F, Raoul H, de Lamballerie X. 2018. Antiviral efficacy of favipiravir against Ebola virus: a translational study in cynomolgus macaques. PLoS Med 15:e1002535. doi: 10.1371/journal.pmed.1002535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bassi MR, Sempere RN, Meyn P, Polacek C, Arias A. 2018. Extinction of Zika virus and Usutu virus by lethal mutagenesis reveals different patterns of sensitivity to three mutagenic drugs. Antimicrob Agents Chemother 62:e00380-18. doi: 10.1128/AAC.00380-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldhill DH, Langat P, Xie H, Galiano M, Miah S, Kellam P, Zambon M, Lackenby A, Barclay WS. 2019. Determining the mutation bias of favipiravir in influenza virus using next-generation sequencing. J Virol 93:e01217-18. doi: 10.1128/JVI.01217-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Borrego B, de Avila AI, Domingo E, Brun A. 2019. Lethal mutagenesis of Rift Valley fever virus induced by favipiravir. Antimicrob Agents Chemother 63:e00669-19. doi: 10.1128/AAC.00669-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westover JB, Sefing EJ, Bailey KW, Van Wettere AJ, Jung KH, Dagley A, Wandersee L, Downs B, Smee DF, Furuta Y, Bray M, Gowen BB. 2016. Low-dose ribavirin potentiates the antiviral activity of favipiravir against hemorrhagic fever viruses. Antiviral Res 126:62–68. doi: 10.1016/j.antiviral.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oestereich L, Rieger T, Lüdtke A, Ruibal P, Wurr S, Pallasch E, Bockholt S, Krasemann S, Muñoz-Fontela C, Günther S. 2016. Efficacy of favipiravir alone and in combination with ribavirin in a lethal, immunocompetent mouse model of Lassa fever. J Infect Dis 213:934–938. doi: 10.1093/infdis/jiv522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sangawa H, Komeno T, Nishikawa H, Yoshida A, Takahashi K, Nomura N, Furuta Y. 2013. Mechanism of action of T-705 ribosyl triphosphate against influenza virus RNA polymerase. Antimicrob Agents Chemother 57:5202–5208. doi: 10.1128/AAC.00649-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beaucourt S, Vignuzzi M. 2014. Ribavirin: a drug active against many viruses with multiple effects on virus replication and propagation. Molecular basis of ribavirin resistance. Curr Opin Virol 8:10–15. doi: 10.1016/j.coviro.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin Z, Smith LK, Rajwanshi VK, Kim B, Deval J. 2013. The ambiguous base-pairing and high substrate efficiency of T-705 (favipiravir) ribofuranosyl 5′-triphosphate towards influenza A virus polymerase. PLoS One 8:e68347. doi: 10.1371/journal.pone.0068347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nystrom K, Wanrooij PH, Waldenstrom J, Adamek L, Brunet S, Said J, Nilsson S, Wind-Rotolo M, Hellstrand K, Norder H, Tang KW, Lagging M. 2018. Inosine triphosphate pyrophosphatase dephosphorylates ribavirin triphosphate and reduced enzymatic activity potentiates mutagenesis in hepatitis C virus. J Virol 92:e01087-18. doi: 10.1128/JVI.01087-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perales C, Beach NM, Gallego I, Soria ME, Quer J, Esteban JI, Rice C, Domingo E, Sheldon J. 2013. Response of hepatitis C virus to long-term passage in the presence of alpha interferon: multiple mutations and a common phenotype. J Virol 87:7593–7607. doi: 10.1128/JVI.02824-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marukian S, Jones CT, Andrus L, Evans MJ, Ritola KD, Charles ED, Rice CM, Dustin LB. 2008. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology 48:1843–1850. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chou TC. 2006. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev 58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 70.Chou TC. 2010. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 71.Zhang N, Fu JN, Chou TC. 2016. Synergistic combination of microtubule targeting anticancer fludelone with cytoprotective panaxytriol derived from Panax ginseng against MX-1 cells in vitro: experimental design and data analysis using the combination index method. Am J Cancer Res 6:97–104. [PMC free article] [PubMed] [Google Scholar]
- 72.Sheldon J, Beach NM, Moreno E, Gallego I, Pineiro D, Martinez-Salas E, Gregori J, Quer J, Esteban JI, Rice CM, Domingo E, Perales C. 2014. Increased replicative fitness can lead to decreased drug sensitivity of hepatitis C virus. J Virol 88:12098–12111. doi: 10.1128/JVI.01860-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moreno E, Gallego I, Gregori J, Lucia-Sanz A, Soria ME, Castro V, Beach NM, Manrubia S, Quer J, Esteban JI, Rice CM, Gomez J, Gastaminza P, Domingo E, Perales C. 2017. Internal disequilibria and phenotypic diversification during replication of hepatitis C virus in a noncoevolving cellular environment. J Virol 91:e02505-16. doi: 10.1128/JVI.02505-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gallego I, Gregori J, Soria ME, García-Crespo C, García-Álvarez M, Gómez-González A, Valiergue R, Gómez J, Esteban JI, Quer J, Domingo E, Perales C. 2018. Resistance of high fitness hepatitis C virus to lethal mutagenesis. Virology 523:100–109. doi: 10.1016/j.virol.2018.07.030. [DOI] [PubMed] [Google Scholar]
- 75.Gallego I, Sheldon J, Moreno E, Gregori J, Quer J, Esteban JI, Rice CM, Domingo E, Perales C. 2016. Barrier-independent, fitness-associated differences in sofosbuvir efficacy against hepatitis C virus. Antimicrob Agents Chemother 60:3786–3793. doi: 10.1128/AAC.00581-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gregori J, Perales C, Rodriguez-Frias F, Esteban JI, Quer J, Domingo E. 2016. Viral quasispecies complexity measures. Virology 493:227–237. doi: 10.1016/j.virol.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 77.Gregori J, Soria ME, Gallego I, Guerrero-Murillo M, Esteban JI, Quer J, Perales C, Domingo E. 2018. Rare haplotype load as marker for lethal mutagenesis. PLoS One 13:e0204877. doi: 10.1371/journal.pone.0204877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol Mol Biol Rev 76:159–216. doi: 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parker WB. 2005. Metabolism and antiviral activity of ribavirin. Virus Res 107:165–171. doi: 10.1016/j.virusres.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 80.Graci JD, Cameron CE. 2006. Mechanisms of action of ribavirin against distinct viruses. Rev Med Virol 16:37–48. doi: 10.1002/rmv.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Domingo E, Perales C. 2018. Quasispecies and virus. Eur Biophys J 47:443–457. doi: 10.1007/s00249-018-1282-6. [DOI] [PubMed] [Google Scholar]
- 82.Sardanyes J, Martinez R, Simo C, Sole R. 2017. Abrupt transitions to tumor extinction: a phenotypic quasispecies model. J Math Biol 74:1589–1609. doi: 10.1007/s00285-016-1062-9. [DOI] [PubMed] [Google Scholar]
- 83.Chao MW, Chang LH, Tu HJ, Chang CD, Lai MJ, Chen YY, Liou JP, Teng CM, Pan SL. 2019. Combination treatment strategy for pancreatic cancer involving the novel HDAC inhibitor MPT0E028 with a MEK inhibitor beyond K-Ras status. Clin Epigenetics 11:85. doi: 10.1186/s13148-019-0681-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Albin JS, Harris RS. 2010. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev Mol Med 12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dapp MJ, Clouser CL, Patterson S, Mansky LM. 2009. 5-Azacytidine can induce lethal mutagenesis in human immunodeficiency virus type 1. J Virol 83:11950–11958. doi: 10.1128/JVI.01406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dapp MJ, Holtz CM, Mansky LM. 2012. Concomitant lethal mutagenesis of human immunodeficiency virus type 1. J Mol Biol 419:158–170. doi: 10.1016/j.jmb.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruiz-Jarabo CM, Ly C, Domingo E, de la Torre JC. 2003. Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV). Virology 308:37–47. doi: 10.1016/s0042-6822(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 88.Harris KS, Brabant W, Styrchak S, Gall A, Daifuku R. 2005. KP-1212/1461, a nucleoside designed for the treatment of HIV by viral mutagenesis. Antiviral Res 67:1–9. doi: 10.1016/j.antiviral.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 89.Mullins JI, Heath L, Hughes JP, Kicha J, Styrchak S, Wong KG, Rao U, Hansen A, Harris KS, Laurent JP, Li D, Simpson JH, Essigmann JM, Loeb LA, Parkins J. 2011. Mutation of HIV-1 genomes in a clinical population treated with the mutagenic nucleoside KP1461. PLoS One 6:e15135. doi: 10.1371/journal.pone.0015135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 76:13001–13014. doi: 10.1128/jvi.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. 2010. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol 28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 93.Gregori J, Esteban JI, Cubero M, Garcia-Cehic D, Perales C, Casillas R, Alvarez-Tejado M, Rodríguez-Frías F, Guardia J, Domingo E, Quer J. 2013. Ultra-deep pyrosequencing (UDPS) data treatment to study amplicon HCV minor variants. PLoS One 8:e83361. doi: 10.1371/journal.pone.0083361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ramírez C, Gregori J, Buti M, Tabernero D, Camós S, Casillas R, Quer J, Esteban R, Homs M, Rodriguez-Frías F. 2013. A comparative study of ultra-deep pyrosequencing and cloning to quantitatively analyze the viral quasispecies using hepatitis B virus infection as a model. Antiviral Res 98:273–283. doi: 10.1016/j.antiviral.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 95.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Quer J, Rodríguez-Frias F, Gregori J, Tabernero D, Soria ME, García-Cehic D, Homs M, Bosch A, Pintó RM, Esteban JI, Domingo E, Perales C. 2017. Deep sequencing in the management of hepatitis virus infections. Virus Res 239:115–125. doi: 10.1016/j.virusres.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 97.Chou TC, Martin N. 2005. CompuSyn for drug combinations: PC software and user’s guide. A computer program for quantitation of synergism and antagonism in drug combinations, and the determination of IC50 and ED50 and LD50 values. ComboSyn, Paramus, NJ. [Google Scholar]
- 98.Chou TC. 2014. Frequently asked questions in drug combinations and the mass-action law-based answers. Synergy 1:3–21. doi: 10.1016/j.synres.2014.07.003. [DOI] [Google Scholar]