This study aimed (i) to compare the performance of the BD Onclarity human papillomavirus (HPV) assay with the Cobas HPV test in identifying cervical intraepithelial neoplasia 2/3 or above (CIN2/3+) in an Asian screening population and (ii) to explore improving the cervical cancer detection specificity of Onclarity by machine learning. We tested 605 stratified random archived samples of cervical liquid-based cytology samples with both assays.
KEYWORDS: cervical cancer, primary screening, HPV test, Onclarity, Cobas
ABSTRACT
This study aimed (i) to compare the performance of the BD Onclarity human papillomavirus (HPV) assay with the Cobas HPV test in identifying cervical intraepithelial neoplasia 2/3 or above (CIN2/3+) in an Asian screening population and (ii) to explore improving the cervical cancer detection specificity of Onclarity by machine learning. We tested 605 stratified random archived samples of cervical liquid-based cytology samples with both assays. All samples had biopsy diagnosis or repeated negative cytology follow-up. Association rule mining (ARM) was employed to discover coinfection likely to give rise to CIN2/3+. Outcome classifiers interpreting the extended genotyping results of Onclarity were built with different underlying models. The sensitivities (Onclarity, 96.32%; Cobas, 95.71%) and specificities (Onclarity, 46.38%; Cobas, 45.25%) of the high-risk HPV (hrHPV) components of the two tests were not significantly different. When HPV16 and HPV18 were used to further interpret hrHPV-positive cases, Onclarity displayed significantly higher specificity (Onclarity, 87.10%; Cobas, 80.77%). Both hrHPV tests achieved the same sensitivities (Onclarity, 90.91%; Cobas, 90.91%) and similar specificities (Onclarity, 48.46%; Cobas, 51.98%) when used for triaging atypical squamous cells of undetermined significance. Positivity in both HPV16 and HPV33/58 of the Onclarity channels entails the highest probability of developing CIN2/3+. Incorporating other hrHPVs into the outcome classifiers improved the specificity of identifying CIN2/3 to up to 94.32%. The extended genotyping of Onclarity therefore can help to highlight patients having the highest risk of developing CIN2/3+, with the potential to reduce unnecessary colposcopy and negative psychosocial impact on women receiving the reports.
INTRODUCTION
Cervical cancer can be prevented effectively by screening, which has been performed traditionally by cytological examination of exfoliated cervical cells (Pap smear) to identify women having cervical intraepithelial neoplasia (CIN). In recent years, primary screening of cervical cancer increasingly relied on high-risk human papillomavirus (hrHPV) detection, as it has been demonstrated that HPV tests generally have higher sensitivity and higher negative predictive value than cytology (1–4).
Most (∼70%) cervical cancers are caused by HPV16 and HPV18 (5). Large trials, such as ATHENA, have demonstrated that patients infected with HPV16 or HPV18 have a higher chance of developing CIN3 or above than patients infected with other hrHPV types (6). It follows that if the HPV test is adopted as the primary screening tool in women aged ≥25 years, women positive for HPV16 and -18 should be immediately referred for colposcopy, whereas those positive for non-16 or -18 HPV types could be further triaged by cytology (Fig. 1A) (7). Our previous studies also found that detection of HPV16 and HPV18 improved the specificity of identifying CIN2/3 or above (CIN2/3+) in our local population (8, 9). The Cobas HPV assay (Cobas) (Roche Molecular Systems, Pleasanton, CA) is an example of the recent generation of an HPV test on the market providing partial genotyping by highlighting HPV16- and HPV18-infected cases among those infected with hrHPV. It is the first HPV test that is approved by the U.S. Food and Drug Administration (FDA) for primary cervical cancer screening. The optimal management of women infected with other hrHPV types, however, is less clear-cut. In addition, while there is evidence suggesting that infection involving multiple high-risk HPV types confers extra risk of cervical cancer (10–12), the clinical significance is unclear and no recommendation has been formulated to specifically address the management of these patients.
FIG 1.
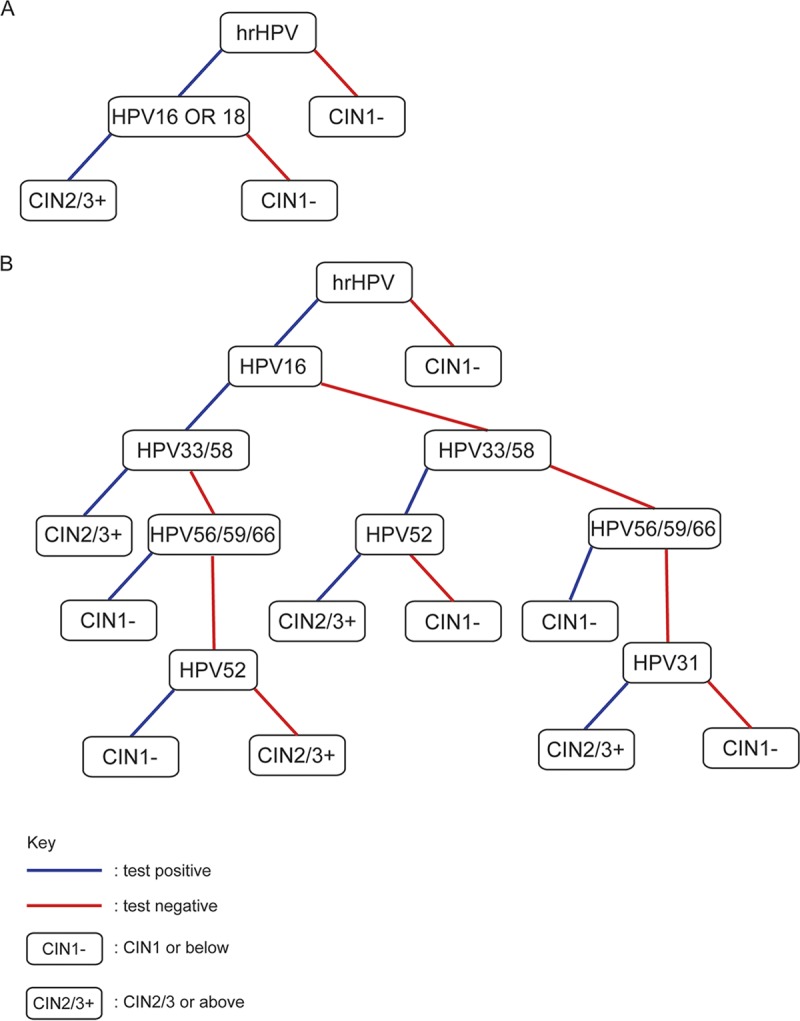
HPV test results can be used to predict the outcome of cervical cancer screening recipients. (A) A classifier using HPV16/18 to triage hrHPV-positive cases. (B) A classifier based on a decision tree model using extended HPV genotyping information.
The BD Onclarity HPV assay (Onclarity) (Becton Dickinson Diagnostics, Sparks, MD) is a multiplex real-time PCR assay detecting the E6 or E7 sequences of 14 hrHPV types. In addition to hrHPV positivity, the assay also provides extended genotyping information by distinguishing the more carcinogenic HPV16, -18, -31, -45, -51, -52 (as six separate channels), and HPV33/58 (combined in one channel) from the remaining 6 hrHPV types reported in two channels of three types each (HPV56/59/66 and HPV35/39/68). It received approval in 2018 from the U.S. FDA for use as a primary screening test, as an adjunct to the Pap test, and for triaging women with atypical squamous cells of undetermined significance (ASC-US) detected by the Pap test (13). Extended genotyping information has been suggested to provide additional indicators of increased cervical cancer risk, as HPV16, -18, -31, -33, -52, and -58 confer higher risk than other hrHPV types (14, 15). Therefore, genotyping information may provide a means to stratify patients positive for hrHPV by signifying which of them would benefit most from immediate colposcopy referral.
In this study, we performed HPV DNA detection with extended genotyping by Onclarity and Cobas in 605 archival cervical cytology samples from Hong Kong women. Using tissue biopsy diagnosis as the standard, we compared the performance of Onclarity and Cobas in identifying high-grade lesions and in triaging equivocal smears. While the hrHPV and HPV16/18 components of Onclarity and Cobas performed similarly, the extended genotyping of Onclarity may provide information for further stratifying patients through machine learning.
MATERIALS AND METHODS
Cervical cytology specimen.
Stratified random samples of 605 archived liquid-based cytology (LBC) residues in PreservCyt solution were selected from the archive of the Cervical Cytology Laboratory, Department of Pathology, The University of Hong Kong. The age of women ranged from 15 to 86 years (median, 39). The selections were made to encompass various cytological diagnostic categories, including 50 negative for intraepithelial lesion (NIL), 238 atypical squamous cells of undetermined significance (ASC-US), 43 atypical glandular cells (AGC), 69 ASC–cannot exclude HSIL (ASC-H), 117 low-grade squamous intraepithelial lesions (LSIL), 58 high-grade squamous intraepithelial lesions (HSIL), and 30 squamous cell carcinomas (SCC) (Table 1). All samples, except NIL and ASC-US samples previously tested HPV negative by Cobas as a management routine, have biopsy specimen-proven diagnoses within 2 years of the initial cytology results. All NIL samples have been confirmed with 2 negative follow-up cytologies. HPV-negative ASC-US cases were also followed up by cytology.
TABLE 1.
Description of the study cohort
| Cyto dxa | No. with worst biopsy specimen finding within 2 yr |
% hrHPV positive by: |
P valued | |||||
|---|---|---|---|---|---|---|---|---|
| No malignancy | CIN1/condylomac | CIN2/3 or abovec | CAc | Total | Onclarity | Cobas | ||
| NILM | 50b | 0 | 0 | 0 | 50 | 16.00 | 18.00 | 1.0000 |
| ASC-US | 112 | 115 | 11 | 0 | 238 | 53.36 | 50.00 | 0.2012 |
| AGC | 6 | 24 | 9 | 4 | 43 | 53.49 | 81.40 | 0.0015 |
| ASCH | 1 | 24 | 44 | 0 | 69 | 92.75 | 91.30 | 1.0000 |
| LSIL | 5 | 105 | 7 | 0 | 117 | 76.07 | 73.50 | 0.6464 |
| HSIL | 0 | 0 | 48 | 10 | 58 | 94.83 | 96.55 | 1.0000 |
| SCC | 0 | 0 | 0 | 30 | 30 | 93.33 | 100.00 | 0.4795 |
| Total | 174 | 268 | 119 | 44 | 605 | 65.12 | 65.79 | 0.7404 |
Cyto dx, cytology diagnosis; NILM, negative for intraepithelial lesion or malignancy; ASC-US, atypical squamous cells of undetermined significance; AGC, atypical glandular cells; ASCH, atypical squamous cells–cannot exclude HSIL; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion; SCC, squamous cell carcinoma.
The 2-year diagnosis of no malignancy was reached by at least 2 consecutive NILM cytology findings in the follow-up period.
CIN1, cervical intraepithelial neoplasia 1; CIN2/3, cervical intraepithelial neoplasia 2/3; CA, SCC/adenocarcinoma.
Determined by McNemar’s test.
Cobas test and Onclarity test were performed on a Cobas 4800 platform and a Viper LT platform, respectively, according to instructions from the manufacturers. All of the genotyping channels of Onclarity were unlocked. A negative result in the hrHPV channel automatically entails negativity in all genotyping channels. The use of archival human cytology specimens for research purposes was approved by the Institutional Review Board of The University of Hong Kong/Hospital Authority Hong Kong West Cluster (HKU HKW/IRB UW 17-249).
Association rule mining (ARM).
Association rules were discovered within a data subset containing only the Onclarity genotyping test results and the binary biopsy specimen-proven outcome (CIN1 or below [CIN1−] and CIN2/3 or above [CIN2/3+]) using the Apriori algorithm (16) within the R extension package arules (17). The parameters were the following: minimum support, 0.009; minimum confidence, 0.01; minimum rule length, 2. Only rules containing CIN2/3+ on the right-hand side (rhs) were retained. The conviction and rule power factor were also calculated with arules.
Classifier construction.
A number of classifiers with different underlying models were built to predict whether a case’s biopsy diagnosis is CIN1− or CIN2/3+ using all results from the ten HPV test channels from Onclarity (hrHPV and HPV16, -18, -31, -45, -51, -52, -33/58, -56/59/66, and -35/39/68). The 605 cases were randomly split into a training set consisting of 485 (80%) of the cases and a test set with the remaining 120 (20%) cases, with proportion of outcomes retained. The training set was used to train 4 classifiers with the model decision tree, random forest, supported vector machine (SVM)-linear, and SVM-nonlinear using the rpart, rf, svmLinear, and svmRadial functions, respectively, of the Caret package of R. Three repeated 10-fold cross validations were performed when building each classifier.
Statistical analysis.
For comparing hrHPV detection by Onclarity and Cobas, the overall percent agreements, positive percent agreements, negative percent agreements, and Cohen’s kappa (κ) values were calculated for the entire set of samples. A kappa value of 0 indicates no agreement better than chance, and a kappa value of 1 indicates perfect agreement. Kappa values from 0 to 0.20, 0.21 to 0.40, 0.41 to 0.60, 0.61 to 0.80, and above 0.81 indicate poor, fair, moderate, good, and very good strength of agreement, respectively (18). For testing the performance in primary screening, sensitivities and specificities of both tests in detecting biopsy specimen-proven CIN2/3+ were calculated. For testing the performance in ASC-US triage, the hrHPV test results were used to highlight biopsy specimen-proven CIN2/3+. McNemar’s test was used to test the difference between the sensitivities or specificities of Onclarity and Cobas HPV tests. A P value of <0.05 was considered statistically significant.
RESULTS
Onclarity and Cobas perform similarly in identifying CIN2/3+.
To evaluate the performance of Onclarity and Cobas in identifying high-grade lesions, we performed the two tests on 605 LBC samples collected in PreservCyt. Although Onclarity test was designed for HPV detection in samples collected in SurePath, a previous study demonstrated that the use of PreservCyt as an alternative transport medium did not lead to remarkable alteration in the performance of the test (19). Stratified random sampling was enforced to ensure inclusion of a significant portion of samples with high-grade cytological findings. The composition of the samples is summarized in Table 1. The overall hrHPV positivities detected by Onclarity and Cobas were similar and were not significantly different (65.12% versus 65.79%; P = 0.7404) (Table 1). Similarly, the hrHPV-positive rates by the two tests in individual cytology diagnosis groups were insignificantly different, except in the AGC group, where the Cobas-positive rate was significantly higher than that of Onclarity (81.40% versus 53.49%; P = 0.0015) (Table 1).
The hrHPV components of the two tests agreed well with each other (86.45% agreement, κ = 0.700) (Table 2). Of the 605 samples, 316 were used in a previous study comparing Aptima and Cobas performance (20). Thus, we were able to gauge some of the discrepant cases with Aptima test results. Out of the 316 samples, 40 yielded discrepant Onclarity and Cobas hrHPV results. As shown in Table S1 in the supplemental material, in the majority (70%) of the samples the hrHPV test results of Aptima agreed with those of Onclarity. The two tests displayed similar sensitivities (Onclarity, 96.32%; Cobas, 95.71%; P = 1.0000) and specificities (Onclarity, 46.38%; Cobas, 45.25%; P = 0.6397) in detecting cases that developed CIN2/3+ within the 2-year follow-up period (Table 3). On the other hand, the two tests showed larger disagreement on HPV16 and HPV18 genotyping. For both HPV types, Cobas yielded more positive results than Onclarity (Table 2). When HPV16 and HPV18 genotyping information was used to interpret hrHPV cases (represented by Fig. 1A), Onclarity was significantly less sensitive (50.92% versus 74.23%; P = 0.0001) but was significantly more specific (87.10% versus 80.77%; P = 0.0003) (Table 3).
TABLE 2.
Concordance between Onclarity and Cobas HPV detection tests
| Onclarity result | All Cobas |
Cobas HPV16 |
Cobas HPV18 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | Positive | Negative | Total | Positive | Negative | Total | |
| All Onclaritya | |||||||||
| Positive | 355 | 39 | 394 | ||||||
| Negative | 43 | 168 | 211 | ||||||
| Total | 398 | 207 | 605 | ||||||
| Onclarity HPV16b | |||||||||
| Positive | 100 | 8 | 108 | ||||||
| Negative | 85 | 412 | 497 | ||||||
| Total | 185 | 420 | 605 | ||||||
| Onclarity HPV18c | |||||||||
| Positive | 26 | 9 | 35 | ||||||
| Negative | 26 | 544 | 570 | ||||||
| Total | 52 | 553 | 605 | ||||||
No. of observed agreements, 523 (86.45% of the observations); kappa = 0.700 (95% CI, 0.640 to 0.760).
No. of observed agreements, 512 (84.63% of the observations); kappa = 0.590 (95% CI, 0.519 to 0.662).
No. of observed agreements, 570 (94.21% of the observations); kappa = 0.568 (95% CI, 0.440 to 0.696).
TABLE 3.
Sensitivity, specificity, PPV, and NPV for diagnoses of biopsy specimen-proven CIN2/3 or above by Onclarity and Cobas tests
| Test | Test resulta
[% (95% CI)] |
|||
|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | |
| Onclarity hrHPV | 96.32 (92.16–98.64) | 46.38 (41.66–51.15) | 39.85 (34.98–44.87) | 97.16 (93.91–98.95) |
| Cobas hrHPV | 95.71 (91.35–98.26) | 45.25 (40.54–50.02) | 39.20 (34.47–44.18) | 96.62 (93.16–98.63) |
| Onclarity hrHPV (HPV16 or HPV18) | 50.92 (42.98–58.82) | 87.10 (83.62–90.08) | 59.29 (50.67–67.50) | 82.80 (79.05–86.12) |
| Cobas hrHPV (HPV16 or HPV18) | 74.23 (66.81–80.76) | 80.77 (76.78–84.34) | 58.74 (51.69–65.53) | 89.47 (86.04–92.31) |
PPV, positive predictive value; NPV, negative predictive value.
Onclarity and Cobas perform similarly in triage of ASC-US.
To evaluate the utilities of the two tests in triaging equivocal abnormal cases, we calculated and compared the sensitivity and specificity of them in highlighting those ASC-US cases which later developed high-grade lesions. Among the 238 ASC-US cases within the cohort, 11 had colposcopic biopsy specimen-proven CIN2/3+ detected within 2 years of the first cytology. Both Onclarity and Cobas hrHPV were positive in 10 of the cases (sensitivity, 90.91%) (Table 4), and their specificities were insignificantly different (48.46% versus 51.98%; P = 0.2012) (Table 4). Similar to the case of primary CIN2/3+ identification, adding HPV16 and HPV18 interpretation increased the specificity. The sensitivities (Onclarity, 54.55%; Cobas, 45.45%) and specificities (Onclarity, 89.43%; Cobas, 88.55%) were insignificantly different (P = 1.000 and 0.6831, respectively) (Table 4). Thus, the two tests performed similarly if they were used for triaging ASC-US.
TABLE 4.
Sensitivity, specificity, PPV, and NPV for triage of ASC-US by Onclarity and Cobas testsa
| Test | Test resulta
[% (95% CI)] |
|||
|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | |
| Onclarity hrHPV | 90.91 (58.72–99.77) | 48.46 (41.79–55.16) | 7.87 (3.84–14.00) | 99.10 (95.08–99.98) |
| Cobas hrHPV | 90.91 (58.72–99.77) | 51.98 (45.27–58.64) | 8.40 (4.10–14.91) | 99.16 (95.41–99.98) |
| Onclarity hrHPV (HPV16 or HPV18) | 54.55 (23.38–83.25) | 89.43 (84.68–93.11) | 20.00 (7.71–38.57) | 97.60 (94.48–99.21) |
| Cobas hrHPV (HPV16 or HPV18) | 45.45 (16.75–76.62) | 88.55 (83.67–92.38) | 16.13 (5.45–33.73) | 97.10 (93.80–98.93) |
PPV, positive predictive value; NPV, negative predictive value.
HPV16, HPV33/58, and HPV31 confer higher RR for CIN2/3 or above than hrHPV.
It has been shown that HPV16 and HPV33/58 detection by Onclarity confers higher 3-year risk for CIN2/3+ than pooled hrHPV detection (21). Here, we also ask whether certain hrHPV genotypes confer higher risk for CIN2/3+ than hrHPV in our cohort. We calculated the relative risk (RR) to develop CIN2/3+ for all the HPV type channels provided by the test relative to results for hrHPV-negative cases. Within the cohort, there were 6 histologically confirmed CIN2/3+ results that were tested hrHPV negative by Onclarity. Thus, the RR of hrHPV-positive cases was 14.01 (Table 5). HPV16-, HPV33/58-, and HPV31-positive cases had higher RR of 24.10, 16.20, and 19.78, respectively (Table 5). Thus, HPV31 showed higher RR than HPV33/58, different from what was observed in reference 21. On the other hand, RR of HPV56/59/66 and -51 were 5.94 and 7.69, respectively, which were much lower than those of hrHPV (Table 5). Hence, our results support the different risk associations of different HPV genotypes besides HPV16 and -18 in different geographic populations.
TABLE 5.
Relative risk for developing CIN2/3 or above by genotype channels of Onclarity
| HPV type | No. of CIN2/3+ cases | No. of positive cases | Relative risk to CIN2/3+ (95% CI) |
|---|---|---|---|
| 16 | 74 | 108 | 24.10 (10.84–53.57) |
| 18 | 11 | 35 | 11.05 (4.37–27.96) |
| 45 | 3 | 9 | 11.72 (3.48–39.50) |
| 33/58 | 47 | 102 | 16.20 (7.16–36.65) |
| 31 | 9 | 16 | 19.78 (8.05–48.62) |
| 56/59/66 | 12 | 71 | 5.94 (2.32–15.25) |
| 51 | 7 | 32 | 7.69 (2.76–21.44) |
| 52 | 30 | 103 | 10.24 (4.40–23.83) |
| 35/39/68 | 22 | 64 | 12.09 (5.12–28.52) |
| 16 and 33/58 | 10 | 11 | 31.97 (14.21–71.90) |
| 33/58 and 35/39/68 | 6 | 9 | 8.79 (2.50–30.93) |
| 33/58 and 52 | 11 | 18 | 21.49 (9.00–51.33) |
| 35/39/68 and 52 | 6 | 10 | 21.10 (8.27–83.86) |
| 16 and 52 | 7 | 13 | 18.94 (7.43–48.26) |
| hrHPV | 157 | 394 | 14.01 (6.31–31.12) |
| Negative | 6 | 211 | Reference group |
A machine learning approach identifies HPV16 and HPV33/58 coinfection as the highest-risk test result.
The Onclarity extended genotyping provides more information based on which prediction of high-grade lesions could be made. Here, we asked whether any of the hrHPV genotypes, when coinfected, entailed higher risk of developing CIN2/3 or above. An association rule mining (ARM) algorithm, Apriori (16), was employed for this purpose. ARM is a machine learning approach originally developed for market basket analysis to reveal rules in the combination of items in retail settings. It can be applied to exhaustively identify relationships between variables within a data set, which can then be ranked according to desired parameters. The results of ARM analysis are rules in the form of {item A, item B, …} → {item X, item Y, …}, with three parameters (support, confidence, and lift) associated with each rule. Lift quantifies the strength of the rule by calculating the ratio of support of the rule to the support of the individual items in the rule. A lift value greater than 1 represents a positive association, i.e., items on the left-hand side (lhs) will increase the probability of the items on the rhs, whereas a lift value of <1 indicates a negative association or substitution (complementary) effect between the lhs and rhs items. Lift equal to or close to 1 suggests the lhs and rhs items are independent. In addition to lift, there are other measures for association rules proposed by various researchers (22). Here, we also calculated the conviction (23) and rule power factor (24) of the rules.
By restricting the resulting rules to contain only CIN2/3 or above on the rhs, 13 rules were identified and ranked by lift (Table 6). Remarkably, all of the rules having single HPV genotyping channels on the lhs were identified except {HPV45} → {CIN2/3 or above}, which was filtered due to insufficient support. Moreover, the ranking by lift mirrored that obtained by RR, e.g., {HPV16} → {CIN2/3 or above} was ranked the highest among rules involving a single channel on the lhs. Therefore, the ARM approach appeared to correctly identify important relationships within the data set.
TABLE 6.
Rules identified by association rule mining
| Rule | Support | Confidence | Rule power factor | Lift | Conviction | Count |
|---|---|---|---|---|---|---|
| 1. {HPV16,HPV33/58} → {CIN2/3 or above} | 0.017 | 0.909 | 0.015 | 3.374 | 8.036 | 10 |
| 2. {HPV16} → {CIN2/3 or above} | 0.122 | 0.685 | 0.084 | 2.543 | 2.321 | 74 |
| 3. {HPV33/58,HPV35/39/68} → {CIN2/3 or above} | 0.010 | 0.667 | 0.007 | 2.474 | 2.192 | 6 |
| 4. {HPV33/58,HPV52} → {CIN2/3 or above} | 0.018 | 0.611 | 0.011 | 2.268 | 1.879 | 11 |
| 5. {HPV35/39/68,HPV52} → {CIN2/3 or above} | 0.010 | 0.600 | 0.006 | 2.227 | 1.826 | 6 |
| 6. {HPV31} → {CIN2/3 or above} | 0.015 | 0.563 | 0.008 | 2.088 | 1.670 | 9 |
| 7. {HPV16,HPV52} → {CIN2/3 or above} | 0.012 | 0.538 | 0.006 | 1.999 | 1.583 | 7 |
| 8. {HPV33/58} → {CIN2/3 or above} | 0.078 | 0.461 | 0.036 | 1.710 | 1.355 | 47 |
| 9. {HPV35/39/68} → {CIN2/3 or above} | 0.036 | 0.344 | 0.013 | 1.276 | 1.113 | 22 |
| 10. {HPV18} → {CIN2/3 or above} | 0.018 | 0.314 | 0.006 | 1.167 | 1.065 | 11 |
| 11. {HPV52} → {CIN2/3 or above} | 0.050 | 0.291 | 0.014 | 1.081 | 1.031 | 30 |
| 12. {HPV51} → {CIN2/3 or above} | 0.012 | 0.219 | 0.003 | 0.812 | 0.935 | 7 |
| 13. {HPV56/59/66} → {CIN2/3 or above} | 0.020 | 0.169 | 0.003 | 0.627 | 0.879 | 12 |
We noticed that a number of rules involving more than one genotyping channel on the lhs ranked high on the list (rule no. 1, 3, 4, 5, and 7) (Table 6). In fact, the rule {HPV16, HPV33/58} → {CIN2/3 or above} appeared to be the most important one within the data set, with the highest confidence, lift, and conviction values. Confidence can be interpreted as an estimate of the conditional probability of finding the rhs under the condition that items on the lhs are found (22). Conviction is similar to lift but also considers the rule’s consequences (23). Thus, a high conviction value of the rule suggests that HPV16-HPV33/58 coinfection and CIN2/3 are unlikely to be independent. Rule power factor indicates how intensely a rule’s items are associated with each other in terms of positive relationships by weighting the rule’s confidence by support (24). The rule {HPV16} → {CIN2/3 or above} exhibited the highest rule power factor, as it also has the highest support (Table 6). However, {HPV16,HPV33/58} → {CIN2/3 or above} remained the third rule with the highest rule power factor, highlighting its importance (Table 6).
We calculated the RR to develop CIN2/3+ for cases with coinfections of HPV16 and HPV33/58, HPV33/58 and HPV35/39/68, HPV33/58 and HPV52, HPV35/39/68 and HPV52, and HPV16 and HPV52. Coinfection by HPV16 and HPV33/58 conferred an RR of 31.97, much higher than those for pooled hrHPV, HPV33/58, or HPV16 alone (Table 5). Notably, coinfection by HPV33/58 and HPV52, HPV35/39/68 and HPV52, and HPV16 and HPV52 also conferred higher RR than pooled hrHPV (Table 5).
Classifiers incorporating extended genotyping results can enhance specificity.
The fact that certain HPV genotype combinations confer even higher risk than HPV16 and pooled hrHPV alone suggested that incorporating the extended genotyping results enhances test specificity. To utilize the additional information, we built a number of classifiers to predict whether a case’s biopsy diagnosis is CIN1 or below or CIN2/3 or above based on all 10 HPV test results (hrHPV and HPV16, -18, -31, -45, -51, -52, -33/58, -56/59/66, and -35/39/68) provided by Onclarity assay. Four hundred eighty-five of the 605 cases (80%; selected randomly) were used as the training set to train the classifiers. As an example of the classifiers, a decision tree using information gain as the split method is shown in Fig. 1B. Three additional classifiers, using random forest, supported vector machine (SVM)-linear, and SVM-nonlinear as the underlying models, were built, and the classifiers were used to make predictions on the test set and compared with the usual way of interpreting HPV test results (represented in Fig. 1A). The performance of the classifiers on predicting the outcome of the test set is shown in Table 7. Of the six ways to interpret Onclarity results, hrHPV has the highest sensitivity of 96.88%, whereas the decision tree model gave the highest specificity of 94.32%. In fact, all of the classifiers incorporating all HPV test results achieved over 90% specificity. However, triaging hrHPV-positive cases with HPV16/18 results gave the best balance of sensitivity and specificity (Table 7).
TABLE 7.
Performance of the classifiers using Onclarity extended genotyping results to predict the outcome of the cases in the test set
| Classifier | Test resulta
[% (95% CI)] |
|||
|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | |
| hrHPV | 96.88 (83.78–99.91) | 51.14 (40.25–61.95) | 41.89 (30.51–53.94) | 97.83 (88.47–99.94) |
| hrHPV (HPV16 + HPV18) | 62.50 (43.69–78.90) | 89.77 (81.47–95.22) | 68.97 (49.17–84.72) | 86.81 (78.10–93.00) |
| Decision treeb | 50.00 (31.89–68.11) | 94.32 (87.24–98.13) | 76.19 (52.83–91.78) | 83.84 (75.09–90.47) |
| Random forest | 53.13 (34.74–70.91) | 90.91 (82.87–95.99) | 68.00 (46.50–85.05) | 84.21 (75.30–90.88) |
| SVMc -linear | 56.25 (37.66–73.64) | 90.91 (82.87–95.99) | 69.23 (48.21–85.67) | 85.11 (76.28–91.61) |
| SVMc -nonlinear | 53.13 (34.74–70.91) | 90.91 (82.87–95.99) | 68.00 (46.50–85.05) | 84.21 (75.30–90.88) |
PPV, positive predictive value; NPV, negative predictive value.
Decision tree is shown in Fig. 1B.
SVM, support vector machine.
DISCUSSION
Summary of principal findings and their significance.
In this study, we demonstrated that the hrHPV component of the Onclarity HPV test performed similarly to that of the Cobas HPV test when used to indicate women at risk of developing CIN2/3+. However, the HPV16 and HPV18 components of the two tests behaved noticeably differently. It follows that when HPV16 and HPV18 genotyping results were incorporated to further triage hrHPV-positive women, Cobas was remarkably more sensitive but Onclarity displayed higher specificity. The specificity of Onclarity could be further enhanced by including the extended genotyping results in the interpretation process. We attempted to derive a decision-making system based on the additional genotyping information available in Onclarity by machine learning. The best classifier achieved a specificity of 94.32%. A specific HPV test can help reduce the unnecessary downstream examinations, such as colposcopy and biopsy tests. Our earlier studies have shown that women receiving positive hrHPV reports suffer from significant anxiety with prolonged psychosocial burden (25).
The higher specificity could partly be attributed to the fact that some of the HPV types are more oncogenic than others (14, 15), and certain HPV genotype combinations may confer higher risks. There is also evidence that coinfection of multiple types of HPV modulates the risk of cervical cancer development. For instance, genotyping analysis of the National Cancer Institute-sponsored Costa Rica HPV Vaccine Trial suggested that coinfection with multiple α9 species was associated with significantly increased risk of CIN2+ or HSIL+ (10). In a study conducted in South America, coinfection of HPV68 with HPV16 increases the risk of CIN2-3+ compared to infection by HPV16 or HPV68 alone (11). Similarly, multiple HPV genotype infection was associated with poorer invasive cervical cancer survival in a Brazilian study (12). On the other hand, the risk of CIN2 progression is reported to be lower in women with combined infection of high-risk HPV and low-risk HPV (26). While the biological effect of multiple HPV infection is unclear, it has been shown that multiple infection entails a larger chance of oncogenic E6/E7 mRNA expression than single infection. Therefore, multiple infection might be a clinically important finding even in low-grade lesions (27).
Using ARM we demonstrated that some infections involving multiple HPV types confer extra risk. While it is possible to calculate the RR of every possible HPV genotype combination, the ARM approach represents a faster and easier-to-implement alternative to highlight the most important combinations, which can then be verified with RR calculations. This attribute is important when correlations were being drawn from larger data sets containing more variables, for example, when the samples were genotyped by HPV line blot assays or chips that cover larger numbers of HPV genotypes (9, 28) or when the samples not only were tested for HPV but also were subjected to quantitative DNA image cytology (29).
Our study suggested that subjects testing positive for the HPV33/58 channel and those positive for HPV16 and HPV33/58 channels of Onclarity were at high risk of developing high-grade lesions. As the Onclarity test cannot distinguish between HPV33 and HPV58 infection, we do not know which of the genotypes is responsible for the elevated risk. However, there were reports that the worldwide uncommon genotype HPV58 was the second most common genotype detected in patients with cervical carcinoma in Hong Kong and displayed a significant association with CIN2/3 and carcinoma (30). HPV58 could be commonly found among squamous cell carcinoma in China and other countries in East Asia, including South Korea and Japan (31). T20I and/or G63S substitution(s) at E7 of HPV58 is common in Asia and is associated with a higher risk for cervical neoplasia (32). HPV58 was the third most common hrHPV within our collection previously evaluated by dot blot assay (9). Therefore, we believe that HPV58 is the more likely genotype to confer increased cancer risk in our cohort.
It is unclear why the two HPV tests performed differently in the AGC group (Table 1), but we noted that discrepant HPV test results tend to happen in samples with equivocal or low-grade lesion cytology diagnosis. In fact, quite a number of ASC-US cases also yielded different Cobas and Onclarity results, but the difference did not reach statistical significance (Table 1). In one of our previous studies, Cobas and Aptima test results were significantly different in ASC-US, AGC, and ASCH samples (20). Tewari et al. also reported that discrepancy between Cobas and Aptima in <CIN2 samples was significantly higher than that in CIN2+ cases (33). Similarly, we have also demonstrated in our earlier study that ASC-US and AGC were the categories yielding the highest percentage of discrepant genotyping results by GenoFlow and Linear Array HPV genotyping assays (9). It has been postulated that compared with samples with apparent high-grade lesions, equivocal or low-grade samples may have lower HPV viral load, which poses challenges to the sensitivities of HPV tests (34, 35).
Limitations of the study.
The study was limited by the small number of samples from which association rules and decision-making models were derived. During ARM many associations were filtered due to insufficient representation within the data set. Similarly, in the resultant decision tree, a number of HPV types are missing, probably because they are underrepresented in the training set.
It did not escape our attention that in the decision tree shown in Fig. 1B, some hrHPV genotypes surprisingly may signify lower risk under certain situations. For example, among hrHPV-positive cases, when a case was HPV16 positive and HPV33/58 negative, HPV56/59/66 positivity prompted CIN1− classification. hrHPV positive, HPV16 negative, HPV33/58 negative, but HPV56/59/66 positive also indicated CIN1− classification. Among our samples, the number of samples that were hrHPV positive, HPV16 positive, HPV33/58 negative, and HPV56/59/66 positive was 12, whereas the number of samples that were hrHPV positive, HPV16 negative, HPV33/58/negative, and HPV56/59/66 positive was 47, totaling 59. In total, there were 292 hrHPV-positive (either HPV16-positive or -negative), HPV33/58-negative cases. Therefore, 59/292 = 20.21% of cases with HPV56/59/66 positivity subsequently developed CIN1−. Among these 59 cases, 51 were indeed diagnosed as CIN1− within 2 years of follow-up, showing this classification is fairly accurate. Thus, the relative risk of these genotype combinations was low (4.32), much lower than those of the pooled hrHPV test covering 14 genotypes (see Table S2 in the supplemental material).
The interpretation of HPV52 was more complex. Among those cases that are hrHPV positive, HPV16 positive, HPV33/58 negative, and HPV56/59/66 negative, HPV52 positivity signified CIN1− classification. However, among hrHPV-positive, HPV16-negative, and HPV33/58-positive cases, HPV52 positivity prompted CIN2/3+ classification. It therefore appears that the interpretation of HPV52 results will depend on the findings for other hrHPV genotypes. However, it should be noted that samples with those genotype combinations were quite rare, and the classifications were not very accurate, as reflected by their similar RR (Table S2).
In addition to the decision tree presented in Fig. 1B, a number of classifiers based on different models were built in order to further explore the potential of the machine learning interpretation of HPV genotyping data, notably a random forest-based classifier which, in effect, classifies using many decision trees. However, at the moment the classifiers built are imperfect, and higher sensitivity is desirable. Furthermore, there was bias in selection of samples, since biopsy and cytology follow-up data originated from retrospective analysis of cytology reports, and prospective study was not involved. However, this study serves to illustrate the potential of utilizing HPV extended genotyping data to stratify risk of subsequent detection of biopsy specimen-proven CIN2/3+. A bigger data set is necessary to build more accurate classifiers. Once available, a classifier can be easily implemented as a web application on which clinical personnel can input a subject’s HPV genotyping test results and derive a prediction of the subject’s outcome. Such information potentially can facilitate prioritizing appointments for colposcopy for effective management of patients in need.
Supplementary Material
ACKNOWLEDGMENTS
The Onclarity HPV test and the Viper LT platform were provided by BD, Ltd., Hong Kong. BD, Ltd., played no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Warrington Hsu for advice on the machine learning analyses.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00997-19.
REFERENCES
- 1.Wright TC, Stoler MH, Behrens CM, Sharma A, Zhang G, Wright TL. 2015. Primary cervical cancer screening with human papillomavirus: end of study results from the ATHENA study using HPV as the first-line screening test. Gynecol Oncol 136:189–197. doi: 10.1016/j.ygyno.2014.11.076. [DOI] [PubMed] [Google Scholar]
- 2.Malagon T, Kulasingam S, Mayrand MH, Ogilvie G, Smith L, Bouchard C, Gotlieb W, Franco EL. 2018. Age at last screening and remaining lifetime risk of cervical cancer in older, unvaccinated, HPV-negative women: a modelling study. Lancet Oncol 19:1569–1578. doi: 10.1016/S1470-2045(18)30536-9. [DOI] [PubMed] [Google Scholar]
- 3.Melnikow J, Henderson JT, Burda BU, Senger CA, Durbin S, Weyrich MS. 2018. Screening for cervical cancer with high-risk human papillomavirus testing: updated evidence report and systematic review for the US Preventive Services Task Force. JAMA 320:687–705. doi: 10.1001/jama.2018.10400. [DOI] [PubMed] [Google Scholar]
- 4.Ogilvie GS, van Niekerk D, Krajden M, Smith LW, Cook D, Gondara L, Ceballos K, Quinlan D, Lee M, Martin RE, Gentile L, Peacock S, Stuart GCE, Franco EL, Coldman AJ. 2018. Effect of screening with primary cervical HPV testing vs cytology testing on high-grade cervical intraepithelial neoplasia at 48 months: the HPV FOCAL Randomized Clinical Trial. JAMA 320:43–52. doi: 10.1001/jama.2018.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control Prevention. 2012. Human papillomavirus-associated cancers–United States, 2004–2008. MMWR Morb Mortal Wkly Rep 61:258–261. [PubMed] [Google Scholar]
- 6.Castle PE, Stoler MH, Wright TC Jr, Sharma A, Wright TL, Behrens CM. 2011. Performance of carcinogenic human papillomavirus (HPV) testing and HPV16 or HPV18 genotyping for cervical cancer screening of women aged 25 years and older: a subanalysis of the ATHENA study. Lancet Oncol 12:880–890. doi: 10.1016/S1470-2045(11)70188-7. [DOI] [PubMed] [Google Scholar]
- 7.The Hong Kong College of Obstetricians and Gynaecologists. 2016. Guidelines for Cervical Cancer Prevention and Screening. The Hong Kong College of Obstetricians and Gynaecologists, Hong Kong, China. [Google Scholar]
- 8.Wong OG, Lo CK, Szeto E, Cheung AN. 2011. Efficacy of Abbott RealTime high risk HPV test in evaluation of atypical squamous cells of undetermined significance from an Asian screening population. J Clin Virol 51:136–138. doi: 10.1016/j.jcv.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Wong OG, Lo CK, Chow JN, Tsun OK, Szeto E, Liu SS, Ngan HY, Cheung AN. 2012. Comparison of the GenoFlow human papillomavirus (HPV) test and the Linear Array assay for HPV screening in an Asian population. J Clin Microbiol 50:1691–1697. doi: 10.1128/JCM.05933-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaturvedi AK, Katki HA, Hildesheim A, Rodriguez AC, Quint W, Schiffman M, Van Doorn LJ, Porras C, Wacholder S, Gonzalez P, Sherman ME, Herrero R, CVT Group. 2011. Human papillomavirus infection with multiple types: pattern of coinfection and risk of cervical disease. J Infect Dis 203:910–920. doi: 10.1093/infdis/jiq139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrillo-García A, Ponce-de-León-Rosales S, Cantú-de-León D, Fragoso-Ontiveros V, Martínez-Ramírez I, Orozco-Colín A, Mohar A, Lizano M. 2014. Impact of human papillomavirus coinfections on the risk of high-grade squamous intraepithelial lesion and cervical cancer. Gynecol Oncol 134:534–539. doi: 10.1016/j.ygyno.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 12.Nogueira Dias Genta ML, Martins TR, Mendoza Lopez RV, Sadalla JC, de Carvalho JPM, Baracat EC, Levi JE, Carvalho JP. 2017. Multiple HPV genotype infection impact on invasive cervical cancer presentation and survival. PLoS One 12:e0182854. doi: 10.1371/journal.pone.0182854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salazar KL, Duhon DJ, Olsen R, Thrall M. 2019. A review of the FDA-approved molecular testing platforms for human papillomavirus. J Am Soc Cytopathol 8:284–292. doi: 10.1016/j.jasc.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Monsonego J, Cox JT, Behrens C, Sandri M, Franco EL, Yap PS, Huh W. 2015. Prevalence of high-risk human papilloma virus genotypes and associated risk of cervical precancerous lesions in a large U.S. screening population: data from the ATHENA trial. Gynecol Oncol 137:47–54. doi: 10.1016/j.ygyno.2015.01.551. [DOI] [PubMed] [Google Scholar]
- 15.Schiffman M, Burk RD, Boyle S, Raine-Bennett T, Katki HA, Gage JC, Wentzensen N, Kornegay JR, Aldrich C, Tam T, Erlich H, Apple R, Befano B, Castle PE. 2015. A study of genotyping for management of human papillomavirus-positive, cytology-negative cervical screening results. J Clin Microbiol 53:52–59. doi: 10.1128/JCM.02116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agrawal R, Srikant R. 1994. Fast algorithms for mining association rules in large databases, abstr 672836. Abstr Proc 20th Int Conf Very Large Data Bases Morgan Kaufmann Publishers Inc, Burlington, MA. [Google Scholar]
- 17.Bettina G, Michael H, Kurt H. 2005. arules–a computational environment for mining association rules and frequent item sets. J Stat Software 14:1–25. doi: 10.18637/jss.v014.i15. [DOI] [Google Scholar]
- 18.Altman DG. 1991. Practical statistics for medical research. Chapman and Hall, London, United Kingdom. [Google Scholar]
- 19.Cuzick J, Ahmad AS, Austin J, Cadman L, Ho L, Terry G, Kleeman M, Ashdown-Barr L, Lyons D, Stoler M, Szarewski A. 2016. A comparison of different human papillomavirus tests in PreservCyt versus SurePath in a referral population-PREDICTORS 4. J Clin Virol 82:145–151. doi: 10.1016/j.jcv.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong OGW, Tsun OKL, Tsui EY, Chow JNK, Ip PPC, Cheung A. 2018. HPV genotyping and E6/E7 transcript assays for cervical lesion detection in an Asian screening population-Cobas and Aptima HPV tests. J Clin Virol 109:13–18. doi: 10.1016/j.jcv.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Wright TC Jr, Stoler MH, Agreda PM, Beitman GH, Gutierrez EC, Harris JM, Koch KR, Kuebler M, LaViers WD, Legendre BL Jr, Leitch SV, Maus CE, McMillian RA, Nussbaumer WA, Palmer ML, Porter MJ, Richart GA, Schwab RJ, Vaughan LM. 2014. Clinical performance of the BD Onclarity HPV assay using an adjudicated cohort of BD SurePath liquid-based cytology specimens. Am J Clin Pathol 142:43–50. doi: 10.1309/AJCP53KMHNRDICBL. [DOI] [PubMed] [Google Scholar]
- 22.Hahsler M. 2015. A probabilistic comparison of commonly used interest measures for association rules. http://michael.hahsler.net/research/association_rules/measures.html. Accessed 1 August 2019.
- 23.Brin S, Motwani R, Ullman JD, Tsur S. 1997. Dynamic itemset counting and implication rules for market basket data. SIGMOD Rec 26:255–264. doi: 10.1145/253262.253325. [DOI] [Google Scholar]
- 24.Ochin S, Kumar N, Joshi N. 2016. Rule power factor: a new interest measure in associative classification. Procedia Computer Sci 93:12–18. doi: 10.1016/j.procs.2016.07.175. [DOI] [Google Scholar]
- 25.Kwan TT, Cheung AN, Lo SS, Lee PW, Tam KF, Chan KK, Ngan HY. 2011. Psychological burden of testing positive for high-risk human papillomavirus on women with atypical cervical cytology: a prospective study. Acta Obstet Gynecol Scand 90:445–451. doi: 10.1111/j.1600-0412.2011.01092.x. [DOI] [PubMed] [Google Scholar]
- 26.Okadome M, Saito T, Tanaka H, Nogawa T, Furuta R, Watanabe K, Kita T, Yamamoto K, Mikami M, Takizawa K, Japanese Gynecologic Oncology Group. 2014. Potential impact of combined high- and low-risk human papillomavirus infection on the progression of cervical intraepithelial neoplasia 2. J Obstet Gynaecol Res 40:561–569. doi: 10.1111/jog.12202. [DOI] [PubMed] [Google Scholar]
- 27.Andersson E, Karrberg C, Radberg T, Blomqvist L, Zetterqvist BM, Ryd W, Lindh M, Horal P. 2012. Type-dependent E6/E7 mRNA expression of single and multiple high-risk human papillomavirus infections in cervical neoplasia. J Clin Virol 54:61–65. doi: 10.1016/j.jcv.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Liu SS, Leung RC, Chan KK, Cheung AN, Ngan HY. 2010. Evaluation of a newly developed GenoArray human papillomavirus (HPV) genotyping assay and comparison with the Roche Linear Array HPV genotyping assay. J Clin Microbiol 48:758–764. doi: 10.1128/JCM.00989-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong OG, Ho MW, Tsun OK, Ng AK, Tsui EY, Chow JN, Ip PP, Cheung AN. 2018. An automated quantitative DNA image cytometry system detects abnormal cells in cervical cytology with high sensitivity. Cytopathology 29:267–274. doi: 10.1111/cyt.12540. [DOI] [PubMed] [Google Scholar]
- 30.Chan PK, Li WH, Chan MY, Ma WL, Cheung JL, Cheng AF. 1999. High prevalence of human papillomavirus type 58 in Chinese women with cervical cancer and precancerous lesions. J Med Virol 59:232–238. doi:. [DOI] [PubMed] [Google Scholar]
- 31.Chan PK. 2012. Human papillomavirus type 58: the unique role in cervical cancers in East Asia. Cell Biosci 2:17. doi: 10.1186/2045-3701-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan PK, Zhang C, Park JS, Smith-McCune KK, Palefsky JM, Giovannelli L, Coutlee F, Hibbitts S, Konno R, Settheetham-Ishida W, Chu TY, Ferrera A, Alejandra Picconi M, De Marco F, Woo YL, Raiol T, Pina-Sanchez P, Bae JH, Wong MC, Chirenje MZ, Magure T, Moscicki AB, Fiander AN, Capra G, Young Ki E, Tan Y, Chen Z, Burk RD, Chan MC, Cheung TH, Pim D, Banks L. 2013. Geographical distribution and oncogenic risk association of human papillomavirus type 58 E6 and E7 sequence variations. Int J Cancer 132:2528–2536. doi: 10.1002/ijc.27932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tewari P, White C, Kelly L, Pilkington L, Keegan H, D'Arcy T, Toole SO, Sharp L, O'Leary JJ, Martin CM. 2018. Clinical performance of the Cobas 4800 HPV test and the Aptima HPV assay in the management of women referred to colposcopy with minor cytological abnormalities. Diagn Cytopathol 46:987–992. doi: 10.1002/dc.24066. [DOI] [PubMed] [Google Scholar]
- 34.Cook DA, Mei W, Smith LW, van Niekerk DJ, Ceballos K, Franco EL, Coldman AJ, Ogilvie GS, Krajden M. 2015. Comparison of the Roche Cobas 4800 and Digene Hybrid Capture 2 HPV tests for primary cervical cancer screening in the HPV FOCAL trial. BMC Cancer 15:968. doi: 10.1186/s12885-015-1959-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rebolj M, Preisler S, Ejegod DM, Rygaard C, Lynge E, Bonde J. 2014. Disagreement between human papillomavirus assays: an unexpected challenge for the choice of an assay in primary cervical screening. PLoS One 9:e86835. doi: 10.1371/journal.pone.0086835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


