Pneumocystis jirovecii pneumonia (PJP) is an important cause of pneumonia in the HIV-negative immunocompromised population, for whom the fungal load is low, the differential diagnosis is difficult, and a bronchoalveolar lavage (BAL) sample is often not readily available. Molecular techniques have improved the microbiological diagnosis in this scenario.
KEYWORDS: HIV negative, Pneumocystis jirovecii, immunocompromised hosts, molecular diagnosis, oral wash
ABSTRACT
Pneumocystis jirovecii pneumonia (PJP) is an important cause of pneumonia in the HIV-negative immunocompromised population, for whom the fungal load is low, the differential diagnosis is difficult, and a bronchoalveolar lavage (BAL) sample is often not readily available. Molecular techniques have improved the microbiological diagnosis in this scenario. The usefulness of two real-time PCR techniques targeting nuclear single-copy and mitochondrial multicopy genes, respectively, applied to oral wash specimens (OW) for PJP diagnosis was assessed, and its accuracy was compared to a BAL fluid-based diagnosis. Immunocompromised patients having PJP in the differential diagnosis of an acute respiratory episode, and from whom OW and BAL or lung biopsy specimens were obtained ≤48 h apart, were retrospectively included. PCRs targeting the dihydropteroate synthase gene (DHPS) and the mitochondrial small-subunit (mtSSU) rRNA gene were performed in paired OW-BAL specimens. Thirty-six patients were included (88.6% HIV negative). Fifteen patients (41.7%) were classified as PJP, and a further 8 were considered P. jirovecii colonized. Quantification of DHPS and mtSSU in BAL fluid showed an accuracy of 96.9% and 93.0%, respectively, for PJP diagnosis, whereas a qualitative approach performed better when applied to OW (accuracy, 91.7%) irrespective of the PCR target studied (kappa = 1). Qualitative molecular diagnosis applied to OW showed an excellent performance for PJP diagnosis regardless of the target studied, being easier to interpret than the quantitative approach needed for BAL fluid.
INTRODUCTION
After the introduction of highly active antiretroviral therapy and specific prophylaxis for patients at risk, the incidence of Pneumocystis jirovecii pneumonia (PJP) has declined in HIV-positive patients from industrialized countries (1, 2), whereas new groups of susceptible patients have emerged (i.e., immunocompromised patients with underlying conditions such as solid-organ or bone marrow transplantation, neoplasms, or autoimmune diseases) (2–5). In the latter, PJP is characterized by a rapidly evolving severe pneumonia with mortality rates up to 40% (6), and its diagnosis poses a challenge both to clinicians and to microbiologists. Signs and symptoms that overlap those of comorbidities frequently seen in these patients (i.e., pharmacological toxicity) make it difficult to establish an accurate clinical diagnosis (2–5). The poor condition of patients at presentation often impedes the immediate performance of a bronchoscopic procedure to collect adequate diagnostic specimens, and the induction of sputum is not always well tolerated. In addition, PJP in immunocompromised HIV-negative patients is typically associated with a low fungal burden, limiting the usefulness of microscopy techniques (6). From the diagnostic perspective, a technique able to provide good sensitivity and accuracy, along with applicability to noninvasively obtained respiratory specimens, would be seen as of great help.
Since their description in the early 1990s (7, 8), molecular assays have gained popularity, and they even have been claimed to be tools of choice for the diagnosis of PJP in standard laboratories (9, 10). Compared to microscopy, they show better sensitivity in settings where low fungal burdens are expected (11). Criticism, nevertheless, has arisen from their limited ability to discriminate infection and colonization, even if the fungal load is measured by means of quantitative real-time PCR (12, 13). Further handicaps of PCR-based techniques are derived from the lack of a universal protocol and from the spectrum of possible genetic targets (14–19).
The diagnostic yield of PCR techniques relies, at least in part, on the appropriate choice of a genetic target. Some targets are present in multiple copies, and even variable numbers of copies, per genome, hindering the interpretation of quantitative results. Others, in contrast, are present as single-copy genes, facilitating the interpretation of results at the expense of a potentially lower sensitivity (20, 21). Recently, the gene encoding the short subunit of the mitochondrial RNA (mtSSU) has been described as a region presenting multiple copies per genome but with reduced variability across physiological states compared to other mitochondrial targets (19), thus becoming a promising candidate for the accurate diagnosis of PJP by using a quantitative approach. Its performance, however, has not been compared to other genes present in a stable but more reduced number of copies per genome.
This study aimed to assess the usefulness of two PCR assays based on the mtSSU and a single-copy nuclear gene, the dihydropteroate synthase (DHPS) gene, for the diagnosis of PJP using two types of respiratory specimens, with particular emphasis on the evaluation of the practicality of oral washes (OWs) for the noninvasive diagnosis of P. jirovecii infection.
MATERIALS AND METHODS
Patients and samples.
A retrospective analysis was carried out between March 2016 and March 2018 at the Vall d’Hebron University Hospital, a 1,000-bed tertiary center in Barcelona, Spain. Immunocompromised patients presenting with signs and symptoms compatible with PJP, and from whom oral wash (OW) and bronchoalveolar lavage (BAL) specimens were obtained ≤48 h apart, were included. One additional patient had a lung biopsy sample with pathological findings compatible with PJP obtained 4 days after the OW. OW specimens were obtained by vigorous gargling of 10 ml of sterile saline for 30 s, whereas lower-airway specimens were collected according to standard bronchoscopy procedures.
Demographics and clinical data were registered, including age, sex, underlying disease, absolute lymphocyte count, immunosuppressive treatment, clinical and radiological features, anti-infective prescriptions (including the use of P. jirovecii-active drugs as prophylaxis or therapy), and outcome of the episode. Final PJP diagnosis was established by consensus between experienced clinical microbiologists and infectious disease practitioners taking into consideration clinical manifestations along with radiological and laboratory findings. In the case that P. jirovecii was detected by PCR but was not considered to be responsible for the acute respiratory episode (i.e., if a proven alternative cause was identified or the patient improved without specific anti-P. jirovecii therapy), patients were categorized as colonized.
Laboratory procedures.
For molecular diagnosis, DNA was extracted from aliquots of OW and BAL fluid using a QIAamp DNA minikit (Qiagen) according to the manufacturer’s instructions. The single-copy nuclear dihydropteroate synthase (DHPS) gene and the mitochondrial small-subunit (mtSSU) rRNA coding region were targeted according to previously described methodologies with minor modifications (19, 22). Reactions were carried out in a Smart Cycler thermal cycler (Cepheid, Sunnyvale, CA) (15 min at 95°C for polymerase activation and DNA denaturation, followed by 45 cycles of 15 s of denaturalization at 95°C and 1 min of annealing and extension at 60°C), in a final volume of 25 μl, with 0.4 μM (each) primer and 0.2 μM probe. Human RNase P target was used as inhibition control (Applied Biosystems, Texas).
An immunofluorescent (IF) staining able to detect cysts and trophic forms (Merifluor Pneumocystis; Meridien Bioscience Inc.) was additionally performed according to the manufacturer’s instructions if the available remnant volume of BAL fluid was ≥5 ml.
Statistical analysis.
PCR results were expressed as cycle threshold (CT) values (mean [95% confidence interval {95% CI}]). The chi-squared or Fisher exact test was used for categorical variables, while the Student t and Mann-Whitney tests were used for quantitative variables as appropriate. Sensitivity, specificity, and positive and negative predictive values (PPV and NPV, respectively) were calculated for each sample-target pair. The kappa index (k) was calculated to evaluate within-sample-type concordance for both targets. A receiver operator characteristic (ROC) curve was constructed to evaluate the diagnostic accuracy of quantitative data. Statistical analysis was performed with the Stata statistical package version 15.0 (StataCorp, College Station, TX, USA). The plot graph was built using GraphPad Prism 8.2.1.
Ethics.
Due to the retrospective nature of the study, requirement for informed consent was waived. The study was approved by the ethical committee of Vall d’Hebron University Hospital [protocol number PR(AG)450/2018].
RESULTS
Thirty-six patients were included in the study; their baseline characteristics and clinical data are shown in Table 1. All but 5 (88.6%) were HIV-negative immunocompromised subjects. The acute respiratory episode was attributed to a P. jirovecii infection in 15 cases (41.7%). An alternative diagnosis was found in the other 21 patients, of whom 8 (22.2% of total) were classified as P. jirovecii colonized.
TABLE 1.
Baseline pathology and clinical characteristics of patients enrolled in the study
| Characteristic | Total (n = 36) | PJP (n = 15) | Other etiologies (n = 21) | P value |
|---|---|---|---|---|
| Mean age, yr (SD) | 60.3 (11.3) | 60.5 (10.4) | 60.1 (12.1) | 0.933 |
| Sex, no. (%) of patients | 0.955 | |||
| Male | 19 (52.8) | 8 (53.3) | 11 (52.4) | |
| Female | 17 (47.2) | 7 (46.7) | 10 (47.6) | |
| Cause of immunosuppression, no. (%) of patients | 0.206 | |||
| Solid-organ malignancies | 11 (30.5) | 6 (40.0) | 5 (23.8) | |
| Hematological malignancies | 10 (27.8) | 3 (20.0) | 7 (33.4) | |
| Bone marrow transplant recipients | 5 (13.9) | 0 (0.0) | 5 (23.8) | |
| HIV | 5 (13.9) | 3 (20.0) | 2 (9.5) | |
| Solid-organ transplant recipients | 3 (8.3) | 1 (6.7) | 2 (9.5) | |
| Autoimmune disease | 1 (2.8) | 1 (6.7) | 0 (0.0) | |
| HIV and solid-organ transplant recipient | 1 (2.8) | 1 (6.6) | 0 (0.0) | |
| Fever, no. (%) of patients | 35 (97.2) | 15 (100.0) | 20 (95.2) | 0.391 |
| Dyspnea, no. (%) of patients | 32 (88.9) | 14 (93.3) | 18 (85.7) | 0.473 |
| Oxygen saturation on room air, % (SD) | 91.2 (5.7) | 91.8 (7.3) | 90.8 (4.4) | 0.609 |
| Interstitial or ground glass radiological pattern, no. (%) of patients | 26 (72.2) | 13 (86.7) | 13 (62.0) | 0.102 |
| Lymphocyte blood count, cells/mm3 (SD) | 911.1 (596.7) | 718.7 (311.8) | 1,048.4 (712.4) | 0.103 |
| Use of previous systemic glucocorticoid therapy, no. (%) of patients | 16 (44.4) | 6 (40) | 10 (47.6) | 0.650 |
OW specimens were obtained on average 1 day (95% CI, 0.71 to 1.30) before the bronchoscopy. None of the patients had been receiving P. jirovecii prophylaxis during the previous 6 months. Pneumocystis-targeted treatment was administered for a mean of 0.75 days (95% CI, 0.21 to 1.29) before OW sampling (11 patients) and 1.63 days (95% CI, 0.97 to 2.30) before bronchoscopy (28 patients). As shown in Table S1 in the supplemental material, these prior short courses of P. jirovecii-active therapy had little or no effect on PCR results in this early stage of disease.
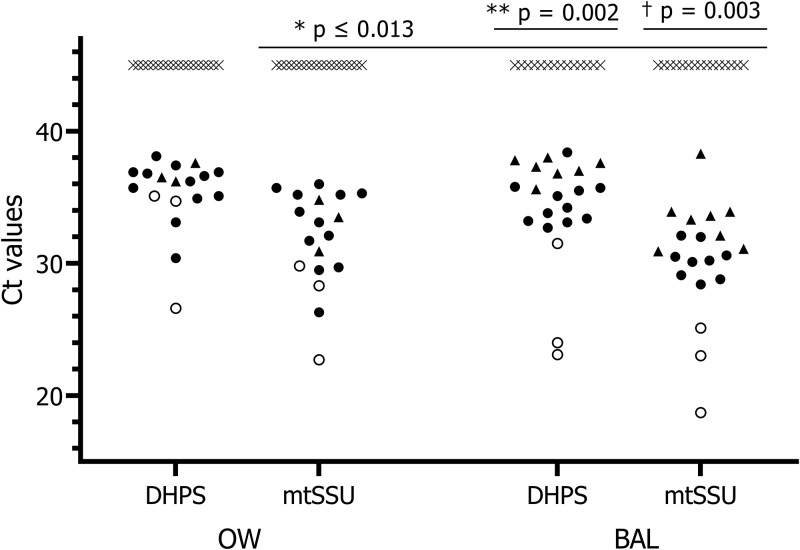
Performance of both PCR techniques is shown in Table 2. Regarding OW specimens, positive results were obtained for both targets in all PJP cases, rendering a sensitivity of 100% for the diagnosis of pneumonia and an excellent concordance (k = 1). Specificity, in contrast, dropped to 84.2% as 3 colonized patients were also OW positive. As depicted in Fig. 1, a huge overlap was noted in the overall load of colonized and pneumonia cases irrespective of the gene targeted (DHPS, colonized 36.8 [35.9 to 37.6] versus PJP 35.6 [34.5 to 36.9], P = 0.407; mtSSU, colonized 33.1 [30.8 to 35.3] versus PJP 32.8 [31.01 to 34.5], P = 0.893) and the HIV status (DHPS, HIV positive 32.1 [26.7 to 37.6] versus HIV negative, 35.7 [34.5 to 36.9], P = 0.064; mtSSU, HIV positive 26.9 [22.7 to 31.2] versus HIV negative 32.8 [31.1 to 34.5], P = 0.013), hampering the establishment of a reliable cutoff. A qualitative PCR result, however, allowed the correct labeling of PJP patients with a 91.7% accuracy irrespective of PCR target.
TABLE 2.
Summary of assay performance of PCR in upper and lower respiratory tract samples for the diagnosis of P. jirovecii pneumoniaa
| Specimen and target | Mean fungal load, CT (95% CI) |
Interpretive criterion | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | |
|---|---|---|---|---|---|---|---|---|
| Colonization | Pneumonia | |||||||
| OW | Pos/Neg | 100 | 85.7 | 83.3 | 100 | 91.7 | ||
| DHPS | 36.8 (35.9–37.6) | 35.6 (34.5–36.9) | ||||||
| mtSSU | 33.1 (30.8–35.3) | 32.8 (31.1–34.5) | ||||||
| BAL fluid | ||||||||
| DHPS | 37.2 (36.6–38.4) | 32.8 (30.6–35.1) | CT 36.8 | 95.2 | 92.9 | 95.2 | 92.9 | 96.9 |
| mtSSU | 33.4 (31.8–35) | 29.1 (26.4–31.9) | CT 30.9 | 100 | 76.9 | 87.5 | 100 | 93.0 |
Abbreviations: PPV, positive predictive value; NPV, negative predictive value; Pos/Neg, positive/negative.
FIG 1.
Fungal load distribution across groups of patients by specimens, assay, and P. jirovecii status. PCR results are shown as follows: white circles, PJP HIV patients; black circles, PJP HIV-negative patients; black triangles, colonized patients; crosses, negative patients. *, fungal load in PJP HIV-positive versus HIV-negative patients; **, fungal load of colonized versus PJP-HIV-negative patients; †, fungal load of colonized versus PJP HIV-negative patients.
BAL specimens obtained from PJP cases were positive at a mean (95% CI) CT of 32.8 (30.6 to 35.1) and 29.1 (26.4 to 31.9) for DHPS and mtSSU, respectively. As seen in OW specimens, larger amounts of fungi were detected in specimens from HIV-positive patients (26.2 [21.0 to 31.4] for DHPS; 22.3 [18.6 to 26.0] for mtSSU) than from HIV-negative subjects (DHPS, 34.6 [33.6 to 35.6]; mtSSU, 31.2 [29.1 to 33.3]) (P ≤ 0.002 and ≤ 0.0055, respectively). Regardless of the HIV status, the ROC curve analysis for BAL fluid (Fig. 2) indicated that a single cutoff of 36.8 for DHPS and 30.9 for mtSSU could distinguish between PJP and non-PJP with an accuracy of 96.9% and 93.0%, respectively (P = 0.525). The fungal load detected in BAL fluid from colonized patients having a positive OW result was not significantly higher than that in OW-negative patients (P = 0.229 and 0.281 for DHPS and mtSSU, respectively). Concordance between techniques was good (k = 0.879).
FIG 2.
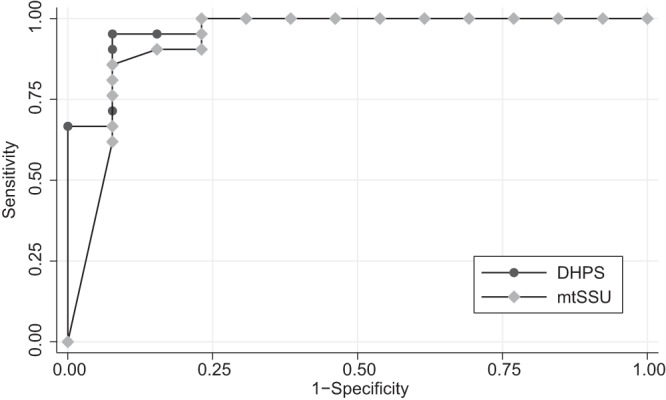
ROC curve analysis of PCR assay performance for PJP diagnosis on BAL specimens.
All the patients classified as PJP on the basis of BAL fluid fungal load were also categorized as pneumonia cases using the qualitative approach in OW irrespective of the gene targeted.
Microscopy allowed the identification of only 48% of PJP cases, with lower sensitivity in HIV-negative than in HIV-positive patients (4/9, 44.4%, versus 3/3, 100%, respectively; P = 0.205).
DISCUSSION
In this retrospective study, the usefulness of two PCR assays in invasive and noninvasive respiratory specimens, as well as the suitability of OW for the diagnosis of PJP, was assessed. Regardless of the Pneumocystis target used on OW, a very good output was found, with 100% sensitivity and negative predictive value, and a 91.7% accuracy for the diagnosis of pneumonia cases, making the use of a noninvasive specimen reliable for the diagnosis of PJP irrespective of the HIV status of the patient.
Two genetic targets showing little variability in the number of copies per genome were chosen (i.e., the 1-copy/genome nuclear gene coding for the dihydropteroate synthase, and a mean of 37 copies/genome for the gene that codes for the short subunit of mitochondrial RNA) (19, 22). The reported stability in the number of copies/genome of targets resulted in a similar performance of PCR techniques in upper- and lower-airway specimens. This limited variability may have been at the root of the reduced overlap found in the lower-airway fungal load of colonized and infected patients, in contrast to published results for targets such as the DNA encoding the large subunit of the mitochondrial RNA (23).
Despite the scant information available on the usefulness of upper respiratory specimens for the diagnosis of PJP in immunocompromised HIV-negative patients, our results are consistent with others (21, 24–28). To et al. (21) reported similar accuracy values in diagnosing PJP in a mixed group of HIV-positive and -negative immunocompromised patients using a multicopy target (the mitochondrial large subunit) on nasopharyngeal specimens. Two PCRs targeting single- and multiple-copy genes (DHPS and the major surface glycoprotein [MSG]) applied to OW were evaluated by Juliano et al. (27) in a cohort of HIV-positive patients. Compared to our findings, they reported poorer individual performance for each individual target (sensitivity, >80%, and specificity, <70%), whereas the diagnostic accuracy was similar to ours when the results of the two PCRs were combined (sensitivity, 100%; specificity, 74%). In our study, excellent concordance between targets in OW specimens allowed the identification of PJP cases with a >90% accuracy regardless of the gene targeted, thus simplifying the diagnosis.
In the case of BAL fluid, our results also compare to data reported from other authors despite differences in the targeted genes (14, 18, 23, 29). A striking difference with respect to OW is that a quantitative approach was needed to differentiate infection and colonization, underscoring the need for an accurate validation of the technique. According to the experience of Fauchier et al. (29) and Louis et al. (30), the yield of both PCRs declined when HIV patients were excluded from the analysis. Nonetheless, the area under the curve (AUC) in the ROC analysis remained above 0.9, supporting their good diagnostic performance in specimens from the lower respiratory tract. Again, the lack of statistically significant difference in the AUCs for DHPS and mtSSU supported the idea of comparable accuracy of the two genes for the microbiological diagnosis of PJP in specimens obtained by bronchoscopy. Historically, setting a cutoff for quantitative results has remained an elusive point due to the variable degree of superposition in the range of fungal load values found in colonized and infected patients (29). However, the results of this study show that a CT-based cutoff of 36.8 for DHPS and 30.9 for mtSSU could reasonably differentiate cases of infection without substantial overlap with values obtained from patients labeled as colonized. The use of CT values instead of the absolute number of copies could have contributed to define groups, as it works in a logarithmic scale.
When the amount of fungi is quantified in terms of absolute number of copies, initiation of treatment has been reported to result in a noticeable reduction of the infectious load (14). Again, the use of CT may be the reason for the lack of differences in fungal load found for specimens collected from naive subjects and from patients given a short course of P. jirovecii-active treatment.
OW specimens were not useful to detect the low fungal loads associated with P. jirovecii-colonized patients, as 5 out of 8 (62.5%) cases were missed. Although slightly higher fungal loads were detected in BAL samples of OW-positive colonized patients, this difference was not large enough to predict an increased risk of OW-positive testing. Thus, OW specimens were found to have a high negative predictive value only to rule out pneumonia. Contrary to what has been previously reported (16), using a target present in multiple copy number per genome did not increase the sensitivity of the test in OW compared to the DHPS, so no additional advantage was provided by the mtSSU in terms of detecting colonized patients at risk of developing pneumonia later on.
This study had several limitations. It was carried out in a single hospital, warranting a more extensive evaluation to extrapolate results to other centers. The number of patients was small, but they all had underlying factors well known to increase the risk of developing PJP. Some patients had received a short course of P. jirovecii-active treatment before sampling, although the results point toward a lack of relevant differences provided that the fungal load was quantified as CT and the treatment was given for no longer than 48 h.
In summary, we assessed the performance of two molecular techniques targeting two genes present as single and multiple copies in the P. jirovecii genome in OW samples in order to evaluate and compare their accuracy for PJP diagnosis in a population where at-risk non-HIV patients predominate over HIV patients. The results of this study suggest that the use of a PCR targeting a gene present in a nonvariable number of copies per genome in OW samples could be a practical approach to the molecular biology-based diagnosis of PJP. A qualitative result obtained in OW seems to offer a reasonably good accuracy for this purpose, thus avoiding the need of interpreting the amount of fungal load, simplifying the diagnostic process. The use of OW can be proposed as a first step for differential diagnosis of acute respiratory distress in patients at risk of developing PJP due to OW being quicker and easier to obtain, presenting a good PJP diagnostic performance, and permitting the establishment of an earlier adequate treatment. When the patient’s status allows, bronchoscopy must be performed since BAL fluid is the appropriate specimen to investigate other etiologies for the clinical episode and to detect P. jirovecii-colonized patients, who are at risk of developing PJP.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andres Anton, M. Antonia Casas, and M. Francisca Madrid for their excellent technical assistance.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01287-19.
REFERENCES
- 1.Maini R, Henderson KL, Sheridan EA, Lamagni T, Nichols G, Delpech V, Phin N. 2013. Increasing Pneumocystis pneumonia, England, UK, 2000–2010. Emerg Infect Dis 19:386–392. doi: 10.3201/eid1903.121151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roux A, Canet E, Valade S, Gangneux-Robert F, Hamane S, Lafabrie A, Maubon D, Debourgogne A, Gal S, Le Dalle F, Leterrier M, Toubas D, Pomares C, Bellanger AP, Bonhomme J, Berry A, Durand-Joly I, Magne D, Pons D, Hennequin C, Maury E, Roux P, Azoulay É. 2014. Pneumocystis jirovecii pneumonia in patients with or without AIDS, France. Emerg Infect Dis 20:1490–1497. doi: 10.3201/eid2009.131668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bienvenu AL, Traore K, Plekhanova I, Bouchrik M, Bossard C, Picot S. 2016. Pneumocystis pneumonia suspected cases in 604 non-HIV and HIV patients. Int J Infect Dis 46:11–17. doi: 10.1016/j.ijid.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 4.Avino L, Roecker A. 2016. A review of Pneumocystis jirovecii pneumonia in the non-HIV-infected population. Ann Pharmacother 50:673–679. doi: 10.1177/1060028016650107. [DOI] [PubMed] [Google Scholar]
- 5.Fily F, Lachkar S, Thiberville L, Favennec L, Caron F. 2011. Pneumocystis jirovecii colonization and infection among non HIV-infected patients. Med Mal Infect 41:526–531. doi: 10.1016/j.medmal.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Salzer HJF, Schäfer G, Hoenigl M, Günther G, Hoffmann C, Kalsdorf B, Alanio A, Lange C. 2018. Clinical, diagnostic, and treatment disparities between HIV-infected and non-HIV-infected immunocompromised patients with Pneumocystis jirovecii pneumonia. Respiration 96:52–65. doi: 10.1159/000487713. [DOI] [PubMed] [Google Scholar]
- 7.Kitada K, Oka S, Kimura S, Shimada K, Serikawa T, Yamada J, Tsunoo H, Egawa K, Nakamura Y. 1991. Detection of Pneumocystis carinii sequences by polymerase chain reaction: animal models and clinical application to noninvasive specimens. J Clin Microbiol 29:1985–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schluger N, Sepkowitz K, Armstrong D, Bernard E, Rifkin M, Cerami A, Bucala R. 1991. Detection of Pneumocystis carinii in serum of AIDS patients with Pneumocystis pneumonia by the polymerase chain reaction. J Protozool 38:240S–242S. [PubMed] [Google Scholar]
- 9.Doyle L, Vogel S, Procop GW. 2017. Pneumocystis PCR: it is time to make PCR the test of choice. Open Forum Infect Dis 4:ofx193. doi: 10.1093/ofid/ofx193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alanio A, Hauser PM, Lagrou K, Melchers WJG, Helweg-Larsen J, Matos O, Cesaro S, Maschmeyer G, Einsele H, Donnelly JP, Cordonnier C, Maertens J, Bretagne S. 2016. ECIL guidelines for the diagnosis of Pneumocystis jirovecii pneumonia in patients with haematological malignancies and stem cell transplant recipients. J Antimicrob Chemother 71:2386–2396. doi: 10.1093/jac/dkw156. [DOI] [PubMed] [Google Scholar]
- 11.Robert-Gangneux F, Belaz S, Revest M, Tattevin P, Jouneau S, Decaux O, Chevrier S, Le Tulzo Y, Gangneux J-P. 2014. Diagnosis of Pneumocystis jirovecii pneumonia in immunocompromised patients by real-time PCR: a 4-year prospective study. J Clin Microbiol 52:3370–3376. doi: 10.1128/JCM.01480-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durand-Joly I, Chabe M, Soula F, Delhaes L, Camus D, Dei-Cas E. 2005. Molecular diagnosis of Pneumocystis pneumonia. FEMS Immunol Med Microbiol 45:405–410. doi: 10.1016/j.femsim.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Jarboui MA, Sellami A, Sellami H, Cheikhrouhou F, Makni F, Ben Arab N, Ben Jemaa M, Ayadi A. 2010. Molecular diagnosis of Pneumocystis jiroveci pneumonia in immunocompromised patients. Mycoses 53:329–333. doi: 10.1111/j.1439-0507.2009.01715.x. [DOI] [PubMed] [Google Scholar]
- 14.Alanio A, Desoubeaux G, Sarfati C, Hamane S, Bergeron A, Azoulay E, Molina JM, Derouin F, Menotti J. 2011. Real-time PCR assay-based strategy for differentiation between active Pneumocystis jirovecii pneumonia and colonization in immunocompromised patients. Clin Microbiol Infect 17:1531–1537. doi: 10.1111/j.1469-0691.2010.03400.x. [DOI] [PubMed] [Google Scholar]
- 15.Tamburrini E, Mencarini P, Visconti E, Zolfo M, Marinaci S, Zinzi D, Margutti P, Ortona E, Siracusano A. 1998. IV. Potential impact of Pneumocystis genetic diversity on the molecular detection of the parasite in human host. FEMS Immunol Med Microbiol 22:37–49. doi: 10.1111/j.1574-695X.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 16.Sasso M, Chastang-Dumas E, Bastide S, Alonso S, Lechiche C, Bourgeois N, Lachaud L. 2016. Performances of four real-time PCR assays for diagnosis of Pneumocystis jirovecii pneumonia. J Clin Microbiol 54:625–630. doi: 10.1128/JCM.02876-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tia T, Putaporntip C, Kosuwin R, Kongpolprom N, Kawkitinarong K, Jongwutiwes S. 2012. A highly sensitive novel PCR assay for detection of Pneumocystis jirovecii DNA in bronchoalveolar lavage specimens from immunocompromised patients. Clin Microbiol Infect 18:598–603. doi: 10.1111/j.1469-0691.2011.03656.x. [DOI] [PubMed] [Google Scholar]
- 18.Fillaux J, Malvy S, Alvarez M, Fabre R, Cassaing S, Marchou B, Linas M, Berry A. 2008. Accuracy of a routine real-time PCR assay for the diagnosis of Pneumocystis jirovecii pneumonia. J Microbiol Methods 75:258–261. doi: 10.1016/j.mimet.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Valero C, Buitrago MJ, Gits-Muselli M, Benazra M, Sturny-Leclère A, Hamane S, Guigue N, Bretagne S, Alanio A. 2016. Copy number variation of mitochondrial DNA genes in Pneumocystis jirovecii according to the fungal load in BAL specimens. Front Microbiol 7:1413. doi: 10.3389/fmicb.2016.01413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linssen CFM, Jacobs JA, Beckers P, Templeton KE, Bakkers J, Kuijper EJ, Melchers WJG, Drent M, Vink C. 2006. Inter-laboratory comparison of three different real-time PCR assays for the detection of Pneumocystis jiroveci in bronchoalveolar lavage fluid samples. J Med Microbiol 55:1229–1235. doi: 10.1099/jmm.0.46552-0. [DOI] [PubMed] [Google Scholar]
- 21.To KKW, Wong SCY, Xu T, Poon RWS, Mok KY, Chan JFW, Cheng VCC, Chan KH, Hung IFN, Yuen KY. 2013. Use of nasopharyngeal aspirate for diagnosis of pneumocystis pneumonia. J Clin Microbiol 51:1570–1574. doi: 10.1128/JCM.03264-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alvarez-Martínez MJ, Miró JM, Valls ME, Moreno A, Rivas PV, Solé M, Benito N, Domingo P, Muñoz C, Rivera E, Zar HJ, Wissmann G, Diehl ARS, Prolla JC, de Anta MTJ, Gatell JM, Wilson PE, Meshnick SR. 2006. Sensitivity and specificity of nested and real-time PCR for the detection of Pneumocystis jiroveci in clinical specimens. Diagn Microbiol Infect Dis 56:153–160. doi: 10.1016/j.diagmicrobio.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Montesinos I, Brancart F, Schepers K, Jacobs F, Denis O, Delforge M-L. 2015. Comparison of 2 real-time PCR assays for diagnosis of Pneumocystis jirovecii pneumonia in human immunodeficiency virus (HIV) and non-HIV immunocompromised patients. Diagn Microbiol Infect Dis 82:143–147. doi: 10.1016/j.diagmicrobio.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Hviid CJ, Lund M, Sørensen A, Ellermann-Eriksen SE, Jespersen B, Dam MY, Dahlerup JF, Benfield T, Jespersen S, Østergaard LJ, Laursen AL. 2017. Detection of Pneumocystis jirovecii in oral wash from immunosuppressed patients as a diagnostic tool. PLoS One 12:e0174012. doi: 10.1371/journal.pone.0174012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guigue N, Alanio A, Menotti J, De Castro N, Hamane S, Peyrony O, LeGoff J, Bretagne S. 2015. Utility of adding Pneumocystis jirovecii DNA detection in nasopharyngeal aspirates in immunocompromised adult patients with febrile pneumonia. Med Mycol 53:241–247. doi: 10.1093/mmy/myu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samuel CM, Whitelaw A, Corcoran C, Morrow B, Hsiao NY, Zampoli M, Zar HJ. 2011. Improved detection of Pneumocystis jirovecii in upper and lower respiratory tract specimens from children with suspected pneumocystis pneumonia using real-time PCR: a prospective study. BMC Infect Dis 11:329. doi: 10.1186/1471-2334-11-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juliano JJ, Barnett E, Parobek CM, Taylor SM, Meshnick SR, Stone S, Chang E, Fong S, Huang L. 2015. Use of oropharyngeal washes to diagnose and genotype Pneumocystis jirovecii. Open Forum Infect Dis 2:ofv080. doi: 10.1093/ofid/ofv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen HH, Huang L, Kovacs JA, Crothers K, Silcott VA, Morris A, Turner JR, Beard CB, Masur H, Fischer SH. 2004. A prospective, blinded study of quantitative touch-down polymerase chain reaction using oral-wash samples for diagnosis of Pneumocystis pneumonia in HIV-infected patients. J Infect Dis 189:1679–1683. doi: 10.1086/383322. [DOI] [PubMed] [Google Scholar]
- 29.Fauchier T, Hasseine L, Gari-Toussaint M, Casanova V, Marty PM, Pomares C. 2016. Detection of Pneumocystis jirovecii by quantitative PCR to differentiate colonization and pneumonia in immunocompromised HIV-positive and HIV-negative patients. J Clin Microbiol 54:1487–1495. doi: 10.1128/JCM.03174-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louis M, Guitard J, Jodar M, Ancelle T, Magne D, Lascols O, Hennequin C. 2015. Impact of HIV infection status on interpretation of quantitative PCR for detection of Pneumocystis jirovecii. J Clin Microbiol 53:3870–3875. doi: 10.1128/JCM.02072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.