Single multiplexed assays could replace the standard 2-tiered (STT) algorithm recommended for the laboratory diagnosis of Lyme disease if they perform with a specificity and a sensitivity superior or equal to those of the STT algorithm.
KEYWORDS: Lyme disease, serodiagnostic, serology, microfluidics, point of care, Borrelia burgdorferi, early Lyme, late Lyme, mChip-Ld
ABSTRACT
Single multiplexed assays could replace the standard 2-tiered (STT) algorithm recommended for the laboratory diagnosis of Lyme disease if they perform with a specificity and a sensitivity superior or equal to those of the STT algorithm. We used human serum rigorously characterized to be sera from patients with acute- and convalescent-phase early Lyme disease, Lyme arthritis, and posttreatment Lyme disease syndrome, as well as the necessary controls (n = 241 samples), to select the best of 12 Borrelia burgdorferi proteins to improve our microfluidic assay (mChip-Ld). We then evaluated its serodiagnostic performance in comparison to that of a first-tier enzyme immunoassay and the STT algorithm. We observed that more antigens became positive as Lyme disease progressed from early to late stages. We selected three antigens (3Ag) to include in the mChip-Ld: VlsE and a proprietary synthetic 33-mer peptide (PepVF) to capture sensitivity in all disease stages and OspC for early Lyme disease. With the specificity set at 95%, the sensitivity of the mChip-Ld with 3Ag ranged from 80% (95% confidence interval [CI], 56% to 94%) and 85% (95% CI, 74% to 96%) for two panels of serum from patients with early Lyme disease and was 100% (95% CI, 83% to 100%) for serum from patients with Lyme arthritis; the STT algorithm detected early Lyme disease in the same two panels of serum from patients with early Lyme disease with a sensitivity of 48.5% and 75% and Lyme arthritis in serum from patients with Lyme arthritis with a sensitivity of 100%, and the specificity was 97.5% to 100%. The mChip-Ld platform outperformed the STT algorithm according to sensitivity. These results open the door for the development of a single, rapid, multiplexed diagnostic test for point-of-care use that can be designed to identify the Lyme disease stage.
INTRODUCTION
Lyme disease (LD), caused by Borrelia burgdorferi and transmitted by the bite of infected Ixodes ticks, is the most common vector-borne disease in the United States (1), with an estimated incidence of ∼300,000 cases per year (2, 3). Lyme disease typically begins with erythema migrans (EM), an expanding skin lesion at the site of the tick bite. If left untreated, spirochetes may disseminate from the site and patients may present with neurologic, cardiac, and/or rheumatologic manifestations (4).
For the laboratory support of Lyme disease diagnosis, the Centers for Disease Control and Prevention (CDC) recommends a standard 2-tiered (STT) approach comprised of a first-tier enzyme immunoassay (EIA) that, if positive, should be followed by a second-tier IgM/IgG immunoblot assay (5). The immunoblot assay is interpreted using standardized criteria, and the IgM immunoblot assay results are used only for disease of ≤30 days’ duration. While the STT approach has worked relatively well when used as recommended, there is plenty of room for improvement. The STT approach requires a complex laboratory infrastructure to perform and has a low sensitivity during early infection, inter- and intralaboratory variability, a long turnaround time, and a high cost because of the high cost for the immunoblot assay. There is also confusion regarding interpretation of the immunoblot assay results (5). Over the last few decades, specific B. burgdorferi epitopes have been mapped. Because only a yes-or-no result is needed for routine cases of suspected Lyme disease, hope has been raised that the STT approach can ultimately be replaced by a single test without the immunoblot assay.
Assays that improve upon the performance of current tests would be most helpful for the laboratory support of Lyme disease diagnosis. While next-generation diagnostic tests are suggested to be at hand (5–7), there remains a need to demonstrate that known epitopes can adequately match the sensitivity and specificity of STT or whether further comprehensive exploration of epitopes is required. Most importantly, it has not been demonstrated that an effective single serodiagnostic test could be offered at the point of care. Rapid assays and point-of-care diagnostic testing could be used in some clinical settings, such as emergency rooms in areas of endemicity and doctors’ private practices (5, 8). Previously, we established a proof of principle for a new rapid test, the mChip-Ld assay, which was developed for point-of-care use (9). Here, we report on the performance of an improved mChip-Ld assay using panels of serum samples from patients rigorously characterized to have confirmed early Lyme disease or Lyme arthritis (a late Lyme disease manifestation) and control serum samples from healthy individuals and individuals with look-alike diseases.
MATERIALS AND METHODS
Ethics statement for human serum panels.
The involvement of human subjects falls under exemption 4, as outlined in HHS regulations (10). A total of 241 deidentified human serum samples (Table 1) were used. The Institutional Review Board (IRB) of IntegReview Inc. (Ethical Review Board Number 2) provided approval under approval number FWA00021769. Serum obtained from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), was collected with written informed consent under IRB-approved protocols. Serum panels from the New York State Department of Health (NYSDOH) were used for assay development under IRB approval number 03-037 of the New York State Department of Health. Serum obtained from the Lyme Disease Biobank (LDB) was collected with informed consent under Advarra IRB approval number Pro00012408.
TABLE 1.
Serum panels, clinical and laboratory definitions, and assay development stagea
| Provider (sample source and characteristics) | Clinical and laboratory definitions | No. of samples | EIA screening for 12 B. burgdorferi antigens | Comparative analysis of 3Ag-EIA vs 3Ag-mChip-Ld |
|---|---|---|---|---|
| NYSDOH (patients with suspected early Lyme disease, 2-tiered serology positive) | C6 and IgM/IgG blotting positive (acute phase, tick exposure, EM for <30 days) | 20 | X | |
| C6 and IgG blotting positive (convalescent phase, clinical signs for >30 days) | 39 | X | ||
| BR (healthy controls from areas of nonendemicity) | Healthy controls from areas of nonendemicity | 20 | X | |
| LDB (patients with lab-confirmed early Lyme disease and healthy controls from areas of endemicity) | Patients with 85% EM and positive by 2-tiered serology, 2-EIA, culture, or PCR | 20 | X | X |
| Healthy controls from areas of endemicity | 20 | X | ||
| NIH | ||||
| Patients with confirmed late Lyme disease | Patients with Lyme arthritis | 20 | X | X |
| Patients with PTLDS | Patients with PTLDS | 20 | X | |
| CDC (patients with confirmed early and late Lyme disease and negative controls) | Patients with early Lyme disease (EM, acute and convalescent phases) | 33 | X | |
| Patients with early disseminated disease with neurologic disease/carditis and Lyme arthritis | 8 | X | ||
| Healthy controls (from areas of endemicity and nonendemicity) and patients with look-alike diseases | 41 | X |
Abbreviations: C6, Immunetics C6 EIA; mChip-Ld, microfluidic rapid assay; EM, erythema migrans. A total of 241 serum samples were tested. For the discovery/screening phase, we used serum from patients with early and late Lyme disease, confirmed Lyme disease (LDB and NIH panels), and suspected Lyme disease with a positive 2-tiered serology (NYSDOH panel). For the comparative analysis, we tested only panels of serum samples from patients with confirmed Lyme disease that are commonly used (early and late Lyme disease, but not PTLDS); the CDC panel was not used for screening, given that panels for which the investigators are blind to the results are unsuitable for use in the discovery phase.
Serum panels.
The panels of serum tested are described in Table 1 and are described in more detail below.
(i) Patients with early Lyme disease (NYSDOH panel). Serum (n = 59 samples) was obtained from patients who presented with an erythematous skin lesion and a history of a recent tick bite or a summer flu-like illness to clinics in parts of New York State where Lyme disease is endemic and who were suspected of having Lyme disease. The samples subsequently tested positive by 2-tiered serology (the STT algorithm) at the New York State Department of Health (NYSDOH). These samples were considered to be from patients (i) suspected of having early Lyme disease (n = 20) and were C6 EIA positive plus IgM/IgG immunoblotting positive (2-tiered algorithm positive for suspected early acute Lyme disease) and (ii) suspected of being in convalescent phase (n = 39) and were C6 EIA positive plus IgG immunoblotting positive (2-tiered algorithm positive for suspected convalescent early Lyme).
(ii) Patients with laboratory-confirmed early acute Lyme disease and healthy controls from areas of endemicity (LDB panel). Serum (n = 40 samples) was obtained from patients with laboratory-confirmed early Lyme disease from areas of endemicity (n = 20 patients with confirmed early acute Lyme disease) and from healthy individuals from areas of endemicity (n = 20). The patients were from eastern Long Island, NY, and Martha’s Vineyard, MA. LD inclusion criteria included presentation in an area of endemicity, physician assessment of erythematous expanding rash (EM), and laboratory confirmation by either 2-tiered serology, 2-EIA (whole-cell extract EIA and C6 peptide EIA), PCR, or culture and PCR.
(iii) Patients with late Lyme disease and PTLDS (NIH panel). Serum (n = 40 samples) was obtained from patients suffering from Lyme arthritis (LA; n = 20) and posttreatment Lyme disease syndrome (PTLDS; n = 20). All patients met the criteria for the diagnosis of Lyme disease (4, 11). Patients with Lyme arthritis had joint swelling in conjunction with serologic evidence of the infection per CDC criteria (12). Patients with PTLDS had Lyme disease, received a minimum of 1 course of recommended therapy (4), and had persistent or relapsing nonspecific symptoms that began within 6 months of treatment and that were severe enough to cause a reduction in activities (4).
(iv) Patients with confirmed early and late Lyme disease and negative controls (CDC panel).
Samples (n = 82 samples) were obtained from patients in areas of endemicity and were clinically characterized by specialized physicians, and the disease was confirmed by laboratory testing (13). The clinical stages of Lyme disease were defined as follows: early acute-phase Lyme disease with EM, in which the patient was at epidemiologic risk, had erythema migrans lesions >5 cm in diameter, and, when possible, was positive by B. burgdorferi culture and/or PCR; early disseminated Lyme disease with neuroborreliosis or Lyme carditis, in which the patient was at epidemiologic risk and had objective clinical manifestations of neuroborreliosis (cranial nerve palsy, lymphocytic meningitis, or radiculopathy) and/or carditis (some patients in this group had single or multiple EM lesions, and some were positive for B. burgdorferi by culture and/or PCR); and late disseminated Lyme disease with arthritis, in which the patient was at epidemiologic risk and had physician-diagnosed arthritis and a positive result by 2-tiered serology. There were 33 samples clinically defined as being from patients with early Lyme disease (some samples were EIA positive, some were positive by IgM blotting, some were positive by IgG blotting, and some were positive by IgM/IgG blotting) and 8 samples clinically defined as being from patients with early disseminated Lyme disease with neuroborreliosis/carditis and late Lyme arthritis. Negative controls (n = 41) included serum from healthy individuals from areas of endemicity and nonendemicity (n = 11) and individuals with syphilis (n = 6), infectious mononucleosis (n=5), fibromyalgia (n = 4), rheumatoid arthritis (n = 4), multiple sclerosis (n = 6), and severe periodontitis (n = 5). This panel was kindly provided by Martin Schriefer from NCID/CDC in 2013, and the investigators were blind to the contents of the panel. The contents of the panel were revealed by Christopher Sexton in 2018 after acquisition of the data.
(v) Healthy controls from areas of nonendemicity (BR panel).
Serum (n = 20 samples) from healthy individuals from an area of nonendemicity was purchased from a commercial source (BioReclammation IVT [BR], MD).
Antigens.
A synthetic peptide, which we denote PepVF (14), was commercially synthesized (GenScript, Piscataway, NJ) and features an N-terminal 17-amino-acid sequence from the IR6 region of B. burgdorferi B31 (15), a glycine linker, and a C-terminal 13-amino-acid proprietary sequence from an internal, non-surface-exposed fragment of FlaB. Eleven genes from B. burgdorferi were cloned in pET28a (GenScript, Piscataway, NJ). We expanded our search for B. burgdorferi antigens by analyzing five new recombinant antigens (Hsp90, ErpB [16, 17], p45, p28, and FlaB), in addition to the best seven markers previously used to establish the proof of principle of this technology (p93/100, BmpA, DbpA, DbpB, recombinant OspC type K [rOspC-K], VlsE, and PepVF [9]). Unlike the first study, we purified recombinant VlsE by affinity chromatography in-house. The following recombinant proteins were purified as described previously (9): p93/100, Hsp90, ErpB/p58, p45, BmpA/p39, VlsE/p35, p28, OspC type K (p23), DbpA/p18, DbpB/p17, and FlaBi (triple fragment of the same internal sequence used in PepVF). Quality control was done by immunoblotting of polyvinylidene difluoride membranes using mouse-generated antigen-specific polyclonal antibodies. (Approval for animal experimentation was obtained from The University of Tennessee Health Science Center Institutional Animal Care and Use Committee [IACUC protocol number 16-154].)
Serologic testing by EIA and on a microfluidic platform.
Serologic testing by EIA was performed as previously described for IgG detection (15). For the microfluidic cassettes, proteins were diluted using 1× EIA coating buffer (Bio-Rad) and spotted on the detection zones at the following concentrations: 100 μg ml−1 for pepVF, 20 μg ml−1 for rOspC-K, and 1 μg ml−1 for recombinant VlsE. Functionalized cassettes had five detection zones, including an internal negative-control zone, an internal positive-control zone spotted with 20 μg ml−1 rabbit anti-goat IgG antibody (Life Technologies), and three antibody detection zones coated with antigen. Protein functionalization and serologic testing for IgM plus IgG were done as described previously (9, 18–22). Serum samples were diluted 10 times in StartingBlock blocking buffer (Thermo Fisher Scientific). Signal measurements were recorded by a benchtop analyzer (Opko Diagnostics) by taking an initial intensity reading (I0) immediately after silver entry into the channel and another intensity reading (I) after 4.5 min of silver development (9). The optical density (OD) was calculated as OD = −log(I/I0).
Statistical analysis and multiplexed algorithm.
For the EIA screening of 12 antigens, we determined the cutoff to be 3 standard deviations above the average OD at 450 nm (OD450) for all 12 antigens tested against the panel of samples from healthy individuals from areas of nonendemicity. For the EIA and the mChip-Ld microfluidic test with the three antigens (3Ag; VlsE and the proprietary synthetic 33-mer peptide [PepVF] to capture sensitivity in all disease stages and OspC for early Lyme disease) (3Ag-EIA and 3Ag-mChip-Ld, respectively), the OD cutoff for each biomarker was determined with the CDC panel by constructing a receiver operating characteristic (ROC) analysis with the area under the curve (AUC) and selecting the OD cutoff value which resulted in the maximum sensitivity given a minimum of 95% specificity.
Our multiplexed signal consisted of a linear sum of weighted ODs for each biomarker, similar to that used in other previous studies with multiplexed markers (23, 24). To determine the optimal weight for each biomarker, we performed ROC analysis for each of the 10,648 different permutations of weights on the CDC panel data and chose the combination of weights that yielded the highest AUC value. We selected the OD cutoff for this weighted multiplexed signal to be the OD value which resulted in the maximum sensitivity given a minimum of 95% specificity. To gain insight into the rough contributions of each biomarker, we also plotted, for a fixed relative weight of one biomarker, the mean AUC score averaged over all combinations of weights for the other two biomarkers (see Fig. S1 in the supplemental material). These steps were applied separately for the mChip and EIA data from the CDC panel to determine the weights and cutoffs, with the same weightings then being used on the other panels. Calculations were performed using GraphPad Prism (version 8) and Python (version 3.0) software.
RESULTS
Screening of diagnostic biomarkers for Lyme disease.
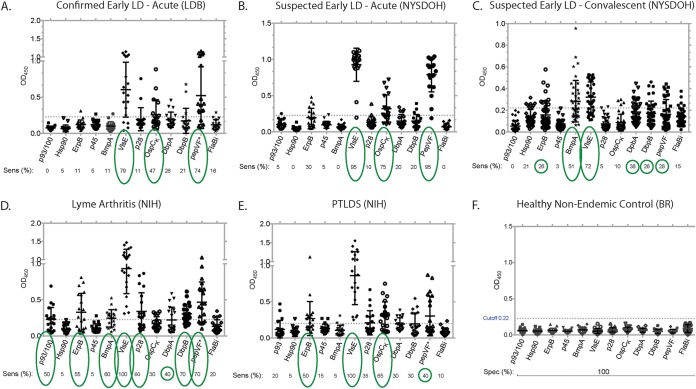
Serum panels (n = 139 samples) classified as being from patients with suspected and confirmed early Lyme with a positive 2-tiered serology (the NYSDOH and LDB panels), patients with Lyme arthritis and PTLDS (the NIH panel), and healthy controls (the BR panel) were used to determine the sensitivity and specificity of the best-in-class antigens by EIA (Fig. 1; Table 1). The cutoff for this EIA screen was 3 standard deviations above the average OD450 for all 12 antigens tested against the panel of samples from healthy individuals from areas of nonendemicity (OD450, ∼0.22); thus, for this phase of test development, we set the specificity of these markers at 100% (Fig. 1F). For the two panels of serum from patients with early acute Lyme disease (the LDB and NYSDOH panels) (Fig. 1A and B), three biomarkers showed considerable sensitivity: VlsE (79% for the LDB panel and 95% for the NYSDOH panel), OspC-K (47% for the LDB panel and 75% for the NYSDOH panel), and PepVF (74% for the LDB panel and 95% for the NYSDOH panel). For the two panels of serum from patients with early convalescent-phase Lyme disease (the NYSDOH panel), two biomarkers (BmpA, VlsE) detected >50% of the samples (Fig. 1C). For the panel of serum from patients with Lyme arthritis, seven biomarkers (p93/100, ErpB, BmpA, VlsE, p28, DbpB, and PepVF) detected >50% of the samples (Fig. 1D). For the panel of serum from patients with PTLDS, three biomarkers (ErpB, VlsE, and OspC-K) detected >50% of the samples (Fig. 1E). The VlsE antigen stood out by detecting all samples in the Lyme arthritis and PTLDS panels (100%) and detected early LD with a >80% sensitivity (79% for the acute-phase serum samples from LDB, 95% for the acute-phase serum samples from NYSDOH, and 72% for the convalescent-phase serum samples from NYSDOH). OspC-K and PepVF were added for sensitivity in the early stage. These three biomarkers (VlsE, OspC-K, and PepVF) were used to further improve the mChip-Ld for Lyme disease serodiagnosis.
FIG 1.
EIA screening of B. burgdorferi biomarkers to select the best in class for the serodiagnosis of the Lyme disease stage. The plots show the specificity (Spec) and the sensitivity (Sens) of the candidate antigens (p93/100, Hsp90, ErpB, p45, BmpA, VlsE, p28, OspC-K, DbpA, DbpB, PepVF, and FlaBi), obtained using IgG EIA, characterized by clinical diagnosis. Sensitivity was determined using samples from patients with laboratory-confirmed (2-tiered serology, 2-EIA, PCR, or culture and PCR) early acute Lyme disease (n = 19 samples; LDB panel) (A), suspected early acute Lyme disease positive by 2-tiered serology (n = 20; NYSDOH panel) (B), suspected early convalescent-phase Lyme disease positive by 2-tiered serology (n = 39; NYSDOH panel) (C), Lyme arthritis (n = 20; NIH panel) (D), and posttreatment Lyme disease syndrome (n = 20; NIH panel) (E) or healthy controls from areas of nonendemicity (n = 20) (F). (F) Specificity was determined using serum from healthy individuals from an area of nonendemicity (n = 20; BR panel). The cutoff was 3 standard deviations above the average OD450 for all 12 antigens tested against the results for healthy controls from areas of nonendemicity (∼OD450, 0.22). Green circles highlight the footprint of the antibody response to B. burgdorferi antigens as Lyme disease progresses from the early to the late stage, large circles represent a sensitivity of ∼50% or greater, and small circles represent a sensitivity of >25%.
Diagnostic performance against three different panels of samples from patients with Lyme disease.
The serum panels from LDB, NIH, and CDC (n = 142) were used to evaluate the three biomarker leads by measuring their sensitivity and specificity by EIA and mChip-Ld, and the results were subsequently compared to those obtained with the STT algorithm. The serum from CDC was tested by a C6 EIA to identify the positive and negative samples and was then used for the ROC analysis to determine the antigen weights and the cutoff values for the three biomarker leads both on the mChip-Ld platform and by EIA (multiplex algorithm). However, this panel was tested by the 3Ag-EIA and the 3Ag-mChip-Ld by a different user who was blind to the results. Panels of serum from patients with Lyme arthritis from LDB and NIH were then used to further analyze the assay. The panel from NYSDOH was not used for comparative analysis because it was not classified as containing samples from patients with confirmed Lyme disease.
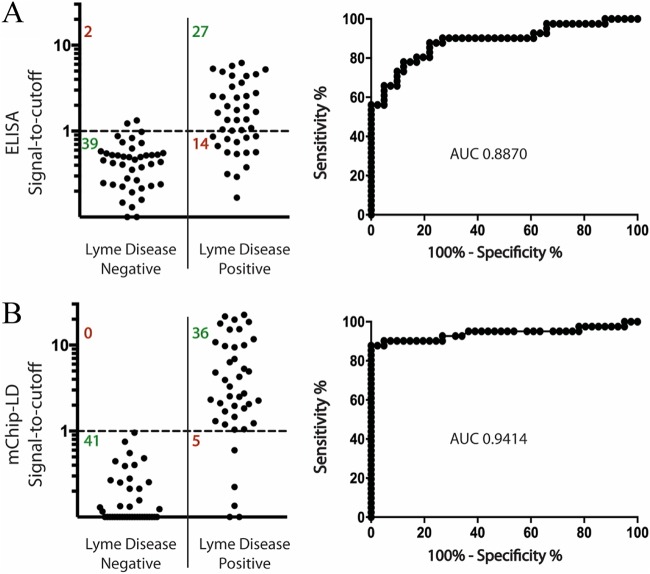
By EIA, of the three antigens tested against the CDC panel, OspC-K had the lowest diagnostic performance (AUC, 0.766) compared to that of PepVF (AUC, 0.849) and VlsE (AUC, 0.842) (Table 2). Combining the three antigens, we used a weighted sum of the three OD signals using the multiplexed algorithm described above and assigned the weights as follows: 1.75, 0.5, and 2.5 for PepVF, OspC-K, and VlsE, respectively. This multiplexed combination (3Ag-EIA) achieved a sensitivity of 65.9% (95% confidence interval [CI], 49.4% to 79.9%) and a specificity of 95.1% (95% CI, 83.5% to 99.4%) with an AUC of 0.887 (Fig. 2A and Table 2). On the microfluidic mChip-Ld platform, we first improved the assay parameters, such as the concentrations of spotted antigens, the concentration and buffers for the secondary antibodies, and the washing conditions, using a small subset of serum samples. Next, we tested the CDC panel. As the CDC panel was the largest panel tested, we used results from this panel to obtain individual cutoff OD signals for each of the three antigens using ROC curve analysis (Table 2). Individually, OspC-K had the lowest diagnostic performance (AUC, 0.862) compared to PepVF (AUC, 0.904) and VlsE (AUC, 0.883). This ranking is similar to the ranking obtained with the EIA results. Using the weight identification strategy described, we obtained weights of 1, 0.125, and 1.25 for PepVF, OspC-K, and VlsE, respectively. The multiplexed combination on the mChip-Ld platform had an AUC of 0.941 and achieved an overall sensitivity of 87.8% (95% CI, 73.8% to 95.9%) and a specificity of 100.0% (95% CI, 91.4% to 100.0%) (Fig. 2B; Table 2).
TABLE 2.
Diagnostic performance with three different panels: comparative analysis between 3Ag-EIA and 3Ag-mCHIP-Lda
| Panel and diagnostic test | Marker | Cutoff | Sensitivity (%) | Specificity (%) | AUC |
|---|---|---|---|---|---|
| CDC (confirmed early and late Lyme disease) | |||||
| EIA | PepVF | >0.2215 | 56.1 (39.8–71.5) | 95.1 (83.5–99.4) | 0.8492 |
| OspC-K | >0.2615 | 26.8 (14.2–42.9) | 95.1 (83.5–99.4) | 0.7662 | |
| VlsE | >0.3110 | 53.7 (37.4–69.3) | 95.1 (83.5–99.4) | 0.8418 | |
| 1.75 PepVF + 0.5 OspC-K + 2.5 VlsE | >1.102 | 65.9 (49.4–79.9) | 95.1 (83.5–99.4) | 0.8870 | |
| mChip-Ld | PepVF | >0.026 | 75.6 (59.7–87.6) | 95.1 (83.5–99.4) | 0.9036 |
| OspC-K | >0.1165 | 65.9 (49.4–79.9) | 95.1 (83.5–99.4) | 0.8623 | |
| VlsE | >0.0208 | 70.8 (54.6–83.9) | 95.1 (83.5–99.4) | 0.8834 | |
| 1 PepVF + 0.125 OspC-K + 1.25 VlsE | >0.0766 | 87.8 (73.8–95.9) | 100.0 (91.4–100.0) | 0.9414 | |
| LDB (laboratory-confirmed early Lyme disease) | |||||
| EIA | PepVF | >0.2215 | 75.0 (50.9–91.3) | 100.0 (83.2–100.0) | 0.9325 |
| OspC-K | >0.2615 | 30.0 (11.9–54.3) | 90.0 (68.0–98.8) | 0.7025 | |
| VlsE | >0.3110 | 75.0 (50.9–91.3) | 100.0 (83.2–100.0) | 0.9038 | |
| 1.75 PepVF + 0.5 OspC-K + 2.5 VlsE | >1.102 | 75.0 (50.9–91.3) | 100.0 (83.2–100.0) | 0.9150 | |
| mChip-Ld | PepVF | >0.026 | 75.0 (50.9–91.3) | 100.0 (83.2–100.0) | 0.9025 |
| OspC-K | >0.1165 | 80.0 (56.3–94.3) | 75.0 (50.9–91.3) | 0.8325 | |
| VlsE | >0.0208 | 80.0 (56.3–94.3) | 90.0 (68.3–98.8) | 0.8750 | |
| 1 PepVF + 0.125 OspC-K + 1.25 VlsE | >0.0766 | 80.0 (56.3–94.3) | 100.0 (83.2–100.0) | 0.8650 | |
| NIH panel (Lyme arthritis) | |||||
| EIA | PepVF | >0.2215 | 70.0 (45.7–88.1) | NA | NA |
| OspC-K | >0.2615 | 25.0 (8.7–49.1) | NA | NA | |
| VlsE | >0.3110 | 95.0 (75.1–99.9) | NA | NA | |
| 1.75 PepVF + 0.5 OspC-K + 2.5 VlsE | >1.102 | 100.0 (83.2–100.0) | NA | NA | |
| mChip-Ld | PepVF | >0.026 | 100.0 (83.2–100.0) | NA | NA |
| OspC-K | >0.1165 | 85.0 (62.1–96.8) | NA | NA | |
| VlsE | >0.0208 | 95.0 (75.1–99.9) | NA | NA | |
| 1 PepVF + 0.125 OspC-K + 1.25 VlsE | >0.0766 | 100.0 (83.2–100.0) | NA | NA |
Abbreviations: AUC, area under the curve; NA, not applicable. Values in parentheses are 95% confidence intervals.
FIG 2.
Performance of the 3Ag-EIA and 3Ag-mChip-Ld using a panel of serum samples for which the investigators were blind to the results. The signal-to-cutoff plots show the sensitivity and the specificity of the multiplexed capture antigen panel (PepVF, OspC-K, and VlsE) with both the EIA (A) and mChip-Ld (B) testing formats. ROC curves are shown on the right with AUC values. The CDC panel was used. It consisted of 41 samples from patients with Lyme disease and 41 samples from controls.
Analysis of the multiplex combination against early and late Lyme disease.
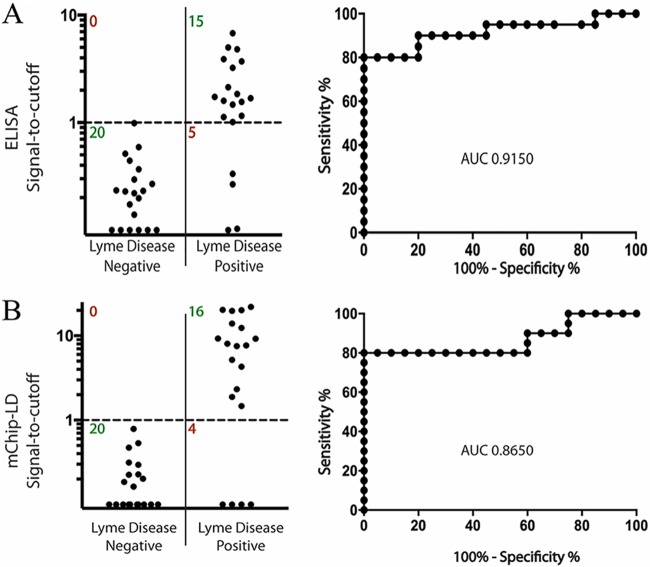
The weights of the antigens and the multiplexing algorithm were then used to analyze the 3Ag-EIA and 3Ag-mChip-Ld assays using two additional panels of serum from patients with Lyme disease: the LDB panel (which contained serum samples from patients with early Lyme disease) and the NIH panel (which contained serum samples from patients with late Lyme disease). With the panel of serum samples from patients with early Lyme disease (the LDB panel), the multiplexed 3Ag-EIA achieved a sensitivity of 75.0% (95% CI, 50.9% to 91.3%) and a specificity of 100% (95% CI, 83.2% to 100%) with an AUC of 0.915 (Fig. 3A; Table 2); the 3Ag-mChip-Ld achieved a sensitivity of 80.0% (95% CI, 56.3% to 94.3%) and a specificity of 100% (95% CI, 83.2% to 100%) with an AUC of 0.865 (Fig. 3B; Table 2). Against the panel of serum samples from patients with Lyme arthritis (the NIH panel), the multiplexed 3Ag-EIA achieved a sensitivity of 100% (95% CI, 83.2% to 100%), which was the same as that of the multiplexed 3Ag-mChip-Ld (Table 2).
FIG 3.
Performance of the 3Ag-EIA and 3Ag-mChip-Ld using the LDB serum panel, consisting of samples from patients with early acute Lyme disease and controls from areas of endemicity. The signal-to-cutoff plots show the sensitivity and the specificity of the multiplexed antigen panel (PepVF, OspC-K, and VlsE) with both the EIA (A) and the mChip-Ld (B) testing formats. ROC curves are shown on the right with AUC values. The same diagnostic cutoff value from the CDC panel was used to analyze the performance of both mChip-Ld and EIA with the LDB panel (n = 40). The LDB panel consisted of samples from patients with early Lyme disease (n = 20) and healthy controls from areas of endemicity (n = 20).
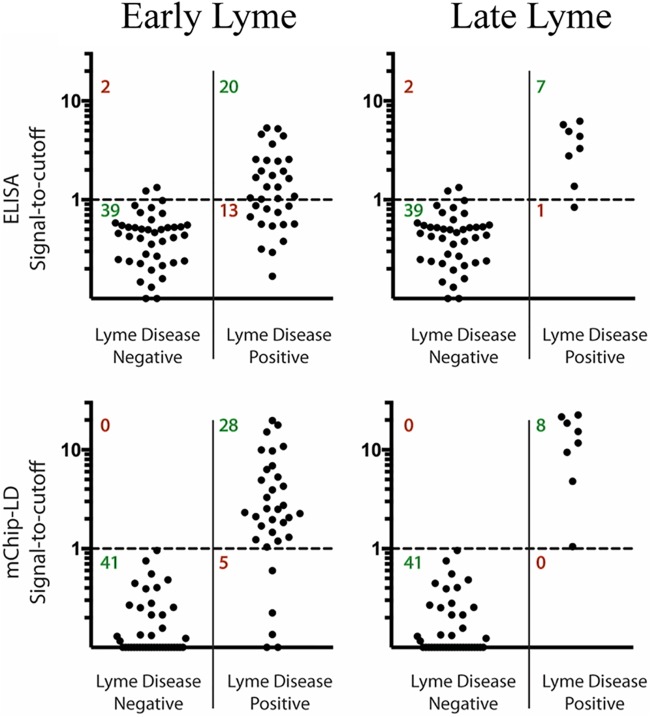
The CDC panel contained samples from patients spanning the range of Lyme disease stages and was used to examine early versus early disseminated plus late Lyme disease detection on both the 3Ag-EIA and 3Ag-mChip-Ld platforms. With samples from patients with early Lyme disease, 3Ag-EIA had a sensitivity of 60.6% (95% CI, 42.1% to 77.1%) and a specificity of 95.1% (95% CI, 83.5% to 99.4%), whereas 3Ag-mChip-Ld had a sensitivity of 84.9% (95% CI, 68.1% to 94.9%) and a specificity of 100% (95% CI, 91.4% to 100.0%) (Fig. 4). With samples from patients with early disseminated and late Lyme disease (n = 8), 3Ag-EIA had a sensitivity of 87.5% (95% CI, 47.4% to 99.7%) and a specificity of 95.1% (95% CI, 83.5% to 99.4%) and 3Ag-mChip-Ld had a sensitivity of 100% (95% CI, 63.1% to 100%) and a specificity of 100% (95% CI, 91.4% to 100%) (Fig. 4). Using the same objective OD cutoff values and relative weights for each antigen, we further confirmed the performance of both assays against the NIH panel of serum samples from patients with Lyme arthritis. Both 3Ag-EIA and 3Ag-mChip-Ld achieved a sensitivity of 100% (Table 2).
FIG 4.
Antibody detection using 3Ag-EIA and 3Ag-mChip-Ld in early and late Lyme disease using a panel of serum samples for which the investigators were blind to the results. Signal-to-cutoff plots of the EIA and mChip-Ld testing formats show a breakdown of the sensitivity and the specificity for the detection of early Lyme disease (n = 33) and early disseminated/late Lyme disease (n = 8). The CDC panel was used. It consisted of 41 samples from patients with Lyme disease and 41 samples from controls.
Comparison of 3Ag-EIA and 3Ag-mChip-Ld with the STT algorithm.
When we analyzed the performance of the three tests using the CDC panel, we observed a marked improvement in the sensitivity of the 3Ag-mChip-Ld (87.8%) and the 3Ag-EIA (65.9%) compared to the results of the 2-tiered test (58.5%). The specificities were 100%, 95.1%, and 97.5%, respectively (Table 3). The increased sensitivity of the 3Ag-mChip-Ld reflects the detection of early Lyme disease in 8/13 patients with early Lyme disease and late Lyme disease in 1/1 patient with late Lyme disease that tested negative or equivocal by EIA (Fig. 4) and the standard 2-tiered (STT) algorithm (Table 3). Remarkably, 3Ag-mChip-Ld performed better than the STT algorithm, and the gain in sensitivity was achieved in the early Lyme disease stage without incurring a loss of specificity. We further confirmed the performance of the three assays against the panels of serum from patients with early Lyme disease (the LDB panel) and Lyme arthritis (the NIH panel) (Table 3). We observed an increase in the sensitivity of the 3Ag-mChip-Ld (80%) versus that of the STT algorithm (75%) with the LDB panel of serum samples from patients with early Lyme disease, whereas no difference was observed for the NIH panel of serum samples from patients with late Lyme disease (all at 100%).
TABLE 3.
Sensitivity and specificity of the multiplex 3Ag-EIA, 3Ag-mCHIP-Ld, and STT algorithm for detection of B. burgdorferi antibody per disease stagea
| Panel | Sensitivity (%) |
Specificity (%) |
||||
|---|---|---|---|---|---|---|
| 3Ag-EIA | 3Ag-mChip-Ld | STT algorithm | 3Ag-EIA | 3Ag-mChip-Ld | STT algorithm | |
| CDC (early LD; n = 33) | 60.6 (42.1–77.1) | 84.9 (68.1–94.9) | 48.5b | NA | NA | NA |
| CDC (ED and Lyme arthritis; n = 8) | 87.5 (47.4–99.7) | 100 (63.1–100) | 100b | NA | NA | NA |
| LDB (early LD; n = 20) | 75 (50.9–91.3) | 80 (56.3–94.3) | 75 | NA | NA | NA |
| NIH (Lyme arthritis; n = 20) | 100 (83.2–100) | 100 (83.2–100) | 100 | NA | NA | NA |
| CDC (controls; n = 41) | NA | NA | NA | 95.1 (83.5 to 99.4) | 100 (91.4 to 100) | 97.5b |
| LDB (controls; n = 20) | NA | NA | NA | 100 (83.2 to 100) | 100 (83.2 to 100) | 100 |
Abbreviations: AUC, area under the curve; ED, early disseminated Lyme disease; LD, Lyme disease; NA, not applicable; EIA, enzyme immunoassay; mChip-Ld, microfluidic rapid assay; Ag, antigen; CDC, Centers for Disease Control and Prevention; LDB, Lyme Disease Biobank; NIH, National Institutes of Health. AUC values for 3Ag-EIA were as follows: 0.8625 for the CDC panel with samples from patients with early LD and controls, 0.9878 for the CDC panel with samples from patients with early disseminated Lyme disease, Lyme arthritis, and controls, and 0.9150 for the LDB panel with samples from patents with early Lyme disease and controls. AUC values for 3Ag-mChip-Ld were as follows: 0.9272 for the CDC panel with samples from patients with early LD and controls, 1.0000 for the CDC panel with samples from patients with early disseminated Lyme disease, Lyme arthritis, and controls, and 0.8650 for the LDB panel with samples from patents with early Lyme disease and controls. Values in parentheses are 95% confidence intervals.
Data were provided by the CDC.
DISCUSSION
The development of assays for the laboratory diagnosis of early Lyme disease remains a challenging unmet need. We improved our microfluidics assay (mChip-Ld) for the rapid detection of B. burgdorferi antibody in serum from Lyme disease patients and analyzed its diagnostic performance against that achieved with samples acquired from three different sources: LDB, NIH, and CDC. The rapid mChip-Ld assay detected early Lyme disease in samples from patients with early Lyme disease with a higher sensitivity and a higher specificity than the STT algorithm.
Currently, CDC recommends the STT algorithm, a two-tiered testing approach comprised of a sensitive first-tier EIA that, if positive, is followed by a second-tier IgM/IgG immunoblot assay if the disease has been present for <30 days or an IgG-only immunoblot assay if the disease has been present for >30 days (25). CDC has updated its recommendations for the serodiagnosis of Lyme disease by deeming those assays that use a second EIA in lieu of the immunoblot assay to be an acceptable alternative for the second tier of the STT algorithm (26). Quantifiable EIA-based methods can provide objective test results, in contrast to the operator-dependent subjective interpretation of immunoblot assay results (5).
Our antigen discovery was done by an EIA screen of old and new B. burgdorferi diagnostic candidates against four panels of serum: three panels consisting of serum from patients in which early and late Lyme disease were confirmed (the LDB and NIH panels) or suspected (the NYSDOH panel) with a positive 2-tiered serology and one panel consisting of serum from negative controls (n = 139 samples) (Table 1 and Fig. 1), providing further evidence that several known antigens (5, 27) can be used to develop sensitive serologic assays for early and early disseminated/late Lyme disease. Our data also show that more antigens become positive with the progression of Lyme disease from the early to the late stage, from VlsE, OspC-K, and PepVF in the early acute phase to p100, ErpB, BmpA, VlsE, p28, DbpA, DbpB, and PepVF in Lyme arthritis. Thus, our data demonstrate the progression of the antibody response to specific antigens of B. burgdorferi per disease stage, which is also seen in immunoblot assays (3 positive bands for IgM, >5 positive bands for IgG). Interestingly, for PTLDS, a protein usually associated with early infection, OspC (27, 28), detected PTLDS in 65% of the samples in the panel of samples from patients with PTLDS. Another interesting observation is that ErpB, which is associated with early disseminated and late Lyme disease (16), detected positive samples in the panel of samples from patients with PTLDS with a 50% sensitivity. The importance of these findings requires further study.
Our screening study identified three markers that performed with a considerably high sensitivity for the detection of anti-B. burgdorferi antibody in serum from patients with early Lyme disease: VlsE (79% and 95%), OspC-K (47% and 75%), and PepVF (74% and 95%). Two of these markers (OspC-K and PepVF) were previously identified in our proof-of-principle study in which we used a small (n = 35) but rigorously characterized panel of serum samples from patients with Lyme disease from the CDC that mostly included samples from patients with convalescent-phase and late Lyme disease (9). Interestingly, subsequent to the proposed use of the PepVF sequence (14), independent large-scale screening efforts identified the same VlsE (29) and FlaB (29, 30) epitopes contained in PepVF. The observed sensitivity of the VlsE antigen for the detection of all cases of Lyme disease may be explained by the association of an immune response toward specific VlsE sequences during early and late stages of the disease (31). Furthermore, anti-VlsE antibody was identified in the anti-Borrelia burgdorferi profiles of PTLDS patients (32), which is also supported by the findings of our study.
Microfluidics offers practical advantages for miniaturizing laboratory-based tests, including portability, multiplexing, speed, and performance (19, 33). Overall, the performance of the mChip-Ld platform largely matched the performance of our laboratory-based 3Ag-EIA IgG functionalized with the same antigens. The improved performance seen in some cases (e.g., samples from patients with early-stage disease, especially those in the CDC and LDB panels) could be due to the mChip-Ld platform detecting IgM antibodies, which peak in the first 2 to 6 weeks after disease onset (34), in addition to IgG, which was the only isotype detected in our EIAs. Traditionally, IgM detection decreases the specificity of an assay (35) and an increase in sensitivity is counterbalanced by a decrease in specificity. Avoiding low specificity was the reason for our exclusion of IgM detection in our antigen discovery phase using EIA. Our IgM/IgG mChip-Ld results showed sensitivities of over 80% to 100% for the panels of serum samples from patients with Lyme disease tested and specificities of 100% with an AUC of 0.865 to 0.941 for the two panels with both positive and negative specimens. These data show that in the microfluidics platform, an increase in sensitivity was not followed by a decrease in specificity, as we predicted for the combined IgM/IgG detection. One possible explanation is that the optimization of parameters specific to an assay and antigen molecules (36) can significantly alter the performance of the microfluidics assay. Here, additional improvement of assay conditions, particularly in reducing the variability of the internal positive- and negative-control signals through buffer modification, was carried out prior to the testing of the specimen panels. Furthermore, for the multiplexed testing, while we previously summed the three OD signals with equal weights (9), here, our final score consisted of a sum of weighted quantitative measurements which were determined empirically (23, 24). Such improvements in assay technology and the multiplexed algorithm contributed to an increased overall sensitivity and specificity of the mChip-Ld. However, further testing in a clinical setting is necessary to confirm whether the mChip-Ld format can consistently perform better than the 3Ag-EIA.
There is no commercially available rapid point-of-care diagnostic test for Lyme disease (19, 37, 38). This work demonstrates an approach that could lead to an objective, point-of-care test for Lyme disease (9) with a diagnostic performance that matches that of current standard laboratory testing or the, in some cases, outperforms current standard laboratory testing with the potential to be a stand-alone replacement for the STT algorithm. The mChip-Ld performed with a sensitivity either similar to or higher than that of the STT algorithm without losing specificity, which remained above 95%. More broadly, this study demonstrates the potential of the microfluidics technology to deliver high performance for multiplexed assays in a portable format in an era when immunodominant epitopes are increasingly being identified for a wide array of infectious organisms.
Additional information.
The cassettes and reagents are from Opko; all reasonable requests for materials sharing will be considered.
Supplementary Material
ACKNOWLEDGMENTS
We thank Martin Schriefer from the National Center for Infectious Diseases, Centers for Disease Control and Prevention, for providing the Lyme disease-characterized serum panels (CDC Lyme disease panel) and Christopher Sexton for revealing the contents of the panel after the data were acquired.
This work was supported by Public Health Service grant R44 AI096551 to M.G.S. via Immuno Technologies Inc. and in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
The content of this publication does not necessarily reflect the views or policies of the U.S. Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. The funders for the study provided support in the form of salaries for authors but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit the manuscript for publication.
M.G.-S. is an employee of Immuno Technologies Inc. and holds a 5% or greater financial interest in Immuno Technologies Inc. V.L. was an employee of Opko Diagnostics LLC while engaged in the research project. M.G.-S. and A.R.M. hold relevant patents. V.L. declares a financial interest in Opko Diagnostics. S.A., S.N., F.S.D.S.M., T.W., R.C.C., M.S.G., S.J.W., and S.K.S. declare no competing financial interests.
S.A., S.N., M.G.-S., and S.K.S. designed the study; S.A., F.S.D.S.M., and R.C.C. performed the microfluidic immunoassays; T.W. and M.S.G. performed the EIA and protein purification; V.L. advised on assay development and provided materials and reagents; S.A., S.N., T.W., M.G.-S., and S.K.S. analyzed the data; and S.A., S.N., M.G.-S, A.R.M., and S.K.S. wrote the paper. E.J.H., S.J.W., and A.R.M. provided Lyme disease-characterized serum panels. All coauthors edited the paper. All figures and tables were created by an author of the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01142-19.
REFERENCES
- 1.Kurtenbach K, Hanincova K, Tsao JI, Margos G, Fish D, Ogden NH. 2006. Fundamental processes in the evolutionary ecology of Lyme borreliosis. Nat Rev Microbiol 4:660–669. doi: 10.1038/nrmicro1475. [DOI] [PubMed] [Google Scholar]
- 2.Hinckley AF, Connally NP, Meek JI, Johnson BJ, Kemperman MM, Feldman KA, White JL, Mead PS. 2014. Lyme disease testing by large commercial laboratories in the United States. Clin Infect Dis 59:676–681. doi: 10.1093/cid/ciu397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson CA, Saha S, Kugeler KJ, Delorey MJ, Shankar MB, Hinckley AF, Mead PS. 2015. Incidence of clinician-diagnosed Lyme disease, United States, 2005–2010. Emerg Infect Dis 21:1625–1631. doi: 10.3201/eid2109.150417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wormser GP, Dattwyler RJ, Shapiro ED, Halperin JJ, Steere AC, Klempner MS, Krause PJ, Bakken JS, Strle F, Stanek G, Bockenstedt L, Fish D, Dumler JS, Nadelman RB. 2006. The clinical assessment, treatment, and prevention of Lyme disease, human granulocytic anaplasmosis, and babesiosis: clinical practice guidelines by the Infectious Diseases Society of America. Clin Infect Dis 43:1089–1134. doi: 10.1086/508667. [DOI] [PubMed] [Google Scholar]
- 5.Branda JA, Body BA, Boyle J, Branson BM, Dattwyler RJ, Fikrig E, Gerald NJ, Gomes-Solecki M, Kintrup M, Ledizet M, Levin AE, Lewinski M, Liotta LA, Marques A, Mead PS, Mongodin EF, Pillai S, Rao P, Robinson WH, Roth KM, Schriefer ME, Slezak T, Snyder J, Steere AC, Witkowski J, Wong SJ, Schutzer SE. 2018. Advances in serodiagnostic testing for Lyme disease are at hand. Clin Infect Dis 66:1133–1139. doi: 10.1093/cid/cix943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porwancher RB, Hagerty CG, Fan J, Landsberg L, Johnson BJ, Kopnitsky M, Steere AC, Kulas K, Wong SJ. 2011. Multiplex immunoassay for Lyme disease using VlsE1-IgG and pepC10-IgM antibodies: improving test performance through bioinformatics. Clin Vaccine Immunol 18:851–859. doi: 10.1128/CVI.00409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradshaw GL, Thueson RK, Uriona TJ. 2017. Improved serodiagnostic performance for Lyme disease by use of two recombinant proteins in enzyme-linked immunosorbent assay compared to standardized two-tier testing. J Clin Microbiol 55:3046–3056. doi: 10.1128/JCM.01004-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nayak S, Blumenfeld NR, Laksanasopin T, Sia SK. 2017. Point-of-care diagnostics: recent developments in a connected age. Anal Chem 89:102–123. doi: 10.1021/acs.analchem.6b04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nayak S, Sridhara A, Melo R, Richer L, Chee NH, Kim J, Linder V, Steinmiller D, Sia SK, Gomes-Solecki M. 2016. Microfluidics-based point-of-care test for serodiagnosis of Lyme disease. Sci Rep 6:35069. doi: 10.1038/srep35069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Code of Federal Regulations. 2009. Title 45. Public welfare. Department of Health Human Services. Protection of human subjects. 45 CFR Part 46 https://www.hhs.gov/ohrp/sites/default/files/ohrp/policy/ohrpregulations.pdf. [PubMed]
- 11.CDC. 2017. Lyme disease (Borrelia burgdorferi) 2017 case definition. CDC, Atlanta, GA: https://wwwn.cdc.gov/nndss/conditions/lyme-disease/case-definition/2017/. Accessed 10 January 2019. [Google Scholar]
- 12.CDC. 1995. Recommendations for test performance and interpretation from the Second National Conference on Serologic Diagnosis of Lyme Disease. MMWR Morb Mortal Wkly Rep 44:590–591. [PubMed] [Google Scholar]
- 13.Molins CR, Sexton C, Young JW, Ashton LV, Pappert R, Beard CB, Schriefer ME. 2014. Collection and characterization of samples for establishment of a serum repository for Lyme disease diagnostic test development and evaluation. J Clin Microbiol 52:3755–3762. doi: 10.1128/JCM.01409-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dattwyler RJ, Gomes-Solecki MJ. 15 February 2011. Peptide diagnostic agent for Lyme disease. US patent 7,887,815.
- 15.Gomes-Solecki MJ, Meirelles L, Glass J, Dattwyler RJ. 2007. Epitope length, genospecies dependency, and serum panel effect in the IR6 enzyme-linked immunosorbent assay for detection of antibodies to Borrelia burgdorferi. Clin Vaccine Immunol 14:875–879. doi: 10.1128/CVI.00122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nowalk AJ, Gilmore RD Jr, Carroll JA. 2006. Serologic proteome analysis of Borrelia burgdorferi membrane-associated proteins. Infect Immun 74:3864–3873. doi: 10.1128/IAI.00189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbour AG, Jasinskas A, Kayala MA, Davies DH, Steere AC, Baldi P, Felgner PL. 2008. A genome-wide proteome array reveals a limited set of immunogens in natural infections of humans and white-footed mice with Borrelia burgdorferi. Infect Immun 76:3374–3389. doi: 10.1128/IAI.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, Wang J, Moore H, Rouse R, Umviligihozo G, Karita E, Mwambarangwe L, Braunstein SL, van de Wijgert J, Sahabo R, Justman JE, El-Sadr W, Sia SK. 2011. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat Med 17:1015–1019. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 19.Chin CD, Cheung YK, Laksanasopin T, Modena MM, Chin SY, Sridhara AA, Steinmiller D, Linder V, Mushingantahe J, Umviligihozo G, Karita E, Mwambarangwe L, Braunstein SL, van de Wijgert J, Sahabo R, Justman JE, El-Sadr W, Sia SK. 2013. Mobile device for disease diagnosis and data tracking in resource-limited settings. Clin Chem 59:629–640. doi: 10.1373/clinchem.2012.199596. [DOI] [PubMed] [Google Scholar]
- 20.Laksanasopin T, Guo TW, Nayak S, Sridhara AA, Xie S, Olowookere OO, Cadinu P, Meng F, Chee NH, Kim J, Chin CD, Munyazesa E, Mugwaneza P, Rai AJ, Mugisha V, Castro AR, Steinmiller D, Linder V, Justman JE, Nsanzimana S, Sia SK. 2015. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci Transl Med 7:273re1. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- 21.Linder V, Sia SK, Whitesides GM. 2005. Reagent-loaded cartridges for valveless and automated fluid delivery in microfluidic devices. Anal Chem 77:64–71. doi: 10.1021/ac049071x. [DOI] [PubMed] [Google Scholar]
- 22.Guo T, Patnaik R, Kuhlmann K, Rai AJ, Sia SK. 2015. Smartphone dongle for simultaneous measurement of hemoglobin concentration and detection of HIV antibodies. Lab Chip 15:3514–3520. doi: 10.1039/c5lc00609k. [DOI] [PubMed] [Google Scholar]
- 23.Salami SS, Schmidt F, Laxman B, Regan MM, Rickman DS, Scherr D, Bueti G, Siddiqui J, Tomlins SA, Wei JT, Chinnaiyan AM, Rubin MA, Sanda MG. 2013. Combining urinary detection of TMPRSS2:ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol Oncol 31:566–571. doi: 10.1016/j.urolonc.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Bryant SE, Xiao G, Barber R, Reisch J, Doody R, Fairchild T, Adams P, Waring S, Diaz-Arrastia R. 2010. A serum protein-based algorithm for the detection of Alzheimer disease. Arch Neurol 67:1077–1081. doi: 10.1001/archneurol.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.CDC. 2018. Lyme disease diagnosis and testing: two-step laboratory testing process. https://www.cdc.gov/lyme/diagnosistesting/labtest/twostep/index.html. Accessed 10 January 2019.
- 26.Mead P, Petersen J, Hinckley A. 2019. Updated CDC recommendation for serologic diagnosis of Lyme disease. MMWR Morb Mortal Wkly Rep 68:703. doi: 10.15585/mmwr.mm6832a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanova L, Christova I, Neves V, Aroso M, Meirelles L, Brisson D, Gomes-Solecki M. 2009. Comprehensive seroprofiling of sixteen B. burgdorferi OspC: implications for Lyme disease diagnostics design. Clin Immunol 132:393–400. doi: 10.1016/j.clim.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engstrom SM, Shoop E, Johnson RC. 1995. Immunoblot interpretation criteria for serodiagnosis of early Lyme disease. J Clin Microbiol 33:419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tokarz R, Mishra N, Tagliafierro T, Sameroff S, Caciula A, Chauhan L, Patel J, Sullivan E, Gucwa A, Fallon B, Golightly M, Molins C, Schriefer M, Marques A, Briese T, Lipkin WI. 2018. A multiplex serologic platform for diagnosis of tick-borne diseases. Sci Rep 8:3158. doi: 10.1038/s41598-018-21349-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahey LJ, Panas MW, Mao R, Delanoy M, Flanagan JJ, Binder SR, Rebman AW, Montoya JG, Soloski MJ, Steere AC, Dattwyler RJ, Arnaboldi PM, Aucott JN, Robinson WH. 2015. Development of a multiantigen panel for improved detection of Borrelia burgdorferi infection in early Lyme disease. J Clin Microbiol 53:3834–3841. doi: 10.1128/JCM.02111-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacek E, Tang KS, Komorowski L, Ajamian M, Probst C, Stevenson B, Wormser GP, Marques AR, Alaedini A. 2016. Epitope-specific evolution of human B cell responses to Borrelia burgdorferi VlsE protein from early to late stages of Lyme disease. J Immunol 196:1036–1043. doi: 10.4049/jimmunol.1501861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandra A, Latov N, Wormser GP, Marques AR, Alaedini A. 2011. Epitope mapping of antibodies to VlsE protein of Borrelia burgdorferi in post-Lyme disease syndrome. Clin Immunol 141:103–110. doi: 10.1016/j.clim.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sackmann EK, Fulton AL, Beebe DJ. 2014. The present and future role of microfluidics in biomedical research. Nature 507:181–189. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 34.Craft JE, Grodzicki RL, Steere AC. 1984. Antibody response in Lyme disease: evaluation of diagnostic tests. J Infect Dis 149:789–795. doi: 10.1093/infdis/149.5.789. [DOI] [PubMed] [Google Scholar]
- 35.Dessau RB. 2013. Diagnostic accuracy and comparison of two assays for Borrelia-specific IgG and IgM antibodies: proposals for statistical evaluation methods, cut-off values and standardization. J Med Microbiol 62:1835–1844. doi: 10.1099/jmm.0.061275-0. [DOI] [PubMed] [Google Scholar]
- 36.Parsa H, Chin CD, Mongkolwisetwara P, Lee BW, Wang JJ, Sia SK. 2008. Effect of volume-and time-based constraints on capture of analytes in microfluidic heterogeneous immunoassays. Lab Chip 8:2062–2070. doi: 10.1039/b813350f. [DOI] [PubMed] [Google Scholar]
- 37.Yager P, Domingo GJ, Gerdes J. 2008. Point-of-care diagnostics for global health. Annu Rev Biomed Eng 10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- 38.Whitesides GM. 2006. The origins and the future of microfluidics. Nature 442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.