Abstract
The arcopallium, a key avian forebrain region, receives inputs from numerous brain areas and is a major source of descending sensory and motor projections. While there is evidence of arcopallial subdivisions, the internal organization or the arcopallium is not well understood. The arcopallium is also considered the avian homologue of mammalian deep cortical layers and/or amygdalar subdivisions, but one-to-one correspondences are controversial. Here we present a molecular characterization of the arcopallium in the zebra finch, a passerine songbird species and a major model organism for vocal learning studies. Based on in situ hybridization for arcopallial-expressed transcripts (AQP1, C1QL3, CBLN2, CNTN4, CYP19A1, ESR1/2, FEZF2, MGP, NECAB2, PCP4, PVALB, SCN3B, SCUBE1, ZBTB20, and others) in comparison with cytoarchitectonic features, we have defined 20 distinct regions that can be grouped into 6 major domains (anterior, posterior, dorsal, ventral, medial and intermediate arcopallium, respectively; AA, AP, AD, AV, AM, and AI). The data also help to establish the arcopallium as primarily pallial, support a unique topography of the arcopallium in passerines, highlight similarities between the vocal robust nucleus of the arcopallium (RA) and AI, and provide insights into the similarities and differences of cortical and amygdalar regions between birds and mammals. We also propose the use of AMV (instead of nucleus taenia/TnA), AMD, AD, and AI as initial steps towards a universal arcopallial nomenclature. Besides clarifying the internal organization of the arcopallium, the data provide a coherent basis for further functional and comparative studies of this complex avian brain region.
Keywords: In situ hybridization; molecularly defined brain regions; oscine songbird; avian; cortical layers; amygdala; taenia; ZEBrA (www.zebrafinchatlas.org, RRID: SCR_012988)
Graphical Abstract
Using in situ hybridization, in comparison with cytoarchitectonic features, the authors identify 6 major domains, and 19 molecularly distinct subdomains within the arcopallium of the adult male Zebra finch. The data provide a basis for further functional and comparative studies of this complex avian brain region.
Introduction
Brain studies in birds offer unique opportunities for understanding the relationship between brain organization and behavior. Birds are increasingly recognized as capable of complex behaviors that require sensorimotor integration, multimodal associations, memory formation and retrieval, temporal reasoning, and other complex integrative functions (Jarvis et al., 2005; Emery, 2005; Gunturkun and Bugnyar, 2016). Accordingly, in comparison with other sauropsids (crocodilians, lizards, turtles), birds generally have an expanded pallium with multiple sensory, motor, and multimodal integrative areas (Butler and Hodos, 2005; Jarvis, 2009). Importantly, pallial areas in birds differ markedly in their cytoarchitectonic organization compared to mammals, but are broadly thought to correspond to pallium-derived brain structures in mammals such as the neocortex, hippocampus, claustrum and amygdalar subdivisions, although exact one-to-one homologies are still a matter of considerable debate (Reiner et al., 2004a; Abellan et al., 2009; Butler et al., 2011; Kuenzel et al., 2011; Puelles, 2011; Dugas-Ford et al., 2012; Belgard et al., 2013; Jarvis et al., 2013; Vicario et al., 2014; Pfenning et al., 2014; Karten, 2015; Vicario et al., 2017; Herold et al., 2018).
The arcopallium, a major avian pallial region, is a complex structure composed of several subdivisions, receives inputs from various brain areas, and originates major descending sensory and motor projections from the telencephalon (Zeier and Karten, 1971; Wild, 1993; Wild et al., 1993; Mello et al., 1998; Cheng et al., 1999; Liao et al., 2011; Mandelblat-Cerf et al., 2014; Herold et al., 2018). These arcopallial projections target a broad range of sub-pallial structures, including multiple nuclei in the brainstem and spinal cord. As such, the arcopallium represents a major output structure through which cortical-like processing circuits are capable of modulating sensory and motor pathways as well as the expression of various behaviors. Accordingly, lesions or inactivation of different regions of the avian arcopallium can have a wide range of effects on behaviors like song learning and production (Nottebohm et al., 1976; Heaton and Brauth, 2000; Mandelblat-Cerf et al., 2014), reproductive behavior (Thompson et al., 1998; Absil et al., 2002), foraging (Xin et al., 2017), fear conditioning (Saint-Dizier et al., 2009) and affective behavior (Cheng et al., 1999) among others.
Despite its central importance, the internal organization of the avian arcopallium is not well understood. Studies based largely on connectivity point to regional specializations (Zeier and Karten, 1971; Wild, 1993; Wild et al., 1993; Mello et al., 1998; Letzner et al., 2016), even though these subdivisions are often difficult to discern with precision based solely on cytoarchitectonics or tract-tracing. The Avian Brain Nomenclature Consortium (Reiner et al., 2004b) proposed several arcopallial subdivisions, consisting of anterior, intermediate, dorsal, and medial parts, as well as the posterior pallial amygdala (PoA), subpallial amygdaloid area (SpA), and nucleus taenia of the amygdala (TnA). Furthermore, some studies provide evidence that discrete arcopallial areas can be identified through immunohistochemical staining for select markers like neuropeptides (e.g. Montagnese et al., 2015), or differential regional binding of ligands for neurotransmitter receptors (Herold et al., 2018). In some cases, discrete arcopallial nuclei that participate in specific circuits and/or functions are readily identifiable. A well-studied example is nucleus robustus arcopallialis (RA), a prominent vocal arcopallial nucleus in songbirds. RA receives projections from other pallial vocal areas, projects to vocal and respiratory centers in the midbrain and medulla, and plays key roles in the encoding and expression of learned vocalizations (Nottebohm et al., 1976; Nottebohm et al., 1982; Vicario, 1991; Wild, 1993; Spiro et al., 1999; Wild et al., 2000; Hahnloser et al., 2002; Long and Fee, 2008; Wild and Botelho, 2015). Analogous nuclei are present in the other bird groups that evolved vocal learning (AAc in parrots and VA in hummingbirds; Brauth et al., 1997; Durand et al., 1997; Gahr, 2000; Jarvis and Mello, 2000; Jarvis et al., 2000), but seem absent in birds that lack this behavioral trait (Kroodsma and Konishi, 1991; Saldanha et al., 2000a, but also see Liu et al., 2013; de Lima et al., 2015). Another example of a discrete arcopallial nucleus is the so-called nucleus taenia, which is thought to mediate telencephalic control of affective state and motivation through projections to the hypothalamus (Zeier and Karten, 1971; Thompson et al., 1998; Cheng et al., 1999). Notwithstanding such examples, most of the arcopallium is still largely unexplored in terms of anatomical, molecular and functional organization, an issue that is further complicated by species differences.
Here we have undertaken a systematic study of the molecular architecture of the arcopallium in the zebra finch. We chose this songbird species as it allows us to assess general features of the arcopallium, as well as specializations of a dedicated nucleus for learned vocalizations (RA). Because a detailed description of arcopallial boundaries, internal laminae and other cytoarchitectonic features was lacking in this species, we initially examined in detail serial Nissl-stained sections, to provide a histological framework for the molecular analysis. We next examined the online zebra finch brain atlas of gene expression (ZEBrA; www.zebrafinchatlas.org, RRID: SCR_012988) and identified a large set of transcripts that were differentially expressed within the arcopallium. In situ hybridization analysis uncovered evidence for 20 discrete arcopallial subdomains, some suggested by Nissl staining while others representing previously unrecognized regional specializations. Comparative analysis points to substantial overlaps in differential markers shared between arcopallial domains and mammalian cortical and amygdalar areas, as well as unique patterns shared with specific cortical or amygdalar subdivisions. We propose a classification of arcopallial subdivisions that brings to light the molecular complexity of this structure, and contributes to designing and interpreting further functional and comparative studies.
Methods
Animals and tissue preparation
Our main goal was to describe the molecular organization of the arcopallium in adult zebra finches (Taeniopygia guttata) by analyzing patterns of constitutively expressed genes. Numerous studies show that sensory stimuli and behavioral state (e.g. sleep, wakefulness, alertness, stress) can modify brain expression patterns (e.g., Mello et al, 1993; Jarvis et al., 1995; Jarvis and Nottebohm, 1998; Dong et al., 2009; Teramitsu et al., 2010; Warren et al., 2010; Whitney et al., 2014), creating significant confounds for searches of neuroanatomical markers. It was therefore very important to make sure that the animals used here consisted of quiet, unstimulated birds. As such, all birds in this study were placed in isolation chambers overnight to minimize singing and hearing of conspecific song. A total of 13 adult male zebra finches were sacrificed by decapitation early the next day following isolation. Their brains were quickly dissected and then blocked in either the sagittal or transverse plane, immersed in Tissue-tek embedding medium (Sakura Finetek, Torrance, CA), frozen in a dry ice-propanol slurry, and stored at −80°C prior to sectioning. The protocol used here was the same as for building the ZEBrA database and website, to ensure that the in situ patterns generated specifically for the present study could be compared with those in ZEBrA. We also followed a nearly identical procedure to obtain brains from 3 two-week post hatch male Leghorn chicks (Gallus gallus domesticus) which were blocked in the transverse plane. We also examined the expression patterns that are available online in the Zebra finch Expression Brain Atlas (ZEBrA; www.zebrafinchatlas.org, RRID: SCR_012988). This database documents the expression of several hundred individual genes by in situ hybridization in a minimum of 2-3 adult male zebra finches, prepared as described above for the current study. All procedures involving live animals were approved by OHSU’s IACUC Committee and are in accordance with NIH guidelines.
Both zebra finch and chick brains were sectioned at 10 μm with a Leica CM1850 cryostat, and mounted on charged microscope slides (Colorfrost Plus; Thermo Fisher Scientific, Waltham, MA). Tissue sections were then fixed in freshly prepared 3% phosphate-buffered paraformaldehyde solution for 5 min, briefly washed twice in 0.1 M PBS, dehydrated in an ascending ethanol series and air dried. For each brain generated for use in this study, every tenth section was stained for Nissl (cresyl violet) using a standard protocol; all other sections were stored at −80°C prior to processing for in situ hybridization.
Probe selection and preparation
For most genes in this study, riboprobes were generated using templates derived from the ESTIMA brain EST/cDNA library (described in Replogle et al., 2008). For each gene, probe specificity was examined by aligning the corresponding EST sequence to the zebra finch genome using UCSC’s genome browser and the BLAT alignment tools. All sequences were confirmed to align to a single locus, or had only very low scoring secondary alignments. In the case of the two probes used for in situ hybridizations with chicken brain tissue, we verified high BLAT genomic alignment scores (91.4% and 95.5% identity at the nucleotide level for C1QL3 and ZBTB20, respectively) and no significant hits to other loci, indicating high specificity and suitability of these probes for cross-species hybridizations. Bacterial stocks for ESTIMA clones of interest were grown overnight, plasmids isolated with a kit (QIAprep spin Miniprep Kit, Qiagen Inc., Valencia, CA), and cDNA inserts were excised by restriction digest (BssHII; New England Biolabs). The templates for riboprobe synthesis of ESR1 and ESR2 were derived from zebra finch cDNAs provided by Art Arnold’s lab (UCLA). In these cases, templates were generated by PCR amplification of an 839 bp fragment of ESR1 (bp275 – 1175 of cDNA L79911 described in Jacobs et al., 1996) and 636 bp fragment of ESR2 (described in Perlman and Arnold, 2003) using primers with T3 and T7 polymerase overhangs. After restriction digest or PCR amplification, DNA templates were cleaned with a purification kit (PureLink Quick PCR Purification Kit, Thermo Fisher Scientific, Waltham, MA). The primary list of probes that were examined and defined a domain or subdomain of the arcopallium are presented in Table 1. We note that a serial set of sections from most brains in this study were hybridized with C1QL3, as preliminary tests determined this gene to be the most reliable and inclusive general marker of the zebra finch arcopallium among the probes tested (see Results), consistent with a previous initial observation about C1QL3 (Fig. S9A in Pfenning et al., 2014).
Table 1†.
Summary of the arcopallial marker expression of genes that provide molecular definitions for the 6 domains and 20 subdomains of the zebra finch arcopallium.
| Whole | Anterior | Medial | Dorsal | Posterior | Intermediate | Ventral | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ventral (AMV) | Dorsal (AMD) | |||||||||||||||||||||
| Gene names | AAc | AArl | AArm | AAv | AMVi | AMVl | AMVm | AMVc | AMVDl | AMDm | AD | APv | APd | RA | AId | AIr | AIrv | AIm | AIv | AV | GenbankID | |
| C1QL3 | + | − | − | − | − | + | − | − | FE726945 | |||||||||||||
| SCUBE1 | + | + | − | − | + | + | + | + | DV947442 | |||||||||||||
| AQP1 | − | − | + | + | + | − | + | + | + | − | + | CK310002 | ||||||||||
| PLPP4 | + | + | − | + | + | CK305549 | ||||||||||||||||
| CRHR2 | + | + | + | CK308437 | ||||||||||||||||||
| ETV1 | + | − | − | + | DV947330 | |||||||||||||||||
| CAMK2N1 | + | + | + | CK308635 | ||||||||||||||||||
| HTR1B | + | + | + | + | + | CK303512 | ||||||||||||||||
| ZBTB20 | + | + | + | − | FE735089 | |||||||||||||||||
| PLS3 | + | + | − | CK302925 | ||||||||||||||||||
| PCP4 | + | + | − | + | + | + | CK311528 | |||||||||||||||
| MGP | + | − | − | − | DV958259 | |||||||||||||||||
| KCNQ5 | + | − | + | + | CK310570 | |||||||||||||||||
| MAP4 | + | − | − | DV945736 | ||||||||||||||||||
| KCND2 | + | − | − | DV956417 | ||||||||||||||||||
| FABP7 | + | + | CK306599 | |||||||||||||||||||
| CYP19A1 | + | + | DV951351 | |||||||||||||||||||
| NECAB2 | − | + | + | + | + | + | CK315373 | |||||||||||||||
| ESR2 | + | PCR temp‡ | ||||||||||||||||||||
| CBLN2 | + | + | FE721173 | |||||||||||||||||||
| CCK | + | − | CK302967 | |||||||||||||||||||
| SV2B | + | + | − | CK302240 | ||||||||||||||||||
| PVALB | + | + | CK305573 | |||||||||||||||||||
| CNTN4 | − | + | + | + | − | DV948150 | ||||||||||||||||
| SCN3B | − | − | FE734016 | |||||||||||||||||||
| SYF2 | + | + | CK306847 | |||||||||||||||||||
| KCNS1 | + | + | FE729668 | |||||||||||||||||||
| HTR2A | + | + | CK306989 | |||||||||||||||||||
| STMN1 | − | CK311233 | ||||||||||||||||||||
| GABRE | + | DV945174 | ||||||||||||||||||||
| CAMKK1 | + | DV947379 | ||||||||||||||||||||
| FEZF2 | + | − | − | DV948042 | ||||||||||||||||||
| SULF2 | + | DV948777 | ||||||||||||||||||||
See Avian Anatomical Abbreviations List for structure abbreviations
Template derived by PCR amplification of a 889 bp fragment of a finch ESR2 cDNA (bp 275-1175) provided by Art Arnold (described in Perlman and Arnold, 2003)
All riboprobes were generated as previously described (Carleton et al., 2014). Briefly, antisense digoxygenin (DIG)-labeled riboprobes were synthesized by incubating the purified DNA template with T3 RNA polymerase (Promega, Madison, WI) and a DIG RNA labeling mix (Roche, Indianapolis, IN) at 37°C for 2 hr, purified on Sephadex G-50 mini-columns, and stored at −20°C until use.
In situ hybridization
We followed a previously described protocol (Carleton et al., 2014) with slight modifications. Briefly, tissue sections were acetylated by incubating slides for 10 min in a freshly prepared solution containing 1.35% triethanolamine and 0.25% acetic anhydride in water. Slides were then briefly washed twice in 2X SSPE (300 mM NaCl, 20 mM NaH2PO4-H2O) and dehydrated in an ethanol series (70%, 95%, 100%). Hybridization solution consisting of 50% formamide, 2X SSPE, 2 μg/μL tRNA, 1 μg/μL BSA, 1 μg/μL Poly A, and 1-4 μL DIG-labeled riboprobe in H2O was freshly prepared and added to slides (16 μL per section). Slides were coverslipped, immersed in a 65°C mineral oil bath, and incubated overnight. The following morning, slides were rinsed in chloroform to remove excess oil and de-coverslipped in 2X SSPE. Slides were then washed at 65°C for 1 hr and 10 min in a 50% formamide 2X SSPE solution followed by two 30 min washes in 0.1X SSPE. During the washes, slides were agitated every 10 min.
Slides were then rinsed in TNT (100 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.3% Triton X-100), covered in blocking buffer (TNB: 100 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.36% w/v BSA, 1% skim milk), and incubated for 30 min at RT in a humidified chamber. Slides were then covered with TNB containing an alkaline phosphatase conjugated anti-DIG antibody (Roche 11 093 274 910; 1:600 in TNB) and incubated for 2 hr in at RT in the humidified chamber. Great care was exercised to prevent drying of solutions over tissue sections during these incubations. Slides were then washed twice for 15 min by gentle shaking in TMN (100 mM Tris-HCl, 150 mM NaCl, 5 mM MgCl2), with vigorous agitation every 5 min. The sections were then transferred to slide mailers containing BCIP/NBT Substrate Solution (PerkinElmer, Waltham, MA) and incubated under gentle shaking at RT for 1-3 days, depending on the target transcript. To minimize artifacts due to chromagen oxidation, slide mailers were filled to the top with freshly filtered chromagen solution and sealed prior to incubation. Slides were then rinsed in DI water, fixed for 20 min in 3% phosphate-buffered paraformaldehyde solution, rinsed in DI water again, and coverslipped with VectaMount permanent mounting medium (Vector, Burlingame, CA). Additional positive (GAD2 probe) and negative (no probe) control slides were included with each hybridization. These slides were used to confirm hybridization quality and monitor background signal during chromagen incubation.
Image Acquisition, Reference Sections, Cell Mapping, and Figure Preparation
Hybridized sections and the reference Nissl-stained sections from brains processed for in situ were photographed at 10X magnification under brightfield optics using a Lumina HR camera mounted on a Nikon E600 microscope yoked to a PC running Neurolucida version 2017 (MBF Bioscience, Williston, VT). For each photograph, the area of interest was selected, background correction was applied, and the image was obtained using the scan slide feature. Images were saved as Tiff files.
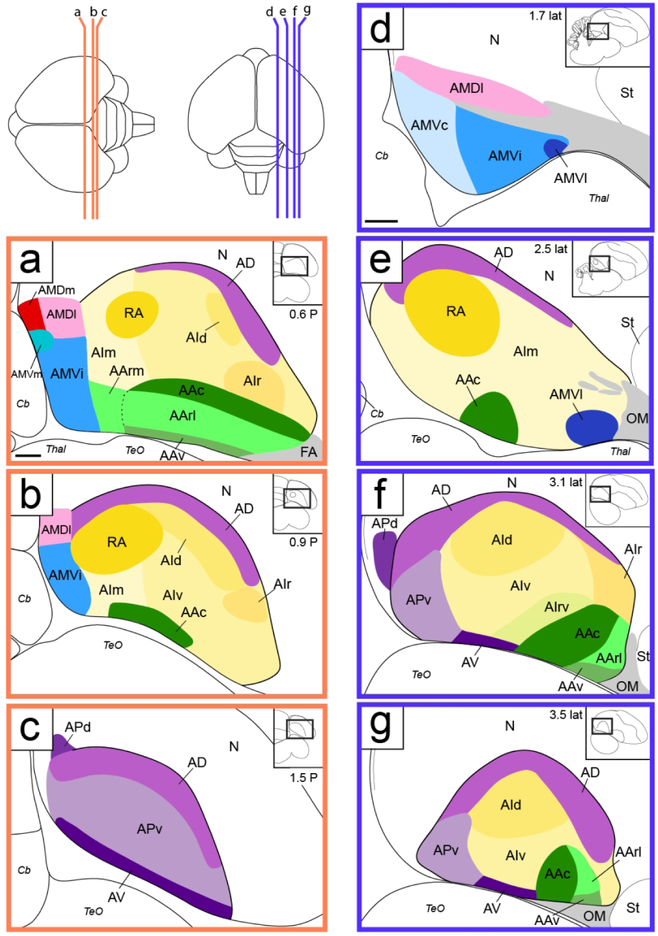
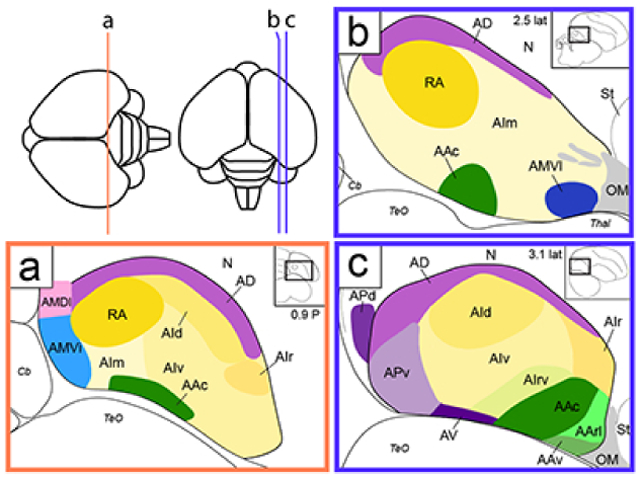
An overview of the range of sections analyzed by in situ hybridization in the transverse and sagittal planes of sections is shown in Figure 1. To provide a general reference of cytoarchitectonic features of the zebra finch arcopallium (Figures 2 and 3), we utilized the high resolution digital images of Nissl-stained serial 30 μm sections from the online histological atlas database (http://zebrafinch.brainarchitecture.org, RRID:SCR_004277), described in Karten et al., 2013. These images allow for clearer visualization of cytoarchitectonic features than the 10 μm sections routinely processed for in situ hybridization (ISH). Representative sections from this collection containing the arcopallium and other structures of interest were identified from one hemisphere in the sagittal plane (brain #0812) and one brain in the transverse plane (brain #0821). Sagittal plane images and schematics based on this series were rotated and aligned to the original block faces as described in Karten et al., 2013. Transverse images and schematics were aligned such that the midline was vertically oriented. We note that this transverse series was prepared in the Frankfurt plane, which is obtained by tilting the anterior pole of the brain slightly downward prior to vertical blocking. As such, the sections are at a more inclined plane than the transverse brains used for in situ hybridization in this and other zebra finch studies.
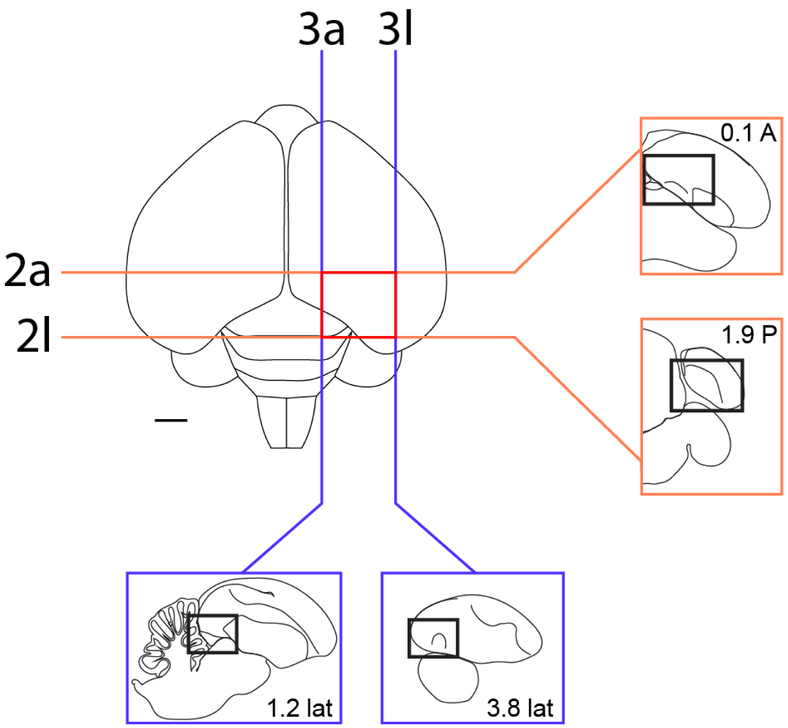
Figure 1.
Localization of the arcopallium in zebra finches. The position and range of sections that contain the arcopallium in adult male zebra finches are shown relative to a dorsal view of the brain. The lines indicate the approximate position of the first and last section in the transverse (orange) and sagittal (blue) series that contain arcopallium and shown in Figs. 2/4 and Figs. 3/5, respectively. Sections containing the arcopallium are located within the area indicated by the red box. The schematic drawings in the blue and orange boxes indicate major structures seen at these brain levels in both planes, while the small rectangles depict the areas of interest containing the arcopallium and shown in Figs. 2-5. Scale: 1mm.
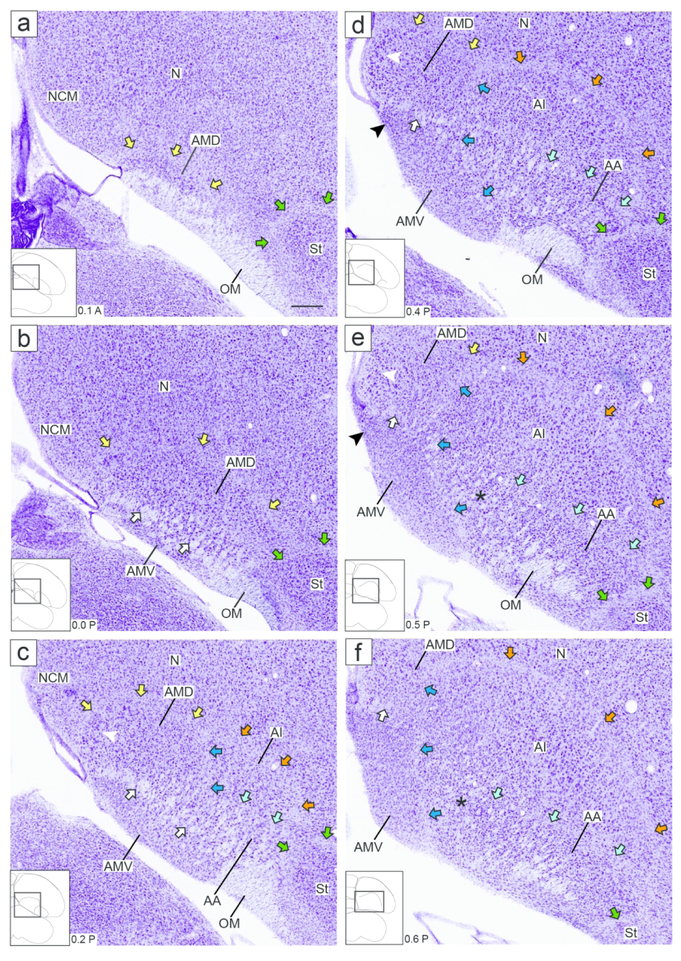
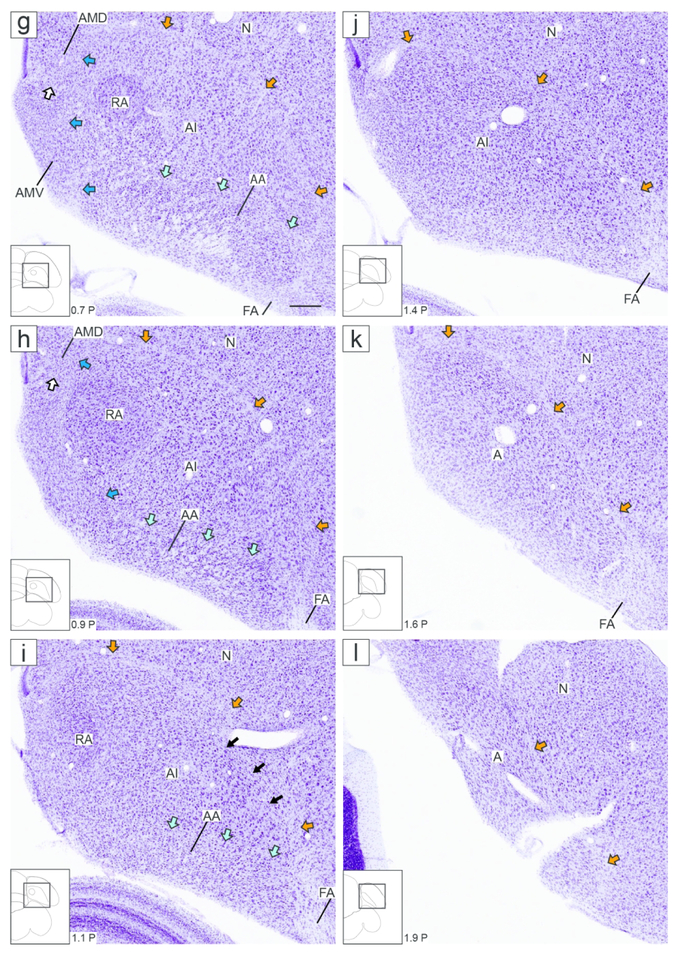
Figure 2.
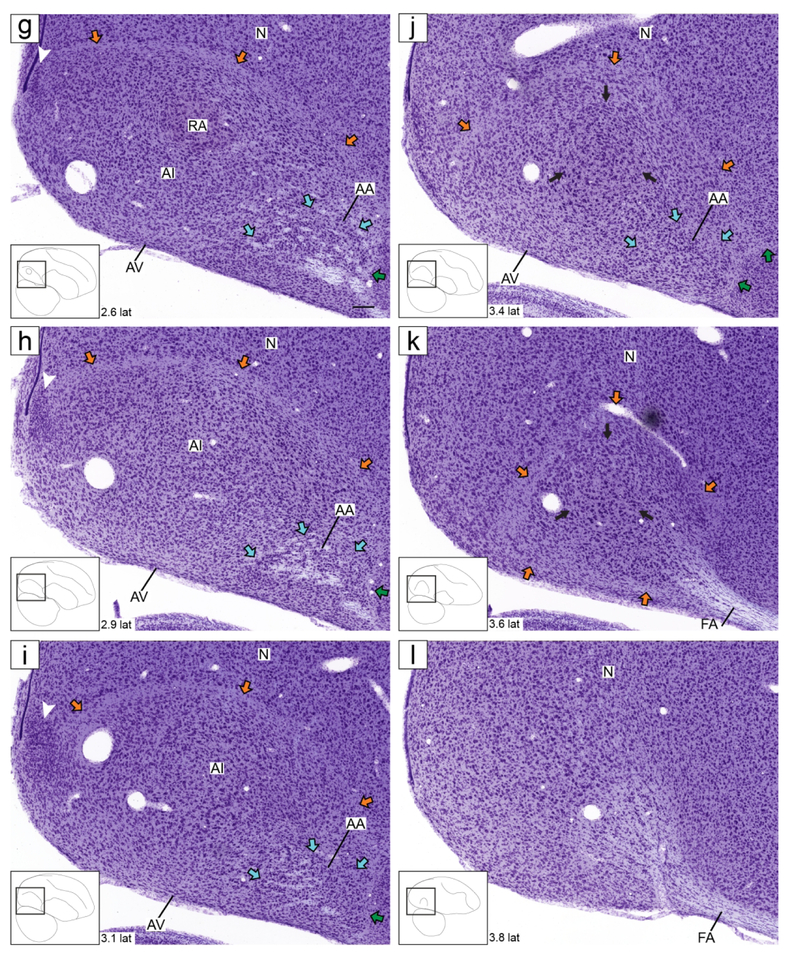
Zebra finch arcopallium on Nissl-stained transverse sections. The images (from Karten et al., 2013) depict the arcopallium and major neighboring structures, with boundaries, internal laminae and cytoarchitectonic domains indicated. The small rectangles in the drawings on the lower left indicate the locations of the areas shown in the images, and the coordinates (in mm) indicate the position of the images along the A-P axis relative to the stereotaxic zero (see also Fig. 1, in orange). Yellow arrows: mLAD; orange arrows: lLAD; green arrows: LPS; white arrows: mOM fiber bundles; dark blue arrows: LAV; light blue arrows: LAO; white arrowhead in (c-e): medial part of AMD with large, darkly staining cells; black arrowhead in (d-e): medial part of AMV with small, tightly packed cells; asterisks in (e-f): medial part of AA rich in fiber bundles; black arrows in (i): dorso-lateral part of AI with darkly staining large cells. For abbreviations, see list. Scale: 250 μm.
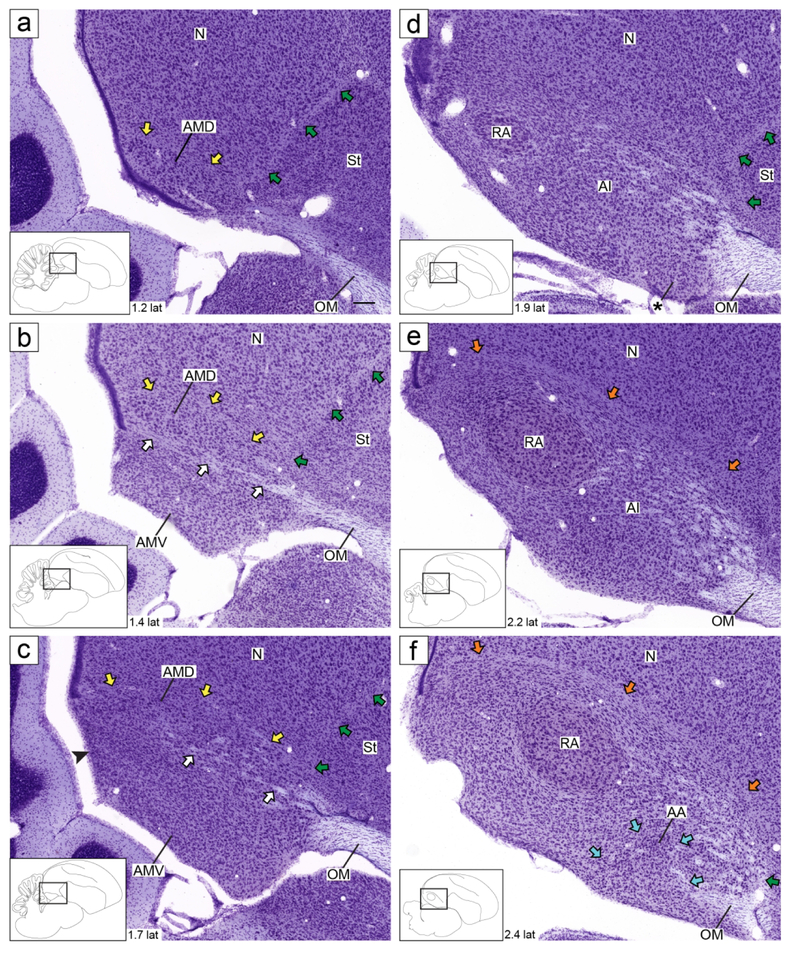
Figure 3.
Zebra finch arcopallium on Nissl-stained sagittal sections. The images (from Karten et al., 2013) depict the arcopallium and major neighboring structures, with boundaries, internal laminae and cytoarchitectonic domains indicated. The small rectangles in the drawings on the lower left indicate the locations of the areas shown in the images, and the coordinates (in mm) indicate the position of the images relative to the midline (see also Fig. 1, in blue). Yellow arrows: mLAD; orange arrows: lLAD; green arrows: LPS; white arrows: mOM fiber bundles; light blue arrows: LAO; black arrowhead in (c): caudal part of AMV with small, tightly packed cells; asterisk in (d): rostro-lateral distinct part of AMV; white arrowhead in (g-i): nucleus with tightly packed cells at the caudal end of the arcopallium; black arrows in (j-k): dorso-lateral part of AI with darkly staining large cells. For abbreviations, see list. Scale: 250 μm.
Additional drawings of whole sections or of areas of interest were generated using Neurolucida/Lucivid. These drawings were based on major features like section borders and borders of major structures like cerebellum and optic tectum, as well as features that are readily identifiable under Nissl staining, including ventricles, major tracts and laminae, and some individual nuclei and regions that have clear cytoarchitectonic borders. We note here that particular attention was paid to laminae, which are thin bands of tissue that delineate brain divisions or subdivisions, and that compared to surrounding tissues have lower density of neuronal cells and higher density of fibers and glial cells. According to this definition, boundaries of individual nuclei are not considered laminae. Laminae were very helpful to define internal subdivisions of the arcopallium as seen under Nissl, and served as important guides for interpreting the in situ images. Drawings were aligned to the Karten finch atlas as discussed above. Illustrator (CC; San Jose, CA) was used to modify the thickness, scaling, and alignment of the contours in all drawings.
For the reconstructions in the transverse and sagittal planes (Figures 4 and 5), every 10th slide (200 μm intervals) in the range that spans the arcopallium was stained for Nissl, and the consecutive slides in the sagittal series were hybridized with probes for C1QL3, SCUBE1, CBLN2, and ZBTB20, and the transverse with C1QL3, ZBTB20, SCUBE2, and CBLN2. The digital images from C1QL3 at each level in the series were aligned with the corresponding drawings from the adjacent Nissl-stained sections in Illustrator (Adobe systems, San Jose, CA), using the borders of the section and of structures like the tectum and cerebellum as cues. The images from all other genes at each level were then similarly aligned relative to C1QL3. The same rectangular window containing the arcopallium and adjacent structures of interest was then applied to crop all aligned images at each level in both series so that comparable panels could be generated for each level. Thus, all images at each level in both series represent the same location in the aligned adjacent sections (depicted as a rectangle containing the arcopallium in the insets of Figures 4 and 5). Images were then scaled down to the desired figure size, without resampling to maintain maximum resolution. Photoshop CC 2018 (Adobe Systems, San Jose, CA) was used to adjust contrast, brightness, and greyscale balance of images, and to correct for artifacts introduced during slide processing (specifically, scratches and chromogen artifactual precipitate). Final figures were constructed in Illustrator CC 2018 (Adobe Systems, San Jose, CA). All general patterns presented in the serial reconstructions were replicated in at least 2 brains and were observed to be largely consistent across both hemispheres in the case of transverse sections.
Figure 4.
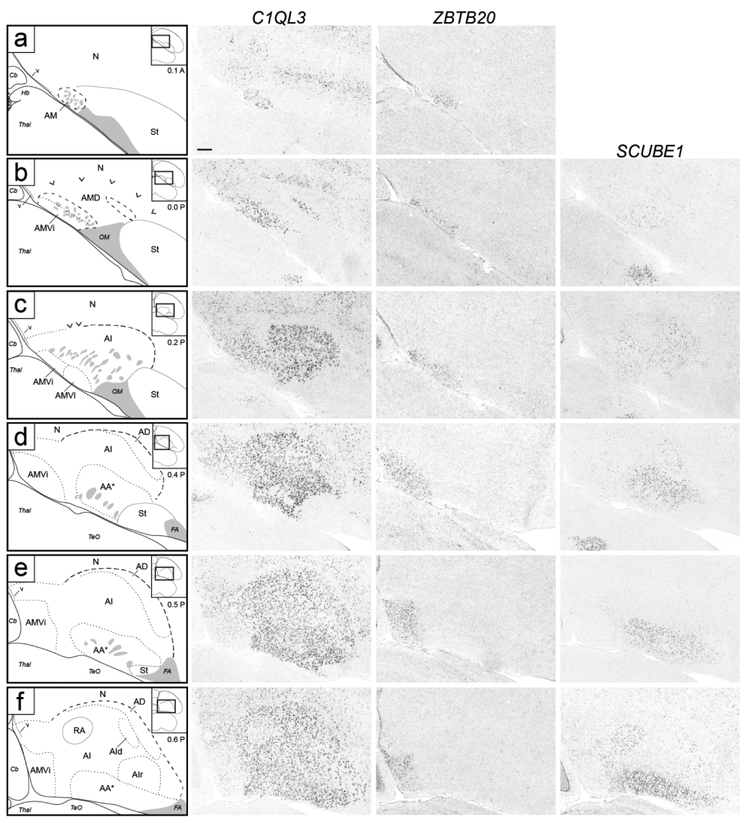
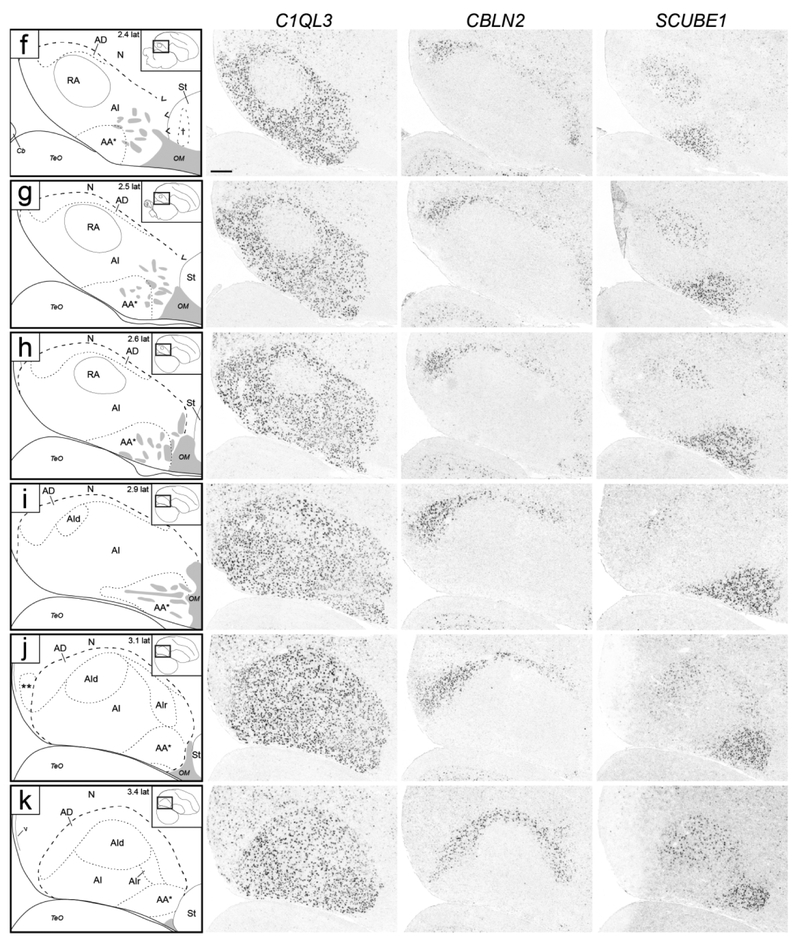
Molecular definition of arcopallial boundaries and major domains on transverse sections. Shown are in situ hybridization images for a general arcopallium marker (C1QL3) and for markers of arcopallial divisions AMV (subdomains AMVi and AMVl; ZBTB20), AD (CBLN2), and AA*, RA and AId (SCUBE1). ZBTB20 is only shown for rostral levels (a-f), CBLN2 is only shown for caudal levels (f-k), and level (f) is repeated for ZBTB20 and SCUBE1. Left column: drawings depicting the structures shown on the in situ images. Solid lines represent tissue borders; dashed lines represent gene expression boundaries drawn after alignment and superposition of in situ images from adjacent sections (thick dashes: dorsal C1QL3 boundary; small dashes: internal domains based on boundaries of expression for regional markers, and medial dorsal border of low C1QL3 expression). Carets in (b-c) depict the dorsal border of the rostral arcopallium that is devoid of C1QL3 expression, grey patches indicate fiber bundles, double asterisks in (j-k) indicate small caudal nucleus of low C1QL3 and high CBLN2 expression. The small rectangles in the drawings on the upper right indicate the locations of the areas shown relative to reference sections, coordinates (in mm) indicate the position along the A-P axis relative to the stereotaxic zero (for full range of sections, see Fig. 1, in orange). For abbreviations, see list. Scale: 250 μm.
Figure 5.
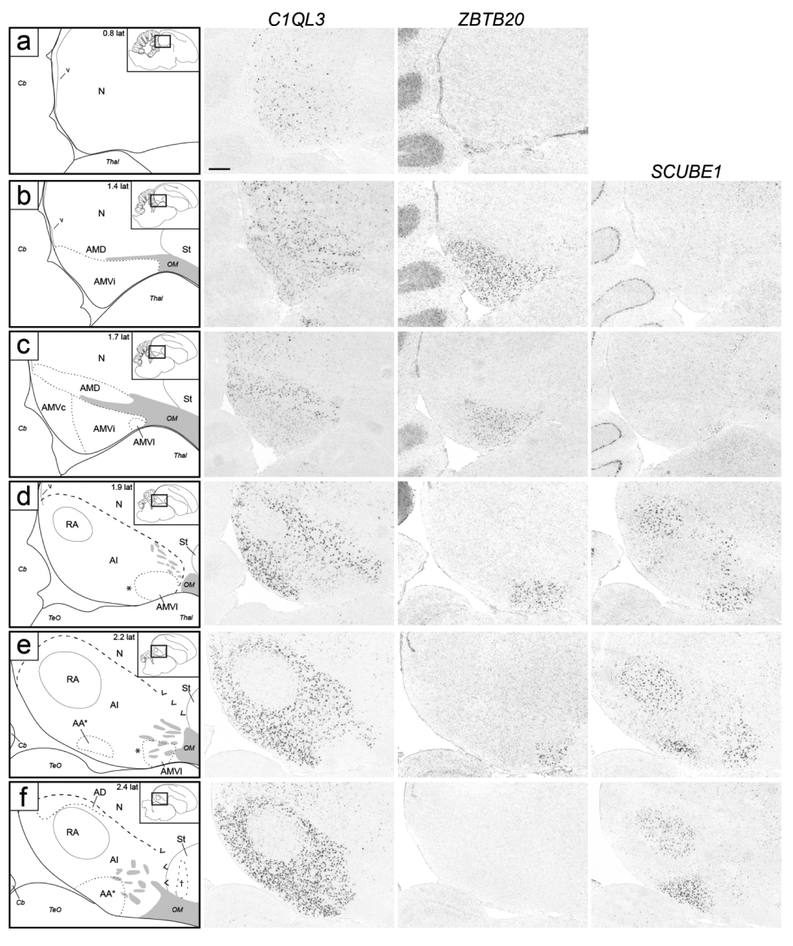
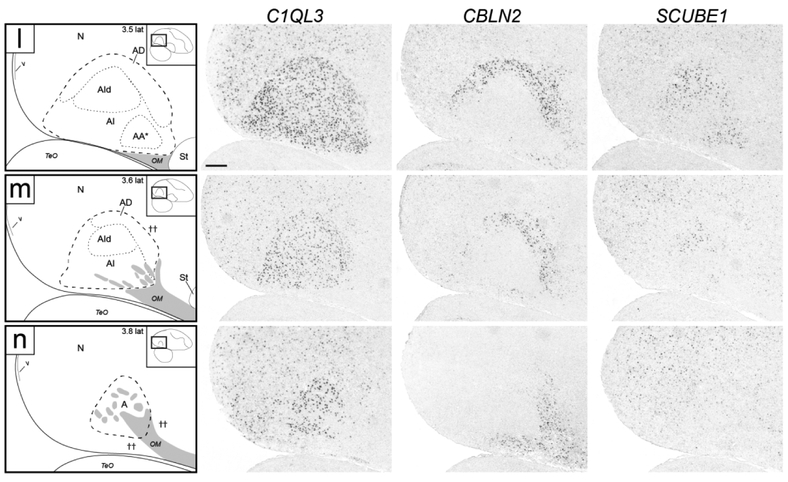
Molecular definition of arcopallial boundaries and major domains on sagittal sections. Shown are in situ hybridization images for a general arcopallium marker (C1QL3) and for markers of AMV (ZBTB20), AD (CBLN2), and AA*, RA, AId and AIr (SCUBE1). ZBTB20 is only shown for medial levels (a-f), CBLN2 is only shown for lateral levels (f-n), and level f is repeated for ZBTB20 and SCUBE1. Left column: drawings depicting the structures shown on the in situ images. Dashed lines represent gene expression boundaries drawn after alignment and superposition of in situ images from adjacent sections (thick dashes: dorsal C1QL3 boundary; small dashes: internal domains based on boundaries of expression for regional markers, and in panel c also the dorsal border of the medial region of moderate C1QL3 expression). Grey patches indicate fiber bundles, asterisk in (d-e) indicates region of low C1QL3 expression just caudal to AMVl, carets in (e-g) depict dorsal border of area of no C1QL3 expression in rostro-dorsal arcopallium, cross in (f) represents a region of high CBLN2 in the caudal striatum rostral to the arcopallium, double asterisk in (j) indicates small caudal nucleus of low C1QL3 and high CBLN2 expression, double crosses in (m-n) indicate region of high CBLN2 expression just rostral and ventral to the lateral arcopallium. The small rectangles in the drawings on the upper right indicate the locations of the areas shown relative to reference sections, coordinates (in mm) indicate the position relative to the midline (for full range of sections, see Fig. 1, in blue). For abbreviations, see list. Scale: 250 μm.
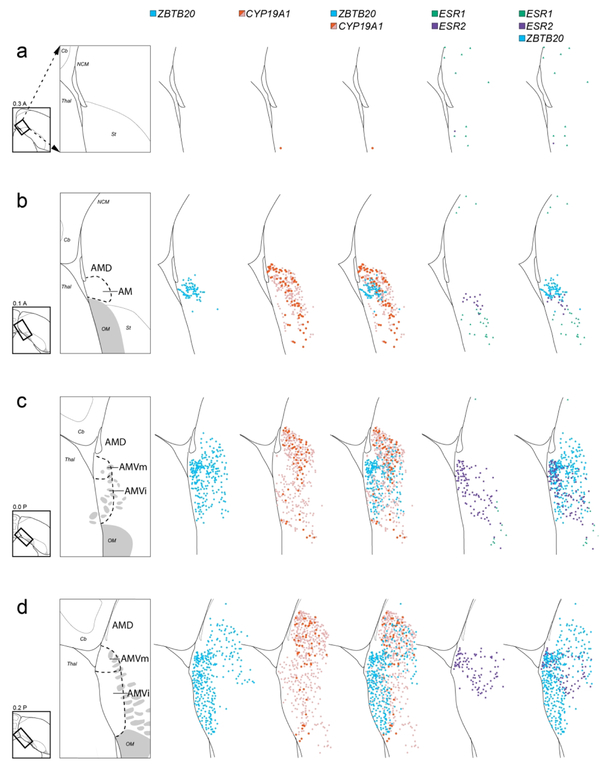
For the mappings presented in Figure 9, every 10th slide (200 μm intervals) in the range that spans the medial arcopallium was stained for Nissl, and the consecutive slides at each level were hybridized with probes for ZBTB20, CYP19A1, ESR1, and ESR2. The hybridized sections were mapped for labeled cells using Neurolucida. Contours representing borders of the tissue and of major structures were initially drawn based on cytoarchitectonics visible in Nissl-stained sections. These drawings were then aligned to consecutive sections processed for in situ, and the respective labeled cells mapped onto the initial drawings using Neurolucida. Using this strategy, all labeled cells from the consecutive hybridized sections at each examined level in the series could be mapped onto the same initial drawings of the Nissl-stained sections. These detailed mappings were performed in two males, one case is presented in Figure 9.
Figure 9.
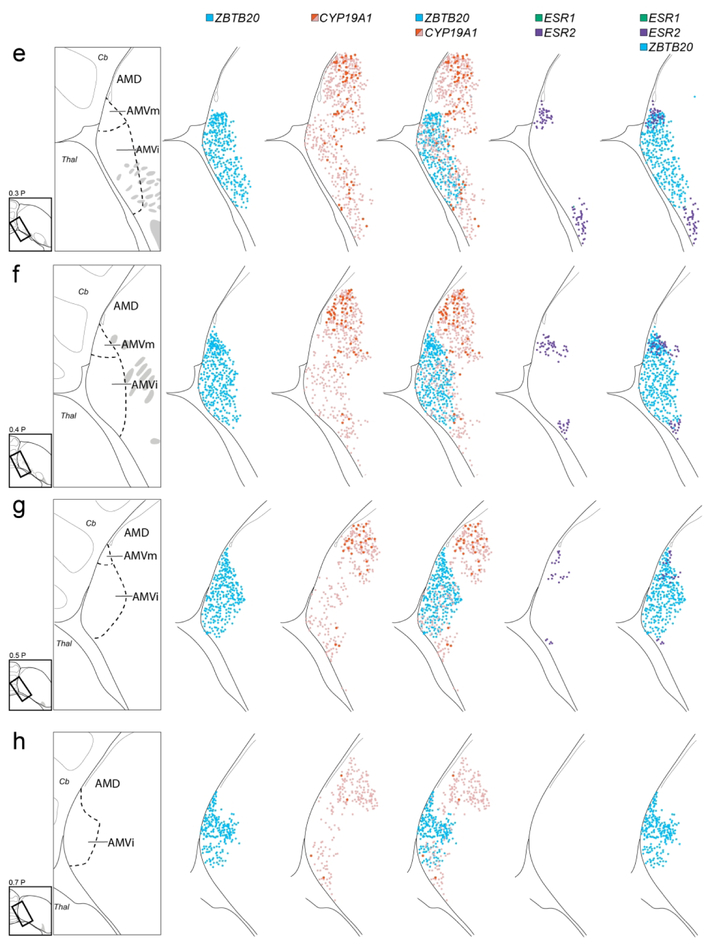
Cellular mapping of differential AM markers. Shown from left to right for each level examined are maps of labeled cells in serial in situ hybridization transverse sections processed for: ZBTB20 (blue), CYP19A1 (red for high label cells, pink for low label cells), ZBTB20 and CYP19A1 combined, ESR2 (purple) and ESR1 (green) combined, and ZBTB20 and ESR1/2 combined. The drawings on the left indicate the region and structures shown on the maps, providing a guide for which specific structures contain the various cell types mapped in this analysis. The diagrams to the bottom left of the drawings indicate levels analyzed, coordinates (in mm) indicate the position along the A-P axis and the rectangles indicate the specific areas mapped relative to the reference sections, grey patches depict major fiber bundles. For clarity, we did not distinguish AMD subdivisions in this Figure. Arrows in (a) indicate how the mapped rectangle areas have been rotated to facilitate comparisons across panels. For abbreviations, see list. Scale: 250 μm.
Analysis of arcopallial domain markers in ZEBrA and the Allen Mouse Brain Atlas.
After completing the initial part of the study, where we identified 20 molecularly-defined arcopallial domains and subdomains, we returned to the ZEBrA database and inspected in detail the patterns for ~200 genes that included sections covering the medial-to-lateral extent of the arcopallium. These genes were evaluated for expression in the arcopallial domains and subdomains described in Table 1, and scored as a positive (+) or negative (−) marker if expression in that subdomain was high or low compared to adjacent domains or subdomains based on visual inspection. Total counts of positive and negative markers for each domain/subdomain were tallied and are presented in Table 2. The lists of all specific genes in ZEBrA that were scored as markers of each domain/subdomain are presented in Table S1.
Table 2†.
Breakdown of the numbers of positive (+) and negative (−) markers in ZEBrA that provide further support for the molecularly defined arcopallial domains in Table 1.
| Whole | Anterior | Medial (AM) | Dorsal | Posterior | Intermediate | Ventral | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ventral(AMV) | Dorsal (AMD) | ||||||||||||||||||||
| Marker Regulation |
AAc | AArl | AArm | AAv | AMVi | AMVl | AMVm | AMVc | AMDl | AMDm | AD | APv | APd | RA | AId | AIr | AIrv | AIm | AIv | AV | |
| − | 30 | 3 | 0 | 0 | 4 | 8 | 4 | 0 | 1 | 1 | 0 | 2 | 4 | 1 | 83 | 34 | 11 | 4 | 0 | 0 | 0 |
| + | 6 | 23 | 1 | 0 | 3 | 14 | 9 | 2 | 6 | 9 | 0 | 23 | 10 | 12 | 46 | 17 | 5 | 6 | 0 | 0 | 8 |
See Avian Anatomical Abbreviations List tor structure abbreviations
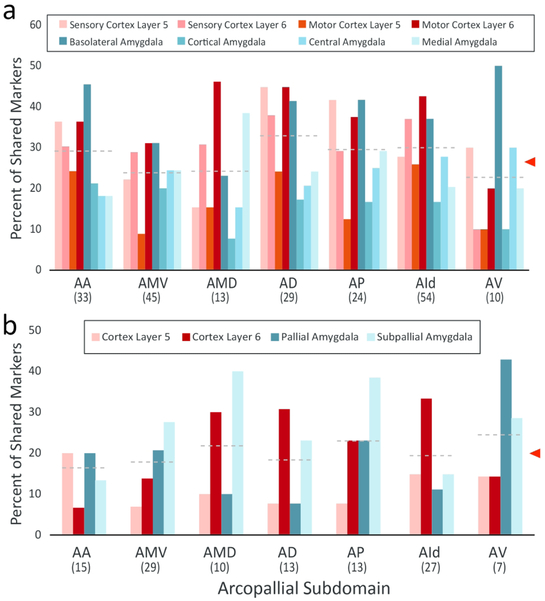
We next conducted a comparative analysis with mammalian brain structures based on the Allen Mouse Brain Atlas (MBA; mouse.brain-map.org, SCR_002978). Specifically, we examined whether the genes identified as markers of at least one arcopallial domain or subdomain in the zebra finch (Table 1 and Table S1; n=129 genes, RA-specific markers excluded) could also be classified as markers of specific layers of the sensory or motor cortices, or of major amygdalar subdivisions. RA-specific markers were excluded, since our goal was to seek for correspondences between avian and mammalian structures rather than vocal learning circuitry analogs. For the MBA cortical analysis we examined: (a) layers 5, 6a and 6b in the primary motor cortex; and (b) layers 5, 6a and 6b in the primary sensory cortex. For the primary sensory cortex we examined primary auditory cortex (MBA’s reference plate 74), or primary visual cortex (MBA’s reference plate 82) when sections containing primary auditory cortex were unavailable for a given gene. For the amygdalar structures we examined the sub-pallial central (CEA; MBA’s reference plates 63-75) and medial amygdala (MEA MBA’s reference plates 63-76), as well as the pallial basolateral (BLA; MBA’s reference plates 63-79) and cortical amygdala (COA; MBA’s reference plates 63-79). We note that the majority of genes included in this analysis were part of the Anatomic Gene Expression Atlas (AGEA) subset within MBA, thus constituted high quality gene sets in this database, but some cases where the expression was difficult to assess with confidence in the MBA had to be removed. For each gene, we used the “Expression Energy” masking feature while scoring expression patterns to help determine the expression contrast across structures annotated in the MBA reference atlas. Genes were scored as markers of a cortical layer if expression in that layer differed compared to the other layers examined in the cortical area of interest (motor or sensory). Genes were scored as markers of an amygdalar structure if the majority of subdivisions within the amygdalar structure were differential compared to the other amygdalar structures. To simplify the comparison with mammalian structures we collapsed the set of genes that defined 18 arcopallial subdomains (RA excluded) into 7 domain/subdomains consisting of AA, AMV, AMD, AD, AP, AId, and AV. We note that while AMV and AMD are subdivisions of AM, they were analyzed separately because many markers in ZEBrA differentiate them. In the case of AI, we only considered AId as there are only very few markers for other AI subdivision. To determine the degree of similarity between arcopallial domains and mammalian structures we calculated the percent of shared markers for each pairwise comparison. Specifically, we calculated the percentages of molecular markers for each of the 7 arcopallial subdomains examined that were also markers of a cortical layer and/or amygdalar structure.
Results
We have conducted a large scale examination of the molecular architecture of the zebra finch arcopallium. While the bulk of the effort is based on gene expression, it was helpful to first identify cyto- and myeloarchitectonic features (e.g., cell groups, laminae, and fiber tracts) that could be readily identifiable under Nissl and myelin staining, to provide a general framework and guidance for the molecular analysis. This was accomplished by examining high-resolution digital images of serial sagittal and transverse sections from the online histological zebra finch brain atlas (http://zebrafinch.brainarchitecture.org, RRID:SCR_004277; Karten et al., 2013). Below we present first the Nissl-based and in situ observations for the whole arcopallium, followed by analyses of arcopallial subdivisions.
Defining the Arcopallium in zebra finches
Under Nissl, the zebra finch arcopallium appears as a large structure in the caudal telencephalon (Fig. 1), spanning ~1.8 mm along the A-P axis (from 0.1 A to 1.9 P; Fig. 1, orange lines) and ~2.6 mm along the M-L axis (1.2 to 3.8 from the midline; Fig. 1, blue lines). Its boundaries and nearby structures change along the rostro-caudal axis. As best seen in transverse sections, the rostral arcopallium (from 0.1 A to ~0.6 P) is bound ventro-laterally by the pallial-subpallial lamina (LPS; green arrows in Fig. 2a-f), which separates it from the striatum. Caudally, as the striatum ends, the arcopallium is bound ventro-laterally by the fronto-arcopallial tract (FA; Fig. 2g-k). Dorsally, the arcopallium is bound by the dorsal arcopallial lamina (LAD), which separates it from the nidopallium; a smaller medial part (mLAD) starts rostrally (yellow arrows in Fig. 2a-e), whereas the larger, more arching lateral part (lLAD) is more prominent at caudal levels (orange arrows in Fig. 2c-l). In sagittal sections, the arcopallium is seen as bound rostrally by the LPS (green arrows in Fig. 3a-d and 3f-j), which separates it from the caudal striatum, while at the lateral-most level it is bound rostrally by the FA (Fig. 3k-l). Dorsally, mLAD and lLAD are seen respectively in medial (yellow arrows in Fig. 3a-c), and lateral sections (orange arrows in Fig. 3e-k), but the LAD is difficult to visualize at the transition zone (Fig. 3d, not drawn).
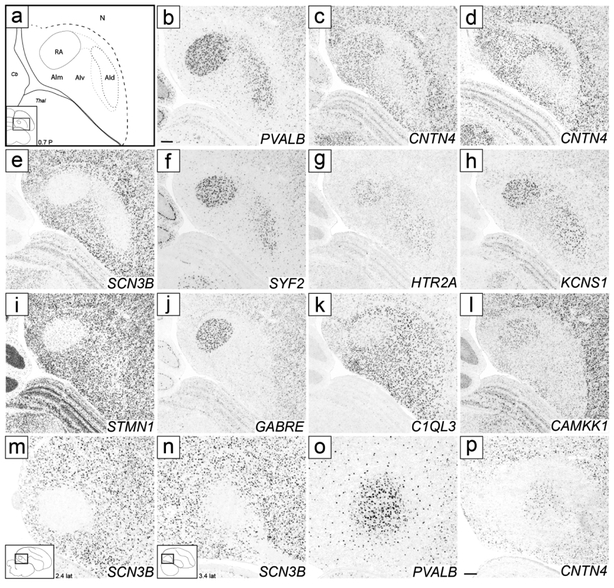
We next sought to identify molecular markers to help define the arcopallium and clarify its borders. The ZEBrA in situ hybridization database includes patterns for numerous transcripts reported as differentially expressed in the arcopallium compared to other telencephalic regions (see arcopallium markers in ZEBrA:www.zebrafinchatlas.org, RRID: SCR_012988). We conducted a detailed in situ evaluation of a subset of these genes, including both up- (C1QL3, ETV1, SV2B) and down-regulated (AQP1, HPCAL1, PLPP4) markers. We note that C1QL3 and ETV1 were previously suggested as good candidate arcopallial markers (Pfenning et al., 2014), and that most of the genes we examined were found to be good general markers of the arcopallium, often identifying boundaries not clearly seen by Nissl. C1QL3, however had the most consistent and inclusive pattern, providing the strongest contrast between the arcopallium and the adjacent striatum and nidopallium. As seen on both the transverse and sagittal series, most of the area of high C1QL3 expression (Figs. 4c-k and 5d-n) matched the Nissl-defined arcopallium (Figs. 2 and 3). C1QL3 was thus chosen as a general arcopallial marker and routinely hybridized to serial sections for most brains in this study.
A closer look revealed that the rostral-most arcopallium, located medio-dorsally to OM (thus corresponding to the rostral portion of Nissl-defined AMD, as seen in Fig. 2b) was negative for C1QL3 (Fig. 4b). In fact, the dorsal border of this region could be defined by the higher C1QL3 expression in the overlying nidopallium (carets in Fig. 4b). This C1QL3-negative region is immediately caudal to the medial striatum (the region defined by carets in Fig. 4b is at the same position as the medial extent of the striatum in Fig. 4a). High C1QL3 expression started rostrally as small islands of labeled cells (thick dashed lines in Fig. 4a-b) within this C1QL3-negative region. Slightly more caudally, the C1QL3-positive clusters became elongated, merged (Fig. 4c), and quickly expanded to eventually occupy almost the entire arcopallium (Fig. 4d-k). The rostral region lacking C1QL3 expression waned caudally, but could still be seen as a small dorsal domain, at the transition between AM and AI (carets in Fig. 4c). C1QL3 expression was also low medially, so that the C1QL3-defined dorsal boundary was less distinct medially (thin dorso-medial dashed line in Fig. 4c-i) than laterally (thick dorso-lateral dashed line in Fig. 4c-k). C1QL3 expression was particularly low medio-ventrally (Fig. 4e-g) and in RA (Fig. 4f-i). A similar pattern was seen in sagittals, a clear dorsal boundary seen laterally (Fig. 5c) but not medially (Fig. 5b), as well as very low expression in AMV (Figs. 5b-c), the rostral-most region (boundary indicated by carets in Fig. 5e-g), rostroventrally (Figs. 5d-e), and in RA (Fig. 5d-h). C1QL3 was also distinctively low in a discrete caudal domain (double asterisks in Fig. 5j) that corresponded to a caudal nucleus with tightly packed cells next to the lateral ventricle tip, as clearly seen in Nissl-stained sagittal sections (white arrowhead in Fig. 3h-i).
Defining subdivisions of the Arcopallium
Before describing the regional in situ data, we note that some prominent features seen under Nissl provided indications of arcopallial subdivisions and helped build a framework for molecular analysis. These included, in the transverse series: (a) the occipito-mesencephalic tract (OM), which appears as a compact ventral tract in rostral sections (Fig. 2a-b) and whose medial part (mOM) breaks into thick fiber bundles that are gradually displaced medially (white arrows in Fig. 2b-h); (b) an arching vertical arcopallial lamina that we named LAV runs roughly parallel to the medial surface of the telencephalon and is most clearly seen rostrally and ventrally (dark blue arrows in Fig. 2c-h); and (c) an oblique arcopallial lamina that that we named LAO that runs parallel to the ventral surface of the telencephalon (light blue arrows in Fig. 2c-i). Whereas LAV is not readily visible in the sagittal series, LAO and OM are prominent, the latter appearing rostrally as a compact tract (Fig. 3a-f) that at medial levels breaks into thick fiber bundles along the rostro-caudal axis of the arcopallium (white arrows in Fig. 3b-c).
We next evaluated patterns for transcripts identified in the ZEBrA database as differentially expressed in specific arcopallial domains, and identified ZBTB20, CBLN2 and SCUBE1 as having the most restricted distributions within the arcopallium. Their analysis, compared with the Nissl reference sections (Figs. 2-3), C1QL3 (Figs. 4 and 5), and other spatially restricted markers (Figs. 6-13) provided a basis for molecularly defining several arcopallial domains and subdomains.
Figure 6.
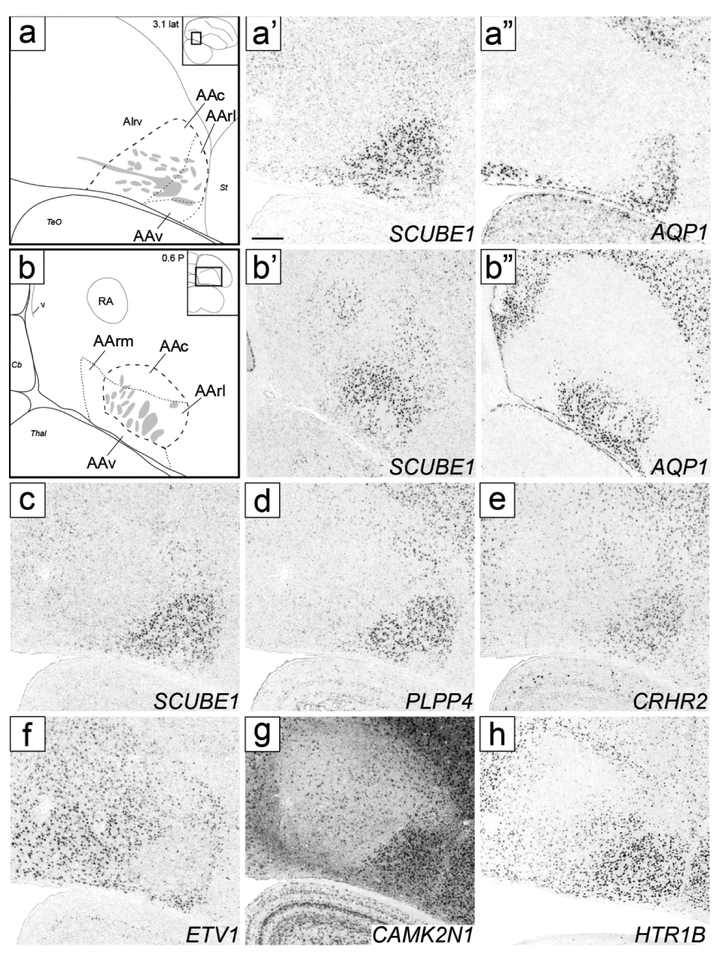
Molecular definition of AA and its subdomains. (a) Drawing depicts structures in sagittal images (a’-a” and c-h). (a’-a’’) In situ hybridization images define AA and its subdomains AAc, AArl and AAv based on SCUBE1 (a’) and AQP1 (a’’); AIrv in (a) indicates region of moderate HTR1B expression caudal to AAc (seen in h). (b) Drawing depicts structures in transverse images (b’-b’’). (b’-b’’) In situ hybridization images define AA and its subdomains AAc, AArm, AArl and AAv based on SCUBE1 (b’) and AQP1 (b’’). In both (a) and (b), thick dashed lines represent SCUBE1 expression, thin dashed lines represent AQP1 expression, grey patches indicate fiber bundles, diagrams on upper right indicate positions of the drawn areas relative to reference sections, and coordinates (in mm) indicate the position along the respective axes. (c-h) In situ hybridization sagittal images for positive (c-e/g-h: SCUBE1, PLPP4, CRHR2, CAMK2N1 and HTR1B) and negative (f: ETV1) markers of AA and its subdomains; (c) is a repeat of (a’) for contrast with other genes. For abbreviations, see list. Scale: 250 μm.
Figure 13.
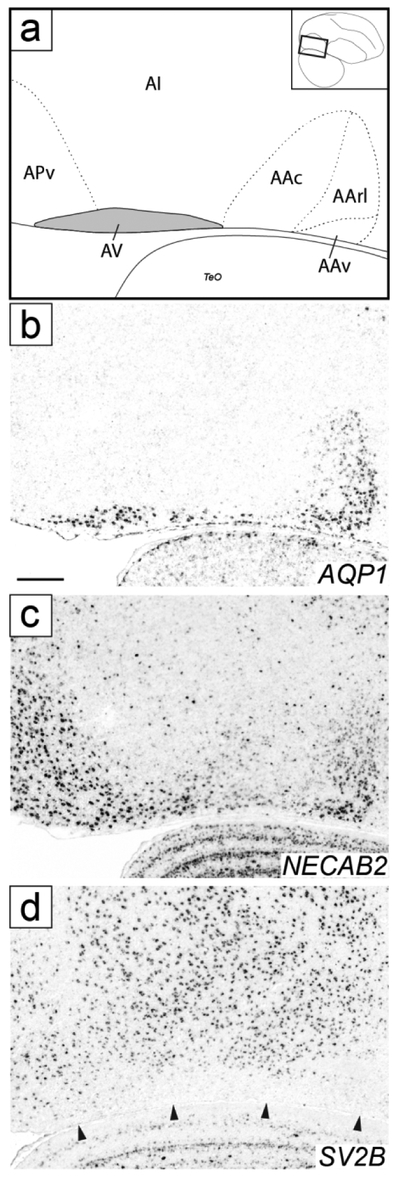
Molecular definition of AV. (a) Drawing depicting structures shown in the sagittal in situ images in (b-d); area in grey shows molecularly defined AV, dashed lines indicate boundaries of AA and AP, as detailed further in Figs. 6 and 11, rectangle in the inset on the top right indicates location of area shown relative to a reference section, coordinate (in mm) indicates the position along the A-P axis. (b-d) Sagittal in situ hybridization images show positive (b-c; AQP1 and NECAB2) and negative (d; SV2B) AV markers; arrowheads in (d) indicate ventral border of the arcopallium. We note that (b) replicates Fig. 6a’’, although partially and at a different angle. For abbreviations, see list. Scale: 250 μm.
Anterior arcopallium (AA) complex
Under Nissl, a distinct arcopallial division starts rostroventrally in the transverse series, named here anterior arcopallium (AA; Fig. 2c-d). AA is bound laterally by the LPS (green arrows in Fig. 2c-d), medially by the LAV (dark blue arrows in Fig. 2d), dorsally by the LAO (light blue arrows in Fig. 2d), and ventrally by OM. More caudally, AA becomes wider with a trapezoid shape and bound ventrally by the ventral border of the telencephalon (Fig. 2e-f), but tapers off at a level where RA is large and OM fiber bundles break up (Fig. 2h-i). The rostro-medial part of AA is rich in fine fiber bundles (asterisks in Fig. 2e-f). In medial sagittal sections, AA is seen rostro-ventral to lateral RA, has a triangular shape, and encompasses the rostral-most fiber bundles that coalesce to form the OM tract (Fig. 3f-i). Laterally, AA becomes flattened dorsally (Fig. 3h-j), and ventrally, AA is devoid of fiber bundles.
SCUBE1 was strongly expressed in a domain (indicated as AA* in Figs. 4d-I and 5e-m) that covered most of the Nissl-defined AA (Figs. 2c-i and 3f-j). On rostral transverse sections, this SCUBE1 domain appeared as a rectangular area close to the ventral border of the telencephalon and medial to the St (Fig. 4d-g) that tapered off caudally (Fig. 4h-i), although it did not extend as far medially as Nissl-defined AA. On sagittals, this SCUBE1 domain started as a small ventral area rostral to intermediate RA (Fig. 5e) that laterally became larger and triangular, occupying the rostro-ventral most arcopallium (Fig. 5f-g). Laterally, the SCUBE1 domain became flattened dorsally (Fig. 5i-k), and then waned (Fig. 5l-m). The central part of this domain included many thick fiber bundles associated with OM rostro-ventrally.
In contrast, AQP1 was selectively expressed in the rostral part of the SCUBE1 domain, but also extended medially and ventrally beyond this domain. Taken together, the sum of the area defined by both markers (Fig. 6a-b) closely matched the Nissl-defined AA (Figs. 2c-i and 3f-i). These markers also provide evidence for AA subdomains. Specifically, a caudal subdomain (AAc) was positive for SCUBE1 only, whereas a rostro-lateral subdomain (AArl) was positive for both SCUBE1 and AQP1 (Figs. 6a- a’’ and 6b-b’’) and contained some of the largest fiber bundles as they coalesce to form the OM tract. SCUBE1- and AQP1-positive cells in AArl (Figs. 6b’ and b’’ respectively) were interspersed among these fiber bundles. In contrast, rostro-medial (AArm) and ventral (AAv) subdomains were positive for AQP1, but negative for SCUBE1 (Fig. 6a-a’’ and 6b-b’’). The distinction between AArm and AArl was best seen in transverse sections (Fig. 6b-b’’). AArm matched the medial-most AA subregion rich in small fiber bundles as seen in Nissl (asterisks in Fig. 2e-f).
Other genes further support AA and subdomains. The up-regulated expression of PLPP4 and CRHR2 (Fig. 6d-e), and down-regulated expression of ETV1 (Fig. 6f) matched closely the SCUBE1 pattern. CAMK2N1 appeared as a positive AA marker due to down-regulation throughout the arcopallium compared with AA and adjacent forebrain areas (Fig. 6g). HTR1B showed upregulation in AA, but also moderate expression that extended caudally beyond AA into rostroventral AI (Fig. 6h, labeled AIrv in Fig. 6a; see AI below). PLPP4 and ETV1 were also differentially expressed in AAv (Fig. 6d and f), further differentiating AAv from the rest of the AA. NECAB2 matched closely the AQP1 pattern, with low expression in AAc but high expression in AArl and AAv (Fig. 13a-c) as well as in AArm (not shown, but see NECAB2 and AQP1 patterns in ZEBrA).
Medial arcopallium (AM)
Under Nissl, we defined AM as a medial division composed of distinct dorsal and ventral subdivisions, named AMD and AMV, respectively. In the transverse series (Fig. 2a-h), these subdivisions are bound laterally by the LAV (dark blue arrows in Fig. 2c-h), and separated from each other by the fiber bundles from mOM (white arrows in Fig. 2b-h). AMD, the rostral-most region of the arcopallium (Fig. 2a), is bound ventrally by mOM and dorsally by mLAD (yellow arrows in Fig. 2a-e). AMV starts rostrally as an elongated thin band at the ventral border of the telencephalon, below mOM (Fig. 2b). More caudally it becomes thicker and vertically elongated (Fig. 2c-g), tapering off by the level of caudal RA. At its thickest, it occupies the medio-ventral-most corner of the telencephalon (Fig. 2d). Both AMD and AMV seem to shift medially going from rostral to caudal levels (Fig. 2a-d), due to the medial border of the telencephalon shifting laterally as the cerebellum expands and the caudomedial nidopallium (NCM) wanes. In medial sagittal sections, AMV and AMD are separated from each other by mOM fiber bundles (white arrows in Fig. 3b-c). Further specializations within AM include: a) the medial-most AMD has darkly staining large cells that are distinct from the rest of the AMD and from the nidopallium above (white arrowhead in Fig. 2c-e); b) the medial-most AMV has smaller and more densely-packed cells than the rest of the AMV (black arrowhead in Fig. 2d-e); c) the caudal-most AMV has a higher density of small, compact cells than more rostral AMV (black arrowhead in Fig. 3c); and d) a rostral region in lateral AMV, bound rostrally by OM and dorsally by mOM appears as a distinct subdomain in sagittal sections (asterisk in Fig. 3d). As detailed below, several in situ patterns differentiate Nissl-defined AM from other arcopallial regions and/or support the differentiation of AMV and AMD and their subdomains.
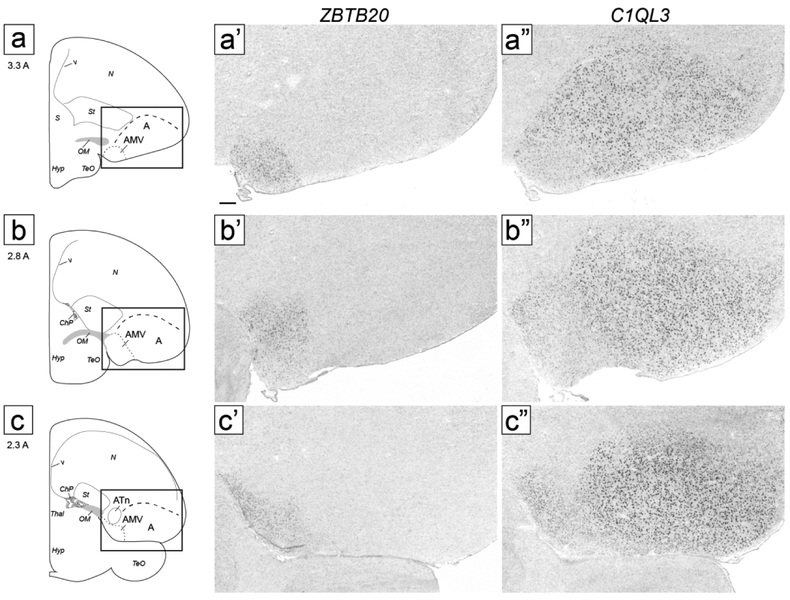
AMV:
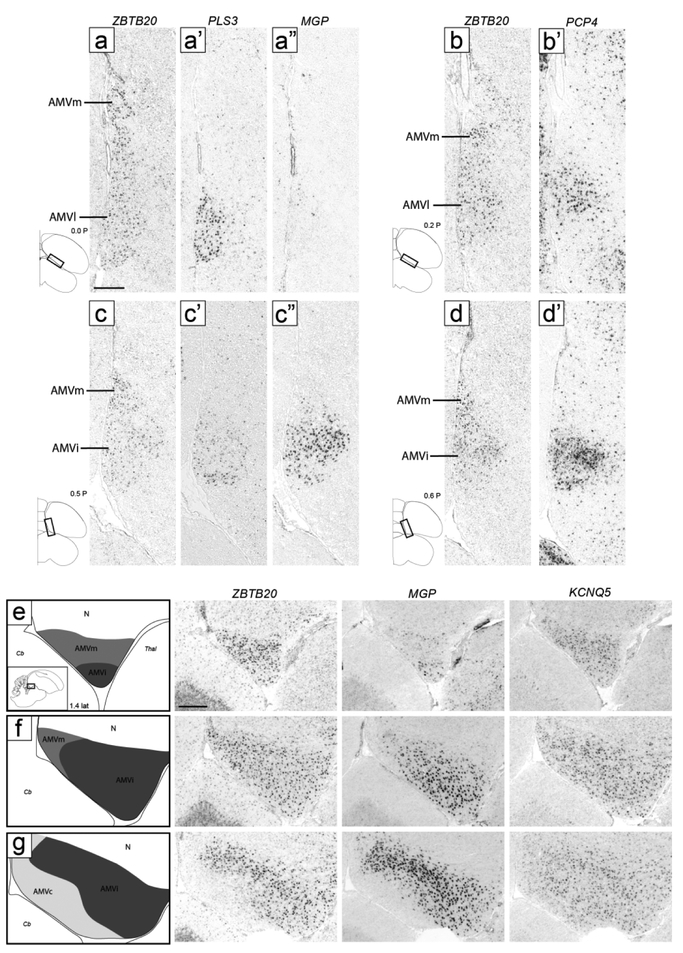
ZBTB20 expression corresponded closely to the Nissl-defined AMV, but our molecular data revealed a prominent central subdomain, which we named intermediate AMV (AMVi), as well as distinct subdomains, which we detail below. On transverse sections, AMVi started rostrally as a small elongated region medial to OM and ventral to the medial OM fiber bundles (Fig. 4b-c). It then appeared to shift medially (Fig. 4d) and became elongated vertically, occupying the entire medio-ventral corner of the telencephalon (Fig. 4e-f). At more caudal levels it waned and terminated as a small round region just medial to caudal RA (Fig. 4g-h; ZBTB20 images not shown but indicated as AMV in the drawings on the left). On sagittals, the ZBTB20 domain we defined as AMVi was seen ventral to mOM medially (Fig. 5b-c).
At rostral levels, the lateral part of the ZBTB20-expressing domain overlapped with the ventral part of a broad region of moderate SCUBE1 expression. This lateral AMV subdomain (AMVl) is best seen on sagittals (Fig. 5c-e), and matched closely the distinct rostro-lateral part of AMV seen under Nissl (compare ZBTB20 and SCUBE1 in Fig. 5d-e with the region indicated by an asterisk in Fig. 3d). The overlap of ZBTB20 and SCUBE1 in AMVl could also be seen on transverse sections (Fig. 4c). AMVl could be distinguished from the larger AMVi based on MGP, which was expressed in AMVi but not in AMVl (compare Fig. 7c’’ with Fig. 7a’’), whereas other markers like PLS3 and PCP4 were highly expressed in both AMVi and AMVl (compare Fig. 7c’/d’ to Fig. 7a’/b’). We also noted a medial AMV subdomain (AMVm) that could be defined by expression of ZBTB20 (Fig. 7a) but lack of expression of PLS3 (Fig. 7a’), MGP (Fig. 7a”), and PCP4 (Fig. 7b’). This molecularly defined AMVm matched closely the medial-most AMV seen under Nissl as having smaller and more densely-packed cells than other AMV subdomains (black arrowhead in Fig 2d-e). Further supporting AMVm and AMVl, we found that MAP4 and KCND2 behaved similarly to MGP, with high expression in AMVi but not in AMVl or AMVm, whereas FABP7 was expressed highly both in AMVi and AMVm (not shown). Lastly, a caudal subdomain (AMVc) best seen in sagittals was defined by lack of expression of ZBTB20 and C1QL3 (Fig. 5c). AMVc corresponds to the caudal area of high density of small cells seen ventral to mOM under Nissl (black arrowhead in Fig. 3C).
Figure 7.
Molecular definition of AMV subdomains. (a-d) Transverse in situ hybridization images define AMV subdomains based on ZBTB20 and other markers. AMVl is positive for PLS3 (a’), negative for MGP (a’’), and positive for PCP4 (b’); AMVi shows moderate expression of PLS3 (c’) and high expression of MGP (c’’) and PCP4 (d’); AMVm only expresses ZBTB20. Diagrams on bottom left show location of regions shown in all panels relative to reference sections, coordinates (in mm) indicate the position along the A-P axis. (e-g) In situ hybridization images show AMV subdomains on medial sagittal sections. Drawings on the left indicate structures shown in the in situ images, diagram on bottom left of (e) indicates location of areas shown in panels (e-g), coordinate (in mm) indicates the position along the M-L axis. AMVm (medium grey) shows high expression of ZBTB20 and KCNQ5 but not of MGP, AMVi (dark grey) shows high expression of all three markers, and AMVc (light grey) shows high expression of KCNQ5 but not of ZBTB20 or MGP. For abbreviations, see list. Scale: 250 μm.
Examination of medial sagittal sections at close intervals further clarified relationships across AMV subdomains (Fig. 7e-g). AMVm was seen very medially as a ZBTB20- and KCNQ5-positive but MGP-negative domain, contrasting with the more ventral AMVi, positive for all 3 markers (Fig. 7e). At more lateral levels, AMVi expanded, while AMVm shifted caudally and waned (Fig. 7f). As AMVm ended, AMVc appeared as a caudo-ventral region that was KCNQ5-positive but ZBTB20- and MGP-negative (Fig. 7g). More laterally, the ZBTB20-defined AMVi shifted rostrally, AMVc expanded caudally, and AMVl (SCUBE1-positive) started as a small domain rostrally (Fig. 5c). More laterally, we observed an abrupt transition after which strong C1QL3 expression started, AMVi ended, and the ZBTB20-expressing domain became restricted to the now larger AMVl (Fig. 5d). At this level, the area of low C1QL3 expression extended caudal to the ZBTB20-positive domain, such that a small C1QL3- and ZBTB20-negative domain was present just caudal to the ZBTB20-defined AMVl (asterisk in Fig. 5d-e). Lastly, we note that at the rostral-most level of the arcopallium, a small round area of strong ZBTB20 expression medial to St and OM matched a similar area of strong C1QL3 expression (AM in Fig. 4a). We labeled this rostral area AM because we were unable to establish whether it was dorsal or ventral to mOM, and thus whether it corresponded to AMV, AMD, or an area where these two subdivisions merge.
AMD:
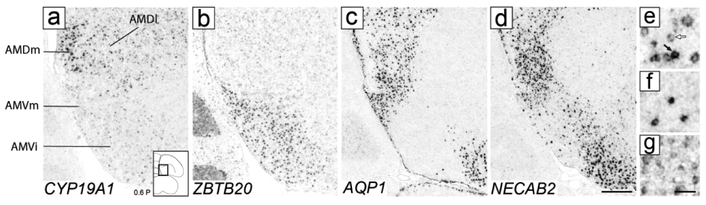
CYP19A1 was most highly expressed in a domain within AM located dorsal to AMV and ventral to the medial nidopallium and that resembled AMD as defined by Nissl (e.g., compare CYP19A1 in Fig. 8a to Nissl in Fig. 2d). AMD was notably devoid of high ZBTB20 expression that defines AMV (Fig. 8b). CYP19A1 expression was most conspicuous in the medial-most part of AMD named AMDm, identifiable under Nissl as a medial region of darkly stained large cells (white arrowhead in Fig. 2c-e), and less marked in lateral AMD (AMDl). On sagittals, the high CYP19A1-expressing AMDm corresponded to the medial-most AMD (dorsal to the OM fibers), representing the first arcopallial domain seen medially (not shown, but see ZEBrA, CYP19A1, level 5). At more lateral and rostral levels, however, while CYP19A1 expression was still enriched in AMD, it was not restricted to this domain and thus less useful to molecularly define it (details below). We also note that while other genes like AQP1 and NECAB2 were not specific markers of AMD, they were substantially enriched in this subdomain, contrasting sharply with their low expression in AMV (Fig. 8c-d). Interestingly, these AMD-enriched genes also showed strong expression in AMVm, further establishing the distinct molecular identity of this subdomain.
Figure 8.
Molecular definition of AMD. (a-b) In situ hybridization images on adjacent transverse sections show high CYP19A1 expression in AMDl and AMDm (a) and ZBTB20 expression in AMVi and AMVm (b). (c-d) In situ hybridization images on transverse sections show high expression of AQP1 (c) and NECAB2 (d) in AMD and AMVm, and low expression in AMVi. The diagram on the bottom right of (a) shows the approximate location of panels (a-d) relative to a reference section, coordinate (in mm) indicates the position along the A-P axis. (e-g) High power views depicting examples of labeled cells that were mapped on serial sections (as shown in Fig. 9). (e) CYP19A1-expressing cells in AMDm, including high label (black arrow) and low label (white arrow) types. (f) ESR1-expressing cells in NCM. (g) ESR2-expressing cells in the ventro-medial arcopallium. For abbreviations, see list. Scale: 250 μm for (a-d); 50 μm for (e-g).
Differential cell type mapping in AM:
To better characterize and distinguish AMV and AMD, we examined closely the distribution of individual ZBTB20- and CYP19A1-expressing cells on serial transverse sections. ZBTB20-labeled cells (blue diamonds in Fig. 9) had a discrete distribution that matched closely the definition of AMV, especially at intermediate to caudal levels, and were particularly dense in AMVm (Fig. 9e-h). At the rostral-most levels, ZBTB20 cells had a somewhat broader distribution, and while still more numerous medio-ventrally, some cells were also seen more dorsally and laterally (Fig. 9c-d).
CYP19A1-expressing cells could be divided into two populations, a sparser high expression cell-type and a more numerous low expression cell-type (black and white arrows, respectively, in Fig. 8e), which we mapped separately. The high label cells (red circles in Fig. 9) had a spatially more restricted distribution that largely corresponded to AMD (Fig. 9c-h), and were seen at several rostro-caudal levels just dorsal to and largely non-overlapping with the ZBTB20-expressing cells. Some of these cells were also seen more laterally and ventrally, although rarely within AMV subdivisions. In contrast, low label cells (pink circles in Fig 9) had a broader distribution than AMD and were also seen within AMV subdivisions (Fig. 9c-h). Overall, CYP19A1 expression was more marked in AMD but also occurred in AMV (Fig. 9, compare the CYP19A1 and ZBTB20 distributions in the overlay column). Thus, in contrast with the more discrete distribution of ZBTB20 in AMV, CYP19A1 was not as distinct a marker for distinguishing these subdivisions. We also note that CYP19A1-expressing cells in AMD were continuous in their distribution with labeled cells in NCM, thus this gene cannot be used as a marker to distinguish the AMD border with NCM. For clarity, CYP19A1-expressing cells in NCM were not included in the Fig. 9 maps.
We also mapped the expression of estrogen receptors alpha (ER-alpha, a.k.a. ESR1) and beta (ER-beta, a.k.a. ESR2) on sections adjacent to ZBTB20 and CYP19A1 from the same series as above, as these receptors have been previously described as enriched in medio-ventral arcopallium and as defining nucleus taenia. ESR2-expressing cells (Fig. 8g) formed two distinct and densely packed clusters (purple squares in Fig. 9e-g). The medial ESR2 cluster corresponded to the ZBTB20-positive AMVm, and the lateral cluster was located just lateral to the ZBTB20-expressing AMVi (Fig. 9e-g, overlay of ZBTB20 and ESR2). More rostrally, ESR2-expressing cells had a somewhat broader distribution that extended beyond AMVm (Fig. 9c-d). Interestingly, ESR2 cells were distinctly absent in the areas of high CYP19A1 expression, especially AMD, as well as in most of the ZBTB20-positive AMV. In contrast to ESR2, ESR1-expressing cells (Fig. 8f, green triangles in Fig. 9) were not observed in the arcopallium; instead, these cells were seen within NCM medially (Fig. 9a-b), or within the St laterally (Fig. 9a-b).
Dorsal arcopallium (AD)
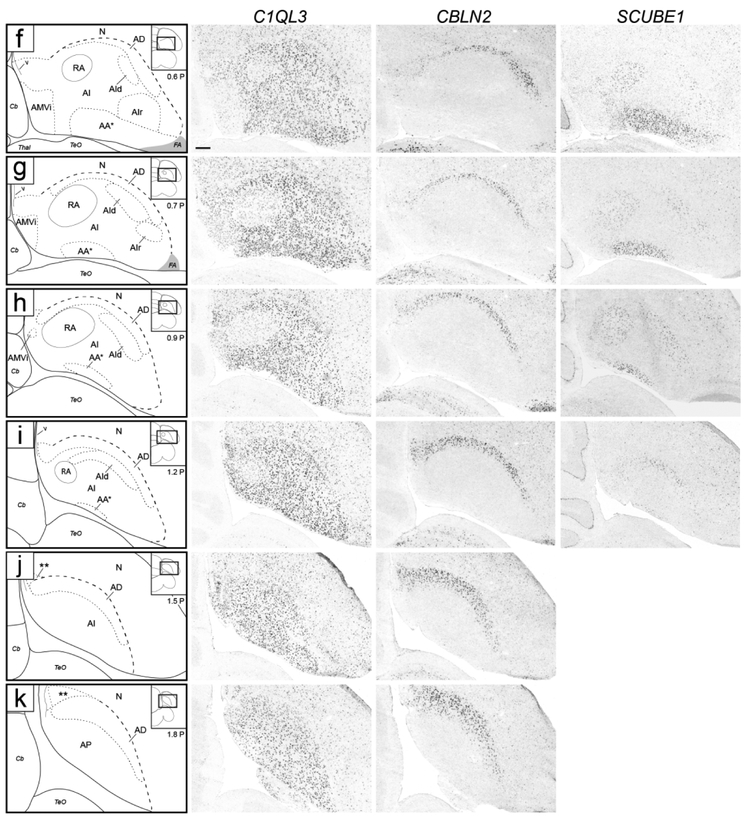
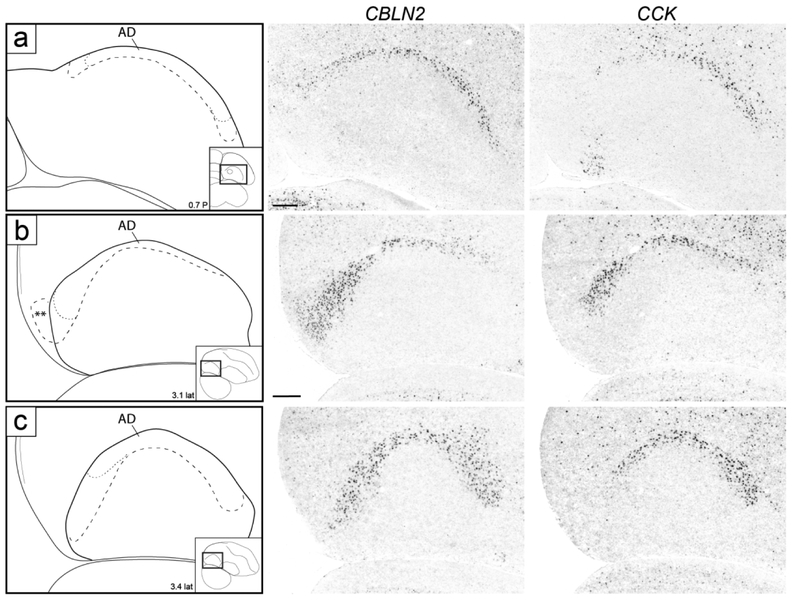
While not readily defined under Nissl, this domain could be defined molecularly. CBLN2 was prominently expressed in a dorsal domain just underneath the dorsal border of the arcopallium as defined by C1QL3 (Figs. 4f-k and 5f-m). On transverse serial sections, this domain was seen as an arching region dorsal to RA that became more distinct laterally, and waned medially over RA (Fig. 4f). Caudal to RA this CBLN2-expressing domain became thicker medially (Fig. 4i-k). On sagittals, a full dorsal arching domain was clearly seen at lateral levels (Fig. 5k-l), whereas medially this CBLN2 dorsal domain waned rostrally and expanded caudally (Fig. 5f-j). CBLN2 was also strongly expressed in a discrete medial region within the caudal striatum rostral to the arcopallium (cross in Fig. 5f), and in a lateral region rostral and ventral to the lateral arcopallium (double crosses in Figs. 5m-n).
To examine AD more closely, we compared CBLN2 expression with CCK, also identified as expressed in a discrete dorsal arcopallial region (Fig. 10). CCK expression largely matched CBLN2 in the dorsal domain but did not extend as medially or laterally (Fig. 10a), and while CCK also showed some caudal expansion (Fig. 10b), it was more restricted than CBLN2 (Fig. 10b-c). Because CBLN2 extended caudally beyond CCK and overlapped with genes that helped to define a posterior arcopallial domain (see below), we concluded that CCK was a more specific marker of dorsal arcopallium (AD). Of note, AD was not distinguishable on Nissl-stained sections, except for levels where a distinct dorsal AI suggested a more dorsal narrow region just underneath LAD (dorsal to the black arrows in Figs. 2i and 3j). This dorsal region appears to correspond closely to the CBLN2-positive/CCK-positive domain. Lastly, other genes like HTR1B (Fig. 6h) and CNTN4 (see Fig. 12c-d below) also defined AD by differential expression compared to the underlying AI.
Figure 10.
Molecular definition of AD. (a-c) In situ hybridization images of CBLN2 and CCK in transverse (a) and sagittal (b-c) sections. Both markers are expressed in AD, but CCK expression is restricted to AD, whereas CBLN2 expression extends further medially and laterally (a) as well as caudally (b-c), beyond AD. Drawings on the left indicate structures shown on in situ panels; larger dashed line indicates AD boundary by CBLN2, thinner dashed line indicates limits of CCK expression; insets on bottom right of drawings indicate location of area of interest on reference sections, coordinates (in mm) indicate the position along the respective axes, double asterisks in (b) indicates region of CBLN2 expression that extends beyond the caudal arcopallium boundary based on C1QL3 expression, as detailed in Figure 11. For abbreviations, see list. Scale: 250 μm.
Figure 12.
Molecular definition of RA, AId, and other AI subdomains. (a) Drawing depicting structures shown in the transverse in situ images in (b-l); dashed lines ventro-medial to AId and between AId and RA indicate regions of variable expression of AId markers; AIm indicates region of high expression of CNTN4 in medial AI; rectangle in the inset on bottom left indicates location of area shown relative to a reference section, coordinate (in mm) indicates the position along the A-P axis. (b-h) In situ hybridization images of positive and negative markers of RA and AId. (i-l) In situ hybridization images of positive and negative markers of RA. (m-p) In situ hybridization images of RA and AId markers on sagittal sections, at the level of RA (m) and AId (n-p); rectangles in the insets on bottom left indicate location of areas shown relative to reference sections, coordinates (in mm) indicate the position along the M-L axis. For abbreviations, see list. Scale: 250 μm.
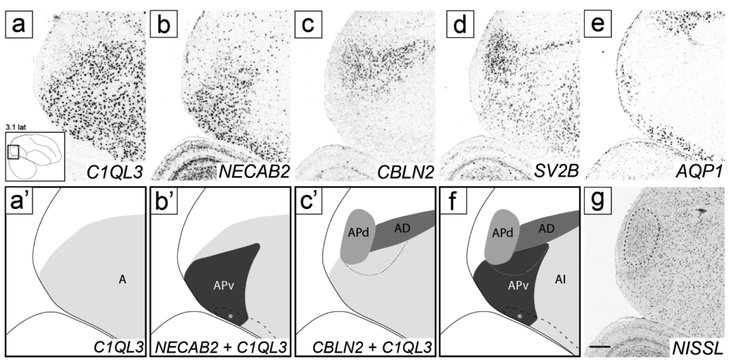
Posterior arcopallium (AP)
This region could not be readily defined under Nissl, and its molecular definition required an examination of multiple genes. The caudal-most expansion of the CBLN2 expression domain was most prominent on sagittal sections lateral to RA (Figs. 5i-j and 10b-c), but could also be seen caudal to RA (Fig. 5f-h). While continuous with AD, this caudal expansion of the CBLN2 expression domain appeared to extend even beyond the C1QL3 boundary (e.g. double asterisks in Figs. 4k and 5j). To explore this region further, we compared CBLN2 with other markers that were prominently differential in this arcopallial region.
While C1QL3 was uniformly expressed throughout the caudal arcopallium and helped to define a distinct caudal boundary (Fig. 11a-a’), NECAB2 was highly expressed within the posterior part of the C1QL3-positive domain, ventrally but not dorsally. We named the NECAB2- and C1QL3-positive domain ventral posterior arcopallium (APv; Fig. 11b-b’). NECAB2 expression contrasted sharply with CBLN2, which was markedly expressed dorsally (Fig. 11c-c’). The CBLN2-expressing domain included AD, whose caudal border is defined by the caudal boundary of CCK expression (as shown in Fig. 10a) and which was contained within the C1QL3-expressing region (Fig. 11c’). We noted, though, a small ventral region of weaker CBLN2 expression that extended caudoventrally beyond AD (thin dashed line in Fig. 11c’). Upon overlaying the CBLN2 and NECAB2 expression domains, this ventral CBLN2-expressing domain overlapped partially with NECAB2-defined APv (thin dashed line in Fig. 10f). CBLN2 also extended caudally and dorsally beyond the C1QL3 boundary, to include a caudal nucleus that showed distinctly higher SV2B expression (Fig. 11d) compared to other parts of the arcopallium or the overlying nidopallium. The caudal boundary of AQP1, a general negative arcopallial marker, also extended beyond the caudal C1QL3 boundary to include the SV2B-positive nucleus (Fig. 11e).
Figure 11.
Molecular definition of AP and subdomains. (a-e) In situ hybridization sagittal images of the caudal arcopallium region processed for C1QL3, a general arcopallial marker (a), NECAB2, a marker of APv (b), CBLN2, a marker of AD and APd (c), SV2B, a marker of APd (d), and AQP1, a general negative marker of the arcopallium and of APd (e). The lower left inset in (a) indicates the location of the region shown in all panels relative to a reference section, coordinates (in mm) indicate the position along the M-L axis. (a’-c’) Schematic drawings depicting the AP subdomains defined by the in situ images in (a-c). (f) Summary diagram including all AP subdomains in (a’-c’). (g) Nissl-stained section close to the in situ processed sections in (a-e), depicting APc (see also Fig. 3g-i). Thick dashed line (b’, f) indicates continuation of NECAB2 expression with AV rostrally; asterisk indicates overlap of AP and AQP1-defined AV, further detailed in Figure 13; medium dashed line (c’, f, g) indicates APd boundary, and thin dashed line (c’ and f) indicates region of overlap between NECAB2 and CBLN2. For abbreviations, see list. Scale: 250 μm.
This SV2B-defined nucleus corresponded to a compact distinct nucleus of small tightly packed cells located immediately rostral to the caudo-ventral tip of the lateral ventricle, most readily seen on sagittal sections just lateral to RA (Fig. 11g and white arrowhead in Fig. 3g-i). It obscures the caudal-ventral end of LAD, thus it is difficult to conclusively determine whether it is within the arcopallium based on Nissl alone. We interpret the molecular data as evidence that this nucleus corresponds to a caudal-most and dorsal extension of the posterior arcopallium, and thus named it APd (Fig. 11f, also double asterisks in Figs. 4j-k, 5j and 10b).
Interestingly, the smaller rostro-ventral part of APd shows C1QL3/NECAB2 expression, but the larger caudo-dorsal part is C1QL3/NECAB2-negative (Fig. 11f). The ventral part of the NECAB2 expression domain overlapped with AQP1, thus defining a distinct ventral subdomain within APv (white asterisk in Fig. 6b’ and f). Taken together, we have identified 4 components of the AP (three subdivisions of APv plus APd), as drawn in Fig. 11f. Lastly, NECAB2 expression also extended rostro-ventrally beyond APv (Fig. 6b and black dashed line in Fig. 6b’), defining a domain discussed below under AV.
Intermediate arcopallium (AI)
This large and complex region and its internal subdivisions could not be fully defined based on Nissl staining alone, but some cytoarchitectonic features provided useful guides for in situ analysis. At rostral levels in the transverse series it appears as a prominent subdivision bound medially by LAV (dark blue arrows in Fig. 2c-d) and ventrally by LAO (light blue arrows in Fig. 2c-d). It expands rapidly more caudally, but keeps the same boundaries along the rest of the transverse series (Fig. 2e-i). Within medial AI, just lateral to the caudal AMD and AMV, lies the robust arcopallial nucleus (RA; Fig. 2g-i). Consistent with the previous literature on this nucleus (e.g., Arnold 1980, Konishi and Akutagawa, 1985), RA has distinct neurons with large somata and a darkly-stained neuropil. Lateral to RA is an arching AI region containing cells that appear larger and more darkly staining than in the surrounding arcopallium, though the exact boundaries are not clearly discernible (e.g., see black arrows Fig. 2i). In sagittals, this AI region appears as a round structure reminiscent of nucleus RA (black arrows Fig. 3j-k), however it is located laterally to RA and lacks the clear borders and prominent features that distinguish RA from its surrounds. This lateral-dorsal arching AI region, where present, suggests a thin arcopallial region dorsal to AI and underneath the LAD. As detailed next, while we did not find markers that consistently labeled the entire AI, some markers defined AI subdomains.
AId and RA:
Besides being an AA marker, SCUBE1 was moderately but consistently upregulated in RA and in a dorso-lateral AI subdomain lateral to RA and immediately ventral to AD that we named dorsal AI (AId; Fig. 4f-i), compared to other AI domains. On caudal transverse sections (e.g. Fig. 4i), RA and AId formed an almost continuous dorsal arching domain extending from RA to ventro-lateral AI. At more rostral levels RA and the AId were separated by an intervening SCUBE1-negative region (Fig. 4f-h). This dorso-lateral SCUBE1-defined AId could also be followed on the sagittal series, starting lateral to RA and extending to more lateral levels of the arcopallium (Fig. 5i-m). The location of this SCUBE1-defined AId resembled closely the AId as seen under Nissl on both transverse (Fig. 2i) and sagittal (Fig. 3j-k) sections. Under Nissl, this domain appears to have somewhat larger cells and lower cell density compared with more ventral parts of the AI. The combined RA and AId arching region helps define the ventral border of the AD, as well as provide a dorsal boundary for other domains within the AI.
Other markers were more variable in defining AId, suggesting AId may possess subdomains. For example, PVALB-defined AId (Fig. 12b) appeared thicker than for SCUBE1 and connected to RA without an evidence for a non-differential intervening region (Fig. 12b; see fine dashed lines in Fig. 12a). In contrast, CNTN4-defined AId at the same level was thinner (Fig. 12c) and less connected with RA, connecting only at more caudal levels (Fig. 12d), thus resembling SCUBE1 more than PVALB (although CNTN4 differed from SCUBE1 by high expression medially). All other identified markers of both structures had the same direction of differential expression in RA and AId, including both negative (SCN3B; Fig. 11e) and positive (SYF2, HTR2A, KCNS1; Figs 11f-h) markers, and none of these were exclusively differential in AId. These other genes also varied in their AId definition, the ventral boundary being sharper for SCN3B and SYF2 and graded for KCNS1 and HTR2A, and the degree of connection between RA and AId varied among transcripts. While the entire AId was clearly seen on transverse sections, the differential expression in RA and AId could also be seen on sagittal sections, with the pattern in AId (Fig. 12n-p) closely resembling that in RA as seen in medial sections (Fig. 12m). Lastly, a smaller set of genes was exclusively differential in RA, including negative (STMN1, C1QL3; Fig. 11i/k) and positive (GABRE, CAMKK1; Fig. 11j/l) RA markers. These exclusive RA-specific markers respected closely the Nissl-defined cytoarchitectonic boundaries of RA.
Other AI subregions:
Other parts of AI showed differential expression, but in most cases they were not as clearly defined by a single gene, requiring a subtraction across markers and/or subregions. SCUBE1, for example, was also expressed in a rostral domain within AI located dorsal to AA at lateral levels. This region appeared as a rostral extension of the AI, and we thus named it rostral AI (AIr; Fig. 5j-k; also see Fig. 4f-g). In contrast to SCUBE1, other markers that define AId did not show any evidence of expression in AIr, indicating that this region is not simply a rostral extension of AId. For HTR1B, besides strong expression in AA, we observed moderate but distinct expression in a rostro-ventral region just caudal to AA which we named rostro-ventral AI (AIrv; Fig. 6a and h). CNTN4, besides its high expression in RA and AId, was also strongly expressed in a broad medial domain that included RA and AM. This CNTN4 expression domain had a sharp border ventral to RA, defining a region that we named medial AI (AIm; Fig. 12a). Lastly, CNTN4 also helped to define a CNTN4 negative region ventral to AId and lateral to AIm that we named ventral AI (AIv; Fig. 12a). Further support for AI subdomains came from other ZEBrA data (see below).
Ventral arcopallium (AV)
Under Nissl, we identified a region along the ventral border of the telencephalon and caudal to AA whose cells appear to be arranged parallel to the tissue border and thus distinct from the overlying A, especially on sagittals. We have termed this region ventral arcopallium (AV; Fig. 3g-j). Besides their high expression in AArl and AAv, AQP1 and NECAB2 also showed high expression in a band of tissue at the ventral edge of the arcopallium that resembled Nissl-defined AV. This ventral AQP1- and NECAB2-expressing domain continued rostrally with AAv and overlapped caudally with the ventral part of NECAB2-defined APv (Fig. 13a-c). It was also defined by low expression of SV2B, which was otherwise generally enriched throughout the arcopallium (Fig. 13d). Also in contrast to AQP1 and NECAB2, this ventral band did not express SCUBE1, thus SCUBE1 expression in AA served as a rostral boundary for this ventral domain (Fig. 6a-a’ and 6c).
Other markers and overall summary of zebra finch data
As summarized in Table 1, the genes analyzed in this study provided supporting evidence for 6 distinct major arcopallial domains (AA, AM - subdivided into AMV and AMD, AD, AP, AI, and AV), altogether comprising 20 subdomains. Seeking additional support for these findings, we next re-examined the ZEBrA database and identified another 171 genes that were general markers of the entire arcopallium or differentially expressed in the various arcopallial domains and/or subdomains defined in the present study (Table 2; details in SuppInfo: SuppTable1.xlsx). A large subset of these genes (n=124) represent RA markers, since they derived from microarray screenings for song nuclei markers (Lovell et al., 2008; Pfenning et al., 2014; Lovell et al, 2018). Because of differences in cutting angle and/or covered regions in the brains used to generate ZEBrA, the detailed definitions of arcopallial subdomains differs somewhat across genes. Nonetheless, the numerous differentially expressed genes available in ZEBrA for most arcopallial subdivisions provide substantial further support for our proposed molecular architecture. Some noteworthy examples (see patterns in ZEBrA) include general markers of the arcopallium besides C1QL3 (e.g. CADPS2, FOSL2, LMO3), prominent markers of AA (e.g. GRIK3, KCTD20, LMO1, RHOB, ZEB2), AMVl (e.g. SLC24A2, KCTD12) and AMD (e.g. PLPP4), and general markers of AI (LGI1, NTS).
Comparative analyses
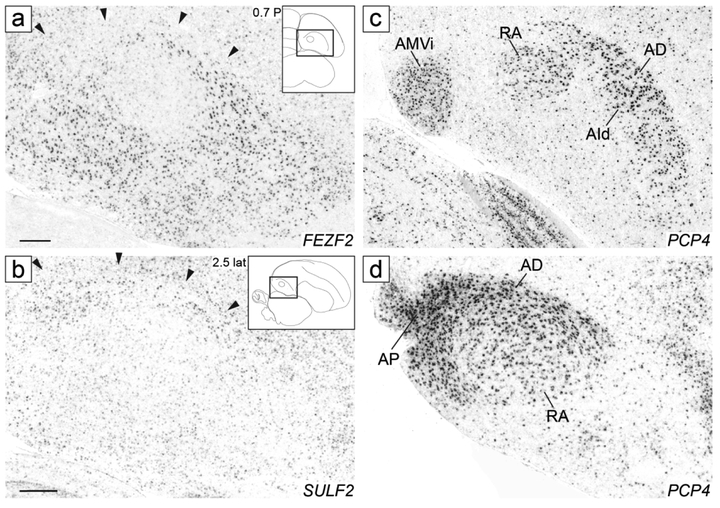
We next examined in zebra finches the expression patterns of several genes previously described as arcopallial markers in chicken (Dugas-Ford et al., 2012), some of which expressed in pyramidal cells in layers 5/6 of the mammalian cortex (e.g. Allen Mouse Brain Atlas; MBA; mouse.brain-map.org, SCR_002978; Lein et al., 2007), and found marked similarities in the patterns across species. For example, FEZF2, which in chick showed broad expression throughout the arcopallium except for the dorsal region (Fig. S5B’’ in Dugas-Ford et al., 2012), also showed broad expression in the zebra finch arcopallium, but was distinctly low in RA as well as in AD, including the caudal portion of AD just caudal to RA (Fig. 14a). SULF2, which in chick was largely restricted to a portion of the dorsal arcopallium (Fig S5E’’ in Dugas-Ford et al., 2012), showed low expression throughout the finch arcopallium but was distinctly enriched in AD (Fig. 14b), and thus was an almost mirror image of FEZF2 in both species. PCP4, which in chick was highly expressed in a ventro-medial region and in a large region including both the dorsal arcopallium and the dorsal part of the intermediate arcopallium (Figs. 2H and S5D’’ in Dugas-Ford et al., 2012), in finch showed strong expression in AMV, in RA, in a dorsal band that included AD and AId (Fig. 14c), and in AP (Fig. 14d). We also confirmed that ETV1 is generally high in the arcopallium but negative in AA (as in Fig 2I in Dugas-Ford et al., 2012, and in Fig. 12a in Jarvis et al., 2013), noting that it also showed weak expression in AD and AP (not shown, but see ETV1 in the ZEBrA website).
Figure 14.
Analysis of chicken arcopallial domain markers in zebra finch. In situ hybridization images of FEZF2 in the transverse plane (a), SULF2 in the sagittal plane (b), and PCP4 in both transverse (c) and sagittal (d) planes. FEZF2 is a negative marker of AD (a), SULF2 is a positive marker of AD (b), and PCP4 is a positive marker of AMV, RA, AId, AD, and AP (c-d). Black arrowheads in (a-b) indicate the position of the dorsal arcopallial lamina. Inset drawings and rectangles indicate the approximate anterior-posterior (a, c) and medial-lateral levels in mm (b, d), and the position of the in situ images in (a-d). For abbreviations, see list. Image scalebar = 250 μm.
To further probe into molecular similarities of the arcopallium between zebra finches and chicken, which is positioned very differently in these two species, we next examined in chicken the expression of some of the arcopallial markers described above for finches. Using crosshybridization with finch probes we obtained data for C1QL3 and for ZBTB20. Similar to zebra finch, C1QL3 was generally high throughout the chicken arcopallium compared to adjacent brain structures (Fig. 15a’’-c’’), matching well the location and boundaries of this structure in chicken (overall lateral position, ventro-lateral to the striatum; see drawings in Fig. 15a-c). The ventro-medial region showed lower expression than the larger, more lateral part of the arcopallium. Thus, the C1QL3 expression in chicken matched well that in finch as a robust marker of the arcopallium. ZBTB20 was selectively expressed in the ventro-medial region, appearing more ventral at rostral levels and shifting dorsally and medially at more caudal levels (Fig. 15a’-c’). The overall shape and location of the ZBTB20 expression domain relative to the rest of the arcopallium resembled closely the pattern in finch (compare with Fig. 4d-f), thus we suggest that this marker reveals the AMV domain of chicken. This region has been identified as the hippocampal-amygdalar transition zone in the chick atlas (AHi in Plate/Figure 18 in Puelles, 2007). We note that this region of ZBTB20 expression excludes the large nucleus identified as Tn (ATn) in the chick atlas (Puelles, 2007).
Figure 15.
Analysis of zebra finch arcopallial domain markers in chicken. In situ hybridization images of ZBTB20 (a’-c’) and C1QL3 (a”-c”) expression in two-week old chick brain in transverse sections corresponding to interaural 3.3, 2.8, and 2.3 mm based on Puelles et al., 2007. Left column (a-c): Drawings depicting major structures and lamina present in in situ images (middle and right columns) defined in adjacent Nissl-stained sections. Coordinates (in mm) indicate the approximate interaural anterior-posterior position. Dashed lines define gene expression boundaries drawn obtained after aligning and superimposing adjacent in situ images (thick dashes: dorsal C1QL3 boundary; thin dashes: boundary of AMV as defined by ZBTB20). For abbreviations, see list. Scale: 250 μm.
We next examined expression patterns in ZEBrA and MBA as the basis for comparing gene expression patterns in avian arcopallial domains and specific mammalian brain areas. Previous studies indicate that the avian arcopallium may correspond to mammalian infra-granular cortical layers and/or amygdalar subdivisions (Abellan et al., 2009; Kuenzel et al., 2011; Puelles, 2011; Dugas-Ford et al., 2012; Jarvis et al., 2013; Vicario et al., 2014; Karten, 2015; Vicario et al., 2017; Herold et al., 2018), thus we focused our analysis on those structures. We initially identified 129 genes whose expression could be assessed in areas of interest in both ZEBrA (7 arcopallial domains: AA, AMV and AMD within AM, AD, AP, AId, and AV) and the MBA (layers 5 and 6 of sensory or motor cortex, and central, medial, basolateral, and cortical amygdala; see Methods for details). Out of these, 26 genes were exclusive markers of arcopallial structures (i.e., not differentially expressed in areas of interest in mouse), and 103 were shared markers between avian arcopallial and mouse cortical and/or amygdalar subdivisions. Upon a closer look at the 103 shared markers, avian arcopallial domains shared 71 markers with layer 5 and/or 6 of the motor cortex, 68 markers with layer 5 and/or 6 of sensory cortex, and 84 markers with at least one amygdalar subdivision. We next examined the degree to which markers were shared between specific arcopallial subdomains and specific cortical layers and amygdalar subdivisions. To facilitate comparisons, we calculated the percent of shared markers for each pairwise comparison across species, as well as the average proportion of markers that each arcopallial domain shared with all mammalian structures examined (see Methods). We found that all arcopallial domains shared considerable proportions of markers with cortical infragranular layers in both sensory and motor areas (Fig. 16a, orange/red hues), as well as with both pallial (Fig. 16a, dark blue) and sub-pallial (Fig. 16a, light blue) amygdalar structures. Several variations were noteworthy though. For instance, all arcopallial domains (AMD, in particular) shared more markers with layer 6 than with layer 5 of motor cortex, whereas some domains (most prominently AV and AP) shared more markers with layer 5 than layer 6 of the sensory cortex. In fact, all domains (AMV and AP, in particular) were found to share fewer markers with layer 5 of motor cortex than the average proportion shared across all mammalian structures. As for amygdalar structures, most arcopallial domains (most prominently AA and AV) shared more markers with BLA compared with other regions and with the average per domain. A notable exception was AMD, the only arcopallial domain that shared more markers than average with the medial amygdala. Most domains (most noticeably AMD and AV) also shared noticeably fewer markers than average with the cortical amygdala.
Figure. 16.
Analysis of marker correspondences between arcopallial subdomains and mouse cortical and amygdalar subdivisions. The expression patterns of 129 genes were analyzed in parallel in finch (ZEBrA) and mouse (MBA; mouse.brain-map.org, SCR_002978) and scored based upon whether they were makers of at least one of 7 major arcopallial subdomains (AA, AMV and AMD within AM, AD, AP, AId, and AV), 4 cortical layers (layers 5 and 6 of sensory and motor cortex) or 4 major amygdalar nuclei (central, medial, basolateral, and cortical amygdala; see Methods for details). (a) Graph showing the proportions of molecular markers shared between each arcopallial domain and layers 5 and 6 of motor and sensory cortices, as well as subpallial (central and medial) and pallial (basal lateral and cortical) amygdalar nuclei. (b) Graph showing the proportion of genes that each arcopallial domain shares with cortical layers or amygdalar nuclei after removing genes found to be markers of both cortical and amygdalar structures. In (a) and (b), dotted lines indicate the mean proportion of shared markers for each arcopallial domain compared with all mammalian structures analyzed; red triangle indicate the overall average proportion of markers shared across all comparisons; numbers shown beneath the arcopallial domains represent the numbers of genes that were shared uniquely with at least one cortical layer or amygdalar nucleus. For abbreviations, see list.
Despite these trends, ~80% of the shared markers were found to be markers of both cortical and amygdalar structures, thus limiting their usefulness for identifying properties uniquely shared between arcopallial subdivisions and specific cortical or amygdalar structures. We therefore removed genes that were shared cortical and amygdalar markers in the MBA, and reanalyzed the remaining core set of 66 genes. To simplify, we collapsed the sets of markers of specific sensory and motor layers, and amygdalar structures into general markers of layer 5 and 6, and pallial and subpallial amygdala. Of the 66 genes analyzed, 19 were found to be shared between an arcopallial domain and one or more cortical layers, and 21 were shared between one arcopallial domain and one or more amygdalar nuclei (Table 3). After examining the distribution of markers per structure, we found that all arcopallial subdomains still shared proportions of unique markers with both cortical and amygdalar structures (Fig. 16b). Nonetheless, we noted a trend towards a large proportion of markers being shared between cortical layer 6 and AId, and between amygdalar structures and AMV and AV, which shared more markers with the subpallial and pallial amygdala respectively. AMD, AD, and AP also shared more markers with the subpallial compared to pallial amygdala, but also shared numerous markers with cortical layer 6.
Table 3†.
Summary of the expression 40 markers shared between finch arcopallial domains and either mouse cortical or amygdalar structures.
| Cortical Subdomains | Amygdalar Subdomains |
Arcopallial Domains/Subdomains | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Names |
MOp5 | MOp6 | S5 | S6 | CEA | MEA | BLH | COA | AA | AMV | AMD | AD | AP | AId | AV |
| NRXN1 | − | − | − | ||||||||||||
| SYNGR3 | − | + | + | ||||||||||||
| IL1RAPL2 | + | + | − | ||||||||||||
| SCN1A | + | − | − | + | |||||||||||
| GAP43 | + | ||||||||||||||
| ARHGDIB | − | − | − | + | + | + | + | + | |||||||
| ADRA1D | − | − | + | − | |||||||||||
| HTR2A | − | − | + | + | |||||||||||
| ADAM23 | − | − | + | ||||||||||||
| CABP1 | − | − | + | ||||||||||||
| PVALB | − | − | + | ||||||||||||
| CRHR1 | − | − | − | + | + | + | |||||||||
| CBLN2 | − | − | + | + | |||||||||||
| VIP | − | − | + | ||||||||||||
| PAK6 | − | − | |||||||||||||
| FAM163B | + | + | + | − | − | ||||||||||
| KCTD20 | − | + | |||||||||||||
| KCTD3 | − | − | − | ||||||||||||
| SLC6A6 | − | + | |||||||||||||
| ID2 | − | + | + | + | |||||||||||
| CCK | − | − | + | ||||||||||||
| PRKAR1B | − | − | |||||||||||||
| NTS | + | + | + | + | + | + | − | + | |||||||
| ZBTB20 | + | + | + | + | |||||||||||
| DAPK1 | + | + | + | ||||||||||||
| CAMTA1 | + | + | |||||||||||||
| RCAN2 | + | + | |||||||||||||
| KCTD12 | + | + | + | + | |||||||||||
| NECAB2 | + | − | − | + | + | + | + | ||||||||
| CALB2 | + | + | + | ||||||||||||
| GLRA3 | + | + | − | ||||||||||||
| CDH4 | + | + | + | − | |||||||||||
| CYP19A1 | + | + | |||||||||||||
| ESR2 | + | + | |||||||||||||
| KCMF1 | − | + | |||||||||||||
| CNTN4 | + | + | + | + | |||||||||||
| ZEB2 | + | + | + | ||||||||||||
| WASF1 | + | + | − | ||||||||||||
| SV2B | + | + | + | + | |||||||||||
| ABCG4 | + | − | |||||||||||||
| GABRB3 | − | ||||||||||||||
| LMO1 | + | + | |||||||||||||
| AQP1 | + | − | + | − | + | ||||||||||
| HTR1B | + | + | − | ||||||||||||
| PLPP4 | + | + | + | ||||||||||||
| FNDC9 | + | + | |||||||||||||
| CRHR2 | + | ||||||||||||||
| GLRA4 | + | ||||||||||||||
| PLD1 | − | + | + | − | |||||||||||
| PLXNC1 | − | − | − | ||||||||||||
| CACNB2 | − | − | |||||||||||||
| KIF26B | − | ||||||||||||||
| PTPRZ1 | − | ||||||||||||||
| MAOB | + | + | + | ||||||||||||
| FABP7 | + | ||||||||||||||
| KCND2 | + | ||||||||||||||
| MAOA | + | ||||||||||||||
| MGAT4C | + | ||||||||||||||
| MGP | + | ||||||||||||||
| ATP2A3 | + | − | |||||||||||||
| QSOX1 | + | + | |||||||||||||
| SEPT12 | − | ||||||||||||||
| SYF2 | − | ||||||||||||||
| CALN1 | + | ||||||||||||||
| CXCL14 | |||||||||||||||
| DCX | |||||||||||||||
Abbreviations: MOp5, primary motor cortex layer 5; MOp6, primary motor cortex layer 6; S5, sensory cortex layer 5; S6, sensory cortex layer 6; CEA, central amygdala; MEA, medial amygdala; BLH, basolateral amygdala; COA, cortical amygdalar area; For other abbreviations see Avian Anatomical Abbreviations List.
Discussion
We have studied the molecular architecture of the arcopallium in comparison with its cytoarchitectonic organization in zebra finches. This study adds to previous efforts to molecularly characterize this complex avian brain region (e.g. Jarvis et al., 2013, Vicario et al., 2017), by presenting a comprehensive molecular dataset that is supported by a histological framework. The gene expression level analysis also contrasts with the ligand binding assays previously used to characterize this region in the pigeon (Herold et al., 2018). As summarized in Tables 1 and 2, SuppInfo:SuppTable1.xlsx, and Fig. 17, the data provide definitions of most boundaries and internal divisions of the arcopallium, and reveal multiple domains and subdomains with unique molecular signatures. The main implications are discussed below, including proposed correspondences between major arcopallial subdivisions in zebra finch compared with previous studies in other songbirds (summarized in SuppInfo:SuppTable2.xlsx) and other avian groups (summarized in Table 4), as well as in mammals.
Figure 17.
Summary of major arcopallial domains and subdomains, identified based on their molecular signatures. Depicted are the various areas identified in this study, on representative transverse (a-c; orange) and sagittal (d-g; blue) sections. We note the transverse plane corresponds to the Frankfurt plane (as discussed in Karten et al., 2013), and thus the diagrams are not exactly orthogonal to those presented in sagittal plane. The levels of the sections for each series are indicated on the dorsal brain views on the upper left. Rectangles in the insets on the upper right of each panel indicate location of area shown relative to a reference section. Coordinates (in mm) indicate the position along the A-P axis for transverse sections (a-c) and position along the M-L axis for sagittal sections (d-g). For abbreviations, see list. Scale bar: 250 μm.
Table 4.
Proposed correspondence between major arcopallial subdivisions in passerines and non-passerines.
| Anterior (AA) | Medial (AM) Medial Ventral (AMV) Medial Dorsal (AMD) |
Dorsal (AD) |
Posterio r (AP) |
Intermediate (AI) | Ventral (AV) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Passerines |
Proposed Zebra Finch |
AAc | AArl | AArm | AAv | AMVi | AMVl | AMVm | AMVc | AMDl | AMDm | AD | APv | APd | RA | AId | AIr | AIrv | AIm | AIv | AV |
|
Previous Oscines1 |
AA | nd | nd | Tn or TN | Tn | Ad | Ap | RA | Ad, AId, Ai | nd | nd | AIV, RAcup | nd | ||||||||
| Ai† | |||||||||||||||||||||
| Suboscines2 | RA | nd | RA | nd | nd | nd | LAI | nd | ni | RA, LAI | nd | nd | nd | nd | nd | ||||||
| Ai | |||||||||||||||||||||
| Non-passerines |
Previous Pigeon/Dove3 |
? (AMp?) | Tn, TnA | AMm+AMp | AD | ? | ni | AId | nd | nd | nd | nd | ? (PoAb?) | ||||||||
| AM | AI | ||||||||||||||||||||
| Chicken4 | ? (AHil?, AIA?, AIPo?) | AHi + AHil + AHilCo, Tn | ATn | ADo, AD | ? (APir?) | ni | AD, (ACo3?) | nd | nd | nd | nd | ? | |||||||||
| AHi + AHil + AHilCo + ATn | ACo1-4 | ||||||||||||||||||||
| Quail5 | ? | nd | nd | Tn‡ | nd | Tn? | nd | ? | ni | nd | nd | nd | nd | nd | ? | ||||||
| AI | |||||||||||||||||||||
| Proposed | ? | AMV | AMD | AD | ? | ni | AI | ? | |||||||||||||
nd = structure not previously described in the literature but likely present in that species or group.
ni = structure not indicated, suggesting it is not present in that species or group.
? = unclear whether the structure has been described in the literature or is present in that species or group.
Oscines: Proposed correspondences between zebra finch AM (described here) and nucleus Taenia (Tn/TN/TnA) in oscines presented in SupportingInfo_SuppTable2.xlsx. AA: Dugas-Ford et al., 2012; Jarvis et al., 2013; Ad: Jarvis et al., 2013; Ap: Jarvis et al., 2013; Ai†: Jarvis et al., 2013; Ad:Johnson et al., 1995; Iyengar et al., 1999; Bottjer et al., 2000; AId: Paterson et al., 2017; Ai: Wang et al., 2015; AIV: Mandelblat-Cerf et al., 2014; RAcup: Vates et al., 1996; Mello et al., 1998.
Suboscines: RA=AA/AMVl: de Lima et al., 2015; RA=AId: Liu et al., 2103; LAI=AD/AId: Feenders et al., 2008; Ai: Fusani et al., 2014.
Pigeon/Dove: Abbreviations defined in Herold et al., 2018; Atoji et al., 2006; Martinez-Vargas et al., 1976; Cheng et al., 1999; Karten and Hodos, 1967; den Boer-Visser, 2004, Reiner et al., 2004a. Note: AA as defined in Herold et al., 2018 likely does not correspond to finch AA.
Chicken: AD: Dugas-Ford et al., 2012; Tn: Balthazart et al., 1998; Thompson et al., 1998; Absil et al., 2002; Ball and Balthazart, 2004; Ball and Balthazart, 2010. All other definitions from the chick atlas (Puelles et al., 2007).
Quail: Tn‡: medial part of Tn in Ball et al., 1999; Tn?: Foidart et al., 1995.
Defining and locating the arcopallium in zebra finches and other songbirds
The location of the zebra finch arcopallium, largely caudal to the striatum, as well as its main cytoarchitectonic features, are similar to those in other songbirds. This includes canaries, starlings, song sparrows and white-throated sparrows, for which a brain atlas (Stokes et al., 1974) or histological data on the arcopallium are available (Jarvis et al., 1997; Bernard et al., 1999; Cheng et al., 1999; Leung et al., 2009). While major laminae like LAD and LPS are seen under Nissl, mLAD is less distinct than the arching lLAD, appearing on sagittals as a faint lamina that separates AMD from NCM (yellow arrows in Fig. 3a-c). Thus, it is easy to mistake the medial-most arcopallium (AMDm) for the ventral-most part of the overlying NCM, especially on thin sections. This misidentification was present in an early version of the annotated histological reference atlas in the ZEBrA website (www.zebrafinchatlas.org, RRID:SCR_012988), an error that has now been corrected, and we suggest it might also have affected previous analyses of NCM. Markers that do not distinguish AMD from NCM (e.g. AQP1 and NECAB2 in Fig. 8c-d) suggest that in some respects AMD is more similar to NCM than to the arcopallium. Furthermore, the general arcopallial marker C1QL3 shows low expression in AMV, which might suggest AMV as not being part of the arcopallium. Interestingly, the patterns of some developmentally expressed genes appear to include parts of the caudal nidopallium and medial arcopallium (e.g., Figs. 7, 11, and 18 in Chen et al, 2013), suggesting that these sub-regions may share similar developmental origins and/or cell types. Nonetheless, mLAD is clearly seen on well-stained thick transverse sections as separating AM from the NCM (yellow arrows in Fig. 2a-e), and the cytoarchitectonic features of AMD and AMV are distinct from those of the nidopallium. Combined with their overall molecular profiles, we conclude that both AMD and AMV are integral parts of the arcopallium and not the nidopallium. AM also shows a pallial-like pattern for GAD2 (see ZEBrA), indicating that AM is not part of the striatum.
The caudal arcopallial boundary posed another challenge, as LAD is not clearly seen caudal to and slightly lateral to RA, due to the presence of APc at the caudo-ventral tip of the lateral ventricle (white arrowhead in Fig. 3g-i; Fig. 11g). While the general arcopallial marker C1QL3 largely excluded this nucleus from the arcopallium, other evidence favored its inclusion, namely the expression of other general (AQP1, SV2B) and domain-specific (CBLN2) markers, and the fact that at least one portion of that nucleus falls within C1QL3-defined arcopallium.
Major arcopallial subdivisions
We obtained evidence for 20 arcopallial domains and subdomains, based on molecular criteria and partially from cytoarchitectonics (Table 1 and Fig. 17). Several subdivisions were not discernible under Nissl and could only be identified based on molecular criteria. Below we discuss the main regions in the context of previous studies. Little is known on the function of most of markers studied, thus functional inferences are limited. We note that some domains have been previously proposed by Jarvis et al., 2013, but specific boundaries were not always clearly defined (see proposed correspondences in Table 4).
AA:
AA is distinguishable by Nissl, and was previously identified as a region of low ETV1 (aka ER81) expression (Dugas-Ford et al., 2012; Jarvis et al., 2013). Here we provide a more complete definition of AA, supported by additional markers that define subdomains. AArl is rich in fiber bundles, thus markers like AQP1 and NECAB2 could be related to the organization or fasciculation of projections originating from the arcopallium. The enrichment of HTR1B, a G-protein coupled serotonin receptor, suggests local high sensitivity to serotonin modulation. We note that a clone previously identified as SEMA6A (CK308635) was also described as an AA marker (Jarvis et al., 2013). We recently found that this clone is a fusion of the 5’-end of SEMA6A and the 3’-end of CAMK2N1 (see CAMK2N1 in ZEBrA). Based on comparisons with other gene-specific clones, we have concluded that CK308635 probes CAMK2N1, which makes it an AA marker, whereas SEMA6A is not an AA marker (see SEMA6A in ZEBrA).
AM:
The medial-most part of the arcopallium has a complex shape and several subdomains (Figs. 3a, 5b, 7e-g). Interestingly, the rostral-most part of the arcopallium, which was not clearly delineated in the canary atlas, also lies within AM and not in AA (in Fig. 4a-d, compare C1QL3 to ZBTB20, which labels AMV, and to SCUBE1, which labels AA). Importantly, AMD and AMV are respectively dorsal and ventral to the OM fibers. The canary atlas incorrectly used the same label (nucleus taenia, Tn) to indicate these two structures: the ventral-most Tn-labeled region is ventral to the OM (plates 25-29 in Stokes et al., 1974) and corresponds to the subdomain we have named AMV, the more dorsal Tn-labeled structure is located dorsal to the OM (plates 23-24 in Stokes et al., 1974), and corresponds to part of the region we have named AMD. Our data indicates that AMD continues more caudally, also including an unlabeled dorso-medial arcopallial area located between Tn and LAD in plates 25-26 of the canary atlas (Stokes et al., 1974). It continues even further caudally in plates 27-30, where it is located medial to nucleus RA, but at these levels the LAD was drawn obliquely toward the midline. Based on cytoarchitectonic features (Fig. 2c-g) and C1QL3 expression (Fig. 4d-g), our interpretation is that this medial part of the LAD was drawn erroneously in plates 27-30, and that the AMD is an integral part of the arcopallium. Unfortunately, these errors have been perpetuated in the literature on songbird Tn.
AMV and AMD are molecularly distinct, particularly at intermediate to caudal levels, where ZBTB20 and strong CYP19A1 (aromatase) are clearly segregated. While ZBTB20 was not previously studied, the CYP19A1 data are largely consistent with previous papers, although our interpretation differs. For example, an early study showed high CYP19A1 mRNA expression in a medial structure dorsal to the OM in zebra finches (labeled Tn in Fig. 5D in Shen et al., 1995), which we interpret as corresponding to the region of strong labeling that we have named AMD (e.g., maps of dark red cells in Fig. 9c-f). However, the authors presented their expression data schematically as corresponding to a large area (Tn in Fig. 1K in Shen et al., 1995) which seems to encompass both AMV and the ventral part of AMD. They also presented schematically the narrow region of aromatase expression medial to nucleus RA as part of the nidopallium, thus outside the arcopallium (Fig. 1L in Shen et al., 1995), following the schematics in Stokes et al., 1974. As discussed above, this region medial to RA corresponds to AMD and is part of the arcopallium, not nidopallium. Another study in zebra finches showed CYP19A1-immunolabeled cells in two different arcopallial structures labeled Tn in schematic diagrams (CYP19A1 distribution in Fig. 1F and G, in Balthazart et al., 1996), which we interpret as corresponding to AMV and AMD, respectively. In yet another study, aromatase expression mapping was shown schematically as occurring in a large Tn-labeled area that likely includes both AMV and part of AMD, and where the more caudal part of AMD is excluded from the arcopallium (Fig. 3 in Saldanha et al., 2000b). Our high resolution cellular mapping on serial sections in zebra finches shows, however, that while some expression of CYP19A1 occurs within AMV, the highest CYP19A1 expression actually occurs within AMD, thus dorso-medial to AMV. Our data are also consistent with that in song sparrows, where we interpret the area of high CYP19A1 expression (labeled Tn in Fig. 2D in Soma et al., 2003) as corresponding to AMD as defined here, and in white-throated sparrows, where expression is higher in AMD than in AMV (Maney, D., personal communication). However, we note that the data in canary and house sparrows suggest comparable CYP19A1 expression in what we interpret to be AMV and AMD (labeled TN and A respectively in Figs. 2E and F, and combined as structure 5 in Fig. 3 in Metzdorf et al., 1999), suggesting species differences in aromatase expression between AM subdomains. Overall, ZBTB20 is a more robust domain-specific marker (restricted to AMV), and while CYP19A1 is more highly expressed in AMD, this gene does not respect Nissl-defined borders as well as ZBTB20.
Other markers label AMV or portions of it, reflecting the complexity of this AM subdivision. For example, ZBTB20-defined AMV seems to correspond to the ventro-medial GPR30-expressing arcopallial domain in finches (Acharya and Veney, 2012), although it is not clear whether the high-immunoreactivity corresponds to soma or terminals. In white-throated sparrows, the AMV shows high mRNA expression of GPR30 and HTR1E (Maney, D., personal communication), as well as high immunoreactivity for the serotonin transporter (SLC6A4A; Fig. 4C in Matragrano et al., 2012), the latter suggesting AMV as an important target for serotonergic modulation.
Our data on ESR1/2 (ERalpha/beta) expression in songbirds, while partially consistent with the existing literature, provides a more accurate account of the distribution of ESR1/2. Consistent with findings in starlings (Figs. 9 and 10A, Bernard et al., 1999), we found no evidence of ESR1 expression within the arcopallium, but did find strongly labeled cells dorso-ventrally along the medial wall of the nidopallium (Fig. 8f) and in lateral areas within the striatum (not shown). In contrast, ESR1-positive cells were previously reported in what appears to be AMVm within the finch arcopallium (schematic diagram in Fig. 4D in Jacobs et al., 1996). We suggest that the data in Jacobs et al., 1996, which were only presented schematically, do not reflect an accurate mapping of ESR1-expressing cells. A similar problem may have occurred in previous studies that defined the distribution of ER-immunolabeled cells in canary and zebra finch, such as in Gahr et al., 1987 (a schematic diagram in Fig. 2c-d, but no primary data presented for the medial area labeled Tn) and in Gahr et al., 1993 (the structure labeled Tn in Fig. 1b, which corresponds to structure 7 in Fig. 5A is clearly not in the arcopallium, and appears to correspond to a striatal site). In contrast, ESR2 is clearly expressed in discrete sites in the finch arcopallium, and is an exquisite marker of AMVm (best seen in Fig. 9E), further supporting a molecular distinction for this subdomain. This observation is consistent with the high ESR2 expression reported in starlings, which we interpret as being restricted to AMVm in that species (Fig. 10C in Bernard et al., 1999). Overall, we found very little evidence of overlapping expression of ESR1/2 and CYP19A1, suggesting that for the most part ESRs are not co-expressed with CYP19A1 at the cellular level within the finch arcopallium. This contrasts markedly with the largely overlapping distributions of ESRs and aromatase in areas like NCM.
Although not explicitly addressed in our study, cells immunoreactive for androgen receptor (AR) have been reported in a presumed songbird Tn, with possible differences between zebra finches and canaries (Balthazart et al., 1992). The lack of primary data presentation in that study limits our interpretation, but the schematic diagrams presented depict labeled cells along a ventral strip in the arcopallium, which in finch could be interpreted as corresponding to ventral parts of AMD and AMV (labeled Tn in Fig. 3E in Balthazart et al., 1992), whereas in canary the cells appear to be located in a ventral region lateral to AM, possibly corresponding to AV (labeled Tn in Fig. 5D in Balthazart et al., 1992). The canary data seem consistent with a subsequent study that reports no AR-labeled cells in Tn (Table 1 in Metzdorf et al., 1999), defined in this study as the medio-ventral areas above and below the OM combined, thus corresponding to what we defined here as AMD and AMV combined. Further comparative analysis is needed before establishing whether AR expression can define AM subdomains and/or whether it shows real species differences.
In sum, different studies on aromatase and ESRs have inconsistently labeled different arcopallial domains as Tn, an error originating from the canary atlas (Stokes et al., 1974). This imprecision, combined with incomplete mapping and lack of presentation of primary data, has led to confusion and conflicts across studies, sometimes even in the same species. Our proposed nomenclature and detailed mapping avoid this confusion, and allow us to conclude that: 1) AMV and AMD have distinct molecular profiles, and can be distinguished by ZBTB20 expression (AMV) and high density of strongly labeled CYP19A1 cells (AMD); 2) arcopallial ESR2 expression is restricted to AMVm, helping define this discrete subdomain; 3) ESR1 is not expressed in the finch arcopallium; 4) we recommend against using CYP19A1 data alone to define AM subdivisions; and 5) we recommend against using Tn to name medial arcopallial structures because: 5a) the term is fraught with errors from previous atlases and studies; 5b) AMD/AMV offer a more precise terminology and 5c) it is unclear what exact arcopallial subdomain in songbirds corresponds to the Tn defined in non-passeriformes. A summary of proposed correspondences between subdomains proposed here and structures labeled nucleus Tn in various songbird studies is presented in SuppInfo:SuppTable2.xlsx.
Connectivity and lesion data for AM in songbirds amount to one study in starlings that focused on the area we defined here as AMV (named Tn in Cheng et al., 1999). The data suggest that AMV is connected to hypothalamic targets as well as pallial areas, which would support an amygdaloid nature of AMV. Accordingly, bilateral lesions to AMV resulted in apparent disruption of social behaviors but not of locomotion. The study is not conclusive, however, as some projections were not confirmed with retrograde tract-tracing, there are fibers of passage concerns, and the behavioral analysis was limited.
AI:
AI represents the bulk of the arcopallium, and while it was proposed in finches by Jarvis et al., 2013, no AI boundaries were determined in that study. No markers studied here uniformly labeled the entire AI, and cytoarchitectonics alone does not differentiate it from all other arcopallial subdivisions. However, we could distinguish it by subtracting other molecularly defined subdivisions.
Although RA lies in close proximity to AM, the molecular and cytoarchitectonic data point to RA’s closer similarity to AId (Table 1), including the fact that all AId markers were also RA markers. Supporting this relationship, RA and AId show similar alpha2-adrenergic receptor ligand binding (Riters and Ball, 2002). Moreover, while LMAN sends projections to RA, the shell of LMAN sends projections to AId (labeled Ad in Johnson et al., 1995, Iyengar et al., 1999 and Bottjer et al., 2000). AId also receives projections from the dorso-lateral caudal nidopallium (dNCL), an area lateral to HVC (Bottjer et al., 2000), whereas HVC itself projects to RA. Such parallel projections argue that RA represents a specialized target within a broader projection system from the nidopallium to dorsal AI. Evidence of neuronal activation within AId associated with wing beats and/or flight in finches (LAI in Figs. 6 and 7 in Feenders et al., 2008; see also Stetner and Fee, 2017) suggests a general involvement of AId in movement control. These observations support the notion that RA represents a specialized region of AId that is dedicated to vocal-motor control, although a role for AId in integrating multimodal sensory and limbic projections has also been suggested (e.g., Bottjer et al., 2000).
The large set of RA markers in ZEBrA derives from microarray screenings (Lovell et al., 2008; Pfenning et al., 2014). While recently validated through microarray re-analysis and confirmatory in situs (Lovell et al., 2018), the contrasting microarray sample was ventro-lateral to RA (Fig. S17A in Pfenning et al., 2014), likely within AIv (see Fig. 16B). Our present analysis reveals that several RA markers are shared markers with AId, rather than RA-specific. The true set of RA-specific markers has thus been overestimated, and a more adequate contrasting sample would have been AId, since it shares more features with RA. Nonetheless, several markers like STMN1, GABRE, C1QL3, and CAMKK1 (Fig. 12i-l) are RA-specific and not differential in AId, reflecting unique properties of RA, although how they modulate RA physiology is unknown. Some markers (e.g. C1QL3) were previously described as shared molecular specializations of RA and human laryngeal cortex, representing convergent features of vocal-motor cortical neurons in vocal learners (Pfenning et al., 2014). However, given the similarities between RA and AId, it remains to be tested whether other markers shared between humans and vocal learning birds are truly RA-specific or represent broader specializations of cortical motor areas.
The so-called RA cup is an AI region immediately adjacent to RA, especially rostro-ventrally, (Kelley and Nottebohm, 1979), receives projections from auditory areas field L and the shelf adjacent to HVC (Vates et al., 1997; Mello et al., 1998), sends descending projections to auditory thalamic and midbrain nuclei (Mello et al., 1998), and shows evidence of activation in response to song stimulation (Mello and Clayton, 1995). It occupies a similar position in the central auditory circuitry as the arcopallial nuclei Aivm and Aidm in non-oscines (Wild et al., 1993). While we did not find markers that specifically labeled the ‘RA cup’, it seems to be included in the CNTN4-defined AIm. The presence of this nucleus supports the notion that AI contains neuronal groups that originate long descending sensory projections.
Mandelblat-Cerf et al (2014) defined AIV as an AI region that excludes RA and AId. We believe this region corresponds largely to what we define here as AIv and AIm, but may also have impinged on AA and AV. The connectivity data in Mandelblat-Cerf et al., 2014, confirm major auditory inputs to AIv and descending auditory projections from AIv, consistent with previous tract tracing data for the auditory system of finches (Mello et al., 1998), but also provide evidence of descending projections from AIv to midbrain dopaminergic areas, which among other targets project to vocal striatal areas. This descending projection from AIv was suggested as a pathway through which cortical auditory processing of song could affect vocal learning through dopaminergic modulation. Evidence from lesions indeed suggests that parts of AI other than RA may be involved in vocal learning. Bottjer and Altenau (2010) provided evidence that lesions of Ad (Fig. 1 in Bottjer and Altenau, 2010), which we interpret as including portions of AId and AD as defined here, prevented the development of stable vocal sequences, but did not affect vocal production. In contrast, Mandelblat-Cerf et al., 2014 showed that lesions of AIV (their nomenclature) affect vocal learning. However, we interpret these lesions as including AIv, AV, and possibly the caudal part of AA (compare their Fig. 4a with our Fig. 13). Furthermore, their control lesions that do not impinge on AIv do not affect vocal learning. We interpret those control lesions (labeled Ad in Fig. 5a, right panel in Mandelblat-Cerf et al., 2014) as including both AId and AD as defined in the present study (our AId corresponding to Ad in Mandelblat-Cerf et al., 2014). We note, however that larger lesions that targeted Ad and extended laterally (Mandelblat-Cerf et al., 2014), did lead to severe motor deficits. While these larger lesions still likely included AId and AD as defined here, the deficits are consistent with the notion that one or both of these subdivisions may correspond to a general motor representation area. We also suggest that apparent discrepancies in how AIv/AId/AD lesions affect vocal learning may be explained in part by the relatively large lesion sizes and likelihood that multiple domains were targeted. Overall, there is considerable heterogeneity in terms of AI projections and functions. How the molecularly defined subdomains correspond to areas of distinct connectivity and function remains a future goal.
Other domains:
AD is a narrow band dorsal to AId and ventral to LAD, representing the dorsal-most part of the arcopallium. It is distinct from the region named Ad in previous studies (Johnson et al., 1995; Iyengar et al., 1999; Bottjer et al., 2000; Mandelblat-Cerf et al., 2014), but which we have named AId. An area labeled LAI that may have included AId and portions of AD shows ZENK induction with hopping behavior in finches (Figs. 7D, 9B and S3Bb in Feenders et al., 2008). Jarvis et al. (2013) sampled a domain that may correspond to the AD described here, although the boundaries were not defined. The fact that several AD markers are neuropeptides (e.g. CCK, CBLN2, UTS2B) or receptors for neuropeptides (CRHR1) or serotonin (HTR1B) suggests that AD is a major target of neuromodulation. We also provide a clear definition and subdivisions for AP. An AP was also proposed by Jarvis et al. (2013), but no clear boundaries were identified, and a distinct nucleus equivalent to APv was not acknowledged. Interestingly, the caudal-most portion of LAD may be more clearly seen with thionin stain (Fig. 15E and F in Jarvis et al., 2013), which supports the inclusion of the entire region containing APc as part of the arcopallium. While an AV was not explicitly named, ZENK is upregulated by flight in warblers in a region that we interpret as including AV and AA (Fig. 3F in Feenders et al., 2008). The pattern of upregulation of ZENK is reminiscent of AQP1 expression (our Fig. 6a”), suggesting a functional connection between these domains. AV receives ipsilateral projections from LMAN shell and dNCL, and sends contralateral projections back to AV, LMAN shell and dNCl, suggesting that an inter-hemispheric pathway involving AV coordinates vocal behavior (Paterson and Bottjer, 2017).
Comparison with sub-oscines
Sub-oscines are a distinct Passeriformes sub-order that for the most part seems to lack vocal learning and a full-fledged forebrain song circuitry (Kroodsma and Konishi, 1991; Gahr, 2000; Saldanha et al., 2000a), although there is suggestive evidence of limited vocal learning and/or rudimentary telencephalic vocal areas in some species (Liu et al., 2013; de Lima et al., 2015). General features of the arcopallium in finches appear similar to those in sub-oscines, including the location caudal to medial St and OM, and the rostro-caudal orientation of OM fiber bundles. However, the nuclei described as a putative RA in two sub-oscines, the scale-backed antbird (de Lima et al., 2015) and the Eastern phoebe (Liu et al., 2013) more likely correspond to other arcopallial domains. Specifically, the probes used to define a “rudimentary RA” (i.e. RGS4) in the antbird seem to have identified AMVl and/or AA, and not RA (e.g., compare Fig. 2E-F in de Lima, 2015, to RGS4, level 8 in ZEBrA), and the domain described as “RA-like” in phoebe defined by PVALB, GRIK1, and GRIA1 (Fig.1 in Liu et al., 2013) likely corresponds to an intermediate arcopallial domain that encompasses or includes AId. Nevertheless, since RA in finches may be a specialization of AId, it will be important to examine whether AId is also present in sub-oscines, and whether there is any trace of either by cytoarchitectonic or molecular criteria.
The golden-collared manakin, a sub-oscine that shows elaborate courtship displays under the control of sex steroids, shows broad expression of androgen receptor in the intermediate arcopallium (Fig. 1H in Fusani et al., 2014). This presumably includes the movement-activated region named LAI in finches (Feenders et al., 2008) that we interpret as corresponding to AId (and possibly AD). Based on the shape and position similarities of the arcopallium in manakin and finch, we conclude that this AR-expressing region in manakin likely includes AId. However, the AR distribution appears much more widespread than finch AId, suggesting that either AId is much enlarged or that male steroids exert more widespread actions on the arcopallium in this sub-oscine group. Because of the strong action of androgen on male courtship displays, these observations suggest that a cortical-like region may be involved in the motor control of courtship, and that courtship may have a learned component.
Comparison with other avian orders
C1QL3 is a robust arcopallial marker in both zebra finches and chicken, and these two distant species share other arcopallial markers with more restricted patterns (e.g. PCP4, FEZF3, SULF2, ETV1; details below). Thus, major molecular properties observed in finches appear to be conserved features of the avian arcopallium. Furthermore, the classification of arcopallial domains that we propose for the finch appears to match that described based on ligand binding densities for neurotransmitter receptors in pigeon (Herold et al., 2018). Upon a closer look, however, significant differences in arcopallial topography between finches and non-oscine species pose challenges for comparative analysis. In passerines, the arcopallium is located medial and caudal to the striatum, an arrangement that we believe is unique to this avian order. In contrast, as detailed in atlases, the arcopallium in chicken and other galloanseriformes (quail, duck) and columbiformes (pigeon, dove) is located mostly lateral, ventral, and only partially caudal to the lateral striatum (Karten and Hodos, 1967; Kuenzel and Masson, 1988; den Boer-Visser, 2004; Puelles, 2007). Furthermore, the characteristic arcopallial fiber bundles course along the medial-lateral axis and merge to form the OM, which is located medial to the arcopallium. A topography similar to that in galloanseres and columbiformes is seen in other neoaves like hummingbirds, as well as in parrots, considered a psittaco-passerifomes sub-order, and thus the closest sister taxa to songbirds (Suh et al., 2011; Jarvis et al., 2014), in falcons (e.g. Bagnoli and Francesconi, 1984), the closest outgroup to parrots and passerines (Suh et al., 2011; Jarvis et al., 2014), and in owls (e.g. Tyto alba in Brain-maps.org).
This non-passeriformes organization seems widespread across avian orders, thus it may have been the ancestral condition in birds, but it differs markedly from that in songbirds. Here the arcopallium is clearly caudal to, not lateral or ventral to the striatum, and occupies the space that in galliformes and other non-oscines lies over the tectum and rostral to the cerebellum. Furthermore, the OM fibers in songbirds are oriented along the AP axis, thus clearly different from the ML orientation in non-oscines. We also note that nucleus RA is located caudally in the songbird arcopallium, whereas the analogous vocal nuclei are positioned laterally in hummingbirds and parrots (VA and AAc, respectively; Jarvis and Mello, 2000; Jarvis et al., 2000), both representing non-oscines with an overall arcopallial layout more similar to that in galliformes than in songbirds.
These topographic differences suggest that as the arcopallium shifted to a caudal position in songbirds it also underwent a rotation, whereby the ML axis seen in chicken and other non-oscines may correspond to the AP axis in songbirds. According to this hypothesis, medial and lateral structures in chicken may have shifted respectively to rostral and caudal positions in finches, whereas the dorsal, central (intermediate) and ventral structures would have retained their position relative to each other. Thus, structures with the same topography along the ML and AP axes would not be the same across species. The hypothesis also predicts that a nonrotating pivot point could have retained the same position in songbirds as in other avian groups. Alternative possibilities cannot be ruled out but appear less likely, namely that the same domains show different molecular profiles across species, or that these domains and their molecular specializations are lineage-specific and thus absent in the other lineage. Under any of these scenarios, simple topographical correspondences of arcopallial domains between passeriformes and other avian orders are likely incorrect. We also note the suggestion by Wang et al. (2015) that the arcopallium in all vocal learning bird groups seems more medially placed compared to vocal non-learners due to the presence of nidopallium lateral to the arcopallium only in the former. We agree that, similar to songbirds, part of the nidopallium in hummingbirds and parrots is lateral to the arcopallium, possibly due to a lateral expansion of the nidopallium in these two vocal learning groups. However, the bulk of the arcopallium in these bird groups is mostly lateral to the striatum, and in that regard resembles more closely the arrangement present in non-vocal learners such as chicken, quail, and dove.
While comparative molecular data are scarce, some observations support this axis rotation hypothesis. For example, AA in finches is located just caudal to the OM and rostral to the AI, contains many OM-related fiber bundles (e.g., Fig. 6a), and shows marked downregulation of ETV1 (as well as low expression of PCP4; see pattern in ZEBrA). In chicken, ETV1 and PCP4 are sharply downregulated in a medial region (Fig. S5A’’ and S5D’’ in Dugas-Ford et al., 2012) just lateral to the OM (named vaf in Puelles, 2007), where this fiber tract breaks into smaller fiber bundles. In the chick atlas, this region seems to include parts of the area named the Hilar region of the amygdala (AHil), as well as the intermedioanterior and intermedioposterior amygdala (AIA and AIPo; Figures 14-15 in Puelles, 2007). In pigeon, this region seems to correspond to the lateral portion of the area named arcopallial mediale pars parvocellularis (AMp), as in Fig. 2i in Herold et al., 2018, and Fig. 1A in Atoji et al., 2006. Thus, an arcopallial domain comparable to finch AA seems to be located medially in chicken and pigeon, and one can follow the progression from OM to a domain of ETV1 downregulation to the AI along the AP axis in finches and along the ML axis in non-passeriformes. In contrast, whereas the region named AA in pigeon can be grouped together with AI and AD in terms of similarity in connectivity (Shanahan et al., 2013), the region named AA in finches does not group with AD and AI when comparing patterns of mRNA expression across different parts of the arcopallium (Jarvis et al., 2013); also see discussion in Herold et al., 2018). Similarly, pigeon AA shows high GABAA binding, but we did not find evidence for enhanced expression of GABA-related receptor subunits in finch AA. Thus, AA in non-passeriformes does not seem to correspond to finch AA. Also according with the ML to AP axis rotation hypothesis, the AA in non-passeriformes would occupy a lateral position in songbirds, lateral domains in chicken (such as the transition zone into the piriform cortex (named APir in Puelles, 2007) would occupy a posterior position in songbirds, possibly corresponding to finch APv and/or APd, and posterior domains in chicken (such as chicken APM/APL in Puelles, 2007) might occupy a more medial or posterior medial position in finch. Although these predictions are largely untested, the discrepancy of dopamine receptor expression between finch and pigeon APs (as noted in Herold et al., 2018), would be consistent with the rotation hypothesis.
This hypothesis also predicts that AD should be similar in finch and chicken, and indeed SULF2 and FEZF2 are markers of AD (positive and negative, respectively) in both species. PCP4 is also a positive marker of AD in both species, but in finches PCP4 is also strongly expressed in AId, which is just ventral to AD, and in chicken PCP4 is highly expressed in a dorsal band which appears thicker than AD (Fig. S5D’’ in Dugas-Ford et al., 2012) and appears to include the dorsal part of the AId. This suggests that chicken also has a PCP4-positive AId located just ventral to AD. The existence of a distinct AId in non-passeriformes is supported by ligand binding densities in the pigeon (Herold et al., 2018). Interestingly, pigeon AId has higher binding for 5-HT1A, whereas finch AId can be defined by higher expression of HTR2A, both consistent with a high sensitivity for serotonergic modulation, but further comparative data are needed to further establish a correspondence of AI subdivisions. The rotation hypothesis also predicts that the domain we have defined as finch AV, a thin strip of tissue at the ventral-most border of the arcopallium, may correspond to the PoAB in pigeon (and not the more internally located AV in Herold et al., 2018). However, we do not have molecular data to test this hypothesis. Lastly, GRIK3 mRNA is high in finch AA, and kainate binding is high in pigeon AA (Herold et al., 2018), which would seem inconsistent with the rotation hypothesis. However, it is difficult to compare gene expression with autoradiographic ligand binding data, and we had few probes for receptors, limiting our ability to draw strong conclusions across these studies.
In both finches and chicken ZBTB20 is selectively expressed in a well-defined ventro-medial arcopallial domain that we named AMV (compare Fig. 4d-f with Fig. 15a’-c’). This region includes structures that in the chick atlas have been named the amygdalo-hippocampal area, and the medial and core parts of the amygdalar hilar area (AHi, AHiL and AHiLCo in Figures 14-19; Puelles, 2007), and excludes the so-called ATn, a distinct nucleus both in the chick atlas (Fig. 17-21 in Puelles, 2007) and in our own data (ATn in Fig. 15c’). We interpret the ZBTB20-expressing domain as including the ventro-medial region that in quail is named Tn, expresses AR (Balthazart et al., 1998b) and is thought to be part of a circuit that regulates sexual behavior (Thompson et al., 1998; Absil et al., 2002; reviewed in Ball and Balthazart, 2004; 2010). While we do not have data in columbiformes, we suggest that the ZBTB20-expressing area seen in finch and chicken likely corresponds to a ventro-medial area that includes the structure labeled Tn and/or TnA in pigeons and doves (Martinez-Vargas et al., 1976; Cheng et al., 1999; Herold et al., 2018), following the conventions in atlases (Karten and Hodos, 1967; den Boer-Visser, 2004), and from the avian nomenclature consortium (Reiner et al., 2004a). Electrolytic lesions indicate that Tn in dove plays a role of in social behaviors and affective state (e.g. Cheng et al., 1999). Receptor autoradiographic studies indicate that compared to other arcopallial domains, TnA is enriched in muscarinic cholinergic and α2 adrenergic receptors (Fernandez-Lopez et al., 1997; Herold et al., 2018), suggesting a prominent role for cholinergic and adrenergic modulation. Overall, we propose that the AMV we have described in finch and chicken corresponds to, or at least includes the structure that has been called Tn and/or TnA in both galliformes and columbiformes, noting that the so-called ATn in the chick atlas (Puelles, 2007) is excluded.
We also found that ER-beta (ESR2) expressing cells are highly enriched in the medial-most subdomain of finch AMV, named here AMVm. In quail, ESR2 expression is also highly enriched in the medial-most part of the arcopallium, and even though this ESR2-expressing region has been labeled Tn (Fig. 2A-C in Ball et al., 1999) or TnA (Fig. 1A-C in Halldin et al., 2006), it seems smaller than the Tn as defined in other quail studies (Balthazart et al., 1998a,b; Thompson et al., 1998; Absil et al., 2002). We thus suggest that the region defined by ESR2 expression in quail is just the medial-most part of quail Tn, possibly corresponding to AMVm as defined here in finch. We are unaware of studies assessing ESR2 in Tn in columbiformes. In our finch study we did not detect ER-alpha (ESR1) in the arcopallium, although we observed strong expression in sparse cell populations in the medial nidopallium and striatum (not shown). In contrast, a sparse population of cells expressing ESR1 mRNA has been reported in quail arcopallium, within a region that we interpret as the medial part of Tn (TnA in Fig. 1A-C in Halldin et al., 2006). Thus, there seems to be a species difference in arcopallial ESR1 expression. While the ESR1 mRNA data in quail is consistent with earlier autoradiographic and immunohistochemical studies of ER expression in galliformes and dove, the very sparse ESR1 mRNA distribution differs from the presumably more widespread ER expression in Tn in these earlier studies (Martinez-Vargas et al., 1976; Balthazart et al., 1989; Watson and Adkins-Regan, 1989; Metzdorf et al., 1999). We note, though, that the earlier studies did not differentiate between ER receptor subtypes, and the data were summarized in tables or schematically, preventing an accurate assessment of the brain distribution of labeled cells.
The highest expression of aromatase (CYP19A1) in finch occurred in an area dorso-medial to the ZBTB20 domain that we have defined as AMD. In quail, aromatase immunoreactive cells have been reported in a structure referred to as N. taenia (structure 7 in Fig. 2D-G in Foidart et al., 1995). However, in other quail studies (Balthazart et al., 1998a,b; Thompson et al., 1998; Absil et al., 2002), aromatase positive cells were mapped to a region dorso-medial to a structure labeled Tn, which likely corresponds to AMV based on the chicken data from the present study. Aromatase expression has also been reported in a structure referred to as Tn in partridge (TN label ventral to OM in Fig. 2D in Metzdorf et al., 1999). However, because the data are from a medial sagittal plane and because of the different orientation of the arcopallium in songbirds and galliformes, we interpret that labeled cell group as being the same as in Foidart et al., (1995), whereas the more laterally placed AMV (i.e. most of the Tn) was likely not assessed (or reported). Based on our detailed mapping in finches, the region of high aromatase expression reported in galliformes (quail/partridge) likely corresponds to a structure dorso-medial to Tn in those species, which we interpret as corresponding to AMD as defined in the present study. Alternatively, the area of high aromatase expression in galliformes could correspond to the dorso-medial portion of Tn/AMV. Altogether, because of possible species differences, the regulation of aromatase expression by hormone levels and reproductive state, and a brain distribution that tends to not respect cytoarchitectonic boundaries, we recommend against using aromatase (or estrogen receptors) alone to define specific nuclei or domains in the avian arcopallium. Lastly, because of the marked lack of ZBTB20 expression, we suggest that the dorso-medial part of AM in chick, which includes ATn, might correspond to the region we defined as AMD in finch, a possibility that can be tested in the future with markers.
Our comparative analysis is summarized in Table 4. We believe that for a subset of the arcopallial domains we have proposed for zebra finches, namely AM (AMV and AMD), AD, and AI, equivalent areas have been identified in other birds groups. We therefore propose that this set of terms be used instead of terms like nucleus taenia, which are not anatomically based and that have been used inconsistently in the literature. In contrast, due to different arcopallial topographies in passerines vs. non-passerines, it is currently unclear how finch AA, AP, and AV correspond to structures identified in other avian orders. Settling these issues and devising a universal nomenclature will require further efforts.
Comparison with mammals
A long-standing contention in comparative neuroanatomy is whether the arcopallium, or portions therein, correspond to avian homologues of cortical versus amygdalar structures in mammals. While settling this issue will require a more in-depth understanding of developmental trajectories and molecular profiles of related areas in both lineages, the evidence presented here contributes to this debate in significant ways. For instance, most major arcopallial domains in adult zebra finches were found to share several differential markers with both deep cortical layers and amygdalar subdivisions. In fact, many of the genes that are markers of cortical layers in mammals are also markers of amygdalar structures, and thus their usefulness in establishing exact correspondences between brain areas in birds and mammals is limited. This includes some markers previously used to argue for molecular similarities between the avian arcopallium and mammalian pyramidal cells in cortical layers 5/6 (e.g. FEZF2, PCP4, and ETV1; Dugas-Ford et al., 2012), as they also label amygdalar structures (e.g. MEA, CEA, and BLA). However, when these and other shared markers of cortical and amygdalar structures were removed from the analysis, we found that the ventral and ventromedial arcopallial subdomains (AMV and AV) shared a predominance of markers with amygdalar structures over cortical layers, whereas intermediate and dorsal arcopallial subdomains (AI/AId/AD), as well as AMD shared a predominance of markers with cortical layers over amygdalar structures. Nonetheless, nearly every arcopallial subdivision still shared markers with both cortical and amygdalar structures. One possible explanation is that most or all arcopallial subdivisions are composed of a mixture of cell populations derived from both cortical layers and amygdalar subdivisions in mammals. Alternatively, individual arcopallial neurons may express genes that are markers of both cortical and amygdalar subdivisions. In either case, the interpretation of molecular similarities and homologies across these avian and mammalian brain structures is clearly highly influenced by the choice of markers studied. Similar observations on molecular similarities in pallial areas of birds and mammals were made by Jarvis et al (2103). Based on a more limited set of genes, those authors suggested a variant of the field homology hypothesis initially proposed by Butler and Molnar (Butler and Molnar, 2002), whereby avian areas like the arcopallium would be homologous to both cortical layers and parts of the claustrum/amygdalar complex in mammals. To some extent similar conclusions were reached by Herold et al. (2018) based on ligand binding data in pigeon, although these authors focused their discussion mostly on the need to further characterize the diversity of avian arcopallial subdomains before attempting to propose molecular-based homologies.
Below we discuss further some specific domains.
AM:
ZBTB20 is a specific marker of AMV in finch, and of a corresponding medio-ventral domain named the amygdalo-hippocampal area in the chick atlas (Puelles, 2007). ZBTB20-defined AMV also likely corresponds to (or at least contains) the area referred to as Tn/TnA in several studies in columbiformes and chicken. In mouse (MBA;mouse.brain-map.org, SCR_002978), differential expression of ZBTB20 is most prominent in the cortical amygdalar area, including the medial transition zone into the dentate gyrus, as well as in striatal CEA and less prominently in the MEA. Consistent with the possibility that AMV/Tn corresponds to the avian homologue of the cortical amygdalar area, and not the striatal CEA or MEA, the adult finch AMV is clearly pallial, located dorsal to the LPSP, and has a pallial-like GAD2 expression pattern. An amygdaloid nature of AMV is also consistent with data showing robust connections of a medial arcopallial domain (labeled Tn) with the hypothalamus in songbirds and columbiformes (Zeier and Karten, 1971; Cheng et al., 1999). In finches, this region appears to lack expression of striatal-related developmental markers (i.e. PAX6, ISL1, LHX5/ 6, NKX2-1; Vicario et al., 2017). Interestingly, we have identified a separate area of high ZBTB20 expression within the ventral striatum of finches (not mapped). This sub-pallial area might correspond to the striatal region suggested to represent the core nuclei of the CEA and MEA in zebra finches, based on the developmental expression of striatal-related markers (Vicario et al., 2017), but this remains to be tested.
An alternative overall scenario is that AMV/Tn, or at least part of it, represents the avian homologue of core nuclei of the mammalian MEA and/or CEA. This scenario is supported by a striatal-like high expression of GAD2 in the medial part of Tn in chicks (Sun et al., 2005; Yamamoto et al., 2005), and evidence that this nucleus plays a role in adaptive patterns of social behavior (Cheng et al., 1999). Moreover, in at least some avian species, Tn appears to express CYP19A1 and ESR2 (e.g. Foidart et al., 1995; Foidart et al., 1999), which are both markers of core nuclei in the MEA and CEA of mammals (see MBA). However, this interpretation would require evidence that the avian arcopallium contains the avian homologues of sub-pallial portions of the amygdala, which would be inconsistent with the analyses and conclusions from Vicario et al. (2013, 2017).
Based on current evidence it is not possible to resolve these apparent discrepancies, however several factors should be considered: 1) AMV/Tn may not be identical in songbirds and nonsongbirds, and AMV/Tn in the latter may include a sub-pallial component not present in songbirds; 2) In zebra finches, the highest arcopallial expression of CYP19A1 occurs in AMD, a region dorso-medial to AMV that is clearly pallial, which weakens the argument that CYP19A1 expression reveals a sub-pallial part of the amygdala in songbirds; 3) The pallial expression of CYP19A1, ESR1, and ESR2 is quite variable across avian species, thus these genes do not appear to reliably define conserved domains and probably should not be used to establish homologies with mammals; 4) As with ZBTB20, we have also observed ventral striatal expression of CYP19A1, ESR1, and ESR2, suggesting the existence of sub-pallial cell populations that might be more consistent with a sub-pallial central or medial amygdalar identity; 5) Lastly, we suggest that the high expression of ESR2 in finch AMVm could represent a pallial specialization characteristic of songbirds (or birds in general, although still untested) that is absent in mammals.
RA, AId, and AD:
Our data indicate that AI/AId and AD share more molecular features with deep cortical layers than with amygdalar structures, consistent with several lines of evidence suggesting that AI, and more specifically AId, corresponds to pyramidal cells in layer 5/6 of motor cortex. In pigeons, for example, while medial parts of the arcopallium (including Tn) project to the hypothalamus and are amygdaloid-like, intermediate and dorsal parts of the arcopallium project to thalamus, midbrain (tectum, tegmentum), lateral reticular formation, and brain stem nuclei, indicating they are somatic-like (Zeier and Karten, 1971). Similar to pigeon AI and consistent with a somatic function, finch AId projects to a myriad of targets, including striatum, thalamus, and nuclei of the midbrain reticular formation (Bottjer et al., 2000), and shows evidence of neuronal activation associated with wing movements (Feenders et al., 2008), suggesting a general involvement in movement control. AId (and AD) is labeled by PCP4, which in the mammalian cortex is a marker of pyramidal cells in layers 5/6 (Dugas-Ford et al., 2012). One should note, however, that finch AId is also projects to the ventral tegmental area and the lateral hypothalamus (Bottjer et al., 2000), suggesting an amygdaloid nature to this region of the AI and highlighting possible species differences in the connectivity of this arcopallial domain. We also note that several other markers of cortical pyramidal cells that were examined (Dugas-Ford et al., 2012) turned out not to be general markers of the finch arcopallium, but instead showed differential expression in AD. The overall molecular data also seems to favor a general correspondence between AD and deep cortical layers, most prominently cortical layer 6. Thus, our data support the overall interpretation that intermediate and dorsal parts of the avian arcopallium may represent cell populations homologous to neuronal cells in deep cortical layers in mammals.
In sum, we have performed an extensive characterization of the organization of the arcopallium in zebra finches, based on analysis of in situ hybridization patterns of a large collection of transcripts and close examination of cytoarchitectonic features. The data provides evidence for 20 distinct regions, grouped into six major domains, as well as novel insights into molecular convergences and homologies with areas like cortical layers and amygdala subdivisions in mammals. The study also provides a roadmap for further detailed connectivity and functional studies of this key but complex avian brain region.
Supplementary Material
Acknowledgments
We thank members of the avian arcopallial discussion group (Donna Maney, Erich Jarvis, Loretta Medina, Anton Reiner, Luke Remage-Healey, as well as other members of these labs) for the stimulating and helpful discussions on the medial arcopallium anatomy and nomenclature, and Donna Maney, in particular, for granting us permission to discuss unpublished gene expression data in white-throated sparrows. We also thank the Art Arnold lab for providing cDNA clones for ESR1 and 2.
Avian Anatomical Abbreviations List
- A
arcopallium
- AA
anterior arcopallium
- AAc
caudal part of the anterior arcopallium
- AArl
rostro-lateral part of the anterior arcopallium
- AArm
rostro-medial part of the anterior arcopallium
- AAv
ventral part of the anterior arcopallium
- AD
dorsal arcopallium
- AI
intermediate arcopallium
- AId
dorsal part of the intermediate arcopallium
- AIm
medial part of the intermediate arcopallium
- AIr
rostral part of the intermediate arcopallium
- AIrv
rostro-ventral part of the intermediate arcopallium
- AIv
ventral part of the intermediate arcopallium
- AM
medial arcopallium
- AMV
ventro-medial arcopallium
- AMVc
caudal part of the ventro-medial arcopallium
- AMVi
intermediate part of the ventro-medial arcopallium
- AMVl
lateral part of the ventro-medial arcopallium
- AMVm
medial part of the ventro-medial arcopallium
- AMD
dorso-medial arcopallium
- AMDl
lateral part of the dorso-medial arcopallium
- AMDm
medial part of the dorso-medial arcopallium
- AP
posterior arcopallium
- APd
dorsal part of the posterior arcopallium
- APv
ventral part of the posterior arcopallium
- AV
ventral arcopallium
- Cb
Cerebellum
- ChP
choroid plexus
- FA
Fronto-arcopallial tract
- Hb
habenula
- Hyp
hypothalamus
- LAD
dorsal arcopallial lamina
- LAV
vertical arcopallial lamina
- lLAD
lateral part of the dorsal arcopallial lamina
- LPS
pallial-subpallial lamina
- mLAD
medial part of the dorsal arcopallial lamina
- mOM
medial part of the occipito-mesencephalic tract
- N
nidopallium
- NCM
caudomedial nidopallium
- OAL
oblique arcopallial lamina
- OM
occipito-mesencephalic tract
- RA
robust nucleus of the arcopallium
- S
septum
- St
Striatum
- TeO
optic tectum
- Thal
thalamus
- Tn
nucleus taenia
Footnotes
Data Availability
All of the data supporting the findings of this study are presented in this article or in SupportingInfo:SuppTable1 and SupportingInfo:SuppTable2. High-resolution photomicrographs of in situ hybridization images for most of the genes analyzed in this study in the sagittal plane are also available online publicly on ZEBrA (www.zebrafinchatlas.org; RRID: SCR_012988).
References
- Abellan A, Legaz I, Vernier B, Retaux S, Medina L. 2009. Olfactory and amygdalar structures of the chicken ventral pallium based on the combinatorial expression patterns of LIM and other developmental regulatory genes. J Comp Neurol 516(3):166–186. [DOI] [PubMed] [Google Scholar]
- Absil P, Braquenier JB, Balthazart J, Ball GF. 2002. Effects of lesions of nucleus taeniae on appetitive and consummatory aspects of male sexual behavior in Japanese quail. Brain Behav Evol 60(1):13–35. [DOI] [PubMed] [Google Scholar]
- Acharya KD, Veney SL. 2012. Characterization of the G-protein-coupled membrane-bound estrogen receptor GPR30 in the zebra finch brain reveals a sex difference in gene and protein expression. Dev Neurobiol 72(11):1433–1446. [DOI] [PubMed] [Google Scholar]
- Arnold AP. 1980. Quantitative analysis of sex differences in hormone accumulation in the zebra finch brain: methodological and theoretical issues. J Comp Neurol 189(3):421–436. [DOI] [PubMed] [Google Scholar]
- Atoji Y, Saito S, Wild JM. 2006. Fiber connections of the compact division of the posterior pallial amygdala and lateral part of the bed nucleus of the stria terminalis in the pigeon (Columba livia). J Comp Neurol 499(2):161–182. [DOI] [PubMed] [Google Scholar]
- Bagnoli P, Francesconi W. 1984. Mapping of functional activity in the falcon visual system with [14C] 2-deoxyglucose. Exp Brain Res 53(2):217–222. [DOI] [PubMed] [Google Scholar]
- Ball GF, Balthazart J. 2004. Hormonal regulation of brain circuits mediating male sexual behavior in birds. Physiol Behav 83(2):329–346. [DOI] [PubMed] [Google Scholar]
- Ball GF, Balthazart J. 2010. Japanese quail as a model system for studying the neuroendocrine control of reproductive and social behaviors. ILAR J 51(4):310–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball GF, Bernard DJ, Foidart A, Lakaye B, Balthazart J. 1999. Steroid sensitive sites in the avian brain: does the distribution of the estrogen receptor alpha and beta types provide insight into their function? Brain Behav Evol 54(1):28–40. [DOI] [PubMed] [Google Scholar]
- Balthazart J, Absil P, Foidart A, Houbart M, Harada N, Ball GF. 1996. Distribution of aromatase-immunoreactive cells in the forebrain of zebra finches (Taeniopygia guttata): implications for the neural action of steroids and nuclear definition in the avian hypothalamus. J Neurobiol 31(2):129–148. [DOI] [PubMed] [Google Scholar]
- Balthazart J, Foidart A, Baillien M, Harada N, Ball GF. 1998a. Anatomical relationships between aromatase and tyrosine hydroxylase in the quail brain: double-label immunocytochemical studies. J Comp Neurol 391(2):214–226. [PubMed] [Google Scholar]
- Balthazart J, Foidart A, Houbart M, Prins GS, Ball GF. 1998b. Distribution of androgen receptor-immunoreactive cells in the quail forebrain and their relationship with aromatase immunoreactivity. J Neurobiol 35(3):323–340. [PubMed] [Google Scholar]
- Balthazart J, Foidart A, Wilson EM, Ball GF. 1992. Immunocytochemical localization of androgen receptors in the male songbird and quail brain. J Comp Neurol 317(4):407–420. [DOI] [PubMed] [Google Scholar]
- Balthazart J, Gahr M, Surlemont C. 1989. Distribution of estrogen receptors in the brain of the Japanese quail: an immunocytochemical study. Brain Res 501(2):205–214. [DOI] [PubMed] [Google Scholar]
- Belgard TG, Montiel JF, Wang WZ, Garcia-Moreno F, Margulies EH, Ponting CP, Molnar Z. 2013. Adult pallium transcriptomes surprise in not reflecting predicted homologies across diverse chicken and mouse pallial sectors. Proc Natl Acad Sci U S A 110(32):13150–13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard DJ, Bentley GE, Balthazart J, Turek FW, Ball GF. 1999. Androgen receptor, estrogen receptor alpha, and estrogen receptor beta show distinct patterns of expression in forebrain song control nuclei of European starlings. Endocrinology 140(10):4633–4643. [DOI] [PubMed] [Google Scholar]
- Bottjer SW, Altenau B. 2010. Parallel pathways for vocal learning in basal ganglia of songbirds. Nat Neurosci 13(2):153–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottjer SW, Brady JD, Cribbs B. 2000. Connections of a motor cortical region in zebra finches: relation to pathways for vocal learning. J Comp Neurol 420(2):244–260. [PubMed] [Google Scholar]
- Brauth SE, Heaton JT, Shea SD, Durand SE, Hall WS. 1997. Functional anatomy of forebrain vocal control pathways in the budgerigar (Melopsittacus undulatus). Annals of the New York Academy of Science 807:368–385. [DOI] [PubMed] [Google Scholar]
- Butler AB, Hodos W. 2005. Comparative vertebrate neuroanatomy : evolution and adaptation. Hoboken, N.J.: Wiley-Interscience; xxi, 715 p. p. [Google Scholar]
- Butler AB, Molnar Z. 2002. Development and evolution of the collopallium in amniotes: a new hypothesis of field homology. Brain Res Bull 57(3–4):475–479. [DOI] [PubMed] [Google Scholar]
- Butler AB, Reiner A, Karten HJ. 2011. Evolution of the amniote pallium and the origins of mammalian neocortex. Ann N Y Acad Sci 1225:14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton JB, Lovell PV, McHugh A, Marzulla T, Horback KL, Mello CV. 2014. An optimized protocol for high-throughput in situ hybridization of zebra finch brain. Cold Spring Harb Protoc 2014(12):1249–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-C, Winkler CM, Pfenning AR, Jarvis ED. 2013. Molecular profiling of the developing avian telencephalon: Regional timing and brain subdivision continuities. Journal of Comparative Neurology. Vol 521 p 3666–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M, Chaiken M, Zuo M, Miller H. 1999. Nucleus taenia of the amygdala of birds: anatomical and functional studies in ring doves (Streptopelia risoria) and European starlings (Sturnus vulgaris). Brain Behav Evol 53(5–6):243–270. [DOI] [PubMed] [Google Scholar]
- de Lima JL, Soares FA, Remedios AC, Thom G, Wirthlin M, Aleixo A, Schneider MP, Mello CV, Schneider PN. 2015. A putative RA-like region in the brain of the scale-backed antbird, Willisornis poecilinotus (Furnariides, Suboscines, Passeriformes, Thamnophilidae). Genet Mol Biol 38(3):249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boer-Visser AM, Brittijn ML, Dubbeldam JL, editor. 2004. A stereotaxic atlas of the brain of the collared dove, Streptopelia decaocto: using the old and new nomenclature: Shaker Publishing B.V. [Google Scholar]
- Dong S, Replogle KL, Hasadsri L, Imai BS, Yau PM, Rodriguez-Zas S, Southey BR, Sweedler JV, Clayton DF. 2009. Discrete molecular states in the brain accompany changing responses to a vocal signal. Proc Natl Acad Sci U S A 106(27):11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas-Ford J, Rowell JJ, Ragsdale CW. 2012. Cell-type homologies and the origins of the neocortex. Proc Natl Acad Sci U S A 109(42):16974–16979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand SE, Heaton JT, Amateau SK, Brauth SE. 1997. Vocal control pathways through the anterior forebrain of a parrot (Melopsittacus undulatus). Journal of Comparative Neurology 377:179–206. [DOI] [PubMed] [Google Scholar]
- Emery NJ. 2005. Cognitive ornithology: the evolution of avian intelligence. Phil. Trans. R. Soc. B 361: 23–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenders G, Liedvogel M, Rivas M, Zapka M, Horita H, Hara E, Wada K, Mouritsen H, Jarvis ED. 2008. Molecular Mapping of Movement-Associated Areas in the Avian Brain: A Motor Theory for Vocal Learning Origin. PLoS ONE. Vol 3 p e1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Lopez A, Revilla V, Candelas MA, Gonzalez-Gil J, Diaz A, Pazos A. 1997. A comparative study of alpha2- and beta-adrenoceptor distribution in pigeon and chick brain. Eur J Neurosci 9(5):871–883. [DOI] [PubMed] [Google Scholar]
- Foidart A, Lakaye B, Grisar T, Ball GF, Balthazart J. 1999. Estrogen receptor-beta in quail: cloning, tissue expression and neuroanatomical distribution. J Neurobiol 40(3):327–342. [PubMed] [Google Scholar]
- Foidart A, Reid J, Absil P, Yoshimura N, Harada N, Balthazart J. 1995. Critical re-examination of the distribution of aromatase-immunoreactive cells in the quail forebrain using antibodies raised against human placental aromatase and against the recombinant quail, mouse or human enzyme. J Chem Neuroanat 8(4):267–282. [DOI] [PubMed] [Google Scholar]
- Fusani L, Donaldson Z, London SE, Fuxjager MJ, Schlinger BA. 2014. Expression of androgen receptor in the brain of a sub-oscine bird with an elaborate courtship display. Neurosci Lett 578:61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahr M 2000. Neural song control system of hummingbirds: comparison to swifts, vocal learning (Songbirds) and nonlearning (Suboscines) passerines, and vocal learning (Budgerigars) and nonlearning (Dove, owl, gull, quail, chicken) nonpasserines. J Comp Neurol 426(2):182–196. [PubMed] [Google Scholar]
- Gahr M, Flugge G, Guttinger HR. 1987. Immunocytochemical localization of estrogen-binding neurons in the songbird brain. Brain Res 402(1):173–177. [DOI] [PubMed] [Google Scholar]
- Gahr M, Guttinger HR, Kroodsma DE. 1993. Estrogen receptors in the avian brain: survey reveals general distribution and forebrain areas unique to songbirds. J Comp Neurol 327(1):112–122. [DOI] [PubMed] [Google Scholar]
- Gunturkun O, Bugnyar T. 2016. Cognition without Cortex. Trends Cogn Sci 20(4):291–303. [DOI] [PubMed] [Google Scholar]
- Hahnloser RH, Kozhevnikov AA, Fee MS. 2002. An ultra-sparse code underlies the generation of neural sequences in a songbird. Nature 419(6902):65–70. [DOI] [PubMed] [Google Scholar]
- Halldin K, Axelsson J, Holmgren C, Brunstrom B. 2006. Localization of estrogen receptor-alpha and -betamRNA in brain areas controlling sexual behavior in Japanese quail. J Neurobiol 66(2):148–154. [DOI] [PubMed] [Google Scholar]
- Heaton JT, Brauth SE. 2000. Effects of lesions of the central nucleus of the anterior archistriatum on contact call and warble song production in the budgerigar (Melopsittacus undulatus). Neurobiol Learn Mem 73(3):207–242. [DOI] [PubMed] [Google Scholar]
- Herold C, Paulitschek C, Palomero-Gallagher N, Gunturkun O, Zilles K. 2018. Transmitter receptors reveal segregation of the arcopallium/amygdala complex in pigeons (Columba livia). J Comp Neurol 526(3):439–466. [DOI] [PubMed] [Google Scholar]
- Iyengar S, Viswanathan SS, Bottjer SW. 1999. Development of topography within song control circuitry of zebra finches during the sensitive period for song learning. Journal of Neuroscience 19(14):6037–6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs EC, Arnold AP, Campagnoni AT. 1996. Zebra finch estrogen receptor cDNA: cloning and mRNA expression. J Steroid Biochem Mol Biol 59(2):135–145. [DOI] [PubMed] [Google Scholar]
- Jarvis ED. 2009. Evolution of the Pallium in Birds and Reptiles In: Binder MD, Hirokawa N, Windhorst U, eds. Encyclopedia of Neuroscience. Berlin, Heidelberg: Springer Berlin Heidelberg; p 1390–1400. [Google Scholar]
- Jarvis ED, Gunturkun O, Bruce L, Csillag A, Karten H, Kuenzel W, Medina L, Paxinos G, Perkel DJ, Shimizu T, Striedter G, Wild JM, Ball GF, Dugas-Ford J, Durand SE, Hough GE, Husband S, Kubikova L, Lee DW, Mello CV, Powers A, Siang C, Smulders TV, Wada K, White SA, Yamamoto K, Yu J, Reiner A, Butler AB. 2005. Avian brains and a new understanding of vertebrate brain evolution. Nat Rev Neurosci 6(2):151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Mello CV. 2000. Molecular mapping of brain areas involved in parrot vocal communication. J Comp Neurol 419(1):1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Mello CV, Nottebohm F. 1995. Associative learning and stimulus novelty influence the song-induced expression of an immediate early gene in the canary forebrain. Learn Mem. 2(2):62–80. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SY, Faircloth BC, Nabholz B, Howard JT, Suh A, Weber CC, da Fonseca RR, Li J, Zhang F, Li H, Zhou L, Narula N, Liu L, Ganapathy G, Boussau B, Bayzid MS, Zavidovych V, Subramanian S, Gabaldon T, Capella-Gutierrez S, Huerta-Cepas J, Rekepalli B, Munch K, Schierup M, Lindow B, Warren WC, Ray D, Green RE, Bruford MW, Zhan X, Dixon A, Li S, Li N, Huang Y, Derryberry EP, Bertelsen MF, Sheldon FH, Brumfield RT, Mello CV, Lovell PV, Wirthlin M, Schneider MP, Prosdocimi F, Samaniego JA, Vargas Velazquez AM, Alfaro-Nunez A, Campos PF, Petersen B, Sicheritz-Ponten T, Pas A, Bailey T, Scofield P, Bunce M, Lambert DM, Zhou Q, Perelman P, Driskell AC, Shapiro B, Xiong Z, Zeng Y, Liu S, Li Z, Liu B, Wu K, Xiao J, Yinqi X, Zheng Q, Zhang Y, Yang H, Wang J, Smeds L, Rheindt FE, Braun M, Fjeldsa J, Orlando L, Barker FK, Jonsson KA, Johnson W, Koepfli KP, O'Brien S, Haussler D, Ryder OA, Rahbek C, Willerslev E, Graves GR, Glenn TC, McCormack J, Burt D, Ellegren H, Alstrom P, Edwards SV, Stamatakis A, Mindell DP, Cracraft J, Braun EL, Warnow T, Jun W, Gilbert MT, Zhang G. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346(6215):1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Nottebohm F. 1997. Motor-driven gene expression. Proc Natl Acad Sci U S A 94(8):4097–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Ribeiro S, da Silva ML, Ventura D, Vielliard J, Mello CV. 2000. Behaviourally driven gene expression reveals song nuclei in hummingbird brain. Nature 406(6796):628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Schwabl H, Ribeiro S, Mello CV. 1997. Brain gene regulation by territorial singing behavior in freely ranging songbirds. Neuroreport 8(8):2073–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Yu J, Rivas MV, Horita H, Feenders G, Whitney O, Jarvis SC, Jarvis ER, Kubikova L, Puck AE, Siang-Bakshi C, Martin S, McElroy M, Hara E, Howard J, Pfenning A, Mouritsen H, Chen CC, Wada K. 2013. Global view of the functional molecular organization of the avian cerebrum: mirror images and functional columns. J Comp Neurol 521(16):3614–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F, Sablan MM, Bottjer SW. 1995. Topographic organization of a forebrain pathway involved with vocal learning in zebra finches. J Comp Neurol 358(2):260–278. [DOI] [PubMed] [Google Scholar]
- Karten HJ. 2015. Vertebrate brains and evolutionary connectomics: on the origins of the mammalian 'neocortex'. Philos Trans R Soc Lond B Biol Sci 370(1684). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karten HJ, Brzozowska-Prechtl A, Lovell PV, Tang DD, Mello CV, Wang H, Mitra PP. 2013. Digital atlas of the zebra finch (taeniopygia guttata) brain: A high resolution photo atlas. J Comp Neurol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karten HJ, Hodos W. 1967. A stereotaxic atlas of the brain of the pigeon (Columba livia). Baltimore, MD: Johns Hopkins Press. [Google Scholar]
- Kelley DB, Nottebohm F. 1979. Projections of a telencephalic auditory nucleus-field L-in the canary. J Comp Neurol 183(3):455–469. [DOI] [PubMed] [Google Scholar]
- Konishi M, Akutagawa E. 1985. Neuronal growth, atrophy and death in a sexually dimorphic song nucleus in the zebra finch brain. Nature 315(6015):145–147. [DOI] [PubMed] [Google Scholar]
- Kroodsma DE, Konishi M. 1991. A suboscine bird (eastern phoebe, Sayornis phoebe) develops normal song without auditory feedback. Animal Behavior 42:477–487. [Google Scholar]
- Kuenzel WJ, Masson M. 1988. A stereotaxic atlas of the brain of the chick (Gallus domesticus). Baltimore, MD: Johns Hopkins University Press. [Google Scholar]
- Kuenzel WJ, Medina L, Csillag A, Perkel DJ, Reiner A. 2011. The avian subpallium: new insights into structural and functional subdivisions occupying the lateral subpallial wall and their embryological origins. Brain Res 1424:67–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445(7124):168–176. [DOI] [PubMed] [Google Scholar]
- Letzner S, Simon A, Gunturkun O. 2016. Connectivity and neurochemistry of the commissura anterior of the pigeon (Columba livia). J Comp Neurol 524(2):343–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CH, Goode CT, Young LJ, Maney DL. 2009. Neural distribution of nonapeptide binding sites in two species of songbird. J Comp Neurol 513(2):197–208. [DOI] [PubMed] [Google Scholar]
- Liao S-q, Hou G-q, Liu X-l, Long C, Li D-f. 2011. Electrophysiological properties of neurons in the robust nucleus of the arcopallium of adult male zebra finches. Neuroscience Letters. Vol 487 p 234–239. [DOI] [PubMed] [Google Scholar]
- Liu WC, Wada K, Jarvis E, Nottebohm F. 2013. Rudimentary substrates for vocal learning in a suboscine. Nat Commun 4:2082. [DOI] [PubMed] [Google Scholar]
- Long MA, Fee MS. 2008. Using temperature to analyse temporal dynamics in the songbird motor pathway. Nature 456(7219):189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Clayton DF, Replogle KL, Mello CV. 2008. Birdsong "transcriptomics": neurochemical specializations of the oscine song system. PLoS ONE 3(10):e3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Huizinga NA, Friedrich SR, Wirthlin M, Mello CV. 2018. The constitutive differential transcriptome of a brain circuit for vocal learning. BMC Genomics 19(1):231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelblat-Cerf Y, Las L, Denisenko N, Fee MS. 2014. A role for descending auditory cortical projections in songbird vocal learning. eLife Sciences Vol 3 p e02152–e02152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vargas MC, Stumpf WE, Sar M. 1976. Anatomical distribution of estrogen target cells in the avian CNS: a comparison with the mammalian CNS. J Comp Neurol 167(1):83–103. [DOI] [PubMed] [Google Scholar]
- Matragrano LL, Sanford SE, Salvante KG, Beaulieu M, Sockman KW, Maney DL. 2012. Estradiol-dependent modulation of serotonergic markers in auditory areas of a seasonally breeding songbird. Behav Neurosci 126(1):110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CV, Vicario DS, Clayton DF. 1993. Song presentation induces gene expression in the songbird forebrain. Proc Natl Acad Sci U S A 89(15):6818–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CV, Clayton DF. 1995. Differential induction of the ZENK gene in the avian forebrain and song control circuit after metrazole-induced depolarization. J Neurobiol 26(1):145–161. [DOI] [PubMed] [Google Scholar]
- Mello CV, Vates GE, Okuhata S, Nottebohm F. 1998. Descending auditory pathways in the adult male zebra finch (Taeniopygia guttata). J Comp Neurol 395(2):137–160. [PubMed] [Google Scholar]
- Metzdorf R, Gahr M, Fusani L. 1999. Distribution of aromatase, estrogen receptor, and androgen receptor mRNA in the forebrain of songbirds and nonsongbirds. J Comp Neurol 407(1):115–129. [PubMed] [Google Scholar]
- Montagnese CM, Szekely T, Csillag A, Zachar G. 2015. Distribution of vasotocin- and vasoactive intestinal peptide-like immunoreactivity in the brain of blue tit (Cyanistes coeruleus). Front Neuroanat 9:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottebohm F, Kelley DB, Paton JA. 1982. Connections of vocal control nuclei in the canary telencephalon. J Comp Neurol 207(4):344–357. [DOI] [PubMed] [Google Scholar]
- Nottebohm F, Stokes TM, Leonard CM. 1976. Central control of song in the canary, Serinus canarius. J Comp Neurol 165(4):457–486. [DOI] [PubMed] [Google Scholar]
- Paterson AK, Bottjer SW. 2017. Cortical inter-hemispheric circuits for multimodal vocal learning in songbirds. J Comp Neurol 525(15):3312–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman WR, Arnold AP. 2003. Expression of estrogen receptor and aromatase mRNAs in embryonic and posthatch zebra finch brain. J Neurobiol 55(2):204–219. [DOI] [PubMed] [Google Scholar]
- Pfenning AR, Hara E, Whitney O, Rivas MV, Wang R, Roulhac PL, Howard JT, Wirthlin M, Lovell PV, Ganapathy G, Mouncastle J, Moseley MA, Thompson JW, Soderblom EJ, Iriki A, Kato M, Gilbert MT, Zhang G, Bakken T, Bongaarts A, Bernard A, Lein E, Mello CV, Hartemink AJ, Jarvis ED. 2014. Convergent transcriptional specializations in the brains of humans and song-learning birds. Science 346(6215):1256846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles L 2011. Pallio-pallial tangential migrations and growth signaling: new scenario for cortical evolution? Brain Behav Evol 78(1):108–127. [DOI] [PubMed] [Google Scholar]
- Puelles L, Paxinos G, Watson C, Martinez S, Martinez-de-la-Torre M 2007. The Chick Brain in Stereotaxic Coordinates: An Atlas Based on Neuromeres: Elsevier Science & Technology Books. [Google Scholar]
- Reiner A, Perkel DJ, Bruce LL, Butler AB, Csillag A, Kuenzel W, Medina L, Paxinos G, Shimizu T, Striedter G, Wild M, Ball GF, Durand S, Guturkun O, Lee DW, Mello CV, Powers A, White SA, Hough G, Kubikova L, Smulders TV, Wada K, Dugas-Ford J, Husband S, Yamamoto K, Yu J, Siang C, Jarvis ED. 2004a. The Avian Brain Nomenclature Forum: Terminology for a New Century in Comparative Neuroanatomy. J Comp Neurol 473:E1–E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Perkel DJ, Mello CV, Jarvis ED. 2004b. Songbirds and the revised avian brain nomenclature. Ann N Y Acad Sci 1016:77–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replogle KL, Arnold AP, Ball GF, Band M, Bensch S, Brenowitz EA, Dong S, Drnevich J, Ferris M, George JM, Gong G, Hasselquist D, Hernandez AG, Kim R, Lewin HA, Liu L, Lovell PV, Mello CV, Naurin S, Rodriguez-Zas S, Thimmapuram J, Wade J, Clayton DF. 2008. The Songbird Neurogenomics (SoNG) Initiative: community-based tools and strategies for study of brain gene function and evolution. BMC Genomics 9(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riters LV, Ball GF. 2002. Sex differences in the densities of alpha 2-adrenergic receptors in the song control system, but not the medial preoptic nucleus in zebra finches. J Chem Neuroanat 23(4):269–277. [DOI] [PubMed] [Google Scholar]
- Saint-Dizier H, Constantin P, Davies DC, Leterrier C, Levy F, Richard S. 2009. Subdivisions of the arcopallium/posterior pallial amygdala complex are differentially involved in the control of fear behaviour in the Japanese quail. Brain Res Bull 79(5):288–295. [DOI] [PubMed] [Google Scholar]
- Saldanha CJ, Schultz JD, London SE, Schlinger BA. 2000a. Telencephalic aromatase but not a song circuit in a sub-oscine passerine, the golden collared manakin (Manacus vitellinus). Brain Behav Evol 56(1):29–37. [DOI] [PubMed] [Google Scholar]
- Saldanha CJ, Tuerk MJ, Kim YH, Fernandes AO, Arnold AP, Schlinger BA. 2000b. Distribution and regulation of telencephalic aromatase expression in the zebra finch revealed with a specific antibody. J Comp Neurol 423(4):619–630. [DOI] [PubMed] [Google Scholar]
- Shanahan M, Bingman VP, Shimizu T, Wild M, Gunturkun O. 2013. Large-scale network organization in the avian forebrain: a connectivity matrix and theoretical analysis. Front Comput Neurosci 7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P, Schlinger BA, Campagnoni AT, Arnold AP. 1995. An atlas of aromatase mRNA expression in the zebra finch brain. J Comp Neurol 360(1):172–184. [DOI] [PubMed] [Google Scholar]
- Soma KK, Schlinger BA, Wingfield JC, Saldanha CJ. 2003. Brain aromatase, 5 alpha-reductase, and 5 beta-reductase change seasonally in wild male song sparrows: relationship to aggressive and sexual behavior. J Neurobiol 56(3):209–221. [DOI] [PubMed] [Google Scholar]
- Spiro JE, Dalva MB, Mooney R. 1999. Long-range inhibition within the zebra finch song nucleus RA can coordinate the firing of multiple projection neurons. J Neurophysiol 81(6):3007–3020. [DOI] [PubMed] [Google Scholar]
- Stetner ME, Fee MS. 2017. Quantification of single unit activity related to head and body movements in the intermediate arcopallium of the zebra finch Soc for Neuroscience. Washington, D.C. [Google Scholar]
- Stokes TM, Leonard CM, Nottebohm F. 1974. The telencephalon, diencephalon, and mesencephalon of the canary, Serinus canaria, in stereotaxic coordinates. J Comp Neurol 156(3):337–374. [DOI] [PubMed] [Google Scholar]
- Suh A, Paus M, Kiefmann M, Churakov G, Franke FA, Brosius J, Kriegs JO, Schmitz J. 2011. Mesozoic retroposons reveal parrots as the closest living relatives of passerine birds. Nat Commun 2:443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Wang HB, Laverghetta A, Yamamoto K, Reiner A. 2005. The distribution and cellular localization of glutamic acid decarboxylase-65 (GAD65) mRNA in the forebrain and midbrain of domestic chick. J Chem Neuroanat 29(4):265–281. [DOI] [PubMed] [Google Scholar]
- Teramitsu I, Poopatanapong A, Torrisi S, White SA. 2010. Striatal FoxP2 is actively regulated during songbird sensorimotor learning. PLoS One 5(1):e8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RR, Goodson JL, Ruscio MG, Adkins-Regan E. 1998. Role of the archistriatal nucleus taeniae in the sexual behavior of male Japanese quail (Coturnix japonica): a comparison of function with the medial nucleus of the amygdala in mammals. Brain Behav Evol 51(4):215–229. [DOI] [PubMed] [Google Scholar]
- Vates GE, Vicario DS, Nottebohm F. 1997. Reafferent thalamo- "cortical" loops in the song system of oscine songbirds. J Comp Neurol 380(2):275–290. [PubMed] [Google Scholar]
- Vicario A, Abellan A, Desfilis E, Medina L. 2014. Genetic identification of the central nucleus and other components of the central extended amygdala in chicken during development. Front Neuroanat 8:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario A, Mendoza E, Abellan A, Scharff C, Medina L. 2017. Genoarchitecture of the extended amygdala in zebra finch, and expression of FoxP2 in cell corridors of different genetic profile. Brain Struct Funct 222(1):481–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario DS. 1991. Organization of the zebra finch song control system: II. Functional organization of outputs from nucleus Robustus archistriatalis. J Comp Neurol 309(4):486–494. [DOI] [PubMed] [Google Scholar]
- Wang R, Chen C-C, Hara E, Rivas MV, Roulhac PL, Howard JT, Chakraborty M, Audet J-N, Jarvis ED. 2015. Convergent differential regulation of SLIT-ROBO axon guidance genes in the brains of vocal learners. Journal of Comparative Neurology. Vol 523 p 892–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Kunstner A, Searle S, White S, Vilella AJ, Fairley S, Heger A, Kong L, Ponting CP, Jarvis ED, Mello CV, Minx P, Lovell P, Velho TA, Ferris M, Balakrishnan CN, Sinha S, Blatti C, London SE, Li Y, Lin YC, George J, Sweedler J, Southey B, Gunaratne P, Watson M, Nam K, Backstrom N, Smeds L, Nabholz B, Itoh Y, Whitney O, Pfenning AR, Howard J, Volker M, Skinner BM, Griffin DK, Ye L, McLaren WM, Flicek P, Quesada V, Velasco G, Lopez-Otin C, Puente XS, Olender T, Lancet D, Smit AF, Hubley R, Konkel MK, Walker JA, Batzer MA, Gu W, Pollock DD, Chen L, Cheng Z, Eichler EE, Stapley J, Slate J, Ekblom R, Birkhead T, Burke T, Burt D, Scharff C, Adam I, Richard H, Sultan M, Soldatov A, Lehrach H, Edwards SV, Yang SP, Li X, Graves T, Fulton L, Nelson J, Chinwalla A, Hou S, Mardis ER, Wilson RK. 2010. The genome of a songbird. Nature 464(7289):757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JT, Adkins-Regan E. 1989. Neuroanatomical localization of sex steroid-concentrating cells in the Japanese quail (Coturnix japonica): autoradiography with [3H]-testosterone, [3H]-estradiol, and [3H]-dihydrotestosterone. Neuroendocrinology 49(1):51–64. [DOI] [PubMed] [Google Scholar]
- Whitney O, Pfenning AR, Howard JT, Blatti CA, Liu F, Ward JM, Wang R, Audet J-N, Kellis M, Mukherjee S, Sinha S, Hartemink AJ, West AE, Jarvis ED. 2014. Core and region-enriched networks of behaviorally regulated genes and the singing genome. Science 346:1256780–1256780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild JM. 1993. Descending projections of the songbird nucleus robustus archistriatalis. J Comp Neurol 338:225–241. [DOI] [PubMed] [Google Scholar]
- Wild JM, Botelho JF. 2015. Involvement of the avian song system in reproductive behaviour. Biol Lett 11(12):20150773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild JM, Karten HJ, Frost BJ. 1993. Connections of the auditory forebrain in the pigeon (Columba livia). J Comp Neurol 337(1):32–62. [DOI] [PubMed] [Google Scholar]
- Wild JM, Williams MN, Suthers RA. 2000. Neural pathways for bilateral vocal control in songbirds. Journal of Comparative Neurology 423(3):413–426. [DOI] [PubMed] [Google Scholar]
- Xin Q, Ogura Y, Uno L, Matsushima T. 2017. Selective contribution of the telencephalic arcopallium to the social facilitation of foraging efforts in the domestic chick. Eur J Neurosci 45(3):365–380. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sun Z, Wang HB, Reiner A. 2005. Subpallial amygdala and nucleus taeniae in birds resemble extended amygdala and medial amygdala in mammals in their expression of markers of regional identity. Brain Res Bull 66(4–6):341–347. [DOI] [PubMed] [Google Scholar]
- Zeier H, Karten HJ. 1971. The archistriatum of the pigeon: organization of afferent and efferent connections. Brain Res 31(2):313–326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.