Summary
The CRISPR-associated protein Cas12a (Cpf1), which has been repurposed for genome editing, possesses two distinct nuclease activities: endoribonuclease activity for processing its own guide RNAs, and RNA-guided DNase activity for target DNA cleavage. To elucidate the molecular basis of both activities, we determined crystal structures of Francisella novicida Cas12a in a binary complex with a guide RNA, and in a R-loop complex containing a non-cleavable guide RNA precursor and full-length target DNA. Corroborated by biochemical experiments, these structures elucidate the mechanisms of guide RNA processing and pre-ordering of the seed sequence in the guide RNA that primes Cas12a for target DNA binding. The R-loop complex structure furthermore reveals the strand displacement mechanism that facilitates guide-target hybridization and suggests a mechanism for double-stranded DNA cleavage involving a single active site. Together, these insights advance our mechanistic understanding of Cas12a enzymes that may contribute to further development of genome editing technologies.
Keywords: CRISPR-Cas, Cpf1, Cas12a, Cas9, CRISPR RNA, crRNA processing, seed sequence, R-loop, target DNA cleavage, nuclease
Introduction
Prokaryotic CRISPR-Cas systems (clustered regularly interspaced short palindromic repeats and CRISPR-associated proteins) function as interference mechanisms that utilize CRISPR RNAs (crRNAs) as a guide to target invading nucleic acids such as viruses and plasmids (Marraffini, 2015; Sorek et al., 2013; van der Oost et al., 2014; Wiedenheft et al., 2012). While Class 1 systems (comprising types I, III and IV) typically form multi-subunit protein-crRNA effector complexes, Class 2 systems (comprising types II, V and VI) rely on a single, crRNA-guided protein for target interference (Mohanraju et al., 2016). In most DNA-targeting CRISPR-Cas systems, target DNA binding is dependent on the initial recognition of a Protospacer Adjacent Motif (PAM), a short sequence juxtaposed to the protospacer (target) site that is complementary to the crRNA spacer segment (Mojica et al., 2009). Additionally, target specificity is strongly influenced by complementarity between the target and a “seed” sequence in the crRNA (Jinek et al., 2012; Künne et al., 2014; Semenova et al., 2011; Wiedenheft et al., 2011), although mismatches to seed sequence nucleotides are sometimes tolerated (Fu et al., 2013; Hua Fu et al., 2014; Kleinstiver et al., 2016; Pattanayak et al., 2013). These molecular recognition events result in the formation of an R-loop structure in which the crRNA forms an RNA-DNA heteroduplex with the complementary DNA strand (the target strand), thereby displacing the non-complementary DNA strand (the non-target strand) (Jinek et al., 2012; 2014; Jore et al., 2011).
RNA-guided nucleases belonging to Class 2 CRISPR-Cas systems have emerged as powerful tools for precision genome editing and gene expression control (Charpentier and Marraffini, 2014; Doudna and Charpentier, 2014; Hsu et al., 2014; Mali et al., 2013a). Cas9, the hallmark protein of type II systems, associates with a dual guide RNA structure composed of a processed crRNA and a trans-activating crRNA (tracrRNA). Cas9 utilizes the 20-nucleotide segment at the 5’ end of the crRNA to bind cognate DNA sequences, and deploys its HNH and RuvC nuclease domains to cleave the double-stranded DNA target (Jinek et al., 2012; Karvelis et al., 2013). By programming Cas9 with artificial crRNA-tracrRNA guides or with chimeric single-molecule guide RNAs (sgRNAs), double-strand DNA breaks (DSBs) can be generated in genomic DNA of eukaryotic cells (Cho et al., 2013; Cong et al., 2013; Hwang et al., 2013; Jinek et al., 2013; Mali et al., 2013b). More recently, the type V CRISPR-associated protein Cpf1, recently renamed as Cas12a (Shmakov et al., 2017), has been repurposed for genome editing applications (Endo et al., 2016; Hur et al., 2016; Kim et al., 2016a; 2016b; Kleinstiver et al., 2016; Tóth et al., 2016; Xu et al., 2016; Zetsche et al., 2015; 2016). While Cas9 requires both a crRNA and a tracrRNA for DNA targeting, Cas12a requires only a single crRNA guide. In contrast to Cas9, Cas12a processes its own crRNAs (Fonfara et al., 2016), which has been exploited for multiplex genome editing (Zetsche et al., 2016). Like Cas9, Cas12a has a conserved RuvC nuclease domain; however, it lacks the HNH domain that Cas9 requires for target strand cleavage (Zetsche et al., 2015). Cas12a generates staggered-end breaks in the target DNA, whereas Cas9-mediated DSBs are blunt-ended (Zetsche et al., 2015). Furthermore, all Cas12a orthologs characterized to date recognize a T-rich PAM at the 5’ end of the target site (5’-(T)TTN), while Streptococcus pyogenes Cas9, the Cas9 ortholog most widely used in genome engineering applications, recognizes GG-containing PAMs at the 3’ end of the target (Zetsche et al., 2015). These distinguishing features of Cas12a make it a useful addition to the CRISPR-Cas genome editing toolkit, broadening the spectrum of targetable genomic sites.
Recent structural studies of Cas12a have revealed its bilobed molecular architecture consisting of a recognition lobe (REC) and a nuclease lobe (NUC) (Dong et al., 2016; Gao et al., 2016; Yamano et al., 2016). The structure of a binary complex of Lachnospiraceae bacterium Cas12a (LbCas12a) bound to a crRNA has revealed that the repeat-derived part of the crRNA forms an pseudoknot structure coordinated by a magnesium ion (Dong et al., 2016). Structures of ternary complexes of Acidaminococcus sp. Cas12a (AsCas12a) bound to a crRNA and a partially double-stranded DNA target uncovered the mechanism of PAM recognition and crRNA-target DNA hybridization (Gao et al., 2016; Yamano et al., 2016). However, many aspects of Cas12a mechanisms remain poorly understood. Pre-crRNA processing by Cas12a is dependent on the presence of divalent metal ions, but it is currently unclear whether the metal ions are directly involved in the catalytic mechanism (Fonfara et al., 2016). Biochemical studies of Cas12a suggested that, like for Cas9, target DNA recognition is dependent on a seed sequence in the crRNA; however, the LbCas12a binary complex structure did not provide structural evidence for seed-dependent DNA targeting. Moreover, it is thought that Cas12a has two distinct DNA nuclease domains (Yamano et al., 2016), yet the mechanisms for R-loop formation and target DNA cleavage remains elusive.
To shed light on the molecular mechanisms of pre-crRNA processing and crRNA-guided target binding and cleavage by Cas12a enzymes, we have determined crystal structures of Francisella novicida U112 Cas12a (FnCas12a) in a binary complex with a crRNA, as well as in a ternary complex with an uncleavable pre-crRNA mimic and a full-length double-stranded DNA target. These structures reveal the structural basis for seed-dependent DNA targeting, and, together with biochemical assays, demonstrate that pre-crRNA processing proceeds via a divalent cation-independent catalytic mechanism. Importantly, the structure of the ternary complex reveals the structural basis for crRNA-dependent R-loop formation and provides clues about the mechanism of target DNA cleavage. Together, our structural observations and corroborating biochemical experiments advance our understanding of Cas12a endonucleases, and establish a mechanistic foundation for their use in genome engineering applications.
Results
Cas12a pre-orders the crRNA seed sequence for target DNA recognition
Previous biochemical studies suggested that Cas12a-mediated DNA targeting relies on the presence of a seed sequence in the crRNA guide (Fonfara et al., 2016; Zetsche et al., 2015). However, the recent crystal structure of LbCas12a in complex with a processed crRNA revealed that, while the repeat-derived pseudoknot in the 5’ handle of the crRNA is ordered, the entire spacer-derived part of the crRNA is disordered (Dong et al., 2016). This contrasts with other RNA-guided systems, in which a seed sequence of the guide is typically pre-ordered in a helical conformation to reduce the entropic penalty of guide-mediated target binding (Elkayam et al., 2012; Jiang et al., 2015; Nakanishi et al., 2012; Schirle and Macrae, 2012; Wang et al., 2008). To reconcile these observations, it has been suggested that PAM-binding by Cas12a induces conformational changes that result in pre-ordering of the crRNA seed sequence (Gao et al., 2016).
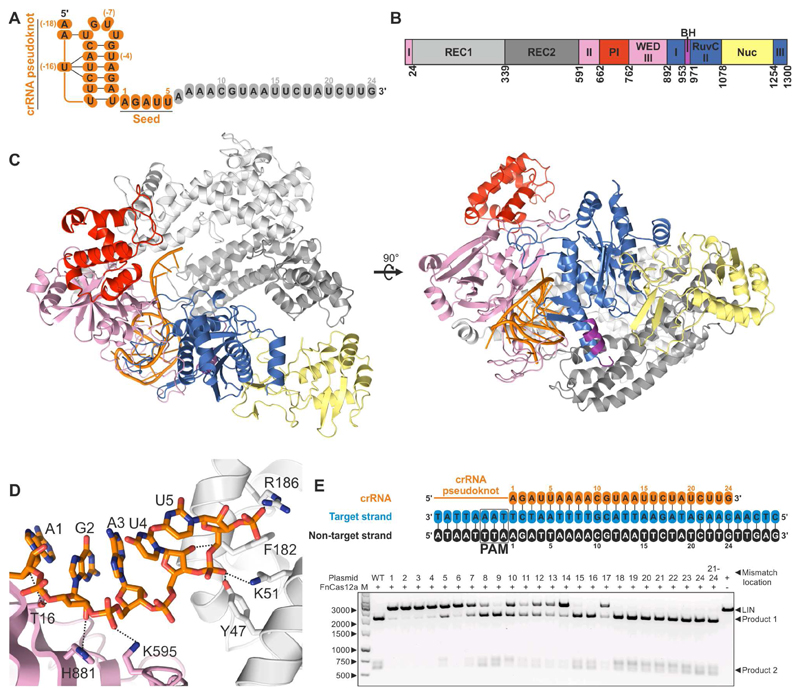
To obtain further insights into the mechanism of RNA-guided target DNA recognition, we co-crystallized purified FnCas12a (Figure S1) with a 43-nt crRNA and determined the structure of the complex at 3.4 Å resolution (Figure 1 and Table 1). The structure reveals a bilobed architecture similar to other Cas12a structures (Figure S2), with the two lobes connected by the Wedge (WED) domain (Dong et al., 2016; Gao et al., 2016; Yamano et al., 2016). The N-terminal recognition (REC) lobe consists of two α-helical domains (REC1 and REC2) that have been shown to coordinate the crRNA-target DNA heteroduplex (Gao et al., 2016; Yamano et al., 2016). The C-terminal nuclease (NUC) lobe consists of the C-terminal RuvC and Nuc domains involved in target cleavage, the Arginine-rich Bridge Helix (BH), and the PAM interacting (PI) domain. As observed in the LbCas12a and AsCas12a structures, the repeat-derived segment of the crRNA forms a pseudoknot (Figure S3A) stabilized by intramolecular base pairing and hydrogen bonding interactions (Figure S3B). The pseudoknot is coordinated by residues from the WED, RuvC and REC2 domains (Figure S3C), as well as by two hydrated magnesium cations (Figure S3A and S3C). Notably, nucleotides 1–5 of the crRNA are ordered in the central cavity of FnCas12a and adopt an A-form-like helical conformation (Figure 1D and Figure S3D). Conformational ordering of the seed sequence is facilitated by multiple interactions between the ribose and phosphate moieties of the crRNA backbone and FnCas12a residues in the WED and REC1 domains. These include residues Thr16, Lys595, His804 and His881 from the WED domain, and residues Tyr47, Lys51, Phe182 and Arg186 from the REC1 domain (Figure 1D and Figure S3C), all of which are strongly conserved in Cas12a orthologs (Figure S4). Consistent with a seed sequence-dependent mechanism of DNA targeting and in broad agreement with previous analyses of AsCas12a and LbCas12a activities in vivo (Kleinstiver et al., 2016; Kim et al., 2016a), cleavage of DNA substrates with single-nucleotide mismatches to the seed segment (positions 1-6) was almost completely impaired (Figure 1E and Table S2), while mismatches in the PAM-distal region of the DNA target were mostly tolerated. The structure of the FnCas12a-crRNA complex further reveals that the bases of the seed sequence are solvent-exposed and poised for hybridization with target DNA (Figure S3E). To corroborate this observation, we performed an electrophoretic mobility shift assay (EMSA) using 8-nucleotide single-stranded DNAs complementary to different segments of a crRNA complexed with a FnCas12a protein (FnCas12aDM) containing mutations in the RuvC (E1006Q) and Nuc (R1218A) domains that impair DNA cleavage (Figure S5). An oligonucleotide complementary to crRNA nucleotides 1–8 was bound by the FnCas12aDM-crRNA complex, while oligonucleotides complementary to the non-seed segment of the crRNA or only partially overlapping with the seed sequence did not bind. These results show that the seed sequence in the FnCas12a-crRNA complex is accessible for DNA binding. Collectively, these results demonstrate that FnCas12a pre-orders the crRNA seed sequence independent of PAM binding and indicate that pre-ordering of the seed sequence enhances base-pairing with the target DNA strand.
Figure 1. Structure of FnCas12a in complex with crRNA guide.
(A) Sequence of the crRNA guide (see also Table S1). Nucleotides that are ordered in the structure are colored orange, unordered nucleotides are colored grey.
(B) Schematic diagram of the domain organization of FnCas12a. REC: Recognition. PI: Protospacer Adjacent Motif (PAM) interacting. WED: Wedge. BH: Bridge Helix. Nuc: Nuclease. Both the WED and RuvC domains are formed by three discontinuous segments of the protein sequence. For the genomic context and purification details of FnCas12a, see Figure S1.
(C) Overall structure of the FnCas12a-crRNA complex. Domains are colored according to the scheme in panel B. For alignments of the structures of the binary complexes of FnCas12a and LbCas12a, see Figure S2.
(D) Close-up view of the crRNA seed sequence. Hydrogen bonds between FnCas12a residues and the seed sequence are displayed as dashed lines. For details of FnCas12a-crRNA binding interactions, see Figure S3. For a sequence alignment of Cas12a orthologs in which the conserved residues involved in crRNA seed sequence pre-ordering are indicated, see Figure S4.
(E) Target DNA complementarity to the crRNA seed sequence is critical for cleavage. Top: Schematic representation of the crRNA (orange) and target DNA sequences used in the nuclease activity assays (see also Table S1 and Table S2). Numbering indicates the position of the mismatches between the crRNA and DNA targets used in the nuclease activity assays. Bottom: FnCas12a-crRNA complexes were incubated with pre-linearized plasmid DNA substrates containing a wild type (WT) target sequence or sequences with mismatches in the target site. Cleavage products were resolved by agarose gel electrophoresis. LIN: linearized plasmid substrate. M: 1kb DNA Ladder. For EMSA binding assays demonstrating the importance of the crRNA seed sequence in target binding, see Figure S5.
Table 1. Crystallographic data collection and refinement statistics.
| Dataset | Binary complex | Ternary complex |
|---|---|---|
| FnCas12a | WT | E1006Q/R1218A |
| RNA | crRNA1 | pre-crRNAX |
| DNA | — | oDS142 (TS) + oDS141 (NTS) |
| X-ray source | SLS PXIII | SLS PXIII |
| Space group | P21212 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 214.7, 277.1, 135.6 | 82.4, 143.7, 84.6 |
| a, b, g (°) | 90.0, 90.0, 90.0 | 90.0, 94.2, 90.0 |
| Wavelength (Å) | 1.00768 | 1.00768 |
| Resolution (Å)* | 169.70–3.34 (3.54–3.34) | 44.59–2.50 (2.65–2.50) |
| Rsym (%)* | 19.5 (117.9) | 6.2 (87.8) |
| CC1/2 (%) | 99.7 (59.5) | 99.9 (58.9) |
| I/sI* | 10.9 (1.73) | 14.6 (1.49) |
| Completeness (%)* | 99.7 (98.1) | 98.5 (96.5) |
| Redundancy* | 7.0 (7.0) | 3.5 (3.5) |
| Refinement | ||
| Resolution (Å) | 49.24–3.34 | 44.59–2.50 |
| No. reflections | 117207 | 131823 |
| Rwork / Rfree | 0.233 / 0.255 | 0.209 / 0.236 |
| No. atoms | ||
| Protein | 41724 | 10380 |
| Nucleic acid | 2028 | 2095 |
| Ion/ligand | 8 | 48 |
| Water | 45 | 41 |
| B-factors | ||
| mean | 82.4 | 70.4 |
| Protein | 83.1 | 69.9 |
| Nucleic acid | 67.8 | 72.5 |
| Ion/ligand | 65.7 | 70.8 |
| Water | 80.7 | 47.9 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.011 | 0.014 |
| Bond angles (°) | 0.73 | 0.82 |
| Ramachandran plot | ||
| % favoured | 98.2 | 97.5 |
| % allowed | 1.8 | 2.5 |
| % outliers | 0.0 | 0.0 |
| Molprobity | ||
| Clashscore | 10.1 | 13.8 |
Values in parentheses denote highest resolution shell
Pre-crRNA processing proceeds via a metal-independent mechanism
In contrast to other CRISPR-Cas systems that rely on standalone ribonucleases for pre-crRNA processing (Charpentier et al., 2015), Cas12a proteins harbor an intrinsic endoribonuclease activity that enables it to cleave pre-crRNA directly upstream of the crRNA pseudoknot (Fonfara et al., 2016; Zetsche et al., 2016). Biochemical studies have indicated that divalent cations are required for pre-crRNA processing by FnCas12a (Fonfara et al., 2016). Although Mg-dependence for pre-crRNA processing has not been established for LbCas12a, it displays ~50-fold higher affinity to crRNA in the presence of magnesium (Dong et al., 2016). Corroborating this observation, a single hydrated magnesium ion is coordinated by the crRNA pseudoknot in the LbCas12a-crRNA structure (Dong et al., 2016), whereas two hydrated magnesium ions are observed in the FnCas12a-crRNA complex (Figure S3). We therefore hypothesized that the previously observed inhibition of pre-crRNA processing by FnCas12a in the absence of divalent cations was due to impaired substrate binding and is not indicative of a divalent cation-dependent catalytic mechanism.
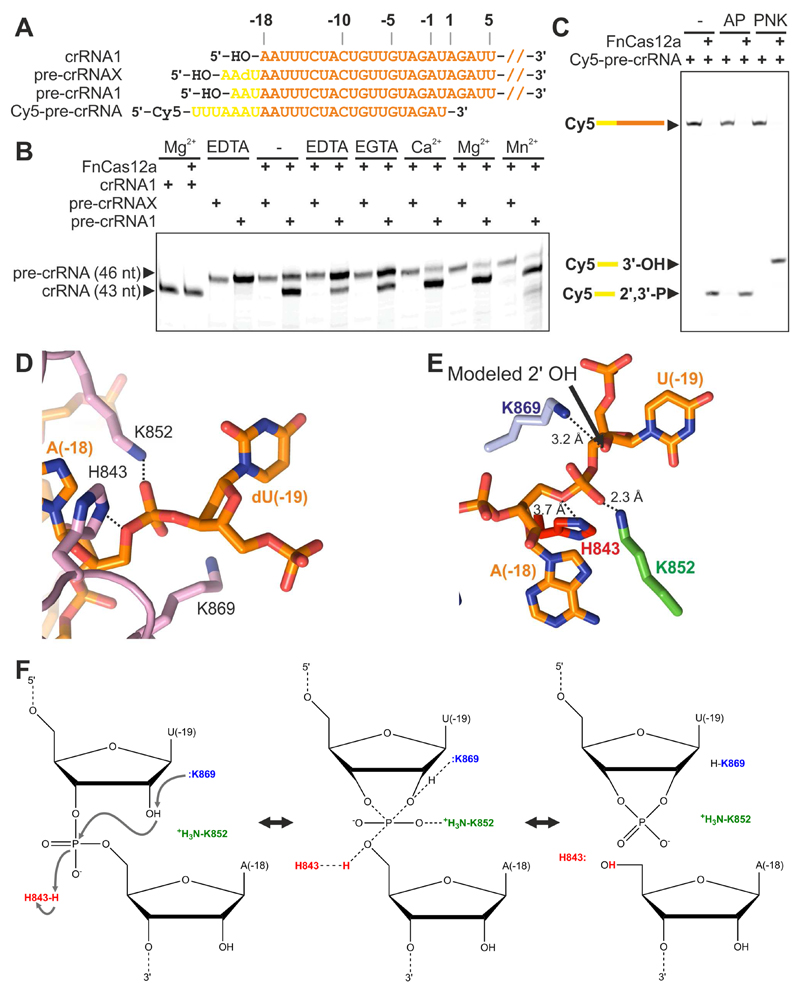
To test this hypothesis, we performed pre-crRNA processing assays at substrate concentrations exceeding the reported dissociation constant of the LbCas12a-crRNA complex, to ensure enzyme saturation by the substrate (Figure 2). To test whether the 2’-hydroxyl group directly upstream of the processing site plays a role in the cleavage mechanism, the assays were performed with an all-ribose pre-crRNA substrate (pre-crRNA1) or a pre-crRNA mimic (pre-crRNAX) containing a 2’-deoxynucleotide directly upstream of the scissile phosphate. Both pre-crRNAs were incubated with FnCas12a in the presence of divalent cations (Ca2+, Mg2+, or Mn2+) or chelating agents (EDTA or EGTA). Pre-crRNA1 was processed both in the presence of divalent cations and in the presence of chelating agents (Figure 2B), indicating that pre-crRNA processing is not strictly divalent cation-dependent but more efficient in presence of cations. In light of earlier observations (Dong et al., 2016; Fonfara et al., 2016), this suggests that cations stimulate pre-crRNA processing, either by increasing the binding affinity of FnCas12a to the pre-crRNA substrate, or by ordering the crRNA pseudoknot. In contrast, pre-crRNAX was not processed under any of these conditions (Figure 2B), demonstrating that the 2’-hydroxyl group of nucleotide U(-19) is essential for pre-crRNA processing. RNA cleavage reactions dependent on the presence of the 2’-hydroxyl group in the ribonucleotide upstream of the scissile phosphate typically proceed via acid-base (rather than metal-dependent) catalysis and yield products containing 5’-hydroxyl and 2’,3’-cyclic phosphate groups. To test whether Cas12a processes its crRNA guide by such mechanism, we investigated the fate of the scissile phosphate using a truncated pre-crRNA substrate mimic containing a 5’-end fluorophore. The FnCas12a-generated 5’ cleavage product was treated with Alkaline Phosphatase (AP), which specifically removes 3’ (or 2’) phosphate groups, but not 2’,3’-cyclic phosphates (Morse and Bass, 1997), or T4 Polynucleotide Kinase (PNK), which can remove 2’,3’-cyclic phosphates (Das and Shuman, 2013). Treatment with PNK resulted in a reduction in mobility of the 5’ cleavage product, while treatment with AP had no effect, indicating that the 5’ cleavage product contains a 2’,3’-cyclic phosphate group (Figure 2C). Combined, these results demonstrate that pre-crRNA processing by FnCas12a proceeds through a metal-independent catalytic mechanism involving a nucleophilic attack on the scissile phosphate by the 2’-hydroxyl of the upstream ribonucleotide.
Figure 2. Pre-crRNA processing by FnCas12a.
(A) Schematic representation of pre-crRNA substrates. Pre-crRNAX contains a deoxyuridine nucleotide at position (-19). Cy5-pre-crRNA is covalently labeled with a Cy5 fluorophore at the 5’ end of a truncated pre-crRNA sequence.
(B) FnCas12a-mediated pre-crRNA processing depends on the presence of a ribose 2'-hydroxyl group in the nucleotide upstream of the scissile phosphate. FnCas12a was incubated with pre-cRNA1 or pre-crRNAX in the presence of chelating agents or divalent cations. Processing products were analyzed by denaturing polyacrylamide gel electrophoresis.
(C) FnCas12a-mediated pre-crRNA processing generates a 5' product containing a 2',3'-cyclic phosphate. FnCas12a was incubated with Cy5-pre-crRNA, after which processing products were treated with Alkaline Phosphatase (AP; removes 3’-end phosphates) or T4 Polynucleotide Kinase (PNK; removes 2’,3’-cyclic phosphates). Reaction products were analyzed by denaturing polyacrylamide gel electrophoresis and fluorescence detection.
(D) Close-up view of the pre-crRNA processing site in FnCas12a bound to pre-crRNAX. For a sequence alignment of Cas12a orthologs in which the conserved residues involved in pre-crRNA processing are indicated, see Figure S4.
(E) Structural model of bona fide pre-crRNA substrate in which nucleotide U(-19) contains a (modeled) 2’-hydroxyl group, based on panel D. The 2’-hydroxyl group is positioned in close proximity of Lys869, suggesting that Lys869 acts as a general base in the catalytic mechanism.
(F) Proposed model for the acid-base catalytic mechanism of pre-crRNA processing by FnCas12a.
Structural basis of pre-crRNA processing
To obtain structural insights into the mechanisms of pre-crRNA processing and R-loop formation, we reconstituted a ternary complex containing FnCas12a lacking DNA endonuclease activity (FnCas12aDM; double mutant E1006Q/R1218A), pre-crRNAX (Figure 2A), and a substrate DNA comprising full-length target and non-target strands (henceforth referred to as the FnCas12a R-loop complex). To facilitate R-loop formation in the substrate DNA, the target and non-target DNA strands were mismatched at positions 1–20. We succeeded in crystallizing the resulting complex and determined its structure at 2.5 Å resolution (Table 1). In the R-loop complex structure, pre-crRNAX nucleotide dU(-19) is ordered (Figure 2D). The scissile phosphate connecting residues A(-18) and dU(-19) is surrounded by the side chains of His843, Lys852 and Lys869, which are strictly conserved in Cas12a orthologs (Figure S4) and essential for crRNA processing (Fonfara et al., 2016). The 2’-deoxyribose moiety of dU(-19) is distorted and adopts a C2’-endo geometry. Modeling a ribose moiety in its place results in a geometry that would favor an in-line nucleophilic attack by the 2’ hydroxyl on the scissile phosphate (Figure 2E). Lys869 is located ~4 Å away from the C2’ atom of dU(-19), which suggests that it would be positioned within hydrogen-bonding distance of the 2’-hydroxyl group of U(-19) in the pre-crRNA substrate. The π-amino group of Lys852 is hydrogen-bonded to the pro-Sp non-bridging oxygen atom of the scissile phosphate, while the side chain of His843 is in close proximity to the 5’ oxygen of A18. Notably, the two hydrated Mg2+ ions bound to the RNA are located ~6 and ~20 Å away from the scissile phosphate, suggesting that they do not directly participate in catalysis. These structural observations, together with biochemical experiments are consistent with an acid-base catalytic mechanism, in which Lys869 acts as the general base catalyst to deprotonate the attacking 2’ hydroxyl group of U(-19), while His843 acts as a general acid to protonate the 5’-oxygen leaving group of A(-18) (Figure 2F). In turn, the side chain of Lys852 is involved in charge stabilization of the transition state. Collectively, these interactions facilitate the intramolecular attack of the 2’-hydroxyl group of U(-19) on the scissile phosphate and promote the formation of the 2’,3’-cyclic phosphate product.
Mechanism of R-loop formation in Cas12a
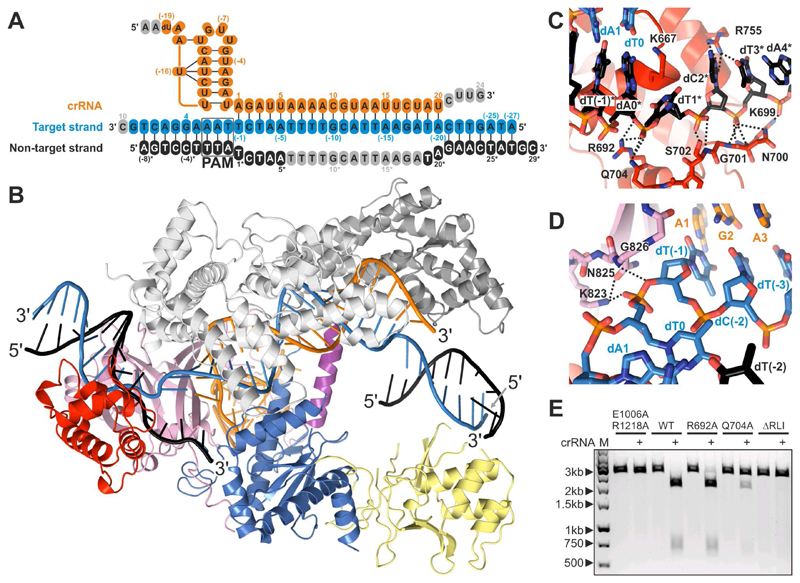
The FnCas12a R-loop complex structure reveals the conformational rearrangements occurring in FnCas12a upon target DNA binding as well as the mechanism of R-loop formation (Figure 3, Figure S6, and Figure S7). Crucially, the structure of the R-loop bound complex reveals that the DNA target and non-target strands remain base-paired to each other in both flanking regions of the R-loop. The crRNA and the target DNA strand form a 20-bp heteroduplex comprising crRNA nucleotides A1–U20 and target strand nucleotides dT(-1)–dA(-20), while the non-target DNA strand is unpaired in the region spanning dT1*–dA20*, with nucleotides dT6*–dT19* (including the scissile phosphate) lacking ordered electron density (Figure 3A and 3B). The crRNA-target DNA strand heteroduplex is enclosed in a central cavity formed by the REC and NUC lobes, and interacts extensively with the REC1 and REC2 domains (Figure S7). Consistently, mismatches in the crRNA-target DNA heteroduplex at positions 1-8, 10-14, and 17, which are directly contacted by FnCas12a residues (Figure S7), resulted in reduced cleavage activity (Figure 1E). The R-loop structure also reveals that nucleotides 21-24 of the crRNA do not contribute to target DNA binding. This is consistent with previous studies demonstrating that 3’-terminal truncations of the crRNA do not impair FnCas12a-mediated DNA cleavage or genome editing (Kleinstiver et al., 2016; Zetsche et al., 2015). Corroborating this, FnCas12a-catalyzed cleavage of a target DNA is not perturbed when nucleotides 21–24 of the crRNA are mismatched to the target DNA (Figure 1E).
Figure 3. Structure of FnCas12a-crRNA in complex with a dsDNA target.
(A) Sequences of the crRNA guide and DNA target (see also Table S1). Structurally disordered nucleotides are colored grey.
(B) Overall structure of the FnCas12a R-loop complex. The nucleic acid strands are colored as in panel A. For alignments of the structures of the ternary complexes of FnCas12a and AsCas12a, see Figure S2. For conformational rearrangements in FnCas12a upon transition from the binary to the ternary complex, see Figure S6. For details of FnCas12a-crRNA-DNA interactions, see Figure S7.
(C) Close-up view of the interactions with the non-target DNA strand at the PAM-proximal end of the R-loop.
(D) Close-up view of the interactions with the target DNA strand at the PAM-proximal end of the R-loop.
(E) Mutations of the FnCas12a residues interacting with the displaced non-target DNA strand impair substrate DNA cleavage. Wild-type and mutated FnCas12a was incubated with crRNA and a pre-linearized plasmid DNA substrate. Cleavage products were analyzed by agarose gel electrophoresis. LIN: linearized plasmid substrate. M: 1kb DNA Ladder. ΔRLI: Ala-Gly3 substitution of R-loop interacting residues Thr698–Ser702. 1 and 2: products of linear DNA cleavage.
The PAM-containing DNA duplex comprises target strand nucleotides dT0–dT8 and non-target strand nucleotides dA(-8)*–dA0* and is contacted by the PI, WED, and REC1 domains (Figure S7). As in AsCas12a, the 5’-TTN-3’ PAM is recognized in FnCas12a by a mechanism combining the shape-specific recognition of a narrowed minor groove, with base-specific recognition of the PAM bases by two invariant residues, Lys671 and Lys613 (Figure S7). Directly downstream of the PAM, the duplex of the target DNA is disrupted by the side chain of residue Lys667, which is inserted between the DNA strands and forms a cation-π stacking interaction with the dA0–dT0* base pair (Figure 3C). The phosphate group linking target strand residues dT(-1) and dT0 is coordinated by hydrogen-bonding interactions with the side chain of Lys823 and the backbone amide of Gly826. Target strand residue dT(-1) bends away from residue T0, allowing the target strand to interact with the seed sequence of the crRNA (Figure 3D). The non-target strand nucleotides dT1*–dT5* interact with the Arg692–Ser702 loop in FnCas12a through hydrogen-bonding and ionic interactions between backbone phosphate groups and side chains of Arg692, Asn700, Ser 702 and Gln704, as well as main chain amide groups of Lys699, Asn700 and Ser702 (Figure 3C). Alanine substitution of Q704 or replacing residues Thr698–Ser702 in FnCas12a with the sequence Ala-Gly3 substantially reduced DNA cleavage activity, suggesting that these residues contribute to R-loop formation by stabilizing the displaced conformation of the non-target DNA strand (Figure 3E).
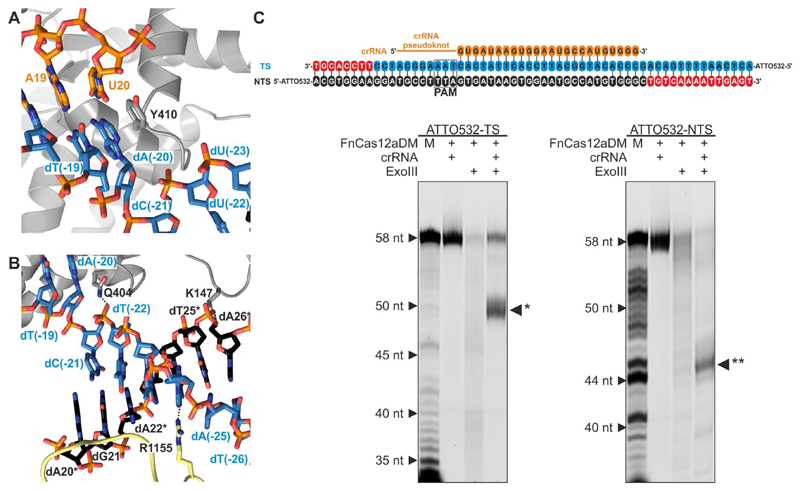
In the FnCas12a R-loop complex, the crRNA-target strand heteroduplex is terminated by a stacking interaction with a conserved aromatic residue (Tyr410, Figure 4A and Figure S4), as previously observed in the structures of DNA-bound AsCas12a complexes (Gao et al., 2016; Yamano et al., 2016). This prevents base pairing between the crRNA and the target strand beyond nucleotides U20 and dA(-20), respectively. Beyond this point, the target DNA strand nucleotides re-engage the non-target DNA strand, forming a PAM-distal DNA duplex comprising nucleotides dC(-21)-dA(-27) and dG21*-dT27*, respectively (Figure 4B). The duplex is confined between the REC2 and Nuc domains at the end of the central channel formed by the REC and NUC lobes. To validate these structural observations, we probed the extent of the R-loop structure by footprinting with the dsDNA-specific exonuclease III (ExoIII). Incubation of the R-loop complex with ExoIII resulted in the formation of well-defined shorter fragments of both the target and non-target strands (Figure 4C), indicating FnCas12a protects approximately 8 bp of duplex DNA upstream of the PAM and 5 bp downstream of the RNA-DNA hybrid. These results corroborate our structural insights, confirming that a substantial part of the DNA downstream of the crRNA-bound target site is shielded by the enzyme in its central channel.
Figure 4. The PAM-distal DNA rehybridizes within the confines of FnCas12a.
(A) Close-up view of the PAM-distal end of the crRNA-target DNA heteroduplex. Tyr410 stacking caps the crRNA-target DNA heteroduplex at base pair U20-dA(-20) and prevents duplex formation beyond nucleotide U20 of the crRNA.
(B) Close-up view of the PAM-distal flank of the R-loop. The PAM-distal segments of the target and non-target DNA strands hybridize in the central cavity of FnCas12a, between the REC2 and Nuc domains.
(C) Exonuclease III footprinting analysis of the FnCas12a–R-loop complex. Top: schematic representation of the crRNA sequence (orange) and target DNA sequences used in the footprinting assay (see also Table S1). FnCas12aDM-crRNA was bound to target DNA duplexes in which either the target strand (TS) or the non-target strand (NTS) was 5’-labeled with ATTO532. The resulting complexes were treated with Exonuclease III (ExoIII) and cleavage products were analyzed by denaturing polyacrylamide gel electrophoresis and fluorescence detection. M: Marker generated by S1 treatment of the labeled DNA strand. * and ** indicate the TS and NTS products formed by ExoIII when the dsDNA is protected by the FnCas12aDM-crRNA1 complex. Nucleotides removed from the TS and from the NTS by ExoIII are colored red in the schematic diagram at the top of the panel.
Implications for target DNA cleavage
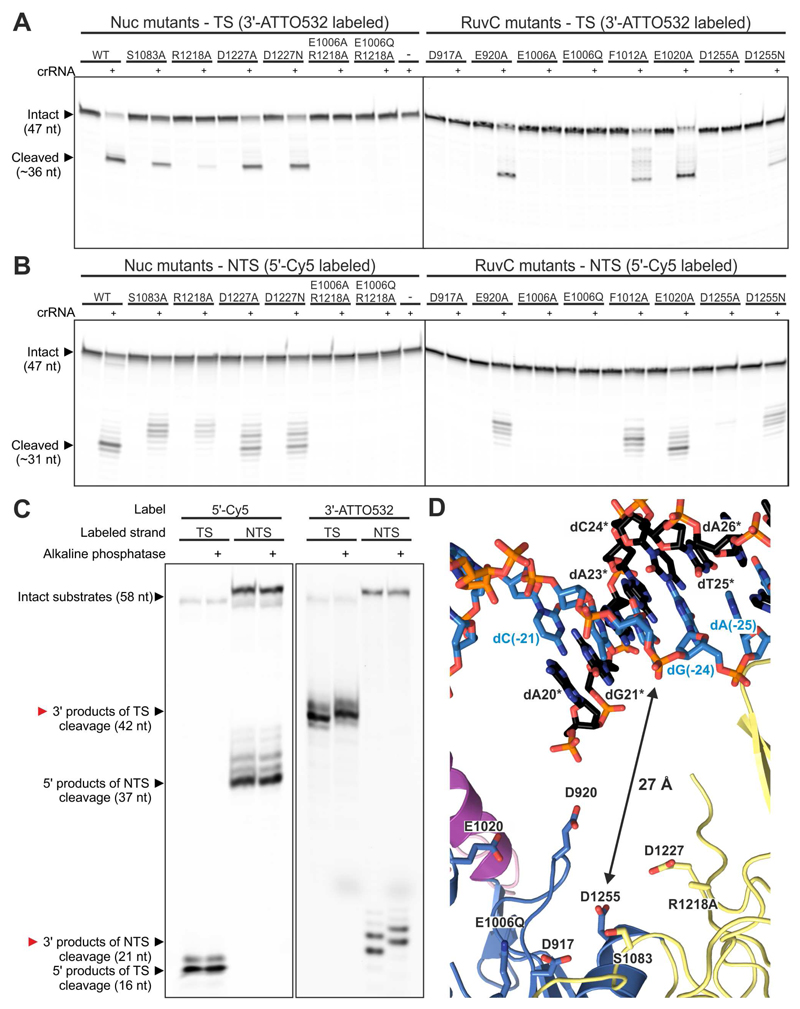
Previous biochemical studies of Cas12a showed that mutations in the RuvC domain impair cleavage of both strands in a target DNA duplex, while a mutation in the neighboring Nuc domain impaired target strand cleavage only (Yamano et al., 2016). This led to the suggestion that Cas12a contains two juxtaposed DNA nuclease active sites located at the interface of the RuvC and Nuc domains, and that cleavage of the non-target DNA strand by the RuvC domain is prerequisite for target strand cleavage by the Nuc domain. A recent structural study of Alicyclobacillus acidoterrestritis Cas12b (AacCas12b, previously known as AacC2c1), which is distantly related to Cas12a, however uncovered that the enzyme can independently accommodate both the target and non-target DNA strand in the catalytic pocket of the RuvC domain, implying that both strands are cleaved by the RuvC domain, while the Nuc contributes to DNA binding domain but lacks catalytic activity (Yang et al., 2016). Although the Nuc domains of Cas12a and Cas12b have a different fold, we hypothesized that Cas12a also cleaves both the target and non-target DNA strands using a single active site.
To verify this hypothesis, we performed extensive mutational analysis of the putative active site in FnCas12a (Figure 5A and 5B). Consistent with previous studies, alanine substitutions of the residues predicted to coordinate Mg2+ ions in the RuvC active site (D917A, E1006A and D1255A) impaired cleavage of both DNA strands. Based on similarities with other RNaseH-superfamily nucleases (Nowotny et al., 2005), substitution of Glu1006 with glutamine is predicted to impair substrate hydrolysis while preserving magnesium coordination in the RuvC active site and hence substrate DNA binding. The complete loss of target strand DNA cleavage observed for the E1006Q mutant (Figure 5B) suggests that the RuvC domain is not simply required for proper coordination of target strand, but that instead it directly catalyzes target strand cleavage. In agreement with this hypothesis, none of the mutations in the Nuc domain resulted in complete loss of cleavage of either DNA strand. Alanine substitution of the strictly conserved Arg1218 had the strongest effect, but affected both target and non-target strand cleavage equally. This is contrary to previous data suggesting that this residue is involved in target strand cleavage only (Yamano et al., 2016). We additionally analyzed the cleavage products of DNA cleavage by FnCas12a and determined that the cleavage of both DNA strands generates products containing 5’-phosphate and 3’-hydroxyl groups (Figure 5C). Together, these observations strongly suggest that both the target and non-target DNA strands are cleaved by the same catalytic mechanism in a single active site in Cas12a enzymes.
Figure 5. FnCas12a cleaves the target and non-target DNA strands with the same catalytic site.
(A) Target strand DNA cleavage is inhibited by specific mutations in the RuvC domain (right panel), but not by any of the mutations in the Nuc domain (left panel). Wild-type and mutated FnCas12a were incubated with crRNA and a dsDNA oligonucleotide substrate in which the target strand was labeled with 3’-ATTO532. Cleavage products were analyzed by denaturing polyacrylamide gel electrophoresis and fluorescence detection.
(B) Non-target strand DNA cleavage is completely inhibited by specific mutations in the RuvC domain (right panel), but not by any of the mutations in the Nuc domain (left panel). Wild-type and mutated FnCas12a were incubated with crRNA and a dsDNA oligonucleotide substrate in which the non-target strand was labeled with 5’-Cy5. Cleavage products were analyzed as in (A).
(C) Cleavage of both target and non-target DNA strands generates products containing a 5’-phosphate and 3’-hydroxyl groups. dsDNA oligonucleotide substrates, labeled as indicated with covalently attached fluorophores, were cleaved with the FnCas12a-crRNA complex. Cleavage products were further incubated in the presence or absence of alkaline phosphatase and analyzed by denaturing polyacrylamide gel electrophoresis and fluorescence detection.
(D) Close-up view of the RuvC and Nuc domain residues involved in DNA cleavage and the PAM-distal part of the R-loop. Arrow indicates the displacement of the scissile phosphate in the target DNA strand from the RuvC domain active site. For a modeled non-target strand in the RuvC catalytic site of FnCas12a, see Figure S8.
The structure of the FnCas12a R-loop complex provides clues about the putative mechanism of DNA cleavage. The scissile phosphate in the target strand is located within the PAM-distal DNA duplex of the R-loop structure, approximately 27 Å away from the RuvC catalytic triad (Asp917, Glu1006, Asp1255) (Figure 5D). Although the cleavage site is located in the disordered part of the non-target strand, the phosphate group of dA20* is located in the vicinity of the RuvC domain and projects toward the RuvC active cleft. Together, these observations suggest that the R-loop complex structure represents a pre-cleavage state in which the non-target DNA strand is poised to enter the active site at the RuvC-Nuc domain interface. To delineate the path of the non-target strand, we modeled the binding of a DNA oligonucleotide in the FnCas12a RuvC active cleft based on structural superposition of the R-loop complex with the structure of AacCas12b containing a fragment of the non-target DNA strand in the RuvC cleft (Figure S8, PDB accession code: 5U33). This places the 3’-terminal nucleotide of the modeled DNA fragment (corresponding to nucleotide dT19* immediately downstream of the scissile phosphate) and the 5’-terminal nucleotide (dA20*) in the PAM-distal part of the non-target strand in close proximity. However, this distance (~13 Å) is considerably larger than what can be spanned by a single phosphodiester linkage. We postulate that conformational rearrangements within the REC and NUC lobes of Cas12a, possibly in combination with additional repositioning of the PAM-distal DNA duplex, facilitate binding of the non-target DNA strand in the active cleft. In one of the structures of AacCas12b (PDB accession code: 5U30), the target strand adopts a sharply bent conformation which allows it to bind the RuvC catalytic site with the same 5’-3’ polarity as the non-target DNA strand (Yang et al., 2016). Consistent with this, we observe that target strand cleavage by FnCas12a yields a 5’-phosphorylated cleavage product, pointing to a similar mechanism of target strand binding and cleavage by the RuvC active site in FnCas12a. We speculate that this process involves further unwinding of the PAM-distal DNA duplex beyond the target strand nucleotide 24, since the target strand is expected bind to the RuvC active site in a single-stranded form.
Discussion
By visualizing the structures of Cas12a complexes, our studies shed light on the dual catalytic activities of Cas12a responsible for crRNA processing and for crRNA-guided DNA targeting and cleavage. These insights not only advance our understanding of the function of Cas12a in type V CRISPR-Cas systems, but also provide a framework for further development of Cas12a-based genome editing tools and applications. FnCas12a has been used for genome editing in plants (Endo et al., 2016). Moreover, given the high degree of both sequence and structural homology between FnCas12a and LbCas12a (43% sequence identity) and AsCas12a (37%) (Figure S4), the Cas12a orthologs that function efficiently in mammalian cells (Zetsche et al., 2015), mechanistic insights derived from our work are directly applicable to these enzymes. The structure of FnCas12a bound to a processed crRNA guide reveals that nucleotides 1-5 of the guide segment are pre-ordered in a helical conformation and poised to mediate target DNA recognition. In line with the results of biochemical experiments described in this and previous studies (Fonfara et al., 2016; Zetsche et al., 2015), the structure confirms that Cas12a proteins utilize a seed sequence for target binding similar to other RNA-guided nucleases such as Cas9 and Argonaute proteins.
Cas12a is one of only three RNA-guided nucleases (the other being Cas13a and Cas13b, both of which display RNA-guided RNase activity) that have been demonstrated to process their own guide RNAs from precursor transcripts (East-Seletsky et al., 2016; Smargon et al., 2017). This property is an integral part of its functional mechanism in vivo and has been exploited in multiplexed genome editing (Zetsche et al., 2016). Our results further demonstrate that FnCas12a catalyzes the processing of its own crRNAs by an divalent cation-independent catalytic mechanism, contravening earlier studies (Fonfara et al., 2016). Consistent with data showing that the affinity of LbCas12a for its cognate crRNA is increased in the presence of Mg2+ (Dong et al., 2016), we observe two hydrated Mg2+ ions bound to the crRNA pseudoknot in the FnCas12a structure. We therefore conclude that divalent cations are required for efficient crRNA binding rather than for the crRNA processing mechanism per se.
The structure of FnCas12a bound to a guide RNA precursor and a target DNA pinpoints specific FnCas12a residues involved in facilitating strand separation in the duplex DNA target and R-loop formation. In Cas9, local melting of the target DNA in the proximity of the PAM is followed by directional RNA-DNA hybridization (Anders et al., 2014; Jiang et al., 2016; 2015). The mechanism of PAM-dependent R-loop formation in Cas12a is likely to be analogous, although it has not been examined in detail. Building on recent structural studies of the distantly related Cas12b (C2c1) enzyme (Yang et al., 2016), our structural and biochemical insights provide compelling evidence that Cas12a enzymes contain a single deoxyribonuclease catalytic site responsible for the alternative cleavage of both the target and non-target strands in a duplex DNA substrate.
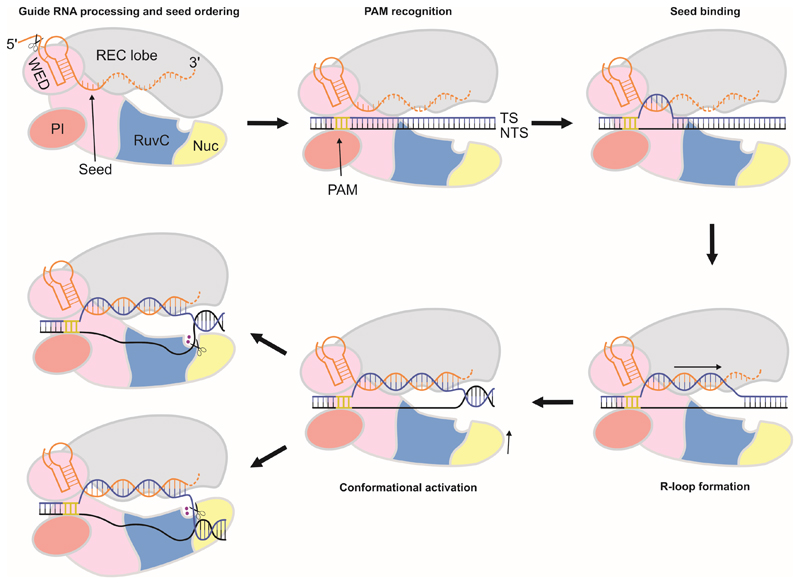
Based on our work, we propose a model for crRNA-mediated target binding and cleavage by Cas12a proteins (Figure 6). Upon crRNA binding and processing, Cas12a adopts a conformation in which it is primed for target DNA recognition, with the seed sequence of the bound crRNA pre-ordered to mediate base-pairing with the target DNA strand. Notably, the RuvC active site is blocked by the REC lobe in the FnCas12a–crRNA complex, implying that the enzyme is unable to catalyze DNA cleavage in the absence of a cognate DNA target. Cas12a subsequently recognizes the 5’-TTN-3’ PAM in a target DNA (Yamano et al., 2016), positioning the target DNA sequence in register with the crRNA and facilitating localized strand separation of the DNA in the proximity of the PAM. This allows the target DNA strand to be captured by the pre-ordered seed sequence of the crRNA, leading to directional DNA strand separation and crRNA-target DNA hybridization. This is accompanied by a conformational rearrangement of the REC lobe, which results in the opening of the central cleft of the enzyme to accommodate the RNA-DNA heteroduplex and exposes the RuvC catalytic site. A conserved aromatic residue in the REC lobe caps the crRNA-target DNA heteroduplex, limiting its length to twenty base pairs. As a result, the scissile phosphate groups in the target and non-target DNA strands are located within the ssDNA-dsDNA junction at the PAM-distal end of the R-loop, and are oriented toward the NUC lobe. We posit that the conformational dynamics of Cas12a allows the R-loop complex to sample two alternative conformations that lead to the cleavage of either the target or non-target DNA strand by a single active site located at the interface of the RuvC and Nuc domains. Whether the two cleavage events are independent of each other or occur in a defined sequential order is currently unclear. The precise mechanism of target DNA cleavage by Cas12a thus awaits further investigation. In conclusion, our studies provide critical insights into the molecular function of type V CRISPR effector nucleases and set the stage for their further exploitation in genome editing.
Figure 6. Schematic model of crRNA-guided target DNA binding and cleavage by Cas12a enzymes.
Cas12a catalyzes the processing of its own crRNA guide and pre-orders crRNA nucleotides 1-5 in the seed sequence. PAM recognition by a preformed binding site in Cas12a positions the target DNA in register with the pre-ordered crRNA seed sequence. Local strand separation in the vicinity of the PAM allows the target strand to base pair to the crRNA seed sequence to initiate R-loop formation. The R-loop is formed by directional guide-target hybridization. The RNA-DNA heteroduplex is terminated after 20 base pairs at the PAM-distal end. The resulting R-loop structure can adopt two alternative conformations that result in the insertion of either the target or the non-target DNA strand in a single catalytic site located at the interface of the RuvC and Nuc domains.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
- EXPERIMENTAL MODEL AND SUBJECT DETAILS
-
○E. coli Rosetta (DE3)
-
○NEB® 5-alpha Competent E. coli (High Efficiency)
-
○Mach1™ Competent Cells (E. coli)
-
○
- METHOD DETAILS
-
○FnCas12a expression and purification
-
○Crystallization and structure determination
-
○Plasmid cleavage experiments
-
○EMSA experiments
-
○Pre-crRNA processing experiments
-
○Exonuclease III protection assay
-
○Oligonucleotide dsDNA cleavage experiment
-
○
DATA AND SOFTWARE AVAILABILITY
Star★Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Martin Jinek (jinek@bioc.uzh.ch).
Experimental Model and Subject Details
E. coli Rosetta (DE3)
For protein expression, E. coli Rosetta (DE3) was grown in LB at 37 °C in a shaker incubator at 120 rpm until an OD600 nm of 0.6 was reached after the temperature was switched to 20 °C. After 30 minutes, expression was induced by addition of isopropyl-1-thio-β-D-galactopyranoside (IPTG) to a final concentration of 0.1 mM. Expression took place at 20 °C for 14 hours.
NEB® 5-alpha Competent E. coli (High Efficiency)
For cloning, NEB® 5-alpha competent E. coli was transformed according to the manual provided by the manufacturer and grown on LB agar plates at 37 °C overnight.
Mach1™ Competent Cells (E. coli)
For cloning, Mach1™ Competent Cells were transformed according to the manual provided by the manufacturer and grown on LB agar plates at 37 °C overnight.
Method Details
FnCas12a expression and purification
FnCas12a was heterologously expressed in Escherichia coli from a codon-optimized synthetic gene and purified by a combination of Ni2+-affinity, ion exchange and gel filtration chromatographic steps. The gene sequence was synthesized and inserted into plasmid pML-1B (obtained from the UC Berkeley MacroLab, Addgene #29653) by ligation-independent cloning using oligonucleotides oDS027 and oDS028 (Table S1) to generate a protein expression construct encoding the FnCas12a polypeptide sequence (residues 1-1300) fused with an N-terminal tag comprising a hexahistidine sequence, a maltose binding protein (MBP) and a Tobacco Etch Virus (TEV) protease cleavage site (Table S2). To express catalytically inactive FnCas12a protein (FnCas12aDM), the E1006Q and R1218A point mutations were inserted by QuikChange site-directed mutagenesis and verified by DNA sequencing. The proteins were expressed in E. coli Rosetta™ 2 (DE3) strain. Cultures were grown to an OD600 nm of 0.5-0.6; expression was induced by the addition of IPTG to a final concentration of 0.2 mM and incubation was continued at 18 °C overnight. Cells were harvested by centrifugation and the cell pellet was resuspended in 50 ml Buffer I (20 mM Tris-HCl pH8, 500 mM NaCl, 5mM imidazole, supplemented with protease inhibitors AEBSF at 10 mg/ml and Pepstatin A at 0.1 mg/ml). Cells were lysed by sonication and the lysates was centrifuged for 45 min at 4 °C at 30,000x g to remove insoluble material. The clarified lysate was applied to a 5 ml Ni-NTA Superflow (Qiagen) column. The column was washed with 10 column volumes of Buffer II (20 mM Tris/HCl pH 8, 250 mM NaCl, 15 mM Imidazole) and bound protein was eluted in Buffer III (20 mM Tris/HCl pH 8, 250 mM NaCl, 250 mM Imidazole). Fractions containing FnCas12a were pooled and TEV protease was added in a 1:100 (w/w) ratio. The sample was dialyzed against Buffer IV (10 mM HEPES-KOH pH 7.5, 250 mM KCl) at 4 °C overnight. For further purification the protein was diluted 1:1 with 20 mM HEPES KOH (pH 7.5) and loaded on a HiTrap Heparin HP column (GE Healthcare). The column was washed with IEX Buffer A (20 mM HEPES-KOH pH 7.5, 100 mM KCl) and eluted with IEX Buffer B (20 mM HEPES-KOH pH 7.5, 2 M KCl) by applying a gradient from 0% to 50% over a total volume of 60 ml. Peak fractions were analyzed by SDS-PAGE and fractions containing FnCas12a were combined, and DTT was added to a final concentration of 1 mM. The protein was fractionated on a HiLoad 16/600 Superdex 200 gel filtration column (GE Healthcare), eluting with SEC buffer (20mM HEPES-KOH pH 7.5, 500mM KCl, 1mM DTT). Peak fractions were combined, concentrated to 10 mg/ml, flash frozen in liquid nitrogen and stored at -80 °C until further use.
Point mutations were inserted in the FnCas12a coding sequence by QuikChange site-directed mutagenesis and verified by DNA sequencing. For DNA cleavage assays, FnCas12a and its mutants were heterologously expressed in Escherichia coli Rosetta 2 (DE3) as described above. Cells were harvested by centrifugation and the cell pellet was resuspended in 20 ml Buffer I. Cells were lysed by sonication and the lysate was centrifuged for 45 min at 4 °C at 30,000 x g to remove insoluble material. The clarified lysate was applied to 500 μl Ni-NTA Superflow (Qiagen) resin. The column was washed with 2x 10 column volumes of Buffer II (20 mM Tris/HCl pH 8, 250 mM NaCl, 15 mM Imidazole) and bound protein was eluted in 1 ml Buffer III (20 mM Tris/HCl pH 8, 250 mM NaCl, 250 mM Imidazole). Purified protein was diluted to 5 μM in Buffer III.
Crystallization and structure determination
The binary FnCas12a-crRNA complex was reconstituted by combining purified apo-FnCas12a at a concentration of 10 mg/ml with synthetic crRNA1 (obtained from Integrated DNA Technologies, Table S1) in a 1:1.2 molar ratio (FnCas12a:crRNA1) and incubating for 10 min at RT. The complex was crystallized at 20 °C using the hanging drop vapor diffusion method by mixing equal volumes of protein and reservoir solution. Initial crystals were obtained at a complex concentration of 10 mg/ml from 50 mM HEPES-NaOH pH 6.5, 20% Polyethylene glycol 3,350, 1% (w/v) Tryptone and 1 mM Sodium azide. Crystal growth was optimized by iterative microseeding. Data was collected from crystals grown obtained using a complex concentration of 9 mg/ml and reservoir solution containing 50 mM HEPES-NaOH, pH 6.5, 20% (w/v) Polyethylene glycol 3,400, 1.5% (w/v) Tryptone (Hampton Research), and 1 mM Sodium azide for 10 weeks at 20 °C. MgCl2 was added to the hanging drop to a final concentration of 1 mM. For cryoprotection, crystals were transferred to cryoprotectant A (50mM HEPES-NaOH pH 7, 20% (w/v) polyethylene glycol 3,400, 1% crystallization grade Tryptone (Hampton Research), 15% (v/v) glycerol) and flash-cooled in liquid nitrogen.
To reconstitute the FnCas12a-crRNAX-DNA ternary complex, purified Apo-FnCas12a at a concentration of 5 mg/ml was mixed with crRNAX (synthesized by Integrated DNA Technologies, Table S1) in a 1:1.4 molar ratio (FnCas12a:crRNA1) and incubated for 10 min at 37 °C. Target strand DNA oligonucleotide (oDS142, obtained from Sigma-Aldrich, Table S1) was added in a 1:1.6 molar ratio (FnCas12a:oDS142) and the sample was incubated for 10 min at 37 °C. Non-target strand DNA oligonucleotide (oDS141, Table S1) was subsequently added in a 1:1.8 molar ratio (FnCas12a:oDS142), followed by further incubation for 10 min at 37 °C. The ternary complex was crystallized at 20 °C using the hanging drop vapor diffusion method by mixing equal volumes of protein and reservoir solution. Initial crystallization hits were obtained at a protein concentration of 4.25 mg/ml in 0.1 M Bis-Tris propane (BTP, pH 6.5), 0.2 M KSCN, 20% (w/v) polyethylene glycol 3,350. Diffraction data was obtained using crystals grown in refinement screens with a final protein concentration of 3.3 mg/ml protein with reservoir solution containing 0.1M BTP pH 6.5, 0.1 M KSCN, and 12.5% (w/v) polyethylene glycol 3,400, for 5 weeks at 20 °C. Crystals were transferred to cryoprotectant solution B (0.1M BTP pH 6.5, 0.2M KSCN, 20% polyethylene glycol 3,400, 15% (v/v) ethylene glycol, and 5 mM MgCl2) and flash-cooled in liquid nitrogen.
X-ray diffraction data were measured at beamline X06DA (PXIII) of the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland). Data were indexed, integrated, and scaled using XDS. Crystals of the FnCas12a-crRNA binary complex diffracted to a resolution of 3.4 Å and belonged to space group P21212, with four copies of the complex in the asymmetric unit. The structure of the binary complex was solved by molecular replacement using Phaser (McCoy et al., 2007), as implemented within the CCP4 package. The search model was generated using the Phyre2 modeling server (2015) based on the binary LbCas12a complex (PDB accession code: 5ID6 (Dong et al., 2016)) and modified by removing long loops and truncating amino acid side chains. Phases obtained using the initial molecular replacement solution were improved by density modification using phenix.resolve (2004) and phenix.morph_model (2013). The atomic model was built manually in Coot (2010) and refined using phenix.refine (Afonine et al., 2012). The final binary complex model contains crRNA1 residues (-18)–(+5) and FnCas12a residues 1–1300, except for residues 333–339, 430–437, 962–967, 1134–1136 and 1154–1161, which lack ordered electron density.
Ternary complex crystals belonged to space group P21 (with one copy per asymmetric unit) and diffracted to a resolution of 2.5 Å. The structure was solved by molecular replacement in phenix.phaser, using the WED-PI-RuvC domains (residues 591-1300) and the REC1–2 domains (residues 24-591) of FnCas12a, as well as the crRNA pseudoknot (nucleotides (-18)–0) from the binary complex structure as separate search models. The atomic model was completed manually in Coot and refined using phenix.refine. The final ternary complex model contains crRNAX residues (-19)–20, target DNA strand nucleotides (-27)–10, and non-target strand nucleotides (-8*)–5* and 19*–29*, and FnCas12a residues 1–1300, except for disordered residues 133–135, 424–443, 1009–1018, 1157–1163, and 1223–1226.
Plasmid cleavage experiments
Target DNA sequence complementary to crRNA1 was generated by annealing complementary synthetic oligonucleotides (Table S1) and inserting the productinto pUC19 between at the EcoRI and HindIII restriction sites. Mutations in the target region were introduced by Quikchange Site Directed Mutagenesis (Table S1 and Table S2). Plasmids were linearized by incubating 640 ng of plasmid DNA with 15 units of SspI-HF (New England Biolabs) in CutSmart buffer (New England Biolabs) for 1 h at 37 °C, followed by heat-inactivation for 20 min at 65 °C.
For cleavage assays, FnCas12a (10 μM in SEC buffer) was mixed with crRNA1 (10 μM in H2O) in the presence of 7.1 mM MgCl2 and incubated at 37 °C for 10 minutes to allow binary complex formation, followed by the additional of linearized plasmid. The final 20 μl reaction contained final concentrations of 0.5x SEC buffer, 2.5 μM FnCas12a, 1 μM crRNA1, 5 mM MgCl2, 255 ng linearized plasmid DNA and 0.3x CutSmart buffer (New England Biolabs), and was incubated for 30 min at 37 °C. Reactions were stopped by adding EDTA and Proteinase K (Thermo Fisher Scientific) to final concentrations of 80 mM and 0.8 mg/ml, respectively, and incubating for 30 min at 37 °C after which 6X DNA loading dye (Thermo Fisher Scientific) was added. The DNA was resolved on 0.8% agarose gels stained with GelRed Nucleic Acid Gel Stain (Biotum) and visualized using a ChemiDoc Touch gel imager (Bio-Rad).
EMSA experiments
FnCas12aDM (10 μM SEC buffer), or only SEC buffer, was mixed with crRNA1 (10 μM in H2O) in the presence of 5 mM MgCl2 and incubated at 37 °C for 10 minutes to allow binary complex formation. Samples were serially diluted in reaction buffer (0.5x SEC buffer, 5 mM MgCl2) and 3’-Cy5 labeled single stranded DNA (oDS205-211, Table S1) was added to a final concentration of 100 nM. Binding reactions contained 0.5x SEC buffer, 5 μM-5 nM FnCas12a-crRNA complex (or the same concentrations of only RNA for negative controls), 5 mM MgCl2 and 100 nM Cy5-labeled ssDNA in a total volume of 20 μl. Samples were incubated for 1 h at 37 °C after which native loading dye (0.5X SEC buffer, 5 mM MgCl2, 50% glycerol) was added in a 4:1 ratio (sample:native loading dye). Samples were resolved at 4 °C on an 8% native polyacrylamide gel supplemented with 5 mM MgCl2 using 0.5X TBE running buffer supplemented with 5 mM MgCl2. Fluorescence of the Cy5-labeled ssDNA was detected using a Typhoon FLA 9500 gel imager (GE Healthcare).
Pre-crRNA processing experiments
For pre-crRNA processing experiments, FnCas12a was incubated with crRNA substrates (crRNA1, crRNAX, or pre-crRNA1, Table S1) and divalent cations as indicated. Processing reactions (total volume of 20 μl) contained 2.5 μM FnCas12a, 0.5x SEC buffer, 1 μM crRNA substrate and 5 mM chelating agent or divalent cation (EDTA, EGTA, CaCl2, MgCl2, MnCl2 or NiCl2) and incubated for 10 min at 37 °C. Reactions were stopped by adding EDTA and proteinase K to final concentrations of 80 mM and 0.8 mg/ml, respectively, and incubated for a further 30 min at 37 °C. Samples were mixed with equal volumes of 2x RNA loading dye and incubated for 10 min at 95 °C. Reactions were resolved on a 20% denaturing (7M Urea) polyacrylamide gel. Nucleic acids were stained with GelRed Nucleic Acid Gel Stain (diluted 1:15,000 in 05x TBE) for 30 min. Fluorescence was detected using a Typhoon FLA 9500 gel imager.
To determine the terminal groups of crRNA processing products, FnCas12a was mixed with Cy5-pre-crRNA (Table S1), and MgCl2 to final concentrations of 2.5 μM FnCas12a, 0.5x SEC buffer, 1 μM Cy5-pre-crRNA substrate and 5 mM MgCl2 in a total volume of 20 μl. Samples were incubated for 30 min at 37 °C. The samples were mixed with DEPC H2O, FastAP Thermosensitive Alkaline Phosphatase (1 U/μl; Thermo Fisher Scientific), or Polynucleotide Kinase (PNK; 10 U/μl; Thermo Fisher Scientific) in a 9:1 ratio (v/v), and incubated for 30 min at 37 °C. Reactions were terminated by adding EDTA and Proteinase K to final concentrations of 80 mM and 0.8 mg/ml, respectively and incubating for 30 min at 37 °C. Samples were mixed with equal volumes of 2x RNA loading dye, incubated for 10 min at 95 °C, and resolved on a 20% denaturing (7M Urea) polyacrylamide gel. Fluorescence of the Cy5-labeled 5’-product was detected using a Typhoon FLA 9500 gel imager.
Exonuclease III protection assay
10 μl FnCas12aDM (5μM in 0.5x SEC buffer) was mixed with 5 μl crRNA-λ (10 μM, Table S1) and 2 μl MgCl2 (50 mM). The mixture was incubated for 15 min at 37 °C to allow binary complex formation. 2 μl double stranded λ-DNA (0.5 μM; generated by mixing 10 μM λ-TS and λ-NTS (Table S1) in a 1:1 ratio, incubating for 5 min at 95 °C, slowly cooling down to room temperature and diluting to 0.5 μM in H2O) was added and the sample was incubated for 30 min at 37 °C to allow ternary complex formation (the molar ratio FnCas12a:crRNA:dsDNA was 50:50:1). 1 μl of Exonuclease III (100 units/μl; Thermo Scientific) was added and the sample was incubated for 1h at 37 °C. For the marker, λ-TS and λ-NTS were incubated with S1 nuclease (1:100 dilution; Thermo Scientific) for 1 min at 37 °C. The reactions were terminated by adding EDTA and Proteinase K to final concentrations of 80 mM and 0.8 mg/ml, respectively and incubating for 30 min at 37 °C. Samples were mixed with equal volumes of clear 2x RNA loading dye, incubated for 10 min at 95 °C, and resolved on an 11% denaturing (7M Urea) polyacrylamide gel. Fluorescence of the Cy5-labeled 5’-product was detected using a Typhoon FLA 9500 gel imager.
Oligonucleotide dsDNA cleavage experiment
Target strand oligonucleotides (100μM) and non-target strand oligonucleotides (100μM) were annealed by incubation at 95 °C for 5 minutes, followed by slow cooling to room temperature. The formed dsDNA (50μM) was diluted in water to 3.33 μM. For the FnCas12a mutant analysis, the ratio of labelled vs non-labelled strands was 1:1.5. For experiments investigating the fate of the scissile phosphate groups, the ratio of TS and NTS was 1:1.
For mutant analysis experiments, 10 μl FnCas12a (5μM in Buffer III) was mixed with 2 μl crRNA1 (10 μM, Table S1), 2 μl MgCl2 (50 mM), and 4 μl H2O. The sample was incubated at 37 °C for 10 minutes to allow complex formation, after which 2 μl dsDNA target (3.33 μM) was added. Samples were incubated for 10 min at room temperature. The reactions were terminated by adding EDTA and Proteinase K to final concentrations of 80 mM and 0.8 mg/ml, respectively and incubating for 30 min at 37 °C.
For the scissile phosphate fate experiments, 10 μl FnCas12a (5μM in 1 x SEC buffer) was mixed with 2 μl crRNA1 or crRNA-λ (10 μM, Table S1), 2 μl MgCl2 (50 mM) and 4 μl H2O. The sample was incubated at 37 °C for 15 minutes to allow complex formation, after which 2 μl dsDNA target (3.33 μM) was added. Samples were incubated for 60 min at 37 °C. The sample was heated for 15 min at 95 °C to promote FnCas12a denaturation and dissociation from the DNA. Samples were centrifuged for 30 minutes at 20.000 x g. 15 μl of the supernatant was mixed with 2 μl 10X FastAP buffer (ThermoFisher) and 3 μl Thermosensitive Alkaline Phosphatase (1 U/μl, ThermoFisher) or 3 μl H2O (negative controls) and incubated at 37 °C for 90 minutes. Thermosensitive Alkaline Phosphatase was heat-inactivated by heating at 75 °C for 10 minutes. Samples were mixed with equal volumes of clear 2x RNA loading dye, incubated for 10 min at 95 °C, and resolved on 14% denaturing (7M Urea) polyacrylamide gels. Fluorescence of the Cy5-labeled and ATTO532-labeled products was detected using a Typhoon FLA 9500 gel imager.
Data and Software Availability
The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank (PDB) under accession numbers XXX and XXX. The unprocessed image files used to prepare the figures in this manuscript are deposited in Mendeley Data and are available at http://dx.doi.org/10.17632/fwbdxjzvcs.1.
Supplementary Material
Supplemental Information includes eight figures and tables and can be found with this article online.
Highlights.
Crystal structures of binary (crRNA) and ternary (R-loop) Cas12a complexes
Cas12a pre-orders the seed sequence of the crRNA to facilitate target binding
crRNA is processed via acid-base catalysis, generating a 2’,3’-cyclic phosphate
Cleavage of target and non-target DNA strands is catalyzed by the same active site
eTOC Blurb.
Swarts et al. solved the structures of Cas12a bound to a guide RNA and in complex with a guide RNA and double stranded target DNA to reveal the mechanisms of guide RNA processing and R-loop formation, and provide insights into the mechanism of DNA cleavage.
Acknowledgments
We are grateful to Meitian Wang, Vincent Olieric and Takashi Tomizaki at the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) for assistance with X-ray diffraction measurements. We thank S. Chardia, P. Mohanraju, R. Pritchard and S. Halter for technical assistance. We thank M. Clerici for help with X-ray data processing and members of the Jinek group for discussions and critical reading of the manuscript. This work was supported by a Swiss National Science Foundation (SNSF) Project Grant to M.J. (SNSF 31003A_149393), by a Netherlands Organization for Scientific Research (NWO) CW-TOP grant (714.015.001) to J.v.d.O., and by a long-term postdoctoral fellowship from the European Molecular Biology Organization (EMBO) to D.C.S. (EMBO ALTF 179-2015).
Footnotes
Author Contributions
D.C.S., J.v.d.O. and M.J. designed experiments. D.C.S. prepared and crystallized the Cas12a complexes, collected X-ray data, determined crystal structures and carried out biochemical assays. M.J. assisted with X-ray structure determination and supervised the project. D.C.S. and M.J. wrote the manuscript, with input from J.v.d.O.
References
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier E, Marraffini LA. Harnessing CRISPR-Cas9 immunity for genetic engineering. Curr Opin Microbiol. 2014;19C:114–119. doi: 10.1016/j.mib.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier E, Richter H, van der Oost J, White MF. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev. 2015;39:428–441. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim J-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U, Shuman S. Mechanism of RNA 2“,3-”cyclic phosphate end healing by T4 polynucleotide kinase-phosphatase. Nucleic Acids Res. 2013;41:355–365. doi: 10.1093/nar/gks977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Ren K, Qiu X, Zheng J, Guo M, Guan X, Liu H, Li N, Zhang B, Yang D, et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016;532:522–526. doi: 10.1038/nature17944. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346 doi: 10.1126/science.1258096. 1258096. [DOI] [PubMed] [Google Scholar]
- East-Seletsky A, O'Connell MR, Knight SC, Burstein D, Cate JHD, Tjian R, Doudna JA. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn C-D, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A, Masafumi M, Kaya H, Toki S. Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci Rep. 2016;6 doi: 10.1038/srep38169. 38169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Yang H, Rajashankar KR, Huang Z, Patel DJ. Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 2016;26:901–913. doi: 10.1038/cr.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Fu BX, Hansen LL, Artiles KL, Nonet ML, Fire AZ. Landscape of target:guide homology effects on Cas9 mediated cleavage. Nucleic Acids Res. 2014;42:13778–13787. doi: 10.1093/nar/gku1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JK, Kim K, Been KW, Baek G, Ye S, Hur JW, Ryu S-M, Lee YS, Kim J-S. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat Biotechnol. 2016 doi: 10.1038/nbt.3596. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh J-RJ, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016 doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343 doi: 10.1126/science.1247997. 1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biology. 2013;10:841–851. doi: 10.4161/rna.24203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim J, Hur JK, Been KW, Yoon S-H, Kim J-S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016a;34:863–868. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- Kim Y, Cheong S-A, Lee JG, Lee S-W, Lee MS, Baek I-J, Sung YH. Generation of knockout mice by Cpf1-mediated gene targeting. Nat Biotechnol. 2016b;34:808–810. doi: 10.1038/nbt.3614. [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP, Tsai SQ, Prew MS, Nguyen NT, Welch MM, Lopez JM, McCaw ZR, Aryee MJ, Joung JK. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016;34:869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künne T, Swarts DC, Brouns SJJ. Planting the seed: target recognition of short guide RNAs. Trends Microbiol. 2014;22:74–83. doi: 10.1016/j.tim.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. 2013a;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013b;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Morse DP, Bass BL. Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry. 1997;36:8429–8434. doi: 10.1021/bi9709607. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013 doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, Macrae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci US A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smargon AA, Cox DBT, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, Abudayyeh OA, Essletzbichler P, Shmakov S, Makarova KS, et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol Cell. 2017;65:618–630.e7. doi: 10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- Tóth E, Weinhardt N, Bencsura P, Huszár K, Kulcsár PI, Tálas A, Fodor E, Welker E. Cpf1 nucleases demonstrate robust activity to induce DNA modification by exploiting homology directed repair pathways in mammalian cells. Biol Direct. 2016;11:46. doi: 10.1186/s13062-016-0147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Bultema J, Waghmare SP, Waghmare S, Zhou K, Barendregt A, Westphal W, Heck AJR, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci US A. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Qin R, Li H, Li D, Li L, Wei P, Yang J. Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol J. 2016 doi: 10.1111/pbi.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Nishimasu H, Zetsche B, Hirano H, Slaymaker IM, Li Y, Fedorova I, Nakane T, Makarova KS, Koonin EV, et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell. 2016;165:949–962. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Gao P, Rajashankar KR, Patel DJ. PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease. Cell. 2016;167:1814–1828.e12. doi: 10.1016/j.cell.2016.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015:1–14. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2016 doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Ren K, Qiu X, Zheng J, Guo M, Guan X, Liu H, Li N, Zhang B, Yang D, Ma C, et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016;532:522–526. doi: 10.1038/nature17944. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phasercrystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–4. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology. 2011;7:539–539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Read RJ, Adams PD, Brunger AT, Afonine PV, Hung L-W. Model morphing and sequence assignment after molecular replacement. Acta Crystallogr D Biol Crystallogr. 2013;69:2244–2250. doi: 10.1107/S0907444913017770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank (PDB) under accession numbers XXX and XXX. The unprocessed image files used to prepare the figures in this manuscript are deposited in Mendeley Data and are available at http://dx.doi.org/10.17632/fwbdxjzvcs.1.