Abstract
Ricin undergoes retrograde transport to the endoplasmic reticulum (ER), and ricin toxin A chain (RTA) enters the cytosol from the ER. Previous reports indicated that RTA inhibits activation of the unfolded protein response (UPR) in yeast and in mammalian cells. Both precursor (preRTA) and mature form of RTA (mRTA) inhibited splicing of HAC1u (u for uninduced) mRNA, suggesting that UPR inhibition occurred on the cytosolic face of the ER. Here, we examined the role of ribosome binding and depurination activity on inhibition of the UPR using mRTA mutants. An active-site mutant with very low depurination activity, which bound ribosomes as WT RTA, did not inhibit HAC1u mRNA splicing. A ribosome-binding mutant, which showed reduced binding to ribosomes but retained depurination activity, inhibited HAC1u mRNA splicing. This mutant allowed separation of the UPR inhibition by RTA from cytotoxicity because it reduced the rate of depurination. The ribosome-binding mutant inhibited the UPR without affecting IRE1 oligomerization or cleavage of HAC1u mRNA at the splice site junctions. Inhibition of the UPR correlated with the depurination level, suggesting that ribosomes play a role in splicing of HAC1u mRNA. We show that HAC1u mRNA is associated with ribosomes and does not get processed on depurinated ribosomes, thereby inhibiting the UPR. These results demonstrate that RTA inhibits HAC1u mRNA splicing through its depurination activity on the ribosome without directly affecting IRE1 oligomerization or the splicing reaction and provide evidence that IRE1 recognizes HAC1u mRNA that is associated with ribosomes.
Keywords: ribosome, ribosome-inactivating protein (RIP), RNA processing, RNA splicing, endoplasmic reticulum stress (ER stress), unfolded protein response (UPR), ribosomal ribonucleic acid (rRNA) (ribosomal RNA), ricin, rRNA depurination, alpha-sarcin/ricin loop, HAC1/XBP1 splicing
Introduction
Ricin is a type II ribosome-inactivating protein (RIP)2 produced by the castor bean plant Ricinus communis. Due to its toxicity and ease of isolation, ricin has been classified as a category B select agent (1). Ricin is more toxic to cancer cells than healthy cells and has been used as the toxic component of immunotoxins (2–4). The holotoxin is composed of an enzymatically active ricin toxin A chain (RTA) linked to ricin toxin B chain (RTB) by a disulfide bond. RTB facilitates entry of the holotoxin into the cell by binding to glycoproteins or glycolipids on the cell surface. After endocytosis, a small fraction of ricin is transported to the endoplasmic reticulum (ER) via retrograde transport. Upon entry into the ER, the disulfide bond of the holotoxin is reduced, allowing RTA to separate from RTB. Reduction of the disulfide bond partially unfolds RTA and allows it to cross the ER membrane. RTA is thought to exploit the ER-associated degradation (ERAD) pathway to enter the cytosol (5, 6). A fraction of RTA escapes the ubiquitin-mediated degradation in the cytosol, ultimately reaching ribosomes (7). RTA inhibits protein synthesis by removing an adenine from the sarcin/ricin loop of the 28S rRNA. This depurination event halts translation at the elongation step, leading to cell death.
Because RTA partially unfolds in the ER, it encounters the unfolded protein response (UPR), an ER stress response pathway, which is activated when unfolded proteins accumulate in the ER (8). The UPR reduces protein translation, increases expression of ER chaperones, and targets misfolded proteins for degradation (8). Accumulation of misfolded proteins in the ER is sensed by a transmembrane protein kinase/RNase, IRE1 (inositol requiring enzyme 1), that transmits a signal from the ER to the nucleus. The N-terminal domain of IRE1 is located in the ER lumen, whereas its C-terminal cytosolic portion contains Ser/Thr protein kinase and endoribonuclease domains (9). Upon accumulation of unfolded proteins in the ER, IRE1 oligomerizes and autophosphorylates (9). The cytosolic endoribonuclease domain of IRE1 excises an intron from the HAC1 mRNA in yeast and XBP1 mRNA in mammalian cells (9, 10). A tRNA ligase, Rlg1p, rejoins the cleaved ends of HAC1 mRNA in yeast (11).
The unspliced form of HAC1 mRNA, HAC1u (u for uninduced) is not translated because base-pairing interaction between the intron and the 5′-untranslated region (UTR) represses its translation (12, 13). However, after splicing, HAC1i (i for induced) is translated very efficiently. HAC1 activates transcription of the genes encoding the ER-resident chaperones and ERAD components by binding to the unfolded protein response element (UPRE) (14). Although removal of the intron relieves post-transcriptional silencing of HAC1u mRNA, how base pairing between the 5′-UTR and the intron prevents translation of the HAC1u mRNA in the absence of the UPR is not well-understood. It is not clear whether ribosomes play a role in the unconventional splicing of HAC1u mRNA in the cytosol. According to one model, the substrate for splicing is HAC1u mRNA stuck on stalled ribosomes (13). Another model proposes that the substrate for IRE1 splicing is untranslated HAC1u mRNA rather than polysomal mRNA containing stalled ribosomes (15).
We showed that the precursor form of an inactive RTA mutant with a point mutation at its active site (preE177K) (16) accumulated in the ER and induced the UPR pathway, whereas the precursor form of WT RTA (preRTA) inhibited tunicamycin (Tm), and DTT induced UPR by blocking splicing of the HAC1u mRNA in yeast (17). Because ER trafficking delayed the entry of preRTA to the cytosol, the inhibitory effect of preRTA on HAC1 mRNA processing could be separated from translation inhibition and cell death (17). Treatment of mammalian cells with ricin holotoxin or RTA led to inhibition of Tm-induced splicing of XBP1u mRNA (18), indicating that yeast is a relevant model to investigate the effect of RTA on the IRE1α-XBP1 arm of the UPR pathway.
We recently characterized mutations in RTA that affected RTA-ribosome interaction but not the enzymatic activity of RTA (19). These mutations lie on the opposite side of the active site at arginine residues critical for ribosome binding (19, 20). Here, we explore the mechanism by which RTA inhibits the UPR using two RTA mutants with reduced cytotoxicity: a ribosome-binding mutant, which binds ribosomes poorly but retains depurination activity, and an active site mutant, which binds ribosomes but has defective depurination activity. We show that ribosome depurination by RTA results in the inhibition of HAC1u mRNA splicing in the cytosol and present evidence that IRE1 recognizes HAC1u mRNA that is associated with ribosomes. These results provide unique insight into the mechanism of UPR inhibition by RTA and the regulation of HAC1u mRNA splicing.
Results
An RTA mutant with reduced depurination activity inhibits the UPR
To determine whether slowing the rate of ribosome depurination affects RTA-mediated inhibition of the UPR, we transformed yeast with R193A/R235A, which has an intact active site but shows greatly reduced ribosome binding in vitro and delayed ribosome depurination in vivo; G212E, which binds ribosomes like WT mRTA in vitro but has very low depurination activity due to a mutation near the active site (19); and the mature form of WT RTA (mRTA) (Table S1). The RTA constructs lacked the 35-amino acid signal sequence that targets RTA to the ER (16) so that they were only expressed in the cytosol. Expression was regulated by the galactose-inducible GAL1 promoter because of the high cytotoxicity of mRTA. Cells grown in dextrose showed no sign of cytotoxicity and displayed similar growth as the vector control (VC) (Fig. S1A). Expression of mRTA resulted in the most dramatic decrease in cell viability. Yeast expressing R193A/R235A also showed a decrease in cell viability relative to VC, whereas expression of G212E had no effect on cell viability compared with cells harboring the VC plasmid (Fig. S1A).
In the absence of RTA (VC), yeast growing in galactose media exhibited a doubling time of 3.3 h (Figs. S1, B and C). Expression of all forms of RTA increased the doubling time relative to the VC, with the greatest effect observed with mRTA, where the doubling time was 19 h. Yeast expressing R193A/R235A had a doubling time of 14 h, whereas G212E had a doubling time closer to that of yeast harboring the VC but still increased to 5.7 h. Despite the reduction in enzymatic activity caused by the G212E mutation, sufficient activity remained to elicit moderate cytotoxic effects, which were consistent with the lag in growth (Fig. S1B).
Ribosome depurination by mRTA was observed even during growth in dextrose media (0 h) due to the leakiness of the GAL1 promoter and high activity of mRTA (Fig. S1D). Ribosome depurination by mRTA showed a steady increase up to 8 h postinduction (hpi). Yeast expressing R193A/R235A showed an increase in depurination, but the rate of depurination was slower compared with mRTA (19). Only at 6 hpi did R193A/R235A reach the same level of depurination as mRTA. Depurination by G212E never reached the WT levels (Fig. S1D).
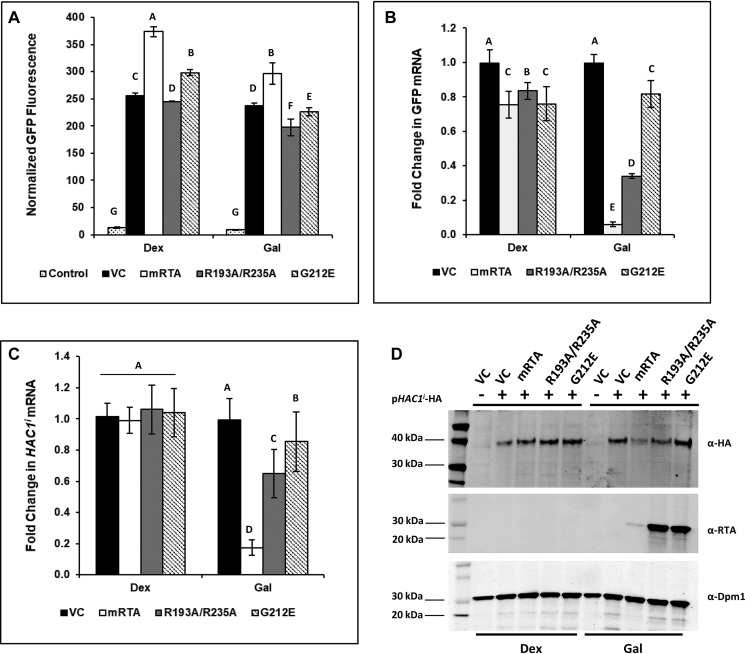
To monitor activation of the UPR, we co-transformed yeast harboring mRTA, R193A/R235A, G212E, or VC with a UPRE-GFP reporter plasmid. ER stress was induced at 6 hpi on galactose by adding 2 mm DTT, which induces UPR by preventing disulfide bond formation. There was no detectable RTA protein in cells grown under non-RTA-inducing conditions (dextrose) (Fig. 1A). Cells grown in galactose showed RTA protein at a level inversely proportional to the level of cytotoxicity (19, 20). RTA protein levels did not change in cells undergoing ER stress (Fig. 1A).
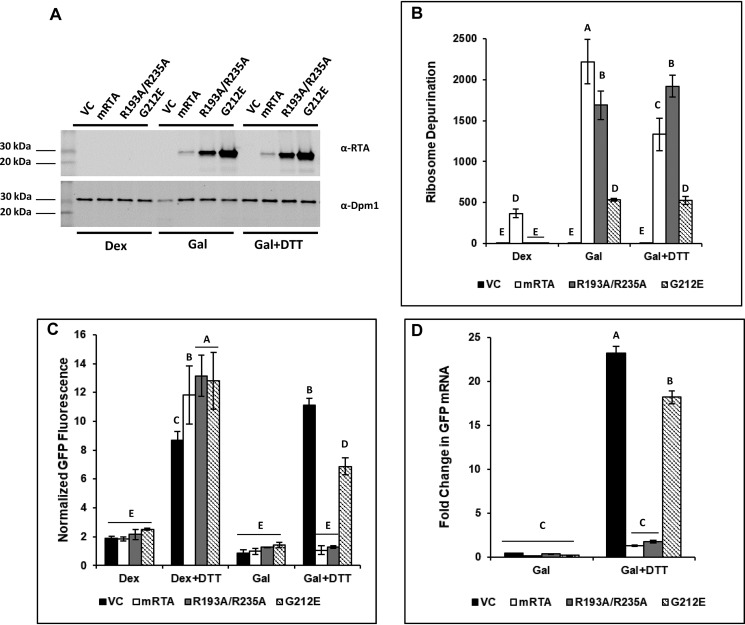
Figure 1.
Depurination of rRNA correlates with inhibition of the UPR in yeast expressing RTA mutants during ER stress. Yeast transformed with VC, WT (mRTA), or mutant RTA expression plasmids were grown in dextrose or galactose for 6 h to induce RTA expression, followed by an additional 90-min growth in the absence or presence of 2 mm DTT to induce ER stress. Aliquots were taken to prepare protein lysates for Western blotting or flow cytometry analysis and total RNA for qRT-PCR analysis. A, protein prepared from yeast carrying VC, WT, or mutant RTA expression plasmids grown in Dex or Gal or in the presence of ER stress (Gal + DTT) was subjected to Western blot analysis with monoclonal antibodies against RTA (top) and Dpm1 (bottom) as a loading control. Uncropped Western blots are shown in Fig. S4. B, ribosome depurination in yeast carrying VC, WT, or mutant RTA expression plasmids quantified by qRT-PCR using total RNA. The y axis shows the average -fold change in ribosome depurination compared with VC, with error bars representing the range of depurination from three biological replicates using three technical replicates for each. Means with different letters show significant differences according to the LSD test (p < 0.01). C, GFP fluorescence from a UPRE-GFP reporter measured by flow cytometry in yeast carrying VC, WT, and mutant RTA expression plasmids grown in dextrose in the absence (Dex) or presence of ER stress (Dex + DTT) and with RTA expression in the absence (Gal) or presence (Gal + DTT) of ER stress. The y axis shows the GFP signal normalized to yeast lacking the UPRE-GFP reporter from a minimum of three biological replicates along with the S.E. (n = 3). Means with different letters show significant differences according to the LSD test (p < 0.01). D, -fold change in GFP mRNA expressed from the UPRE-GFP reporter in yeast carrying VC, WT, or mutant RTA expression plasmids was quantified by qRT-PCR using total RNA prepared from cells grown in dextrose or during RTA expression in the absence (Gal) or presence of ER stress (Gal + DTT). The y axis shows the average -fold change in GFP mRNA compared with the same cells grown in dextrose, with error bars representing the range of GFP expression from two biological replicates using three technical replicates for each. Means with different letters show significant differences according to the LSD test (p < 0.001).
Depurination activity of RTA on ribosomes was monitored using qRT-PCR (21). The level of depurinated rRNA remained relatively low in dextrose except for yeast harboring mRTA. The depurination increased >2000-fold in mRTA and ∼1700-fold in R193A/R235A compared with yeast carrying the VC at 6 hpi (galactose (Gal)) (Fig. 1B). Despite reduced activity expression of G212E resulted in a >500-fold increase in depurinated rRNA. When ER stress was induced by the addition of 2 mm DTT, ribosome depurination remained largely unchanged at ∼1900-fold and ∼500-fold in R193A/R235A and G212E, respectively. A slight decrease in rRNA depurination was seen in mRTA (∼1300-fold). This drop may be due to cytotoxicity of mRTA coupled with inhibition of the UPR, resulting in cell death.
To show that the UPRE-GFP reporter was responsive to ER stress in the absence of RTA, we treated cells grown in dextrose with DTT and measured the increase in GFP fluorescence by flow cytometry. The normalized GFP fluorescence increased 9-, 12-, 13-, and 13-fold for VC, WT, R193A/R235A, and G212E, respectively, demonstrating that the UPRE-GFP reporter was functional (Fig. 1C, Dex + DTT). The normalized GFP fluorescence remained relatively low when RTA expression was induced in galactose for 6 h (Gal). When ER stress was induced by the addition of DTT to the galactose medium for 90 min, we observed an 11-fold increase in GFP fluorescence in yeast carrying the VC plasmid, indicating that the UPR has been induced (Gal + DTT). There was no increase in GFP fluorescence in yeast expressing mRTA, which was consistent with previous data showing that mRTA inhibits the UPR (17). Yeast expressing R193A/R235A showed complete inhibition of the UPRE-GFP reporter expression, suggesting that it inhibits the UPR at a similar level as mRTA. We observed a 7-fold increase in the GFP signal in yeast expressing G212E during ER stress, compared with the 11-fold increase in the VC, suggesting that the UPR was not induced at a similar level, possibly due to the reduced level of ribosome depurination.
To determine whether the decrease in fluorescence is due to reduced transcription of GFP from the UPRE-GFP reporter, we measured the GFP mRNA levels by qRT-PCR. There was no difference in GFP mRNA levels when cells were grown in Gal, indicating that expression of RTA does not influence expression of the reporter (Fig. 1D). When ER stress was induced by DTT, a 23-fold increase in GFP mRNA level was observed in cells harboring the VC. This increase in GFP mRNA was inhibited by mRTA and R193A/R235A, demonstrating that inhibition of the UPRE-GFP reporter by RTA is occurring at the level of transcription. Yeast expressing G212E showed an 18-fold increase in GFP mRNA, compared with the 23-fold increase in the VC, suggesting that G212E could partially inhibit activation of the UPR.
The decreased rate of ribosome depurination leads to specific inhibition of the UPR pathway genes
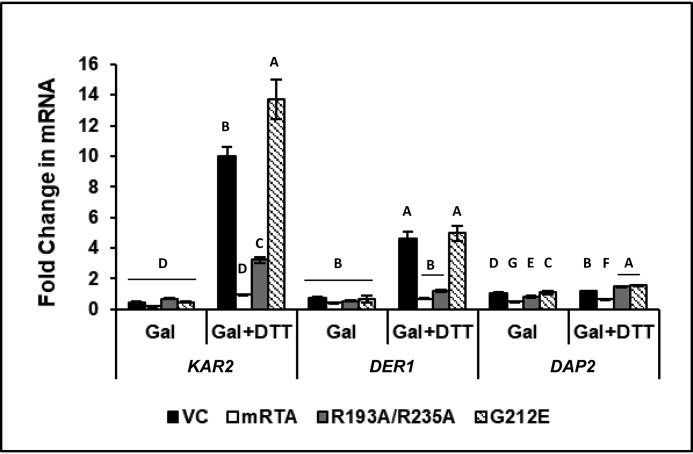
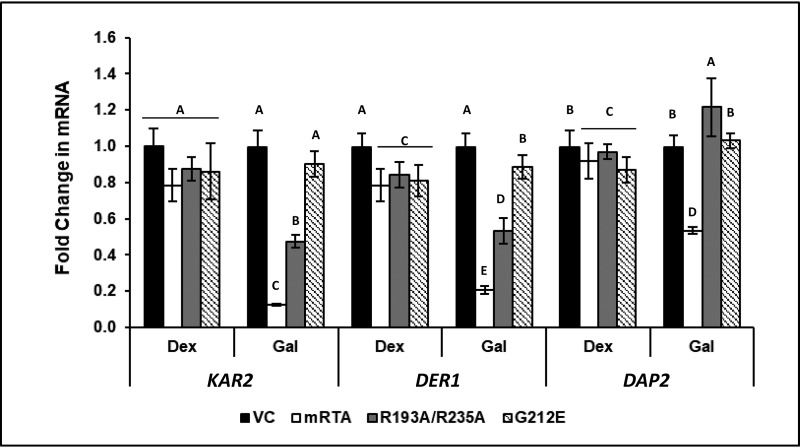
To determine whether expression of R193A/R235A attenuates signaling downstream of HAC1 induction, we examined expression of biological targets of UPR, such as KAR2, which is directly associated with the UPR, and DER1, which is closely linked to the UPR through the ERAD pathway. As a control, we monitored expression of DAP2, a dipeptidyl aminopeptidase that is involved in vacuolar sorting and does not have a known role in the ER stress. Expression of all genes remained unchanged when RTA was induced by growth in galactose relative to growth in dextrose (Fig. 2). When ER stress was induced in the VC, we observed 10- and 5-fold increases in the expression of KAR2 and DER1, respectively, whereas DAP2 showed no change. Similar results were observed for yeast expressing G212E, where KAR2 expression increased 14-fold and DER1 increased 5-fold. Once again, DAP2 expression remained unchanged. Expression of KAR2 and DER1 was completely inhibited in the presence of mRTA, and expression of DAP2 was also affected. The R193A/R235A inhibited expression of KAR2 and DER1 and had no effect on DAP2 expression. These results showed that R193A/R235A specifically inhibits transcriptional activation of the UPR pathway genes.
Figure 2.
Expression of genes associated with the UPR is specifically inhibited by R193A/R235A under conditions of ER stress. -Fold change in mRNA level of KAR2, DER1, and DAP2 in yeast carrying VC, mRTA, or mutant RTA expression plasmids was quantified by qRT-PCR using total RNA prepared from cells grown in dextrose or galactose in the absence (Gal) or presence of ER stress (Gal + DTT). The y axis shows the average -fold change in mRNA compared with the same cells grown in dextrose, with error bars representing the range of expression from two biological replicates using three technical replicates for each. Statistical analysis was conducted separately for each gene. Means with different letters (for each gene tested) show significant differences (p < 0.001). When one-way analyses of variance and Fisher's LSD statistical analysis were applied to the DAP2 expression data, we observed significant differences across all variants due to low mean values and low S.E., such that the slightest differences were perceived as significant.
HAC1u mRNA processing is inhibited in yeast expressing R193A/R235A during ER stress
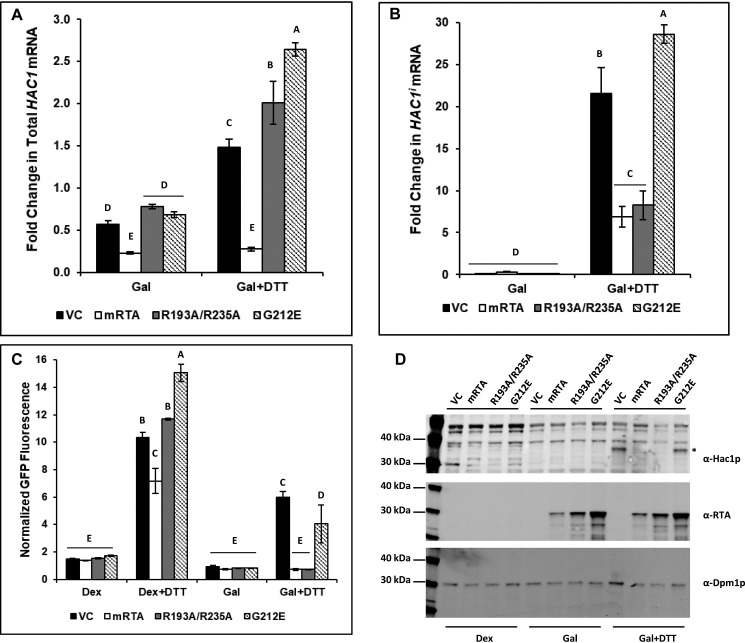
To determine whether inhibition of the UPR by R193A/R235A occurred at the HAC1u mRNA processing step, we developed a qRT-PCR assay that specifically detects HAC1i in the presence of both HAC1 mRNA species. The reverse primer spans the splice site, where the last 2 bp anneal 5′ to the splice site and the remainder of the primer anneals 3′ to the splice site. The specificity for amplification of HAC1i but not HAC1u mRNA was achieved by introducing a secondary mutation at the penultimate base in the primer. As a control, we quantified the total HAC1 mRNA. We did not observe a significant change (<2-fold) in total HAC1 RNA levels in yeast carrying the VC or mutant RTA plasmids after shifting cells from dextrose to galactose (Fig. 3A). Yeast expressing mRTA showed a ∼3-fold decrease in total HAC1 RNA. When ER stress was induced, there was a modest increase (1.5–2.5-fold) in total HAC1 mRNA in cells carrying the VC, R193A/R235A, and G212E. In contrast, total HAC1 RNA levels remained ∼3-fold lower in yeast expressing mRTA compared with growth in dextrose (Fig. 3A). Similar levels of HAC1i mRNA were present in all cells grown in galactose regardless of the expression vector (Fig. 3B). However, when ER stress was induced with DTT, HAC1i mRNA increased >20-fold in the VC and ∼30-fold in G212E, indicating that the UPR has been activated. In contrast, HAC1i mRNA increased only 7-fold in mRTA (Fig. 3B). Similarly, only an 8-fold increase in HAC1i mRNA was observed in R193A/R235A. The total HAC1 mRNA level increased under ER stress (Fig. 3A), whereas the amount of HAC1i mRNA remained at a similar level as in mRTA in yeast expressing the ribosome-binding mutant (Fig. 3B), indicating that it specifically inhibits the splicing of HAC1u mRNA during ER stress.
Figure 3.
Processing of HAC1u to HAC1i is specifically inhibited in yeast expressing R193A/R235A during ER stress. -Fold change in total HAC1 RNA level (A) and HAC1i mRNA level (B) in yeast carrying VC, WT (mRTA), or mutant RTA expression plasmids was quantified by qRT-PCR using total RNA prepared from cells grown in dextrose or galactose in the absence (Gal) or presence of ER stress (Gal + DTT). The y axis shows the average -fold change in mRNA compared with the same cells grown in dextrose, with error bars representing the range of HAC1 mRNA from two biological replicates using three technical replicates for each. Statistical analysis was conducted separately for each. Means with different letters show significant differences (p < 0.001). C, GFP fluorescence from a UPRE-GFP reporter measured by flow cytometry is shown. The y axis shows the GFP signal normalized to yeast lacking the UPRE-GFP reporter from a minimum of three biological replicates along with the S.E. (n = 3). Means with different letters show significant differences (p < 0.01). D, protein prepared from yeast carrying VC, WT, or mutant RTA expression plasmids grown in dextrose or galactose in the absence (Gal) or presence of ER stress (Gal + DTT) was analyzed on SDS-PAGE followed by Western blot analysis with polyclonal antibodies against HAC1 (top), RTA (middle), and Dpm1 (bottom). Uncropped Western blots are shown in Fig. S5.
The normalized GFP fluorescence from the reporter showed that the UPR was induced under ER stress–inducing conditions in yeast harboring the VC and mutant RTA plasmids in the absence of RTA (Dex + DTT) (Fig. 3C). However, the reporter activity was induced only in yeast harboring the VC or expressing G212E after ER stress and was inhibited in yeast expressing mRTA or R193A/R235A (Gal + DTT) (Fig. 3C).
Inhibition of the processing of HAC1u to HAC1i would result in the loss of HAC1 protein. Immunoblot analysis showed HAC1 protein migrating at ∼38 kDa (*) specific to yeast carrying the VC plasmid or expressing G212E grown under ER stress conditions but absent in yeast expressing mRTA or R193A/R235A (Gal + DTT) (Fig. 3D). HAC1 migrates more slowly on SDS-polyacrylamide gels than its predicted size of ∼27 kDa at a size of just under 40 kDa (22). The immunoblot analysis was consistent with the splicing results (Fig. 3B) and showed that mRTA and R193A/R235A inhibited processing of HAC1u mRNA, which in turn reduced HAC1 protein levels. We did not observe HAC1 in the absence of ER stress in cells grown in dextrose or in galactose. These results showed that HAC1u mRNA processing occurs in yeast expressing G212E when ER stress is induced. However, expression of the ribosome-binding mutant has a specific inhibitory effect on HAC1u mRNA processing during ER stress.
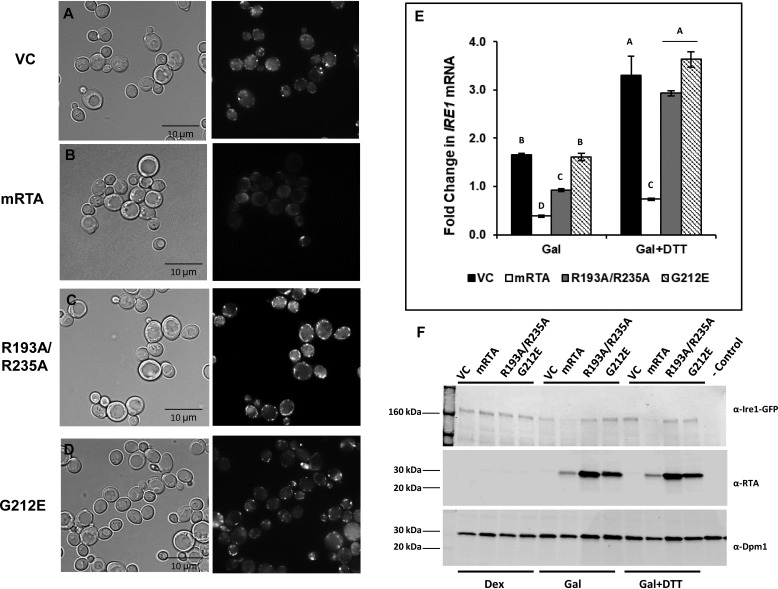
Inhibition of HAC1u mRNA processing by R193A/R235A occurs after IRE1 oligomerization
Oligomerization is critical for autophosphorylation and activation of IRE1, which is required for HAC1u mRNA processing. Therefore, a potential target for RTA-mediated inhibition of HAC1u mRNA processing is IRE1 oligomerization. To determine whether RTA inhibits the UPR by disrupting this process, we monitored IRE1 oligomerization in vivo by epifluorescence microscopy using an Δire1 yeast strain in which GFP-tagged IRE1 was integrated at the LEU2 locus (23). The phase-contrast (left) and GFP images (right) of this strain harboring the RTA plasmids after treatment with 5 mm DTT to induce ER stress are shown in Fig. 4 (A–D). We observed discrete brightly fluorescing foci in yeast carrying the VC (Fig. 4A), indicating that IRE1 oligomerization had occurred. However, these foci were absent in yeast expressing mRTA (Fig. 4B). During ER stress, the appearance of brightly fluorescing foci could readily be seen when R193A/R235A was expressed (Fig. 4C). Yeast expressing G212E (Fig. 4D) did not inhibit IRE1 oligomerization and appeared just like VC, which is consistent with previous data showing that this mutant does not inhibit the UPR.
Figure 4.
IRE1 oligomerization is observed in yeast expressing R193A/R235A, but not mRTA. Yeast (Δire1, IRE1-GFP::LEU2) transformed with VC, WT (mRTA), or mutant RTA expression plasmids were grown in dextrose or galactose for 6 h, to induce RTA expression, followed by an additional 90-min growth in the absence or presence of 5 mm DTT to induce ER stress. Aliquots were removed for epifluorescent microscopy, preparation of total RNA for qRT-PCR analysis, and protein lysate preparation for Western blot analysis. A–D, bright field (left) and fluorescence images (right) of yeast carrying VC (A), mRTA (B), R193A/R235A (C), and G212E (D) expression plasmids grown under conditions of RTA expression and ER stress. Scale bars, 10 μm. E, -fold change in IRE1 mRNA quantified by qRT-PCR using total RNA prepared from cells grown in the absence (Gal) or presence of ER stress (Gal + DTT). The y axis shows the average -fold change in IRE1 mRNA compared with the same cells grown in dextrose, with error bars representing the range of expression from two biological replicates using three technical replicates for each. Means with different letters show significant differences according to the LSD (p < 0.001) test. F, protein prepared from yeast carrying VC, WT, or mutant RTA expression plasmids grown in dextrose or during RTA expression in the absence (Gal) or presence of ER stress (Gal + DTT) subjected to Western blot analysis with antibodies against GFP to detect IRE1-GFP (top), RTA (middle), and Dpm1 (bottom). Uncropped Western blots are shown in Fig. S6.
A possible explanation for the inhibition of IRE1 oligomerization by mRTA is that the expression of IRE1 may be affected. To address this, we monitored the IRE1 mRNA level compared with the same cells grown in dextrose. The IRE1 mRNA level was slightly higher (<2-fold) in VC or in yeast expressing G212E. Expression of mRTA reduced IRE1 mRNA level, whereas IRE1 expression in cells expressing R193A/R235A was only slightly lower compared with growth in dextrose (Fig. 4E). When ER stress was induced, we observed a similar increase in IRE1 mRNA level in yeast expressing the RTA mutants, except in mRTA, where IRE1 mRNA levels remained low (Fig. 4E).
To determine whether the reduction in IRE1 expression leads to a loss of IRE1 protein, we monitored IRE1-GFP protein levels using antibodies against GFP. The lysates from all cells grown in dextrose and containing the integrated IRE1-GFP construct showed a high-molecular weight band that was absent from the negative control lysate (−control) prepared from yeast without GFP-tagged IRE1 (Fig. 4F). When RTA expression was induced, there was a loss of IRE1-GFP protein in mRTA, whereas yeast harboring VC or either mutant showed little change in IRE1-GFP protein levels. RTA protein was not detectable when cells were grown in dextrose but was clearly visible upon RTA induction and during ER stress (Fig. 4F, central panel). The UPR inhibition by mRTA was likely the result of a reduction in the IRE1 mRNA level possibly due to the cytotoxicity of mRTA. IRE1 oligomerization was not affected in yeast expressing the ribosome-binding mutant, although UPRE-GFP reporter activity and expression of UPR genes were inhibited, suggesting that the inhibitory effect of this mutant on UPR was downstream of IRE1 oligomerization.
HAC1 RNA is not a direct target for RTA-mediated UPR inhibition
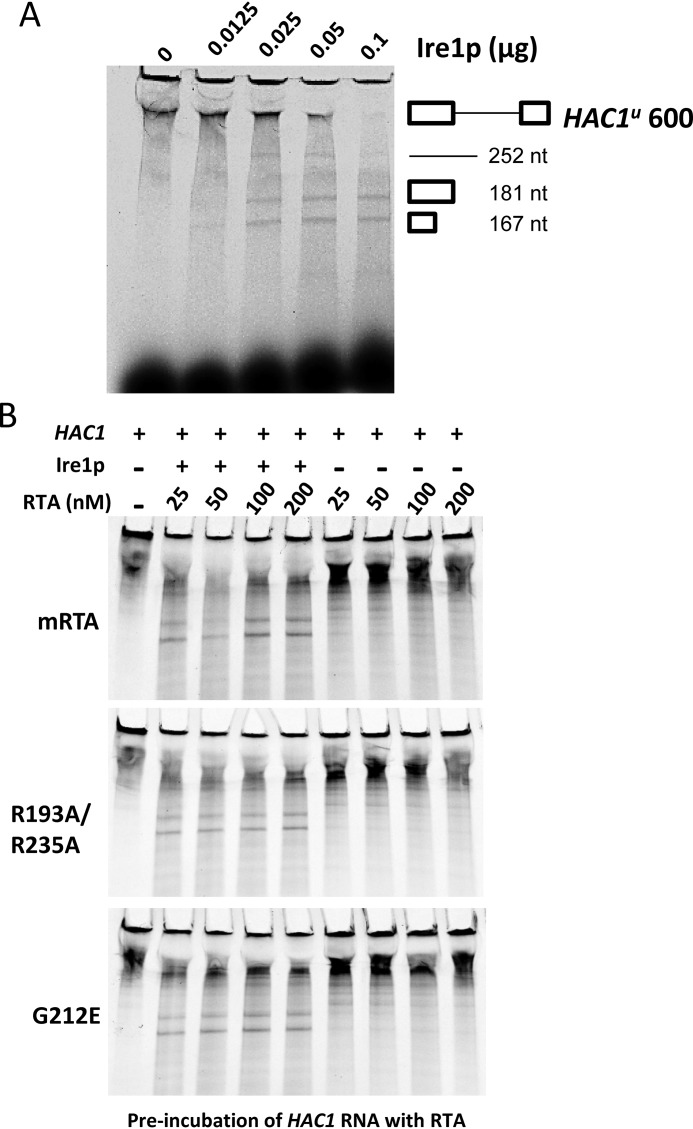
We showed that R193A/R235A inhibits HAC1 mRNA processing after IRE1 oligomerization, raising the possibility that HAC1 mRNA splicing is a direct target for RTA. HAC1u mRNA splicing has been reconstituted in vitro using recombinant GST-tagged IRE1 consisting of the cytoplasmic kinase and RNase domains (24). To determine whether RTA inhibits HAC1u mRNA splicing, an in vitro transcribed and Cy5-labeled 600-nucleotide HAC1u RNA transcript consisting of 181- and 167-nucleotide 5′ and 3′ exons and the 252-nucleotide intron was incubated with GST-tagged IRE1 (Ire1KR32) containing the cytoplasmic kinase and RNase domains (amino acids 641–1115) (25). Incubation of HAC1u RNA transcript with increasing amounts of IRE1 resulted in complete processing of the transcript into the 252-nucleotide intron and 181- and 167-nucleotide exon fragments (Fig. 5A). To determine whether mRTA and RTA mutants inhibited processing of the HAC1u RNA transcript, we preincubated the HAC1 transcript with increasing amounts of recombinant mRTA (25–200 nm final concentration) for 30 min prior to the addition of IRE1. Even at the highest concentration of mRTA (200 nm), the transcript was processed into the two-exon and single-intron fragments (Fig. 5B). Similar results were observed when increasing amounts of recombinant R193A/R235A or G212E were added to the splicing reaction (Fig. 5B). Reactions set up in the absence of IRE1 demonstrated that the presence of RTA alone had no effect on the stability of the transcript. The same results were obtained when RTA was preincubated with IRE1. Therefore, inhibition of HAC1u mRNA processing by RTA is not a result of direct inhibition of the splicing reaction or inhibition of the RNase activity of IRE1.
Figure 5.
In vitro processing of HAC1u to HAC1i is not affected by mRTA or mutant forms. A, incubation of in vitro transcribed Cy5-UTP–labeled 600-nt HAC1u RNA transcript with increasing amounts of IRE1 results in processing of HAC1u into 181- and 167-nt 5′ and 3′ exons and a 252-nt intron. B, HAC1u processing in the presence of increasing amounts of mRTA (top), R193A/R235A (middle), and G212E (bottom). HAC1u transcript was preincubated with RTA for 30 min prior to the addition of 0.05 μg of recombinant IRE1 and then incubated for an additional 30 min.
Activation of UPR correlates with the depurination activity of RTA mutants when HAC1i is provided in trans
The HAC1i mRNA level markedly decreased and HAC1 protein was not detected in yeast expressing R193A/R235A, suggesting that the turnover of HAC1i mRNA may be affected. To determine whether R193A/R235A affects the stability of HAC1i mRNA, yeast containing the UPRE-GFP reporter integrated at the URA3 locus was co-transformed with RTA expression plasmids along with the pHACi-HA plasmid. The normalized GFP fluorescence from yeast carrying the RTA expression plasmids in the presence of pHACi-HA is shown in Fig. 6A. As a control, we included the normalized fluorescence from the UPRE-GFP reporter carrying the pHACu-HA plasmid (Control) to demonstrate that activation of the reporter depended upon the presence of mature HAC1-HA (Fig. 6A). Yeast harboring the pHACi-HA plasmid showed a 250–350-fold increase in normalized GFP fluorescence, whereas fluorescence in the control yeast carrying the pHAC1u-HA plasmid remained very low, demonstrating that HACi-HA activates the UPR in the absence of ER stress. After 6 h of RTA induction, fluorescence remained unchanged for yeast carrying the VC and showed a minimal decrease in yeast carrying the RTA expression plasmids. These data were consistent with our previous results (17) and suggested that RTA-mediated inhibition of the UPR occurs at the point of HAC1u mRNA processing.
Figure 6.
The UPR is partially induced in the presence of HAC1i in yeast expressing R193A/R235A. Yeast containing the UPRE-GFP reporter integrated at URA3 were co-transformed pHAC1i-HA and VC, WT (mRTA), or mutant RTA expression vectors. Cells were grown in dextrose and then back-diluted into galactose medium to induce expression of RTA and incubated for 6 h. Aliquots were removed for flow cytometry, total RNA preparation for qRT-PCR analysis, and protein lysate preparation for Western blot analysis. A, GFP fluorescence from UPRE-GFP reporter was measured by flow cytometry in yeast constitutively expressing HAC1i and carrying VC, mRTA, and mutant RTA expression plasmids grown in dextrose or galactose. Yeast cells carrying the pHAC1u and VC plasmids are used as controls. The y axis shows the GFP signal normalized to yeast lacking the UPRE-GFP reporter from a minimum of three biological replicates along with the S.E. (n = 3). Means with different letters show significant differences (p < 0.01). B, -fold change in GFP mRNA expressed from the UPRE-GFP reporter quantified by qRT-PCR in yeast carrying WT or mutant RTA expression plasmids compared with VC using total RNA from cells grown in dextrose or galactose. The y axis shows the average -fold change in GFP mRNA compared with the VC, with error bars representing the range of expression from two biological replicates using three technical replicates for each. Means with different letters show significant differences (p < 0.001). C, -fold change in HAC1i mRNA expressed from the UPRE-GFP reporter quantified by qRT-PCR in yeast carrying mRTA or mutant RTA expression plasmids compared with VC using total RNA from cells grown in Dex and Gal. The y axis shows the average -fold change in HAC1i mRNA, with error bars representing the range of expression from two biological replicates using three technical replicates for each. Means with different letters show significant differences (p < 0.001). D, protein prepared from yeast carrying VC, mRTA, or mutant RTA expression plasmids was subjected to SDS-PAGE followed by Western blot analysis with monoclonal antibodies against HA (top), RTA (middle), and Dpm1 (bottom). Uncropped Western blots are shown in Fig. S7.
Because GFP is a highly stable protein, fluorescence observed during growth in galactose may be due to accumulation of GFP protein during growth in dextrose. To address this, we monitored GFP mRNA levels in cells carrying the RTA expression vectors relative to yeast carrying the VC. The GFP mRNA levels were similar, yet slightly reduced, for yeast transformed with the expression vectors on dextrose (Fig. 6B). However, when RTA was expressed by growth in galactose, we observed a ∼17-fold reduction in GFP mRNA in yeast expressing mRTA and a ∼3-fold decrease in yeast expressing R193A/R235A. Expression of G212E caused a slight decrease compared with VC. These data suggested that mRTA and R193A/R235A may be affecting transcription and/or the stability of the GFP mRNA. To investigate this, we quantified the HAC1i mRNA levels in cells grown in dextrose and galactose. The HAC1i mRNA levels were similar in cells grown in dextrose (Fig. 6C). Expression of mRTA resulted in a >5-fold reduction in the HAC1i mRNA, which is consistent with the decrease in expression of the GFP reporter. Expression of R193A/R235A and G212E led to a slight (<2-fold) but significant reduction in HAC1i mRNA levels.
To determine whether R193A/R235A affects translation of the HAC1i mRNA, we analyzed HAC1-HA protein level by immunoblot analysis using anti-HA antibodies. When cells were grown in dextrose, HAC1-HA was detected in lysates prepared from yeast harboring the pHACi-HA plasmid but not in the lysate prepared from yeast carrying pHAC1u-HA (−VC, Fig. 6D). When RTA is expressed, we observed a measurable decrease in HAC1-HA in yeast expressing mRTA and a <2-fold reduction in cells expressing R193A/R235A, which correlated with the <2-fold reduction in HAC1i mRNA levels. Yeast expressing G212E showed little effect on HAC1-HA level and appeared similar to VC (Fig. S2). These results indicate that the reduction in HAC1i mRNA and protein levels correlates with the depurination activity of RTA mutants on the ribosome.
To determine whether the ribosome-binding mutant and mRTA affect expression of genes associated with the UPR when HAC1-HA was provided in trans, we examined expression of KAR2, DER1, and DAP2. The fold increase in expression of each gene in dextrose in the presence of the pHACi-HA plasmid compared with control carrying the pHACu-HA plasmid is shown in Fig. S3. KAR2 and DER1 showed average increases of 4.8 ± 0.5- and 3.1 ± 0.5-fold, whereas DAP2 did not change in the presence of the pHACi-HA plasmid. Expression of KAR2, DER1, and DAP2 were similar to VC on dextrose (Fig. 7). However, expression of mRTA resulted in 10- and 5-fold decreases in KAR2 and DER1, respectively, and ∼2-fold reduction in DAP2. R193A/R235A caused a 2-fold decrease in KAR2 and DER1 and a slight increase (1.2-fold) in DAP2 compared with the VC, whereas G212E did not have an appreciable effect. These data indicate that whereas mRTA causes a general reduction in the mRNA levels of all genes analyzed, R193A/R235A causes a lower reduction and G212E has the least effect, indicating that the inhibitory effect of RTA mutants on UPR responsive genes downstream of HAC1i mRNA correlates with their depurination activity.
Figure 7.
Expression of the UPR genes is partially induced in yeast expressing R193A/R235A in the presence of HAC1i. Fold change in KAR2, DER1, and DAP2 mRNA in yeast carrying VC, mRTA, or mutant RTA expression plasmids was quantified by qRT-PCR using total RNA prepared from cells grown in dextrose or during RTA expression (Gal) compared with the VC. The y axis shows the average -fold change in mRNA, with error bars representing the range of expression from two biological replicates using three technical replicates for each. Statistical analysis was conducted separately for each gene. Means with different letters show significant differences according to the LSD (p < 0.001) test for each gene.
HAC1u mRNA is associated with ribosomes and does not get processed on depurinated ribosomes during the UPR
The ribosome-binding mutant specifically inhibited the UPR by blocking processing of HAC1u mRNA. The UPR inhibition correlated with the level of ribosome depurination, suggesting that ribosomes play a role in splicing of HAC1u mRNA. To investigate this, we assessed ribosome occupancy of HAC1u, HAC1i, and total HAC1 mRNA in the absence or presence of ER stress by qRT-PCR using RNA extracted from purified ribosomes.
We first monitored induction of UPR in cells that will be used for ribosome isolation by measuring expression of the integrated UPRE-GFP reporter. Low GFP levels were observed when cells were grown under non-UPR-inducing conditions (Dex and Gal) (Fig. S8A). GFP fluorescence increased between ∼15- and 30-fold upon the addition of DTT in the absence of RTA (Dex + DTT), demonstrating induction of the UPR.
The mRTA and R193A/R235A inhibited activation of the UPRE-GFP reporter in the presence of DTT (Gal + DTT), whereas yeast carrying the VC or G212E showed a similar level (∼15-fold) of UPRE-GFP induction (Fig. S8A). RTA expression was induced >25-fold when yeast carrying the different expression plasmids were grown on galactose (Fig. S8B).
Ribosomes were purified by high-speed centrifugation, and ribosome purity was assessed using equivalent amounts of cytoplasmic (Cyto.) and ribosomal (Rb) fractions by Western blot analysis with antibodies against ribosomal protein L3 (RPL3) and cytoplasmic protein phosphoglycerate kinase 1 (PGK1) (Fig. S8C). The ∼44 kDa RPL3 band was present only in the ribosomal fraction, whereas the ∼45 kDa PGK1 band was present in the cytoplasmic fraction. Less than ∼5% of PGK1 in the cytosolic fraction was observed in the ribosomal fraction.
As a control for detection of ribosome associated RNAs by qRT-PCR, we examined the association of ACT1 pre-mRNA, which contains an intron and ACT1 mRNA without the intron with ribosomes using primers specific for the exon or intron sequences after normalization of each to 25S rRNA. The normalized level of ACT1 pre-mRNA and mRNA in the ribosome fraction was compared with the normalized level in the total RNA. The level of ACT1 mRNA (exon) was very similar in total RNA and RNA extracted from ribosomes, whereas the level of ACT1 pre-mRNA (intron) decreased ∼3-fold in purified ribosomes (Fig. S8D) compared with the level in total RNA, indicating that qRT-PCR detects specifically ribosome-associated RNAs.
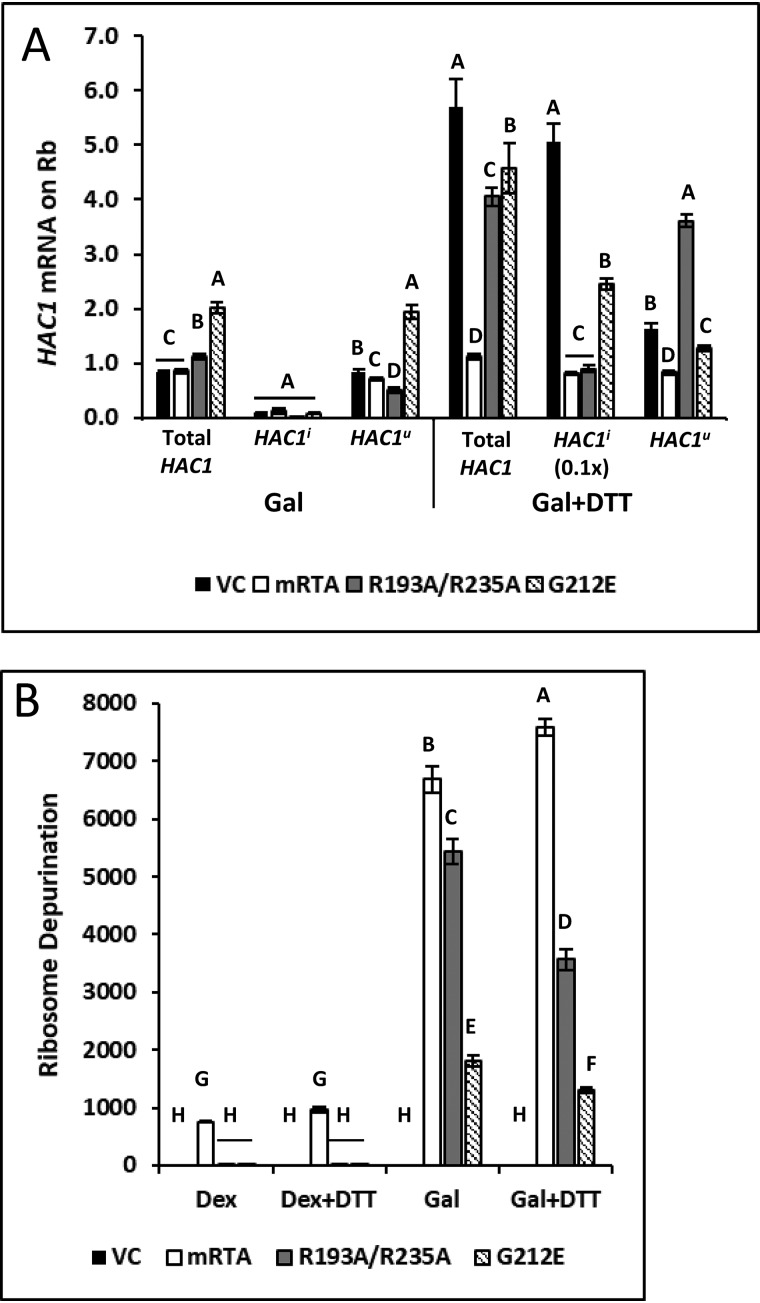
We measured the change in total HAC1, HAC1i, and HAC1u mRNA associated with the ribosome in yeast grown in galactose or Gal + DTT compared with the same cells grown in dextrose. The total and HAC1u mRNA were present on the ribosome in similar amounts in VC, mRTA, and R193A/R235A, whereas G212E showed a slight increase (<2-fold) when cells were grown on Gal (Fig. 8A). The amount of HAC1i mRNA found on the ribosome remained low regardless of the expression plasmid. Induction of the UPR by the addition of DTT caused ∼6-fold increase in the total HAC1 mRNA on the ribosome in the VC and ∼4- and 5-fold increase in R193A/R23A and G212E, respectively. There was no increase in total HAC1 mRNA in mRTA due to an overall decrease in transcription. We observed ∼50-fold increase in HAC1i mRNA in the VC (10× reduced scale in Fig. 8A). Likewise, in G212E, HAC1i mRNA on the ribosome increased ∼25-fold, whereas mRTA or R193A/R235A showed only a slight increase in HAC1i mRNA on the ribosome. These results show that HAC1i mRNA accumulates on the ribosome during ER stress in yeast expressing either mutant, but processing of that message is specifically inhibited by the ribosome-binding mutant. HAC1u mRNA level on the ribosome was ∼3.5-fold higher in R193A/R235A in the presence of DTT compared with in the absence of DTT, whereas in VC and G212E, HAC1u mRNA was similar to that observed in the absence of DTT. The HAC1u mRNA level associated with ribosomes decreased and the HAC1i mRNA level increased in the VC and G212E compared with the total HAC1 mRNA due to processing. However, the level of HAC1u mRNA associated with ribosomes was similar to the level of total HAC1 mRNA in R193A/R235A because HAC1u mRNA did not get processed. We conclude that the ribosome-binding mutant does not inhibit loading of HAC1u mRNA on the ribosome but inhibits splicing of HAC1u mRNA on the ribosome during the UPR.
Figure 8.
Ribosome-binding mutant traps HAC1u on the depurinated ribosome. A, analysis of total HAC1, HAC1u, and HAC1i on the ribosome by qRT-PCR using RNA extracted from ribosomes purified from yeast grown in the absence (Gal) or presence of ER stress (Gal + DTT) compared with the same cells grown in dextrose. Note that values shown for HAC1i are reduced by 10-fold for presentation on the same bar graph as total and HAC1u mRNA. The y axis shows mean fold change in HAC1 mRNA, with error bars representing the range of abundance from three biological replicates using three technical replicates for each. Statistical analysis was done separately for each HAC1 mRNA. Means with different letters show significant differences according to the LSD test (p < 0.01). B, depurination level of ribosomes isolated from yeast carrying VC, WT, or mutant RTA grown in dextrose and galactose in the absence (Gal) or presence of ER stress (Gal + DTT) quantified by qRT-PCR. The y axis shows mean fold change in ribosome depurination compared with VC with S.E. representing range of depurination from three biological replicates using three technical replicates for each. Statistical analysis was done using the means from each HAC1 mRNA. Means with different letters show significant differences according to the LSD test (p < 0.01).
The depurination state of purified ribosomes was assessed by measuring the -fold increase in depurination relative to the VC (Fig. 8B). Elevated ribosome depurination was observed in yeast carrying mRTA even on dextrose. Depurination increased by ∼6500–7000-fold upon induction of mRTA on galactose in the absence or presence of the UPR induction. Ribosome depurination remained low during growth in dextrose in R193A/R235A and G212E. Depurination increased between ∼5000 and 3000-fold upon expression of R193A/R235A in the absence or presence of the UPR induction. Ribosome depurination by G212E also increased but was ∼3 times lower compared with R193A/R235A, indicating that there is a correlation between processing of HAC1u mRNA on the ribosome and ribosome activity. These results suggest that translation inhibition due to depurination by the ribosome-binding mutant traps HAC1u mRNA on the ribosome in such a way that processing cannot occur.
Discussion
Inhibition of the UPR correlates with the rate of ribosome depurination by RTA
We previously showed that preRTA with its own signal sequence translocated into the ER and inhibited activation of the UPR, whereas an inactive form that translocated into the ER induced the UPR (17). The preRTA inhibited processing of the HAC1u, preventing synthesis of HAC1 (17). Both precursor and the mature form of RTA without the signal sequence inhibited the UPR after treatment with ER stress inducers, suggesting that inhibition of the UPR occurred on the cytosolic face of the ER (17). Because activation of the UPR would induce transcription of ERAD components, which normally translocate misfolded proteins from the ER to the cytosol for degradation, we proposed that inhibition of the UPR may allow ricin to enter the cytosol and avoid degradation (17). Here, we used RTA mutants that lacked the signal sequence to examine the role of the ribosome in UPR inhibition by RTA in the absence of ER trafficking.
Arginine residues at the RTA/RTB interface outside the active site cleft are critical for ribosome interactions of RTA (19, 20). The R193A/R235A double mutation affected the electrostatic interactions of RTA with the ribosome (19). The combination of reduced ribosome binding and intact enzymatic activity led to a reduction in the rate of ribosome depurination by R193A/R235A compared with mRTA (19, 26). The G212E mutation near the active site reduced the depurination activity without affecting the ability to bind to the ribosome (19). The reduced rate of depurination exhibited by R193A/R235A and reduced enzymatic activity by G212E correlated with cytotoxicity and indicated that G212E had lower toxicity than R193A/R235A, which was less toxic than mRTA. As reported previously (19, 20), the level of protein expression showed an inverse correlation with cytotoxicity because a lower rate of depurination allowed the mutant proteins to accumulate with time.
The preRTA used in our previous study showed a specific inhibitory effect on activation of the UPR, which could be separated from the decrease in translation and viability (17). However, mRTA depurinated ribosomes at a faster rate than preRTA because it did not require ER trafficking to enter the cytosol (16, 27). We observed reductions in total HAC1 and IRE1 mRNA and protein levels and a 2-fold decrease in DAP2 mRNA level when mRTA was expressed, indicating that it caused a general decrease in cell viability. In contrast, the ribosome-binding mutant did not reduce the total HAC1, IRE1, or DAP2 mRNA levels but caused a specific reduction in the HAC1i mRNA level, indicating that the effect of this mutant on the UPR could be separated from the decrease in viability. The ribosome-binding mutant offered a unique window into the mechanism of UPR inhibition and showed that RTA inhibits activation of the UPR as a consequence of its depurination activity on the ribosome without directly targeting oligomerization of IRE1 or HAC1u mRNA splicing.
We previously showed that UPR measured by LacZ reporter activity was induced when HAC1 mRNA was provided in trans in yeast expressing preRTA (17). Here, we measured the mRNA level of the UPRE-GFP reporter and HAC1i mRNA and showed that both decreased in yeast expressing the RTA mutants in a manner that correlated with their depurination activity on the ribosome. HAC1i mRNA level decreased at the highest level in yeast expressing mRTA, at a much lower level in R193A/R235A, and at the lowest level in G212E. G212E was able to bind ribosomes in vitro similar to mRTA, indicating that the inhibitory activity on the UPR correlated with the depurination activity of RTA mutants on the ribosome rather than their ability to bind to the ribosome.
RTA inhibits the UPR downstream of IRE1 oligomerization before splicing of the HAC1u mRNA
To determine whether inhibition of the UPR by the ribosome-binding mutant was due to a reduction in the processing of HAC1u mRNA, we developed a qRT-PCR assay that specifically detected HAC1i in the presence of both HAC1 mRNA species. The same strategy was used in the development of the qRT-PCR assay that specifically amplifies depurinated 25S rRNA (21). The total HAC1 RNA level did not change, but the HAC1i mRNA level decreased when R193A/R235A was expressed in yeast, indicating that processing of HAC1u mRNA was specifically inhibited. To address the mechanism, we examined oligomerization of IRE1, which is required for splicing of HAC1u mRNA. Oligomerization of IRE1 proceeded in yeast expressing both mutants. Unlike mRTA, expression of R193A/R235A did not cause a reduction in IRE1 mRNA and protein levels, suggesting that it inhibited the UPR after IRE1 oligomerization, raising the possibility that it may be targeting the HAC1u mRNA. Previous studies indicated that RIPs can target other RNAs besides the rRNA (28). Pokeweed antiviral protein alters splicing of HIV-1 RNAs (29) and can depurinate viral RNAs and mRNA (30, 31). Because splicing of HAC1u mRNA is mediated by a secondary structure at the intron-exon boundaries of the mRNA, we wanted to find out whether RTA has a direct effect on the stability of HAC1u mRNA or its splicing, which occurs in the cytoplasm independent of the spliceosome machinery (13). Using an in vitro HAC1 RNA-processing assay (24), we showed that neither mRTA nor the mutants affected the stability of HAC1u mRNA or the cleavage of HAC1u RNA at either splice junction by IRE1. The mRTA and RTA mutants did not inhibit the nuclease activity of IRE1. It is unlikely that RTA would have an inhibitory effect on RLG1-mediated ligation of the exon fragments, because RIPs do not have any known effects on tRNA synthetases. The ribosome-binding mutant did not directly target the HAC1u mRNA and did not have a direct inhibitory effect on oligomerization of IRE1 or its RNase activity, indicating that RTA inhibits the UPR downstream of IRE1 oligomerization before splicing of HAC1u mRNA.
Model for inhibition of HAC1u mRNA splicing by RTA
Inhibition of the UPR correlated with the extent of ribosome depurination caused by RTA mutants, suggesting that IRE1 recognizes HAC1u mRNA associated with ribosomes. Earlier studies showed that HAC1u and XBP1u are associated with membranes (32). Both RNAs are tethered to the ER membrane to enhance cytoplasmic splicing (23, 33). A portion of HAC1u mRNA co-sediments with polysomes (13). The N terminus of HAC1u was immunoprecipitated with antibody against the N-terminal epitope of HAC1, indicating that it was translated (12, 34). However, there is conflicting evidence regarding the role of the ribosome in the unconventional splicing of HAC1u mRNA. In one model translation of HAC1u, mRNA is initiated as it is exported from the nucleus, but ribosomes stall on the mRNA as the intron is exported from the nucleus base-pairs with the 5′-UTR (13). Translation initiation does not occur on HAC1u mRNA after base pairing is formed (15, 35). Translational repression was shown to be necessary for targeting HAC1u mRNA to IRE1 clusters on the ER membrane (23). Therefore, 5′-UTR–intron interaction is essential not only to repress HAC1u translation, but also to allow efficient splicing and HAC1 synthesis upon UPR induction (13). In another model, the substrate for IRE1 splicing is untranslated mRNA rather than polysome associated HAC1u mRNA containing stalled ribosomes (15). It has been shown that base pairing between the 5′-UTR and the intron prevents ribosome loading, and a combination of inhibited translation initiation and accelerated protein degradation prevents accumulation of HAC1 protein in the absence of UPR (15).
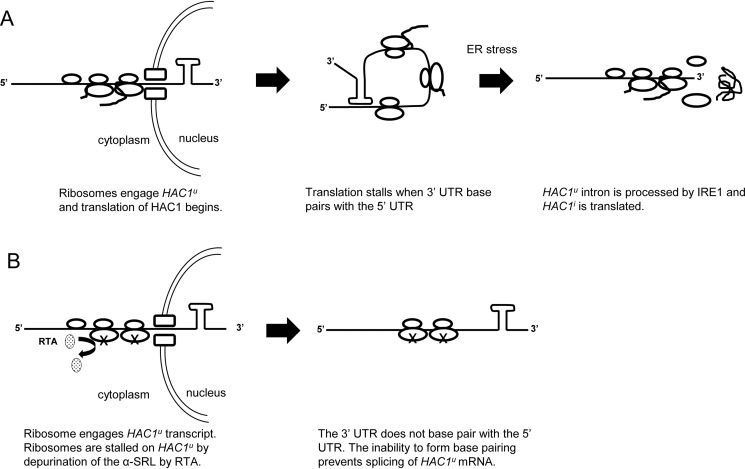
Our results build upon the first model (13), which proposes that the substrate for splicing is HAC1u mRNA that is stuck on translating ribosomes. We show that HAC1u mRNA is engaged with ribosomes and does not get processed on depurinated ribosomes when ribosome-binding mutant is expressed. Because 5′ portions of both unspliced and spliced HAC1 mRNA are identical, we propose that at least one round of translation occurs on HAC1u mRNA before base pairing is established. According to our model (Fig. 9A), as the 5′-end of HAC1 mRNA emerges from the nucleus, it associates with ribosomes and begins to be translated. When the intron at the 3′-UTR enters the cytoplasm, it base-pairs with the 5′-UTR, thereby forming a closed loop that prevents translation initiation. During ER stress, HAC1u mRNA on stalled ribosomes is recruited to IRE1, and IRE1-dependent excision of the intron allows translation of HAC1i mRNA. Because the intron is thought to interact with the 5′-UTR only after 3′-UTR of HAC1u mRNA is exported to the cytosol (13), ribosome depurination by RTA traps ribosomes that are already engaged on HAC1u mRNA and prevents the base-pairing interaction between the 5′-UTR and the intron (Fig. 9B). The inability to form base pairing impedes the process by which HAC1u mRNA can be spliced during ER stress. HAC1u mRNA associated with depurinated ribosomes is not degraded because its level is similar to the level of total HAC1 mRNA associated with ribosomes in yeast expressing R193A/R235A (Fig. 8A).
Figure 9.
Proposed model for inhibition of HAC1u mRNA splicing by RTA. A, HAC1u mRNA associates with ribosomes and begins to be translated as the 5′ end emerges from the nucleus. When the intron at the 3′-UTR enters the cytoplasm, it base-pairs with the 5′-UTR, thereby forming a closed loop that prevents translation initiation. Removal of the intron by IRE1 during ER stress allows translation of HAC1i. B, HAC1u mRNA associates with the ribosome as it is exported from the nucleus. Translation begins, but ribosome depurination by RTA traps ribosomes on HAC1u mRNA, preventing the base-pairing interaction between the 5′-UTR and the intron. The inability to form base pairing impedes the process by which HAC1u can be spliced during ER stress.
RTA inhibits splicing of XBP1u mRNA during ER stress in mammalian cells (18). Although HAC1u 5′-UTR–intron pairing appears not to be conserved in XBP1u mRNA, we believe that our model is applicable to inhibition of XBP1u splicing by RTA. In mammalian cells, translation of XBP1u mRNA is required for ER association (36). A hydrophobic region at the C-terminal region of XBPu protein (HR2) recruits the mRNA–ribosome–nascent chain complex to the ER for processing by IRE1. Translation of XBPu protein pauses to allow sufficient time for recruitment of the complex to IRE1 (33). We propose that ribosome depurination by RTA would inhibit translation of HR2, preventing recruitment of the mRNA–ribosome–nascent chain complex to IRE1 for splicing (18).
Mechanistic data about modulation of the HAC1/XBP1 splicing pathway is important not only for understanding the mechanism of toxicity of ricin, but also for the development of novel therapies against major diseases associated with the UPR pathway, such as cancer and metabolic disorders (37, 38). The IRE1α-XBP1 arm of the UPR pathway has a prosurvival role in cancer, and XBP1 is one of the most promising targets for cancer therapy (39). Inhibition of HAC1/XBP1 splicing by RTA may contribute to the anti-cancer activity of ricin. Although ricin-containing immunotoxins have shown promise in clinical trials against cancer, their use has been limited due to nonspecific toxicity to healthy cells. Small molecules that can activate UPR in the presence of ricin may reduce the cytotoxicity of RTA and may be useful as ricin antidotes. They may also be useful in reducing the nonspecific toxicity of ricin immunotoxins. XBP1 has been shown to mediate a wide range of responses in metabolic disease, and reducing the UPR in obesity has been suggested to have a therapeutic potential in insulin resistance and type 2 diabetes (40). Systemic administration of ricin to animal models lowers blood glucose levels, and at high doses, ricin exposure leads to lethal hypoglycemia (41). Understanding how ricin modulates the HAC1/XBP1 processing may offer novel opportunities for treating metabolic disease and for developing more effective ricin immunotoxins.
Experimental procedures
Cell viability assay, growth curve, and doubling time determination
WT Saccharomyces cerevisiae strain W303a (MATa ade2-1 trp1-1 ura3-1 leu2-3, 112 his3-11, 15 can1-100) was transformed with galactose-inducible yeast expression plasmids containing WT mature RTA (NT1458), R193A/R235A (NT1448), and G212E (NT1468) without the 35-residue signal sequence or the vector control (pRS415). Transformants were grown into mid-log phase in synthetic dropout medium minus leucine, SD−Leu (2% dextrose, 0.67% Bacto-yeast nitrogen base supplemented with amino acids), pelleted, and resuspended at a concentration of ∼0.3 OD600/ml in selective medium containing 2% galactose (SGal−Leu) to induce RTA expression. Aliquots were taken at 0 and 6 hpi, diluted to 0.1 OD600/ml, and diluted in 10-fold series. Five microliters from each dilution were spotted onto S.D.−Leu solid medium and incubated at 30 °C for 36 h and then photographed.
For growth curve analysis, three biological replicates were grown in SD−Leu medium into mid-log phase, pelleted, and resuspended in SGal−Leu medium at a concentration of ∼0.15 OD600/ml in a final volume of 0.75 ml in Costar 24-well plates sealed with Breathe-Easy gas-permeable sealing membranes and incubated at 30 °C with constant shaking in a BioTek Synergy 4 (BioTek Industries) plate reader controlled by Gen5 software. OD600 was measured every 2 h for 24 h. Data were exported into Excel, plotting the average OD600 versus time. Doubling time was calculated for each using the formula, doubling time = ln(2)/growth rate, where growth rate is taken to be the exponent from the line equation of the exponential fit of the curve. The R2 value for each curve was >0.998. The average doubling time for three biological replicates was reported.
Detection of UPR by flow cytometry
WT S. cerevisiae strain W303a (MATa ade2-1 trp1-1 ura3-1 leu2-3, 112 his3-11, 15 can1-100) was co-transformed with LEU2-marked galactose-inducible yeast expression plasmids containing mature RTA (NT1458), R193A/R235A (NT1448), G212E (NT1468), or vector control (pRS415) along with a URA3-marked UPRE-GFP promoter reporter (NT1466) containing four UPREs from the KAR2 promoter driving expression of GFP. Three biological replicates were grown in SD−Leu−Ura medium supplemented with 100 μg/ml myoinositol into mid-log phase, pelleted, and resuspended at a concentration of ∼0.3 OD600/ml in SGal−Leu−Ura medium (supplemented with 100 μg/ml myoinositol) to induce RTA expression. At 6 hpi, the culture was divided in half, and DTT was added to one half at a concentration of 2 mm to induce ER stress. After 90 min, aliquots were taken for flow cytometry, and for total RNA and protein lysate preparation. The median GFP signal from the FL1-A channel was measured using an Accuri C6 flow cytometer (BD Biosciences) and normalized to yeast lacking the reporter to account for background fluorescence. Normalized GFP fluorescence was reported as the average from three biological replicates along with the standard error.
RNA isolation and cDNA synthesis
Total RNA was extracted from ∼2 × 107 cells using the RNeasy Mini Kit (Qiagen) after cell lysis with 425–600-μm glass beads (Sigma) and a 16-tube Mini Bead Beater (BioSpec Products). The optional on-column DNase digestion was carried out as per protocol. The High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) was used for cDNA conversion of 375 ng of total RNA in a 15-μl reaction. rRNA was extracted from purified ribosomes using the RNA cleanup steps found in the RNeasy Mini Kit, including the on-column DNase digestion. The High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) was used for cDNA conversion of ∼150 ng of total RNA in a 20-μl reaction.
qRT-PCR and data analysis
The qRT-PCR assays were performed on a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) using Power SYBR Green Master Mix (Thermo Fisher Scientific) in a total volume of 20 μl with 5 μl of diluted cDNA and forward and reverse primers at a final concentration of 250 nm each. PCR efficiencies for all primer pairs were 90% or greater, and sequences are as follows in the 5′ to 3′ direction: GFP, GTCCTTTTACCAGACAACCATTACC and TTTTCGTTGGGATCTTTCGAA; KAR2, AGACTAAGCGCTGGCAAGCT and ACCACGAAAAGGGCGTACAG; DER1, GCAGCATCACTCGGTGTGTT and TTTCCGTTCTTTTTCAGTTCGTAGT; DAP2, GGCTGCGTGGTGGTCAC and CGCATTTCGGGGTATATATCC; HAC1i, GCGGGAAACAGTCTACCCTTT and TTCAAACCTGACTGCGTTAT; HAC1 total, TTGGCTATCCCTACCAACTTCAA and TTCCTCTTTTGTCTTGGCTCTTTT; HAC1u, TCCTGAACAAATAGAGCCATTCT and TGCGCTTCGGACAGTACAAG; G6PD (yeast ZWF1), CAGCAATGACTTTCAACATCGAA and CCGGCACGCATCATGAT; depurinated rRNA, ACTAATAGGGAACGTGAGCTG and CCGAATGAACTGTTCCACA; 25S, AGACCGTCGCTTGCTACAAT and ATGACGAGGCATTTGGCTAC; IRE1, TGTTCCCCTAGCGCTTCAGA and CACGGAAAGGCGCTATGC; ACT1 (exon), GGATTCCGGTGATGGTGTTACT and TGGCGTGAGGTAGAGAGAAACC; ACT1 (intron), GGATTCTGGTATGTTCTAGCGC and TCTCTCGAGCAATTGGGACC. Quantification by the comparative CT method was used for three technical replicates from a minimum of two biological replicates and analyzed as part of a comparative CT study using StepOnePlus software version 2.3. A comparative CT study allows for analysis across independent qRT-PCR plates utilizing a single threshold cycle as if run on a single plate, which minimizes plate-to-plate and run-to-run variation. The results of a study are reported as a range of the minimum and maximum levels of expression of a gene for all biological replicates (RQmin and RQmax). The means of the RQmin and RQmax are shown as bar graphs in each figure with the error bar representing the range of RNA abundance from all biological replicates. Reference gene normalization used 25S rRNA for depurination assays and for qRT-PCR assays using purified ribosomes and G6PD for expression analysis. Either VC- or dextrose-grown cells were used for comparative normalization, depending on the assay, as stated throughout.
Statistical analysis
SAS software version 12.3 (SAS/STAT User's Guide, version 9.4, SAS Institute, Cary, NC) was used for statistical analyses. One-way analyses of variance and Fisher's least significant difference (LSD) were applied to the analysis of data in Figs. 1 (B–D), 2, 3 (A–C), 4E, 6 (A–C), 7, and 8 (A and C) and Figs. S1D, S3, and S8 (A, B, and D). Means labeled with different letters show significant differences according to the LSD (p < 0.01 or p < 0.001, as indicated in the figure legends).
Protein lysate preparation and Western blot analysis
Protein lysates were prepared as described previously (34). Total protein concentration was determined by A280, diluted to 5 μg/μl in 8 m urea. An equal volume of 2× SDS loading buffer was added and heated to 95 °C for 5 min, and 50 μg of total protein was separated on a 4–20% SDS-polyacrylamide gel (GenScript) run in TRIS-MOPS-SDS buffer at 140 V for ∼60 min. For Western blots relating to purified ribosomes, equivalent cell amount volumes from cytoplasmic and ribosomal fractions were used. Proteins were transferred to a nitrocellulose membrane using the Trans-Blot Turbo system (Bio-Rad) and blocked in Odyssey Blocking Buffer (LI-COR) prior to primary antibody incubation. Primary antibody dilutions were in Odyssey Blocking Buffer plus 0.1% Tween at the following dilutions: 1:5000 for anti-RTA (custom antibody), 1:2000 for anti-Dpm1 (Thermo Fisher Scientific), 1:5000 for anti-GFP (Roche Applied Science), 1:5000 for anti-HA (Covance), 1:1000 for anti-PGK1 (Thermo Fisher Scientific), 1:4000 for anti-RPL3 (gift of Dr. Jonathan Warner), and 1:500 for polyclonal anti-Hac1. Secondary antibodies, IRDye 800CW goat anti-mouse or goat anti-rabbit (LI-COR), were diluted 1:20,000 in blocking buffer plus 0.1% Tween. Blots were imaged on a LI-COR Odyssey CLx system.
Epifluorescence microscopy
Yeast strain YDP007 (MATa ade2-1 trp1-1 ura3-1 leu2-3, 112 his3-11, 15 can1-100, Δire1, IRE1-GFP::LEU2) was transformed with galactose-inducible yeast expression plasmids containing WT mature RTA (NT1425), R193A/R235A (NT1472), G212E (NT1821), or the vector control (pRS416). Transformants were grown into mid-log in SD−Ura (supplemented with 100 μg/ml myoinositol), pelleted, and resuspended at ∼0.2 OD600/ml in SGal−Ura (supplemented with 100 μg/ml myoinositol) and grown at 30 °C for 6 h to induce RTA expression. At 6 hpi, DTT was added at a final concentration of 5 mm and grown for 90 min. Cells were applied to an agar pad and visualized using an Olympus BX41 fluorescence microscope equipped with a CCD camera (Hamamatsu, Bridgewater, NJ) and a ×100 oil objective (1.45 numerical aperture Plan Apo, Olympus). Image acquisition and processing were performed using Metamorph Image software (version 7.0; MDS Analytical Technologies).
IRE1 expression and purification
Ire1p (Ire1KR32) was purified as a glutathione S-transferase fusion protein expressing IRE1 amino acids 641–1115 encompassing the cytoplasmic kinase and RNase domains using pGEX-6P-2 plasmid (GE Healthcare). GST-IRE1 was expressed in Escherichia coli (BL21 CodonPlus RIPL competent cells; Agilent Technologies, Santa Clara, CA). Expression was induced by the addition of 0.7 mm isopropyl 1-thio-β-d-galactopyranoside and incubation overnight at 22 °C. Cell pellets were resuspended in lysis buffer (20 mm HEPES, pH 7.5, 0.5 m NaCl, 10% glycerol, 5 mm MgCl2, 1 mm DTT) and lysed by sonication. Cell debris was pelleted, and the soluble supernatant was applied to a GSH 4B column (GE Healthcare) and washed with 50 ml of lysis buffer. The fusion protein was eluted with 30 mm reduced GSH Elution Buffer, pH 9.0, digested with PreScission Protease (GE Healthcare) to remove the GST tag and dialyzed overnight at 4 °C against lysis buffer. Dialyzed protein was reloaded onto the GSH 4B column, and flow-through was collected and concentrated to a concentration of 1.5 mg/ml.
In vitro HAC1 RNA processing
Cy5-labeled HAC1 RNA was prepared with the AmpliScribe T7 High Yield Transcription Kit (Lucigen) using linearized pCF150 plasmid as template containing a 600-nucleotide fragment of HAC1 sequence with 181 nucleotides of the 5′ exon, the 252-nucleotide intron, and 167 nucleotides of the 3′ exon. The transcript was purified using a Qiagen RNA cleanup procedure that is part of the RNeasy kit (Qiagen). The cleavage reaction was carried out as described previously (24) with 25 ng of HAC1 RNA and recombinant IRE1 ranging from 100 to 12.5 ng/reaction. Assays with RTA ranging from 25 to 200 nm used the same conditions but with preincubation with 50 ng of HAC1 RNA for 30 min at 30 °C followed by the addition of 50 ng of recombinant IRE1 and incubation at 30 °C for 30 min. Reactions were stopped with an equal volume of 2× TBE-urea sample buffer (Thermo Fisher Scientific), heated to 65 °C for 5 min, applied to a 10% TBE-urea polyacrylamide gel (Bio-Rad), and separated at 100 V in 1× TBE. Cy5 fluorescence was detected using a Typhoon FLA 9500 (GE Healthcare).
Ribosome purification
Ribosomes were isolated from ∼5 × 107 cells harvested at early log phase grown in dextrose- or galactose-selective medium in the presence or absence of 2 mm DTT. Cell pellets were stored at −80 °C prior to breakage by bead beating with chilled glass beads in 0.5 ml of 20 mm HEPES, pH 7.6, 5 mm magnesium acetate, 50 mm KCl, 10% glycerol. The supernatant was collected and clarified of debris by centrifugation for 10 min at 16,000 rpm. Triton X-100 was added to 1% final concentration and mixed by inversion for 10 min at 4 °C and then layered onto 0.8 ml of 20 mm HEPES, pH 7.6, 5 mm magnesium acetate, 50 mm KCl, 25% glycerol. Ribosomes were sedimented for 2 h in a Beckman TLA55 rotor at 54,000 rpm. The top 0.5 ml was removed and stored for Western blotting as the cytoplasmic fraction. The ribosome pellets were collected and stored at −80 °C for RNA purification and Western blot analysis.
Author contributions
M. P. and N. E. T. conceptualization; M. P. and N. E. T. formal analysis; M. P., D. V., J. E. M., and J. N. K. investigation; M. P. and J. E. M. methodology; M. P. writing-original draft; J. N. K. and N. E. T. writing-review and editing; N. E. T. supervision; N. E. T. funding acquisition.
Supplementary Material
Acknowledgments
We are grateful to Dr. Xiao-Ping Li for critical reading of the manuscript and for helpful suggestions. We thank Dr. Peter Walter for plasmid pCF150 containing the HAC1u fragment that served as template for the in vitro transcription reaction, Dr. Alexei Korennykh for the Ire1KR32 expression plasmid used for Ire1p purification, Dr. Madhusudan Dey for polyclonal anti-Hac1p antibodies for Western blot analysis, Dr. Karen Chave (Northeast Biodefense Center Protein Synthesis Core) for purification of RTA variants from E. coli, Dr. Jonathan Warner for monoclonal antibodies against RPL3, and the School of Environmental and Biological Sciences (SEBS) Core Facility for assistance with flow cytometry.
This work was supported by National Institutes of Health Grant AI072425 (to N. E. T.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Figs. S1–S8.
- RIP
- ribosome-inactivating protein
- hpi
- hours postinduction
- RTA
- ricin toxin A chain
- RTB
- ricin toxin B chain
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- UPR
- unfolded protein response
- UPRE
- UPR element
- UTR
- untranslated region
- preRTA
- precursor RTA
- mRTA
- mature RTA
- Tm
- tunicamycin
- VC
- vector control
- qRT-PCR
- quantitative RT-PCR
- Gal
- galactose
- Dex
- dextrose
- OD
- optical density
- LSD
- least significant difference
- nt
- nucleotide.
References
- 1. Audi J., Belson M., Patel M., Schier J., and Osterloh J. (2005) Ricin poisoning: a comprehensive review. JAMA 294, 2342–2351 10.1001/jama.294.18.2342 [DOI] [PubMed] [Google Scholar]
- 2. Liu X. Y., Pop L. M., Schindler J., and Vitetta E. S. (2012) Immunotoxins constructed with chimeric, short-lived anti-cd22 monoclonal antibodies induce less vascular leak without loss of cytotoxicity. mAbs 4, 57–68 10.4161/mabs.4.1.18348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weidle U. H., Tiefenthaler G., Schiller C., Weiss E. H., Georges G., and Brinkmann U. (2014) Prospects of bacterial and plant protein-based immunotoxins for treatment of cancer. Cancer Genomics Proteomics 11, 25–38 [PubMed] [Google Scholar]
- 4. Polito L., Djemil A., and Bortolotti M. (2016) Plant toxin-based immunotoxins for cancer therapy: a short overview. Biomedicines 4, E12 10.3390/biomedicines4020012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spooner R. A., and Lord J. M. (2012) How ricin and shiga toxin reach the cytosol of target cells: retrotranslocation from the endoplasmic reticulum. Curr. Top. Microbiol. Immunol. 357, 19–40 10.1007/82_2011_154 [DOI] [PubMed] [Google Scholar]
- 6. Spooner R. A., Lord J. M. (2015) Ricin trafficking in cells. Toxins (Basel) 7, 49–65 10.3390/toxins7010049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deeks E. D., Cook J. P., Day P. J., Smith D. C., Roberts L. M., and Lord J. M. (2002) The low lysine content of ricin a chain reduces the risk of proteolytic degradation after translocation from the endoplasmic reticulum to the cytosol. Biochemistry 41, 3405–3413 10.1021/bi011580v [DOI] [PubMed] [Google Scholar]
- 8. Walter P., and Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- 9. Korennykh A., and Walter P. (2012) Structural basis of the unfolded protein response. Annu. Rev. Cell Dev. Biol. 28, 251–277 10.1146/annurev-cellbio-101011-155826 [DOI] [PubMed] [Google Scholar]
- 10. Gardner B. M., Pincus D., Gotthardt K., Gallagher C. M., and Walter P. (2013) Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb. Perspect. Biol. 5, a013169 10.1101/cshperspect.a013169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sidrauski C., Cox J. S., and Walter P. (1996) tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87, 405–413 10.1016/S0092-8674(00)81361-6 [DOI] [PubMed] [Google Scholar]
- 12. Cox J. S., and Walter P. (1996) A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87, 391–404 10.1016/S0092-8674(00)81360-4 [DOI] [PubMed] [Google Scholar]
- 13. Rüegsegger U., Leber J. H., and Walter P. (2001) Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 107, 103–114 10.1016/S0092-8674(01)00505-0 [DOI] [PubMed] [Google Scholar]
- 14. Bernales S., Papa F. R., and Walter P. (2006) Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22, 487–508 10.1146/annurev.cellbio.21.122303.120200 [DOI] [PubMed] [Google Scholar]
- 15. Di Santo R., Aboulhouda S., and Weinberg D. E. (2016) The fail-safe mechanism of post-transcriptional silencing of unspliced HAC1 mRNA. eLife 5, e20069 10.7554/eLife.20069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yan Q., Li X. P., and Tumer N. E. (2012) N-Glycosylation does not affect the catalytic activity of ricin a chain but stimulates cytotoxicity by promoting its transport out of the endoplasmic reticulum. Traffic 13, 1508–1521 10.1111/j.1600-0854.2012.01404.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parikh B. A., Tortora A., Li X. P., and Tumer N. E. (2008) Ricin inhibits activation of the unfolded protein response by preventing splicing of the HAC1 mRNA. J. Biol. Chem. 283, 6145–6153 10.1074/jbc.M707981200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang C. T., Jetzt A. E., Cheng J. S., and Cohick W. S. (2011) Inhibition of the unfolded protein response by ricin a-chain enhances its cytotoxicity in mammalian cells. Toxins (Basel) 3, 453–468 10.3390/toxins3050453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li X. P., Kahn P. C., Kahn J. N., Grela P., and Tumer N. E. (2013) Arginine residues on the opposite side of the active site stimulate the catalysis of ribosome depurination by ricin A chain by interacting with the P-protein stalk. J. Biol. Chem. 288, 30270–30284 10.1074/jbc.M113.510966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou Y., Li X. P., Chen B. Y., and Tumer N. E. (2017) Ricin uses arginine 235 as an anchor residue to bind to P-proteins of the ribosomal stalk. Sci. Rep. 7, 42912 10.1038/srep42912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pierce M., Kahn J. N., Chiou J., and Tumer N. E. (2011) Development of a quantitative RT-PCR assay to examine the kinetics of ribosome depurination by ribosome inactivating proteins using Saccharomyces cerevisiae as a model. RNA 17, 201–210 10.1261/rna.2375411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kawahara T., Yanagi H., Yura T., and Mori K. (1997) Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell 8, 1845–1862 10.1091/mbc.8.10.1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aragón T., van Anken E., Pincus D., Serafimova I. M., Korennykh A. V., Rubio C. A., and Walter P. (2009) Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature 457, 736–740 10.1038/nature07641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sidrauski C., and Walter P. (1997) The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90, 1031–1039 10.1016/S0092-8674(00)80369-4 [DOI] [PubMed] [Google Scholar]
- 25. Korennykh A. V., Egea P. F., Korostelev A. A., Finer-Moore J., Zhang C., Shokat K. M., Stroud R. M., and Walter P. (2009) The unfolded protein response signals through high-order assembly of ire1. Nature 457, 687–693 10.1038/nature07661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jetzt A. E., Li X. P., Tumer N. E., and Cohick W. S. (2016) Toxicity of ricin A chain is reduced in mammalian cells by inhibiting its interaction with the ribosome. Toxicol. Appl. Pharmacol. 310, 120–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jetzt A. E., Cheng J. S., Tumer N. E., and Cohick W. S. (2009) Ricin A-chain requires c-jun N-terminal kinase to induce apoptosis in nontransformed epithelial cells. Int. J. Biochem. Cell Biol. 41, 2503–2510 10.1016/j.biocel.2009.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barbieri L., Valbonesi P., Bonora E., Gorini P., Bolognesi A., and Stirpe F. (1997) Polynucleotide:adenosine glycosidase activity of ribosome-inactivating proteins: effect on DNA, RNA and poly(A). Nucleic Acids Res. 25, 518–522 10.1093/nar/25.3.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhabokritsky A., Mansouri S., and Hudak K. A. (2014) Pokeweed antiviral protein alters splicing of HIV-1 RNAs, resulting in reduced virus production. RNA 20, 1238–1247 10.1261/rna.043141.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karran R. A., and Hudak K. A. (2011) Depurination of brome mosaic virus RNA3 inhibits its packaging into virus particles. Nucleic Acids Res. 39, 7209–7222 10.1093/nar/gkr383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hudak K. A., Bauman J. D., and Tumer N. E. (2002) Pokeweed antiviral protein binds to the cap structure of eukaryotic mrna and depurinates the mRNA downstream of the cap. RNA 8, 1148–1159 10.1017/S1355838202026638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ron D. (2009) Targeting of mRNAs to their sites of unconventional splicing in the unfolded protein response. Mol. Cell 34, 133–134 10.1016/j.molcel.2009.04.003 [DOI] [PubMed] [Google Scholar]
- 33. Yanagitani K., Kimata Y., Kadokura H., and Kohno K. (2011) Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science 331, 586–589 10.1126/science.1197142 [DOI] [PubMed] [Google Scholar]
- 34. Chapman R. E., and Walter P. (1997) Translational attenuation mediated by an mRNA intron. Curr. Biol. 7, 850–859 10.1016/S0960-9822(06)00373-3 [DOI] [PubMed] [Google Scholar]
- 35. Sathe L., Bolinger C., Mannan M. A., Dever T. E., and Dey M. (2015) Evidence that base-pairing interaction between intron and mrna leader sequences inhibits initiation of HAC1 mRNA translation in yeast. J. Biol. Chem. 290, 21821–21832 10.1074/jbc.M115.649335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yanagitani K., Imagawa Y., Iwawaki T., Hosoda A., Saito M., Kimata Y., and Kohno K. (2009) Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol. Cell 34, 191–200 10.1016/j.molcel.2009.02.033 [DOI] [PubMed] [Google Scholar]
- 37. Hetz C., Chevet E., and Harding H. P. (2013) Targeting the unfolded protein response in disease. Nat. Rev. Drug Discov. 12, 703–719 10.1038/nrd3976 [DOI] [PubMed] [Google Scholar]
- 38. Jiang D., Niwa M., and Koong A. C. (2015) Targeting the IRE1α-XBP1 branch of the unfolded protein response in human diseases. Semin. Cancer Biol. 33, 48–56 10.1016/j.semcancer.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koong A. C., Chauhan V., and Romero-Ramirez L. (2006) Targeting XBP-1 as a novel anti-cancer strategy. Cancer Biol. Ther. 5, 756–759 10.4161/cbt.5.7.2973 [DOI] [PubMed] [Google Scholar]
- 40. Lee J., and Ozcan U. (2014) Unfolded protein response signaling and metabolic diseases. J. Biol. Chem. 289, 1203–1211 10.1074/jbc.R113.534743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roy C. J., Song K., Sivasubramani S. K., Gardner D. J., and Pincus S. H. (2012) Animal models of ricin toxicosis. Curr. Top. Microbiol. Immunol. 357, 243–257 10.1007/82_2011_173 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.