Abstract
Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activase (Rca) is a AAA+ enzyme that uses ATP to remove inhibitors from the active site of Rubisco, the central carboxylation enzyme of photosynthesis. Rca α and β isoforms exist in most higher plant species, with the α isoform being identical to the β form but having an additional 25–45 amino acids at the Rca C terminus, known as the C-terminal extension (CTE). Rca is inhibited by ADP, and the extent of ADP sensitivity of the Rca complex can be modulated by the CTE of the α isoform, particularly in relation to a disulfide bond structure that is specifically reduced by the redox-regulatory enzyme thioredoxin-f. Here, we introduced single point mutations of Lys-428 in the CTE of Rca-α from wheat (Triticum aestivum) (TaRca2-α). Substitution of Lys-428 with Arg dramatically altered ADP inhibition, independently of thioredoxin-f regulation. We determined that the reduction in ADP inhibition in the K428R variant is not due to a change in ADP affinity, as the apparent constant for ADP binding was not altered by the K428R substitution. Rather, we observed that the K428R substitution strongly increased ATP substrate affinity and ATP-dependent catalytic velocity. These results suggest that the Lys-428 residue is involved in interacting with the γ-phosphate of ATP. Considering that nucleotide-dependent Rca activity regulates Rubisco and thus photosynthesis during fluctuating irradiance, the K428R substitution could potentially provide a mechanism for boosting the performance of wheat grown in the dynamic light environments of the field.
Keywords: ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO); ADP; ATP; thioredoxin; plant biochemistry; plant; C-terminal extension (CTE); carbon fixation; light regulation; Rubisco activase (Rca); wheat
Introduction
Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco;2 EC 4.1.1.39) is the enzyme of photosynthesis that catalyzes the fixation of CO2 into sugars (1). Rubisco is susceptible to inhibition by catalytic misfire products in a process sometimes referred to as fallover (2–4). Indeed, the sugar ribulose 1,5-bisphosophate is both a substrate and an inhibitor of Rubisco if the active site is not initially primed by the binding of a Mg2+ ion and carbamylated with a molecule of CO2 separate from the one catalyzed (5, 6). The removal of inhibitors from the Rubisco active site and hence regulation of Rubisco and ultimately photosynthesis are in large part due to its chaperone, Rubisco activase (Rca) (7, 8). Rca is a member of the AAA+ superfamily of enzymes, a group of ATPases with diverse functions, many of which are chaperones and involved in maintaining the function of partner proteins (9). Rca removes tightly bound sugar substrates and inhibitors from the active site of Rubisco through a mechanism that is not fully characterized for higher plants (10). Difficulty in characterizing the interaction with Rubisco arises because the active multimeric Rca complex is dynamic and altered by cofactors such as nucleotides and Mg2+ as well as concentration- and solvent-dependent self-association (11–15). Structural and mutational analyses suggest that the active holoenzyme consists of a hexamer (16); however, dimers, tetramers, dodecamers, and larger complexes are formed under physiologically relevant conditions (12, 13, 17). Two certainties are that ATP is required by Rca to regenerate Rubisco active sites and that ADP inhibits the activity of Rca (18). This provides a tight control over photosynthesis, which depends on ATP produced in illuminated leaves by the chloroplast electron transport chain. Thus, high irradiance leads to a low ADP/ATP ratio and faster Rca activity, whereas low irradiance leads to a higher ADP/ATP ratio and slower Rca activity, maintaining or reducing Rubisco carboxylation activity, respectively.
In most higher plants, there are two isoforms of Rca due to either alternative splicing of pre-mRNA from a single gene or the presence of two separate genes (19). For the spliced variant, the longer polypeptide is referred to as the α isoform, and the shorter is referred to as the β isoform. The primary structural difference between the two is 25–45 additional C-terminal residues in the α isoform. A conserved characteristic of this C-terminal extension (CTE) is the presence of two cysteine residues that form an intradisulfide bond under nonreduced conditions and require specific reduction by the redox-regulated chloroplast protein thioredoxin-f (20). Thioredoxin-f is reduced by electrons generated from the light-dependent reactions of photosynthesis before reducing disulfide bonds in target proteins (21). In Arabidopsis thaliana (Arabidopsis) reduction of the α isoform CTE by thioredoxin-f reduces ADP inhibition, promotes ATP affinity, and thus provides a secondary mechanism for coordinating the regulation of Rca and Rubisco to light conditions (20, 22). Interestingly, it has been shown that the sensitivity of the α and β isoforms to ADP inhibition is species-specific. For example, in Arabidopsis the nonreduced α isoform is sensitive to ADP inhibition, whereas the β isoform has relatively minimal sensitivity (20, 23). Tobacco (Nicotiana tabacum) expresses a β isoform that is sensitive to ADP (23). Irrespective of the sensitivity of the β isoform, it seems that due to the indiscriminate incorporation of both isoforms into the active heterooligomeric Rca complex, changes in ADP inhibition of the α isoform translate to inhibition characteristics of the final active holoenzyme (22, 23). Little is known about the ADP inhibition of Rca from monocot species. Rice (Oryza sativa) and wheat (Triticum aestivum) express both an α and β isoform, with wheat expressing an α isoform (TaRca2-α), the spliced TaRca2-β variant, and a unique β isoform encoded by a separate gene, referred to as TaRca1-β (24, 25). The ADP sensitivity of the α and β isoforms in rice, wheat and between the two wheat β isoforms is not known.
Here, we explore the in vitro ADP inhibition sensitivity of the TaRca2-α, TaRca2-β, and TaRca1-β isoforms of Rca in wheat. We made residue substitutions to the CTE of the TaRca2-α isoform to determine its effect on ADP inhibition. Substitutions were made at position Lys-428, as this residue has recently been identified in Arabidopsis as being a site for ADP regulation through acetylation control (26). We also replaced the cysteine at position 441 to remove the disulfide bond in the nonreduced α isoform as this is also known to affect ADP inhibition in Arabidopsis (20). Substitutions at these two positions of the TaRca2-α CTE show pronounced effects on nucleotide binding and catalysis, providing insight into the C-terminal regulation of Rca–nucleotide interactions.
Results
In this study, we characterized the sensitivity of His-tagged recombinant Rca to ADP inhibition. Initially, Rca from wheat and rice was screened followed by a more in-depth analysis of wheat Rca, including residue substitutions to the TaRca2-α CTE and TaRca1-β C terminus.
The Rca α and β isoforms of wheat and rice were both highly sensitive to ADP inhibition (Fig. 1). At an ADP/ATP fraction of 0.1 (i.e. 10% of the nucleotide content being ADP), the activation velocity of Rca was more than halved relative to velocities in the absence of ADP. The TaRca2-β isoform of wheat was significantly less ADP-sensitive than the α isoform across the entire range of ADP inhibition concentrations. For rice, the β isoform seemed to be less sensitive as well, especially at a fraction of 0.1 ADP/ATP (Fig. 1B). Despite these subtle differences between isoform sensitivities, the β isoform was still highly sensitive to ADP inhibition with both isoforms from both species showing less than 10% of their maximal activity at an ADP/ATP fraction of 0.4.
Figure 1.
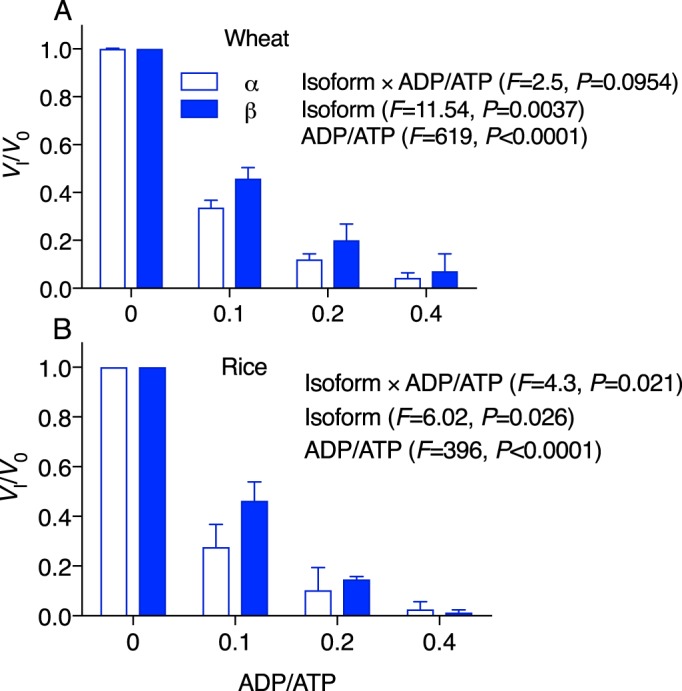
ADP inhibition of Rca for wheat (TaRca2) (A) and rice (OsRca) (B) α and β isoforms. Rca velocity in the presence of 5 mm ATP substrate and varying concentrations of ADP inhibitor was normalized to initial velocity in the absence of inhibitor (Vi/V0) and plotted against the fraction of ADP to ATP. Values are the means ± S.D. (error bars) of three experimental replicates. A two-way ANOVA was performed for each species, with the F and p values of the interaction and each individual factor reported on the graph.
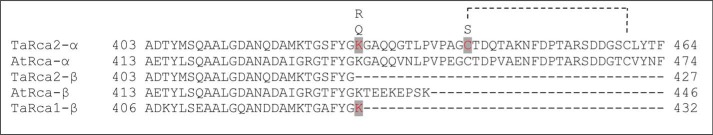
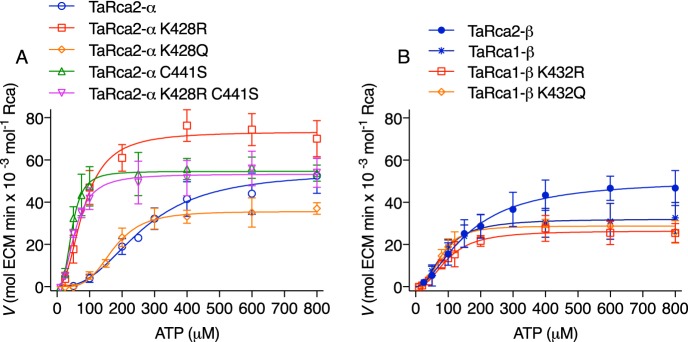
The substitutions K428Q, K428R, and C441S and the double substitution K428R/C441S were introduced to the CTE of the wheat TaRca2-α isoform (Fig. 2). K432Q and K432R substitutions were also made to the corresponding Lys-432 in the TaRca1-β isoform. Substitution to a Gln and Arg were chosen as they are conservative in terms of size or size/polarity and have been used as Lys acetylation and nonacetylated mimics, respectively (27). There is no corresponding Lys residue in the TaRca2-β variant. To determine the importance of these changes on nucleotide binding and catalysis, kinetic curves were generated plotting ATP substrate versus the Rubisco activation velocity of Rca (Fig. 3). For all variants of Rca, an allosteric sigmoidal response of Rca activity to ATP concentration was observed, with a Hill slope ranging from 1.8 to 2.8 but no significant difference between any of the variants (Table 1). Preliminary experiments performed to optimize assay conditions indicated ATP substrate inhibition of Rca at concentrations above 2 mm (Fig. S1). Thus, the ATP-dependent Vmax was determined only at physiologically relevant concentrations to a maximum of 800 μm ATP. Of further note, preliminary experiments demonstrated that inhibited Rubisco (ER) was not at saturating substrate concentrations for Rca, but to increase ER substrate would have sped the reaction above the detection range of the spectrophotometric assay (Fig. S2). To use less Rca in assays was not viable due to the need for concentration-dependent self-association of the Rca enzyme. Thus, the Vmax obtained in this study only relates to the standardized conditions under which the Rca was assayed.
Figure 2.
C terminus alignment of wheat (TaRca) and Arabidopsis (AtRca) α and β Rca isoforms indicating where residue substitutions were made in this study. The amino acid positions 428 for the wheat TaRca2-α isoform and 432 for the wheat TaRca1-β isoform were substituted from a native Lys to either an Arg or Gln. The Cys at position 441 of TaRca2-α isoform was substituted to a Ser to emulate the reduction of the Cys-441–Cys-461 disulfide bond state of the α isoform. The dashed lines indicate where the disulfide bond is formed.
Figure 3.
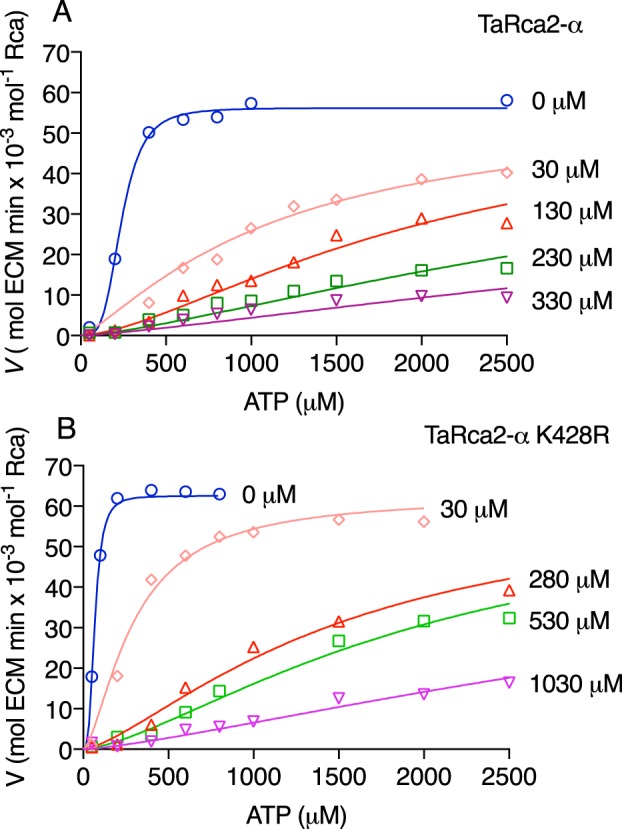
ATP substrate-dependent enzyme kinetic curves of wheat Rca. A, TaRca2-α isoform and derived CTE variants. B, β isoforms and TaRca1-β–derived variants. The enzymatic velocity of Rubisco reactivation by Rca (ECM-regenerated reactions per minute per Rca monomer) were plotted against ATP concentrations added to assays. An ATP-regenerating system using phosphocreatine and creatine phosphokinase was used to limit ADP buildup due to ATP hydrolysis by Rca. Curves are Hill equations (V = Vmax × Sh/(Khalfh + Sh)) where Khalf is the ATP substrate corresponding to half-maximal velocity in μm, h is the Hill slope constant, and S is the ATP substrate concentration in μm. The Vmax, Khalf, and h values were generated from iterative fits using a least-squares model, and values for each curve are presented in Table 1. Values are the means ± S.D. (error bars) of three to five experimental replicates.
Table 1.
ATP- and ADP-dependent kinetic parameters of wheat Rca
Shown are the ATP-dependent Vmax, ATP concentration required to reach half-maximal velocity (Khalf), Hill slopes (h), and IC50 of ADP for the wheat isoforms TaRca2-α, TaRca2-β and TaRca1-β as well as C terminus variants. Superscript letters refer to significant differences in kinetic parameters between Rca variants at p ≤ 0.05 using a one-way ANOVA and Tukey's multiple comparison test. Values are the means and S.D. of three to six experimental replicates.
| Vmax | Khalf | h (constant) | IC50 | |
|---|---|---|---|---|
| ECM min × 10−3 | μm | μm | ||
| TaRca2-α | 55 ± 11a | 269 ± 18a | 2.7 ± 0.4a | 14.2 ± 2a |
| TaRca2-α K428Q | 38 ± 7b,c | 160 ± 33b | 2.8 ± 0.6a | 42 ± 5b |
| TaRca2-α K428R | 72 ± 9d | 72 ± 11c,d | 2.5 ± 0.3a | 57 ± 8b,c |
| TaRca2-α C441S | 55 ± 6a | 48 ± 8c | 2.7 ± 0.6a | 67 ± 10b,c |
| TaRca2-α K428R/C441S | 53 ± 9a | 56 ± 7c | 2.5 ± 0.4a | 65 ± 10b,c |
| TaRca2-β | 51 ± 5a,b | 165 ± 28b | 1.8 ± 0.4a | 29 ± 10a |
| TaRca1-β | 32 ± 8c | 81 ± 13c,d | 2.6 ± 0.7a | 58 ± 6b,c |
| TaRca1-β K428Q | 34 ± 6c | 81 ± 12c,d | 2.7 ± 0.6a | 47 ± 7b,c |
| TaRca1-β K428R | 26 ± 8c | 90 ± 16d | 2.2 ± 0.4a | 66 ± 14c |
The TaRca2-α K428Q variant had a significantly slower Vmax, indicating that a nonbasic amino acid at this position reduces activity. The TaRca2-α K428R variant had a faster Vmax than the TaRca2-α WT (Fig. 3A and Table 1). The two variants TaRca2-α C441S and TaRca2-α K428R/C441S did not have significantly different Vmax values from the TaRca2-α WT. The ATP substrate concentration that corresponded with half-maximal velocity (Khalf) was significantly reduced for all TaRca2-α variants, indicating an increase in ATP affinity. Khalf of TaRca2-α K428Q was intermediate between the TaRca2-α WT and its K428R variant. The K428R variant had a Khalf that was not significantly different from that of the Cys-441 variants, which were similar and had the lowest Khalf values of all TaRca2-α variants.
The TaRca2-β isoform had a greater affinity for ATP than the TaRca2-α splice variant, evident in its significantly lower Khalf (Fig. 3 and Table 1). The TaRca1-β variant, coded by a separate gene and with substantial amino acid sequence differences from the Rca2 spliced variants, had even higher affinity for ATP with a Khalf significantly less than both TaRca2-α and -β isoforms and not significantly different from the TaRca2-α K428R and C441S variants. TaRca1-β had a significantly slower Vmax than the TaRca2-α and -β spliced variants. Gln or Arg substitution at Lys-432 of the TaRca1-β C terminus, which corresponds to the Lys-428 residue of TaRca2-α, did not have any significant effect on Vmax or Khalf relative to the TaRca1-β WT.
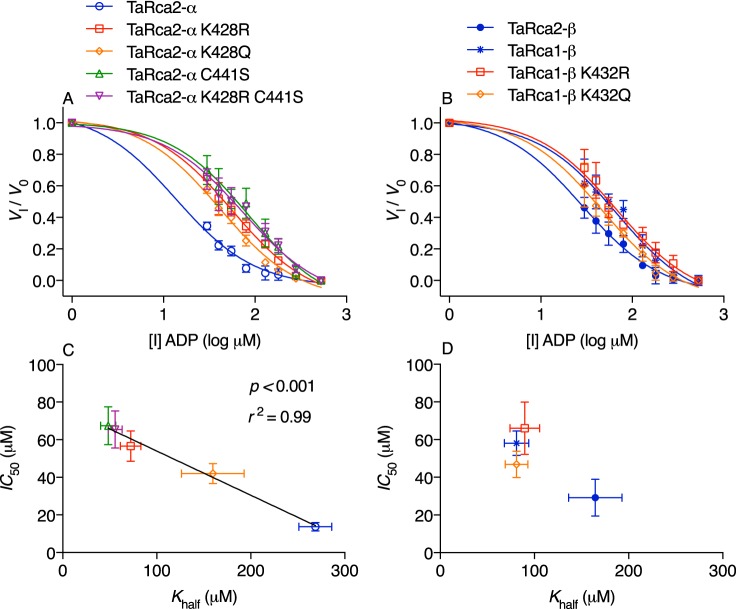
As with Khalf, there were significant differences in ADP inhibition of Rca between the WT isoforms and modified variants (Fig. 4 and Table 1). The TaRca2-α isoform was most susceptible to ADP inhibition with an ADP IC50 significantly below all other variants studied (Fig. 4, A and C). We plotted IC50 against Khalf values and observed a highly significant negative linear correlation between IC50 and Khalf values, indicating that an increase in apparent affinity for ATP (i.e. reduced Khalf) was associated with a reduction in ADP inhibitor sensitivity (i.e. increased IC50) (Fig. 4C). As with Khalf values, Lys-432 substitutions had no significant effect on the IC50 of TaRca1-β. The TaRca2-β WT had an intermediate IC50 among the WT isoforms (Fig. 4D).
Figure 4.
Inhibitor dose-response curves of wheat Rca. A and B, Rca velocity in the presence of varying concentrations of ADP inhibitor (I) was modeled by ordinary least-squares dose-response curves using the following equation: Vi/V0 = Vi/V0 min + (Vi/V0 max − Vi/V0 min)/1 + 10I−logIC50) with the IC50 value corresponding to the ADP inhibitor concentration in μm at which half-inhibitor free velocity is reached. The log-transformed ADP concentrations added to assays are given on the x axis; 500 μm ATP substrate was added in all assays. Calculated IC50 values are presented in Table 1. C and D, scatter plots of IC50 and Khalf for the Rca variants and fit with a linear regression model in the case of the α isoform and CTE variants. Values are the means ± S.D. (error bars) of three to five experimental replicates.
To explore the interaction between nucleotide binding and ADP inhibition further, ATP substrate kinetics at differing ADP inhibitor concentrations were performed for TaRca2-α and its most ADP-insensitive substitution, K428R (Fig. 5). Ordinary least-squares model fitting analysis of the kinetic response to ATP and ADP was used to determine the inhibitor mechanism (i.e. competitive versus noncompetitive) and the apparent inhibition binding constant (Ki) of ADP. A competitive-inhibition model provided a close fit to observations with a global R2 of > 0.98 for both the TaRca2-α and K428R substitution, whereas noncompetitive model analysis gave ambiguous parameter estimations (Table S1). There was no significant difference in Ki values of ADP between the TaRca2-α WT and K428R variant with calculated values of 4.9 ± 1.5 μm for TaRca2-α and 3.4 ± 1.4 μm for TaRca2-α K428R.
Figure 5.
ATP substrate kinetics at differing ADP inhibitor concentrations for TaRca2-α and the ADP-insensitive K428R substitution. Activation velocities of TaRca2-α WT (A) and the K428R substitution (B) with varying concentrations of ADP inhibitor (I) added as indicated on the right of individual curves. A competitive-inhibition model fitted the data best (global R2 > 0.98 for both variants) using the following equations: Khalf-Obs = Khalfh(1 + I/Ki) and V = Vmax × Sh/(Khalf-Obs + Sh) where Khalf is the ATP substrate corresponding to half-maximal velocity in μm, h is the Hill slope, S is the ATP substrate concentration, and I is the ADP inhibitor concentration in μm. The apparent inhibition binding constant (Ki) was determined to be 4.9 ± 1.5 μm for TaRca2-α and 3.4 ± 1.4 μm for K428R through iteration of the curves.
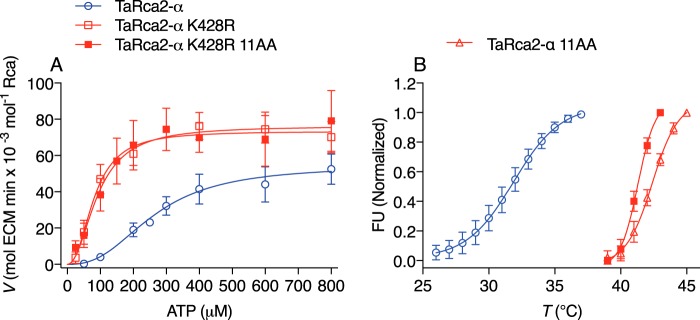
We recently reported on 11 residue substitutions that impart a 7 °C increase in the thermostability of TaRca2-α (28). These 11 residue substitutions were combined with the TaRca2-α K428R substitution we report here, with gains in both thermal stability and ADP insensitivity achieved (Fig. 6). Thus, thermal stability and ADP sensitivity seem to be independent properties of the Rca enzyme.
Figure 6.
The interaction of ATP substrate kinetics and thermostability. A, ATP substrate–dependent enzyme kinetic curves of the TaRca2-α WT, K428R substitution, and the K428R substitution combined with 11 residue substitutions (K428R 11AA) corresponding to a recently reported (28) thermostable isoform. B, thermostability profiling using differential scanning fluorimetry of the TaRca2-α WT, the recently reported thermostable isoform (TaRca2-α 11AA), and the combined K428R 11AA variant. FU refers to fluorescent units, and T refers to temperature. The point of inflection of the fluorescence curves corresponds to the thermal midpoint (50% enzymatic activity) and is 31.9 ± 0.2 °C for TaRca2-α WT, 42.4 ± 0.1 °C for the 11-amino acid (AA) thermostable variant, and 41.3 ± 0.1 °C for the combined K428R 11AA variant. Values are the means ± S.D. (error bars) of three experimental replicates.
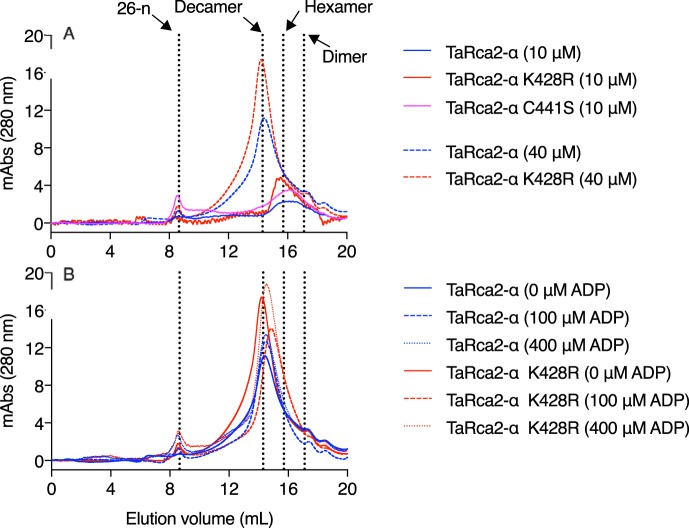
Finally, gel-filtration chromatography was performed to determine whether ADP inhibition sensitivity was associated with changes in oligomerization (Fig. 7). The TaRca2-α K428R and C441S substitutions did not affect oligomer size of TaRca2-α, with a hexamer forming at Rca concentrations of 10 μm. At higher Rca concentrations of 40 μm, the complex took the form of a decamer under the prevailing experimental conditions. Incubation of TaRca2-α or the ADP-insensitive TaRca2-α K428R variant at a concentration of 40 μm and with varying ADP concentrations did not substantially change oligomer size (Fig. 7B). It therefore seems that sensitivity to ADP inhibition was not associated with ADP-dependent changes in oligomer size, at least for the specific conditions analyzed in this study.
Figure 7.
Gel-filtration chromatograms showing the effect of Rca and ADP concentration on the Rca oligomeric complex. Chromatograms are presented as milli-absorbance (mAbs) monitored at a wavelength of 280 nm. A, TaRca2-α and its K428R and C441S substitution variants at a concentration of 10 (solid lines) or 40 μm (dashed lines). B, TaRca2-α and TaRca2-α K428R at a concentration of 40 μm and incubated with no ADP (solid lines) or 100 (dashed lines) or 400 μm (dotted lines) ADP. The elution volumes corresponding to a 26-mer (26-n), decamer, hexamer, and dimer of the TaRca2-α isoform are indicated by the vertical dotted lines.
Discussion
The ADP inhibition of Rca is an important photosynthetic regulatory mechanism in higher plants. The extent of Rca sensitivity to inhibition by ADP seems to be species- and isoform-dependent and can be modulated by the CTE of the α isoform (20, 22, 23), although the extent to which this occurs in monocots has not yet been established. As such, we determined that for both wheat and rice the two isoforms of Rca are sensitive to ADP inhibition based on His-tagged recombinantly purified enzymes. Furthermore, residue substitutions in the wheat α CTE can have a dramatic influence on the extent of ADP sensitivity. We established that ADP inhibition of TaRca2-α depends on ATP but not ADP affinity, and the results we obtained provide a potential mechanism by which photosynthetic performance in wheat might be improved.
In wheat chloroplasts, the ratio of ADP/ATP has been described to vary from 1:3 in the light to 1:1 in the dark (29). Even at the 1:3 expected ratio of ADP/ATP in the light, when photosynthesis is occurring, we observed a substantial inhibition of both Rca α and β activity under in vitro measuring conditions, in the absence of the associated reducing enzyme thioredoxin-f. The results we obtained clearly indicate that for wheat and rice both alternatively spliced isoforms of Rca are sensitive to ADP inhibition (Fig. 1). This sensitivity of both isoforms is in contrast to Arabidopsis, which has been characterized to have an insensitive β isoform (20, 22, 23, 30). However, we show the TaRca1-β isoform of wheat to be significantly less sensitive to ADP inhibition relative to the spliced TaRca2 α and β variants, and as such it may play a role similar to that of the insensitive β isoform of Arabidopsis. A comparison of ADP sensitivity for wheat and rice Rca isoforms presented here and Arabidopsis and tobacco Rca isoforms measured previously (22, 23) is illustrated in Fig. 8.
Figure 8.
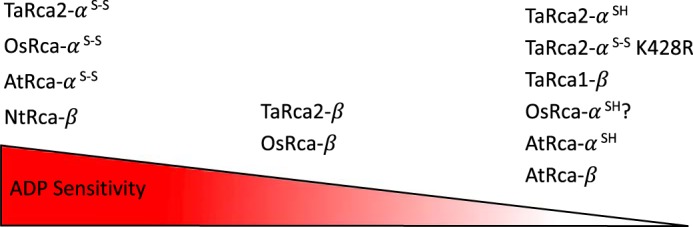
An illustration of ADP sensitivity between species and isoforms of Rca discussed in this report. ADP sensitivity of Rca from wheat (TaRca) and rice (OsRca) measured in this study and from previously reported Arabidopsis (AtRca) and tobacco (NtRca) isoforms are compared. For α isoforms, superscript S–S and SH indicate disulfide bond formation and lack thereof between the conserved cysteine residues of the CTE, respectively. ? indicates an expected but not measured level of ADP sensitivity.
We have recently reported that the wheat TaRca1-β isoform has low gene expression under standard physiological conditions but is induced by heat stress relative to TaRca2 isoforms and is a more thermotolerant variant of wheat Rca (28). We presume the thermal stability of TaRca1-β and its relative insensitivity to ADP inhibition are physiologically linked, as it is likely that heat stress would result in higher ADP/ATP ratios considering that (i) the ATP synthase complex itself seems susceptible to oxidative damage from abiotic stress such as heat (31, 32) and (ii) substrate supply for ATP through the proton-motive force may be reduced as electron transport through the thylakoid membranes is known to be disrupted by heat (33). Furthermore, we demonstrate that improvements to both thermostability and ADP insensitivity can be made to a single Rca isoform, with one property not detrimentally affecting the other (Fig. 6).
There was an allosteric sigmoidal response of Rca activity to ATP concentration, indicating positive allosteric interaction between Rca monomeric subunits driven by ATP (Fig. 3). A similar response is observed in spinach (Spinacia oleracea) and Arabidopsis (13, 22, 34). Positive cooperativity of the ATP hydrolysis of tobacco Rca has also been observed, interestingly with the requirement of both ATP and ADP (35). It is postulated that ATP is required to drive formation of the Rca complex separate from its involvement in Rubisco activation, and indeed multiple studies have shown a positive relationship between the formation of the active complex and ATP concentrations (12, 13, 15). With no significant difference in Hill slopes, it seems alterations to the CTE do not change the extent of this positive interaction between subunits. Interestingly, ATP became a mild substrate inhibitor of Rca activity at ATP concentrations of above 2 mm. Although it is unlikely that such high ATP concentrations have any physiological relevance, this observation is of interest as some in vitro studies use saturating ATP concentrations above 2 mm for the analysis of Rca activity (36, 37), and at such high concentrations the optimal activity of Rca might not be reached. The reduced velocity with higher ATP concentrations is likely linked to the influence ATP and ADP adenylates have over oligomer formation and reassembly (17), and presumably at this higher ATP concentration wheat Rca is in a larger oligomeric conformation with reduced functional capacity. Interestingly, however, the oligomeric sizes of TaRca2-α and TaRca2-α K428R were not responsive to differences in ADP concentration despite differences in ADP sensitivity (Fig. 7).
Under physiologically relevant concentrations and ratios of ATP and ADP, the ADP IC50 was 4-fold greater for the TaRca2-α K428R variant than for TaRca2-α WT. ADP inhibition was associated with ATP binding affinity, evident in a 3-fold lower Khalf of the K428R variant relative to the TaRca2-α WT. Furthermore, an analysis of TaRca2-α and all associated variants in this study shows a strong positive correlation between apparent ATP affinity, extrapolated from reduced Khalf values, and reduced ADP inhibition (Fig. 4C). Similarly, studies of Arabidopsis Rca showed reduced ADP inhibition associated with increased ATP affinity when the α isoform C-terminal disulfide bond was reduced by thioredoxin-f or through cysteine substitution (20, 22). The inhibition kinetic curves we generated for TaRca2-α and its K428R variant indicate that ADP is a competitive inhibitor at the Rca nucleotide-binding site (Fig. 5). Similarly, ADP was found to act as a competitive inhibitor at the ATP-binding site of tobacco Rca, with substantially tighter binding of ADP than ATP (35). Considering ADP acts through competitive inhibition, an improvement in ATP affinity would result in reduced ADP inhibition as observed, but only if ADP affinity did not change correspondingly. Indeed, despite the dramatic difference in ADP inhibition between TaRca2-α and TaRca2-α K428R, the apparent Ki of ADP was not significantly different between these two Rca variants (Fig. 5). In effect, we demonstrate that structural changes arising from the TaRca2-α K428R CTE substitution dramatically influence ATP binding but have no discernable influence on ADP binding. A similar conclusion was drawn from the results pertaining to Arabidopsis CTE Cys substitutions (38). The fact that we observed a dramatic increase in ATP binding affinity, matching the change in binding affinity arising from the loss of a disulfide bond but with only a single residue substitution of a positively charged residue (Lys) to another (Arg), is highly surprising. Lys acetylation can have dramatic influence on protein characteristics (27), and the corresponding Lys-428 residue is acetylated in Arabidopsis (26). However, this post-translational modification has not been reported in monocots. Further experimentation is required to determine whether the changes in catalytic properties arising from the TaRca2-α K428R substitutions are equivalent to deacetylation of Lys-428 in the native protein. Irrespective of the reasoning, considering that ATP and ADP only differ by one Pi, nucleotide interference by the K428R CTE substitution must be highly specific and, in some way, related to the γ-phosphate of ATP. Although not measured, the tight relationship we observed between the ATP Khalf and ADP IC50 across all TaRca2-α variants suggests that ATP affinity and not ADP binding is generally what drives ADP inhibition sensitivity by the CTE of the α isoform of Rca.
The changes in ATP substrate–dependent Vmax for Lys-428 substitutions of TaRca2-α demonstrate that changes at this position not only influence ATP binding to the nucleotide active site but also catalytic properties subsequent to binding. As the C terminus is reported to affect subunit interaction close to the nucleotide-binding pocket of adjacent monomers in the Rca complex (16, 38), we postulate that changes in the catalysis of ATP as a result of a single residue substitution at position Lys-428 might be a consequence of changes in allosteric interactions between adjacent Rca monomers that influence nucleotide active sites in the oligomeric complex. The lack of any difference in Vmax between the TaRca2-α WT and TaRca2-α variants with the C441S substitution suggests that the disulfide bond structure of the CTE is acting through separate mechanisms from that of the TaRca2-α Lys-428 substitutions. The former only imparts effects on the binding of nucleotide, whereas the latter effects binding and catalysis. Furthermore, the nonsignificant difference in Khalf between the TaRca2-α K428R variant containing a nonreduced disulfide bond and the TaRca2-α C441S variant mimicking disulfide bond reduction shows that the impact of the Lys-to-Arg substitution at position 428 on ATP binding matches the effect of disulfide bond reduction on ATP affinity. However, the loss of the greater Vmax of the TaRca2-α K428R single substitution with the TaRca2-α K428R/C441S double substitution (Fig. 3 and Table 1) suggests that there is some cumulative effect of the two sites on ATP kinetics.
Of interest, the Arabidopsis β isoform has a C-terminal Lys residue (Lys-438) that may influence the ADP sensitivity of Rca activity (26), justifying interest in this amino acid in wheat. This is equivalent to the Lys-428 residue in the TaRca2-α CTE but is not present in TaRca2-β (Fig. 2). Hartl et al. (26) found that the ATP hydrolysis activity of an Arabidopsis AtRca-β K438Q substitution had reduced ADP sensitivity, aligning with the reduced activation velocity we observed for K428Q in the α isoform of wheat. However, an AtRca-β K438R substitution resulted in greater ADP inhibition as well as substantially reduced ATP hydrolysis independent of ADP, both inconsistent with the faster activation velocity and ADP insensitivity we observed in the TaRca2-α K428R variant. Furthermore, we substituted the corresponding Lys residue in the TaRca1-β isoform, which in contrast to TaRca2-β does contain a Lys residue at the equivalent TaRca2-α Lys-428 position (Fig. 2), and found neither a Gln nor an Arg substitution to substantially influence ATP-dependent activation velocity, ATP affinity, or ADP inhibition (Figs. 3B and 4, B and D). It therefore seems that the Lys position in question influences nucleotide interaction for the β isoform in Arabidopsis as well as the α isoform in wheat, but differently depending on the residue substitution, and has no influence on TaRca1-β ATP or ADP interaction. Conformational changes in oligomeric structure specific to each Rca variant likely determine the extent to which this C-terminal Lys impacts ATP binding and Rca catalysis. Obtaining a crystal or high-resolution cryo-EM structure of the Rca CTE in future studies is needed for reliable protein structural modeling analysis to answer some of the questions of how the C terminus primary sequence influences ATP binding and catalysis. Another useful experiment in future studies would be to determine whether the improved ATP affinity of TaRca2-α K428R is additive or synergistic to heterooligomers comprising the K428R variant and the TaRca2-α WT isoform.
Finally, in a previous study, an Arabidopsis mutant only expressing the ADP-insensitive β isoform showed faster light induction of nonsteady-state net photosynthesis but had no demonstrable impact on photosynthesis once the steady state had been reached (23). Above-ground biomass for the Arabidopsis mutant increased when irradiance fluctuated from low to high levels on a regular basis. One might therefore expect that under field conditions, with variable light due to factors such as wind-dependent dynamic leaf shading and variable cloud cover, an enhancement of wheat Rca light responsiveness will potentially lead to an increased biomass accumulation and, ultimately, yield. Analysis of wheat expressing Rca variants displaying decreased sensitivity to ADP inhibition over the longer-term, including growth and developmental indicators, would be needed to answer such a question. However, the use of precise gene editing technology to edit wheat genes was recently shown to be feasible (39). Additionally, only one nucleotide mutation (i.e. AAA to AGA) is needed to transform a Lys to an Arg and convert WT wheat Rca into the strong ADP-insensitive variant. The TaRca2-α K428R variant we have characterized is therefore a prime candidate for wheat improvement through gene editing technology.
Experimental procedures
Recombinant Rca protein generation
All Rca genes of interest were synthesized de novo (GENEWIZ, South Plainfield, NJ) with 46 amino acids at the N terminus corresponding to the signal peptide deleted and a 6-amino-acid His tag attached to the C terminus. Genes were ligated into Novagen pET-23d+ vectors (Merck KGaA, Darmstadt, Germany) before being transformed into BL21(DE3) Star Escherichia coli strain following standard procedures. 0.5 liter of lysogeny broth cultures was grown in 2-liter conical flasks at 37 °C and shaken at 180 rpm. Cultures were induced with 0.8 mm isopropyl 1-thio-β-d-galactopyranoside at an OD600 of 0.8–1 and then grown for a further 17 h at 20 °C. All subsequent steps were performed at 4 °C. Cells were removed from culture media by centrifugation at 6000 × g for 20 min and then lysed by sonication for 10 s for five cycles at 16-μm amplitude (Soniprep 150, MSE, London, UK). Purification was performed using an ÄKTA pure protein purification system and 5-ml HisTrap FF columns (GE Healthcare) following the manufacturer's instructions. Final Rca recombinant protein was desalted into a buffer containing 20 mm Tris-HCl, pH 8.0, 0.2 mm EDTA, 7.5 mm MgCl2, 1 mm DTT, and 50 mm KCl at a concentration of 2.2 ± 0.4 mg ml−1; snap frozen in liquid nitrogen; and stored at −80 °C until use. Rca recombinant protein concentration was determined using Protein Assay Dye Reagent Concentrate (Bio-Rad) with a bovine serum albumin (BSA) standard. Final purity of Rca was greater than 80% (Fig. S3).
Rubisco isolation
Rubisco was extracted from the leaves of T. aestivum cv. Fielder (Bread Wheat) grown in a greenhouse in Ghent, Belgium between April and August 2017 with a night temperature of 18 ± 1 °C, day temperature of 20 ± 1 °C, and a 16-h supplemented light period with a photoactive radiation of 375 μmol of photons m2 s−1 under standard physiological conditions. Leaves were harvested between 4 and 6 h into the light period, immediately frozen in liquid N2, and stored at −80 °C until extraction. Frozen leaf tissue was ground into a fine powder using liquid N2 and a mortar and pestle. While on ice, leaf powder was added to and repeatedly vortexed in an extraction buffer consisting of 100 mm Tris-HCl, pH 8.0, 1 mm EDTA, 10 mm MgCl2, 2 mm DTT, 2% (w/v) polyvinylpolypyrrolidone, and plant specific protease inhibitor mixture (Merck KGaA), before being passed through a single layer of Miracloth and fine cotton gauze to remove solid matter. The sample was spun at 24,000 × g for 20 min at 4 °C, and the supernatant was retained. 35% (v/v) saturated ammonium sulfate was added, and the sample was incubated on ice for 30 min before respinning. Saturated ammonium sulfate was added dropwise to the supernatant to a final concentration of 60% (v/v) and slowly stirred at 4 °C for 30 min before being spun again. The resulting pellet was suspended in a sample buffer of 100 mm Tricine-NaOH, pH 8.0, and 0.5 mm EDTA and desalted into the same buffer using PD-10 desalting columns. 20% glycerol was added, and the sample was aliquoted into 50-μl volumes before being snap frozen in liquid nitrogen and stored at −80 °C until use.
Rubisco activation assays
The velocity of Rca in activating Rubisco was measured following the ADP-insensitive coupled-enzyme spectrophotometric method of Scales et al. (36) with the following modifications. All reagents were purchased from Merck KGaA except for d-2,3-phosphoglycerate mutase, which was expressed and purified as outlined previously (36). The assay was scaled down to 100-μl reactions and measured in Costar® 96-well flat-bottom polystyrene plates (Corning), heated to 25 °C using a Thermomixer (Eppendorf). In one set of wells, a reaction solution, with final volume of 80 μl, consisting of N2-sparged Milli-Q H2O, 5% (w/v) PEG-4000, 100 mm Tricine, pH 8, 10 mm MgCl2, 10 mm NaHCO3, 5 mm DTT, 2.4 units of enolase, 3.75 units of phosphoenolpyruvate carboxylase, 6 units of malate dehydrogenase, 0.2 mm 2,3-bisphosphoglycerate, 4 units of d-2,3-phosphoglycerate mutase, 10 units of carbonic anhydrase, and 0.8 mm NADH was added. When ADP inhibition was not being measured, an ATP-regenerating system consisting of 4 mm phosphocreatine and 20 units of creatine phosphokinase was added. ATP and ADP were added at concentrations indicated in the text. In another set of wells, a final volume of 20 μl consisted of 0.2 ± 0.05 μm Rubisco active sites added to either 1) an activation solution (N2-sparged Milli-Q H2O, 20 mm Tricine-NaOH, pH 8, 20 mm NaHCO3, and 10 mm MgCl2) to determine fully carbamylated Rubisco (ECM) activity or 2) 4 mm ribulose 1,5-bisphosphate (99% pure) for Rubisco substrate inhibition (ER). Two minutes prior to measurements, 3.7–4.6 μl (1.6 μm protomer) of Rca was added to ER wells as a separate droplet from the Rubisco solution. Rca was not added to ER samples when measuring spontaneous baseline activity. 10 min after addition of Rubisco, the contents of the reaction solution wells were added to the Rubisco-containing wells by multichannel pipette, and measurements of absorbance at a wavelength of 340 nm were immediately made on an Infinite M200 Pro plate reader (TECAN, Männedorf, Switzerland) every 15 s over an 8-min period. Up to 10 samples were assayed simultaneously. The quantification of ECM-regenerated reactions by Rca per minute (mol of ECM min × 10−3 mol−1 Rca) was calculated by the method outlined by Loganathan et al. (40) over the first 4-min period of measurements (see Fig. S4 for an example). The amount of Rubisco active sites added to the assay was determined from the slope of a linear regression through the data points corresponding to the first 60 s of 3-phosphoglycetic acid product generated from ECM samples and factoring in a wheat Rubisco reaction rate constant (Kcat) of 2.1 at 25 °C (41). Differential scanning fluorimetry was used to determine the conformational thermal stability of Rca as described previously (28).
Gel-filtration chromatography
All gel-filtration chromatography experiments used Rca desalting buffer as described above. Rca was diluted to 10 or 40 μm concentration in a 200-μl volume. The samples were incubated on ice for 2 h with varying ADP concentrations as indicated. Samples were then loaded onto a Superose 6 10/300 GL column (GE Healthcare) equilibrated with 2 column volumes of buffer and eluted at 0.5 ml min−1 at 4 °C. ADP was added to the buffer to match the sample incubation ADP concentrations. Size markers were generated by a standard curve (Fig. S5) using blue dextran 2000 to determine the column void volume, thyroglobulin (669,000 Da), ferritin (440,000 Da), aldolase (158,000 Da), conalbumin (75,000 Da), and ovalbumin (44,000 Da) using a Gel Filtration High Molecular Weight Calibration kit (GE Healthcare) following the manufacturer's instructions.
Statistics and data analysis
All data and statistical analyses were carried out using GraphPad Prism 5.0 software (GraphPad Prism Software Inc., San Diego, CA). Two-way ANOVAs were performed to compare ADP-dependent wheat and rice Rca isoform velocity. For enzyme kinetic analysis, GraphPad Prism ordinary least-squares models with best fit to the data were chosen, with iteration used to determine unknown parameters of interest. For Rca velocity versus ATP substrate concentration, the Hill equation (V = Vmax × Sh/(Khalfh + Sh) was used. It was noted that ATP-dependent velocity of Rca differed when the ATP regeneration system was not included in assays. We attribute this to self-inhibition as we expect hydrolysis of ATP to ADP by Rca over the duration of the assay. We accounted for this by measuring the ATP hydrolysis activity of Rca (37) and including the approximate 30 μm accumulation of ADP over the 4-min measurement period (Fig. S6) into calculations. All experiments were repeated between three and six times, and all values and error bars presented are means and S.D.
Author contributions
A. P. S., D. D. V., B. d. B., A. G., and J. V. R. conceptualization; A. P. S., A. G., and J. V. R. funding acquisition; A. P. S., D. D. V., and N. B. investigation; A. P. S., D. D. V., N. B., M. A. H., B. d. B., A. G., and J. V. R. methodology; A. P. S. writing-original draft; A. P. S., A. G., and J. V. R. project administration; A. P. S., D. D. V., M. A. H., A. G., and J. V. R. writing-review and editing; M. A. H., B. d. B., A. G., and J. V. R. supervision.
Supplementary Material
Acknowledgments
We thank Iris de Beun and Els Vercleyen for vector construction and Dr. Artur Sawicki for comments on the finalized manuscript.
The authors are inventors named on a patent application pertaining to this work.
This article contains Table S1 and Figs. S1–S6.
- Rubisco
- ribulose-1,5-bisphosphate carboxylase/oxygenase
- CTE
- C-terminal extension
- Rca
- Rubisco activase
- Ta
- T. aestivum
- At
- A. thaliana
- Os
- O. sativa
- Nt
- N. tabacum
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- ECM
- fully carbamylated Rubisco activity
- ER
- Rubisco substrate inhibition
- ANOVA
- analysis of variance.
References
- 1. Andersson I. (2008) Catalysis and regulation in Rubisco. J. Exp. Bot. 59, 1555–1568 10.1093/jxb/ern091 [DOI] [PubMed] [Google Scholar]
- 2. Pearce F. G., and Andrews T. J. (2003) The relationship between side reactions and slow inhibition of ribulose-bisphosphate carboxylase revealed by a loop 6 mutant of the tobacco enzyme. J. Biol. Chem. 278, 32526–32536 10.1074/jbc.M305493200 [DOI] [PubMed] [Google Scholar]
- 3. Pearce F. G. (2006) Catalytic by-product formation and ligand binding by ribulose bisphosphate carboxylases from different phylogenies. Biochem. J. 399, 525–534 10.1042/BJ20060430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schrader S. M., Kane H. J., Sharkey T. D., and Caemmerer S. V. (2006) High temperature enhances inhibitor production but reduces fallover in tobacco Rubisco. Funct. Plant Biol. 33, 921–929 10.1071/FP06059 [DOI] [PubMed] [Google Scholar]
- 5. Jordan D. B., and Chollet R. (1983) Inhibition of ribulose bisphosphate carboxylase by substrate ribulose 1,5-bisphosphate. J. Biol. Chem. 258, 13752–13758 [PubMed] [Google Scholar]
- 6. Cleland W. W., Andrews T. J., Gutteridge S., Hartman F. C., and Lorimer G. H. (1998) Mechanism of Rubisco: the carbamate as general base. Chem. Rev. 98, 549–562 10.1021/cr970010r [DOI] [PubMed] [Google Scholar]
- 7. Portis A. R., Jr. (2003) Rubisco activase—rubisco's catalytic chaperone. Photosynth. Res. 75, 11–27 10.1023/A:1022458108678 [DOI] [PubMed] [Google Scholar]
- 8. Salvucci M. E., and Anderson J. C. (1987) Factors affecting the activation state and the level of total activity of ribulose bisphosphate carboxylase in tobacco protoplasts. Plant Physiol. 85, 66–71 10.1104/pp.85.1.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Erzberger J. P., and Berger J. M. (2006) Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 10.1146/annurev.biophys.35.040405.101933 [DOI] [PubMed] [Google Scholar]
- 10. Mueller-Cajar O., Stotz M., and Bracher A. (2014) Maintaining photosynthetic CO2 fixation via protein remodelling: the Rubisco activases. Photosynth. Res. 119, 191–201 10.1007/s11120-013-9819-0 [DOI] [PubMed] [Google Scholar]
- 11. Henderson J. N., Hazra S., Dunkle A. M., Salvucci M. E., and Wachter R. M. (2013) Biophysical characterization of higher plant Rubisco activase. Biochim. Biophys. Acta 1834, 87–97 10.1016/j.bbapap.2012.09.006 [DOI] [PubMed] [Google Scholar]
- 12. Keown J. R., and Pearce F. G. (2014) Characterization of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase activase isoforms reveals hexameric assemblies with increased thermal stability. Biochem. J. 464, 413–423 10.1042/BJ20140676 [DOI] [PubMed] [Google Scholar]
- 13. Wang Z. Y., Ramage R. T., and Portis A. R. Jr. (1993) Mg2+ and ATP or adenosine 5′-[γ-thio]-triphosphate (ATPγS) enhances intrinsic fluorescence and induces aggregation which increases the activity of spinach Rubisco activase. Biochim. Biophys. Acta 1202, 47–55 10.1016/0167-4838(93)90061-U [DOI] [PubMed] [Google Scholar]
- 14. Salvucci M. E. (1992) Subunit interactions of Rubisco activase: polyethylene glycol promotes self-association, stimulates ATPase and activation activities, and enhances interactions with Rubisco. Arch. Biochem. Biophys. 298, 688–696 10.1016/0003-9861(92)90467-B [DOI] [PubMed] [Google Scholar]
- 15. Wang Q., Serban A. J., Wachter R. M., and Moerner W. E. (2018) Single-molecule diffusometry reveals the nucleotide-dependent oligomerization pathways of Nicotiana tabacum Rubisco activase. J. Chem. Phys. 148, 123319 10.1063/1.5005930 [DOI] [PubMed] [Google Scholar]
- 16. Stotz M., Mueller-Cajar O., Ciniawsky S., Wendler P., Hartl F. U., Bracher A., and Hayer-Hartl M. (2011) Structure of green-type Rubisco activase from tobacco. Nat. Struct. Mol. Biol. 18, 1366–1370 10.1038/nsmb.2171 [DOI] [PubMed] [Google Scholar]
- 17. Serban A. J., Breen I. L., Bui H. Q., Levitus M., and Wachter R. M. (2018) Assembly–disassembly is coupled to the ATPase cycle of tobacco Rubisco activase. J. Biol. Chem. 293, 19451–19465 10.1074/jbc.RA118.005047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson S. P., and Portis A. R. Jr. (1989) Adenosine triphosphate hydrolysis by purified rubisco activase. Arch. Biochem. Biophys. 268, 93–99 10.1016/0003-9861(89)90568-7 [DOI] [PubMed] [Google Scholar]
- 19. Nagarajan R., and Gill K. S. (2018) Evolution of Rubisco activase gene in plants. Plant Mol. Biol. 96, 69–87 10.1007/s11103-017-0680-y [DOI] [PubMed] [Google Scholar]
- 20. Zhang N., and Portis A. R. (1999) Mechanism of light regulation of Rubisco: a specific role for the larger Rubisco activase isoform involving reductive activation by thioredoxin-f. Proc. Natl. Acad. Sci. U.S.A. 96, 9438–9443 10.1073/pnas.96.16.9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schürmann P., and Buchanan B. B. (2008) The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid. Redox Signal. 10, 1235–1274 10.1089/ars.2007.1931 [DOI] [PubMed] [Google Scholar]
- 22. Zhang N., Schürmann P., and Portis A. R. Jr. (2001) Characterization of the regulatory function of the 46-kDa isoform of Rubisco activase from Arabidopsis. Photosynth. Res. 68, 29–37 10.1023/A:1011845506196 [DOI] [PubMed] [Google Scholar]
- 23. Carmo-Silva A. E., and Salvucci M. E. (2013) The regulatory properties of Rubisco activase differ among species and affect photosynthetic induction during light transitions. Plant Physiol. 161, 1645–1655 10.1104/pp.112.213348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. To K.-Y., Suen D.-F., and Chen S.-C. (1999) Molecular characterization of ribulose-1,5-bisphosphate carboxylase/oxygenase activase in rice leaves. Planta 209, 66–76 10.1007/s004250050607 [DOI] [PubMed] [Google Scholar]
- 25. Carmo-Silva E., Scales J. C., Madgwick P. J., and Parry M. A. (2015) Optimizing Rubisco and its regulation for greater resource use efficiency. Plant Cell Environ. 38, 1817–1832 10.1111/pce.12425 [DOI] [PubMed] [Google Scholar]
- 26. Hartl M., Füßl M., Boersema P. J., Jost J. O., Kramer K., Bakirbas A., Sindlinger J., Plöchinger M., Leister D., Uhrig G., Moorhead G. B., Cox J., Salvucci M. E., Schwarzer D., Mann M., et al. (2017) Lysine acetylome profiling uncovers novel histone deacetylase substrate proteins in Arabidopsis. Mol. Syst. Biol. 13, 949 10.15252/msb.20177819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hosp F., Lassowskat I., Santoro V., De Vleesschauwer D., Fliegner D., Redestig H., Mann M., Christian S., Hannah M. A., and Finkemeier I. (2017) Lysine acetylation in mitochondria: from inventory to function. Mitochondrion 33, 58–71 10.1016/j.mito.2016.07.012 [DOI] [PubMed] [Google Scholar]
- 28. Scafaro A. P., Bautsoens N., den Boer B., Van Rie J., and Gallé A. (2019) A conserved sequence from heat-adapted species improves Rubisco activase thermostability in wheat. Plant Physiol. 181, 43–54 10.1104/pp.19.00425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stitt M., Lilley R. M., and Heldt H. W. (1982) Adenine nucleotide levels in the cytosol, chloroplasts, and mitochondria of wheat leaf protoplasts. Plant Physiol. 70, 971–977 10.1104/pp.70.4.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen J. B., Orozco E. M. Jr., and Ogren W. L. (1991) Expression of the two isoforms of spinach ribulose 1,5-bisphosphate carboxylase activase and essentiality of the conserved lysine in the consensus nucleotide-binding domain. J. Biol. Chem. 266, 8963–8968 [PubMed] [Google Scholar]
- 31. Buchert F., Schober Y., Römpp A., Richter M. L., and Forreiter C. (2012) Reactive oxygen species affect ATP hydrolysis by targeting a highly conserved amino acid cluster in the thylakoid ATP synthase γ subunit. Biochim. Biophys. Acta 1817, 2038–2048 10.1016/j.bbabio.2012.06.007 [DOI] [PubMed] [Google Scholar]
- 32. Sweetlove L. J., Heazlewood J. L., Herald V., Holtzapffel R., Day D. A., Leaver C. J., and Millar A. H. (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 32, 891–904 10.1046/j.1365-313X.2002.01474.x [DOI] [PubMed] [Google Scholar]
- 33. Sharkey T. D. (2005) Effects of moderate heat stress on photosynthesis: importance of thylakoid reactions, rubisco deactivation, reactive oxygen species, and thermotolerance provided by isoprene. Plant Cell Environ. 28, 269–277 10.1111/j.1365-3040.2005.01324.x [DOI] [Google Scholar]
- 34. Wang Z.-Y., and Portis A. R. Jr. (1991) A fluorometric study with 1-anilinonaphthalene-8-sulfonic acid (ANS) of the interactions of ATP and ADP with rubisco activase. Biochim. Biophys. Acta 1079, 263–267 10.1016/0167-4838(91)90067-A [DOI] [PubMed] [Google Scholar]
- 35. Hazra S., Henderson J. N., Liles K., Hilton M. T., and Wachter R. M. (2015) Regulation of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activase: product inhibition, cooperativity, and magnesium activation. J. Biol. Chem. 290, 24222–24236 10.1074/jbc.M115.651745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scales J. C., Parry M. A., and Salvucci M. E. (2014) A non-radioactive method for measuring Rubisco activase activity in the presence of variable ATP:ADP ratios, including modifications for measuring the activity and activation state of Rubisco. Photosynth. Res. 119, 355–365 10.1007/s11120-013-9964-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barta C., Carmo-Silva A. E., and Salvucci M. E. (2011) Rubisco activase activity assays. Methods Mol. Biol. 684, 375–382 10.1007/978-1-60761-925-3_29 [DOI] [PubMed] [Google Scholar]
- 38. Portis A. R. Jr., Li C., Wang D., and Salvucci M. E. (2008) Regulation of Rubisco activase and its interaction with Rubisco. J. Exp. Bot. 59, 1597–1604 10.1093/jxb/erm240 [DOI] [PubMed] [Google Scholar]
- 39. Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., and Qiu J.-L. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947 10.1038/nbt.2969 [DOI] [PubMed] [Google Scholar]
- 40. Loganathan N., Tsai Y.-C., and Mueller-Cajar O. (2016) Characterization of the heterooligomeric red-type rubisco activase from red algae. Proc. Natl. Acad. Sci. U.S.A. 113, 14019–14024 10.1073/pnas.1610758113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hermida-Carrera C., Kapralov M. V., and Galmés J. (2016) Rubisco catalytic properties and temperature response in crops. Plant Physiol. 171, 2549–2561 10.1104/pp.16.01846 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.