Abstract
The Krüppel-like factor 5 (KLF5) transcription factor is highly expressed in basal type breast cancer and promotes breast cancer cell proliferation, survival, migration, and tumorigenesis. KLF5 protein stability is regulated by ubiquitination. In this study, ubiquitin-specific protease 3 (USP3) was identified as a new KLF5 deubiquitinase by genome-wide siRNA library screening. We demonstrated that USP3 interacts with KLF5 and stabilizes KLF5 via deubiquitination. USP3 knockdown inhibits breast cancer cell proliferation in vitro and tumorigenesis in vivo, which can be partially rescued by ectopic expression of KLF5. Furthermore, we observed a positive correlation between USP3 and KLF5 protein expression levels in human breast cancer samples. These findings suggest that USP3 is a new KLF5 deubiquitinase and that USP3 may represent a potential therapeutic target for breast cancer.
Keywords: breast cancer, cell growth, cell proliferation, ubiquitin, ubiquitin-dependent protease, DUB, KLF5, USP3
Introduction
Breast cancer remains the leading cancer threatening women's health. Breast cancer presents the highest incidence and the second greatest mortality among American women (1). Of those cases, 15–20% of breast cancers are triple-negative breast cancer (TNBC),3 which is characterized as estrogen receptor–negative, progesterone receptor–negative, and human epidermal growth factor receptor 2–negative. TNBC is largely overlapped with basal type breast cancer (2). TNBC is heterogeneous and results in the worst prognosis because of a lack of effective therapeutic targets (3) and distant vital organ metastases (4). In the past 15 years, novel therapeutic options have been developed to target different subsets of TNBC (5, 6), including olaparib for BRCA1/2 mutated breast cancers (7), atezolizumab and nanoparticle albumin–bound paclitaxel for PD-L1–positive TNBC (8), enzalutamide for androgen receptor–positive TNBC (9), and the AKT inhibitor ipatasertib plus paclitaxel (10). Identification of novel specific biomarkers and therapeutic targets for TNBC is important.
Krüppel-like factor 5 (KLF5) is a zinc finger transcription factor that is overexpressed in basal subtype breast cancers (11), and its high mRNA and protein levels are associated with poor prognosis in breast cancer patients (12, 13). Our previous studies demonstrated that KLF5 promotes breast cancer cell proliferation, stemness, survival, migration, tumor growth, and metastasis, partially through up-regulating the transcription of fibroblast growth factor–binding protein 1 (FGF-BP) (14), microsomal prostaglandin E2 synthase 1 (mPGES1) (15), TNFAIP2 (16), and Slug (17) and inhibiting the transcription of p27 (18). Therefore, KLF5 represents an attractive therapeutic target for TNBC. We demonstrated that mifepristone (19), metformin (20), and mithramycin A (21) indirectly inhibited KLF5 expression and TNBC.
As an oncogenic transcription factor, KLF5 is not an ideal candidate target for anti-cancer drug development. Alternatively, the KLF5 upstream positive regulators, especially enzymes, may be better targets for TNBC. Our previous studies showed that KLF5 is an unstable protein that can be ubiquitinated by E3 ubiquitin ligases including WWP1 (22), SCFFbw7 (23), Smurf2 (24), and EFP (25) and degraded by proteasomes (26). KLF5 ubiquitination is a reversible process that can be reversed by deubiquitinating enzymes (DUBs). We previously identified ATXN3L (27) and BAP1 (18) as KLF5 DUBs. However, the endogenous ATXN3L protein has not been detected to date. Whether the expression levels of BAP1 and KLF5 are correlated in breast tumors is unknown, although BAP1 promotes breast cancer cell growth and migration. We suspected that the KLF5 protein is also positively regulated by other DUBs in TNBC.
Given that KLF5 DUBs might be ideal therapeutic targets in TNBC, we screened a DUB siRNA library and identified USP3 as a candidate DUB for KLF5. In this study, we demonstrated that USP3 promotes breast cancer cell proliferation in vitro and tumor growth in vivo. Our results suggest that USP3 is a potential therapeutic target for breast cancer.
Results
USP3 stabilizes the KLF5 protein
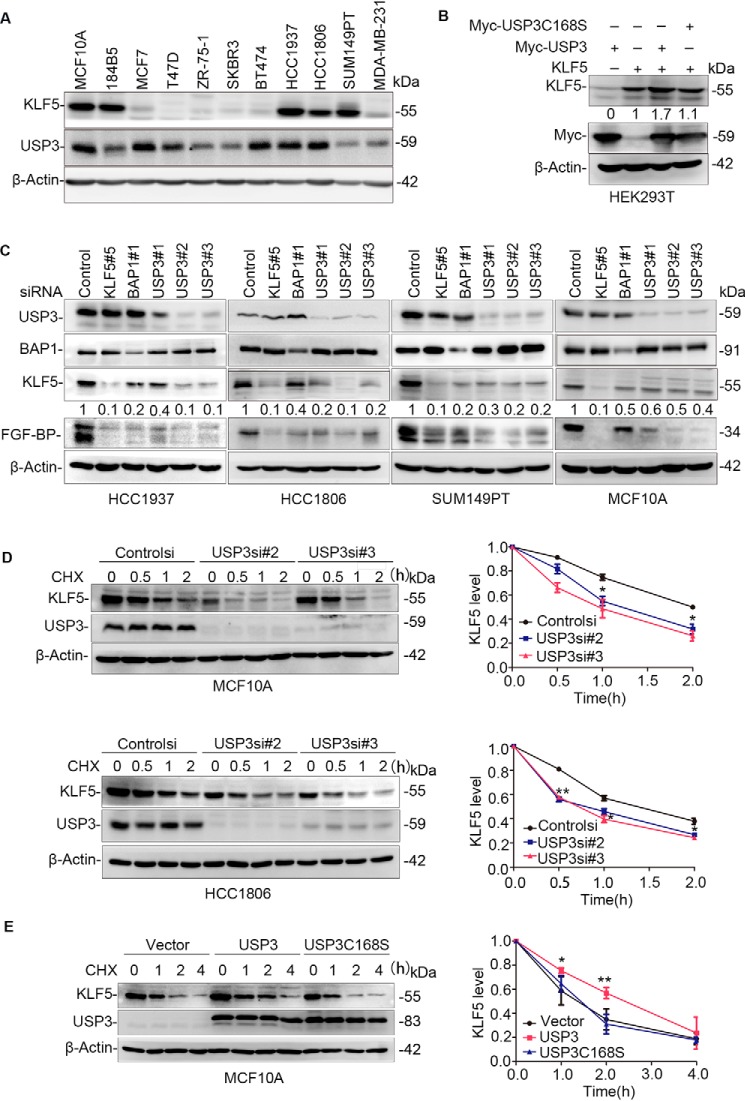
In our previous study, we screened an siRNA library consisting of siRNA pools against 87 human DUBs based on KLF5 protein expression levels in HeLa cells (18). USP3 was identified as a candidate KLF5 DUB. We first examined the protein expression levels of USP3 in two immortalized breast epithelial cell lines (MCF10A and 184B5) and nine breast cancer cell lines. As shown in Fig. 1A, USP3 is detectable in all cell lines and is highly expressed in several KLF5-high cell lines, such as MCF10A, HCC1937, and HCC1806. In addition, we measured the exogenous KLF5 protein level after we overexpressed WT USP3 and the catalytically inactive mutant USP3 C168S (Cys-168 was mutated into Ser so that the deubiquitinase activity was lost) in HEK293T cells. The KLF5 protein level was elevated ∼1.7-fold by USP3 but not by USP3 C168S (Fig. 1B).
Figure 1.
USP3 stabilizes KLF5. A, KLF5 and USP3 protein expression levels in immortalized mammary epithelial cell lines (MCF10A and 184B5) and nine breast cancer cell lines were examined by WB. The experiment was repeated three times, and a representative result is shown. B, USP3 overexpression stabilized the KLF5 protein in a DUB enzyme activity–dependent manner. KLF5 was co-expressed with USP3 or USP3C168S in HEK293T cells. ImageJ was used to measure the band intensities of KLF5 and β-actin. The normalized KLF5 protein levels are shown below the KLF5 blot. The experiment was repeated three times, and a representative result is shown. C, USP3 knockdown decreased KLF5 and FGF-BP protein levels in breast cancer cell lines (HCC1937, HCC1806, and SUM149PT) and a breast epithelial cell line (MCF10A). The cells were transfected with control or three different USP3 siRNAs for 48 h. Control siRNA was used as the negative control. KLF5 and BAP1 siRNAs were used as the positive controls. WB was used to measure the endogenous KLF5 and FGF-BP protein levels. ImageJ was used to measure the band intensities of KLF5 and β-actin. The normalized KLF5 protein levels are shown below the KLF5 blot. The experiment was repeated three times, and a representative result is shown. D, USP3 knockdown shortened the half-life of KLF5. MCF10A and HCC1806 cells were transfected with siRNA and treated with cycloheximide (CHX) for the indicated time points. WB was used to measure the endogenous KLF5 protein level. The graph on the right shows the quantitative results of KLF5 by ImageJ. Error bars represent S.D. The experiment was repeated three times, and a representative result is shown. *, p < 0.05; **, p < 0.01, t test. E, USP3 overexpression extended the half-life of KLF5 protein. USP3 or USP3 C168S were stably overexpressed in MCF10A cells. WB was used to measure the endogenous KLF5 protein levels after the cells were treated with CHX for the indicated time. The graph on the right shows the quantitative results. Error bars represent S.D. The experiment was repeated three times, and a representative result is shown. *, p < 0.05; **, p < 0.01, t test.
To further confirm whether USP3 increases KLF5 protein stability, we knocked down USP3 using three different siRNAs in the MCF10A cell line and breast cancer cell lines (HCC1937, HCC1806, and SUM149PT) with high expression levels of KLF5 (Fig. 1C). Similar to BAP1, USP3 knockdown decreased the endogenous protein levels of KLF5 and its downstream target gene FGF-BP compared with the control in four cell lines, although the down-regulation of KLF5 by USP3 siRNAs in MCF10A was less efficient than three cancer cell lines. However, USP3 knockdown did not affect the KLF5 mRNA level in the HCC1806 and MCF10A cells (Fig. S1, A and B). We further demonstrated that overexpression of WT USP3, but not the catalytically inactive mutant USP3 C168S, rescued the down-regulation of KLF5 protein expression induced by USP3 siRNA#2 in HCC1806 (Fig. S1C). This result indicates that the down-regulation of KLF5 protein expression by USP3 knockdown was not caused by off-target effects of siRNA.
In previous studies, the E3 ubiquitin ligases, such as WWP1 (22), SCFFbw7 (23), and Smurf2 (24), were shown to target the KLF5 protein for ubiquitin-mediated degradation. To test whether USP3 can antagonize E3 ligase-mediated KLF5 degradation, we transfected KLF5, USP3, and E3s, such as WWP1, Myc-FBW7, or FLAG-Smurf2, into HEK293T cells. As expected, USP3 overexpression blocked the KLF5 protein degradation induced by E3 ligases (Fig. S1D).
To further investigate whether endogenous USP3 protects KLF5 protein from degradation, we knocked down USP3 in MCF10A and HCC1806 cells using two different siRNAs and measured the KLF5 protein half-lives with the cycloheximide chase assay. As expected, the KLF5 protein half-life was reduced when USP3 was knocked down compared with control cells (Fig. 1D). Similar results were obtained in SUM149PT breast cancer cells (Fig. S1E).
Furthermore, we found that the KLF5 protein half-life was extended by USP3, but not by USP3 C168S, in MCF10A cells (Fig. 1E). Collectively, these results suggest that USP3 stabilizes the KLF5 protein through its DUB enzyme activity in breast epithelial cells.
USP3 deubiquitinates KLF5
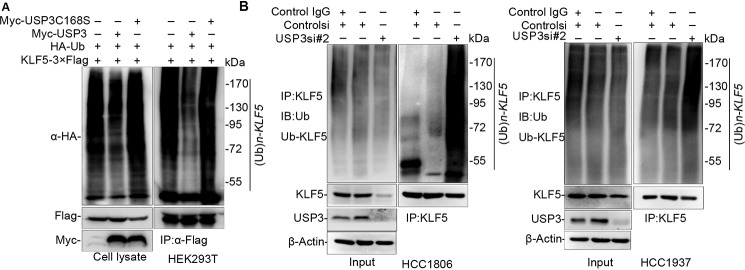
To test whether USP3 increases KLF5 protein stability through deubiquitinating KLF5, we transfected KLF5–3×FLAG, hemagglutinin (HA)-Ub, and USP3 or USP3 C168S into HEK293T cells and treated the cells with the proteasome inhibitor MG132 to block protein degradation. As expected, USP3, but not USP3 C168S, dramatically reduced KLF5 protein polyubiquitination (Fig. 2A). We did not observed the USP3 DUB activity toward p21 (Fig. S2A), suggesting that KLF5 is a specific substrate of USP3.
Figure 2.
USP3 deubiquitinates KLF5. A, USP3 decreased KLF5 ubiquitination in a DUB activity–dependent manner. KLF5–3×FLAG and HA-Ub were co-expressed with USP3 or USP3 C168S in HEK293T cells. WB was used to detect the ubiquitinated KLF5 protein levels after the cells were treated with MG132 for 6 h. The experiment was repeated three times, and a representative result is shown. B, USP3 knockdown in HCC1806 and HCC1937 cells increased the endogenous KLF5 protein ubiquitination. The cells were transfected with control or USP3 siRNAs for 48 h. Control siRNA was used as the negative control. Control IgG refers to goat IgG antibody, which is the negative control for KLF5 antibody. WB was used to detect the ubiquitinated KLF5 protein levels after the cells were treated with MG132 for 6 h. The experiment was repeated three times, and a representative result is shown. IP, immunoprecipitation; IB, immunoblotting.
To test whether USP3 decreases the endogenous KLF5 polyubiquitination, we silenced USP3 by siRNA in HCC1806 and HCC1937 cells. As shown in Fig. 2B, knockdown of USP3 remarkably increased the endogenous KLF5 protein polyubiquitination levels compared with the control cells. Thus, USP3 decreases KLF5 protein ubiquitination in a DUB activity–dependent manner.
USP3 interacts with KLF5
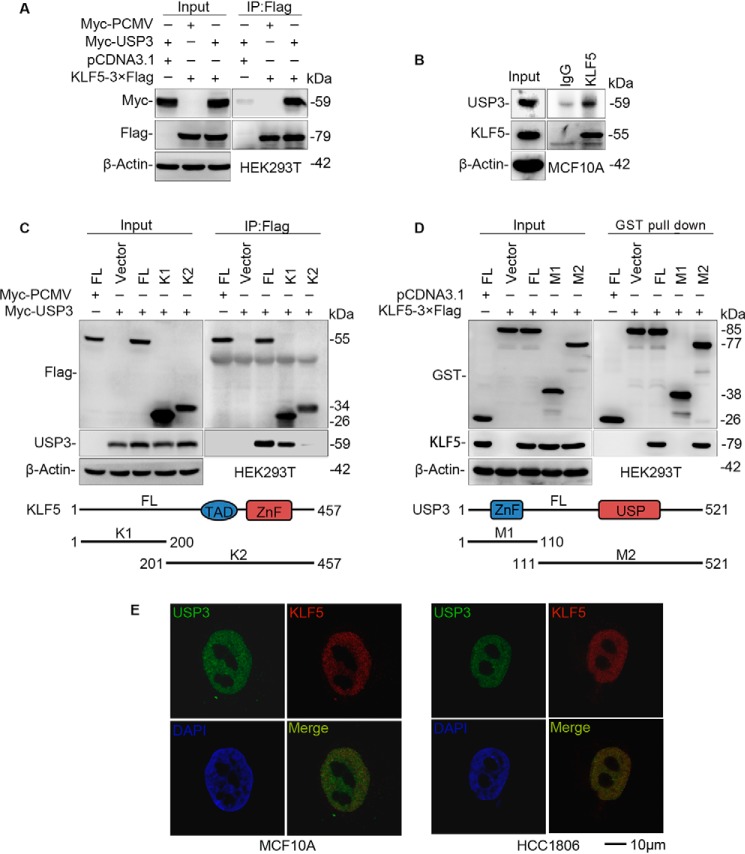
In principle, USP3 and KLF5 proteins should interact with each other if USP3 is a KLF5 DUB. To test this, we cotransfected KLF5–3×FLAG and Myc-USP3 into HEK293T cells and immunoprecipitated KLF5–3×FLAG using an anti-FLAG antibody. As expected, Myc-USP3 was co-immunoprecipitated with KLF5–3×FLAG (Fig. 3A). The catalytically inactive mutant USP3 C168S was still efficiently interacted with KLF5 (Fig. S2B). Next, we demonstrated that endogenous USP3 was also co-immunoprecipitated with KLF5 proteins in MCF10A cells (Fig. 3B).
Figure 3.
USP3 interacts with KLF5. A, exogenous USP3 and KLF5 proteins interact with each other in HEK293T cells. Myc-USP3 and KLF5–3×FLAG were co-expressed in HEK293T cells. KLF5–3×FLAG was immunoprecipitated by FLAG-M2 beads. The experiment was repeated three times, and a representative result is shown. B, endogenous USP3 and KLF5 proteins interact with each other in MCF10A cells. Endogenous USP3 and KLF5 proteins were immunoprecipitated with the anti-KLF5 antibody. IgG serves as the negative control. The experiment was repeated three times, and a representative result is shown. C, mapping the KLF5 domain that interacts with USP3. FLAG-tagged full-length or mutants of KLF5 (a schematic diagram is shown below the panel) and Myc-USP3 were cotransfected into HEK293T cells. Immunoprecipitation was performed with FLAG-M2 beads. The experiment was repeated three times, and a representative result is shown. D, mapping the USP3 domain that interacts with KLF5. GST-tagged full-length or mutants of USP3 (a schematic diagram is shown below the panel) and KLF5–3×FLAG were cotransfected into HEK293T cells. Immunoprecipitation was performed with GSH beads. The experiment was repeated three times, and a representative result is shown. E, endogenous USP3 and KLF5 are co-localized within the nuclei of HCC1806 and MCF10A cells. Immunofluorescence staining was used to detect the subcellular location of USP3 and KLF5. 4,6-Diamidino-2-phenylindole was used to stain the DNA. Scale bar, 10 μm. Endogenous USP3 and KLF5 are co-localized in the nuclei of all nonmitosis cells, and only representative results are shown. IP, immunoprecipitation.
To further map the domain of KLF5 responsible for the interaction, we generated two truncated KLF5 fragments (Fig. 3C). Full-length KLF5 (1–457) and the N terminus of KLF5 (1–200) were shown to interact with USP3, whereas the C terminus of KLF5 (201–457) did not, although they are localized in the nucleus like the full-length KLF5 (Fig. S2C). Furthermore, we tried to map the region of USP3 that is responsible for the interaction by generating two GST-fused USP3 deletion mutants. Using GST immunoprecipitation assays, full-length USP3 (1–521) and the C terminus of USP3 (111–521) interacted with KLF5–3×FLAG; however, the N terminus of USP3 (1–110) did not interact with KLF5–3×FLAG (Fig. 3D). Finally, we demonstrated, by means of immunofluorescence staining, that endogenous USP3 and KLF5 are co-localized in the nuclei of MCF10A and HCC1806 cells (Fig. 3E). Taken together, we concluded that USP3 interacts with KLF5 in the nucleus and that the interaction is mediated by the C terminus of USP3 and N terminus of KLF5.
USP3 promotes breast cancer cell proliferation in vitro partially through KLF5
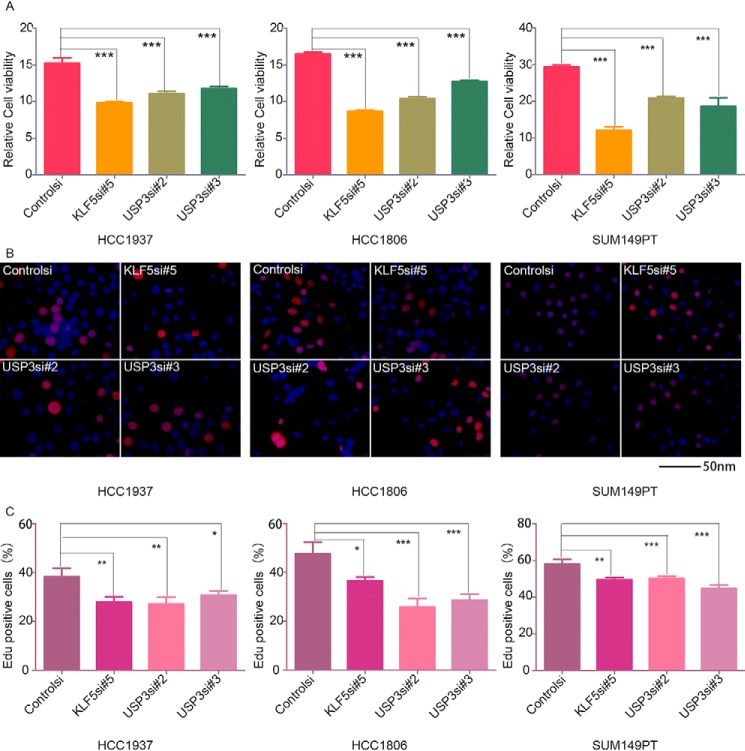
In our previous studies, we demonstrated that KLF5 promoted breast cancer cell proliferation and tumorigenesis (16). However, the function of USP3 in breast cancer has not been studied. We knocked down USP3 using two different siRNAs in HCC1937, HCC1806, and SUM149PT cells and measured cell growth by the SRB assay. As a result, knockdown of USP3 significantly decreased cell viability in the three cell lines compared with the control cells, similar to the knockdown of KLF5 (Fig. 4A). We further confirmed these results by measuring DNA synthesis by the EdU incorporation assay. Consistently, knockdown of USP3 significantly decreased the percentage of EdU-positive cells in the three cell lines compared with the control cells, similar to the knockdown of KLF5 (Fig. 4, B and C).
Figure 4.
USP3 knockdown suppresses breast cancer cell proliferation in HCC1937, HCC1806, and SUM149PT. A, knockdown of USP3 decreased cell viability, as measured by the sulforhodamine B assay at 120 h after transfection. The cells were transfected with siRNA and then implanted into 96-well plates. ***, p < 0.001, t test. Error bars represent S.D. of five samples in parallel. The experiment was repeated three times, and a representative result is shown. B, knockdown of USP3 decreased DNA synthesis, as measured by the EdU incorporation assay at 48 h after transfection. Representative images are shown. Red represents the proliferative cells, and blue represents the total cells. Scale bar, 50 nm. The experiment was repeated three times, and a representative result is shown. C, the quantitative results of DNA synthesis. The percentages of proliferative cells versus total cells from five images were compared by the t test. Graphs are the means ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001, t test. The experiment was repeated three times, and a representative result is shown.
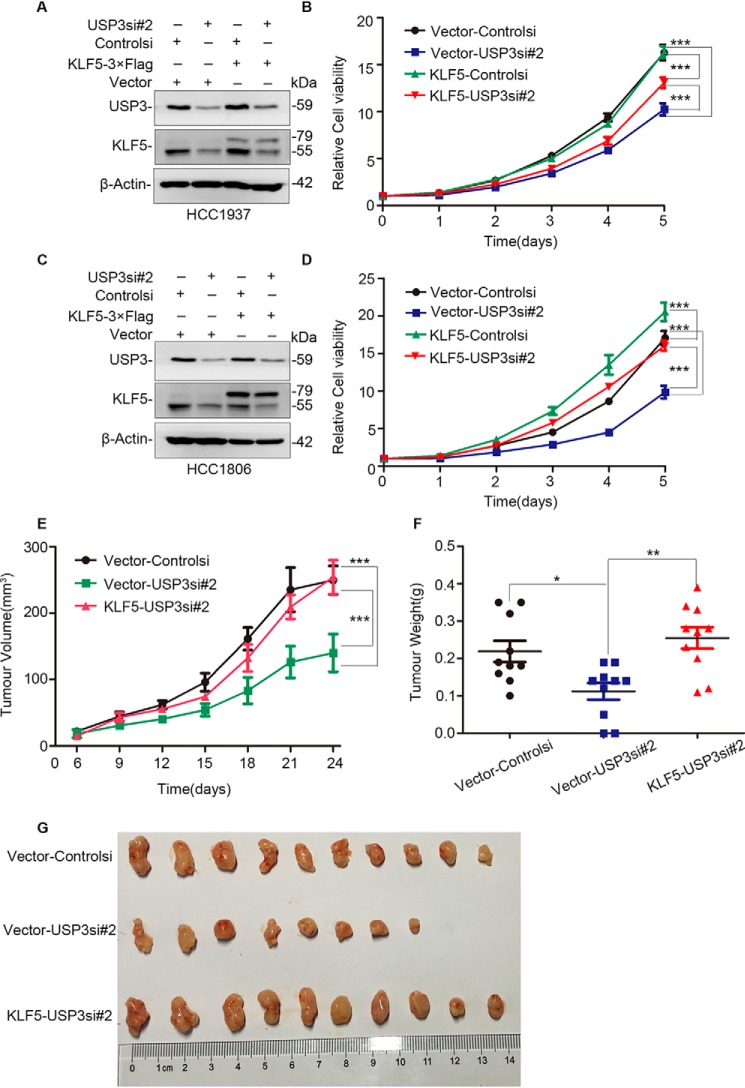
To test whether USP3 promotes breast cell proliferation through KLF5, we stably overexpressed KLF5 in HCC1937 and HCC1806 cells and transiently knocked down USP3 (Fig. 5, A and C). We found that KLF5 overexpression significantly rescued the cell growth arrest induced by USP3 knockdown (Fig. 5, B and D). These results suggest that endogenous USP3 promotes breast cancer cell proliferation partially through stabilizing KLF5.
Figure 5.
USP3 knockdown suppresses cell proliferation partially through KLF5. A, KLF5 stable overexpression and USP3 knockdown in HCC1937 cells were confirmed by WB. Vector alone was used as the negative control for KLF5. B, KLF5 overexpression significantly restored cell proliferation in USP3 knockdown HCC1937 cells. The cells with stable KLF5 overexpression were transfected with siRNA and then implanted into 96-well plates. The cell viability was measured by the sulforhodamine B assay every day. Every experimental group was compared with the control siRNA group. ***, p < 0.001, t test. Error bars represent S.D. of five samples in parallel. Interactions between KLF5 and USP3 were found in HCC1937 (F = 21.051, p < 0.0001) cells by the analysis of variance of factorial design. The experiment was repeated three times, and a representative result is shown. C, KLF5 stable overexpression and USP3 knockdown in HCC1806 cells were confirmed by WB. D, KLF5 overexpression significantly restored cell proliferation in USP3 knockdown HCC1806 cells. ***, p < 0.001, t test. Interaction between KLF5 and USP3 was found in HCC1806 (F = 10.284, p = 0.005) cells by the analysis of variance of factorial design. The experiment was repeated three times, and a representative result is shown. E, stable overexpression of KLF5 restored USP3 knockdown HCC1806 xenograft growth in nude mice. Xenograft tumor growth was measured twice per week. Data points represent the means ± S.D. of five mice per group (10 tumors). ***, p < 0.001, t test. F, stable overexpression of KLF5 significantly increased tumor weight in USP3 knockdown HCC1806 cells (n = 10). Error bars represent S.D. **, p < 0.01, t test. G, stable overexpression of KLF5 generated larger xenografts in USP3 knockdown HCC1806 cells at day 24.
USP3 promotes breast tumor growth in vivo partially through KLF5
To test whether USP3 promotes breast tumor growth in vivo, we knocked down USP3 in KLF5 overexpressed and vector control HCC1806 cells. The cells (1 × 106/spot, n = 6) were subcutaneously injected into fat pads of 6–7-week-old BALB/C nude mice from the Hunan SJA Laboratory Animal Co. Ltd. In a 19-day course, USP3 knockdown HCC1806 breast cancer cells grew significantly more slowly than did the control cells (Fig. 5, E–G, and Fig. S3, A–C). KLF5 overexpression promoted HCC1806 breast cancer cell growth in nude mice (Fig. S3, A–C) and rescued USP3 knockdown-induced growth inhibition (Fig. 5, E–G). The average tumor weights from the different groups also support these conclusions (Fig. 5, F and G, and Fig. S3, B and C). Taken together, these results indicate that USP3 promotes breast cancer tumorigenesis partially by KLF5.
The USP3 and KLF5 protein expression levels are positively correlated in human TNBC specimens
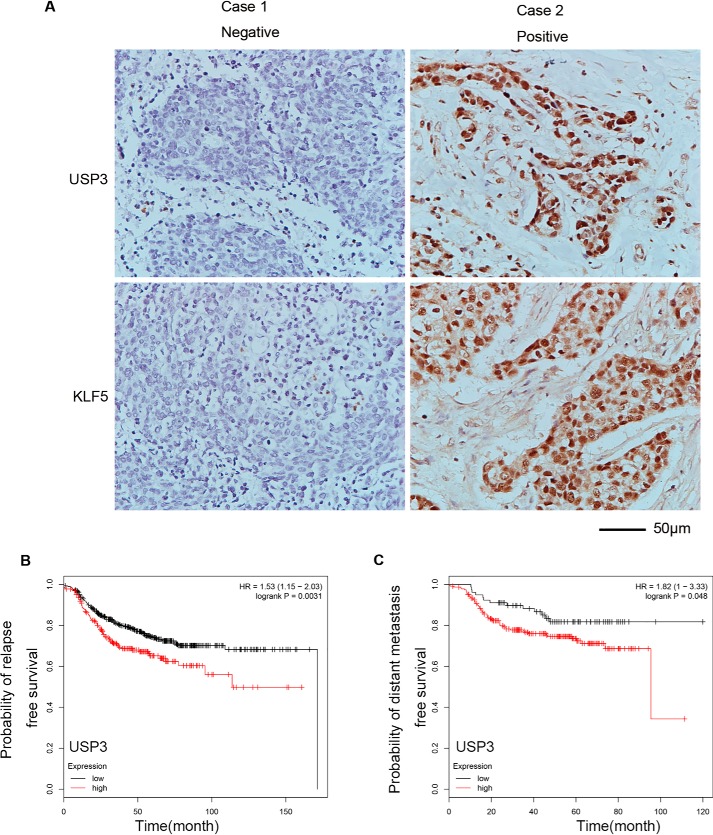
We tested the protein levels of USP3 and KLF5 in human TNBC clinical samples by immunohistochemical staining. Remarkably, USP3 positivity was observed in 46.5% (20 of 43) of TNBC samples. At the same time, KLF5 was positive in 51.2% (22 of 43) of TNBC samples. Moreover, a strong positive correlation (r = 0.6312, p < 0.0001) between USP3 and KLF5 was observed in these TNBC samples (Fig. 6A and Table 1). The original score data were shown in the Table S4. These results suggest that USP3 may positively regulate the expression of KLF5 in TNBC patients.
Figure 6.
USP3 and KLF5 protein expression levels are positively correlated in human TNBC. A, representative IHC results for USP3 and KLF5 in TNBC specimens. The negative results are shown in the left panel, and the positive results are shown in the right panel. Scale bar, 50 μm. B, a high expression level of USP3 mRNA is significantly associated with a short relapse–free survival of breast cancer patients who have received chemoradiotherapy. Kaplan–Meier plotter was used to analyze the breast cancer RNA-sequencing data from TCGA database. **, p < 0.01, t test. C, a high expression level of USP3 mRNA is significantly associated with a short distant metastasis-free survival of breast cancer patients who have received chemoradiotherapy. Kaplan–Meier plotter was used to analyze the breast cancer RNA-sequencing data from TCGA database. *, p < 0.05, t test.
Table 1.
The USP3 and KLF5 protein expression levels are positively correlated in human TNBC specimens
r = 0.6312. p < 0.0001.
| USP3 |
||||
|---|---|---|---|---|
| Negative | Positive | Total | ||
| Negative | 18 | 3 | 21 | |
| KLF5 | Positive | 5 | 17 | 22 |
| Total | 23 | 20 | 43 | |
It has been reported that high KLF5 mRNA and protein levels are associated with poor prognosis in breast cancer patients (12, 13). Based on the TCGA database, we found that a high expression level of USP3 mRNA is significantly associated with a short relapse-free survival and short distant metastasis-free survival in breast cancer patients who have received chemoradiotherapy (Fig. 6, B and C).
USP3 is well-documented to participate in maintaining genome stability (28). Therefore, we tested whether USP3 knockdown sensitized breast cancer cells to genotoxic chemotherapy drugs. As shown in Fig. S4 (A and B), USP3 knockdown significantly sensitized HCC1806 and HCC1937 cells to cisplatin-induced loss of cell viability. However, KLF5 knockdown did not elicit similar effects. We conclude that USP3 confers drug resistance in breast cancer cells independently of KLF5.
Discussion
KLF5 has been implicated as an oncogene and therapeutic target in a number of cancers including breast, colon, bladder, lung, stomach, and ovarian cancer (29). Therefore, it is important to thoroughly understand the upstream regulators of KLF5. At the transcriptional level, progesterone induced the expression of KLF5 in multiple progesterone receptor–positive breast cancer cell lines (30). Sp1 is a crucial transcription factor for KLF5 in breast cancer (21). Recently, KLF5 transcription was reported to be regulated by superenhancers, which could be suppressed by BRD4 inhibitors and CDK7 inhibitors (31, 32). At the post-translational level, we demonstrated that E3 ubiquitin ligases (WWP1 and SCFFbw7) promote KLF5 protein ubiquitination and degradation (22, 23). Metformin inhibited protein kinase A activity to induce glycogen synthase kinase-3β–mediated KLF5 protein phosphorylation and degradation (20). In addition, YAP (33) and TAZ (34) decreased WWP1-mediated KLF5 degradation. We already identified ATXN3L (27) and BAP1 (18) as KLF5 DUBs. In this study, we demonstrated that USP3 is an authentic KLF5 DUB that stabilizes KLF5 protein in TNBC.
Several lines of evidence support the assertion that USP3 is the KLF5 DUB. First, USP3 interacts with KLF5. Second, USP3 decreases KLF5 polyubiquitination and increases KLF5 protein stability in a DUB activity-dependent manner. Additionally, USP3 promotes breast cancer cell proliferation and tumor growth partially through KLF5. Finally, the protein levels of USP3 and KLF5 are positively correlated in human TNBC samples. Our results suggest that USP3 is oncogenic in TNBC because it stabilizes KLF5.
USP3 belongs to the human USP DUB subfamily, which contains two highly conserved sequence regions: a cysteine residue and two histidine residues. USP3 is a single copy gene and is located at chromosome 15q22.3. USP3 is a functional DUB capable of efficiently cleaving a ubiquitin–proline bond (35). USP3 serves a well-recognized role in DNA repair or chromatin remodeling. USP3 was first discovered to be a chromatin modifier required for S phase progression and genome stability by deubiquitinating monoubiquitinated H2A and H2B (28, 36). Further investigation demonstrated that USP3 abrogates the accumulation of BRCA1 and 53BP1 at double-strand breaks by counteracting RNF168- and RNF8-mediated H2A and γH2AX ubiquitination in response to DNA damage (37, 38). Recent studies have shown that USP3 promotes CHK1 activation by removing the K63-linked ubiquitin chain from CHK1 in response to DNA damage (39). It was recently reported that USP3 is a DUB for p53 and suppresses cell proliferation and transformation in U2OS and IMR90 cells (40). Mice deficient in USP3 exhibited increased levels of histone ubiquitination in adult tissues, reduced hematopoietic stem cell reserves over time, and shortened animal lifespan (41, 42). Most importantly, USP3 knockout mice spontaneously developed tumors because of loss of chromosomal integrity (42). Consistently, we found that USP3 knockdown significantly increased cisplatin sensitivity in TNBC cells, although this function was independent of stabilizing KLF5 (Fig. S4).
In addition to its role in DNA repair, USP3 promotes gastric cancer tumorigenesis, and high expression levels of USP3 are associated with poor prognosis in patients with gastric cancer (43). It is likely that USP3 promotes gastric cancer through KLF5, in part because KLF5 also promotes gastric cancer (29).
In conclusion, we identified USP3 as a new KLF5 DUB and demonstrated that USP3 promotes breast cancer cell proliferation and tumor growth partially through stabilizing the KLF5 protein. The expression level of USP3 may serve as a prognostic biomarker in breast cancer. Our study revealed a new mechanism for KLF5 protein regulation. USP3 might be further developed into a therapeutic target for patients with TNBC because depletion of USP3 inhibits tumor growth and sensitizes cancer cells to genotoxic chemotherapeutic drugs.
Materials and methods
DUB siRNA library screening
The siRNA library consisting of 87 human DUBs was purchased from Applied Biosystems (Ambion Silencer siRNA Custom Library, catalog no. 4392425). The screening method was described in our previous study (18).
Antibodies
All primary antibodies were diluted by a factor of 1,000 for Western blotting (WB) unless otherwise specified. The KLF5 rabbit polyclonal antibody was described in a previous study (29). The anti-USP3 rabbit polyclonal antibody was purchased from Abcam (ab101473, 1:3,000 dilution; London, UK). The anti-BAP1 mouse monoclonal (sc-28383), anti-HA (Y11) rabbit polyclonal (sc-805), and anti-Myc (9E10) mouse monoclonal (sc-40) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-FLAG rabbit polyclonal (F7425), anti-GST rabbit polyclonal (G7781), and anti–β-actin mouse monoclonal (A5441, 1:10,000 dilution) antibodies were from Sigma–Aldrich. The goat anti-KLF5 and mouse anti–FGF-BP1 monoclonal antibodies were from R&D Systems (Minneapolis, MN). The anti-WWP1 (H00011059-M01) rabbit polyclonal antibody was from Abnova (Taiwan, China). The mouse anti-ubiquitin (MAB1510) monoclonal antibody was from Millipore.
Cell culture and transfection
All cell lines were purchased from American Type Culture Collection (Manassas, VA) and validated by STR analysis. The HEK293T human embryonic kidney cell line was cultured in Dulbecco's modified Eagle's medium (high glucose) medium containing 5% fetal bovine serum (FBS, HyClone) and 1% penicillin/streptomycin (P/S). The HCC1806 and HCC1937 human breast cancer cell lines were maintained in RPMI 1640 containing 5% FBS and 1% P/S. The SUM149PT breast cancer cell line was grown in HAM/F1 medium supplemented with 10% FBS, 0.005 mg/ml insulin, 1 mg/ml hydrocortisone, 10 mm HEPES, and 1% P/S. The MCF10A immortalized breast epithelial cell line was maintained in Dulbecco's modified Eagle's medium/Ham's F-12 50/50 medium supplemented with 5% horse serum, 0.5 mg/ml hydrocortisone, 10 mg/ml insulin, 20 ng/ml epidermal growth factor, 0.1 mg/ml cholera enterotoxin, 1% P/S, and 2 mm l-glutamine. All of the siRNAs and plasmids were transfected using Lipofectamine 2000, according to the manufacturer's instructions. All siRNA sequences are shown in Table S1. All primers are listed in Table S2.
Deubiquitination assays
HEK293T cells were transiently transfected with HA-Ub, KLF5–3×FLAG, and other plasmids in 6-well plates. Two days after transfection, the cells were treated with 20 μm MG132 for 6 h before harvest in 150 μl of SDS lysis buffer (50 mm Tris-Cl, pH 6.8, 1.5% SDS). The samples were boiled for 15 min. Then 120 μl of protein lysate was diluted with 1.2 ml of BSA buffer (50 mm Tris-Cl, pH 6.8, 180 mm NaCl, 0.5% CA630, 0.5% BSA) for immunoprecipitation with 30 μl of anti-FLAG M2–agarose beads or anti-KLF5 antibody plus protein A/G–agarose (for endogenous KLF5 protein ubiquitination) overnight at 4 °C with rotation. The beads were collected by centrifugation at 1,000 × g for 3 min at 4 °C after being washed three times with 1 ml of ice-cold BSA buffer. The results were analyzed by WB.
Protein–protein interaction
We described immunoprecipitation using anti-FLAG M2–agarose beads (A2220; Sigma) and GST immunoprecipitation using the GSH-Sepharose 4B slurry beads in a previous study (42). Expression plasmids were transfected into HEK293T cells (6-well plates) for 48 h for exogenous KLF5 protein interaction studies. MCF10A cells were seeded in 10-cm plates for endogenous USP3/KLF5 protein interaction analysis.
Cell proliferation in vitro
The proliferation of HCC1937, HCC1806, and SUM149PT cells was measured using Click-iT EdU Alexa Fluor 488 imaging kits (Invitrogen) and the sulforhodamine B assay (42).
Tumorigenesis
Thirty-three 6–7-week-old female nude mice were purchased from SJA Laboratory Animal Co., Ltd. (Hunan, China). The animal protocol was approved by the animal ethics committee of Kunming Institute of Zoology, Chinese Academy of Sciences. Nude mice were randomly distributed into four groups (Vector-Control, Vector-USP3si#2, Vector-KLF5, and KLF5-USP3si#2, with five or six mice/group). One million HCC1806 cells presuspended in Matrigel (BD Biosciences; 1:1 diluted with PBS) were subcutaneously injected into the fourth pair of mammary gland fad pads. Tumor sizes were measured twice a week and calculated using the following equation: ½ × length × width2. Finally, tumors were harvested and weighed according to the standards of the ethics committee.
RT–quantitative PCR assays
Total RNA was isolated from MCF10A and HCC1806 cells using TRIzol reagent (Invitrogen). Reverse transcription was performed using the iScript cDNA synthesis kit (Bio-Rad). Quantitative RT-PCR was performed on an ABI-7900 system using SYBR Green reagents (ABI, Austin, TX). Primers used for USP3, KLF5, and glyceraldehyde-3-phosphate dehydrogenase are shown in Table S3.
Immunohistochemical staining
Human breast cancer specimens were obtained from 42 patients who had undergone surgical resection between 2006 and 2015 at the First Affiliated Hospital of Kunming Medical University (Yunnan, China). Surgical specimens were formalin-fixed, sectioned, stained with hematoxylin and eosin, and examined by microscopy. Diagnoses were made by experienced pathologists and graded using standard histological and modified Scarff–Bloom–Richardson criteria. Human breast cancer specimen slides were incubated at 60 °C for 2 h and subjected to antigen retrieval prior to conventional IHC. Anti-KLF5 and anti-USP3 primary antibodies were used at a 1/1,000 dilution. Staining patterns were interpreted by two pathologists with no prior knowledge of the clinicopathological parameters. Immunostained slides were evaluated under a light microscope. IHC was rated by the Allred score method (43). Briefly, scores were assigned that represented the estimated proportions of positive-staining tumor cells (0, none; 1, 1/100; 2, >1/100 to <1/10; 3, >1/10 to <1/3; 4, >1/3 to <2/3; and 5, >2/3). The average estimated staining intensities in positive cells were assigned intensity scores (0, none; 1, weak; 2, intermediate; and 3, strong). The proportion and intensity scores were added to obtain a total score ranging from 0 to 8. In this study, protein expression levels were classified into four subfamilies: negative (total score, 0), weak positive (total score, 2–3), intermediate positive (total score, 4–5), and strong positive (total score, 6–8). To further exclude the influence of nonspecific staining and facilitate statistics, weak positive cases (total score, 2–3) were included in the negative group, whereas intermediate positive cases (total score, 4–5) and strong positive cases (total score, 6–8) were included in the positive group.
TCGA database information
The identification number that we use in the TCGA database is 221654_s_at, which is for breast cancer patients regardless of hormonal status and human epidermal growth factor receptor 2 status. These patients have received chemotherapy, including adjuvant and neoadjuvant chemotherapy, but the specific regimen is unknown. The patients were segregated into “low” and “high” level of USP3 mRNA by median. There were 798 cases in the RFS analysis and 305 cases in the DMFS analysis.
Statistical analysis
The data are presented as the means ± S.D. Student's t test was used for statistical analysis, unless otherwise indicated. The analysis of variance of factorial design was used for interaction between KLF5 and USP3 (Fig. 5, B and D). GraphPad Prism 6 (GraphPad Software Inc., La Jolla, CA) and SPSS 20.0 Software (IBM Inc., Armonk, NY) were used for all statistical analyses. p values below 0.05 were considered to be statistically significant.
Author contributions
Y. W., J. Q., Q. T., W. C., and C. C. conceptualization; Y. W., R. L., and C. C. data curation; Y. W. and C. C. formal analysis; Y. W. and C. C. validation; Y. W. investigation; Y. W., F. L., C. Y., H. Z., Y. L., and X. W. methodology; Y. W. writing-original draft; Y. W. and C. C. writing-review and editing; Z. L. resources; Z. L., W. C., and C. C. funding acquisition; Z. Z. project administration; R. L., Q. T., W. C., and C. C. supervision; R. L. visualization.
Supplementary Material
This work was supported in part by National Key R&D Program of China Grant 2018YFC2000400; National Nature Science Foundation of China Grants 81660437 (to W. C.), 81830087, U1602221, and 31771516 (to C. C.), 81772847 (to R. L.), and 81672639 (to Z. Z.); and Science and Technology Innovation Team of Yunnan Province Grant 2018HC002. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S4 and Figs. S1–S4.
- DUB
- deubiquitinase
- TNBC
- triple-negative breast cancer
- FGF-BP
- fibroblast growth factor–binding protein 1
- KLF5
- Krüppel-like factor 5
- USP
- ubiquitin-specific protease
- HA
- hemagglutinin
- WB
- Western blotting
- FBS
- fetal bovine serum
- P/S
- penicillin/streptomycin
- IHC
- immunohistochemistry
- GST
- glutathione S-transferase.
References
- 1. Siegel R. L., Miller K. D., and Jemal A. (2018) Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2. Bastien R. R., Rodríguez-Lescure Á., Ebbert M. T., Prat A., Munárriz B., Rowe L., Miller P., Ruiz-Borrego M., Anderson D., Lyons B., Álvarez I., Dowell T., Wall D., Seguí M. Á., Barley L., et al. (2012) PAM50 breast cancer subtyping by RT-qPCR and concordance with standard clinical molecular markers. BMC Med. Genomics 5, 44 10.1186/1755-8794-5-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hurvitz S. A., and Finn R. S. (2009) What's positive about “triple-negative” breast cancer?: Future oncology, future medicine. Future Oncol. 5, 1015–1025 10.2217/fon.09.68 [DOI] [PubMed] [Google Scholar]
- 4. Elias A. D. (2010) Triple-negative breast cancer: a short review. Am. J. Clin. Oncol. 33, 637–645 10.1097/COC.0b013e3181b8afcf [DOI] [PubMed] [Google Scholar]
- 5. Shao F., Sun H., and Deng C. X. (2017) Potential therapeutic targets of triple-negative breast cancer based on its intrinsic subtype. Oncotarget 8, 73329–73344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oualla K., El-Zawahry H. M., Arun B., Reuben J. M., Woodward W. A., Gamal El-Din H., Lim B., Mellas N., Ueno N. T., and Fouad T. M. (2017) Novel therapeutic strategies in the treatment of triple-negative breast cancer. Ther. Adv. Med. Oncol. 9, 493–511 10.1177/1758834017711380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Robson M., Im S. A., Senkus E., Xu B., Domchek S. M., Masuda N., Delaloge S., Li W., Tung N., Armstrong A., Wu W., Goessl C., Runswick S., and Conte P. (2017) Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med. 377, 523–533 10.1056/NEJMoa1706450 [DOI] [PubMed] [Google Scholar]
- 8. Schmid P., Adams S., Rugo H. S., Schneeweiss A., Barrios C. H., Iwata H., Diéras V., Hegg R., Im S. A., Shaw Wright G., Henschel V., Molinero L., Chui S. Y., Funke R., Husain A., et al. (2018) Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 379, 2108–2121 10.1056/NEJMoa1809615 [DOI] [PubMed] [Google Scholar]
- 9. Traina T. A., Miller K., Yardley D. A., Eakle J., Schwartzberg L. S., O'Shaughnessy J., Gradishar W., Schmid P., Winer E., Kelly C., Nanda R., Gucalp A., Awada A., Garcia-Estevez L., Trudeau M. E., et al. (2018) Enzalutamide for the treatment of androgen receptor-expressing triple-negative breast cancer. J. Clin. Oncol. 36, 884–890 10.1200/JCO.2016.71.3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim S. B., Dent R., Im S. A., Espié M., Blau S., Tan A. R., Isakoff S. J., Oliveira M., Saura C., Wongchenko M. J., Kapp A. V., Chan W. Y., Singel S. M., Maslyar D. J., Baselga J., et al. (2017) Ipatasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer (LOTUS): a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 18, 1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ben-Porath I., Thomson M. W., Carey V. J., Ge R., Bell G. W., Regev A., and Weinberg R. A. (2008) An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40, 499–507 10.1038/ng.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tong D., Czerwenka K., Heinze G., Ryffel M., Schuster E., Witt A., Leodolter S., and Zeillinger R. (2006) Expression of KLF5 is a prognostic factor for disease-free survival and overall survival in patients with breast cancer. Clin. Cancer Res. 12, 2442–2448 10.1158/1078-0432.CCR-05-0964 [DOI] [PubMed] [Google Scholar]
- 13. Takagi K., Miki Y., Onodera Y., Nakamura Y., Ishida T., Watanabe M., Inoue S., Sasano H., and Suzuki T. (2012) Krüppel-like factor 5 in human breast carcinoma: a potent prognostic factor induced by androgens. Endocr. Relat. Cancer 19, 741–750 10.1530/ERC-12-0017 [DOI] [PubMed] [Google Scholar]
- 14. Zheng H.-Q., Zhou Z., Huang J., Chaudhury L., Dong J.-T., and Chen C. (2009) Krüppel-like factor 5 promotes breast cell proliferation partially through upregulating the transcription of fibroblast growth factor binding protein 1. Oncogene 28, 3702–3713 10.1038/onc.2009.235 [DOI] [PubMed] [Google Scholar]
- 15. Xia H., Wang C., Chen W., Zhang H., Chaudhury L., Zhou Z., Liu R., and Chen C. (2013) Kruppel-like factor 5 transcription factor promotes microsomal prostaglandin E2 synthase 1 gene transcription in breast cancer. J. Biol. Chem. 288, 26731–26740 10.1074/jbc.M113.483958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jia L., Zhou Z., Liang H., Wu J., Shi P., Li F., Wang Z., Wang C., Chen W., Zhang H., Wang Y., Liu R., Feng J., and Chen C. (2016) KLF5 promotes breast cancer proliferation, migration and invasion in part by upregulating the transcription of TNFAIP2. Oncogene 35, 2040–2051 10.1038/onc.2015.263 [DOI] [PubMed] [Google Scholar]
- 17. Liu R., Shi P., Zhou Z., Zhang H., Li W., Zhang H., and Chen C. (2018) Kruppel-like factor 5 is essential for mammary gland development and tumorigenesis. J. Pathol. 246, 497–507 10.1002/path.5153 [DOI] [PubMed] [Google Scholar]
- 18. Qin J., Zhou Z., Chen W., Wang C., Zhang H., Ge G., Shao M., You D., Fan Z., Xia H., Liu R., and Chen C. (2015) BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat. Commun. 6, 8471 10.1038/ncomms9471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu R., Shi P., Nie Z., Liang H., Zhou Z., Chen W., Chen H., Dong C., Yang R., Liu S., Chen C. (2016) Mifepristone suppresses basal triple-negative breast cancer stem cells by down-regulating KLF5 expression. Theranostics 6, 533–544 10.7150/thno.14315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi P., Liu W., Tala, Wang H., Zhang H., Li F., Wu Y., Kong Y., Zhou Z., Wang C., Chen W., Liu R., and Chen C. (2017) Metformin suppresses triple-negative breast cancer stem cells by targeting KLF5 for degradation. Cell Discovery 3, 17010 10.1038/celldisc.2017.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu R., Zhi X., Zhou Z., Zhang H., Yang R., Zou T., and Chen C. (2018) Mithramycin A suppresses basal triple-negative breast cancer cell survival partially via down-regulating Krüppel-like factor 5 transcription by Sp1. Sci. Rep. 8, 1138 10.1038/s41598-018-19489-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen C., Sun X., Guo P., Dong X. Y., Sethi P., Cheng X., Zhou J., Ling J., Simons J. W., Lingrel J. B., and Dong J. T. (2005) Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J. Biol. Chem. 280, 41553–41561 10.1074/jbc.M506183200 [DOI] [PubMed] [Google Scholar]
- 23. Liu N., Li H., Li S., Shen M., Xiao N., Chen Y., Wang Y., Wang W., Wang R., Wang Q., Sun J., and Wang P. (2010) The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J. Biol. Chem. 285, 18858–18867 10.1074/jbc.M109.099440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Du J. X., Hagos E. G., Nandan M. O., Bialkowska A. B., Yu B., and Yang V. W. (2011) The E3 ubiquitin ligase SMAD ubiquitination regulatory factor 2 negatively regulates Krüppel-like factor 5 protein. J. Biol. Chem. 286, 40354–40364 10.1074/jbc.M111.258707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao K. W., Sikriwal D., Dong X., Guo P., Sun X., and Dong J. T. (2011) Oestrogen causes degradation of KLF5 by inducing the E3 ubiquitin ligase EFP in ER-positive breast cancer cells. Biochem. J. 437, 323–333 10.1042/BJ20101388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen C., Sun X., Ran Q., Wilkinson K. D., Murphy T. J., Simons J. W., and Dong J. T. (2005) Ubiquitin–proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene 24, 3319–3327 10.1038/sj.onc.1208497 [DOI] [PubMed] [Google Scholar]
- 27. Ge F., Chen W., Qin J., Zhou Z., Liu R., Liu L., Tan J., Zou T., Li H., Ren G., and Chen C. (2015) Ataxin-3 like (ATXN3L), a member of the Josephin family of deubiquitinating enzymes, promotes breast cancer proliferation by deubiquitinating Krüppel-like factor 5 (KLF5). Oncotarget 6, 21369–21378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nicassio F., Corrado N., Vissers J. H., Areces L. B., Bergink S., Marteijn J. A., Geverts B., Houtsmuller A. B., Vermeulen W., Di Fiore P. P., and Citterio E. (2007) Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 17, 1972–1977 10.1016/j.cub.2007.10.034 [DOI] [PubMed] [Google Scholar]
- 29. Gao Y., Ding Y., Chen H., Chen H., and Zhou J. (2015) Targeting Kruppel-like factor 5 (KLF5) for cancer therapy. Curr. Top. Med. Chem. 15, 699–713 10.2174/1568026615666150302105052 [DOI] [PubMed] [Google Scholar]
- 30. Liu R., Zhou Z., Zhao D., and Chen C. (2011) The induction of KLF5 transcription factor by progesterone contributes to progesterone-induced breast cancer cell proliferation and dedifferentiation. Mol. Endocrinol. 25, 1137–1144 10.1210/me.2010-0497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang X., Choi P. S., Francis J. M., Gao G. F., Campbell J. D., Ramachandran A., Mitsuishi Y., Ha G., Shih J., Vazquez F., Tsherniak A., Taylor A. M., Zhou J., Wu Z., Berger A. C., et al. (2018) Somatic superenhancer duplications and hotspot mutations lead to oncogenic activation of the KLF5 transcription factor. Cancer Discov. 8, 108–125 10.1158/2159-8290.CD-17-0532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen C. H., Yang N., Zhang Y., Ding J., Zhang W., Liu R., Liu W., and Chen C. (2019) Inhibition of super enhancer downregulates the expression of KLF5 in basal-like breast cancers. Int. J. Biol. Sci. 15, 1733–1742 10.7150/ijbs.35138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhi X., Zhao D., Zhou Z., Liu R., and Chen C. (2012) YAP promotes breast cell proliferation and survival partially through stabilizing the KLF5 transcription factor. Am. J. Pathol. 180, 2452–2461 10.1016/j.ajpath.2012.02.025 [DOI] [PubMed] [Google Scholar]
- 34. Zhao D., Zhi X., Zhou Z., and Chen C. (2012) TAZ antagonizes the WWP1-mediated KLF5 degradation and promotes breast cell proliferation and tumorigenesis. Carcinogenesis 33, 59–67 10.1093/carcin/bgr242 [DOI] [PubMed] [Google Scholar]
- 35. Sloper-Mould K. E., Eyre H. J., Wang X. W., Sutherland G. R., and Baker R. T. (1999) Characterization and chromosomal localization of USP3, a novel human ubiquitin-specific protease. J. Biol. Chem. 274, 26878–26884 10.1074/jbc.274.38.26878 [DOI] [PubMed] [Google Scholar]
- 36. Sharma N., Zhu Q., Wang Q. E., and Wani A. A. (2013) Ubiquitin specific protease 3 is a regulator of histone ubiquitination required for maintenance of genomic stability. Cancer Res. 73, 1785 [Google Scholar]
- 37. Sharma N., Zhu Q., Wani G., He J., Wang Q. E., and Wani A. A. (2014) USP3 counteracts RNF168 via deubiquitinating H2A and γH2AX at lysine 13 and 15. Cell Cycle 13, 106–114 10.4161/cc.26814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raychaudhuri P. (2014) USP3 controls BRCA1 “foci.” Cell Cycle 13, 183–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheng Y. C., and Shieh S. Y. (2018) Deubiquitinating enzyme USP3 controls CHK1 chromatin association and activation. Proc. Natl. Acad. Sci. U.S.A. 115, 5546–5551 10.1073/pnas.1719856115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fu S., Shao S., Wang L., Liu H., Hou H., Wang Y., Wang H., Huang X., and Lv R. (2017) USP3 stabilizes p53 protein through its deubiquitinase activity. Biochem. Biophys. Res. Commun. 492, 178–183 10.1016/j.bbrc.2017.08.036 [DOI] [PubMed] [Google Scholar]
- 41. Lancini C., van den Berk P. C., Vissers J. H., Gargiulo G., Song J. Y., Hulsman D., Serresi M., Tanger E., Blom M., Vens C., van Lohuizen M., Jacobs H., and Citterio E. (2014) Tight regulation of ubiquitin-mediated DNA damage response by USP3 preserves the functional integrity of hematopoietic stem cells. J. Exp. Med. 211, 1759–1777 10.1084/jem.20131436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lancini C., Gargiulo G., van den Berk P. C., and Citterio E. (2016) Quantitative analysis by next generation sequencing of hematopoietic stem and progenitor cells (LSK) and of splenic B cells transcriptomes from wild-type and Usp3-knockout mice. Data in Brief 6, 556–561 10.1016/j.dib.2015.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fang C. L., Lin C. C., Chen H. K., Hseu Y. C., Hung S. T., Sun D. P., Uen Y. H., and Lin K. Y. (2018) Ubiquitin-specific protease 3 overexpression promotes gastric carcinogenesis and is predictive of poor patient prognosis. Cancer Sci. 109, 3438–3449 10.1111/cas.13789 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.