Abstract
Long noncoding RNAs (lncRNAs) modulate gene expression as competing endogenous RNAs (ceRNAs) that sponge regulatory microRNAs (miRNAs). During cellular reprogramming, genes associated with pluripotency establishment need to be up-regulated, and developmental genes need to be silenced. However, how ceRNAs control cellular reprogramming still awaits full elucidation. Here, we used doxycycline-inducible expression of the four transcription factors octamer-binding protein 4 (OCT4), SRY-box 2 (SOX2), Krüppel-like factor 4 (KLF4), and proto-oncogene c-Myc (c-Myc) to generate induced pluripotent stem cells (iPSCs) from mouse embryonic fibroblasts (MEFs). Using RNA-Seq and bioinformatics approaches, we found that the expression levels of miRNAs from MEFs remain high from day 0 to 6 after the doxycycline induction. Many genes targeted by these miRNAs were up-regulated, and long intergenic noncoding RNAs (lincRNAs) and circular RNAs (circRNAs), which have complementary binding sites to these miRNAs, were highly expressed, indicating lincRNAs and circRNAs may function as ceRNAs. Intriguingly, knockdown of the linc/circRNAs that sponge the miRNAs, which target OCT4 down-regulated exogenous OCT4, decreased reprogramming efficiency, and resulted in low-grade iPSCs. Our results suggest that the ceRNA network plays an important role in cellular reprogramming.
Keywords: microRNA (miRNA); long noncoding RNA (long ncRNA, lncRNA); reprogramming; gene regulation; induced pluripotent stem cell (iPS cell) (iPSC); cellular reprogramming; circular RNA (circRNA); competing endogenous RNA (ceRNA); long intergenic noncoding RNA (lincRNA); microRNA (miRNA)
Introduction
The microRNAs (miRNAs)2 (∼22 nucleotide noncoding RNAs) are an abundant class of small noncoding RNAs, which guide the RNA-induced silencing complex to miRNA response elements (MREs) to its targeting transcripts, and can induce mRNA degradation or translational repression (1–3). The efficiency of miRNAs is known related to the abundance of MREs present in the transcripts (4–6). Previous studies have shown that one gene can have multiple MREs for distinct miRNAs, and vice versa, miRNA can target on multiple distinct transcripts (7, 8). A highly expressed MRE-containing transcript can compete for multiple shared miRNAs and lead to observable changes in miRNA activity and thus regulate the expression of one or multiple targeted transcripts. This “sponge” mechanism has been proposed for competing endogenous RNAs (ceRNAs) to regulate other RNA transcripts by competing for shared miRNAs (9, 10).
Such a sponge mechanism involves coding and noncoding transcripts, including long intergenic noncoding RNAs (lincRNAs) (10–13). For example, linc-MD1, a muscle-specific ceRNA, regulates muscle differentiation (13). Recently, another naturally occurring family of noncoding RNAs, the circular RNA (circRNA), from back-spliced exons, has also been reported to act as miRNA sponges to regulate gene expression (14–17), such as the miR-7 inhibitor in the central nervous system (15). Although thousands of lincRNAs have been annotated in eukaryotic genomes and targeted by miRNAs with both computational and experimental evidences (18, 19), so far, only a small number of lincRNAs have been identified to serve as ceRNAs. The circRNAs are also found abundant in eukaryotic transcriptome (20, 21), whether circRNAs can function as miRNA sponges remains completely elusive. Moreover, the biological relevance of ceRNAs has recently been challenged, because the relatively low expression level of most lncRNAs might limit their ability to effectively modulate, in a miRNA-dependent manner, mRNA abundance. For example, the level of one transcript, Aldoa, required to significantly alter the level of one highly abundant miRNA, miR-122, and its targets in adult hepatocytes was found to exceed the changes observed in vivo, even under extreme physiological or disease conditions (5). The discrepancies in the conclusions of the different attempts suggest that substantially more genetic and genomic evidence from diverse cell types will be required to resolve this issue and to establish the general prevalence and physiological relevance of ceRNAs.
Considering that many established ceRNAs neither share an unusual high number of predicted MREs with their mRNA targets nor are especially abundant (10, 22), the ability to modulate mRNA abundance was thought to be limited (4, 6). However, accumulating evidences suggest that ceRNAs might play an important role in regulating key transcription factors during the cell-fate decision process, supported by the observation that the expression of ceRNAs is tightly regulated with spatial and temporal expression patterns. For example, linc-RoR competes for miR-145 binding with key self-renewal transcription factor transcripts, including Nanog, Oct4, and Sox2, and is expressed in undifferentiated embryonic stem cells (ESCs) (11). The specific expression profiles of these ceRNAs might specify the cells in which their activity exerts the greatest effect. Cell-fate decision often involves switch-like responses in the expression levels of key regulatory genes that result in coordinated changes in transcription profiles, suggesting that lowly abundant, but specifically expressed, ceRNAs might function efficiently on regulation of the transition from differentiated to pluripotent cell states.
Cell fate conversion can be induced by over-expressing sets of regulatory transcription factors (23–25). The over-expression of Oct4, Sox2, Klf4, and c-Myc (OSKM) can reprogram mouse embryonic fibroblasts (MEFs) to induced pluripotent stem cells (iPSCs) (26, 27). In this study we performed ribo- minus RNA-seq and small RNA-seq from cells collected at the early reprogramming stages and fully reprogrammed iPSCs, which can generate “all-iPSCs” pups (Fig. 1A), to profile expression patterns of miRNAs, lincRNAs, circRNAs, and mRNAs. We also took advantage of publicly available RNA-seq and small RNA-seq data from Mbd3flox/− (a member of nucleosome remodeling and deacetylase co-repressor complex) reprogramming system, which results in near 100% efficiency of iPSCs reprogramming within 7 days by OSKM induction (28), to confirm our analysis. Our studies ought to determine the possible function of linc/circRNAs as miRNAs sponge during cellular reprogramming.
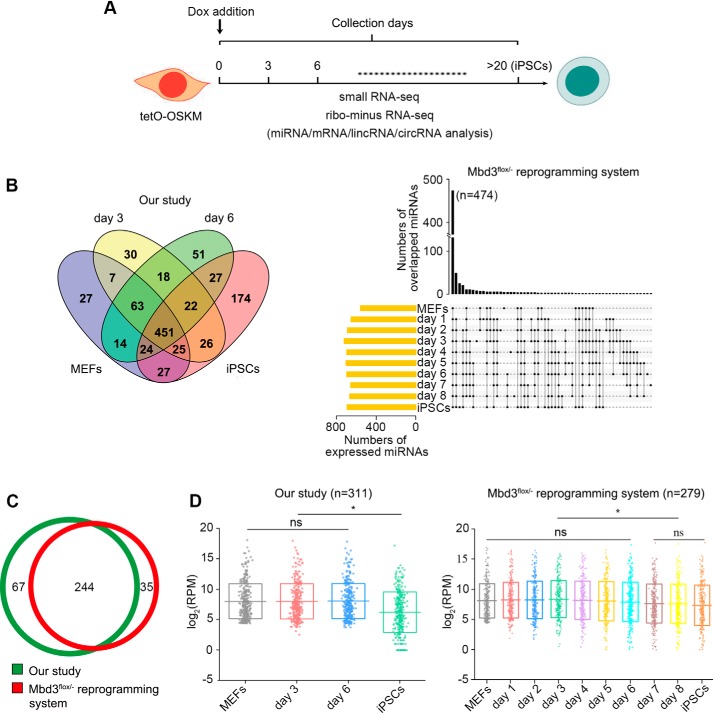
Figure 1.
Profile of miRNAs abundance during reprogramming. A, schematics of iPSCs reprogramming indicating the time points at which samples were collected for library preparations. B, overlap of miRNAs expressed in different reprogramming stages and fully reprogrammed iPSCs. C, highly-expressed miRNAs in MEFs. Only the top 50% abundant and RPM > 20 miRNAs were selected. 244 miRNAs were overlapped between the two reprogramming systems. D, expression patterns of the MEF-high miRNAs over the course of reprogramming and in iPSCs. Each dot represents one MEF-high miRNA. The bottom and top of boxes indicate the 25th and 75th percentiles. The horizontal lines stand for the medians. * indicates p < 0.05.
Results
Profile of miRNAs from MEFs during reprogramming
MEFs, from mice carrying a single Dox-inducible OSKM polycistronic 4F2A cassette (tetO-OSKM), were induced by reprogramming by adding doxycycline to the culture medium. To demonstrate the miRNA-mediated cross-talk during reprogramming, we first profiled small RNAs in MEFs, the reprogramming MEFs (rMEFs) at day 3 and day 6 post-OSKM induction and the fully reprogrammed iPSCs. To confirm our data, profiling of small RNA (GEO accession GSE102518) from the Mbd3flox/− reprogramming system were also analyzed. 451 and 474 miRNAs were co-expressed among MEFs, rMEFs, and iPSCs in our and Mbd3flox/− reprogramming systems, respectively, indicating miRNAs from MEFs are sustainable during reprogramming (Fig. 1B). To figure out the extent of miRNA-mediated interactions between their mRNA targets and lncRNAs, we selected 311 and 279 miRNAs highly expressed in our and Mbd3flox/− MEFs (top 50% abundance and RPM >20), and 244 miRNAs were overlapped (Fig. 1C). The expression levels of the MEF-high miRNAs were not significantly changed over the course of reprogramming, and significantly decreased in reprogrammed cells (at days 7 and 8 in the Mbd3flox/− reprogramming system) and iPSCs (Fig. 1D, Table S1), and the miRNAs were chosen for further analysis.
Activity of miRNAs may be inhibited during reprogramming
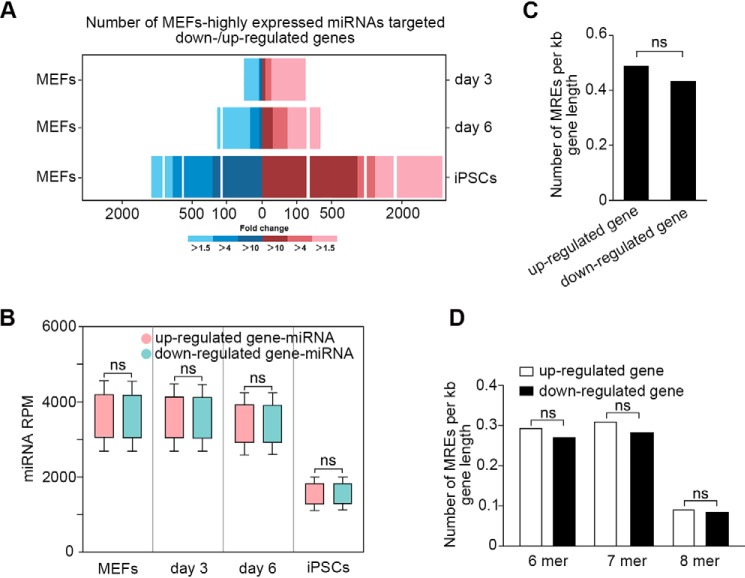
During reprogramming, genes associated with pluripotent establishment need to be activated or up-regulated, whereas developmental genes need to be silenced (28). To determine the role of miRNAs in regulating the transcriptomic changes, we profiled expression of mRNAs on MEFs, rMEFs, and iPSCs as well, and then analyzed the effect of the MEF-high miRNAs on up- and down-regulated genes during reprogramming. We found the miRNAs were not only targeted on the down-regulated genes, but also the up-regulated genes, including key pluripotent transcriptional factors, such as Oct4, Klf4, Nanog, Tbx3, Sall4, and Esrrb. Notably, the number of up-regulated genes targeted by the miRNAs was more than that of the down-regulated genes (Fig. 2A; Table S2). The expression levels of miRNAs targeting on up- and down-regulated genes had no significant difference (p > 0.05), and both were maintained at high levels during reprogramming and decreased in iPSCs (Fig. 2B). Then, we checked the densities of MREs in up- and down-regulated mRNA targets. We found that the MRE density in down-regulated mRNA targets (mean of 0.430 MREs/kb) is not significantly different with up-regulated mRNA targets (mean of 0.487 MREs/kb; p > 0.05) (Fig. 2C). Considering that high-affinity MREs would be more effective to bind miRNAs than low-affinity MREs, resulting in down-regulation of gene expression more efficiently, we analyzed the ratio of 8-mer, 7-mer, and 6-mer MREs in the targets, and found no significant difference between the up- and down-regulated targets (Fig. 2D). Our results show that neither the density nor the affinity of MREs is different between the miRNA-targeted up- and down-regulated genes. During reprogramming, developmental genes should be silenced, so the highly-expressed miRNAs targeting the down-regulated genes may have little effect on reprogramming. However, if expression of the up-regulated genes is influenced by the miRNAs, the reprogramming will fail. Thus, we hypothesize that additional mechanisms may exist to inhibit the activity of the MEF-high miRNAs during reprogramming.
Figure 2.
Activity of miRNAs may be reduced during reprogramming. A, the numbers of differentially expressed genes targeted by MEF-high miRNAs between MEFs and rMEFs or iPSCs. Blue and red bars represent down- and up-regulated genes targeted by the miRNAs, respectively. B, expression patterns of miRNAs targeting on down- and up-regulated genes during reprogramming. There is no significant difference in expression between the two kinds of miRNAs. Up-regulated gene-miRNA: miRNA targeting on up-regulated gene; down-regulated gene-miRNA: miRNA targeting on down-regulated gene. The error bars represent S.D. C, densities of MREs in up-regulated and down-regulated mRNA targets. There is no significant difference for MREs densities between down-regulated (mean of 0.430 MREs/kb) and up-regulated (mean of 0.487 MREs/kb; p > 0.05). D, analysis of MREs affinity in up-regulated and down-regulated mRNA targets. The ratio of 8-mer, 7-mer, and 6-mer MREs in up-regulated targets is 0.092, 0.312, and 0.292 MREs/kb, and 0.084, 0.282, and 0.269 MREs/kb in down-regulated targets, respectively.
Linc/circRNAs may serve as sponges for multiple miRNAs during reprogramming
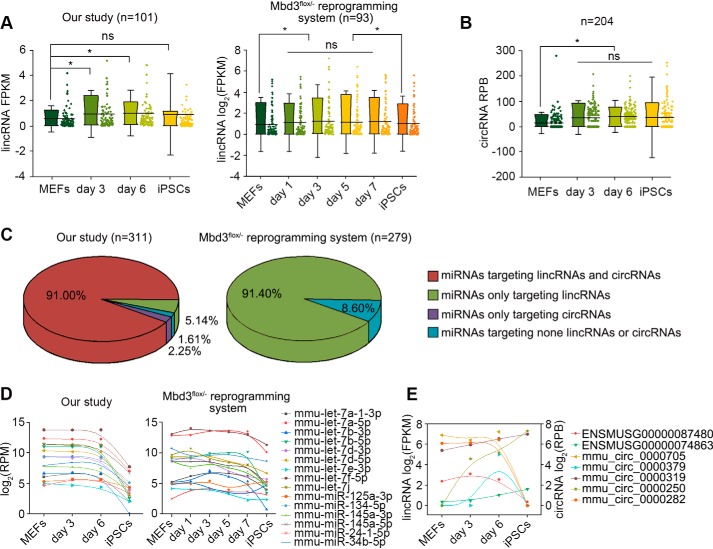
Previous studies have shown that linc/circRNAs can function as miRNA sponges to reduce miRNA activity to regulate its targets (11, 14). We propose that linc/circRNAs may serve as sponges to counteract the activities of the MEF-high miRNAs during reprogramming. Taking advantage of ribo- minus RNA-seq, we characterized linc/circRNAs in rMEFs and iPSCs. 1490 known lincRNAs and 641 known circRNAs were identified (Table S3). Compared to MEFs, 101 lincRNAs (Fig. 3A) and 204 circRNAs (Fig. 3B) were found to express with higher levels in day 3 and 6 rMEFs. The limitation of sequencing libraries from the Mbd3flox/− reprogramming system is the inability to detect nonpolyadenylated (poly(A)−) RNA, so we cannot identify circRNAs using the data (GEO accession GSE102518). However, 93 highly-expressed lincRNAs were characterized during the reprogramming (Fig. 3A). Strikingly, we found that almost all of the MEF-high miRNAs shared complementary binding sites with the highly-expressed linc/circRNAs with only few exceptions (Fig. 3C), indicating that linc/circRNAs might regulate gene expression through inhibiting the activity of miRNAs during reprogramming. In addition, we observed that, in both reprogramming systems, some miRNAs (mmu-let-7f-5p, mmu-let-7a-5p, mmu-let-7i, mmu-let-7d-5p, mmu-let-7b-5p, mmu-let-7d-3p, mmu-let-7b-3p, mmu-let-7e-3p, mmu-let-7a-1–3p, mmu-miR-145a-5p, and mmu-miR-145a-3p), proven to serve as reprogramming barriers, such as let-7 and miR-34 families (29, 30), were relatively high during reprogramming, and low in iPSCs (Fig. 3D). We wonder if there are linc/circRNAs that can counteract the activity of the miRNAs. As expected, we identified some linc/circRNAs, which have complementary binding sites with the miRNAs, and found seven linc/circRNAs were highly expressed in rMEFs at days 3 and/or 6, compared to MEFs (Fig. 3E). The results suggest the general function of linc/circRNAs as sponges on inhibition of activity of miRNAs during reprogramming.
Figure 3.
Linc/circRNAs predicted to sponge multiple miRNAs during reprogramming. A, lincRNAs highly expressed during iPSCs reprogramming. B, circRNAs highly expressed during iPSCs reprogramming. * indicates p < 0.05. The error bars represent S.D. C, MEFs highly expressed miRNAs predicted to target linc/circRNAs. Almost all of the miRNAs shared complementary binding sites with the highly expressed linc/circRNAs with only a few exceptions. D, expression patterns of miRNAs that have been proven to inhibit cellular reprogramming. E, expression patterns of linc/circRNAs predicted to be targeted by the miRNAs inhibiting cellular reprogramming.
Activity of miRNAs targeting on Oct4 is counteracted during iPSCs reprogramming
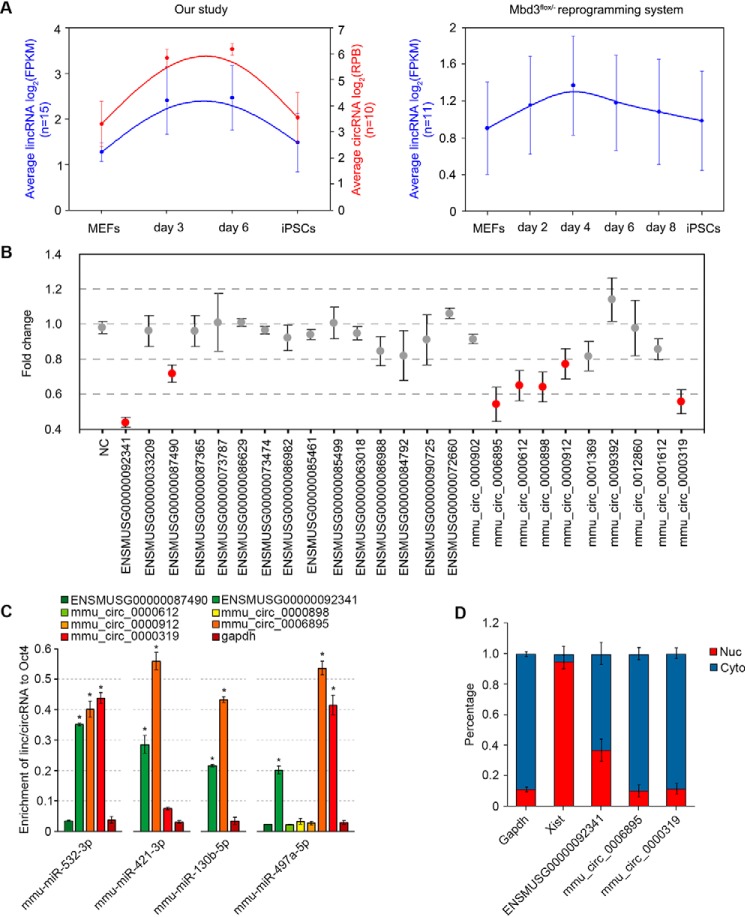
To further demonstrate the ceRNA mechanism during reprogramming, we tested the hypothesis in the context of Oct4, a key pluripotent transcriptional factor that determines the successful reprogramming and is not replaceable in induction of iPSCs (31). We predicted seven MEF-high miRNAs targeting on the CDS region of Oct4 (Oct4-miRNAs) by RNA22 (32) (Fig. 4A). However, definitely, Oct4 was up-regulated (Fig. 4B). During iPSCs reprogramming, endogenous Oct4 activation generally takes place at 7–12 days (33), and, in the study, we found that the expression of endogenous Oct4 was initiated from day 7 post-OSKM induction (Fig. 4C), so the ceRNA mechanism we checked mainly involves the regulation of exogenous Oct4 expression. During reprogramming, the expression of the seven Oct4-miRNAs were sustained in high levels, and significantly decreased in reprogrammed cells (at days 7 and 8 in Mbd3flox/− reprogramming system) and iPSCs (Fig. 4D). This result suggests that the expression of Oct4 is not tightly regulated by the miRNAs or a mechanism that can counteract the function of the miRNAs. To confirm if Oct4-miRNAs can down-regulate the expression of exogenous Oct4 during the process of reprogramming, the increasing concentrations of the mimics of the miRNAs were transfected into MEFs and the expression of Oct4 mRNA was monitored by qRT-PCR at day 3 post-OSKM induction. We found that mmu-miR-130b-5p, mmu-miR-421–3p, mmu-miR-497a-5p, and mmu-miR-532-3p could efficiently down-regulate Oct4 expression at high concentrations, and the dose-dependent manner revealed that activity of the miRNAs might be limited (Fig. 4E). To ensure that the miRNAs can definitely function on Oct4 mRNA, we inserted the native and mutant MREs of the four Oct4-miRNAs into the 3′ end of a standard luciferase reporter after an internal ribosome entry site (Fig. 4F) and transfected into MEFs, respectively. The luciferase assay was detected at day 3 post-OSKM induction and showed that the luciferase activity expressed by the reporters with native MREs was significantly decreased, and by the reporters with mutant MREs had no significant changes, suggesting the miRNAs can indeed repress the expression of Oct4 by targeting on the CDS (Fig. 4G). These results suggest that the Oct4-miRNAs can down-regulate exogenous Oct4 expression, however, their activities are counteracted during reprogramming.
Figure 4.
Activity of Oct4-miRNAs is limited during iPSCs reprogramming. A, MEF-high miRNAs predicted to target on the CDS region of Oct4 (Oct4-miRNAs). Each shape represents a miRNA. B, expression patterns of Oct4 during reprogramming. The expression of Oct4 is shown by red dots. C, expression of exogenous Oct4 (exo-Oct4) and endogenous Oct4 (endo-Oct4) during reprogramming checked by qRT-PCR. D, expression patterns of Oct4-miRNAs during reprogramming. *, indicates p < 0.05. E, fold-change of Oct4 mRNA with increasing concentrations of Oct4-miRNAs as indicated. The increasing concentrations of the miRNA mimics were transfected into MEFs and the expression level of Oct4 mRNA was analyzed by qRT-PCR at day 3 post-OSKM induction. F, luciferase reporter constructs containing the native and mutant MREs in the luciferase 3′ end. The seed sequences of the miRNAs are highlighted in red, and the mutant nucleotides are shown in green. G, luciferase assay results detected at day 3 post-OSKM. The luciferase activity expressed by the reporters with native MREs was significantly decreased, and the reporters with mutant MREs had no significant changes. *, indicates p < 0.01. The error bars represent S.D.
Linc/circRNAs can sustain high expression of exogenous Oct4 by sponging the miRNAs targeting on Oct4
We considered if the linc/circRNAs, which have complementary binding sites with the Oct4-miRNAs could counteract the activity during reprogramming. We indeed found that some linc/circRNAs, which have complementary binding sites with the four Oct4-miRNAs were highly expressed during reprogramming, and decreased in reprogrammed cells and iPSCs (Fig. 5A). We postulate that if we knockdown the expression of the linc/circRNAs, exogenous Oct4 could be down-regulated for the increased activity of the miRNAs. We selected the efficient locked nucleic acid (LNA)-siRNA for each lincRNA and circRNA (LNA-siRNAs targeting the back-splice sequences of circRNAs were used) (Fig. S1). As expected, Oct4 expression was significantly down-regulated at least 20% when some linc/circRNAs were knocked down (Fig. 5B). We supposed that these linc/circRNAs might benefit Oct4 expression through sponging the Oct4-miRNAs. To test the hypothesis, we conducted RNA immunoprecipitation (RIP) using day 3 rMEFs from MEFs transfected with the biotin-labeled miRNAs at a final concentration of 20 nm, and observed that ENSMUSG00000092341, mmu_circ_00006895, and mmu_circ_00000319 were specifically enriched by qRT-PCR analysis normalized to captured Oct4 mRNA (up to 20-fold enrichment compared with Gapdh mRNA), suggesting the three linc/circRNAs are able to interact with the Oct4-miRNAs (Fig. 5C). Further analysis showed multiple complementary binding sites with mmu-miR-130b-5p, mmu-miR-421–3p, mmu-miR-497a-5p, and mmu-miR-532–3p on the sequences of the linc/circRNAs (Fig. S2). LncRNAs in the cytoplasm may function as miRNA sponge. Thus, we analyzed the subcellular localization of the linc/circRNAs. Gapdh and Xist were checked as control. The results showed that the linc/circRNAs were mainly located in the cytoplasm (Fig. 5D). These results show that the linc/circRNAs function as Oct4-miRNAs sponges to sustain high expression of exogenous Oct4 during reprogramming.
Figure 5.
Linc/circRNAs sponge miRNAs targeting on Oct4. A, expression patterns of linc/circRNAs, which have complementary binding sites with the four Oct4-miRNAs. B, fold-change of Oct4 mRNA with knockdown of the linc/circRNAs as indicated. Down-regulation of the linc/circRNAs highlighted in red significantly suppressed Oct4 expression by at least 20% (p < 0.001). C, RIP assay for detecting the interaction of the Oct4-miRNAs with the specific linc/circRNAs. Biotin-labeled miRNAs were transfected into MEFs at a final concentration of 20 nm, and the RIP assay was performed using day 3 rMEFs (n = 3). The enrichment of lincRNAs or circRNAs in bound fractions was evaluated by qRT-PCR analysis normalized to Oct4 mRNA. Gapdh was analyzed as the negative control. D, subcellular localization analysis of the linc/circRNAs. Nuc, nucleoplasm; Cyto, cytoplasm. Gapdh and Xist act as cytoplasmic and nuclear control, respectively.
The linc/circRNAs can improve reprogramming efficiency and the quality of iPSCs
The efficiency to derive alkaline phosphatase (AP)-positive clones was about 1.31% using the secondary reprogramming system (Fig. 6A), and the iPSCs could give rise to chimeric pups and contributed to germline transmission (Table 1). Using the reprogramming system, we investigated whether the linc/circRNAs have any functional effects on reprogramming. To this end, we transfected each of the LNA-siRNAs targeting the linc/circRNAs in MEFs, respectively, and we did not find any significant changes about the formation of AP-positive colonies at day 17 post-OSKM induction. However, when the mixture of the LNA-siRNAs (si-mix) were transfected, we found that colony formation was significantly decreased (Fig. 6A) and only two cell lines were obtained. When additional Oct4 was added back (si-mix+Oct4-addback) by infecting MEFs using lentiviral Oct4, the number of AP-positive colonies was greatly enhanced (Fig. 6A). To test if the overexpression of these lncRNAs can enhance the reprogramming, we cloned a part of the sequence (3000 bp) of ENSMUSG00000092341 (the sequence includes 3, 2, 2, and 1 complementary binding sites of mmu-miR-532–3p, mmu-miR-497a-5p, mmu-miR-421–3p, and mmu-miR-130b-5p, respectively) and overexpressed it in MEFs, and then induced the reprogramming by adding doxycycline. However, after day 17 post-OSKM induction, we did not observe any significant changes in the formation of AP-positive colonies (Fig. S3). To further confirm that low reprogramming efficiency is related to the decreased expression of exogenous Oct4 induced by knockdown of the linc/circRNAs, we tested the expression of the other three reprogramming factors, Sox2, Klf4, and c-Myc, after knockdown of the three linc/circRNAs, and found their expressions were not significantly affected (Fig. S4). Low levels of exogenous Oct4 leads to generation of iPSCs with aberrant DNA methylation of the Dlk1-Dio3 locus, low expression of miRNAs encoded by the locus, and the low capacity in chimeric mice (33). Thus, we checked the methylation status of the Dlk1-Dio3 locus. Control iPSCs showed normal, 50–60% DNA methylation, whereas the linc/circRNA-knockdowned cells, especially the si-mix cells, showed DNA hypermethylation, and Oct4-addback could rescue the methylation defect (Fig. 6B). Consistent with the hypermethylation pattern, the miRNAs in the locus showed significantly lower expression in linc/circRNA-knockdowned cells compared to control iPSCs. Also, the expressions could be rescued by Oct4-addback (Fig. 6C). In addition, we tested the in vivo developmental potency of the iPSCs by injection into blastocysts. In contrast, only one si-mix cell line could produce chimeric mice with the low extent of chimaerism and fail to support germline transmission (Table 1), whereas, all three Oct4-addback cell lines produced chimeric pups and one cell line could contribute to germline transmission (Fig. 6D, Table 1). The results indicate that the linc/circRNAs can improve reprogramming efficiency and the quality of iPSCs by sponging miRNAs targeting on Oct4.
Figure 6.
The linc/circRNAs can improve iPSC reprogramming. A, effect of knockdown of the linc/circRNAs on formation of AP-positive iPSC colonies. MEFs were obtained from E13.5 embryos carrying a single copy of Dox-inducible expression of the four transcription factors. AP staining was performed at day 17 after doxycycline addition. Before Dox induction, 10,000 cells were placed in each well in a 6-well plate. Data are represented as mean ± S.D., n = 3. *, p < 0.001. B, DNA methylation analysis on the Dlk1-Dio3 locus in cell lines. C, expression of the Dlk1-Dio3 locus-encoded miRNAs in cell lines. The 2−ΔΔCt method was used for analysis of the relative expression level of the miRNAs. The relative expression level of the miRNAs in control iPSCs was calculated as 1.00. Expression levels were normalized to control iPSCs. si-mix, cells transfected with a mix of siRNAs targeting on the linc/circRNAs; si-mix+Oct4 addback, cells transfected with mixture of siRNAs targeting on the linc/circRNAs and infected by retroviral Oct4. Each shape represents a cell line. D, images of chimeras obtained from si-mix+Oct4 addback iPSCs and its F1 progeny resulting from germline transmission. *, pup derived from the germline-competent iPSCs.
Table 1.
In vivo developmental potency of iPSCs
The extent of chimaerism was estimated on the basis of coat color.
| Cell lines | Blastocysts injected | Chimeric/total pups | Chimaerism | Germline transmission |
|---|---|---|---|---|
| % | ||||
| Con. line 1 | 44 | 9/16 | 20–80 | Yes |
| Con. line 2 | 48 | 7/22 | 40–70 | Yes |
| si-mix line 1 | 60 | 2/34 | 10, 20 | No |
| si-mix line 2 | 62 | 0/26 | 0 | No |
| si-mix + Oct4 line 1 | 46 | 8/22 | 30–80 | Yes |
| si-mix + Oct4 line 2 | 60 | 9/31 | 10–60 | No |
| si-mix + Oct4 line 3 | 68 | 8/26 | 20–50 | No |
Discussion
Herein we investigated the prevalence and properties of lncRNAs as ceRNA during reprogramming by experimental and bioinformatic approaches. We focused on the analysis of linc/circRNAs with the proposed role in the regulation of cell states from differentiation to pluripotency. Our data reveal the linc/circRNAs can sponge the miRNAs targeting on Oct4 mRNA and regulate expression of exogenous Oct4, and further improve reprogramming efficiency and quality of iPSCs (Fig. 7). In the context of this critical cellular transition when repertoires of miRNAs are limited, the impact of linc/circRNAs on transcriptional programs is likely to be greater than in differentiated cells. Environmental or cellular stress, for example, upon starvation or infection, may also offer similar opportunities for strong effects of ceRNA.
Figure 7.
Model depicting linc/circRNAs as miRNAs sponges sustain high expression of exogenous Oct4 during reprogramming. During reprogramming, the miRNAs targeting Oct4 mRNA from MEFs are highly expressed, however, the expression of exogenous Oct4 is not suppressed. Linc/circRNAs, which have complementary binding sites with the miRNAs targeting on Oct4 are also highly expressed and can counteract the activities of the miRNAs as sponges. The knockdown of the linc/circRNAs results in down-regulation of Oct4 expression, the imprinting defect at the Dlk1-Dio3 locus, decrease of reprogramming efficiency, and low-grade chimera forming iPSCs, and those can be rescued by Oct4-addback. In summary, the linc/circRNAs can improve reprogramming efficiency and quality of iPSCs by sponging miRNAs targeting on Oct4 during reprogramming.
We integrated the data of ribo- minus RNA-seq and small RNA-seq from cells collected during reprogramming and fully reprogrammed iPSCs. During reprogramming, OSKM induction causes drastic changes of transcriptome (23, 27, 34). It leads to inactivation of genes responsible for the identity of the parental cells and the activation of genes that are crucial for the establishment of the lineage of interest (23). Similarly, in our study, we observed initial silencing of fibroblast genes and their transcriptomic activity was gradually replaced with switching-on genes associated with pluripotency. However, we found the pool of miRNAs from MEFs did not changed remarkably during reprogramming, indicating reprogramming of the miRNA expression pattern is a latter event. To ensure the successful reprogramming process, the activity of miRNAs targeting on genes that need to be activated or up-regulated must be reduced. Indeed, we identified many linc/circRNAs, which share miRNA-binding sites with mRNAs of pluripotent transcription factors, and were highly expressed, suggesting that ceRNAs could be beneficial for reprogramming via a miRNA-mediated mechanism.
LncRNAs are proposed to act as sponges for miRNAs to modulate gene expression (4, 35). In mouse ESCs, many lncRNAs have been predicted to compete for miRNAs with mRNAs (4). However, it has not yet been experimentally validated as ceRNAs, especially circRNAs. A previous study has shown that linc-ROR regulates Oct4, Nanog, and Sox2 to sustain self-renewal of human ESCs by sponging miRNAs (11). In the study, we identified that linc/circRNAs could serve as ceRNAs during reprogramming. We evidenced one lincRNA and two circRNAs that could sustain high expression of the core transcription factor Oct4 by sponging miRNAs during reprogramming, and they ultimately improved the reprogramming efficiency and quality of iPSCs. The knockdown of each of the lncRNA could significantly down-regulate Oct4 expression, but the formation of AP-positive colonies was not affected, indicating the lncRNAs as miRNA sponges can mutually compensate and iPSCs reprogramming can tolerate a certain down-regulation of exogenous Oct4 expression. Furthermore, considering that the formation of AP-positive colonies was not significantly changed by knockdown or overexpression of one single lncRNA, we suggest that the combined effect of the lncRNAs as miRNA sponges, rather than a single lncRNA, plays the central role on iPSCs reprogramming. During reprogramming, the activation of endogenous pluripotent genes happens at late stages (27, 36). So, here, the ceRNA mechanism is involved in the regulation of exogenous Oct4 expression. Whether this mechanism regulates endogenous pluripotent gene expression during reprogramming remains to be further determined. Together with previous studies (11), we suggest that the ceRNA network plays an important role on establishment and maintenance of pluripotency during reprogramming and in pluripotent stem cells.
In summary, our results point out a high prevalence of miRNA-mediated interactions between their mRNA targets and lncRNAs, proposing that this mechanism of ceRNAs indeed functions on regulation of gene expression, particularly in the context of cell-fate transitions.
Experimental procedures
Animals
Mice were housed and prepared according to the protocol approved by the Institutional Animal Care and Use Committee of Northeast Agriculture University (protocol number IACUC-02-005) and the IACUC of Weill Cornell Medical College (protocol number 2014-0061).
Procedure of iPSCs reprogramming
For the secondary reprogramming system, MEFs were obtained from E13.5 embryos from C57BL/6 mice carrying a single copy of Dox-inducible expression of OSKM (as a gift from Zev Rosenwaks, Ronald O. Perelman and Claudia Cohen Center for Reproductive Medicine, Weill Cornell Medical College). The MEFs within two passages were split when they reached 80–90% confluence, placed at 5000 cells/cm2. Before Dox induction, 10,000 cells were placed in each well in a 6-well plate. 2 μg/ml of doxycycline was added to induce over-expression of OSKM. Reprogramming was performed in KSR medium (Dulbecco's modified Eagle's medium/F-12 (Gibco) supplemented with 15% Knockout Serum Replacement (Gibco), 2 mm l-glutamine (Gibco), 1× Pen/Strep (Gibco) 100 μm minimal essential medium nonessential amino acids (Gibco), 100 μm β-mercaptoethanol (Gibco), and 1000 units/ml of LIF (ESGRO, Millipore), 3 μm CHIR99021 (Stemgent), 1 μm PD0325901 (Selleck), and 0.5× N-2 supplement (Gibco)). AP staining was performed at day 17 after doxycycline addition.
High throughput RNA-seq
Total RNA was isolated from the cells at days 0, 3, and 6 post-OSKM induction and the fully reprogrammed iPSCs using TRIzol RNA extraction reagent (Thermo Fisher Scientific), following the manufacturer's protocol. The concentration and purity of total RNA were assessed on Nanodrop 2000 (Thermo Fisher Scientific), and the integrity of total RNA was evaluated with 2% agarose gel electrophoresis and a RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA). The cDNA libraries of long chain RNAs (mRNA/lncRNA/circRNA) and small RNAs were generated from 10 and 2 μg of total RNA, respectively. rRNA was removed using a Epicenter Ribo-zero rRNA Kit (Epicenter). Sequencing libraries were generated using the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (number E7530L, New England Biolabs) and NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (number E7300L, New England Biolabs), respectively. Sequencing was performed using Illumina HiSeqTM 2000 platform by the Genomics Facility at Cornell Biotechnology Resource Center. The RNA-seq and small RNA-seq data were uploaded as Gene Expression Omnibus GSE116684 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116684).
RNA-seq data processing and analysis
The qualified reads were aligned with Tophat 2 into the mouse reference genome by default parameters. The reads on the extracted alignment were constructed with Cufflinks to construct transcripts. The transcripts of all samples were combined and reconstructed into a large transcript file (merged.gtf) using Cuffmerge. Then the expression of genes in each sample were quantified to FPKM. For differential expression, we first counted the overlap of reads with genes by htseq-count. Next, we compared the two groups using default parameters in R package DESeq. A gene was considered significant if the Benjamini and Hochberg-adjusted p value (Padj) was less than 5% and the fold-change was greater than 2.
Quantification of miRNA abundance and prediction of miRNA response elements
Clean sequencing reads were obtained by removing from the raw data reads containing more than one low quality (Q-value ≤20) base, with 5′ primer contaminants, without the 3′ primer, without the insert, with poly(A), and shorter than 18 nucleotides. Clean reads were aligned with the reference genome (ftp://ftp.ensembl.org/pub/release-92/fasta/mus_musculus/dna/Mus_musculus.GRCm38.dna.toplevel.fa.gz), the Rfam database 13.0 (37), and the RepBase (38) to identify and remove rRNA, scRNA, snoRNA, snRNA, tRNA, repeat sequences, and fragments from mRNA degradation. The remaining reads were searched against miRBase 22.0 (39) to identify known miRNAs in mouse. The possible MREs of known miRNAs were predicted by RNA22 (32), an miRNA target prediction tool, with default parameter setting and the binding energy cutoff was less than or equal to −20 Kcal/mol. The numbers of MREs per kb length of each up-regulated or down-regulated gene and the average numbers of MREs in each group were calculated. The miRNA expression level was calculated and normalized using RPM (reads per million). We identified significant differentially expressed miRNAs with a false discovery rate <0.05 and fold-change ≥2 threshold by DEGseq (40).
Identification of linc/circRNAs and expression analysis
Pre-processing of raw sequencing data were performed using the FASTX-Toolkit with default parameters by removing low quality reads (more than 20% of the bases qualities are lower than 10), reads with adaptors, and reads with unknown bases (n bases more than 5%). Clean reads were mapped to the mouse reference genome (ftp://ftp.ensembl.org/pub/release-92/fasta/mus_musculus/dna/Mus_musculus.GRCm38.dna.toplevel.fa.gz). Unmapped reads were collected and the software CIRI (41) and find_circ (17) were used to identify circRNAs with default options, and circBase IDs were used to indicate known circRNAs. Counts of identified circRNA reads were normalized by the RPB (junction reads per billion mapped reads) method. For lincRNAs, the reconstruction and identification of transcripts were performed by mapped reads with StringTie (42) and cuffcompare (43). Known lincRNAs were acquired by gene annotations for comparing the assembled transcripts with NCBI, Ensembl, and UCSC mouse known genes. The expression level of lincRNA was calculated using FPKM (fragments per kilobase of transcript per million mapped reads) with the software RSEM (44). Transcripts with a false discovery rate <0.05 and fold-change ≥2 were then identified as significant differentially expressed lincRNA or circRNA using DEGSeq (40).
Experimental validation of miRNA activity and interaction between mRNAs and linc/circRNAs
The miRNAs mimics and inhibitors, and linc/circRNAs siRNAs were synthesized by Exiqon, and transfected in triplicates into MEFs using Lipofectamine RNAi MAX Reagent (Invitrogen) according to the manufacturer's guidelines. rMEFs were collected at day 3 post-OSKM induction, RNAs were extracted and checked by qRT-PCR for gene expressions. RIP was performed using a Magnetic RNA-Protein Pull-down Kit (Pierce, number 20164) according to the manufacturer's protocol. Briefly, the 3′ end-biotinylated miRNAs mimics were transfected into MEFs at a final concentration of 20 nm for 1 day. The biotin-coupled RNA complex was pull downed by incubating the cell lysates with streptavidin-coated magnetic beads. The abundance of linc/circRNAs in bound fractions was evaluated by qRT-PCR analysis normalized to Oct4 mRNA. Gapdh was analyzed as negative control. The TetO-FUW-Oct4 plasmid (Addgene plasmid 20323) was used for Oct4-addback, and 10,000 cells infected with a dose of 5 × 108 cfu of virus for 3 h.
Methylation analysis of the Dlk1-Dio3 locus
Genomic DNA isolation from cells and bisulfite pyrosequencing were performed as follows. Briefly, genomic DNA of iPSCs was extracted with the Gentra Pure gene kit (Qiagen) according to the manufacturer's instructions and quantified using a Nano Drop 2000 spectrophotometer. Bisulfite conversion of 500 ng of genomic DNA per sample was performed with the EpiTect Bisulfite Kit (Qiagen) according to the manufacturer's specifications. Quantification of DNA methylation was carried out by PCR of bisulfite-converted DNA and pyrosequencing. PCR and sequencing primers for bisulfite pyrosequencing were designed using Pyrosequencing Assay Design Software 2.0 (Qiagen). For pyrosequencing, a PyroMark Q96 ID instrument (Qiagen) and PyroMark Gold Q96 reagents (Qiagen) were used. Data were analyzed using the PyroMark CpG Software 1.0.11 (Qiagen).
Statistical analysis
All data are presented as mean ± S.D. Differences between groups were tested for statistical significance using Student's t test or χ2 test. Statistical significance was set at p < 0.01.
Author contributions
X. Z., K. Z., H. Z., and X. P. data curation; X. Z., K. Z., and H. Z. software; X. Z., K. Z., H. Z., and X. P. formal analysis; X. Z., J. Z., and K. Z. methodology; X. Z. and Q. K. writing-original draft; J. Z. resources; J. Z. investigation; J. Z. visualization; Z. Y., D. W., and Q. K. supervision; Z. Y. validation; D. W. and Q. K. conceptualization; Q. K. funding acquisition; Q. K. project administration; Q. K. writing-review and editing.
Supplementary Material
This work was supported by the Natural Science Foundation of Heilongjiang Province Grant C2018032, the National Key Research and Development program of China-Stem Cell and Translational Research Grant 2016YFA0100200, and a Starr Foundation Tri-Institutional Stem Cell Core and Derivation grant. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4 and Tables S1–S3.
The microarray data reported in this paper have been submitted to the Gene Expression Omnibus (GEO) database under GEO accession no. GSE116684.
- miRNA
- microRNA
- MRE
- miRNA response elements
- ceRNA
- competing endogenous RNA
- lincRNA
- long intergenic noncoding RNA
- circRNA
- circular RNA
- ESC
- embryonic stem cell
- MEF
- mouse embryonic fibroblast
- iPSC
- induced pluripotent stem cell
- LNA
- locked nucleic acid
- RIP
- RNA immunoprecipitation
- Dox
- doxycycline
- FPKM
- fragments per kilobase of transcript per million mapped read
- AP
- alkaline phosphatase
- Gapdh
- glyceraldehyde-3-phosphate dehydrogenase
- qRT
- quantitative RT.
References
- 1. Hutvágner G., and Zamore P. D. (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060 10.1126/science.1073827 [DOI] [PubMed] [Google Scholar]
- 2. Zeng Y., Wagner E. J., and Cullen B. R. (2002) Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9, 1327–1333 10.1016/S1097-2765(02)00541-5 [DOI] [PubMed] [Google Scholar]
- 3. Iwakawa H-o., and Tomari Y. (2015) The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 25, 651–665 10.1016/j.tcb.2015.07.011 [DOI] [PubMed] [Google Scholar]
- 4. Tan J. Y., Sirey T., Honti F., Graham B., Piovesan A., Merkenschlager M., Webber C., Ponting C. P., and Marques A. C. (2015) Extensive microRNA-mediated crosstalk between lncRNAs and mRNAs in mouse embryonic stem cells. Genome Res. 25, 655–666 10.1101/gr.181974.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Denzler R., Agarwal V., Stefano J., Bartel D. P., and Stoffel M. (2014) Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 54, 766–776 10.1016/j.molcel.2014.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Denzler R., McGeary S. E., Title A. C., Agarwal V., Bartel D. P., and Stoffel M. (2016) Impact of microRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol. Cell 64, 565–579 10.1016/j.molcel.2016.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Friedman R. C., Farh K. K., Burge C. B., and Bartel D. P. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salmena L., Poliseno L., Tay Y., Kats L., and Pandolfi P. P. (2011) A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell 146, 353–358 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tay Y., Kats L., Salmena L., Weiss D., Tan S. M., Ala U., Karreth F., Poliseno L., Provero P., Di Cunto F., Lieberman J., Rigoutsos I., and Pandolfi P. P. (2011) Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147, 344–357 10.1016/j.cell.2011.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y., Xu Z., Jiang J., Xu C., Kang J., Xiao L., Wu M., Xiong J., Guo X., and Liu H. (2013) Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell 25, 69–80 10.1016/j.devcel.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 12. Laneve P., Po A., Favia A., Legnini I., Alfano V., Rea J., Di Carlo V., Bevilacqua V., Miele E., Mastronuzzi A., Carai A., Locatelli F., Bozzoni I., Ferretti E., and Caffarelli E. (2017) The long noncoding RNA linc-NeD125 controls the expression of medulloblastoma driver genes by microRNA sponge activity. Oncotarget 8, 31003–31015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., and Bozzoni I. (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G., Liang L., Gu J., He X., and Huang S. (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 7, 11215 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., and Kjems J. (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 16. Peng L., Chen G., Zhu Z., Shen Z., Du C., Zang R., Su Y., Xie H., Li H., Xu X., Xia Y., and Tang W. (2017) Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung's disease. Oncotarget 8, 808–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S. D., Gregersen L. H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., le Noble F., and Rajewsky N. (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 18. Fan C., Hao Z., Yan J., and Li G. (2015) Genome-wide identification and functional analysis of lincRNAs acting as miRNA targets or decoys in maize. BMC Genomics 16, 793 10.1186/s12864-015-2024-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shuai P., Liang D., Tang S., Zhang Z., Ye C.-Y., Su Y., Xia X., and Yin W. (2014) Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J. Exp. Bot. 65, 4975–4983 10.1093/jxb/eru256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jeck W. R., and Sharpless N. E. (2014) Detecting and characterizing circular RNAs. Nat. Biotechnol. 32, 453–561 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lasda E., and Parker R. (2014) Circular RNAs: diversity of form and function. RNA 20, 1829–1842 10.1261/rna.047126.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karreth F. A., Reschke M., Ruocco A., Ng C., Chapuy B., Léopold V., Sjoberg M., Keane T. M., Verma A., Ala U., Tay Y., Wu D., Seitzer N., Velasco-Herrera Mdel C., Bothmer A., et al. (2015) The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell 161, 319–332 10.1016/j.cell.2015.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Apostolou E., and Hochedlinger K. (2013) Chromatin dynamics during cellular reprogramming. Nature 502, 462–471 10.1038/nature12749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roy S., and Sridharan R. (2017) Chromatin module inference on cellular trajectories identifies key transition points and poised epigenetic states in diverse developmental processes. Genome Res. 27, 1250–1262 10.1101/gr.215004.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aldiri I., Xu B., Wang L., Chen X., Hiler D., Griffiths L., Valentine M., Shirinifard A., Thiagarajan S., Sablauer A., Barabas M.-E., Zhang J., Johnson D., Frase S., Zhou X., Easton J., et al. (2017) The dynamic epigenetic landscape of the retina during development, reprogramming, and tumorigenesis. Neuron 94, 550–568.e510 10.1016/j.neuron.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takahashi K., and Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 27. Stadtfeld M., Maherali N., Breault D. T., and Hochedlinger K. (2008) Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2, 230–240 10.1016/j.stem.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mor N., Rais Y., Sheban D., Peles S., Aguilera-Castrejon A., Zviran A., Elinger D., Viukov S., Geula S., Krupalnik V., Zerbib M., Chomsky E., Lasman L., Shani T., Bayerl J., Gafni O., et al. (2018) Neutralizing Gatad2a-Chd4-Mbd3/NuRD complex facilitates deterministic induction of naive pluripotency. Cell Stem Cell 23, 412–425.e10 10.1016/j.stem.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Worringer K. A., Rand T. A., Hayashi Y., Sami S., Takahashi K., Tanabe K., Narita M., Srivastava D., and Yamanaka S. (2014) The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 14, 40–52 10.1016/j.stem.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choi Y. J., Lin C.-P., Ho J. J., He X., Okada N., Bu P., Zhong Y., Kim S. Y., Bennett M. J., Chen C., Ozturk A., Hicks G. G., Hannon G. J., and He L. (2011) miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat. Cell Biol. 13, 1353–1360 10.1038/ncb2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen J., Chen X., Li M., Liu X., Gao Y., Kou X., Zhao Y., Zheng W., Zhang X., Huo Y., Chen C., Wu Y., Wang H., Jiang C., and Gao S. (2016) Hierarchical Oct4 binding in concert with primed epigenetic rearrangements during somatic cell reprogramming. Cell Rep. 14, 1540–1554 10.1016/j.celrep.2016.01.013 [DOI] [PubMed] [Google Scholar]
- 32. Miranda K. C., Huynh T., Tay Y., Ang Y.-S., Tam W.-L., Thomson A. M., Lim B., and Rigoutsos I. (2006) A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126, 1203–1217 10.1016/j.cell.2006.07.031 [DOI] [PubMed] [Google Scholar]
- 33. Radzisheuskaya A., and Silva J. C. R. (2014) Do all roads lead to Oct4? the emerging concepts of induced pluripotency. Trends Cell Biol. 24, 275–284 10.1016/j.tcb.2013.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Takahashi K., and Yamanaka S. (2016) A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 17, 183–193 10.1038/nrm.2016.8 [DOI] [PubMed] [Google Scholar]
- 35. Wang H., Niu L., Jiang S., Zhai J., Wang P., Kong F., and Jin X. (2016) Comprehensive analysis of aberrantly expressed profiles of lncRNAs and miRNAs with associated ceRNA network in muscle-invasive bladder cancer. Oncotarget 7, 86174–86185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brambrink T., Foreman R., Welstead G. G., Lengner C. J., Wernig M., Suh H., and Jaenisch R. (2008) Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2, 151–159 10.1016/j.stem.2008.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kalvari I., Argasinska J., Quinones-Olvera N., Nawrocki E. P., Rivas E., Eddy S. R., Bateman A., Finn R. D., and Petrov A. I. (2018) Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46, D335–D342 10.1093/nar/gkx1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bao W., Kojima K. K., and Kohany O. (2015) Repbase update, a database of repetitive elements in eukaryotic genomes. Mobile DNA 6, 11 10.1186/s13100-015-0041-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kozomara A., and Griffiths-Jones S. (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang L., Feng Z., Wang X., Wang X., and Zhang X. (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138 10.1093/bioinformatics/btp612 [DOI] [PubMed] [Google Scholar]
- 41. Gao Y., Wang J., and Zhao F. (2015) CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 16, 4 10.1186/s13059-014-0571-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pertea M., Pertea G. M., Antonescu C. M., Chang T.-C., Mendell J. T., and Salzberg S. L. (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., and Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li B., and Dewey C. N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.