Abstract
Naïve CD4+ T cells in the periphery differentiate into regulatory T cells (Tregs) in which Foxp3 is expressed for their suppressive function. NLRP3, a pro-inflammatory molecule, is known to be involved in inflammasome activation associated with several diseases. Recently, the expression of NLRP3 in CD4+ T cells, as well as in myeloid cells, has been described; however, a role of T cell–intrinsic NLRP3 in Treg differentiation remains unknown. Here, we report that NLRP3 impeded the expression of Foxp3 independent of inflammasome activation in Tregs. NLRP3-deficient mice elevate Treg generation in various organs in the de novo pathway. NLRP3 deficiency increased the amount and suppressive activity of Treg populations, whereas NLRP3 overexpression reduced Foxp3 expression and Treg abundance. Importantly, NLRP3 interacted with Kpna2 and translocated to the nucleus from the cytoplasm under Treg-polarizing conditions. Taken together, our results identify a novel role for NLRP3 as a new negative regulator of Treg differentiation, mediated via its interaction with Kpna2 for nuclear translocation.
Keywords: NLRP3, nuclear translocation, T-cell biology, cell differentiation, forkhead box P3 (FOXP3), protein-protein interaction, gene knockout, transfection, Western blot, immunosuppression, Inflammasome-independent pathway, Kpna2, negative regulator, Treg differentiation
Introduction
Tregs2 are one of the CD4+ T cell subsets, capable of limiting effector CD4+ T cells and immune-mediated inflammation by their suppressive properties (1). The transcription factor Foxp3 controls Treg differentiation and is the key molecule involved in inhibition of the proliferation of conventional T cells (2). The induction of Foxp3 in both peripheral Tregs (pTregs) and in vitro–induced Tregs (iTregs) requires a signaling cascade, involving TCR, IL-2R (CD25), and TGF-βR, even though natural Tregs (nTregs, also known as tTregs) do not require TGF-β signaling to induce the Foxp3 expression in the thymus (3–5). Optimal activating factors of Foxp3 are NFAT, AP-1, c-Rel, NF-κB p65, Foxo, and cAMP-response element-binding protein (by TCR signaling), STAT5 (by IL-2 signaling), and Smad2/3 (by TGF-β signaling) (6), but its suppressive molecules have rarely been reported.
The nod-like receptor family protein 3 (NLRP3) is one of the key sensors of defense against both pathogen-associated molecular patterns by infection of bacteria and endogenous danger-associated molecular patterns generated by tissue damage (7). NLRP3 is activated by various pathogens and endogenous danger signals, such as extracellular ATP, glucose, silica, aluminum salt, and amyloid deposit (8). NLRP3 consists of an N-terminal pyrin domain (PYD), a central nucleotide-binding and oligomerization (NACHT) domain, and a C-terminal leucine-rich repeats (LRR) domain (8). NLRP3 forms the inflammasome together with protease caspase-1 through the adaptor ASC acting as the central hub (9). The assembly of three molecules in the cytoplasm subsequently results in programmed cell death, referred to as pyroptosis (10, 11), and also activates inflammatory cytokines, including IL-1β, IL-18, and IL-33, via cleavage and activation by caspase-1 (12, 13). NLRP3 protein is highly abundant in myeloid cells, such as dendritic cells and macrophages, and it controls adaptive immunity and regulates T cell–mediated autoimmunity (14–17). Recently, lymphoid cells were also reported to express NLRP3 in murine and human CD4+ T cells (15–18). The expression of NLRP3 is related with several diseases, such as autoimmune diseases and cancer in mice, as well as humans (19–23).
The role of T cell–intrinsic NLRP3 has been described in T cell responses, but not in Treg differentiation. In this study, we described that NLRP3-deficient mice generate increased Treg populations. Moreover, NLRP3 knockdown in CD4+ T cells exhibits increased Treg differentiation. Endogenous NLRP3 in CD4+ T cells down-regulated levels of Foxp3 expression levels under Treg-polarizing conditions through its translocation from cytoplasm into the nucleus, mediated by Kpna2. These results demonstrated that pro-inflammatory NLRP3 can repress the differentiation of Tregs by diminishing Foxp3 expression and suggest a role of NLRP3 as a new negative regulator of Treg differentiation.
Results
NLRP3 deficiency increases the generation of Tregs
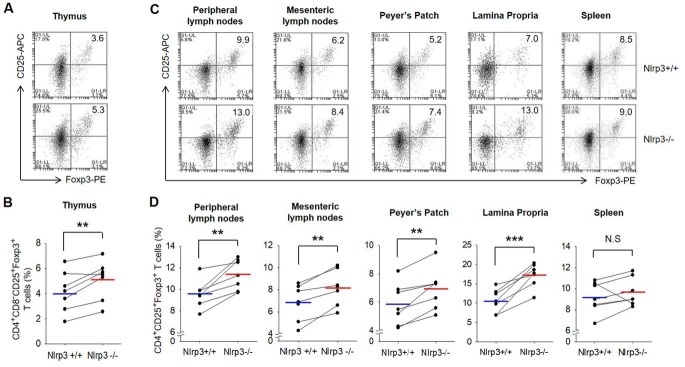
NLRP3 is reported to be expressed in CD4+ T cells as well as myeloid cells. To explore the effect of NLRP3 on Treg differentiation in the de novo pathway, we assessed the level of both pTreg and nTreg populations in Nlrp3+/+ and Nlrp3−/− littermate mice. For identifying pure nTreg populations, CD4+CD8−CD25+Foxp3+ T cells were separated in the thymus and analyzed. For detecting migratory nTregs and generated pTregs in lymphoid tissues, CD4+CD25+Foxp3+ T cells were isolated from various organs, such as the peripheral lymph nodes, mesenteric lymph nodes, Peyer's patch, lamina propria of the small intestine, and spleen. Compared with WT mice, in NLRP3-deficient mice, both nTregs (Fig. 1, A and B) and pTregs (Fig. 1, C and D) were much more abundant in all tissues, except in the spleen. This was not because of a difference in CD62L−CD44+CD4+ effector T cell abundance in Nlrp3−/− mice (Fig. S1A). The level of CD62L+CD44lowCD4+ naïve T cells was similar in both genotypes (Fig. S1B). These results suggest that NLRP3 deficiency elevates Treg generation in various organs in the de novo pathway.
Figure 1.
nTregs and pTregs are more present in Nlrp3−/− mice than Nlrp3+/+ mice. A, the population of nTregs in the thymus of Nlrp3+/+ mice and Nlrp3−/− mice. nTregs were stained with mAbs specific for CD4, CD25, Foxp3, and CD8, after depletion of CD8 cells. B, bar graph indicates the percentage of the cells. C, mixture of migratory nTregs and pTregs isolated from peripheral lymph nodes, mesenteric lymph nodes, Peyer's patch, lamina propria, and spleen was stained with mAbs specific for CD4, CD25, and Foxp3. D, summarized results shown as graph. B and D, blue and red lines represent the mean values of Nlrp3+/+ and Nlrp3−/− from seven independent experiments, respectively. **, p < 0.01; ***, p < 0.001; N.S, not significant (Student's two-tailed paired t test). The lines connected with Nlrp3+/+ and Nlrp3−/− indicate littermates (n = 7).
NLRP3 impedes the expression of Foxp3 in iTregs
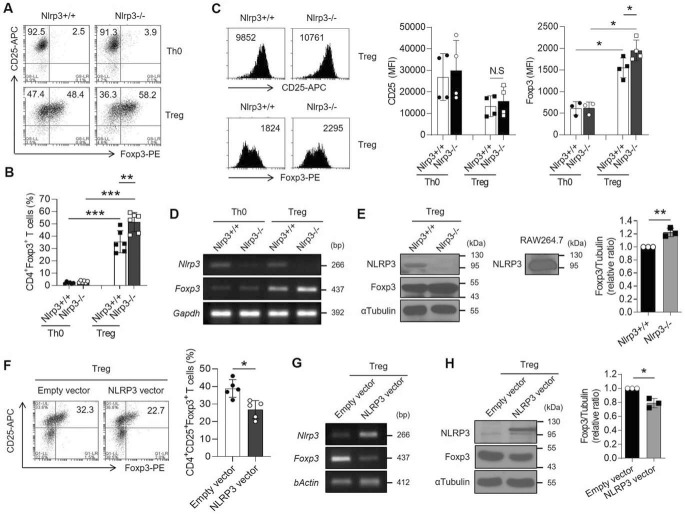
To investigate relevance of our observation regarding the repressive role of endogenous NLRP3 in Treg differentiation, we induced Treg differentiation of naïve CD4+ T cells isolated from either Nlrp3+/+ or Nlrp3−/− mice. Interestingly, the percentage of Foxp3-expressing cells was increased in NLRP3-deficient iTregs compared with the WT iTregs (Fig. 2, A and B). The expression of Foxp3 in single cells was higher in Nlrp3−/− iTregs than Nlrp3+/+ iTregs, although the percentage of CD25-expressing cells remained unaffected (Fig. 2C). NLRP3-deficient iTregs highly expressed Foxp3 at mRNA and protein levels (Fig. 2, D and E). Previous studies showed that Treg differentiation is favored by weak TCR stimulation and the interrupted T cell activation enhances Foxp3 expression (24–26). We investigated whether the activation of Nlrp3−/− naïve T cells was reduced by assessing the expression level of CD69, an early activation marker, after TCR stimulation. Nlrp3−/− naïve T cells were defective in the expression of CD69 after TCR stimulation for 16 h in Treg-polarizing conditions (Fig. S2, A and B). However, the expression levels of Treg-polarizing cytokine receptors (TGF-β and IL-2 receptors) were not significantly different in naïve CD4+ T cells of either genotype (Fig. S2, C and D).
Figure 2.
The expression of Foxp3 is reduced by NLRP3 during iTreg differentiation. A–E, naïve CD4+ T cells isolated from Nlrp3+/+ and Nlrp3−/− mice were polarized into either Th0 or iTregs in the presence of anti-CD3ϵ and anti-CD28 with or without TGF-β and IL-2 for 72 h. A, both CD4+CD25+Foxp3+ cell populations were assessed by flow cytometric analysis. B, bar graph indicates the percentage of Foxp3-expressing cells. C, representative histogram depicts the expression of CD25 and Foxp3 after gated CD4+ cells in either Nlrp3+/+ or Nlrp3−/− iTregs. Bar graph indicates the MFI of CD25 or Foxp3 of each molecule in Th0 and Tregs. D, the expression of Nlrp3 and Foxp3 mRNA was detected by RT-PCR assay. E, NLRP3 and Foxp3 proteins were observed by immunoblot assay. Bar graph indicates the relative ratio of Foxp3 over tubulin in Tregs in independent experiments. F–H, NLRP3 overexpressing virus was transduced to Th0 cells detail described in “Experimental procedures.” As a negative control, the virus with the empty vector was transduced. F, flow cytometry of Tregs expressed both CD25 and Foxp3 in the NLRP3 overexpression condition. Bar graph indicates the percentage of CD4+CD25+Foxp3+ iTregs. G, the expression of Nlrp3 and Foxp3 mRNA. H, the expression of NLRP3 and Foxp3 protein. Bar graph indicates the relative ratio of Foxp3 over tubulin in Tregs. All experiments were independently performed more than three times. Data are represented as mean ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001; N.S, not significant ((B, C) as Kruskal-Wallis test) or ((E, F, H) as Student's two-tailed unpaired t test).
Next, we overexpressed NLRP3 during iTreg differentiation using NLRP3-expressing retroviruses. When CD4+ T cells overexpressed NLRP3, the expression of Foxp3 was significantly down-regulated in iTregs, as observed by flow cytometry, RT-PCR, and Western blotting analyses (Fig. 2, F–H). Taken together, these data demonstrate that NLRP3 inhibits Foxp3 expression during iTreg differentiation.
NLRP3 activity is independent of inflammasome activation in iTreg responses
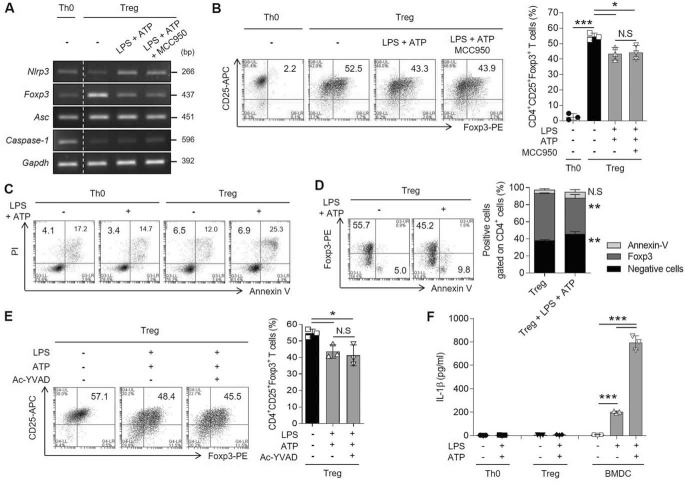
To prove that NLRP3 acts as a disturbing molecule of Treg responses, WT iTregs were stimulated to activate inflammasome, as NLRP3 commonly acts in an inflammasome-dependent manner by interacting with ASC and caspase-1. We speculated that Tregs might also be regulated by NLRP3 during inflammasome activation via danger-associated molecular pattern signals. To determine any involvement of the inflammasome activation in the NLRP3-mediated down-regulation of Treg differentiation, we stimulated iTregs with both LPS and ATP in the presence of MCC950, an inhibitor of NLRP3 inflammasome activation. MCC950 decreases IL-1β expression and secretion in bone marrow-derived dendritic cells (BMDCs) (Fig. S3, A and B). When iTregs were treated with extracellular ATP in the presence of LPS, the Nlrp3 expression was increased and Foxp3 expression was decreased. However, the expression of Foxp3 did not recover in iTregs treated with MCC950 (Fig. 3A). We confirmed that MCC950 inflammasome inhibitor did not affect Foxp3 protein expression, which was down-regulated by ATP (Fig. 3B). As NLRP3 inflammasome regulates both pyroptosis and the secretion of cytokines, especially IL-1β (11), we evaluated the levels of pyroptosis and IL-1β secretion in iTregs under inflammasome-activating conditions. Firstly, we observed that pyroptosis (PI+/annexin V− region) was not induced in iTregs by ATP (Fig. 3C). To investigate the relevance between apoptosis (PI+/annexin V+ region) and Foxp3 expression in Tregs, we assessed the staining of Foxp3 and annexin V. However, the reduction of Foxp3-expressing cells by ATP treatment was not because of apoptosis, with no significant increase in the number of annexin V–positive cells among in Foxp3-negative cells (Fig. 3D). We next investigated whether the activation of caspase-1 and maturation of IL-1β were associated with NLRP3 activation in the iTreg program. We confirmed that although caspase-1 activation was inhibited by Ac-YVAD-CHO, Foxp3 expression was not affected (Fig. 3E). IL-1β levels were low and not affected in iTregs despite stimulation with extracellular ATP, compared with BMDCs (Fig. 3F). Ac-YVAD-CHO also inhibited the secretion of IL-1β in BMDCs (Fig. S3, C and D). These results manifested that T cell–intrinsic NLRP3 inhibits Foxp3 expression and the inhibitory effect of NLRP3 on iTregs is independent of inflammasome activation.
Figure 3.
Treg differentiation is disturbed by NLRP3 without inflammasome activation. During iTreg differentiation, the cells were stimulated with LPS for 24 h, followed by pretreatment with MCC950 (10 μm) for 1 h, before ATP (5 mm) treatment for 6 h. A, the mRNA expression of inflammasome components (Nlrp3, Asc, and caspase-1), as well as Foxp3, was detected in iTregs. B, the population of CD25+Foxp3+ T cells gated on CD4+ T cells as in (A). Bar graph represents the percentage of plots. C, the population of pyroptotic cells was assessed with both PI and annexin V staining in CD4+ T cells. Both Th0 and iTreg cells were treated with ATP in the presence of LPS as in (A). PI+/annexin V− cells describe as the pyroptotic cells; PI+/annexin V+ cells indicate the apoptotic cells. D, Foxp3 and annexin V were detected in live iTregs as in (A). Double negative cells, Foxp3+CD4+ cells, or annexin V+CD4+ cells were quantified in bar graph. E, the population of CD25+Foxp3+CD4+ T cells was observed. The cells were treated with Ac-YVAD-CHO (10 μm) instead of MCC950 according to the culture method described in (A). F, secretory level of IL-1β in both Th0 and iTregs stimulated with LPS and ATP was confirmed by ELISA. Label of BMDCs is a control group about the stimulation of NLRP3 inflammasome activation. Data are representative of three independent experiments (n = 3). The results are represented as mean ± S.D. Statistics was significance by Student's two-tailed unpaired t test (B, D, E) or one-way ANOVA (F). *, p < 0.05; **, p < 0.01; ***, p < 0.001; N.S, not significant.
NLRP3 translocates to the nucleus by interacting with Kpna2 in iTregs
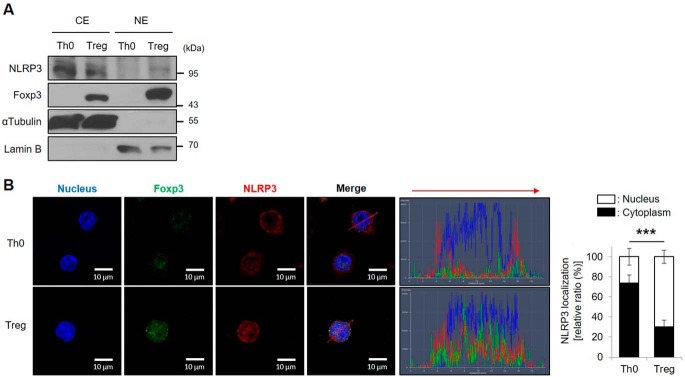
NLRP3 is normally located in the cytoplasm and found in complexes with ASC and pro–caspase-1 in various immune cells, especially in dendritic cells, macrophages, and T lymphocytes (16, 17). We investigated, and compared the localization of NLRP3 in Th0 and iTregs by cytoplasmic/nuclear protein fractions. We found that NLRP3 localized in the nucleus of iTregs (Fig. 4A). By immunocytochemical analysis detecting both NLRP3 and Foxp3, we confirmed that NLRP3 was localized in the cytoplasm of Th0 cells, but translocated to the nucleus in iTregs (Fig. 4B). These results suggested that NLRP3 translocates to the nucleus from the cytoplasm when naïve CD4+ T cells differentiate into iTregs.
Figure 4.
NLRP3 locates in the nucleus of Tregs. A, the cytoplasmic and nuclear protein levels of NLRP3 and Foxp3 were detected in either Th0 or iTregs after cytoplasmic/nuclear fractionation. αTubulin and lamin B were used as markers of cytoplasm and nucleus, respectively. B, immunofluorescence images of Foxp3 (green) and NLRP3 (red) were assessed in WT CD4+ T cells, after TCR stimulation for Th0 cells, or differentiation by TGF-β and IL-2 for 72 h in the presence of TCR stimulation for iTregs. DNA-binding dye DAPI is used for staining the nuclei (blue), and the scale bar is 5 μm. Four images within each field were collected at 630× magnification. Histogram shows the fluorescence intensity corresponding to the red arrow matched the direction. Line colors match with fluorescence colors. Data are representative of independent experiments (n = 4). Bar graph indicates the percentage of NLRP3 localization in either cytoplasm or nucleus of Th0 and Tregs. The results are represented as mean ± S.D. of six cells. Statistics was significance by Student's two-tailed unpaired t test. ***, p < 0.001.
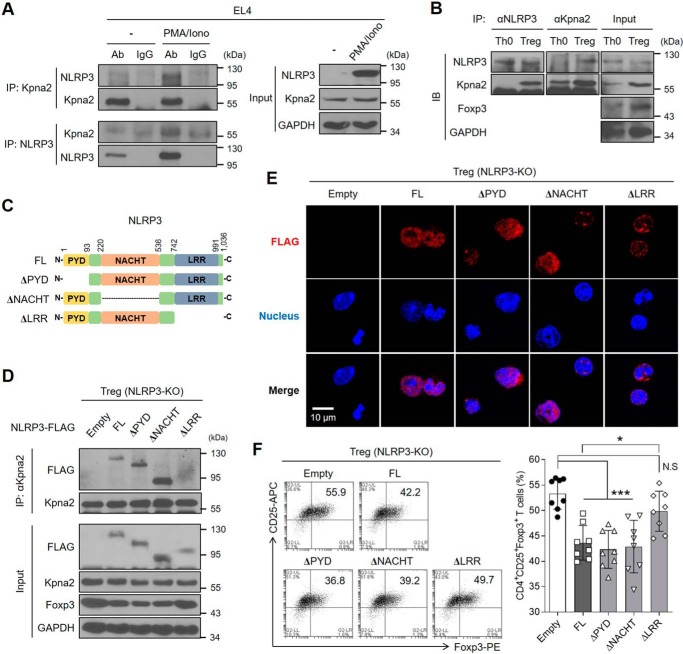
NLRP3 does not contain a nuclear localization signal (NLS)–binding site. To determine how NLRP3 is translocated to the nucleus of iTregs, we focused on importin proteins. Karyopherin α is one of the importin protein groups that recognizes NLS, and it is crucial for nuclear transport (27). As NLRP3 is not translocated into the nucleus when karyopherin α-2 (Kpna2) was silenced in Th2 cells (17), we speculated that Kpna2 is able to mediate the translocation of NLRP3 into the nucleus of Tregs. Kpna2 co-localized with NLRP3 in the nucleus of iTregs, but not Th0 cells (Fig. S4A). We also found co-localization of NLRP3 and Kpna2 in Tregs generated by the de novo pathway (Fig. S4B). Hence, we hypothesized that the sensitivity of interactions between NLRP3 and Kpna2 might be higher in iTregs than Th0 cells. To investigate whether NLRP3 can interact with Kpna2, we first applied Treg differentiation conditions (known to induce high expression of Foxp3) to EL4 mouse lymphoma cells by stimulation with PMA and ionomycin (Fig. S5), as described previously (28). By immunoprecipitation, we confirmed that NLRP3 bound Kpna2 in EL4 cell line under Treg differentiation conditions (Fig. 5A). We next investigated the interaction of NLRP3 and Kpna2 in Th0 and iTregs. Although the expression level of NLRP3 was higher in Th0 cells than iTregs, NLRP3 bound Kpna2 only in iTregs (Fig. 5B). To map the domain of NLRP3 that interacts with Kpna2, we made various deletion mutants of NLRP3 (Fig. 5C). These mutants were ectopically expressed in Nlrp3−/− CD4+ T cells during iTreg differentiation (Fig. 5D). The LRR domain of NLRP3 was required for interacting with Kpna2 (Fig. 5D). Moreover, when the LRR domain of NLRP3 was deleted, NLRP3 did not translocate into the nucleus of iTregs (Fig. 5E). Interestingly, LRR-deleted NLRP3 did not affect the decrease of Foxp3 expression (Fig. 5F). Taken together, the LRR domain of NLRP3 is important for binding to Kpna2 and mediated NLRP3 translocation to the nucleus to diminish Foxp3 expression in iTregs.
Figure 5.
NLRP3 interacts with Kpna2 to translocate into the nucleus of Tregs. A, EL4 cells were stimulated with PMA (50 ng/ml) and ionomycin (1 μg/ml) for 24 h, after which the cells were lysed and cellular proteins were extracted. Afterward, the protein fraction was immunoprecipitated with either rabbit anti-Kpna2 or mouse anti-NLRP3, followed by the immunoblot assay. Immunoglobulins are used as normal controls. B, either Th0 or iTregs was overexpressed with NLRP3, were subjected to the immunoprecipitation assay of NLRP3 and Kpna2, followed by immunoblot assay with mAbs for NLRP3 or Kpna2. C, schematic diagram of the structures of full-length or truncated mutants of NLRP3. D, iTregs isolated from Nlrp3-KO mice were transduced with empty vector, 3×FLAG-NLRP3 or truncated mutants (ΔPYD, ΔNACHT, ΔLRR). Cells were performed by immunoblot assay of FLAG and Kpna2 after immunoprecipitation with anti-Kpna2. E, immunofluorescence imaging of FLAG-tagged (red) NLRP3 was observed in iTregs after overexpression of either NLRP3 full-length or each domain mutant. Nuclei of iTregs were stained as blue. The object glass lens was used at 40× for amplifying the cells, and the scale bar represents 20 μm. F, the populations of iTregs overexpressed by either NLRP3 or each mutant form were detected by flow cytometry. Bar graph represents the percentage of plots. *, p < 0.05; ***, p < 0.001; N.S, not significant (one-way ANOVA). Data (A–E) are representative of two independent experiments (n = 2) and data (F) are representative of eight independent experiments (n = 8), as mean ± S.D.
NLRP3-deficient iTregs have more suppressive ability than WT iTregs
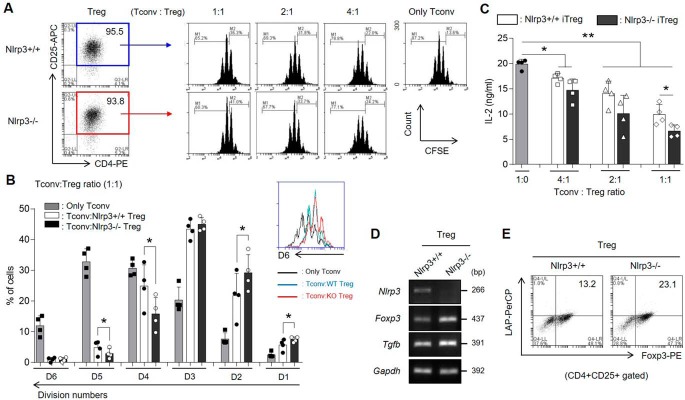
We investigated whether Nlrp3−/− iTreg populations are more suppressive than Nlrp3+/+ iTreg populations. Nlrp3−/− iTregs suppressed proliferation of conventional T (Tconv) cells more significantly than Nlrp3+/+ iTregs, as observed by inhibition of the division of CFSE-stained Tconv cells (Fig. 6, A and B). In addition, more Nlrp3−/− CD4+ T cells from DBA1/J mice were also differentiated into Tregs than Nlrp3+/+ CD4+ T cells in the presence of Treg-polarizing cytokines, and the Nlrp3−/− iTreg population efficiently suppressed the proliferation of Tconv cells (data not shown). Furthermore, Nlrp3−/− iTregs had an increased capacity to inhibit IL-2 production from Tconv cells (Fig. 6C) and exhibited higher expression of TGF-β compared with the Nlrp3+/+ iTregs (Fig. 6, D and E). Collectively, these data demonstrate that lack of NLRP3 in iTregs can effectually suppress Tconv cells by increasing an anti-inflammatory cytokine TGF-β as well as Foxp3.
Figure 6.
Nlrp3−/− iTreg populations are functionally more suppressive than Nlrp3+/+ iTregs. A–C, the iTreg-mediated suppression activity was measured by CFSE dilution of Tconv cells. The CFSE-stained Tconv cells were co-cultured for 3 days with Nlrp3+/+ or Nlrp3−/− iTregs sorted as CD4+CD25+ T cells by at various ratios, or cultured alone. A, proliferation of Tconv cells was determined by CFSE dilution and flow cytometric analysis. B, the percentages of divided Tconv cells were arranged by splitting the number of division in the presence of either Nlrp3+/+ iTregs or Nlrp3−/− iTregs at a 1:1 of Tconv: iTreg ratio. C, the secretory level of IL-2 from Tconv cells was measured by ELISA. D, the mRNA expression levels of Nlrp3, Tgfb as well as Foxp3 were detected in both WT and KO iTregs after Treg differentiation for 72 h. E, the population of Foxp3+LAP (latent TGF-β)+ cells in CD4+CD25+ iTregs were assessed by flow cytometry. Data are representative of four independent experiments (n = 4). The results are presented as mean ± S.D. *, p < 0.05; **, p < 0.01 (Student's two-tailed t test).
Discussion
The expression of Foxp3 is necessary for CD4+ T cells to differentiate into Tregs and homeostasis to maintain in the immune system. Thus, in this study, we focused on the regulation of Foxp3 expression during Treg differentiation. It has been reported that the expression of Foxp3 is suppressed by a few suppressive molecules, such as OX40 (29, 30), Batf/Batf3 (30, 31), Gfi1 (28), and Id2 (32). These suppressive factors are increased in activated T cells; however, they are decreased when activated T cells differentiate to iTregs, except for OX40 (29). We described that the expression of NLRP3 was down-regulated in iTregs compared with that in Th0, as well as other Th subsets (Fig. S6). However, factor(s) that contribute toward limiting the expression of NLRP3 in Tregs are still unclear. Nevertheless, given that the level of NLRP3 decreased in iTregs, we postulated that it may play as a suppressive factor in Treg differentiation.
Intrinsic NLRP3—ASC—caspase-8 inflammasome of Th17 cells, producing mature IL-1β, is associated with pathogenesis of Th17 cell-mediated experimental autoimmune encephalomyelitis (15). NLRP3 inflammasome enhances IFN-γ-producing Th1 cells via complement C5a generated in T cells during intestinal inflammation and viral infection (16). In contrast to several studies exploring the role of NLRP3, recently it was demonstrated that NLRP3 acts as a transcriptional regulator of Th2 differentiation, without the effect on the inflammasome (17). Additionally, NLRP3 is expressed by IL-2/STAT5 pathway in CD4+ T cells (17). IL-2 is sensitized by highly expressed CD25 (IL-2Rα) in most Tregs, indicating that STAT5 binds to the Foxp3 promoter for Treg development (33). We determined that NLRP3 is expressed in TGF-β/IL-2–induced Tregs, even though NLRP3 is an inflammatory factor, and T cell–intrinsic NLRP3 disturbs the Treg differentiation. This effect is not limited to C57BL/6 mice, but is also seen in DBA1/J mice, indicating that the inhibitory effect of NLRP3 on Treg differentiation is not strain-specific. Because Treg populations were more abundant in Nlrp3−/− healthy mice (Fig. 1), the increase of Foxp3+Treg frequency in NLRP3-deficient mice is the endogenous effect of NLRP3 in CD4+ T cells. Tregs up-regulated the NLRP3 expression in response to ATP stimulus, resulting in the NLRP3-mediated inhibition of Foxp3 expression, without inflammasome activation. Moreover, NLRP3 overexpression obstructed the expression of Foxp3. However, this regulation might be achieved through no direct binding of the Foxp3 promoter. Bruchard et al. (17) suggested a consensus motif of 5′-nGRRGGnRGAG-3′ for NLRP3 binding, which does not exist in the Foxp3 promoter. Lack of the NLRP3 did not induce dominant Treg differentiation, suggesting that triggering Foxp3 expression is protected by multiple mechanisms. Future studies would be needed to further elucidate the mechanism of NLRP3-mediated inhibition of Foxp3 expression and the role of NLRP3 in Treg stability.
NLRP3 inflammasome is formed by interacting with ASC adaptor protein and the PYD among the domain of NLRP3 in the cytosol (34). The binding of small heterodimer partner to the PYD of NLRP3 negatively regulates the activation of inflammasome (35). However, we observed that NLRP3 was located in the nucleus of Tregs, but not that of Th0 cells. This unusual nuclear localization of NLRP3 in Tregs occurred via interaction of Kpna2 to the LRR domain of NLRP3. It has been reported that LRRs also provide a versatile structure for the protein-protein interactions (36, 37). The LRR domain of NLRP3 mediates the interaction with promyelocytic leukemia protein 1 (PML-1), but it is not related to inflammasome activation (38). IRF4 is able to directly interact with the LRR domain of NLRP3 to regulate Th2 differentiation via its nuclear translocation by associating with Kpna2 (17). The assistance of Kpna2 is needed because of the lack of a recognizing site of NLS in NLRP3. In this report we demonstrated that Kpna2 interacted with the LRR domain of NLRP3 for facilitating the nuclear translocation of NLRP3, resulting in negative regulation of Foxp3 expression in Tregs.
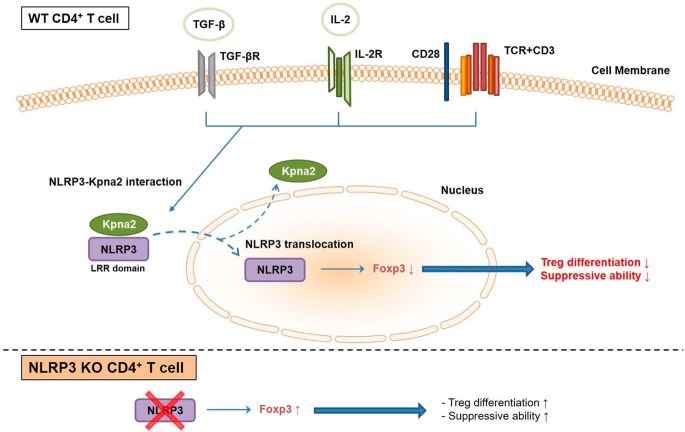
In conclusion (see Fig. 7 for a proposed model), our data suggest that the pro-inflammatory sensor NLRP3 limits the expression of Foxp3, leading to the disruption of Treg differentiation. Although NLRP3 expression is diminished in Tregs, it is presented and located in the nucleus following translocation after association with Kpna2. NLRP3 of Tregs, in the nucleus, can be up-regulated during inflammation followed by repression of Foxp3 expression in an inflammasome-independent manner. These findings provide a rationale for down-regulating NLRP3 in CD4+ T cells in immunotherapy setting for curing autoimmune diseases or graft-versus-host reactions.
Figure 7.
A schematic model for the role of NLRP3 to negatively regulate Treg differentiation. NLRP3 translocates to the nucleus of CD4+ T cells by interacting with Kpna2 during Treg differentiation, followed by down-regulation of Foxp3 expression. However, the deficiency of NLRP3 in CD4+ T cells increases Foxp3 expression, resulting in better suppression of Tconv cells.
Experimental procedures
Mice
Female C57BL/6 mice at 7 weeks of age were obtained from the Orient Bio (Kapyong, Korea). Nlrp3−/− mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All mice were housed under specific pathogen-free conditions. All animal experiments were conducted according to the Korea University Guidelines for the Care and Use of Laboratory Animals (Approval No. KUIACUC-2017–109/KUIACUC-2018–0045). All mice were used at 7–11 weeks of age, but the isolation of nTreg cells from thymus was performed in 5-week-old mice.
Antibodies
For fluorescence study, Abs against FITC- or PerCP-Cy5.5–conjugated CD4 (RM4–5), PE- or APC-conjugated (PC61), and APC-conjugated CD44 (IM7) were purchased from BD Biosciences. Abs against PE-conjugated Foxp3 (NRRF-30), APC-conjugated CD69 (H1.2F3), and FITC-conjugated CD62L (MEL-14) were obtained from eBioscience. PerCP-Cy5.5–conjugated LAP (TW7–16B4) was purchased from BioLegend. Secondary Abs against Alexa Fluor 488–conjugated rabbit IgG H + L, or both Alexa Fluor 594–conjugated mouse IgG H&L and Alexa Fluor 647–conjugated rabbit IgG H&L were purchased from Invitrogen or Abcam, respectively.
For immunoblot study, mouse anti-mouse/human NLRP3/NALP3 (cryo-2) was purchased from AdipoGen. Rabbit anti-mouse/human KPNA2 (polyclonal) was obtained from Abcam. Rabbit anti-mouse FOXP3 (H-190), mouse anti-mouse αtubulin (4G1), goat anti-mouse lamin B (M-20), and mouse anti-mouse GAPDH (6C5) were acquired from Santa Cruz Biotechnology. Mouse anti-FLAG (M2) was purchased from Sigma. HRP-linked anti-mouse and anti-rabbit IgG were obtained from Cell Signaling Technology.
In vitro T cell isolation and differentiation
CD4+ T cells were isolated from peripheral lymph nodes, mesenteric lymph nodes, spleens, Peyer's patches, and thymus of C57BL/6 Nlrp3+/+ or Nlrp3−/− mice by magnetic bead purification (MACS; Miltenyi Biotec). The purity of isolated CD4+ T cell populations was >95% routinely. For in vitro T cell activation, purified CD4+ T cells (1.5 × 106 cells/well) were seeded in anti-CD3ϵ (0.25–2.0 μg/ml)–coated 24-well plate for 16 h. CD4+ T cells (1 × 106 cells/well) were cultured for 3 days with anti-CD3ϵ (1 μg/ml) and anti-CD28 (1 μg/ml) in the presence of TGF-β (2 ng/ml) and IL-2 (2 ng/ml), for the generation of Tregs.
Lamina propria lymphocyte isolation
Lamina propria lymphocytes were isolated from small intestine, as described previously (39). Epithelial cells were lysed by 1 mm EDTA at 37 °C in 150 rpm of shaking incubator for 15 min, followed by washing with warm PBS. This lysis step was performed twice. Then, connective tissues were removed by 0.1 mg/ml collagenase D (Roche) at 37 °C with gentle stirring in 180 rpm. The supernatants were collected after passing through a 40-μm cell strainer. The step was done three times. Lamina propria lymphocytes were collected by Percoll gradient media (Amersham Biosciences) at the 40 and 85% interphase after carefully centrifuge for 20 min at 20 °C at 2000 rpm.
Plasmid construction
DNA fragments corresponding to the coding sequences of the mouse NLRP3 gene were amplified by PCR from pcDNA3-FLAG-NLRP3. 3×FLAG-tagged NLRP3 was cloned into the XhoI and NotI sites in pMXs-IRES-GFP by NEBuilder HiFi DNA Assembly Cloning kit (New England Biolabs, Ipswich, MA). 3×FLAG-tagged truncated mutant constructs of NLRP3 were created by subcloning the PCR products of complementary DNA fragments, containing each domain of the target genes, into pMXs-IRES-GFP. All the constructs were sequenced at Bionics (Seoul, Korea) to verify 100% correspondence with the original DNA sequence.
Retroviral transduction to T cells
Platinum E retroviral packaging cell line was transfected with retroviral expression plasmids based on pMX-IRES-GFP for 48 h by using TransIT-LT1 Transfection Reagent (Mirus Bio LLC, Madison, WI). Virus was collected in culture supernatant through a 0.22-μm syringe filter. Naïve CD4+ T cells isolated from Nlrp3-KO mice were activated for 12 h with anti-CD3ϵ (1 μg/ml) and anti-CD28 (1 μg/ml) followed by spin, infected with retrovirus supernatant in the presence of polybrene (8 μg/ml) at 892 × g for 90 min at 30 °C, as described with minor modification (40). Cells were subsequently cultured with TGF-β (5 ng/ml) and IL-2 (5 ng/ml) for Treg polarization for 72 h, after removing the virus supernatant.
Fluorescent microscopy
For attaching cells, the number of cells (5 × 106 cells/well) was counted and moved at poly-l-lysine–coated 18-mm diameter cover glass for 20 min, after washing with serum-free media. Cells were fixed in 4% paraformaldehyde (pH 7.5) for 10 min at 4 °C, then permeabilized in permeabilization buffer (0.1% Triton X-100 in PBS) for 2 min at room temperature. Before treating the adequate antibodies, cells of nonspecific site were blocked with blocking buffer (0.5% BSA in PBS). Each cell was incubated with both primary Ab (1:50 dilution) overnight at 4 °C, followed by staining with fluorescence-conjugated secondary Ab (1:200 dilution) for 1 h at room temperature. The nuclei of cells were stained with DAPI (Molecular Probes, 5 μg/ml, 3 min, room temperature). After mounting the cells by using Antifade Reagent (Invitrogen), cells were observed with confocal laser scanning microscope (LSM700, Carl Zeiss).
Flow cytometry
For cell surface staining, cells were washed and stained in FACS buffer (0.5% FBS and filtered 0.05% NaN3 in PBS). For intracellular staining, fixation and permeabilization were performed in Cytofix/Cytoperm (BD Biosciences) and then cells were stained in Perm/Wash solutions (BD Biosciences). Especially for staining the Foxp3 or transcription factor, cells were fixed and permeabilized in Foxp3/Transcription Factor Staining buffer set (eBioscience). For detection of intracellular cytokines, cells were activated by PMA (50 ng/ml) and ionomycin (1 μg/ml) in the presence of Golgi Stop (BD Biosciences) for 4 h. All of flow cytometric analyses were performed using FACSCalibur or BD Accuri with BD AccuriC6 software (BD Biosciences).
RT-PCR
Total RNA from T cells were extracted by RiboEX total RNA kit (GeneAll Biotechnology, Seoul, South Korea). The RNA extraction was reverse-transcribed to cDNA library. cDNA was quantified by PCR. After amplification, the PCR products were separated on 1% agarose gels and detected by using Loading STAR (Dyenbio, Kyeongki, Korea).
Cytoplasmic and nuclear fractionation
Protein extracts were washed by PBS, and then the cytoplasmic extract was lysed by CE buffer 1 (0.075% (v/v) Nonidet P-40, 10 mm HEPES, 60 mm KCl, 1 mm EDTA, 1 mm DTT, 1 mm PMSF (pH 7.6) including protease inhibitors) of 5 pellet volumes for 3 min on ice. The cytoplasmic extract was removed from the pellet to a clean tube. The nuclei were gently washed by CE buffer 2 (similar to CD buffer 1 but without the addition of Nonidet P-40). The nuclear pellet was lysed by NE buffer (20 mm Tris-HCl, 420 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 1 mm PMSF, 25% (v/v) glycerol, (pH 8.0) including protease inhibitors) of 1 pellet volume for 10 min on ice. The nuclei extract was removed from the pellet to a clean tube.
Immunoblot and immunoprecipitation assay
Protein extracts were prepared by the lysis buffer (1% SDS, 1 mm sodium orthovanadate, and 10 mm Tris (pH7.4)) in the presence of protease inhibitor mixture (Sigma) for 10 min at 4 °C, followed by sonication, as described in a previous report (17). Protein lysates were separated on 8–12% SDS-PAGE cells, and transferred to 0.45-μm PVDF membranes. The membrane was blocked with 5% skim milk in TBS containing 0.1% Tween 20 (TBS-T). The membrane was incubated in a 1:1000 dilution of a primary antibody in 5% BSA containing TBS-T for overnight at 4 °C. The membrane was incubated in a 1:5000 dilution of a secondary antibody in 2.5% skim milk contained TBS-T for 1 h. Chemiluminescence reagent was Amersham Biosciences ECL Prime.
For immunoprecipitation, protein lysates were prepared at least 400 μg and incubated with 2 μg of primary antibody and Protein G Plus Agarose Beads (Santa Cruz Biotechnology) for 16 h at 4 °C. The beads were washed five times with lysis buffer. Samples were suspended with sample buffer and boiled for 10 min and subsequently were separated on 8% SDS-PAGE gel.
Treg suppression assay
Nlrp3+/+ CD4+ T cells or Nlrp3−/− CD4+ T cells (5 × 104 cells/well) were polarized to Tregs for 3 days at anti-CD3ϵ (1 μg/ml)-coated 96-well plate in the presence of soluble anti-CD28 (1 μg/ml) as triplicate. Either Nlrp3+/+ iTregs or Nlrp3−/− iTregs were restimulated with PMA (50 ng/ml) and ionomycin (1 μg/ml). Both CD25+ Tregs were isolated by FACS Aria and co-cultured with CFSE (1 μm)-labeled CD4+ Tconv cells (5 × 104 cells/well) at different ratio in anti-CD3ϵ (1 μg/ml)-coated 96-well plate in the presence of anti-CD28 (1 μg/ml). The divided aspect of CFSE was examined by flow cytometry and the level of IL-2 was detected by ELISA.
Statistics
Groups were compared by using adequate analytic programs such as GraphPad Prism 8.0 by a two-tailed Student's t test, or SPSS software by Kruskal-Wallis test or one-way ANOVA. A p value of <0.05 was considered significant.
Author contributions
S.-H. P. conceptualization; S.-H. P., A. M., and T. S. K. data curation; S.-H. P. software; S.-H. P. formal analysis; S.-H. P. and T. S. K. funding acquisition; S.-H. P. and S. H. investigation; S.-H. P. and S. H. visualization; S.-H. P., S. H., and A. L. methodology; S.-H. P. writing-original draft; A. M. and T. S. K. supervision; A. M. and T. S. K. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Dr. Ju Han Song (Chonnam National University), Dr. Byung Cheol Lee, and Si Hoon Park (Korea University) for technical assistance for cloning to make NLRP3 overexpression vector or its domain-deleted mutant vector, and Dr. Juliet French and Dr. Jason S. Lee (QIMR) for helpful discussion and comments on this study. We also thank the Gyerim Experimental Animal Resource Center at Korea University for careful maintenance of all animals.
This work was supported by National Research Foundation of Korea (NRF) Grant NRF-2017R1A2B2009442 (to T.S.K) and by the Global Ph.D. Fellowship Program, Project No.: 2015H1A2A1031021, through the National Research Foundation of Korea funded by the Ministry of Education (to S.-H.P). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6.
- Treg
- regulatory T cell
- nTreg
- natural Treg
- pTreg
- peripheral Treg
- iTreg
- induced Treg
- NLRP3
- nod-like receptor family protein 3
- Kpna2
- karyopherin α-2
- TCR
- T cell receptor
- Tconv cells
- conventional T cells
- PYD
- pyrin domain
- LRR
- leucine-rich repeats
- LPS
- lipopolysaccharide
- NLS
- nuclear localization signal
- BMDC
- bone marrow-derived dendritic cell
- PMA
- phorbol 12-myristate 13-acetate
- CFSE
- carboxyfluorescein succinimidyl ester
- Abs
- antibodies
- ANOVA
- analysis of variance.
References
- 1. Josefowicz S. Z., Lu L. F., and Rudensky A. Y. (2012) Regulatory T cells: Mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564 10.1146/annurev.immunol.25.022106.141623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schmidt A., Oberle N., and Krammer P. H. (2012) Molecular mechanisms of Treg-mediated T cell suppression. Front. Immunol. 3, 51 10.3389/fimmu.2012.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shevach E. M., and Thornton A. M. (2014) tTregs, pTregs, and iTregs: Similarities and differences. Immunol. Rev. 259, 88–102 10.1111/imr.12160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shalev I., Schmelzle M., Robson S. C., and Levy G. (2011) Making sense of regulatory T cell suppressive function. Semin. Immunol. 23, 282–292 10.1016/j.smim.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ouyang W., Beckett O., Ma Q., and Li M. O. (2010) Transforming growth factor-β signaling curbs thymic negative selection promoting regulatory T cell development. Immunity 32, 642–653 10.1016/j.immuni.2010.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huehn J., and Beyer M. (2015) Epigenetic and transcriptional control of Foxp3+ regulatory T cells. Semin. Immunol. 27, 10–18 10.1016/j.smim.2015.02.002 [DOI] [PubMed] [Google Scholar]
- 7. Bullon P., and Navarro J. M. (2017) Inflammasome as a key pathogenic mechanism in endometriosis. Curr. Drug Targets 18, 997–1002 10.2174/1389450117666160709013850 [DOI] [PubMed] [Google Scholar]
- 8. Chen S., and Sun B. (2013) Negative regulation of NLRP3 inflammasome signaling. Protein Cell 4, 251–258 10.1007/s13238-013-2128-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elliott E. I., and Sutterwala F. S. (2015) Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunol. Rev. 265, 35–52 10.1111/imr.12286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abe J., and Morrell C. (2016) Pyroptosis as a regulated form of necrosis: PI+/annexin V-/high caspase 1/low caspase 9 activity in cells = pyroptosis? Circ. Res. 118, 1457–1460 10.1161/CIRCRESAHA.116.308699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miao E. A., Leaf I. A., Treuting P. M., Mao D. P., Dors M., Sarkar A., Warren S. E., Wewers M. D., and Aderem A. (2010) Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 11, 1136–1142 10.1038/ni.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lech M., Lorenz G., Kulkarni O. P., Grosser M. O., Stigrot N., Darisipudi M. N., Günthner R., Wintergerst M. W., Anz D., Susanti H. E., and Anders H. J. (2015) NLRP3 and ASC suppress lupus-like autoimmunity by driving the immunosuppressive effects of TGF-β receptor signalling. Ann. Rheum. Dis. 74, 2224–2235 10.1136/annrheumdis-2014-205496 [DOI] [PubMed] [Google Scholar]
- 13. Chen M., Wang H., Chen W., and Meng G. (2011) Regulation of adaptive immunity by the NLRP3 inflammasome. Int. Immunopharmacol. 11, 549–554 10.1016/j.intimp.2010.11.025 [DOI] [PubMed] [Google Scholar]
- 14. Shaw P. J., McDermott M. F., and Kanneganti T. D. (2011) Inflammasomes and autoimmunity. Trends Mol. Med. 17, 57–64 10.1016/j.molmed.2010.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin B. N., Wang C., Zhang C. J., Kang Z., Gulen M. F., Zepp J. A., Zhao J., Bian G., Do J. S., Min B., Pavicic P. G. Jr., El-Sanadi C., Fox P. L., Akitsu A., Iwakura Y., et al. (2016) T cell-intrinsic ASC critically promotes TH17-mediated experimental autoimmune encephalomyelitis. Nat. Immunol. 17, 583–592 10.1038/ni.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arbore G., West E. E., Spolski R., Robertson A. A. B., Klos A., Rheinheimer C., Dutow P., Woodruff T. M., Yu Z. X., O'Neill L. A., Coll R. C., Sher A., Leonard W. J., Köhl J., Monk P., et al. (2016) T helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4+ T cells. Science 352, aad1210 10.1126/science.aad1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bruchard M., Rebé C., Derangère V., Togbé D., Ryffel B., Boidot R., Humblin E., Hamman A., Chalmin F., Berger H., Chevriaux A., Limagne E., Apetoh L., Végran F., and Ghiringhelli F. (2015) The receptor NLRP3 is a transcriptional regulator of TH2 differentiation. Nat. Immunol. 16, 859–870 10.1038/ni.3202 [DOI] [PubMed] [Google Scholar]
- 18. Doitsh G., Galloway N. L., Geng X., Yang Z., Monroe K. M., Zepeda O., Hunt P. W., Hatano H., Sowinski S., Muñoz-Arias I., and Greene W. C. (2014) Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505, 509–514 10.1038/nature12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao C., Gu Y., Zeng X., and Wang J. (2018) NLRP3 inflammasome regulates Th17 differentiation in rheumatoid arthritis. Clin. Immunol. 197, 154–160 10.1016/j.clim.2018.09.007 [DOI] [PubMed] [Google Scholar]
- 20. Vakrakou A. G., Boiu S., Ziakas P. D., Xingi E., Boleti H., and Manoussakis M. N. (2018) Systemic activation of NLRP3 inflammasome in patients with severe primary Sjögren's syndrome fueled by inflammagenic DNA accumulations. J. Autoimmun. 91, 23–33 10.1016/j.jaut.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 21. Gu J., Liu G., Xing J., Song H., and Wang Z. (2018) Fecal bacteria from Crohn's disease patients more potently activated NOD-like receptors and Toll-like receptors in macrophages, in an IL-4-repressible fashion. Microb. Pathog. 121, 40–44 10.1016/j.micpath.2018.05.009 [DOI] [PubMed] [Google Scholar]
- 22. Liu R., Truax A. D., Chen L., Hu P., Li Z., Chen J., Song C., Chen L., and Ting J. P. (2015) Expression profile of innate immune receptors, NLRs and AIM2, in human colorectal cancer: Correlation with cancer stages and inflammasome components. Oncotarget 6, 33456–33469 10.18632/oncotarget.5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang A., Yu J., Yan S., Zhao X., Chen C., Zhou Y., Zhao X., Hua M., Wang R., Zhang C., Zhong C., He N., Ji C., and Ma D. (2018) The genetic polymorphism and expression profiles of NLRP3 inflammasome in patients with chronic myeloid leukemia. Hum. Immunol. 79, 57–62 10.1016/j.humimm.2017.10.013 [DOI] [PubMed] [Google Scholar]
- 24. Sauer S., Bruno L., Hertweck A., Finlay D., Leleu M., Spivakov M., Knight Z. A., Cobb B. S., Cantrell D., O'Connor E., Shokat K. M., Fisher A. G., and Merkenschlager M. (2008) T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc. Natl. Acad. Sci. U.S.A. 105, 7797–7802 10.1073/pnas.0800928105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miskov-Zivanov N., Turner M. S., Kane L. P., Morel P. A., and Faeder J. R. (2013) The duration of T cell stimulation is a critical determinant of cell fate and plasticity. Sci. Signal. 6, ra97 10.1126/scisignal.2004217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schönle A., Hartl F. A., Mentzel J., Nöltner T., Rauch K. S., Prestipino A., Wohlfeil S. A., Apostolova P., Hechinger A. K., Melchinger W., Fehrenbach K., Guadamillas M. C., Follo M., Prinz G., Ruess A. K., et al. (2016) Caveolin-1 regulates TCR signal strength and regulatory T-cell differentiation into alloreactive T cells. Blood 127, 1930–1939 10.1182/blood-2015-09-672428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chook Y. M., and Blobel G. (2001) Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11, 703–715 10.1016/S0959-440X(01)00264-0 [DOI] [PubMed] [Google Scholar]
- 28. Ichiyama K., Hashimoto M., Sekiya T., Nakagawa R., Wakabayashi Y., Sugiyama Y., Komai K., Saba I., Möröy T., and Yoshimura A. (2009) Gfi1 negatively regulates Th17 differentiation by inhibiting RORγt activity. Int. Immunol. 21, 881–889 10.1093/intimm/dxp054 [DOI] [PubMed] [Google Scholar]
- 29. Vu M. D., Xiao X., Gao W., Degauque N., Chen M., Kroemer A., Killeen N., Ishii N., and Li X. C. (2007) OX40 costimulation turns off Foxp3+ Tregs. Blood 110, 2501–2510 10.1182/blood-2007-01-070748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang X., Xiao X., Lan P., Li J., Dou Y., Chen W., Ishii N., Chen S., Xia B., Chen K., Taparowsky E., and Li X. C. (2018) OX40 Costimulation inhibits Foxp3 expression and Treg induction via BATF3-dependent and independent mechanisms. Cell Rep. 24, 607–618 10.1016/j.celrep.2018.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee W., Kim H. S., Hwang S. S., and Lee G. R. (2017) The transcription factor Batf3 inhibits the differentiation of regulatory T cells in the periphery. Exp. Mol. Med. 49, e393 10.1038/emm.2017.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hwang S. M., Sharma G., Verma R., Byun S., Rudra D., and Im S. H. (2018) Inflammation-induced Id2 promotes plasticity in regulatory T cells. Nat. Commun. 9, 4736 10.1038/s41467-018-07254-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fan M. Y., Low J. S., Tanimine N., Finn K. K., Priyadharshini B., Germana S. K., Kaech S. M., and Turka L. A. (2018) Differential roles of IL-2 signaling in developing versus mature Tregs. Cell Rep. 25, 1204–1213.e1204 10.1016/j.celrep.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chae J. J., Cho Y. H., Lee G. S., Cheng J., Liu P. P., Feigenbaum L., Katz S. I., and Kastner D. L. (2011) Gain-of-function pyrin mutations induce NLRP3 protein-independent interleukin-1β activation and severe autoinflammation in mice. Immunity 34, 755–768 10.1016/j.immuni.2011.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang C. S., Kim J. J., Kim T. S., Lee P. Y., Kim S. Y., Lee H. M., Shin D. M., Nguyen L. T., Lee M. S., Jin H. S., Kim K. K., Lee C. H., Kim M. H., Park S. G., Kim J. M., Choi H. S., and Jo E. K. (2015) Small heterodimer partner interacts with NLRP3 and negatively regulates activation of the NLRP3 inflammasome. Nat. Commun. 6, 6115 10.1038/ncomms7115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kobe B., and Kajava A. V. (2001) The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11, 725–732 10.1016/S0959-440X(01)00266-4 [DOI] [PubMed] [Google Scholar]
- 37. Gay N. J., Packman L. C., Weldon M. A., and Barna J. C. (1991) A leucine-rich repeat peptide derived from the Drosophila Toll receptor forms extended filaments with a β-sheet structure. FEBS Lett. 291, 87–91 10.1016/0014-5793(91)81110-T [DOI] [PubMed] [Google Scholar]
- 38. Lo Y. H., Huang Y. W., Wu Y. H., Tsai C. S., Lin Y. C., Mo S. T., Kuo W. C., Chuang Y. T., Jiang S. T., Shih H. M., and Lai M. Z. (2013) Selective inhibition of the NLRP3 inflammasome by targeting to promyelocytic leukemia protein in mouse and human. Blood 121, 3185–3194 10.1182/blood-2012-05-432104 [DOI] [PubMed] [Google Scholar]
- 39. Kim M. S., and Kim T. S. (2014) IgA+ plasma cells in murine intestinal lamina propria as a positive regulator of Treg differentiation. J. Leukoc. Biol. 95, 461–469 10.1189/jlb.0613310 [DOI] [PubMed] [Google Scholar]
- 40. Singh Y., Garden O. A., Lang F., and Cobb B. S. (2016) Retroviral transduction of helper T cells as a genetic approach to study mechanisms controlling their differentiation and function. J. Vis. Exp. 117, e54698 10.3791/54698 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.