Abstract
Anthocyanin and proanthocyanidin biosynthesis pathways are believed to overlap. This study examined proanthocyanidin accumulation in seed coats of morning glories (Ipomoea nil and I. purpurea) carrying mutations in CHS-D, CHI, and ANS genes encoding chalcone synthase, chalcone isomerase, and anthocyanidin synthase, respectively. Chemical staining revealed that mutants accumulate proanthocyanidin normally. Thus, the tested genes are not essential to proanthocyanidin biosynthesis, but are essential to anthocyanin biosynthesis in flowers and stems. Based on the results and the I. nil draft genome sequence, the genes involved in proanthocyanidin biosynthesis, including a new copy of the flavanone 3-hydroxylase gene could be predicted. Moreover, the genome has no homologs for known enzymes involved in producing flavan-3-ols, the starter and extension units of proanthocyanidin. These results suggested that I. nil produces flavan-3-ols through an undiscovered biosynthesis pathway. To characterize proanthocyanidin pigmentation further, we conducted mutant screening using a large I. nil population. We discovered that the brown mutant lines (exhibiting brown seeds and normal anthocyanin pigmentation) do not accumulate proanthocyanidin in their seed coats. Thus, the brown mutation should be useful for further investigations into the various mechanisms controlling anthocyanin and proanthocyanidin pathways.
Keywords: Anthocyanin, Ipomoea nil, Ipomoea purpurea, proanthocyanidin, seed coat
Introduction
Proanthocyanidins (or condensed tannins) are flavan-3-ol polymers and a class of flavonoid pigments. Distributed in a wide range of plant species, proanthocyanidins play an important role in defense against UV radiation, microbial pathogens, insect pests, and herbivore predation (Barbehenn and Peter Constabel 2011; Dixon et al. 2005). Their oxidization in seed coats confers the typical brown color indicating seed maturation (Marles et al. 2003; Tanner et al. 2003). Proanthocyanidins also have economic importance as the source of astringency in fruit, wines, and teas, while also being powerful antioxidants with potential protective effects against cancers, reactive oxygen species, and cholesterol accumulation (Dixon et al. 2005). Thus, considerable effort has been expended on increasing proanthocyanidin quantity in fruits and legumes (Dixon et al. 2013).
Proanthocyanidins and anthocyanins are thought to be more recently evolved flavonoids (Koes et al. 1994). The early steps of their biosynthesis pathway involve the same enzymes (Figure 1). In Arabidopsis, both anthocyanin and proanthocyanidin are absent if genes encoding any one of the following enzymes are defective: chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydoroxylase (F3′H), dihydroflavonol 4-reductase (DFR), and anthocyanidin synthase (ANS) (Feinbaum and Ausubel 1988; Koornneef 1990; Shirley et al. 1992). However, it is unclear if these genes mediate both anthocyanin and proanthocyanidin production in other plants.
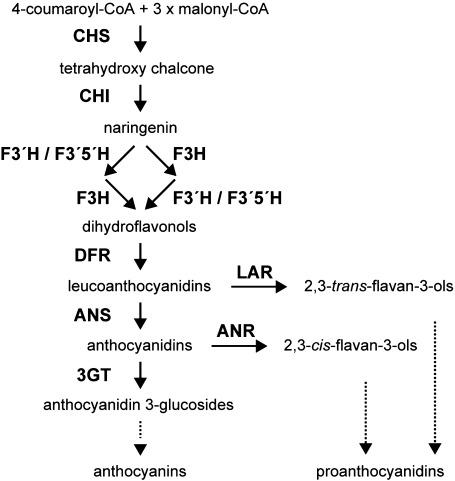
Figure 1. Schematic diagram representing the flavonoid pathway of anthocyanin and proanthocyanidin biosynthesis in plants. Enzymes are shown in boldface. CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydoroxylase; F3′5′H, flavonoid 3′,5′-hydoroxylase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; 3GT, UDP-glucose: flavonoid 3-O-glycosyltransferase; LAR, leucoanthocyanidin reductase; ANR, anthocyanidin reductase.
Leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR) are two enzymes involved in the late steps of proanthocyanidin biosynthesis (Devic et al. 1999; Tanner et al. 2003; Xie et al. 2003). Respectively, LAR and ANR catalyze 2,3-trans-flavan-3-ol formation from leucoanthocyanidins and 2,3-cis-flavan-3-ol from anthocyanidins (Figure 1) (Winkel-Shirley 2006). In addition, 2,3-trans-flavan-3-ols (catechins) and 2,3-cis-flavan-3-ols (epicatechins) are the starter and extension units of proanthocyanidin. Arabidopsis produces proanthocyanidin based on 2,3-cis-flavan-3-ols, whereas many other plants base their proanthocyanidin production on both flavan-3-ol types (Abrahams et al. 2003; Harborne and Williams 2000; Routaboul et al. 2006; Tanner et al. 2003). No LAR genes have been found in Arabidopsis (Lepiniec et al. 2006).
The expression of proanthocyanidin and anthocyanin biosynthesis genes is controlled by transcriptional activators containing the R2R3-MYB domain, basic helix-loop-helix (bHLH) domain, and WD40 repeats (WDR). These activators interact to regulate anthocyanin/proanthocyanidin pigmentation and other epidermal traits, including root hair and trichome formation, as well as seed coat mucilage production (Hichri et al. 2011; Koes et al. 2005; Lepiniec et al. 2006; Ramsay and Glover 2005).
Morning glory species, the Japanese morning glory (Ipomoea nil) and the common morning glory (I. purpurea) are commercially important horticultural plants (Figure 2A–L). Wild type I. nil and I. purpurea have blue and purple flowers, respectively (Figure 2A, J), through the production of anthocyanins. Several mutations resulting in flower-color alterations have been isolated in these species (Chopra et al. 2006; Iida et al. 2004), allowing for characterization of the structural and regulatory genes for anthocyanin biosynthesis (Koes et al. 2005; Petroni and Tonelli 2011). Structural genes, CHS-D, CHI, DFR-B, and ANS in I. nil, as well as CHS-D in I. purpurea, are essential for flower and stem anthocyanin production (Figure 2B–F, K) (Chopra et al. 2006; Habu et al. 1998; Hoshino et al. 2009; Iida et al. 2004; Inagaki et al. 1994), but their exact roles in proanthocyanidin production are unclear. Ipomoea seed coats accumulate proanthocyanidins, but seeds of mutants for the above genes are dark brown and indistinguishable from wild-type seeds (Park and Hoshino 2012; Park et al. 2007), likely because phytomelanins comprise the dark-brown pigment along with proanthocyanidins. Thus, proanthocyanidin accumulation cannot be characterized using seed appearance alone. In a recent study, we used chemical staining to show that the DFR-B gene is not essential for proanthocyanidin synthesis in I. nil (Park and Hoshino 2012). Previous work has also demonstrated that InMYB1 and InWDR1 in I. nil and IpMYB1 and IpbHLH2 in I. purpurea all activate anthocyanin pigmentation in flowers (Chang et al. 2005; Morita et al. 2006; Park and Hoshino 2012; Park et al. 2007). Furthermore, InWDR1 and IpbHLH2 are also involved in activating both proanthocyanidin and phytomelanin pigmentation in seed coats, as evidenced by the lack of brown seed pigmentation among mutants (Figure 2F, L). However, InWDR1 does not regulate CHS-D, CHI, or ANS, whereas IpbHLH2 activates DFR-B and ANS expression, but not CHS-D expression in seed coats (Park and Hoshino 2012; Park et al. 2007).
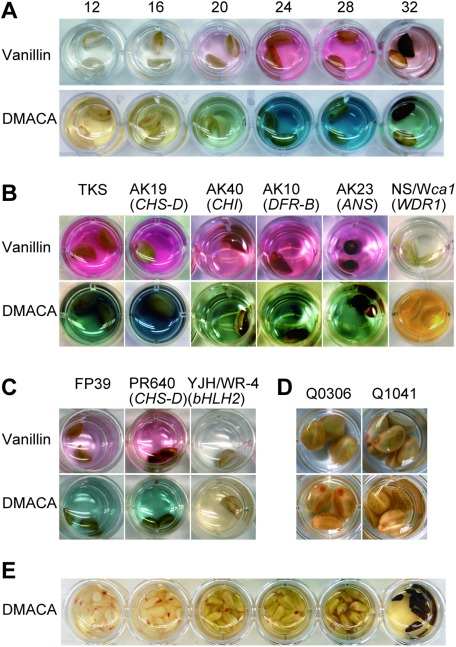
Figure 2. Flower and seed phenotypes of Ipomoea species. (A–G) Ipomoea nil lines. (A) TKS (wild-type), (B) AK19 (CHS-D mutant), (C) AK40 (CHI mutant), (D) AK10 (DFR-B mutant), (E) AK23 (ANS mutant), (F) NS/W1ca1 (InWDR1 mutant), and (G) Q0306 (brown mutant). (H) Seed pigmentation and trichome formation in NS/W1ca1 (left) and Q0306 (right). The photograph was taken using a digital microscope (VHX-100, Keyence, Osaka, Japan), and the scale bar represents 1 mm. (I) SEM image of seed trichome in Q0306; the scale bar represents 200 µm. (J–L) Phenotypes of I. purpurea lines. (J) FP39 (wild-type), (K) PR640 (CHS-D mutant), and (L) YJH/DR-4 (bHLH2 mutant). (M) Wild-type I. quamoclit line, Q0055.
From these observations, we hypothesized that CHS-D, CHI, and ANS are not involved in proanthocyanidin production, similar to our findings for DFR-B. To test this hypothesis, we measured proanthocyanidin accumulation in CHS-D, CHI, and ANS loss-of-function morning glory mutants. We then predicted genes involved in proanthocyanidin biosynthesis using the published I. nil draft genome sequence (Hoshino et al. 2016). Finally, we screened a large collection of I. nil lines and related Ipomoea species to isolate novel mutants exhibiting altered proanthocyanidin pigmentation in seed coats.
Materials and methods
Plant materials
Wild-type I. nil (Tokyo-kokei standard, TKS; Kawasaki and Nitasaka 2004), I. purpurea (YO/FP-39, FP39; Habu et al. 1998), and I. quamoclit (Q0055; Sakata Nursery, Yokohama, Japan) all have dark brown seeds, but produce blue, purple, and red flowers, respectively. Negative controls for the proanthocyanidin assay were recessive InWDR1 and IpbHLH2 mutants of I. nil (NS/W1ca1) and I. purpurea (YJH/WR-4); both produce ivory seeds that do not accumulate proanthocyanidins (Morita et al. 2006; Park et al. 2007). The study also used lines AK19 (Ginsekai; Takii & Co., Ltd., Kyoto Japan), AK40, AK10, and AK23 (r3) that were, respectively, I. nil CHS-D, CHI, DFR-B, and ANS mutants (Chopra et al. 2006; Habu et al. 1998; Hoshino et al. 2001, 2009; Iida et al. 2004; Inagaki et al. 1994). The I. purpurea CHS-D mutant (PR640) was also included. All mutant lines produced white flowers and green stems. Drs. Keiichi Shimizu, Norio Saito, and Caitilin Corberly provided AK10, AK23, and PR640, respectively. Line AK40 was from our own collection. Finally, proanthocyanidin mutants were screened using 205 lines (Table 1) from the National BioResource Project (NBRP) morning glory (http://www.shigen.nig.ac.jp/asagao/index.jsp).
Table 1. Summary of proanthocyanidin mutant screening.
| Seed color | Line | PAs* |
|---|---|---|
| Black or dark-brown | Q0114, Q0160, Q0188, Q0191, Q0205, Q0240, Q0243, Q0254, Q0255, Q0270, Q0273, Q0303, Q0304, Q0312, Q0313, Q0316, Q0321, Q0325, Q0330, Q0333, Q0335, Q0336, Q0337, Q0339, Q0342, Q0343, Q0344, Q0346, Q0347, Q0349, Q0352, Q0353, Q0355, Q0357, Q0370, Q0371, Q0373, Q0374, Q0375, Q0410, Q0415, Q0426, Q0438, Q0441, Q0442, Q0448, Q0449, Q0459, Q0464, Q0465, Q0466, Q0467, Q0468, Q0470, Q0471, Q0515, Q0525, Q0537, Q0538, Q0539, Q0575, Q0584, Q0626, Q0635, Q0640, Q0644, Q0645, Q0652, Q0661, Q0663, Q0664, Q0666, Q0667, Q0668, Q0669, Q0670, Q0671, Q0672, Q0673, Q0677, Q0679, Q0703, Q0726, Q0751, Q0771, Q0783, Q0789, Q0821, Q0829, Q0830, Q0837, Q0840, Q0889, Q0893, Q0895, Q0933, Q0943, Q0961, Q0962, Q0963, Q0964, Q0965, Q1055, Q1057, Q1058, Q1065, Q1071, Q1072, Q1075, Q1083, Q1094, Q1095, Q1096, Q1097, Q1098, Q1099, Q1202, Q1214, Q1243, Q1245, Q1246, Q1247, Q1248, Q1249, Q1250, Q1251, Q1252, Q1255, Q1256, Q1257, Q1258, Q1259, Q1261, Q1263, Q1267, Q1268, Q1270, Q1271, Q1272, Q1273, Q1274, Q1275, Q1276, Q1277, Q1278, Q1279, Q1280, Q1281, Q1282, Q1283, Q1284, Q1285, Q1286, Q1287, Q1288, Q1289, Q1290, Q1291, Q1292, Q1293, Q1294, Q1295, Q1297, Q1298, Q1299, Q1300, Q1301, Q1302, Q1303, Q1304, Q1305, Q1306, Q1308, Q1309, Q1310, Q1312, Q1313, Q1314, Q1315, Q1316, Q1317, Q1319, Q1320, Q1322 | + |
| Light-brown | Q0306, Q0314, Q0332, Q0338, Q0341, Q0350, Q0356, Q0359, Q0545, Q0607, Q0674, Q0790, Q0810, Q1041, Q1264, Q1296, Q1307, Q1311 | − |
| Ivory | Q0646, Q0660, Q1059 | − |
*Proanthocyanidins detected using chemical staining.
Proanthocyanidin analysis
Seed proanthocyanidins were quantified using vanillin and DMACA staining, as described previously (Abrahams et al. 2002; Debeaujon et al. 2000; Park et al. 2007). Immature seeds were soaked in either 5% vanillin solution (MeOH : HCl=2 : 1, v/v) or 0.15% DMACA (MeOH : HCl=3 : 1, v/v) solutions for 10 min at room temperature. Proanthocyanidin presence resulted in pink-reddish (vanillin) or bluish (DMACA) staining.
Sequence and expression analysis
To identify proanthocyanidin biosynthesis genes, Arabidopsis proteins were searched against the I. nil draft genome sequence (Hoshino et al. 2016) using BLASTP. Protein databases from NCBI and NBRP (morning glory, http://viewer.shigen.info/asagao/) were used. Distribution of LAR and ANR were tested using the KEGG database that includes protein sequences predicted from the whole genome sequences of multiple plant species (Kanehisa et al. 2016).
The wild-type I. nil (TKS) was used for reverse transcription-polymerase chain reaction (RT-PCR). Total RNA from immature seed coats at 4, 8, 12, 16, 20, 24, and 28 days after pollination (DAP) was extracted using a Get pureRNA Kit (Dojindo Molecular Technologies, Kumamoto, Japan). RNA extraction from flower buds was performed as described previously (Park et al. 2007). First-strand cDNA was synthesized using the SuperScript III reverse transcriptase kit (Thermo Fisher Scientific, Waltham, MA, USA). The internal control was glyceraldehyde 3-phosphate dehydrogenase 2 (GAPDH2) (Park and Hoshino 2012). Amplification of F3H-A, F3H-B, and F3H-C cDNA was performed with the following primer sets, respectively: F3HA-F2/R1 (5′-AAG GGC ATT GAC GAC GTC CT-3′, 5′-CAG ATG GAA ATA GCA GCC GA-3′); F3HB-F2/R1 (5′-GGC ATT GAC GAC GAC GTC CA-3′, 5′-CTC CAC ATC CCT CAT CCT TG-3′); and F3HC-F2/R1 (5′-CAT CGA CGA CGG CGG CGT TA-3′, 5′-GAA ATT ATG GAG CGG GCT AG-3′). Thermocycling conditions were as follows: initial denaturation at 94°C for 2 min, followed by 30 cycles of denaturation (98°C for 10 s), annealing (60°C for 15 s), and extension (68°C for 1 min).
Scanning electron microscopy
Mature seeds were glued to a stage, frozen in liquid nitrogen for 1 min, and examined using a scanning electron microscope (XL30, Philips, Amsterdam, Netherlands) at 10 kV.
Results
Proanthocyanidin accumulation in I. nil seed coats over time
Proanthocyanidins were previously observed in immature I. nil seeds at 24 DAP (Park and Hoshino 2012), but the pigment’s temporal accumulation during seed development has not been characterized. Therefore, we assayed proanthocyanidin accumulation every 4 day from 12 to 32 DAP (Figure 3A). Proanthocyanidin accumulation was undetectable at 12 DAP but increased from 16 to 24 DAP, peaking at 24–28 DAP, and then decreasing at 32 DAP, when seeds turned black (Figure 3A). As seeds mature, proanthocyanidin content likely decreases through forming insoluble, oxidized complexes with other phenolics and cell wall polysaccharides, thus causing seed color to darken (Marles et al. 2003).
Figure 3. Proanthocyanidin accumulation in Ipomoea seeds. (A) Temporal accumulation of proanthocyanidins in wild-type I. nil (TKS). Numerals on photos indicate days after pollination (DAP); seeds were soaked in a vanillin or DMACA solution for staining. (B) Proanthocyanidin accumulation in immature (28 DAP) seeds of anthocyanin-deficient I. nil mutants. The lines used were TKS (wild-type), AK19 (CHS-D mutant), AK40 (CHI mutant), AK10 (DFR-B mutant), AK23 (ANS mutant), NS/W1ca1 (InWDR1 mutant). Flower and seed phenotypes of these lines are shown in Figure 2. (C) Proanthocyanidin accumulation in seed coat of the CHS-D mutant I. purpurea line (PR640) at 28 DAP. Lines FP39 (wild-type) and YJH/WR-4 (bHLH2 mutant) were used as positive and negative controls, respectively. (D) Proanthocyanidin mutant screening revealed that brown mutant lines Q0306 and Q1041 were proanthocyanidin mutants. (E) Immature seeds at various developmental stages of wild-type I. quamoclit (Q0055) remained unstained after being soaked in a DMACA solution.
Proanthocyanidin accumulation in anthocyanin mutants
We examined proanthocyanidin production in seed coats of CHS-D, CHI, and ANS mutants. Proanthocyanidin accumulation was highest at 28 DAP in wild-type plants (Figure 3A). When immature mutant seeds of I. nil and I. purpurea were treated with vanillin-HCl and DMACA-HCl solutions at 28 DAP, all exhibited reddish and bluish staining (Figure 3B, C), indicating proanthocyanidin accumulation. Overall, staining results were indistinguishable across mutants and the wild type. Corroborating previous studies showing a lack of proanthocyanidin accumulation in the seed coats of InWDR1 and IpbHLH2 mutants (Park and Hoshino 2012; Park et al. 2007), we did not observe staining in these lines (Figure 3B, C). Our data suggested that proanthocyanidins accumulate normally in seed coats of CHS-D, CHI, and ANS mutants. Therefore, despite being essential for anthocyanin production in flower petals, these three genes (along with DFR-B) in I. nil, as well as CHS-D in I. purpurea, are not required for seed-coat proanthocyanidin accumulation.
Identification of additional copies of proanthocyanidin biosynthesis genes from the I. nil draft genome
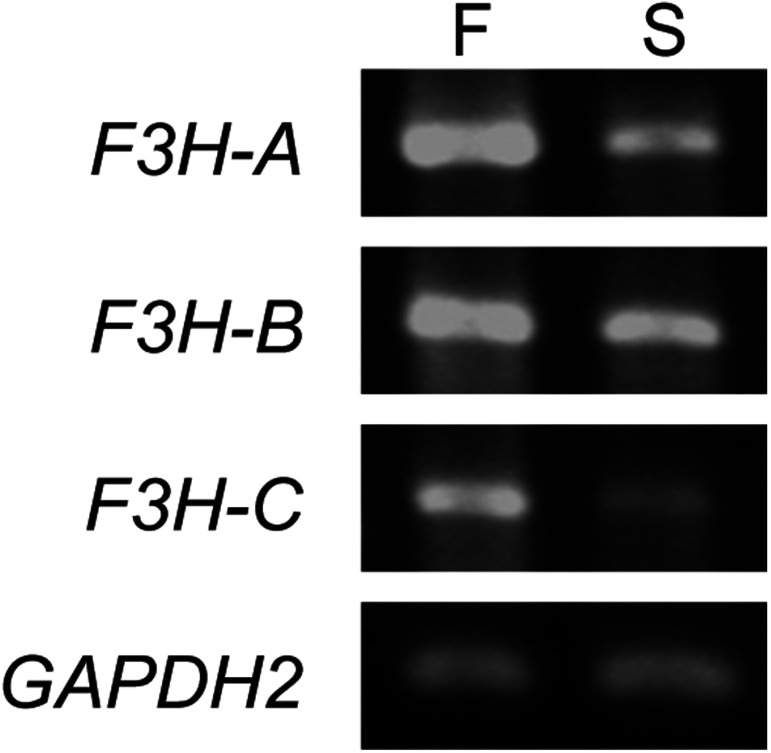
Although flavonoid biosynthesis genes have been extensively characterized in I. nil, their genomic copy numbers remain unknown. We therefore employed BLASTP searches with Arabidopsis flavonoid biosynthesis enzyme sequences against I. nil protein databases to confirm copy number. The results are summarized in Table 2. No additional gene copies were found, except for F3H. Previous studies have shown that F3H-A is likely responsible for anthocyanin pigmentation in flowers because its expression is activated by InWDR1 and InMYB1 (Morita et al. 2006). The I. nil genome carries two additional F3H genes: F3H-B and F3H-C (Table 2). F3H-B seems to be a pseudogene because its deduced protein sequence is shorter at the 3′ end than either F3H-A (36 amino acids) or F3H-C (30 amino acids). Publicly available RNA-seq data (Hoshino et al. 2016) and RT-PCR analysis indicated that all three F3H are transcribed in both flower and seed coats (Table 2, Figure 4). As no F3H mutants have been isolated, we do not know the exact roles of these three F3H genes in anthocyanin and proanthocyanidin biosynthesis.
Table 2. Flavonoid biosynthesis genes found in the Ipomoea nil genome.
| Gene | Accession No. | Gene ID | RNA expression | RNA-seq | Mutation | Necessity** | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Original* | NCBI | Petal | Seed coat | Flower | Seed coat | Petal | Seed coat | |||
| CHS-D | AB001818 | INIL12g08537 | 109176547 | + | − | + | − | transposon | essential | unnecessary |
| CHS-E | AB001819 | INIL14g35461 | 109161594 | + | + | + | + | essential*** | essential | |
| CHI | INIL11g18443 | 109186979 | + | + | + | + | transposon | essential | not essential | |
| F3H-A | D83041 | INIL02g39928 | 109167724 | + | + | + | + | (redundant) | (redundant) | |
| F3H-B | INIL02g39929 | 109167725 | + | + | + | + | pseudo gene | unnecessary | unnecessary | |
| F3H-C | INIL05g09512 | 109177773 | + | + | + | + | (redundant) | (redundant) | ||
| F3′H | AB113261 | INIL04g34064 | 109160581 | + | + | + | + | substitution | not essential | not essential |
| DFR-A | INIL05g09558 | 109177611 | + | + | + | − | unnecessary | redundant | ||
| DFR-B | AB006792 | INIL05g09559 | 109177619 | + | + | + | − | transposon | essential | not essential |
| DFR-C | INIL05g09560 | 109177620 | + | − | + | − | unnecessary | unnecessary | ||
| ANS | AB073920 | INIL13g40254 | 109168138 | + | − | + | − | transposon | essential | unnecessary |
| 3GT | LC019108 | INIL10g12321 | 109182028 | + | + | + | + | transposon | essential | (unnecessary) |
*The original gene ID is from a previous publication (Hoshino et al. 2016). **Predictions without mutant analyses are presented in parentheses. ***CHS-E expression is responsible for flower tube pigmentation in some I. nil lines (Hoshino et al. 2009).
Figure 4. RT-PCR analysis for the F3H genes in Ipomoea nil. “F” and “S” indicate flower petals and seed coats, respectively.
Among the flavonoid biosynthesis genes given in Table 2, CHS-D, CHS-E, F3′H, ANS, and 3GT genes have a single intron, while CHI carries two introns. F3H and DFR genes contain three and five introns, respectively. Two of three F3H (F3H-A and F3H-B) and three DFR genes were duplicated in tandem on chromosomes 2 and 5, respectively. The publicly available RNA-seq data (Hoshino et al. 2016) indicated that all the genes are transcribed in leaves and stems in addition to flowers. The data also indicated that all genes, except for F3H-B and 3GT, are expressed in roots. Only CHI, F3H-B, F3H-C, and F3′H transcripts were found in the RNA-seq data from embryos.
Moreover, BLASTP searches indicated that I. nil does not possess genes related to flavan-3-ol production, given that the best-match sequences using Medicago truncatula LAR and Arabidopsis ANR as queries were a putative isoflavone reductase (XP_019174833) and DFR-B, respectively. To test whether LAR and ANR absence is unique to I. nil, we evaluated putative flavonoid biosynthesis pathways using the KEGG database. We found that LAR and ANR are absent from the genomes of Cucumis sativus (cucumber), Cucumis melo (muskmelon), Momordica charantia (bitter melon), Cucurbita maxima (winter squash), Capsicum annuum (red pepper), Sesamum indicum (sesame), and Dendrobium officinale.
Screening of I. nil proanthocyanidin mutants
We screened I. nil proanthocyanidin mutants to further examine the genetic mechanisms underlying proanthocyanidin synthesis. Most mutant lines exhibited morphological alterations in flower and leaf shape or pigmentation. Of the 205 screened lines, 184 had black or dark-brown seeds, 18 had light-brown seeds, and 3 had ivory-colored seeds (Table 1, Figure 2). Classical genetic studies showed that brown and ca-white (ca) mutations confer light-brown and ivory-colored seeds, respectively (Hagiwara 1931, 1937; Miyake and Imai 1920; Miyazawa 1923). The gene Ca (encoding InWDR1) regulates both flower and seed-coat pigmentation (Morita et al. 2006). Immature seeds from all 205 lines were subjected to DMACA-HCl assays. Every line with black or dark-brown seeds exhibited staining characteristic of proanthocyanidin accumulation. Lines Q0114, Q0353, and Q0830 were F3′H mutants (Morita et al. 2005), suggesting that F3′H is not essential for proanthocyanidin biosynthesis.
In contrast, the 18 light-brown and ivory lines did not exhibit vanillin-HCl and DMACA-HCl staining (Table 1, Figure 3D), indicating that proanthocyanidin accumulation did not occur in their seed coats.
Dark-brown seed pigmentation in I. quamoclit without proanthocyanidin accumulation
We also investigated proanthocyanidin accumulation in the cypress vine (I. quamoclit), a species related to I. nil. Although mature I. quamoclit seeds were dark brown (Figure 2M), blue staining after DMACA-HCl treatment was not observed in seeds at any developmental stage (Figure 3E). This outcome strongly suggests that proanthocyanidins are not the source of dark-brown pigmentation in I. quamoclit seeds.
Discussion
In this study, we demonstrated that CHS-D, CHI, and ANS in I. nil, as well as CHS-D in I. purpurea, are not essential for proanthocyanidin accumulation in seed coats (Figure 3B, C). These genes, however, are essential to anthocyanin production in flowers and stems (Table 2) (Chopra et al. 2006; Habu et al. 1998; Hoshino et al. 2009; Iida et al. 2004; Inagaki et al. 1994). Our previous study similarly showed that DFR-B in I. nil is not essential for proanthocyanidin accumulation in seed coats, but essential for anthocyanin pigmentation (Park and Hoshino 2012). Together, the data suggest that different genes are involved in the early steps of anthocyanin and proanthocyanidin biosynthesis. In contrast, Arabidopsis CHS, CHI, F3H, F3′H, DFR, and ANS are essential for both anthocyanin and proanthocyanidin biosynthesis; their mutants do not accumulate proanthocyanidin in seed coats and thus present a transparent test of phenotype (Abrahams et al. 2003; Shikazono et al. 2003; Shirley et al. 1992, 1995; Wisman et al. 1998).
Both I. nil and I. purpurea have at least two active CHS genes (CHS-D and CHS-E), but only the latter is expressed in seed coats (Johzuka-Hisatomi et al. 1999; Park and Hoshino 2012; Park et al. 2007). This finding coincides with our conclusion that CHS-D is unnecessary for proanthocyanidin accumulation. Instead, CHS-E is likely to be responsible for proanthocyanidin biosynthesis in I. nil and I. purpurea.
We had previously shown that among the three active DFR genes in I. nil, DFR-A and DFR-B transcripts accumulate in the seed coats (Park and Hoshino 2012). Because DFR-B mutants nevertheless accumulate proanthocyanidin in seed coats (Figure 3B) (Park and Hoshino 2012), we conclude that DFR-A and DFR-B are functionally redundant, at least in terms of proanthocyanidin production.
We found a single gene copy of CHI, F3′H, and ANS in I. nil (Table 2). This outcome appears to be inconsistent with the observation of normal proanthocyanidin accumulation in CHI mutants. The most plausible explanation is that CHI activity is not essential for proanthocyanidin synthesis in the I. nil seed coats. Indeed, chalcones can spontaneously isomerize into flavanones without CHI activity (Davies and Schwinn 2005). The reaction is nonstereospecific, resulting in (2S)- and (2R)-flavanone; CHI activity simply guarantees generation of the former. Additionally, F3′H is also non-essential for proanthocyanidin production because its mutants (Q0114, Q0353, and Q0830; Morita et al. 2005) accumulate proanthocyanidins (Table 1). This result suggests that both flavan-3-ols with and without the 3′-hydroxyl group are proanthocyanidin precursors in I. nil. Our current study examining publicly available RNA-seq data (Table 2) further confirmed that ANS is not expressed in I. nil seed coats (Park and Hoshino 2012). Because ANS activity is essential for 2,3-cis-flavan-3-ol but not 2,3-trans-flavan-3-ol production (Park and Hoshino 2012), we suggest that I. nil uses the latter as starter and extension units of proanthocyanidins. This hypothesis is further supported by the observation that ANS mutants accumulate proanthocyanidin normally. Taken together, existing data allow us to predict the roles of proanthocyanidin-production genes in I. nil seed coats (Table 2). Specifically, CHS-E, CHI, and the two F3H (F3H-A, F3H-C) and DFR (DFR-A and DFR-B) genes are involved in the biosynthesis pathway, but CHI and DFR-B are not essential.
Previous work has suggested that LAR converts leucoanthocyanidins to 2,3-trans-flavan-3-ols in the proanthocyanidin pathway (Figure 1) (Xie and Dixon 2005). Unexpectedly, our BLASTP search revealed that the I. nil genome does not contain LAR or ANR genes. Moreover, the absence of LAR or ANR in some plant species, especially plants belonging Cucurbitaceae, was supported by the KEGG database survey. Among Cucurbitaceae plants, proanthocyanidins were found in the seeds and whole fruits of cucumber and bitter melon, respectively (Tan et al. 2014; Zhu et al. 2016). Thus, some plants may produce proanthocyanidins via undiscovered biosynthesis pathways without LAR and ANR involvement. Further analysis of I. nil proanthocyanidins will benefit the identification of such pathways.
Our screening of I. nil lines revealed that brown and InWDR1 (ca) mutant lines did not accumulate proanthocyanidin in their seed coats. Previously, we used another InWDR1 mutant line (NS/W1ca1) to show that InWDR1 activates proanthocyanidin pigmentation in seeds (Park and Hoshino 2012), and our current findings confirm those results. Although InWDR1 mutation removes anthocyanin, resulting in white flowers and green stems (Hagiwara 1931, 1937; Miyake and Imai 1920; Miyazawa 1923), the effects of the brown mutation on anthocyanin pigmentation have not been reported until now. Here we demonstrate that brown mutants show normal anthocyanin accumulation. This study is the first to describe mutations that result in alterations to proanthocyanidin but not anthocyanin pigmentation among Ipomoea species. Based on our results, we propose that brown mutants are a useful material for detailed examinations of differential mechanisms underlying anthocyanin and proanthocyanidin production. We observed that brown mutants have slightly darker seeds than InWDR1 mutants (Figure 2F–H). The latter accumulate phytomelanins at approximately 20% of wild-type levels. Therefore, pigmentation differences between brown and InWDR1 mutants may be attributable to differing phytomelanin content in seed coats. More research is necessary to determine whether the brown gene controls phytomelanin in addition to proanthocyanidin. Notably, brown mutants produce normal seed trichomes (Figure 2H–I). InWDR1 and IpbHLH2 control seed trichome formation in I. nil and I. purpurea, respectively, besides anthocyanin biosynthesis in flowers and proanthocyanidin and phytomelanin pigmentation in seeds (Morita et al. 2006; Park and Hoshino 2012; Park et al. 2007). These observations suggest that the Brown gene product is not a component of the transcriptional activator complex including InWDR1 and InbHLH2, which is the product of IpbHLH2 ortholog in I. nil.
Finally, our screening revealed that despite having dark-brown seed coats, I. quamoclit does not accumulate proanthocyanidins (Figure 3E). We suggest that the brown pigmentation of I. quamoclit seed coats may comprise phytomelanins. Additionally, previous studies have shown that ANS is expressed in I. purpurea but not I. nil seed coats, indicating that their respective flavan-3-ols units are 2,3-trans-flavan-3-ols and 2,3-cis-flavan-3-ols (Park and Hoshino 2012; Park et al. 2007). Together, the data suggest that Ipomoea species exhibit considerable diversity in the compositions of their seed coat pigments.
In conclusion, our anthocyanin mutant analyses suggested that I. nil uses a different set of genes for the early steps in anthocyanin and proanthocyanidin biosynthesis pathways, although Arabidopsis uses the same set of genes for these steps. This allows normal proanthocyanidin accumulation in seed coats of anthocyanin mutants of I. nil. The lack of LAR and ANR genes in I. nil implied the possibility that some plant species produce proanthocyanidins via unelucidated biosynthesis pathways. The brown mutants and I. quamoclit are useful materials for further elucidation of anthocyanin and proanthocyanidin production.
Acknowledgements
The authors wish to thank Dr. Shigeru Iida for his participation in valuable discussions; Drs. Norio Saito, Keiichi Shimizu, and Caitilin Corberly for providing Ipomoea lines; and the NIBB Model Plant Research Facility and the NIBB Functional Genomics Facility for technical support. This study was funded in part by the Center for the Promotion of Integrated Sciences (CPIS) of SOKENDAI and the 2015 Yeungnam University research grant.
References
- Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR (2003) The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J 35: 624–636 [DOI] [PubMed] [Google Scholar]
- Abrahams S, Tanner GJ, Larkin PJ, Ashton AR (2002) Identification and biochemical characterization of mutants in the proanthocyanidin pathway in Arabidopsis. Plant Physiol 130: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbehenn RV, Peter Constabel C (2011) Tannins in plant-herbivore interactions. Phytochemistry 72: 1551–1565 [DOI] [PubMed] [Google Scholar]
- Chang SM, Lu Y, Rausher MD (2005) Neutral evolution of the nonbinding region of the anthocyanin regulatory gene Ipmyb1 in Ipomoea. Genetics 170: 1967–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra S, Hoshino A, Boddu J, Iida S (2006) Flavonoid pigments as tools in molecular genetics. In: Grotewold E (ed) The Science of Flavonoids. Springer, New York, pp 147–173
- Davies KM, Schwinn KE (2005) Molecular biology and biotechnology of flavonoid biosynthesis. In: Andersen OM, Markham KR (eds) Flavonoids Chemistry, Biochemistry and Applications. CRC Press, Boca Raton, pp 143–218
- Debeaujon I, Leon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devic M, Guilleminot J, Debeaujon I, Bechtold N, Bensaude E, Koornneef M, Pelletier G, Delseny M (1999) The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J 19: 387–398 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Liu C, Jun JH (2013) Metabolic engineering of anthocyanins and condensed tannins in plants. Curr Opin Biotechnol 24: 329–335 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Xie DY, Sharma SB (2005) Proanthocyanidins: A final frontier in flavonoid research? New Phytol 165: 9–28 [DOI] [PubMed] [Google Scholar]
- Feinbaum RL, Ausubel FM (1988) Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol Cell Biol 8: 1985–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habu Y, Hisatomi Y, Iida S (1998) Molecular characterization of the mutable flaked allele for flower variegation in the common morning glory. Plant J 16: 371–376 [DOI] [PubMed] [Google Scholar]
- Hagiwara T (1931) Genetico-physiological studies on the seed-colour of Japanese morning glory. Jpn J Genet 7: 1–16 (in Japanese) [Google Scholar]
- Hagiwara T (1937) On a mutable gene of the seed coat colour in Pharbitis nil. Jpn J Genet 13: 185–192 (in Japanese) [Google Scholar]
- Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55: 481–504 [DOI] [PubMed] [Google Scholar]
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62: 2465–2483 [DOI] [PubMed] [Google Scholar]
- Hoshino A, Jayakumar V, Nitasaka E, Toyoda A, Noguchi H, Itoh T, Shin-I T, Minakuchi Y, Koda Y, Nagano AJ, et al. (2016) Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nat Commun 7: 13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Johzuka-Hisatomi Y, Iida S (2001) Gene duplication and mobile genetic elements in the morning glories. Gene 265: 1–10 [DOI] [PubMed] [Google Scholar]
- Hoshino A, Park KI, Iida S (2009) Identification of r mutations conferring white flowers in the Japanese morning glory (Ipomoea nil). J Plant Res 122: 215–222 [DOI] [PubMed] [Google Scholar]
- Iida S, Morita Y, Choi JD, Park KI, Hoshino A (2004) Genetics and epigenetics in flower pigmentation associated with transposable element in morning glories. Adv Biophys 38: 141–159 [PubMed] [Google Scholar]
- Inagaki Y, Hisatomi Y, Suzuki T, Kasahara K, Iida S (1994) Isolation of a Suppressor-mutator/Enhancer-like transposable element, Tpn1, from Japanese morning glory bearing variegated flowers. Plant Cell 6: 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka-Hisatomi Y, Hoshino A, Mori T, Habu Y, Iida S (1999) Characterization of the chalcone synthase genes expressed in flowers of the common and Japanese morning glories. Genes Genet Syst 74: 141–147 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44(D1): D457–D462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S, Nitasaka E (2004) Characterization of Tpn1 family in the Japanese morning glory: En/Spm-related transposable elements capturing host genes. Plant Cell Physiol 45: 933–944 [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Koes RE, Quattrocchio F, Mol JNM (1994) The flavonoid biosynthetic pathway in plants: Function and evolution. BioEssays 16: 123–132 [Google Scholar]
- Koornneef M (1990) Mutations affecting the testa color in Arabidopsis. Arabidopsis Inf Serv 27: 1–4 [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Marles MA, Ray H, Gruber MY (2003) New perspectives on proanthocyanidin biochemistry and molecular regulation. Phytochemistry 64: 367–383 [DOI] [PubMed] [Google Scholar]
- Miyake K, Imai Y (1920) Genetic studies of Japanese morning glory. Bot Mag Tokyo 34: 1–26 (in Japanese) [Google Scholar]
- Miyazawa B (1923) Genetic studies on the seed color on Japanese morning glory. Jpn J Genet 2: 1–11 (in Japanese) [Google Scholar]
- Morita Y, Hoshino A, Kikuchi Y, Okuhara H, Ono E, Tanaka Y, Fukui Y, Saito N, Nitasaka E, Noguchi H, et al. (2005) Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose: Anthocyanidin 3-O-glucoside-2″-O-glucosyltransferase, due to 4-bp insertions in the gene. Plant J 42: 353–363 [DOI] [PubMed] [Google Scholar]
- Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S (2006) Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol 47: 457–470 [DOI] [PubMed] [Google Scholar]
- Park KI, Hoshino A (2012) A WD40-repeat protein controls proanthocyanidin and phytomelanin pigmentation in the seed coats of the Japanese morning glory. J Plant Physiol 169: 523–528 [DOI] [PubMed] [Google Scholar]
- Park KI, Ishikawa N, Morita Y, Choi JD, Hoshino A, Iida S (2007) A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. Plant J 49: 641–654 [DOI] [PubMed] [Google Scholar]
- Petroni K, Tonelli C (2011) Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci 181: 219–229 [DOI] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10: 63–70 [DOI] [PubMed] [Google Scholar]
- Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, Einhorn J, Lepiniec L (2006) Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224: 96–107 [DOI] [PubMed] [Google Scholar]
- Shikazono N, Yokota Y, Kitamura S, Suzuki C, Watanabe H, Tano S, Tanaka A (2003) Mutation rate and novel tt mutants of Arabidopsis thaliana induced by carbon ions. Genetics 163: 1449–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Hanley S, Goodman HM (1992) Effects of ionizing radiation on a plant genome: Analysis of two Arabidopsis transparent testa mutations. Plant Cell 4: 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Tan SP, Vuong QV, Stathopoulos CE, Parks SE, Roach PD (2014) Optimized aqueous extraction of saponins from bitter melon for production of a saponin-enriched bitter melon powder. J Food Sci 79: E1372–E1381 [DOI] [PubMed] [Google Scholar]
- Tanner GJ, Francki KT, Abrahams S, Watson JM, Larkin PJ, Ashton AR (2003) Proanthocyanidin biosynthesis in plants. Purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J Biol Chem 278: 31647–31656 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2006) The biosynthesis of flavonoids. In: Grotewold E (ed) The Science of Flavonoids. Springer, New York, pp 71–95
- Wisman E, Hartmann U, Sagasser M, Baumann E, Palme K, Hahlbrock K, Saedler H, Weisshaar B (1998) Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc Natl Acad Sci USA 95: 12432–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DY, Dixon RA (2005) Proanthocyanidin biosynthesis: Still more questions than answers? Phytochemistry 66: 2127–2144 [DOI] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299: 396–399 [DOI] [PubMed] [Google Scholar]
- Zhu LJ, Deng XG, Zou LJ, Wu JQ, Zhang DW, Lin HH (2016) Proanthocyanidins accelerate the germination of cucumber (Cucumis sativus L.) seeds. J Plant Biol 59: 143–151 [Google Scholar]