Abstract
Recently, two rice genes, OsAPETALA2 (OsAP2) and OsWRKY24 have been reported to be positive regulators involved in increased lamina inclination and grain size through cell elongation. Here, we found that the two genes have tightly linked expression patterns and functional convergence in rice, and are also likely to play an opposite role in Arabidopsis. Overexpression of the two rice transcription factors in Arabidopsis caused smaller plant size with reduced cell size, and the expression of a series of genes encoding expansins and xyloglucan endotransglucosylase/hydrolases (XTHs) involved in cell elongation was reduced. However, transgenic Arabidopsis expressing OsWRKY24-SRDX as a synthetic chimeric repressor displayed indistinguishable phenotypes from wild-type plants. Moreover, the subcellular localization pattern of OsWRKY24 in Arabidopsis was different from that in rice. Thus, we demonstrate an example of transcription factors from one species playing distinct roles in different plant species.
Keywords: APETALA2 transcription factor, cell size, RNA expression, transgenic Arabidopsis, WRKY transcription factor
Transcription factors (TFs) are master regulators that control the expression of many other genes. Recently, two rice TFs, Oryza sativa APETALA 2 (OsAP2) and OsWRKY24 have been isolated as positive regulators in cell elongation in rice (Jang and Li 2017). The AP2 family TFs characterized by the AP2/ERF DNA-binding domain are plant-specific (Riechmann and Meyerowitz 1998) and known to be involved in regulation of diverse processes in plant development and stress responses (Nakano et al. 2006; Sharoni et al. 2011; Zhao et al. 2015).
In the progression of plant growth and development, AINTEGUMENTA and AINTEGUMENTA-LIKE6 are related to flower organ growth and ovule development in Arabidopsis (Elliott et al. 1996; Jofuku et al. 1994; Klucher et al. 1996; Krizek 2009; Mizukami and Fischer 2000). Moreover, mutations in AP2 increased the seed size because of increased cell number and size in the Arabidopsis embryo (Jofuku et al. 2005; Ohto et al. 2005). Other AP2 TFs, AtERF1, AtDREB1, and TINY were able to dwarf plant height of Arabidopsis (Liu et al. 1998; Solano et al. 1998; Wilson et al. 1996).
In rice, several AP2 genes including SUBMERGENCE1A (SUB1A), SNORKEL1 (SK1), SK2, Oryza sativa ERF protein associated with tillering and branching (OsEATB) and REDUCED LEAF ANGLE1 (RLA1)/SMALL ORGAN SIZE1 (SMOS1) have been reported to play roles in regulating internode elongation, which is primarily post-mitotic expansion of differentiating cells (Aya et al. 2014; Hattori et al. 2009; Qi et al. 2011; Xu et al. 2006). In particular, RLA1/SMOS1 was revealed to be a positive regulator in the brassinosteroid (BR) signaling pathway and also an auxin-dependent regulator of cell expansion (Hirano et al. 2014; Qiao et al. 2017).
The WRKY family TFs are only found in higher plants and the family is one of the largest TF families in plants. Since the first member was isolated in sweet potato, many WRKY TFs have been identified, including 74 in Arabidopsis and 102 in rice (Rushton et al. 2010). WRKY TFs are defined by their WRKY domain, a conserved DNA-binding domain containing 60 amino acid residues, which binds to a W-box (TTG ACC/T) in the regulatory DNA sequences. Additionally, WRKY TFs possess a zinc-finger motif at their C-terminus, either Cx4–5Cx22–23HxH (CCHH) or Cx7Cx23HxC (CCHC). Members of the WRKY family TFs are involved in a wide range of developmental and physiological processes including trichome and seed development, senescence, biotic and abiotic stress response (Jiang et al. 2017). In terms of cell elongation/enlargement, a loss-of-function mutant of the maternally expressed WRKY gene, ttg-2 produced small seeds displaying reduced cell elongation in integuments, and precocious endosperm cellularization (Garcia et al. 2005). Recently, new roles of WRKY TFs in BR signaling have been revealed (Chen et al. 2017) and BR regulates a wide range of plant growth and development, such as cell elongation, cell division, xylem differentiation and stress responses. In rice, OsWRKY78 is an example of a WRKY TF that functions in regulating cell elongation (Zhang et al. 2011). Both RNA interference (RNAi) and T-DNA insertion lines provided evidence showing that OsWRKY78 may act as a regulator in stem elongation and seed development of rice. Recently, a foxtail millet mutant, loose-panicle 1 (lp1) impaired in a WRKY gene, has been reported to have an interesting phenotype, reduced internode length but increased panicle length (Xiang et al. 2017).
Previously, we reported that two rice TFs, OsAP2 (Os10g41330) and OsWRKY24 (Os01g61080) are positive regulators for cell elongation acting downstream of Oryza sativa BUL1 Downstream Gene1 (OsBDG1) in rice (Jang and Li 2017). Increased expression of OsAP2 and OsWRKY24 in lamina joint and panicle of rice caused increased lamina inclination and grain size through the cell elongation in those parts. In this study, we tested whether the functional roles of the two rice TFs are conserved in a heterologous system, Arabidopsis. To our surprise, transgenic Arabidopsis plants overexpressing the two rice TFs exhibited smaller plant body size with reduced epidermal cell size, which is an opposite phenotype to that of transgenic rice (Jang and Li 2017). Thus, we show that TFs, master regulators that control gene expression in one plant species may produce distinct phenotypes in heterologous systems. This information may provide more possibilities for application of genetic modification (GM) technology in plants.
First, we conducted co-expression analysis of the two rice genes, OsAP2 and OsWRKY24 by using RiceFREND database (Sato et al. 2013; http://ricefrend.dna.affrc.go.jp). The database includes 815 pieces of microarray data from various tissues at different developmental stages and phyto-hormone treatment conditions with single or multiple guide gene searches. The expression of the two genes, OsAP2 and OsWRKY24 are closely linked based on the analysis result through the database (Figure 1A) supporting our previous spatiotemporal expression analyses (Jang and Li 2017). Moreover, the two genes are also involved in stress response in rice: OsAP2 transcript is known to be more abundant in the submergence tolerant rice, M202 (Sub1) related to control M202 at 1 day after submergence (Jung et al. 2010) and OsWRKY24 was previously shown to enhance resistance to salt and drought through the cross-talk of gibberellic acid and abscisic acid signaling pathways under stress conditions (Zhang et al. 2009). As shown in Figure 1B, the expression patterns of the two genes by treatment of chemicals inducing abiotic stresses such as drought and salt stresses are also similar. Results of lamina inclination assay with brassinolide (Sigma-E1641) treatments demonstrated that induced expression of the two genes in the lamina joint under the control of the OsBUL1 promoter confers increased sensitivity to brassinolide resulting in increased lamina angles confirming the expression hierarchy among OsBUL1, OsBDG1 and OsAP2 and OsWRKY24 with brassinosteroid response (Figure 1C; Jang and Li 2017; Jang et al. 2017a).
Figure 1. Expression patterns with functional convergence of OsAP2 and OsWRKY24 in rice. (A) In the RiceFREND database, OsAP2 (Os10g41330) marked as 1 is presented to be co-expressed with OsWRKY24 (Os01g61080) marked as 2. The red and blue circles represent plant-pathogen interaction and the phosphatidylinositol signaling system, respectively (in KEGG pathway). (B) Expression of OsAP2 and OsWRKY24 under different abiotic stress conditions. Rice seedlings were germinated and incubated on 1/2 MS media for 10 days and transferred to test tubes containing water (mock), ABA (60 µM; Sigma-A4906), PEG6000 (15%; Alfa Aesar-A17541) and NaCl (150 mM; Amresco-0241) solution, respectively. The plants were grown for 2 days under 12 h-light photoperiod at a constant temperature of 28°C, and each sample was harvested at 3 h, 12 h and 48 h time points. Actin (Act) was used as standard for quantification of gene expression. Data are represented as mean±SD of three biological replicates. W, water control; ABA, Abscisic acid 60 µM; PEG, Polyethylene glycol 6000 15%; NaCl, NaCl 150 mM. (C) Lamina inclination assay (Li et al. 2017) with exogenous brassinolide treatment (10−8 and 10−6 M; Sigma-E1641). Leaves of transgenic rice plants expressing OsAP2 and OsWRKY24 under the control of OsBUL1 promoter (Jang and Li 2017) were used together with those of WT control, TNG67. Values represent mean±SD (n≥15). Data that is significantly different from the corresponding control is indicated (*p<0.05; students’ t-test).
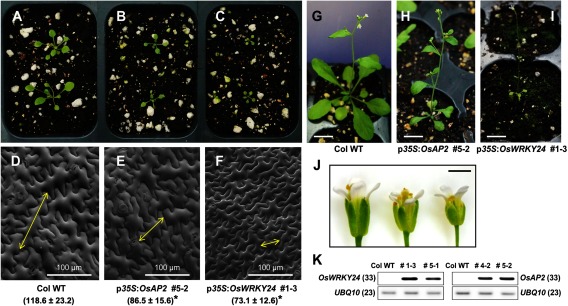
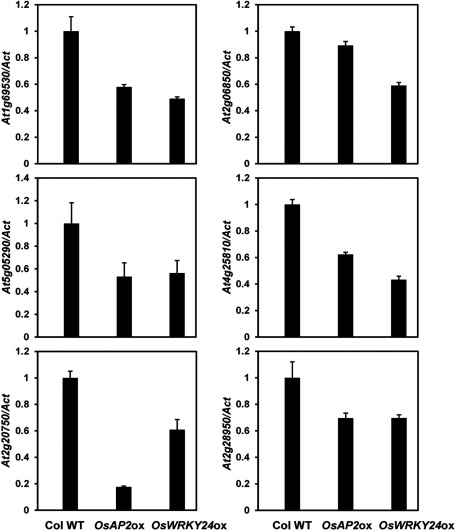
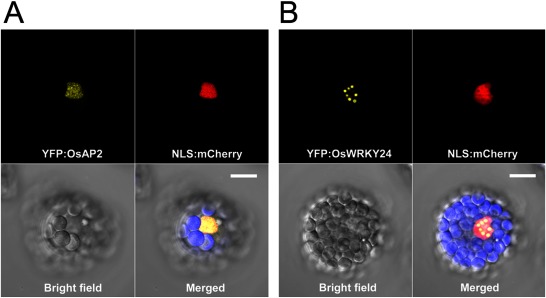
To examine whether the functional activities of OsAP2 and OsWRKY24 are conserved in Arabidopsis, the two genes were ectopically expressed in Arabidopsis using CaMV35S promoter. Compared with the WT, transgenic Arabidopsis showed much smaller plant body size starting from the seedling stages (Figure 2A–C, 2G–I, 2K). Moreover, the flower size of the transgenic plants was reduced (Figure 2J). Particularly, transgenic Arabidopsis overexpressing OsWRKY24 exhibited an extremely tiny body and produced a small number of short seeds (Supplementary Figure S1). To test whether cell size is also affected in those plants, epidermal cells of the first true leaf from each transgenic plant were examined using a scanning electron microscope (SEM) and indeed, smaller size epidermal cells were observed in the transgenic plants (Figure 2D–F). In order to examine the phenotype by molecular analyses, expression of several representative genes involved in cell elongation was investigated by quantitative RT-PCR (Figure 3). The expression level of Expansin genes including Expansin-A1 (At1g69530), Expansin-A2 (At5g05290), Expansin-A6 (At2g28950) and xyloglucan endo-transglycosylase/hydrolase (XTH) genes such as XTH4 (At2g06850) and XTH23 (At4g25810) was lower in transgenic plants showing smaller epidermal cells than the WT control. Subcellular localization of OsAP2 and OsWRKY24 was also examined using Arabidopsis mesophyll protoplasts. Yellow florescence signals are exclusively detected in the nucleus from both Yellow Florescence Protein (YFP):OsAP2 and YFP:OsWRKY24 fusion proteins (Figure 4), which is similar to the results in rice mesophyll protoplasts (Jang and Li 2017). Interestingly, however, the YFP:OsWRKY24 protein forms nuclear-speckles only in Arabidopsis mesophyll cells (Figure 4B) implying the distinct characteristics between mesophyll cells of the two different plant species, rice and Arabidopsis. Nuclear speckles are believed to serve as an active hub coordinating transcription of genes with the processing of their pre-mRNAs (Hall et al. 2006). Thus, OsWRKY24 is likely to participate in active transcription of downstream genes in Arabidopsis. Furthermore, in order to test whether OsWRKY24 acts as a transcriptional activator in plants, a dominant repressor version of OsWRKY24 [OsWRKY24-SUPERMAN repressive domain X (SRDX)] was generated by the fusion of SRDX (LDLDLELRLGFA) repressor domain to the carboxyl terminal of OsWRKY24. It was found that fusion of this motif to transcriptional activators converts them into dominant repressors (Hiratsu et al. 2003). Transgenic plants overexpressing OsWRKY24-SRDX exhibited no significant phenotypic alterations compared to the WT (Figure 5) indicating OsWRKY24 may act as a transcriptional activator in Arabidopsis.
Figure 2. Transgenic Arabidopsis plants overexpressing OsAP2 and OsWRKY24. Twenty-four-day-old seedlings of wild-type Columbia (A, D), p35S:OsAP2 (B, E) and p35S:OsWRKY24 (C, F) plants. Epidermal cells of the first leaves from each plant are shown in D, E and F. Yellow opposing arrows on D, E and F indicate the length of a representative epidermal cell from each genotype. The cell length of each genotype is shown as mean±SD (µm, n≥15). Student’s t-test was used for statistical analysis (*p<0.05). The cell length was measured with ImageJ software (https://imagej.nih.gov/ij/). At the flowering stage, the tiny body size of transgenic Arabidopsis plants (H, I) can be observed compared to wild-type plant (G). Bar=1 cm (G–I). (J) Flower size is also reduced in transgenic plants for p35S:OsAP2 (middle) and p35S:OsWRKY24 (right) compared to that of wild-type plants (left). Bar=1 mm. (K) Expression of transgenes, OsAP2 and OsWRKY24 in transgenic plants. The numbers of cycles used in the RT-PCR are in parenthesis.
Figure 3. Expression of genes encoding cell-wall loosening proteins in Arabidopsis. Expression level of AtEXPA1 (At1g69530), AtEXPA2 (At5g05290), AtEXPA6 (At2g28950), AtEXPB1 (At2g20750), AtXTH4 (At2g06850) and AtXTH23 (At4g25810) was investigated in 2-week-old seedlings of wild-type Columbia, p35S:OsAP2 and p35S:OsWRKY24 plants. qPCR was performed on a CFX96TM real-time system (Bio-Rad) using Maxima SYBR Green qPCR Master Mix (Thermo). The primers used for quantification are listed in Supplementary Table S1. Actin (Act) was used as a standard for quantification of gene expression. Data are represented as mean±SD of three biological replicates.
Figure 4. Subcellular localization of OsAP2 and OsWRKY24 in Arabidopsis mesophyll cells. (A) YFP:OsAP2 and nuclear localization signal (NLS):mCherry were co-transformed into Arabidopsis protoplasts. YFP signals from YFP:OsAP2 are exclusively localized in the nucleus. (B) YFP:OsWRKY24 and NLS:mCherry were co-transformed into Arabidopsis protoplasts. YFP signals from YFP:OsWRKY24 form nuclear speckles. Red florescence is from a nuclear marker, NLS:mCherry (Lee et al. 2012). Isolation and transfection of Arabidopsis protoplasts followed, as described by Wu et al. (2009). Bar=10 µm.
Figure 5. Transgenic Arabidopsis plants overexpressing a chimeric repressor of OsWRKY24 (OsWRKY24-SRDX). The OsWRKY24-SRDX DNA fragment for an entry clone was amplified by PCR using the following primers, 5′GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT CAT GAC AAC CTC GTC GTC3′ and 5′GGG GAC CAC TTT GTA CAA GAA AGC TGG GTG CTA TGC GAA TCC TAG TTC CAG TTC GAG ATC CAG GTC TAG GTA GAG CGA GTT CTG3′. Transgenic plants containing p35S:OsWRKY24-SRDX construct with high expression level of the transgene exhibit an indistinguishable phenotype to WT.
Since TFs are master controllers of gene expression through the formation of homo/hetero-multimeric complexes, overexpression of TFs can be anticipated to produce distinct phenotypes in different plant species. For instance, although overexpression of a key flowering gene of Arabidopsis, FD encoding a basic leucine zipper (bZIP) TF, causes early flowering in Arabidopsis, transgenic rice overexpressing FD exhibits no significant alteration in heading date but has smaller grain size with semi-dwarfism (Abe et al. 2005; Jang et al. 2017b). Previously, we identified two rice genes, OsAP2 and OsWRKY24 as putative downstream genes of OsBDG1, a positive regulator controlling rice leaf angle through cell elongation of the lamina joint (Jang and Li 2017). The spatiotemporal expression of the two genes using various organs at different developmental stages displayed similar patterns and the induced expression of OsAP2 and OsWRKY24 was observed by exogenous treatment of phytohormones such as gibberellin and/or brassinosteroid (Jang and Li 2017). Furthermore, ectopic expression of OsAP2 and OsWRKY24 under the control of OsBUL1 promoter, which is preferentially active in lamina joints and spikelets caused increased leaf angle and grain size with elongated epidermal cells in lamina joint and spikelet lemma (Jang and Li 2017).
In this study, we surveyed the co-expression network of rice genes through the RiceFREND database (Sato et al. 2013) with OsAP2 and OsWRKY24 and found their expression patterns to be closely linked. Since some genes of the AP2 family and WRKY family are known to play pivotal roles in the adaptation of plants to abiotic stresses including drought stress (Bakshi and Oelmüller 2014; Phukan et al. 2017), we also investigated expression patterns of the two genes under several abiotic stress-inducing conditions using abscisic acid (60 µM ABA; Sigma-A4906), polyethylene glycol (15% PEG6000; Alfa Aesar-A17541) and NaCl (150 mM; Amresco-0241). Of note, the two genes showed similar expression patterns under different stress conditions. Functional study using transgenic rice plants expressing OsAP2 and OsWRKY24 under the control of OsBUL1 promoter demonstrated that the two genes confer increased sensitivity to brassinolide supporting the notion that genes with similar expression patterns tend to be functionally related across several experimental conditions. Transgenic Arabidopsis overexpressing OsAP2 and OsWRKY24 also showed similar phenotypes such as small plant body with decreased cell size, and expression of expansin and XTH genes involved in cell expansion/elongation through cell-wall loosening was also reduced in the transgenic plants (Cosgrove 2000; Eklöf and Brumer 2010). Notably, the phenotype of smaller cells in Arabidopsis caused by OsAP2 and OsWRKY24 is opposite to that of transgenic rice with increased expression of those genes (Jang and Li 2017) suggesting that the two TFs may control the expression of distinct target genes in Arabidopsis and rice. Nuclear speckled localization of YFP signals from YFP:OsWRKY24 in Arabidopsis mesophyll protoplasts, which is different from that in rice mesophyll protoplasts (Jang and Li 2017) may support this notion. Previously, overexpression of BOLITA, an Arabidopsis AP2 gene caused small and compact flowers in Arabidopsis while it produced bigger flowers with broader petals in tobacco (Marsch-Martinez et al. 2006) supporting the notion that ectopic expression of TFs in heterologous systems may cause novel phenotypes, which may be due to divergent cis-elements in the regulatory region of gene expression in genomes of distinct species and/or modification of transcription machinery for downstream genes through interaction with other TFs. Thus, we believe that the two genes, OsAP2 and OsWRKY24 have potential to control cell size in plants, and it would be also interesting to identify their target genes in Arabidopsis and rice.
Acknowledgements
We thank members of core facility laboratories of Academia Sinica for plant transformation and confocal microscopy. We are also grateful to Ms. Miranda Loney for help with English editing. This work was supported in part by grants from MOST, Taiwan (106-2313-B-001-005- to SJ) and BCST of ABRC, Academia Sinica.
Abbreviations
- ABA
Abscisic acid
- BR
Brassinosteroid
- PEG
Polyethylene glycol
- RT-PCR
Reverse transcription-polymerase chain reaction
- SEM
Scanning electron microscope
- SRDX
SUPERMAN repressive domain X
- TF
Transcription factor
- XTH
Xyloglucan endotransglucosylase/hydrolase
- YFP
Yellow florescence protein
Supplementary Data
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Aya K, Hobo T, Sato-Izawa K, Ueguchi-Tanaka M, Kitano H, Matsuoka M (2014) A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ size downstream of an auxin signaling pathway. Plant Cell Physiol 55: 897–912 [DOI] [PubMed] [Google Scholar]
- Bakshi M, Oelmüller R (2014) WRKY transcription factors: Jack of many trades in plants. Plant Signal Behav 9: e27700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Nolan TM, Ye H, Zhang M, Tong H, Xin P, Chu J, Chu C, Li Z, Yin Y (2017) Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29: 1425–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ (2000) Loosening of plant cell walls by expansins. Nature 407: 321–326 [DOI] [PubMed] [Google Scholar]
- Eklöf JM, Brumer H (2010) The XTH gene family: An update on enzyme structure, function, and phylogeny in xyloglucanremodeling. Plant Physiol 153: 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Fitz Gerald JN, Berger F (2005) Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Smith KP, Byron M, Lawrence JB (2006) Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol 288: 664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hirano K, Yoshida H, Aya K, Kawamura M, Hayashi M, Hobo T, Sato-Izawa K, Kitano H, Ueguchi-Tanaka M, Matsuoka M (2017) SMALL ORGAN SIZE 1 and SMALL ORGAN SIZE 2/DWARF AND LOW-TILLERING form a complex to integrate auxin and brassinosteroid signaling in rice. Mol Plant 10: 590–604 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Jang S, An G, Li HY (2017a) Rice leaf angle and grain size are affected by the OsBUL1 transcriptional activator complex. Plant Physiol 173: 688–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Li HY (2017) Oryza sativa BRASSINOSTEROID UPREGULATED1 LIKE1 induces the expression of a gene encoding a small leucine-rich-repeat protein to positively regulate lamina inclination and grain size in rice. Front Plant Sci 8: 1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Li HY, Kuo ML (2017b) Ectopic expression of Arabidopsis FD and FD PARALOGUE in rice results in dwarfism with size reduction of spikelets. Sci Rep 7: 44477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Ma S, Ye N, Jiang M, Cao J, Zhang J (2017) WRKY transcription factors in plant responses to stresses. J Integr Plant Biol 59: 86–101 [DOI] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, Omidyar PK, Gee Z, Okamuro JK (2005) Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc Natl Acad Sci USA 102: 3117–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Seo YS, Walia H, Cao P, Fukao T, Canlas PE, Amonpant F, Bailey-Serres J, Ronald PC (2010) The submergence tolerance regulator Sub1A mediates stress-responsive expression of AP2/ERFtranscription factors. Plant Physiol 152: 1674–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8: 137–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B (2009) AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol 150: 1916–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LY, Wu FH, Hsu CT, Shen SC, Yeh HY, Liao DC, Fang MJ, Liu NT, Yen YC, Dokládal L, et al. (2012) Screening a cDNA library for protein-protein interactions directly in planta. Plant Cell 24: 1746–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HY, Wang HM, Jang S (2017) Rice lamina joint inclination assay. Bio Protoc 7: e2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsch-Martinez N, Greco R, Becker JD, Dixit S, Bergervoet JH, Karaba A, de Folter S, Pereira A (2006) BOLITA, an Arabidopsis AP2/ERF-like transcription factor that affects cell expansion and proliferation/differentiation pathways. Plant Mol Biol 62: 825–843 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ (2005) Control of seed mass by APETALA2. Proc Natl Acad Sci USA 102: 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phukan UJ, Jeena GS, Tripathi V, Shukla RK (2017) Regulation of Apetala2/Ethylene response factors in plants. Front Plant Sci 8: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Sun F, Wang Q, Chen M, Huang Y, Feng YQ, Luo X, Yang J (2011) Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol 157: 216–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao S, Sun S, Wang L, Wu Z, Li C, Li X, Wang T, Leng L, Tian W, Lu T, et al. (2017) The RLA1/SMOS1 Transcription factor functions with OsBZR1 to regulate brassinosteroid signaling and rice architecture. Plant Cell 29: 292–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379: 633–646 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Sato Y, Namiki N, Takehisa H, Kamatsuki K, Minami H, Ikawa H, Ohyanagi H, Sugimoto K, Itoh J, Antonio BA, et al. (2013) RiceFREND: A platform for retrieving coexpressed gene networks in rice. Nucleic Acids Res 41(D1): D1214–D1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharoni AM, Nuruzzaman M, Satoh K, Shimizu T, Kondoh H, Sasaya T, Choi IR, Omura T, Kikuchi S (2011) Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol 52: 344–360 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K, Long D, Swinburne J, Coupland G (1996) A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS (2009) Tape-Arabidopsis Sandwich: A simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J, Tang S, Zhi H, Jia G, Wang H, Diao X (2017) Loose Panicle1 encoding a novel WRKY transcription factor, regulates panicle development, stem elongation, and seed size in foxtail millet, Setaria italica, L., P. Beauv. PLoS One 12: e0178730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Zhang CQ, Xu Y, Lu Y, Yu HX, Gu MH, Liu QQ (2011) The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 234: 541–554 [DOI] [PubMed] [Google Scholar]
- Zhang ZL, Shin M, Zou X, Huang J, Ho TH, Shen QJ (2009) A negative regulator encoded by a rice WRKY gene represses both abscisic acid and gibberellinssignaling in aleurone cells. Plant Mol Biol 70: 139–151 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Cheng S, Song Y, Huang Y, Zhou S, Liu X, Zhou DX (2015) The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell 27: 2469–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.