Abstract
The WUSCHEL-RELATED HOMEOBOX1 (WOX1) transcription factor and its homolog PRESSED FLOWER (PRS) are multifunctional regulators of leaf development that act as transcriptional repressors. These genes promote cell proliferation under certain conditions, but the related molecular mechanisms are not well understood. Here, we present a new function for WOX1 in cell proliferation. To identify the WOX1 downstream genes, we performed a microarray analysis of shoot apices of transgenic Arabidopsis thaliana lines harboring [35Sp::WOX1-glucocorticoid receptor (GR)] in which the WOX1 function was temporarily enhanced by dexamethasone. The downregulated genes were significantly enriched for the Gene Ontology term “response to auxin stimulus”, whereas the significantly upregulated genes contained auxin transport-associated PIN1 and AUX1 and the auxin response factor MP, which are involved in formation of auxin response maxima. Simultaneous treatments of synthetic auxin and dexamethasone induced the formation of green compact calli and the unorganized proliferation of cells in the hypocotyl. A microarray analysis of 35Sp::WOX1-GR plants treated with indole-3-acetic acid and dexamethasone revealed that WOX1 and auxin additively influenced their common downstream genes. Furthermore, in the presence of an auxin-transport inhibitor, cell proliferation during leaf initiation was suppressed in the prs mutant but induced in a broad region of the peripheral zone of the shoot apical meristem in the ectopic WOX1-expressing line FILp::WOX1. Thus, our results clarify the additive effect of WOX1/PRS and auxin on their common downstream genes and highlight the importance of the balance between their functions in controlling cell proliferation.
Keywords: auxin, cell proliferation, microarray, PRESSED FLOWER (PRS), WUSCHEL-RELATED HOMEOBOX1 (WOX1)
Introduction
Plant-specific WUSCHEL RELATED-HOMEOBOX (WOX) family transcription factors play fundamental roles for plant development, such as body axis-dependent patterning and the maintenance of shoot, root and vascular stem cells (van der Graaff et al. 2009). The members of this family diverged during plant evolution and are conserved (Lian et al. 2014; Nardmann et al. 2009). Among WOX family members, WOX1 and its paralog PRESSED FLOWER (PRS; also called WOX3), which act as transcriptional repressors (Ikeda et al. 2009; Lin et al. 2013; Zhang et al. 2014), play crucial roles in the leaf and floral organ development of seed plant species. WOX1 and PRS promote cell proliferation during leaf initiation and leaf lamina growth (Alvarez et al. 2016; Cho et al. 2013; Ishiwata et al. 2013; Lin et al. 2013; McHale 1993; McHale and Marcotrigiano 1998; Nakata et al. 2012; Nakata and Okada, 2013; Nardmann et al. 2004; Scanlon and Freeling 1997; Scanlon et al. 1996; Tadege et al. 2011; Vandenbussche et al. 2009; Zhuang et al. 2012). WOX1 and PRS are specifically expressed at the margins and the region between the adaxial and abaxial domains (hereafter, the middle domain) in leaf primordia and contribute to margin cell differentiation and the maintenance of adaxial–abaxial patterning (Honda et al. 2018; Ishiwata et al. 2013; Matsumoto and Okada 2001; Nakata and Okada 2012; Nakata and Okada, 2013; Nakata et al. 2012; Nardmann et al. 2004; Tadege et al. 2011; Vandenbussche et al. 2009; Yoshikawa et al. 2016; Zhang et al. 2014; Zhuang et al. 2012).
In grasses, relatively high WOX1 and PRS expression levels lead to the enlargement of leaves and stems, and increased biomass production (Ishiwata et al. 2013; Wang et al. 2017). In addition, overexpression in tobacco caused unorganized cell proliferation to form callus-like tumors (Tadege et al. 2011). However, extremely high expression levels also caused dwarfism in both grasses and eudicot species. In Arabidopsis thaliana, the overexpression of WOX1 inhibits plant growth and negatively affects shoot meristem development (Zhang et al. 2011). The ectopic high expression of WOX1 by the FIL promoter and the constitutive expression of PRS caused local abnormal outgrowths, which formed protrusions or ridges, but did not appear to globally increase the biomass (Matsumoto and Okada 2001; Nakata et al. 2012). Unorganized cell proliferation, such as calli formation in tobacco, did not occur when WOX1 was overexpressed in A. thaliana. Such context-dependent phenotypes have made it difficult to determine the roles of WOX1 and PRS in cell proliferation and have delayed the development of industrial applications.
In this study, we aimed to understand the function of WOX1 in cell proliferation in more detail. To improve our knowledge of events occurring downstream of WOX1, we examined gene expression profiles of early responses to the transient activation of WOX1 by glucocorticoid receptor (GR) and dexamethasone (DEX) in A. thaliana. The microarray analysis identified hundreds of WOX1-upregulated and downregulated genes. A Gene Ontology (GO) analysis revealed that auxin-responsive genes were significantly enriched among both the upregulated and downregulated genes. We then focused on the relationship between WOX1 and the auxin pathway and performed further microarray and pharmacological analyses. The data allows a deeper understanding of the link between WOX1 and the auxin pathway as related to cell proliferation.
Materials and methods
Plant materials and growth conditions
The A. thaliana accession Columbia-0 was used as wild type. Among the mutants and transgenic lines used in this study, prs-2, wox1-101, prs-2 wox1-101, FILp::WOX1 (Nakata et al. 2012) and DR5::GFP (Ottenschläger et al. 2003) have been previously described. The generation of 35Sp::WOX1-GR and 35Sp::GFP-GR is described below.
Surface-sterilized seeds were sown and stored at 4°C in the dark for 2–3 day. Plants were grown under continuous white fluorescent light at 22°C on solid or in liquid media. Solid and liquid media contained 0.5×Murashige and Skoog salts, 1% sucrose and 0.05% MES-KOH (w/v) pH 5.7 with or without 1.2% purified agar, respectively. For pharmacological analyses, the plants were incubated on solid or in liquid medium with or without DEX, 2,4-dichlorophenoxyacetic acid (2,4-D), indole-3-acetic acid (IAA) and 1-N-naphtylphtalamic acid (NPA).
Microarray analyses
For microarray analyses, 35Sp::WOX1-GR and 35Sp::GFP-GR were generated using the GR–DEX system that was established in Aoyama and Chua 1997. For the construction of 35Sp::WOX1-GR and 35Sp::GFP-GR, a partial DNA fragment of Rattus norvegicus GR protein was amplified by PCR and cloned into the multiple cloning site of pBlueScript SKII(+) using the restriction enzymes BamHI and KpnI. Full-length sequences of WOX1 and G3GFP (Kawakami and Watanabe 1997) were amplified by PCR and cloned into pBlueScript SKII(+) containing the GR fragment using the restriction enzymes SacII and BamHI, generating WOX1-GR and GFP-GR, respectively, DNA fragments. These were transferred separately to pDONR221 (Thermo Fisher Scientific, USA) and to pFAST-G02 by the GATEWAY reaction (Shimada et al. 2010). These constructs were introduced into Columbia-0 by vacuum infiltration using Agrobacterium tumefaciens strain ASE. The first generation of transgenic (T1) plants was screened for GFP fluorescent seeds (35Sp::WOX1-GR and 35Sp::GFP-GR) and obtained homozygous T3 lines. Primer sequences are shown in Supplemental Table S1.
For the transient application of DEX and/or IAA before harvesting, plants were grown on solid medium for 6 day and transferred to liquid medium with or without 10 µM DEX, 10 µM cycloheximide (CHX) and 20 µM IAA. DEX, CHX and IAA treatments were started 6, 6 and 3 h, respectively, before sample collection.
For the microarray analysis, RNA was extracted from shoot apices, including leaf primordia, using a Plant RNeasy Mini Kit (QIAGEN, Germany), and their quality was checked using a Bioanalyzer 2100 (Agilent, USA). For both DEX and CHX treatments of 35Sp::WOX1-GR, sampling was performed twice in two independent lines (n=4). For the analysis of 35Sp::GFP-GR and a combined application of DEX and IAA, sampling was performed from two independent lines (n=2). Microarray analyses were performed using an Arabidopsis Whole Genome Microarray 4X44K Ver. 4 and the one-color method (Agilent). The labelled-cRNA preparation, hybridization and scanning were performed by Miltenyi Biotec Inc. (Germany). The signal intensities from the raw data obtained from individual experiments were normalized by dividing them by their median values. For the identification of WOX1 downstream genes, p-values, as determined by Student’s t-test, and q-values of the false discovery rate (FDR), as determined by the Benjamini-Hochberg method, were calculated with Microsoft Excel and the R stats package (https://stat.ethz.ch/R-manual/R-devel/library/stats/html/00Index.html). GO analyses were performed with GOEAST (Zheng and Wang 2008). For the analysis of simultaneous applications of DEX and IAA, genes with a fold change of more than 1.5-fold in both of the independent 35Sp::WOX1-GR lines were collected as differentially expressed genes.
The effects of interactions between two factors can be estimated using linear modeling (Brady et al. 2015). Therefore, to investigate the contribution of the interaction on individual gene expression levels, the following general linear model was fitted to log2-transformed signal values of the given probe:
| S~µ+D+I+D:I+ε | (1) |
where S, µ, ε, D, I and D : I are log2-transformed signal values, intercept, residuals, effects of DEX, IAA, and the interaction between DEX and IAA, respectively. The t- and p-values were obtained for DEX, IAA, and DEX : IAA coefficients, and the q-values were calculated using the Benjamini-Hochberg method. The probes showing signal values less than −3 in all eight samples were eliminated to satisfy the normal distribution assumption required for the linear model.
The graphical presentations of the results were performed with Microsoft Excel, the R gplots package (https://www.rdocumentation.org/packages/gplots/versions/3.0.1), the R beeswarm package (http://www.cbs.dtu.dk/~eklund/beeswarm/) and the Python Matplotlib pyplot library (https://matplotlib.org/api/pyplot_api.html). Raw data from the microarray analyses were deposited to the Gene Expression Omnibus (GSE79647: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=evihssyaddqdfmj&acc=GSE79647).
Reverse transcriptase-quantitative PCR (RT-qPCR)
For the RT-qPCR analysis, seedlings of two independent lines were grown on a solid medium for 6 day and were treated transiently with DEX and IAA under the same conditions as for the microarray analyses. Total RNA was extracted from the aerial plant parts using a Plant RNeasy Mini Kit. cDNA was synthesized using a QuantiTect reverse transcription kit (QIAGEN). Quantitative PCR was performed on a 7500 fast Real-Time PCR Systems (Thermo Fisher Scientific) using the QuantiTect SYBR Green PCR kit (QIAGEN). Relative expression levels were analyzed by the comparative CT (ΔΔCT) method using ACT2 as an endogenous control. The experiment was performed with biological triplicates. Primer sequences are shown in Supplemental Table S1.
Observation and staining
The stereoscopic observation was performed with a LEICA MZ FLIII (Leica, Germany). For scanning electron microscopy, samples frozen in liquid nitrogen for 1 min were observed with a XL30 scanning electron microscope (FEI, USA). For the observation of the DR5::GFP pattern, leaves were stained with 50 µg ml−1 FM4-64 (Thermo Fisher Scientific) for 5 min and observed with a TCS SP8 (Leica) equipped with a white light laser and the time-gating detector HyD. For the analysis of plants treated with DEX and/or 2,4-D, hypocotyls were stained by the modified pseudo-Schiff–propidium iodide staining method (Truernit et al. 2008) with some changes (Nakata et al. 2018), and observed with a LSM710 (Zeiss, Germany). For observation with a confocal laser scanning microscope, GFP, FM4-64 and propidium iodide fluorescence were detected with the excitation/emission values of 488/495–540 nm, 488/580–625 nm and 514/566–719 nm, respectively. Obtained images were processed using ImageJ (https://imagej.nih.gov/ij/).
Results
Enhancement of the WOX1 function by the GR–DEX system
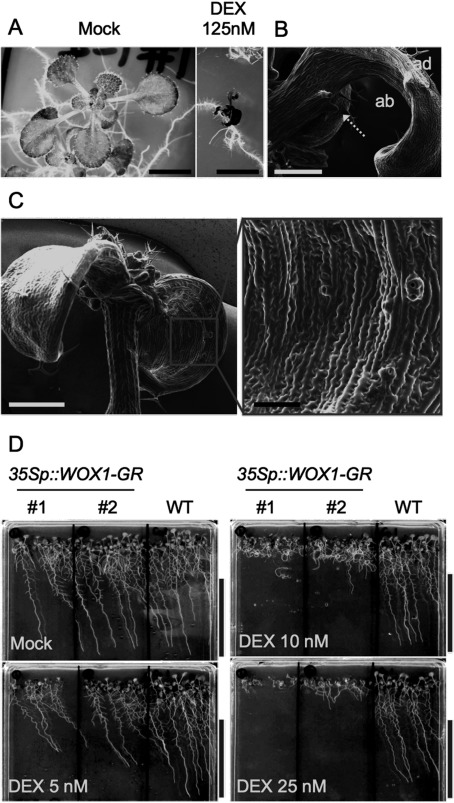
To confirm the enhancement of the WOX1 function by the GR–DEX system, we investigated the phenotypes of 35Sp::WOX1-GR transgenic lines treated with DEX. Of 24 independent T2 transformants, 13 lines showed growth defects when grown on the medium containing 125 nM DEX (example shown in Figure 1A). In these lines, the numbers of leaves decreased, and the leaves were narrow. We found adventitious protrusions on the abaxial sides of leaves in 5 of the 13 lines (Figure 1B). The leaf epidermises were covered with elongated oblong cells resembling leaf margin cells (Figure 1C). This phenotype occurs with the ectopic expression of WOX1, i.e. FILp::WOX1 (Nakata et al. 2012). Using two of these five transgenic lines, we examined the effects of lower concentrations of DEX (5, 10, 25 nM DEX). The lower the DEX concentration, the milder the phenotype (Figure 1D). The relatively weak dwarf phenotype was consistent with the phenotype of wox1-D in a previous study (Zhang et al. 2011). We also found the growth inhibition of roots in addition to shoots. No positive effect of WOX1 on biomass was found in the tested concentration range. These phenotypes indicate that the WOX1 function was strongly enhanced by the DEX treatment in these two 35Sp::WOX1-GR lines.
Figure 1. Phenotype of the DEX-treated 35Sp::WOX1-GR Arabidopsis line. (A) Phenotype of 35Sp::WOX1-GR plants treated with mock or 125 nM DEX as observed by stereomicroscopy. (B–C) Leaves of 35Sp::WOX1-GR plants as observed by scanning electron microscopy. The arrow in (B) indicates an abaxial protrusion. “ad” and “ab” in (B) indicate the adaxial and abaxial sides, respectively. Almost all epidermis cells other than stomata have changed to an elongated oblong form. (D) The dose-dependent effects of DEX on the growth of 35Sp::WOX1-GR plants 13 day after germination. Scale bars, 5 mm (A), 500 µm (B, C left), 100 µm (C right), 5 cm (D).
Transient enhancement of WOX1 function alters auxin-related gene expression levels
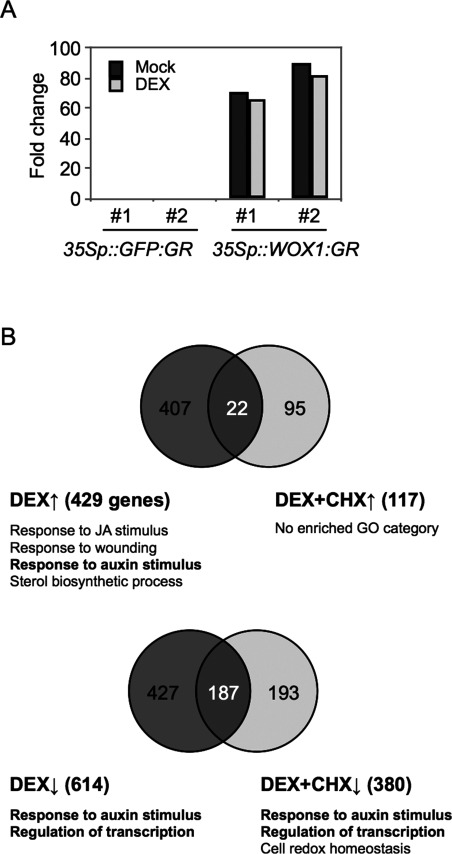
To identify WOX1-downstream genes, we performed a microarray analysis of shoot apices that included leaf primordia harvested immediately after a 5–6-h DEX treatment. WOX1 expression increased more than 60 times in these two lines (Figure 2A). We found that 429 and 614 genes were upregulated and downregulated by the DEX treatment, respectively [Figure 2B; FDR <0.05, log2 fold change (log2 FC)>1]. A GO analysis revealed the upregulated genes were enriched with GO terms, including “response to jasmonic acid” (17 genes, p-value=7.33e-6), “response to wounding” (13 genes, p-value=2.77e-3), “sterol biosynthetic process” (5 genes, p-value=6.84e-3) and “response to auxin stimulus” (16 genes, p-value=4.62e-3). The downregulated genes were enriched with other GO terms, including “response to auxin stimulus” (37 genes, p-value=3.25e-14) and “regulation of transcription, DNA dependent” (72 genes, p-value=2.03e-6) (Figure 2B).
Figure 2. Genes altered in the DEX-treated 35Sp::WOX1-GR plants with or without CHX. (A) Venn diagram illustrating the numbers of genes upregulated (upward-pointing arrow) and downregulated (downward-pointing arrow) by DEX and/or CHX. The enriched GO categories and the gene numbers are indicated beside the diagram. GO category numbers: GO:0009753 (response to jasmonic acid), GO:0009611 (response to wounding), GO:0009733 (response to auxin stimulus), GO:0016126 (sterol biosynthetic process), GO:0006355 (DNA-dependent regulation of transcription) and GO:0045454 (cell redox homeostasis). (B) A comparison between the expression level of WOX1 in the control (35Sp:GFP-GR) and 35Sp::WOX1-GR plants.
To clarify which genes have the potential to be directly affected by WOX1, we performed the same transcriptome analysis in the presence of CHX, which inhibits protein synthesis. Under these conditions, 117 and 380 genes were upregulated and downregulated by the DEX treatment, respectively (Figure 2B; FDR <0.05, log2 FC >1). Because WOX1 acts as transcriptional repressor (Lin et al. 2013), it is plausible that more genes are downregulated than upregulated. A GO analysis revealed the upregulated genes were not significantly enriched with any GO terms. In contrast, the downregulated genes were enriched with GO terms, including “response to auxin stimulus” (25 genes, p-value=7.93e-10), “regulation of transcription, DNA dependent” (48 genes, p-value=6.56e-5) and “cell redox homeostasis” (10 genes, p-value=3.84e-4). Among genes involved in “response to auxin stimulus”, 14 SAUR genes, IAA16, DFL2, AIR3 and ACS4 were downregulated regardless of the presence of CHX (Table 1). Genes involved in “regulation of transcription” contained 10 transcription factor families, including bHLH, NAC, ERF/AP2 and GRAS (Table 2). Interestingly, five abscisic acid-receptor genes were downregulated by DEX regardless of the presence of CHX (Table 3). Among adaxial- and abaxial-specific genes, ZPR1 and ETT were upregulated, while AS2 and YAB5 were significantly downregulated by the DEX treatment (Table 3). However, the changes in the expression levels of these genes, except for YAB5, were subtle in the presence of CHX (Table 3). Thus, transcriptional regulatory genes, auxin-responsive genes and abscisic acid-receptor genes were identified as potential direct targets of WOX1 repression.
Table 1. “Response to auxin stimulus” genes downregulated by WOX1 with or without CHX.
| Description | Gene name | AGI code | Fold change (D/M)*1 | p-value*2 | q-value*3 | Fold change (D/M+C)*1 | ProbeName | ||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | ||||||
| Aux/IAA | IAA16 | AT3G04730 | −1.47 | 0.06 | 0.000 | 0.019 | −1.06 | 0.04 | A_84_P23996 |
| GH3 | DFL2 | AT4G03400 | −1.10 | 0.04 | 0.000 | 0.018 | −1.86 | 0.14 | A_84_P20399 |
| SAUR | SAUR61 | AT1G29420 | −4.11 | 0.19 | 0.000 | 0.020 | −3.22 | 0.25 | A_84_P537376 |
| SAUR63 | AT1G29440 | −4.10 | 0.12 | 0.000 | 0.017 | −5.28 | 0.19 | A_84_P279980 | |
| SAUR64 | AT1G29450 | −3.17 | 0.20 | 0.001 | 0.024 | −4.25 | 0.20 | A_84_P19713 | |
| SAUR65 | AT1G29460 | −3.16 | 0.17 | 0.001 | 0.021 | −2.14 | 0.20 | A_84_P15930 | |
| SAUR68 | AT1G29490 | −3.06 | 0.15 | 0.000 | 0.020 | −1.15 | 0.09 | A_84_P843096 | |
| SAUR66 | AT1G29500 | −1.58 | 0.11 | 0.001 | 0.025 | −3.26 | 0.14 | A_84_P11207 | |
| SAUR68 | AT1G29510 | −1.69 | 0.09 | 0.000 | 0.021 | −2.67 | 0.16 | A_84_P22555 | |
| SAUR56 | AT1G76190 | −1.63 | 0.11 | 0.001 | 0.025 | −1.56 | 0.15 | A_84_P19956 | |
| SAUR59 | AT3G60690 | −1.13 | 0.11 | 0.003 | 0.035 | −1.19 | 0.10 | A_84_P11862 | |
| SAUR15 | AT4G38850 | −3.68 | 0.33 | 0.002 | 0.031 | −4.53 | 0.29 | A_84_P16734 | |
| AT5G18030 | −1.28 | 0.13 | 0.004 | 0.037 | −2.42 | 0.18 | A_84_P152798 | ||
| SAUR22 | AT5G18050 | −1.28 | 0.16 | 0.006 | 0.046 | −1.60 | 0.09 | A_84_P141269 | |
| SAUR23 | AT5G18060 | −1.39 | 0.10 | 0.001 | 0.026 | −1.56 | 0.12 | A_84_P94979 | |
| SAUR24 | AT5G18080 | −1.48 | 0.09 | 0.001 | 0.024 | −2.12 | 0.14 | A_84_P272980 | |
| Others | AIR3 | AT2G04160 | −2.63 | 0.20 | 0.001 | 0.027 | −2.51 | 0.12 | A_84_P93389 |
| ACS4 | AT2G22810 | −3.11 | 0.31 | 0.003 | 0.035 | −3.33 | 0.17 | A_84_P17344 | |
*1 Means of log2 fold changes for four experiments; D/M indicates DEX/Mock; +C indicates “with CHX”. *2 p-values as assessed by a two-sided t-test. *3 False discovery rate as assessed by the Benjamini–Hochberg method.
Table 2. “Regulation of transcription “ genes downregulated by WOX1 with or without CHX.
| Description | Gene name | AGI code | Fold change (D/M)*1 | p-value*2 | q-value*3 | Fold change (D/M+C)*1 | ProbeName | ||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | ||||||
| bZIP | bZIP3 | AT5G15830 | −3.34 | 0.44 | 0.007 | 0.048 | −4.42 | 0.29 | A_84_P15068 |
| bZIP44 | AT1G75390 | −1.46 | 0.16 | 0.004 | 0.038 | −1.30 | 0.12 | A_84_P817973 | |
| −1.48 | 0.13 | 0.002 | 0.030 | −1.58 | 0.09 | A_84_P817969 | |||
| −1.46 | 0.12 | 0.002 | 0.028 | −1.52 | 0.08 | A_84_P22038 | |||
| C2H2 | GIS | AT3G58070 | −1.85 | 0.24 | 0.007 | 0.048 | −2.10 | 0.10 | A_84_P23199 |
| ERF/AP2 | ERF106 | AT5G07580 | −3.11 | 0.24 | 0.002 | 0.027 | −2.22 | 0.16 | A_84_P821249 |
| −3.13 | 0.25 | 0.002 | 0.028 | −2.21 | 0.11 | A_84_P12054 | |||
| ERF107 | AT5G61590 | −3.28 | 0.13 | 0.000 | 0.018 | −5.80 | 0.09 | A_84_P21661 | |
| RAP2.1 | AT1G46768 | −1.67 | 0.06 | 0.000 | 0.018 | −1.13 | 0.31 | A_84_P108742 | |
| −1.70 | 0.12 | 0.001 | 0.026 | −1.56 | 0.07 | A_84_P786987 | |||
| −1.80 | 0.10 | 0.001 | 0.021 | −1.49 | 0.08 | A_84_P844167 | |||
| HB | HAT1 | AT4G17460 | −2.56 | 0.18 | 0.001 | 0.026 | −2.51 | 0.17 | A_84_P19589 |
| HAT22 | AT4G37790 | −1.51 | 0.20 | 0.008 | 0.049 | −1.36 | 0.05 | A_84_P18621 | |
| MYB | AT5G56840 | −2.10 | 0.21 | 0.003 | 0.035 | −1.49 | 0.09 | A_84_P11239 | |
| RL3 | AT4G36570 | −3.86 | 0.40 | 0.004 | 0.037 | −3.01 | 0.19 | A_84_P789861 | |
| NAC | NAC020 | AT1G54330 | −1.08 | 0.34 | 0.002 | 0.032 | −1.23 | 0.25 | A_84_P23710 |
| NAC028 | AT1G65910 | −1.23 | 0.10 | 0.000 | 0.021 | −1.22 | 0.09 | A_84_P11441 | |
| NAC003 | AT1G02220 | −2.81 | 0.06 | 0.006 | 0.044 | −2.17 | 0.05 | A_84_P799508 | |
| WRKY | WRKY7 | AT4G24240 | −1.88 | 0.09 | 0.000 | 0.020 | −1.06 | 0.03 | A_84_P10055 |
| GRAS | AT3G49950 | −1.75 | 0.20 | 0.004 | 0.040 | −1.57 | 0.03 | A_84_P13705 | |
| SCL7 | AT3G50650 | −2.40 | 0.13 | 0.001 | 0.021 | −1.37 | 0.02 | A_84_P17490 | |
| SCL15 | AT4G36710 | −1.29 | 0.17 | 0.007 | 0.047 | −1.16 | 0.01 | A_84_P13917 | |
| bHLH | AKS2 | AT1G05805 | −1.22 | 0.07 | 0.001 | 0.023 | −1.15 | 0.04 | A_84_P104016 |
| AIF4 | AT1G09250 | −1.38 | 0.13 | 0.003 | 0.034 | −1.35 | 0.14 | A_84_P820734 | |
| −1.45 | 0.12 | 0.002 | 0.028 | −1.32 | 0.10 | A_84_P23695 | |||
| bHLH030 | AT1G68810 | −1.37 | 0.06 | 0.000 | 0.019 | −2.10 | 0.12 | A_84_P19990 | |
| bHLH129 | AT2G43140 | −2.80 | 0.22 | 0.002 | 0.028 | −1.82 | 0.09 | A_84_P838362 | |
| −3.21 | 0.25 | 0.002 | 0.028 | −2.59 | 0.08 | A_84_P150068 | |||
| SPCH | AT5G53210 | −1.91 | 0.15 | 0.001 | 0.027 | −1.11 | 0.10 | A_84_P576326 | |
| PIF7 | AT5G61270 | −2.65 | 0.23 | 0.002 | 0.030 | −1.58 | 0.02 | A_84_P827848 | |
| −2.85 | 0.10 | 0.000 | 0.018 | −1.55 | 0.02 | A_84_P553165 | |||
| ZF | ZF3 | AT5G43170 | −2.15 | 0.11 | 0.000 | 0.021 | −2.25 | 0.10 | A_84_P21591 |
| −2.20 | 0.11 | 0.000 | 0.021 | −2.20 | 0.09 | A_84_P826434 | |||
*1 Means of log2 fold changes for four experiments; D/M indicates DEX/Mock; +C indicates “with CHX”. *2 p-values as assessed by a two-sided t-test. *3 False discovery rate as assessed by the Benjamini–Hochberg method.
Table 3. Other genes significantly upregulated or downregulated by WOX1 with DEX.
| Description | Gene name | AGI code | Fold change (D/M)*1 | p-value*2 | q-value*3 | Fold change (D/M+C)*1 | ProbeName | ||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | ||||||
| ABA receptor | PYR1 | AT4G17870 | −2.44 | 0.10 | 0.000 | 0.019 | −1.66 | 0.15 | A_84_P120122 |
| PYL4 | AT2G38310 | −2.74 | 0.09 | 0.000 | 0.018 | −2.62 | 0.09 | A_84_P197484 | |
| PYL5 | AT5G05440 | −1.94 | 0.15 | 0.001 | 0.027 | −1.95 | 0.08 | A_84_P146199 | |
| −1.89 | 0.14 | 0.001 | 0.027 | −1.97 | 0.11 | A_84_P817892 | |||
| −1.85 | 0.08 | 0.000 | 0.019 | −1.98 | 0.11 | A_84_P852047 | |||
| PYL6 | AT2G40330 | −4.35 | 0.25 | 0.001 | 0.022 | −3.69 | 0.11 | A_84_P174151 | |
| RCAR3 | AT5G53160 | −1.36 | 0.06 | 0.000 | 0.020 | −1.02 | 0.01 | A_84_P94069 | |
| Adaxial gene | AS2 | AT1G65620 | −1.14 | 0.08 | 0.001 | 0.025 | −0.52 | 0.09 | A_84_P15163 |
| ZPR1 | AT2G45450 | 1.26 | 0.10 | 0.001 | 0.027 | 0.41 | 0.07 | A_84_P62200 | |
| Abaxial gene | YAB5 | AT2G26580 | −1.15 | 0.06 | 0.000 | 0.020 | −0.83 | 0.06 | A_84_P516292 |
| −1.14 | 0.10 | 0.002 | 0.031 | −0.94 | 0.06 | A_84_P788325 | |||
| ETT | AT2G33860 | 1.31 | 0.06 | 0.000 | 0.019 | 0.43 | 0.13 | A_84_P18296 | |
*1 Means of log2 fold changes for four experiments; D/M indicates DEX/Mock; +C indicates “with CHX”. *2 p-values as assessed by a two-sided t-test. *3 False discovery rate as assessed by the Benjamini-Hochberg method.
Because significant numbers of genes categorized into “response to auxin stimulus” were upregulated and downregulated, we examined the expression profiles of genes involved in the auxin pathway (Table 4). Among the auxin biosynthetic genes, TAA1, YUC1, YUC4, NIT1 and NIT3 were upregulated, while TAR2, YUC2, YUC5, NIT4 and CYP83B1 were downregulated. Among transport-associated genes, AUX1, LAX1 and PIN1 were upregulated, while PIN3 and PIN7 were downregulated. The expression of the activator-type auxin response factor MP increased. The changes in all of these genes were statistically significant (Table 4). Regulation on AUX1, LAX1, PIN1 and MP appeared to be mainly indirect because changes in the expression levels of these four genes are limited to the absence of CHX (Table 4).
Table 4. Expression changes in auxin-pathway genes significantly upregulated or downregulated by WOX1 (FDR <0.05).
| Description | Gene name | AGI code | Fold change (D/M)*1 | p-value*2 | q-value*3 | Fold change (D/M+C)*1 | ProbeName | ||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | ||||||
| Biosynthesis | TAA1 | AT1G70560 | 1.03 | 0.06 | 0.001 | 0.024 | 0.10 | 0.05 | A_84_P19052 |
| TAR2 | AT4G24670 | −1.41 | 0.11 | 0.001 | 0.027 | −0.70 | 0.12 | A_84_P10057 | |
| YUC1 | AT4G32540 | 0.50 | 0.02 | 0.000 | 0.020 | 0.02 | 0.08 | A_84_P22386 | |
| YUC2 | AT4G13260 | −2.01 | 0.17 | 0.002 | 0.030 | −0.91 | 0.37 | A_84_P16644 | |
| YUC4 | AT5G11320 | 0.77 | 0.08 | 0.004 | 0.038 | −0.10 | 0.47 | A_84_P767924 | |
| 0.76 | 0.05 | 0.001 | 0.025 | −0.59 | 0.07 | A_84_P18684 | |||
| YUC5 | AT5G43890 | −3.74 | 0.15 | 0.000 | 0.019 | −3.03 | 0.23 | A_84_P23485 | |
| NIT1 | AT3G44310 | 0.71 | 0.05 | 0.001 | 0.025 | 0.22 | 0.13 | A_84_P808866 | |
| 0.64 | 0.07 | 0.004 | 0.037 | 0.15 | 0.09 | A_84_P21247 | |||
| NIT3 | AT3G44320 | 1.74 | 0.11 | 0.001 | 0.025 | 1.23 | 0.06 | A_84_P22193 | |
| NIT4 | AT5G22300 | −1.42 | 0.09 | 0.001 | 0.024 | −1.21 | 0.16 | A_84_P10196 | |
| CYP83B1 | AT4G31500 | −0.45 | 0.04 | 0.002 | 0.028 | −0.13 | 0.06 | A_84_P16706 | |
| Transport | AUX1 | AT2G38120 | 0.74 | 0.09 | 0.005 | 0.041 | 0.12 | 0.05 | A_84_P107652 |
| LAX1 | AT5G01240 | 0.75 | 0.03 | 0.000 | 0.019 | 0.28 | 0.10 | A_84_P859674 | |
| 0.94 | 0.10 | 0.003 | 0.036 | 0.45 | 0.05 | A_84_P20541 | |||
| PIN1 | AT1G73590 | 1.03 | 0.08 | 0.001 | 0.027 | 0.10 | 0.05 | A_84_P11175 | |
| PIN3 | AT1G70940 | −0.50 | 0.06 | 0.005 | 0.041 | −0.19 | 0.06 | A_84_P13326 | |
| PIN7 | AT1G23080 | −1.08 | 0.08 | 0.001 | 0.027 | −0.69 | 0.25 | A_84_P67534 | |
| Activator ARF | MP | AT1G19850 | 1.42 | 0.04 | 0.000 | 0.017 | 0.23 | 0.06 | A_84_P21913 |
| Repressor ARF | ARF1 | AT1G59750 | 0.16 | 0.00 | 0.000 | 0.017 | 0.01 | 0.01 | A_84_P18037 |
| ARF6 | AT1G30330 | −0.29 | 0.03 | 0.005 | 0.041 | −0.37 | 0.06 | A_84_P21873 | |
| ARF16 | AT4G30080 | 0.36 | 0.01 | 0.000 | 0.017 | 0.29 | 0.06 | A_84_P101526 | |
*1 Means of log2 fold changes for four experiments; D/M indicates DEX/Mock; +C indicates “with CHX”. *2 p-values as assessed by a two-sided t-test. *3 False discovery rate as assessed by the Benjamini–Hochberg method.
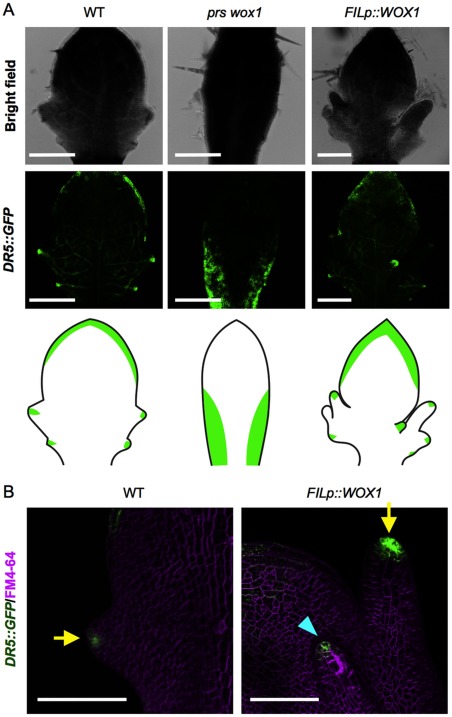
To investigate the importance of WOX1 expression in the regulation of auxin transport-associated genes and MP in developing leaves, auxin response patterns in the wild type, prs wox1 and plants harboring the FILp::WOX1 construct were examined by confocal laser scanning microscopy using the DR5::GFP marker. In the wild-type leaves, spots on the marginal tip showed the maximum fluorescence, but in the leaves of prs wox1, the spot pattern disappeared, and the auxin response was detected in a broad region of the leaf base (Figure 3A). In the leaves of the FILp::WOX1 plants, there was no apparent change in the global pattern of auxin at the margins (Figure 3A), but the spots with the maximum fluorescence were also found at the tips of the ectopically developed abaxial protrusions (Figure 3B; an arrow head). The DR5::GFP pattern in FILp::WOX1 leaves was consistent with the PIN1p::PIN1-GFP pattern in the same genotype (Alvarez et al. 2016). Thus, WOX1 may affect auxin patterning at the leaf margins probably through the regulation of auxin transport.
Figure 3. DR5::GFP fluorescent pattern in prs wox1 and the FILp::WOX1 plants. (A) Confocal laser scanning microscope images of developing leaves. The bottom columns show the leaf shape and the GFP pattern in the individual lines. (B) The GFP spots on a growing tip. The arrows indicate the marginal tip, and the arrow head indicates an abnormal abaxial tip. Scale bars, 200 µm (A), 100 µm (B).
In conclusion, the microarray data suggested that WOX1 had bifurcated effects on the auxin pathway: the downregulation of auxin responsive genes and the upregulation of auxin transport-associated genes and MP.
The combination of WOX1 and auxin induces unorganized cell proliferation
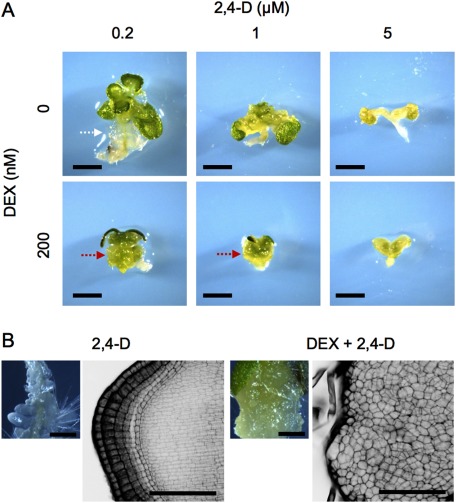
To clarify whether the complicated relationship between auxin and WOX1 is responsible for the phenotypic variation in cell proliferation, we analyzed the phenotypes of plants treated with a synthetic auxin analog, 2,4-D, together with enhanced WOX1 activity. In the liquid medium, a relatively low 2,4-D concentration alone resulted in the formation of white and friable tissues on hypocotyls (Figure 4A, a white arrow), while green compact calli were formed on hypocotyls by the simultaneous application of DEX with a relatively low 2,4-D concentration (Figure 4A). The higher the concentration of 2,4-D was, the slower the cell proliferation was, regardless of the presence of DEX (Figure 4A). Green compact calli on hypocotyls were also formed when plants were grown on the solid medium containing both DEX and 2,4-D (Figure 4B). In the green compact calli, unorganized cell proliferation, with random division planes, was observed (Figure 4B). This phenotype was clearly different from the root tissue-like layered structures formed on hypocotyls treated with 2,4-D alone (Figure 4B). Thus, the combination of WOX1 and auxin stimulated unorganized cell proliferation but the balance between the functions of the two factors is important in determining the rate of cell proliferation and the characteristics of the cells.
Figure 4. Effects of a simultaneous application of DEX and 2,4-D on Arabidopsis seedling phenotypes. (A) The 35Sp::WOX1-GR plants grown on liquid media containing various concentrations of DEX and/or 2,4-D for three weeks. The relatively low concentration of 2,4-D led to white friable tissue formation (white arrow) in the absence of DEX but green compact calli in the presence of DEX (red arrows). (B) The 35Sp::WOX1-GR plants grown on solid media containing 125 nM DEX and/or 1 µM 2,4-D for 12 day. The left panels are stereomicroscopic images, and the right ones are black-and-white reversal images observed with confocal laser scanning microscope after modified pseudo-Schiff–propidium iodide staining. Scale bars, 2 mm (A), 0.5 mm (B, left panels), 100 µm (B, right panels).
WOX1 and auxin additively influence the expression of their common downstream genes
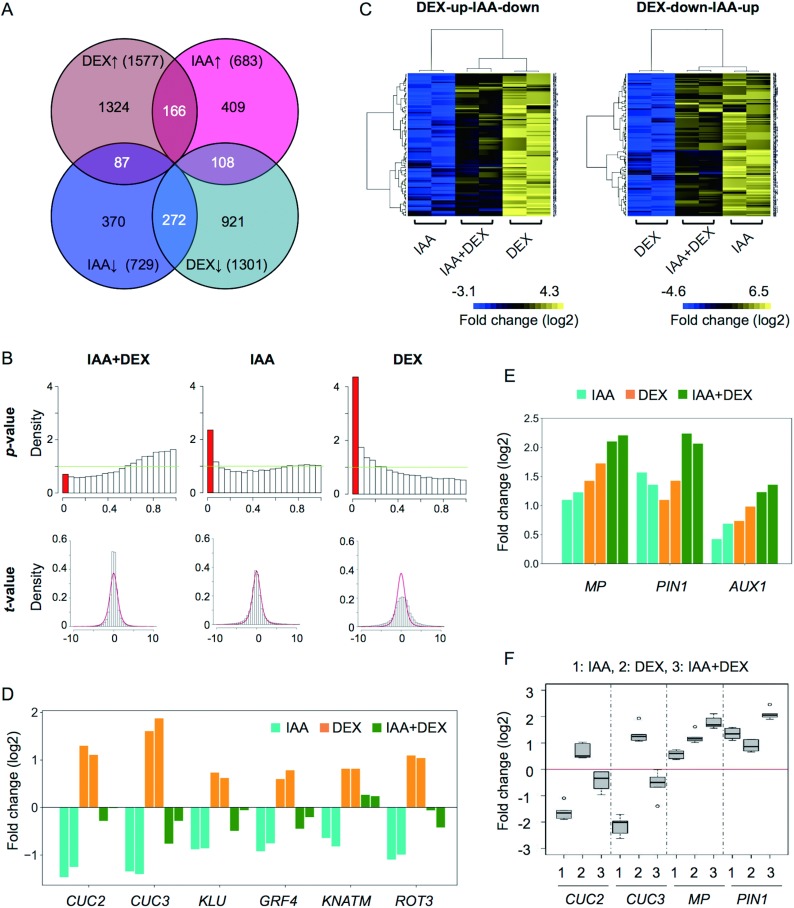
To investigate the effects of simultaneous auxin and DEX treatments on gene expression profiles, we performed microarray analyses (Figure 5). As shown in Figure 5A, 166 genes were upregulated commonly by DEX and by IAA (DEX-up-IAA-up; Supplemental Table S2), 87 genes were upregulated by DEX but downregulated by IAA (DEX-up-IAA-down; Supplemental Table S3), 108 genes were downregulated by DEX but upregulated by IAA (DEX-down-IAA-up; Supplemental Table S4), and 272 genes were downregulated by both DEX and IAA (DEX-down-IAA-down; Supplemental Table S5). MP, PIN1 and AUX1 were found in DEX-up-IAA-up. DEX-up-IAA-down included TAA1 and regulators of shoot apical meristem/leaf development, including CUC2, CUC3 and KLU. The Aux/IAA genes, the SAUR genes, ARGOS and CKX1, which is a member of the CKX genes involved in cytokinin degradation, were included in DEX-down-IAA-up, while CKX7 and ARR7, which is a negative regulator of cytokinin signaling, and the abscisic acid-receptor genes (PYR1, PYL4-6 and RCAR3) were found in DEX-down-IAA-down (Supplemental Table S2).
Figure 5. Genes altered in the 35Sp::WOX1-GR treated with DEX and IAA. (A) Venn diagram illustrating the numbers of genes upregulated (upward-pointing arrow) and downregulated (downward-pointing arrow) with or without DEX and IAA. (B) Histograms showing the distributions of calculated t-values and p-values for the linear model’s analysis of the interaction between DEX and IAA. See Materials and Methods for details. (C) Heat maps of fold changes in the expression levels of DEX-up-IAA-down and DEX-down-IAA-up genes compared with the mock treatment. Expression levels of DEX-up-IAA-down genes treated with IAA+DEX were greater than with IAA alone and lower than with DEX alone (left), while those of DEX-down-IAA-up genes treated with IAA+DEX were lower than with IAA alone and greater than with DEX alone (right). Data from different probes for a single gene are shown in different rows in the heat map. (D) Log2FCs in the expression levels of the DEX-up-IAA-down genes involved in shoot meristem and leaf development in comparison with the mock treatment. Two bars of the same color represent the results of two independent lines. (E) Log2FCs in the expression levels of MP, PIN1 and AUX1 in comparison with the mock treatment. Two bars of the same color represent the results of two independent lines. (F) Boxplots showing the results of the RT-qPCR analysis of CUC2, CUC3, MP and PIN1. Data were normalized by the ACT2 expression level.
To examine the global interaction between WOX1 and IAA, the expression levels of both genes under each condition were fitted to the statistical model. The density of genes potentially affected by a single treatment of DEX or IAA (p<0.05) was much higher than the average, while the density affected depending on the interaction between DEX and IAA was lower (Figure 5B). Based on these results, even though >600 genes were affected by both DEX and IAA, the effects of DEX and IAA are likely to be additive at the transcriptome level. In particular, in both the DEX-up-IAA-down and DEX-down-IAA-up categories, changes in gene expression by the simultaneous DEX plus IAA treatment (IAA+DEX) were intermediate between those of single DEX and IAA treatments (Figure 5C). As with the global trend, changes in the expression levels of regulators of shoot apical meristem/leaf development that were categorized into DEX-up-IAA-down in the presence of IAA+DEX were offset between the changes after treatments with IAA and DEX alone (Figure 5D). MP, PIN1 and AUX1 were further upregulated by IAA+DEX (Figure 5E). The expression changes of CUC2, CUC3, MP and PIN1 were confirmed by RT-qPCR (Figure 5F). These expression analyses indicate that WOX1 and auxin additively control the expression of several genes responsible for leaf development.
The WOX1/PRS is responsible for leaf initiation when normal auxin patterning is disrupted
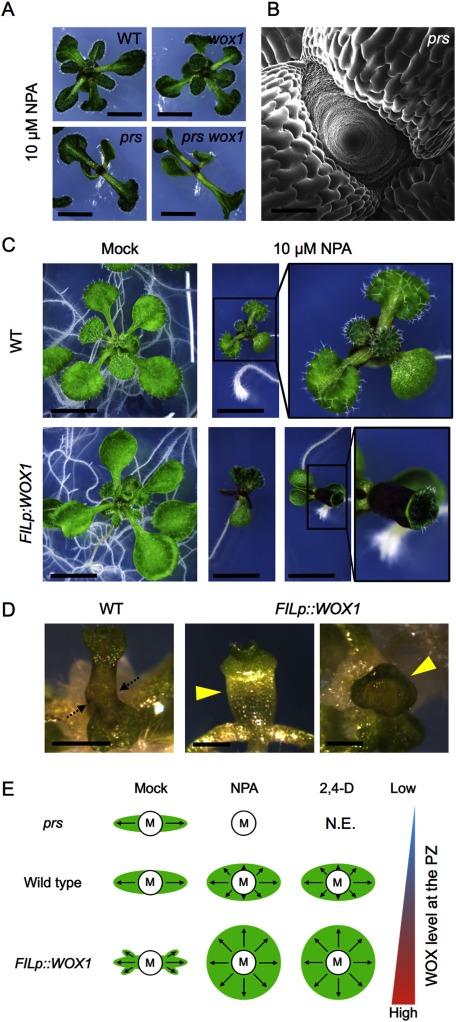
The phenotype of 35Sp::WOX1-GR simultaneously treated with DEX and 2,4-D suggested that the balance between the auxin and WOX1 functions is important for unorganized cell proliferation. To clarify whether the function of WOX1/PRS in leaf development is also closely related to that of the auxin pathway, we disrupted the auxin pattern in seedlings of loss-of-function mutants by treatment with NPA, an inhibitor of polar auxin transport (Thomson et al. 1973) and analyzed the phenotype. In A. thaliana, leaf initiation phenotypes of all prs, wox1 and prs wox1 mutants had no, or only slight, phenotypic differences from the wild type in the absence of NPA (Nakata et al. 2012). NPA caused a severe inhibition of leaf initiation in the prs mutant background unlike the wild type (Figure 6A, B). The wox1 mutation did not have any effect (Figure 6A). The phenotype of prs indicated that PRS plays an important role in leaf initiation when the normal auxin pattern is disturbed. For further characterization, we examined the phenotype of the FILp::WOX1 plants treated with NPA. These seedlings frequently formed fused leaves or cup-shaped leaves in the presence of 10 µM NPA (Figure 6C). Fused or cup-shaped leaves were not formed in the wild-type seedling treated with 10 µM NPA (Figure 6C). The similar cup-shaped leaf phenotype was also observed in the 2,4-D-treated FILp::WOX1 plants (Figure 6D). The leaf primordia are initiated by cell proliferation along periclinal division planes from the peripheral region of the shoot apical meristem. Thus, the cup-shaped leaf phenotype is considered to be a result from cell proliferation that occurred in the whole peripheral zone of the shoot apical meristem (Figure 6E). These findings indicate that PRS and WOX1 are involved in cell proliferation in coordination with the auxin pathway during leaf initiation.
Figure 6. Roles of WOX1/PRS and auxin in leaf development. (A) Phenotype of the aerial part of wild type, prs, wox1 and prs wox1 in the presence of 10 µM NPA. Leaf initiation defects in 10 µM NPA-treated prs and prs wox1 plants. (B) Short pin-like phenotype of the 10 µM NPA-treated prs shoot apex. (C) Phenotype of wild type and the FILp::WOX1 plants treated with 10 µM NPA. (D) Leaf phenotype of 1 µM 2,4-D-treated FILp::WOX1 plants. The arrows indicate the fusion of petioles between two leaves observed in wild type. The arrow heads indicate cup-shaped leaves observed in the FILp::WOX1 plants. Plants shown in (A–D) were grown on a solid medium. (E) Summary of the leaf phenotypes of mock-, NPA- or 2,4-D-treated plants. Schematic diagram of the vicinity of the shoot apical meristem observed from above. “M” represents the center of the meristem, “PZ” represents the peripheral zone, and “N.E”. represents “not examined”. Arrows indicate the degree and direction of growth. Scale bars, 4 mm (A), 200 µm (B), 1 cm (C), 1 mm (D).
Discussion
This study provides a new insight into the relationship between the function of auxin and WOX1 at the gene expression level. In addition, it also suggested that the relationship is important for cell proliferation.
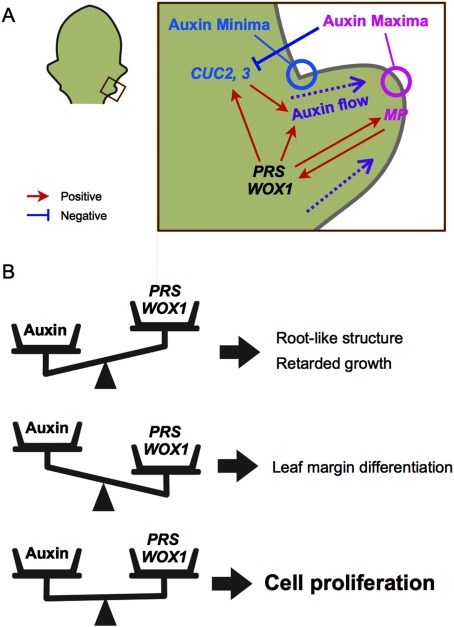
Here, we identified hundreds of genes affected by the activation of WOX1 using a microarray analysis of the shoot apices of 35Sp::WOX1-GR plants after DEX treatments. Both the upregulated and downregulated genes were enriched for the GO term “response to auxin stimulus”, which suggests that the effects of WOX1 on the auxin pathway are complex. In experiments with CHX, WOX1 indirectly activated and directly repressed auxin responsive genes. The effects of DEX and IAA were globally additive, and changes in the expression levels of both the DEX-up-IAA-down and the DEX-down-IAA-up genes did not occur when plants were treated with IAA+DEX. In addition, MP, PIN1, AUX1 and LAX1 were upregulated by DEX. The expression sites of MP, PIN1, AUX1 and LAX1 overlap with those of WOX1 and PRS at the leaf margins (Kasprzewska et al. 2015; Qi et al. 2014; Scarpella et al. 2006). A role for WOX1 in auxin transport is indicated by the observation that the DR5::GFP signal is broadly distributed in the leaf base of prs wox1 mutant and the auxin maxima were ectopically formed at the abaxial protrusions of the FILp::WOX1 plants leaves. Based on these findings, we concluded that WOX1 positively regulates auxin response and transport but suppresses the expression of some auxin downstream genes independent of auxin. Furthermore, the results inferred that WOX1 stimulates only particular events downstream of auxin, while preventing others. The microarray analysis using a mutant of the WOX1 homolog of Medicago truncatula (stenofolia) revealed the decreased expression of the PIN1 homolog and the increased expression of auxin-responsive SAUR homologous genes (Tadege et al. 2011), which is consistent with our results. However, in A. thaliana prs wox1, the DR5::GFP signal remained in the basal region, which was different from the extremely depressed signal of the DR5 marker in the tobacco mutant of WOX1 homolog (lam1) (Tadege et al. 2011). Therefore, the role of WOX1 in the auxin pathway suggested by our study may be basically, but not fully, conserved among eudicot plants.
Recent studies have revealed that the expression levels of WOX1 and PRS in leaf primordia are induced by auxin (Caggiano et al. 2017; Shi et al. 2017) and that the induction is directly mediated by MP (Guan et al. 2017). These findings, together with our analysis, indicate that there is a positive feedback between auxin and WOX1/PRS (Figure 7A). WOX1 and PRS act as the middle-domain regulators (Nakata et al. 2012) and mutually regulate the adaxial and abaxial genes in A. thaliana, Petunia x hybrida, M. truncatula and rice (Guan et al. 2017; Honda et al. 2018; Nakata and Okada 2012; 2013; Nakata et al. 2012; Vandenbussche et al. 2009; Zhang et al. 2014; Table 3). However, the adaxial/abaxial pattern contributes to dynamic auxin pattern formation (Caggiano et al. 2017; Izhaki and Bowman 2007). The adaxial/abaxial genes affect auxin responsive genes (Huang et al. 2014; Merelo et al. 2013), while auxin is important for the adaxial/abaxial patterning (Qi et al. 2014). Besides adaxial/abaxial associated genes, our data show that the expression levels of CUC2 and CUC3 were oppositely regulated by auxin and WOX1 (Figure 7A). CUC2, which functions as a dimer with CUC3 (Rubio-Somoza et al. 2014), modulates the growth patterning along the medial-lateral axis (Hasson et al. 2011; Kawamura et al. 2010; Nikovics et al. 2006) and cooperatively works with PIN1, AUX1 and LAX1 (Bilsborough et al. 2011; Kasprzewska et al. 2015). Thus, WOX1/PRS, MP-auxin, adaxial/abaxial genes and CUC2/CUC3 establish a complicated regulatory network, and this network organizes leaf developmental events based on the adaxial–abaxial and the medial–lateral axes.
Figure 7. Models of the functions of WOX1/PRS and auxin. (A) The spatio-temporal model of the fuctions of WOX1/PRS and auxin in leaf development of wild type. (B) The model of the role of the balance between WOX1/PRS and auxin in controlling cell proliferation.
In addition to the role in leaf patterning, the combination of WOX1 and auxin contributed to cell proliferation under certain conditions. The simultaneous application of DEX and 2,4-D to plants harboring 35Sp::WOX1-GR construct induced unorganized cell proliferation. However, the application of DEX without 2,4-D caused dwarfism and the differentiation of the leaf-margin cells, while a high concentration of 2,4-D retarded cell proliferation, even in the presence of DEX. Based on these results, we propose a model in which the balance between the activity levels of auxin and WOX1 is a key factor in stimulating unorganized cell proliferation (Figure 7B). In cells where the level of auxin is low and WOX1/PRS is highly expressed, cell differentiation is triggered, resulting in the suppression of cell proliferation. Also, in a tissue where auxin is abundant but the expression level of WOX1/PRS is not high enough, root tissues are formed or cell proliferation is slowed or another tissue identities such as root are acquired. Cell proliferation occurs only when the amount of auxin and the expression level of WOX1 are well balanced. In this model, the additive effect of WOX1 and auxin at the gene expression level potentially explains why the balance between them is important for cell proliferation because only a subset of genes are strongly upregulated and downregulated whereas modulation of other WOX1 and auxin downstream genes are offset.
This model can also explain the phenotypes of NPA-treated prs and NPA-treated the FILp::WOX1 plants. The phenotype of the NPA-treated prs mutant indicated that PRS has the most important role in leaf initiation among the redundantly acting WOX family members. In vegetative shoots, PRS is expressed in the peripheral zone of the shoot apical meristem (Caggiano et al. 2017) as well as in early leaf primordia, in contrast with the faint or non-expression of WOX1 (Caggiano et al. 2017; Matsumoto and Okada 2001; Nakata et al. 2012; Yu et al. 2017). Because NPA causes the broad distribution of the auxin response throughout the peripheral zone (Caggiano et al. 2017), the reduced total activity level of the WOX genes in the prs mutant might disrupt the balance between the activity levels of WOX and auxin, resulting in retarded cell proliferation during leaf initiation. However, because the FIL promoter induces relatively strong expression levels throughout the emerging primordia (Tameshige et al. 2013), in the FILp::WOX1 plants, WOX1 can be expressed in a broad region of the peripheral zone. In addition, the NPA treatment on the FILp::WOX1 plants caused the retention of auxin, which may have generated a proper balance between WOX1 and auxin throughout the peripheral zone, resulting in the formation of cup-shaped or fused leaves. 2,4-D is a substrate of AUX1 but is not moved by an efflux carrier (Delbarre et al. 1996; Yang et al. 2006). Therefore, 2,4-D could trigger the cup-shaped phenotype similar to that induced by NPA. Although this model is consistent with the phenomenon found in this study, our observations were limited to mature leaves. Consequently, it is necessary to further verify this model by conducting detailed analyses of the leaf primordial at the cellular level.
In this study, unorganized cell proliferation was caused by the combined actions of WOX1 and auxin but how these actions can be used to improve biomass is unclear. Because the balance between the actions of WOX1 and auxin could be important, changes in the amount of auxin that are dependent on leaf development may make it difficult to continue triggering cell proliferation using the combination of WOX1 and auxin. In addition, the constitutive expression of WOX1 strongly inhibited root growth in A. thaliana, which might be a negative factor that interferes with biomass improvement. Continuous cell proliferation in developing leaves can be caused by the prolonged expression of PRS and WOX1 that results from the suppression of the TCP/NGA function (Alvarez et al. 2016). This report suggests that under particular cellular environments the increased expression levels of PRS and WOX1 could result in biomass improvement, even in A. thaliana. Studies of grass species have suggested that the cytokinin pathway is responsible for WOX1-triggered biomass improvement (Wang et al. 2017). Here, both WOX1 and auxin downregulated CKX7 and ARR7, which negatively affect the cytokinin pathway (Kölmer et al. 2014; To et al. 2007). Therefore, WOX1 may improve the biomass together with auxin through the activation of the cytokinin pathway. Interestingly, our microarray analysis also implied that both WOX1 and auxin suppress the abscisic acid pathway, which negatively affects leaf growth (Tardieu et al. 2010), through the downregulation of abscisic-acid receptors. To elucidate the regulatory mechanism of cell proliferation, it is necessary to understand the balance between auxin and WOX1, and the relationships with the cytokinin and with the abscisic-acid pathways. Fully understanding how the balance of WOX1/PRS and auxin controls cell proliferation could lead to their manipulation, which could improve biomass and allow for the modification of plant morphology in the future.
Acknowledgements
We thank Prof. M. Nishimura (NIBB) for the use of the Agilent Bioanalyzer, Dr. T. L. Shimada (Chiba Univ.) and Prof. I. Hara-Nishimura (Konan Univ.) for providing the pFAST-G02 vector, Prof. K. Palme (Univ. of Freiburg) for providing DR5::GFP seeds, the Arabidopsis Biological Resource Center and Dr S. Tabata (Kazusa DNA Res. Inst.) for providing the mutant seeds, Dr. K. Tatematsu (NIBB), Dr. G Horiguchi (Rikkyo Univ.) and Ms. Y. Tsuzuki (NIBB) for technical supports, Dr. R. Tsugeki (Kyoto Univ.), Dr. K. Toyokura (Kobe Univ.), Dr. M. Ikeuchi (RIKEN) and other members of Okada’s lab for fruitful discussions, and Dr. M. Sato (Keio Univ.) and Dr. A.J. Nagano (Ryukoku Univ.) for helpful comments. We thank Lesley Benyon, PhD, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry for Education, Culture, Sports, Science, and Technology of Japan (Grant number 19060004 to K.O.), a Grant-in-Aid for Creative Scientific Research from the Japan Society for the Promotion of Science (Grant number 19GS0315 to K.O.) and a fellowship from the Japan Society for the Promotion of Science (Grant numbers 20-2203 and 21-1024 to M.N. and T.T., respectively). This work was also supported by the Model Plant Research Facility, NIBB Bioresource Center. The microscopic work was partly supported by Live Imaging Center (WPI-ITbM) in Nagoya University and the Japan Advanced Plant Science Network.
Abbreviations
- WOX1
WUSCHEL-RELATED HOMEOBOX1
- PRS
PRESSED FLOWER
- GR
glucocorticoid receptor
- DEX
dexamethasone
- CHX
cycloheximide
- GO
Gene Ontology
- 2,4-D
2,4-dichlorophenoxyacetic acid
- IAA
indole-3-acetic acid
- NPA
1-N-naphtylphtalamic acid
Supplementary Data
References
- Alvarez JP, Furumizu C, Efroni I, Eshed Y, Bowman JL (2016) Active suppression of a leaf meristem orchestrates determinate leaf growth. eLife 5: e15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua NH (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M (2011) Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci USA 108: 3424–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Burow M, Busch W, Carlborg Ö, Denby KJ, Glazebrook J, Hamilton ES, Harmer SL, Haswell ES, Maloof JN, et al. (2015) Reassess the t Test: Interact with All Your Data via ANOVA. Plant Cell 27: 2088–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggiano MP, Yu X, Bhatia N, Larsson A, Ram H, Ohno CK, Sappl P, Meyerowitz EM, Jönsson H, Heisler MG (2017) Cell type boundaries organize plant development. eLife 6: e27421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Yoo SC, Zhang H, Pandeya D, Koh HJ, Hwang JY, Kim GT, Paek NC (2013) The rice narrow leaf2 and narrow leaf3 loci encode WUSCHEL-related homeobox 3A (OsWOX3A) and function in leaf, spikelet, tiller and lateral root development. New Phytol 198: 1071–1084 [DOI] [PubMed] [Google Scholar]
- Delbarre A, Muller P, Imhoff V, Guern J (1996) Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198: 532–541 [DOI] [PubMed] [Google Scholar]
- Guan C, Wu B, Yu T, Wang Q, Krogan NT, Liu X, Jiao Y (2017) Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis. Curr Biol 27: 2940–2950.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson A, Plessis A, Blein T, Adroher B, Grigg S, Tsiantis M, Boudaoud A, Damerval C, Laufs P (2011) Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell 23: 54–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda E, Yew C, Yoshikawa T, Sato Y, Hibara K, Itoh J (2018) LEAF LATERAL SYMMETRY1, a member of the WUSCHEL-RELATED HOMEOBOX3 gene family, regulates lateral organ development differentially from other paralogs, NARROW LEAF2 and NARROW LEAF3 in rice. Plant Cell Physiol 59: 376–391 [DOI] [PubMed] [Google Scholar]
- Huang T, Harrar Y, Lin C, Reinhart B, Newell NR, Talavera-Rauh F, Hokin SA, Barton MK, Kerstetter RA (2014) Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis-element and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIPIII factors. Plant Cell 26: 246–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M (2009) Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21: 3493–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwata A, Ozawa M, Nagasaki H, Kato M, Noda Y, Yamaguchi T, Nosaka M, Shimizu-Sato S, Nagasaki A, Maekawa M, et al. (2013) Two WUSCHEL-related homeobox genes, narrow leaf2 and narrow leaf3, control leaf width in rice. Plant Cell Physiol 54: 779–792 [DOI] [PubMed] [Google Scholar]
- Izhaki A, Bowman JL (2007) KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19: 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzewska A, Carter R, Swarup R, Bennett M, Monk N, Hobbs JK, Fleming A (2015) Auxin influx importers modulate serration along the leaf margin. Plant J 83: 705–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami S, Watanabe Y (1997) Use of green fluorescent protein as a molecular marker tag of protein movement in vivo. Plant Biotechnol 14: 127–130 [Google Scholar]
- Kawamura E, Horiguchi G, Tsukaya H (2010) Mechanisms of leaf tooth formation in Arabidopsis. Plant J 62: 429–441 [DOI] [PubMed] [Google Scholar]
- Köllmer I, Novák O, Strnad M, Schmülling T, Werner T (2014) Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J 78: 359–371 [DOI] [PubMed] [Google Scholar]
- Lian G, Ding Z, Wang Q, Zhang D, Xu J (2014) Origins and evolution of WUSCHEL-related homeobox protein family in plant kingdom. Sci World J 2014: 534140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Niu L, McHale NA, Ohme-Takagi M, Mysore KS, Tadege M (2013) Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants. Proc Natl Acad Sci USA 110: 366–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto N, Okada K (2001) A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev 15: 3355–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale NA (1993) LAM-1 and FAT genes control development of the leaf blade in Nicotiana sylvestris. Plant Cell 5: 1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale NA, Marcotrigiano M (1998) LAM1 is required for dorsoventrality and lateral growth of the leaf blade in Nicotiana. Development 125: 4235–4243 [DOI] [PubMed] [Google Scholar]
- Merelo P, Xie Y, Brand L, Ott F, Weigel D, Bowman JL, Heisler MG, Wenkel S (2013) Genome-wide identification of KANADI1 target genes. PLoS One 8: e77341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Matsumoto N, Tsugeki R, Rikirsch E, Laux T, Okada K (2012) Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 24: 519–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Okada K (2012) The three-domain model: A new model for the early development of leaves in Arabidopsis thaliana. Plant Signal Behav 7: 1423–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Okada K (2013) The Leaf adaxial–abaxial boundary and lamina growth. Plants 2: 174–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata MT, Sato M, Wakazaki M, Sato N, Kojima K, Sekine A, Nakamura S, Shikanai T, Toyooka K, Tsukaya H, et al. (2018) Plastid translation is essential for lateral root stem cell patterning in Arabidopsis thaliana. Biol Open 7: bio028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardmann J, Ji J, Werr W, Scanlon MJ (2004) The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 131: 2827–2839 [DOI] [PubMed] [Google Scholar]
- Nardmann J, Reisewitz P, Werr W (2009) Discrete shoot and root stem cell-promoting WUS/WOX5 functions are an evolutionary innovation of angiosperms. Mol Biol Evol 26: 1745–1755 [DOI] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P (2006) The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschläger I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K (2003) Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci USA 100: 2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Wang Y, Yu T, Cunha A, Wu B, Vernoux T, Meyerowitz E, Jiao Y (2014) Auxin depletion from leaf primordia contributes to organ patterning. Proc Natl Acad Sci USA 111: 18769–18774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I, Zhou CM, Confraria A, Martinho C, von Born P, Baena-Gonzalez E, Wang JW, Weigel D (2014) Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr Biol 24: 2714–2719 [DOI] [PubMed] [Google Scholar]
- Scanlon MJ, Freeling M (1997) Clonal sectors reveal that a specific meristematic domain is not utilized in the maize mutant narrow sheath. Dev Biol 182: 52–66 [DOI] [PubMed] [Google Scholar]
- Scanlon MJ, Schneeberger RG, Freeling M (1996) The maize mutant narrow sheath fails to establish leaf margin identity in a meristematic domain. Development 122: 1683–1691 [DOI] [PubMed] [Google Scholar]
- Scarpella E, Marcos D, Friml J, Berleth T (2006) Control of leaf vascular patterning by polar auxin transport. Genes Dev 20: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Dong J, Xue J, Wang H, Yang Z, Jiao Y, Xu L, Huang H (2017) Model for the role of auxin polar transport in patterning of the leaf adaxial–abaxial axis. Plant J 92: 469–480 [DOI] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Hara-Nishimura I (2010) A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J 61: 519–528 [DOI] [PubMed] [Google Scholar]
- Tadege M, Lin H, Bedair M, Berbel A, Wen J, Rojas CM, Niu L, Tang Y, Sumner L, Ratet P, et al. (2011) STENOFOLIA regulates blade outgrowth and leaf vascular patterning in Medicago truncatula and Nicotiana sylvestris. Plant Cell 23: 2125–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameshige T, Fujita H, Watanabe K, Toyokura K, Kondo M, Tatematsu K, Matsumoto N, Tsugeki R, Kawaguchi M, Nishimura M, et al. (2013) Pattern dynamics in adaxial–abaxial specific gene expression are modulated by a plastid retrograde signal during Arabidopsis thaliana leaf development. PLoS Genet 9: e1003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu F, Parent B, Simonneau T (2010) Control of leaf growth by abscisic acid: Hydraulic or non-hydraulic processes? Plant Cell Environ 33: 636–647 [DOI] [PubMed] [Google Scholar]
- Thomson KS, Hertel R, Müller S, Tavares JE (1973) 1-N-naphthylphthalamic acid and 2,3,5-triiodobenzoic acid : In-vitro binding to particulate cell fractions and action on auxin transport in corn coleoptiles. Planta 109: 337–352 [DOI] [PubMed] [Google Scholar]
- To JP, Deruère J, Maxwell BB, Morris VF, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ (2007) Cytokinin regulates type-A Arabidopsis Response Regulator activity and protein stability via two-component phosphorelay. Plant Cell 19: 3901–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J, Palauqui JC (2008) High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M, Horstman A, Zethof J, Koes R, Rijpkema AS, Gerats T (2009) Differential recruitment of WOX transcription factors for lateral development and organ fusion in Petunia and Arabidopsis. Plant Cell 21: 2269–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Laux T, Rensing SA (2009) The WUS homeobox-containing (WOX) protein family. Genome Biol 10: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Niu L, Fu C, Meng Y, Sang D, Yin P, Wu J, Tang Y, Lu T, Wang ZY, et al. (2017) Overexpression of the WOX gene STENOFOLIA improves biomass yield and sugar release in transgenic grasses and display altered cytokinin homeostasis. PLoS Genet 13: e1006649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E (2006) High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol 16: 1123–1127 [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Tanaka SY, Masumoto Y, Nobori N, Ishii H, Hibara K, Itoh J, Tanisaka T, Taketa S (2016) Barley NARROW LEAFED DWARF1 encoding a WUSCHEL-RELATED HOMEOBOX 3 (WOX3) regulates the marginal development of lateral organs. Breed Sci 66: 416–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Guan C, Wang J, Sajjad M, Ma L, Jiao Y (2017) Dynamic patterns of gene expression during leaf initiation. J Genet Genomics 44: 599–601 [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang Y, Li G, Tang Y, Kramer EM, Tadege M (2014) STENOFOLIA recruits TOPLESS to repress ASYMMETRIC LEAVES2 at the leaf margin and promote leaf blade outgrowth in Medicago truncatula. Plant Cell 26: 650–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu R, Qin G, Chen Z, Gu H, Qu LJ (2011) Over-expression of WOX1 leads to defects in meristem development and polyamine homeostasis in Arabidopsis. J Integr Plant Biol 53: 493–506 [DOI] [PubMed] [Google Scholar]
- Zheng Q, Wang XJ (2008) GOEAST: A web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res 36(suppl_2): W358-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang LL, Ambrose M, Rameau C, Weng L, Yang J, Hu XH, Luo D, Li X (2012) LATHYROIDES, encoding a WUSCHEL-related Homeobox1 transcription factor, controls organ lateral growth, and regulates tendril and dorsal petal identities in garden pea (Pisum sativum L.). Mol Plant 5: 1333–1345 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.