Abstract
Abscisic acid (ABA) plays an important role in plant growth, development, and stress responses. ABA regulates many aspects of plant growth and development, including seed maturation, dormancy, germination, the transition from vegetative to reproductive growth, leaf senescence and responses to environmental stresses, such as drought, high salinity and cold. It is also known that mitogen-activated protein kinase (MAPK) cascades function in ABA signaling. Recently, we and another group have identified the ABA-inducible MAP3Ks MAP3K17 and MAP3K18 as the upstream MAP3Ks of MKK3, implicating the MAP3K17/18-MKK3-MPK1/2/7/14 cascade in ABA signaling. It has also been reported that overexpression of MAP3K18 in Arabidopsis causes an early leaf senescence phenotype, ABA hypersensitive stomata closing, and drought tolerance. In this study, we generated transgenic plants overexpressing MAP3K17 (35S:MAP3K17) and its kinase-inactive form (35S:MAP3K17KN). The bolting of 35S:MAP3K17 was earlier than WT, and the fresh weights of the seedlings were smaller, whereas 35S:MAP3K17KN showed the opposite phenotype. These results indicate that the transition from vegetative to reproductive growth can be regulated by overexpression of MAP3K17 and its kinase-inactive form. Moreover, 35S:MAP3K17 showed lower sensitivity to ABA during post-germinated growth, whereas 35S:MAP3K17 KN showed the opposite phenotype, suggesting the negative roles of MAP3K17 in the response to ABA. Our work provides the possibility to regulate plant growth and development by the genetic manipulation of ABA-induced MAPK cascades, leading to improved crop growth and productivity.
Keywords: ABA response, Arabidopsis, floral transition, MAP3K, post-germinated growth
As sessile organisms, plant growth is limited by many aspects of external stress, such as cold, heat, drought, and biotic attacks. To survive them, plants have developed various signal transduction pathways to modulate cellular responses to environmental changes. Abscisic acid (ABA) is one of the major plant hormones that play important roles in plant growth and stress responses. ABA regulates many aspects of plant growth and development, including seed maturation, dormancy, germination, the transition from vegetative to reproductive growth, leaf senescence and responses to environmental stresses, such as drought, high salinity and cold (Finkelstein 2013). ABA signaling comprises the cellular events of ABA perception and subsequent downstream pathways that regulate ABA responses. Various signaling components, for example, second messengers, including Ca2+ and reactive oxygen species (ROS), SNF1-related protein kinase 2 (SnRK2), protein phosphatase 2C (PP2C) pathway and G-protein, have been identified in ABA signaling (Mori et al. 2006; Munemasa et al. 2013; Wang et al. 2001). Mitogen-activated protein kinase (MAPK) cascades have also been implicated in this signaling pathway (Liu et al. 2012).
The MAPK cascade is a highly conserved signaling system in eukaryotic cells that converts signals generated from receptors/sensors into cellular responses. In plants, MAPK pathways have been implicated in the responses to various biotic and abiotic stresses, plant hormones, cell division and developmental processes (MAPK Group 2002; Nakagami et al. 2005; Takahashi et al. 2004). A MAPK cascade is composed of three classes of enzymes: MAPK, MAPK kinase (MAPKK) and MAPKK kinase (MAP3K). In the genome of the model plant Arabidopsis thaliana, there are at least 80 MAP3K, 10 MAPKK (MKK1–MKK10) and 20 MAPK (MPK1–MPK20) genes (Colcombet and Hirt 2008). Several MAPK cascades have been identified in stress and developmental signal transduction pathways (Nakagami et al. 2005; Takahashi et al. 2004). We have previously reported that the MEKK1-MKK1-MPK4 cascade is stimulated following wounding stress (Hadiarto et al. 2006; Matsuoka et al. 2002). Other studies have also indicated that these cascades are involved in several other stress signaling pathways: the MEKK1-MKK2-MPK4/MPK6 cascade in salt and cold stress signaling (Teige et al. 2004) or the MEKK1-MKK4/MKK5-MPK3/MPK6 cascade following pathogen infection (Asai et al. 2002).
Regarding the involvement of the MAPK cascade in ABA signaling, several works have been reported as follows. MKK1-MPK6 regulates the ABA-dependent expression of CAT1 and H2O2 production (Xing et al. 2008). MPK9/12 positively regulates stomatal closure via ROS-mediated ABA signaling. MPK4/6 are activated by ABA (Ichimura et al. 2000), and MKK3-MPK1/2 mediates ABA signaling and is involved in salt stress tolerance (Hwa et al. 2008). Recently, we and another group have identified MAP3K17 and MAP3K18 as the upstream MAP3Ks of MKK3, implicating the MAP3K17/18-MKK3-MPK1/2/7/14 cascade in ABA signaling (Danquah et al. 2015; Matsuoka et al. 2015). Moreover, it has been shown that MAP3K18 directly interacts with ABI1 protein phosphatase, which composes the core signaling module of ABA and is regulated by the ubiquitin-proteasome pathway (Mitula et al. 2015). Overexpression of MAP3K18 in Arabidopsis causes an early leaf senescence phenotype (Matsuoka et al. 2015), ABA hypersensitive stomata closing (Mitula et al. 2015), and drought tolerance (Li et al. 2017). The transcripts of MAP3K17 and MAP3K18 are induced by ABA (Menges et al. 2008), and these genes seem to have redundant functions in ABA signaling. In this study, we generated transgenic Arabidopsis overexpressing MAP3K17 and kinase-inactive MAP3K17 (MAP3K17KN) to analyze the function of MAP3K17 in ABA signaling. We analyzed their growth characteristics and the response to ABA in post-germinated growth.
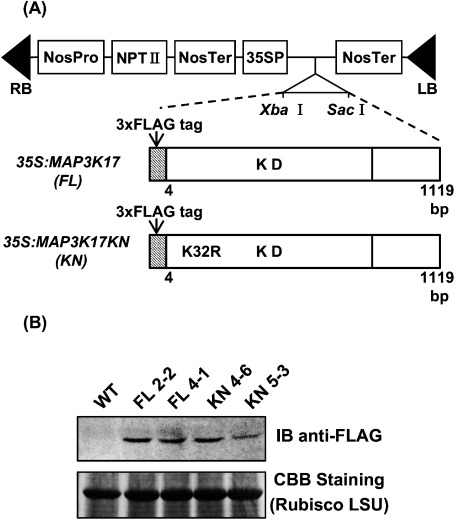
Molecular cloning of the full-length cDNA for MAP3K17 and production of the transgenic plants overexpressing MAP3K17 and the kinase-inactive form of MAP3K17 (MAP3K17 KN) were carried out as follows. Arabidopsis thaliana (Col-0) seeds were surface-sterilized with 70% (v/v) ethanol for 3 min, followed by a solution of NaClO (1% w/v) containing Triton X-100 (0.1% v/v) for 7 min. The seeds were subsequently washed five times with sterile water, plated onto Gamborg’s B5 agar (0.8% w/v) medium and incubated for two days at 4°C before germination at 22°C, and the seedlings were grown at 22°C under continuous light conditions. Total RNA was extracted from two-week-old Arabidopsis plants using an RNeasy Plant Mini Kit (Qiagen) and treated with DNaseI (Invitrogen) to remove residual DNA contamination. The cDNA was synthesized from 0.5 µg of Arabidopsis total RNA using a PrimeScript 1st strand cDNA Synthesis Kit (TAKARA). The cDNA for Arabidopsis MAPKKK17 was isolated using RT-PCR with the forward primer 5′-GAA AGA ATT CAT GGA ATG GAC TAG AGG AAG-3′ and the reverse primer 5′-GTT TCT CGA GTT ACA ATT CCC CCA CCA ATA-3′. PCR was performed with KOD-Plus- Neo DNA polymerase (TOYOBO) at 98°C for 2 min, followed by 30 cycles of 94°C for 10 s, 50°C for 30 s and 68°C for 1 min. The PCR product was cloned into a cloning vector, pBluescript II SK (−). The plasmid clones were verified using DNA sequencing. The kinase negative mutation of MAP3K17 (K32R), designated MAP3K17 KN, was created using a Quick-Change site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions and verified by DNA sequencing. The cDNA for MAP3K17 and MAP3K17 KN genes without the start codon were translationally fused to the coding sequence of the 3xFLAG-tag in the N-terminus (Figure 1A). The 3xFLAG-tag fused MAP3K17 and the 3xFLAG-tag fused MAP3K17 KN, designated 3xFLAG-MAP3K17 and 3xFLAG-MAP3K17 KN, respectively, were inserted between the CaMV 35S promoter and the NOS terminator of the plant expression vector pBI121 (Clontech). The resulting constructs were introduced into Agrobacterium tumefaciens (strain C58) by triparental mating with Escherichia coli (strain DH5α) containing pRK2013 (Wise et al. 2006) and transferred into wild-type Arabidopsis (Columbia ecotype) by the vacuum infiltration method (Bechtold et al. 1998). The transgenic plants were germinated on 0.5× Murashige and Skoog medium with 20 µg ml−1 kanamycin. Eight and seven independent lines of the transgenic plant overexpressing 3xFLAG-MAP3K17 and 3xFLAG-MAP3K17 KN, respectively, were generated. All transgenic lines used in this study were T4 homozygous plants. Two-week-old seedlings of WT and transgenic plants overexpressing 3xFLAG-MAP3K17 and 3xFLAG-MAP3K17 KN were ground in liquid nitrogen and then thawed in an extraction buffer (100 mM Tris-HCl (pH 8.0), 1 mM EDTA, 1% Triton X-100, 150 mM NaCl, 1 mM PMSF, 1 µg ml−1 leupeptin, 2 mM DTT, 1 mM sodium vanadate, 25 mM sodium fluoride and 50 mM β-glycerophosphate). After centrifugation, the supernatants were used for immunoblot analysis. The aliquots of the crude extracts from the seedlings of the WT and transgenic plants overexpressing 3xFLAG-MAP3K17 and 3xFLAG-MAP3K17 KN were resolved using SDS-PAGE, and immunoblot analysis was performed as previously described (Matsuoka et al. 2002). An anti-FLAG-tag antibody was used as the primary antibody. After extensive washing of the membrane with TBS-T buffer, an alkaline phosphatase-conjugated anti-mouse secondary antibody (Promega) was employed, and the color reaction was conducted using 5-bromo-4-chloro-3-indolyl-phosphate and nitro-blue tetrazolium as substrates. Each immunoblot analysis was repeated at least three times, and the results from one representative experiment are shown. Equal amounts of the sample were resolved on SDS-PAGE and stained with CBB for a loading control. The immunoblot results showed that both 3xFLAG-MAP3K17 and 3xFLAG-MAP3K17 KN proteins were expressed in each transgenic plant (Figure 1B). We used both lines of transgenic plants overexpressing 3xFLAG-MAP3K17 (FL 2-2 and FL 4-1) and 3xFLAG-MAP3K17 KN (KN4-6 and KN 5-3) for further analysis.
Figure 1. Overexpression of MAP3K17 and its kinase-inactive mutant in Arabidopsis. (A) Schematic representation of the constructs for the overexpression of 3xFLAG-MAP3K17 (FL) and 3x-FLAG MAP3K17KN (K32R, KN). The 3xFLAG-tag was translationally fused to the N-terminus of the coding sequence, without an initiation codon, of MAP3K17 and MAP3K17 KN and then inserted between the CaMV 35S promoter (35SP) and the NOS terminator (NOS Ter) of the plant expression vector pBI121 (Clontech) using XbaI and SacI restriction enzyme sites. RB: right border, LB: left border, NOS Pro: NOS promoter, NPTII: neomycin phosphotransferase II gene, NOS Ter: NOS terminate, KD: kinase domain (B) Detection of the 3xFLAG-MAP3K17 and 3xFLAG-MAP3K17KN proteins. Total protein extracts from WT and each transgenic plant were resolved by SDS-PAGE. Immunoblot analysis was conducted using the anti-Flag tag antibody. Equal amounts of the samples were resolved by SDS-PAGE and stained with CBB, and the large subunit (LSU) of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) is shown as the loading control for equal protein amounts in the WT and transgenic plants.
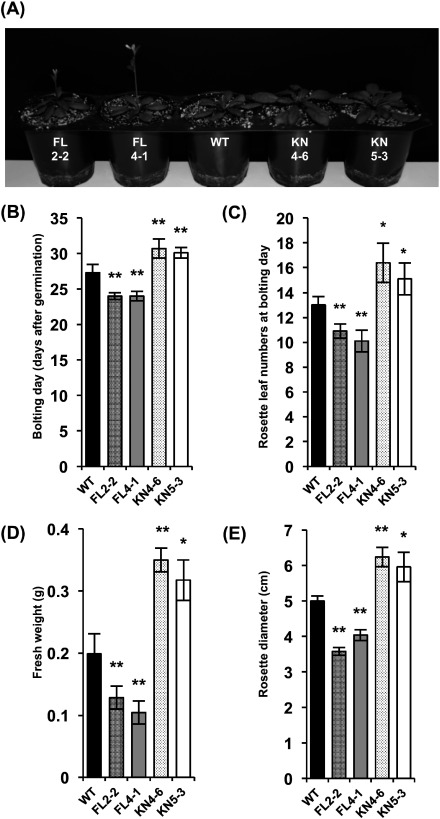
To analyze the effects of overexpressing 3xFLAG-MAP3K17 and 3xFLAG-MAP3K17 KN on plant growth, WT and both transgenic plants were germinated on soil and grown at 22°C under continuous light. Bolting time was monitored daily and calculated as the number of days from sowing to the first elongation of the floral stem at 1 mm height. Rosette leaf numbers were counted at bolting day. Representative plants (28 days after germination) are shown in Figure 2A. The bolting of both FL lines preceded that of WT by approximately 3 days (Figure 2B). On the other hand, the bolting of both KN lines was approximately 3 days later than that of WT. Similarly, the numbers of rosette leaf at bolting day were smaller in both FL lines and bigger in both KN lines (Figure 2C). To evaluate plant growth, the shoots were harvested at 4 weeks after germination and immediately measured the fresh weights and the diameter of rosette leaves. The fresh weights and the size (rosette diameter) of both FL plants were significantly lower than those of WT plants at 4 weeks after germination, on the other hand, those of both KN plants were clearly greater to WT (Figure 2D, E).
Figure 2. Growth characteristics of transgenic Arabidopsis plants overexpressing MAP3K17 and MAP3K17KN. WT and the transgenic plants were germinated on soil and grown at 22°C under continuous light conditions. (A) Representative (28-day-old) image of WT and transgenic plants. Bolting days (B), rosette leaf numbers at bolting day (C), fresh weights at 4 weeks (D), and rosette diameters at 4 weeks (E) of WT and transgenic plants. Bolting time was monitored daily and determined as the number of days from sowing to the first elongation of the floral stem at 1 mm height. All results are presented as the means. The bars indicate the standard errors from ten (B, C) and five (D, E) replicates. Asterisks indicate significant differences (Student’s t-test, * p<0.05, ** p<0.01) between WT and each transgenic line.
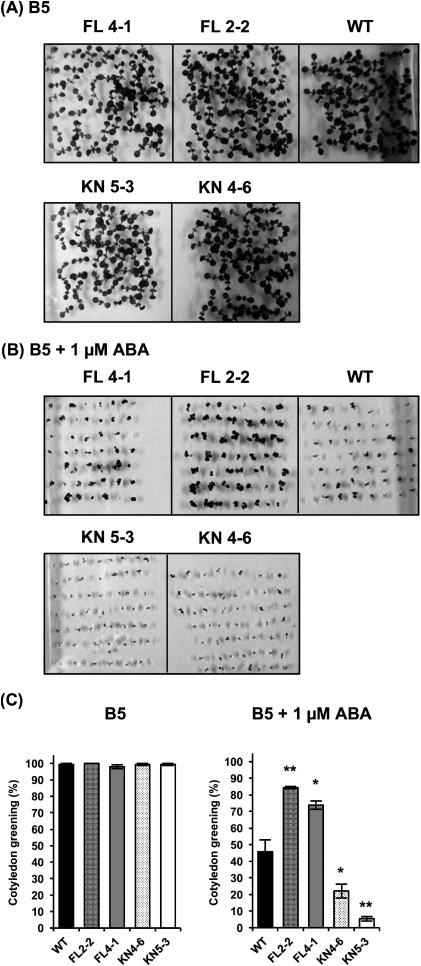
Next, we analyzed the ABA-related response of transgenic Arabidopsis plants overexpressing MAP3K17 and MAP3K17KN. The sterilized seeds of WT and transgenic Arabidopsis plants overexpressing MAP3K17 and MAP3K17KN were sown in Gamborg’s B5 medium supplemented with or without 1 µm ABA. The cotyledon greening was recorded at 7 days after transfer to 22°C. Every experiment was repeated three times (50 seeds for each repeat). Representative pictures are shown in Figure 3A (Gamborg’s B5 medium) and Figure 3B (Gamborg’s B5 medium plus 1 µm ABA). On Gamborg’s B5 medium plates, all plants germinated and grew without significant differences among them (Figure 3C). In the presence of ABA, the cotyledon greening rates of FL 2-2 and FL 4-1 were 84.4±0.7% and 73.9±2.5%, respectively, which was significantly higher than that of WT (45.8±7.2%). The cotyledon greening rates of KN 4-6 and KN 5-3 were 22.1±4.2% and 5.4±1.4%, respectively, indicating a sensitive phenotype of both KN plants to ABA (Figure 3C).
Figure 3. Effect of ABA on the post-germinated growth of WT and transgenic Arabidopsis plants overexpressing MAP3K17 and MAP3K17KN. The seeds of WT and transgenic Arabidopsis plants overexpressing MAP3K17 (FL 2-2, FL 4-1) and MAP3K17KN (KN 4-6, KN 5-3) were sown onto Gamborg’s B5 medium plates with or without 1 µM ABA. Images of 7-day-old germination on B5 medium plates (A) and B5 medium plates containing 1 µM ABA (B) are shown. (C) The cotyledon greening rates were scored at 7 days after cultivation. All results are presented as the means. The bars indicate the standard errors from three independent experiments (50 seeds for each repeat). Asterisks indicate significant differences (Student’s t-test, * p<0.05, ** p<0.01) between WT and each transgenic line.
In this study, we generated transgenic plants overexpressing ABA-inducible MAP3K, MAP3K17 (35S:MAP3K17) and its kinase-inactive form (35S:MAP3K17KN). The bolting of 35S:MAP3K17 was earlier than WT, and the fresh weights of the seedlings were smaller, whereas 35S:MAP3K17KN showed the opposite phenotype (Figure 2). These results indicate that the transition from vegetative to reproductive growth can be regulated by overexpression of MAP3K17 and its kinase-inactive form. Floral transition is one of the most important decisions in the plant life cycles and four regulatory pathways, the photoperiod, vernalization, autonomous pathways, and gibberellic acid, have been identified in Arabidopsis (Mouradov et al. 2012). The inhibitory role of ABA in regulating the floral transition was initially proposed based on the early-flowering phenotype of ABA-deficient mutant (Martinez-Zapater et al. 1994). It has been also reported that the inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis (Wang et al. 2013). 35S:MAP3K17 showed an early bolting phenotype and a lower sensitivity to ABA in cotyledon greening (Figure 3), suggesting the negative roles of MAP3K17 in the response to ABA. Transcriptome and proteome analysis of 35S:MAP3K17 and 35S:MAP3K17KN may lead to the identification of the downstream signaling pathways of the MAP3K17/18-MKK3-MPK1/2/7/14 cascade and the regulatory components of floral transition. As described above, ABA regulates many aspects of plant growth, development, and responses to environmental stresses. (Finkelstein 2013). It has been reported that several MAP3K genes were upregulated by ABA treatment (Menges et al. 2008). Previously, we reported that one of the Arabidopsis MAPKKK genes, MAP3Kδ4 (At4g23050), was induced and activated by ABA treatment (Shitamichi et al. 2013). Overexpression of MAP3Kδ4 in Arabidopsis enhanced tolerance to high salinity and showed vigorous growth (Sasayama et al. 2011). Recently, it has been reported that transgenic tobacco overexpressing cotton GhMKK3 enhanced drought tolerance (Wang et al. 2016), and two recessive mutations in MKK3 orthologs have been identified in both wheat and barley using QTL mapping for seed dormancy (Nakamura et al. 2016; Torada et al. 2016). These results suggest that ABA-induced MAP3Ks, including MAP3K17, and the components of their downstream pathways are targets of genetic modification for regulating plant growth and enhancing stress tolerance.
Acknowledgements
This research was supported by JSPS KAKENHI Grant Numbers JP19770031, JP17K08196 (to D.M.).
Abbreviations
- ABA
abscisic acid
- MAPK
mitogen-activated protein kinase
- MAPKK
MAPK kinase
- MAP3K
MAPKK kinase
References
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Bögre L, Ligterink W, Meskiene I, Barker PJ, Heberle-Bors E, Huskisson NS, Hirt H (1997) Wounding induces the rapid and transient activation of a specific MAP kinase pathway. Plant Cell 9: 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J, Hirt H (2008) Arabidopsis MAPKs: A complex signaling network involved in multiple biological processes. Biochem J 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Danquah A, de Zélicourt A, Boudsocq M, Neubauer J, Frei Dit Frey N, Leonhardt N, Pateyron S, Gwinner F, Tamby JP, Ortiz-Masia D, et al. (2015) Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J 82: 232–244 [DOI] [PubMed] [Google Scholar]
- Dóczi R, Brader G, Pettkó-Szandtner A, Rajh I, Djamei A, Pitzschke A, Teige M, Hirt H (2007) The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 19: 3266–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R (2013) Abscisic acid synthesis and response. Arabidopsis Book 11: e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadiarto T, Nanmori T, Matsuoka D, Iwasaki T, Sato K, Fukami Y, Azuma T, Yasuda T (2006) Activation of Arabidopsis MAPK kinase kinase (AtMEKK1) and induction of AtMEKK1-AtMEK1 pathway by wounding. Planta 223: 708–713 [DOI] [PubMed] [Google Scholar]
- Hwa CM, Yang XC (2008) The AtMKK3 pathway mediates ABA and salt signaling in Arabidopsis. Acta Physiol Plant 30: 277–286 [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2000) Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J 24: 655–665 [DOI] [PubMed] [Google Scholar]
- Jammes F, Song C, Shin D, Munemasa S, Takeda K, Gu D, Cho D, Lee S, Giordo R, Sritubtim S, et al. (2009) MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc Natl Acad Sci USA 106: 20520–20525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Cai H, Liu P, Wang C, Gao H, Wu C, Yan K, Zhang S, Huang J, Zheng C (2017) Arabidopsis MAPKKK18 positively regulates drought stress resistance via downstream MAPKK3. Biochem Biophys Res Commun 484: 292–297 [DOI] [PubMed] [Google Scholar]
- Liu Y (2012) Roles of mitogen-activated protein kinase cascades in ABA signaling. Plant Cell Rep 31: 1–12 [DOI] [PubMed] [Google Scholar]
- MAPK Group (2002) Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater JM, Coupland G, Dean C, Koornneef M (1994) The transition to flowering in Arabidopsis In: EM Meyerowitz, CR Somerville (eds) Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, pp 403–433
- Matsuoka D, Nanmori T, Sato K, Fukami Y, Kikkawa U, Yasuda T (2002) Activation of AtMEK1, an Arabidopsis mitogen-activated protein kinase kinase, in vitro and in vivo: Analysis of active mutants expressed in E. coli and generation of the active form in stress response in seedlings. Plant J 29: 637–647 [DOI] [PubMed] [Google Scholar]
- Matsuoka D, Yasufuku T, Furuya T, Nanmori T (2015) An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol Biol 87: 565–575 [DOI] [PubMed] [Google Scholar]
- Menges M, Dóczi R, Okrész L, Morandini P, Mizzi L, Soloviev M, Murray JA, Bögre L (2008) Comprehensive gene expression atlas for the Arabidopsis MAP kinase signalling pathways. New Phytol 179: 643–662 [DOI] [PubMed] [Google Scholar]
- Mitula F, Tajdel M, Cieśla A, Kasprowicz-Maluśki A, Kulik A, Babula-Skowrońska D, Michalak M, Dobrowolska G, Sadowski J, Ludwików A (2015) Arabidopsis ABA-activated kinase MAPKKK18 is regulated by protein phosphatase 2C ABI1 and the ubiquitin-proteasome pathway. Plant Cell Physiol 56: 2351–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Ichimura K, Shinozaki K (1997) Environmental stress response in plants: The role of mitogen-activated protein kinases. Trends Biotechnol 15: 15–19 [DOI] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, et al. (2006) CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol 4: e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: Interacting pathway as a basis for diversity. Plant Cell 14(Suppl): S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Muroyama D, Nagahashi H, Nakamura Y, Mori IC, Murata Y (2013) Regulation of reactive oxygen species-mediated abscisic acid signaling in guard cells and drought tolerance by glutathione. Front Plant Sci 4: 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAP kinase pathways in plant stress signaling. Trends Plant Sci 10: 339–346 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Pourkheirandish M, Morishige H, Kubo Y, Nakamura M, Ichimura K, Seo S, Kanamori H, Wu J, Ando T, et al. (2016) Mitogen-activated Protein Kinase Kinase 3 regulates seed dormancy in barley. Curr Biol 26: 775–781 [DOI] [PubMed] [Google Scholar]
- Sasayama D, Matsuoka D, Oka M, Shitamichi N, Furuya T, Azuma T, Itoh K, Nanmori T (2011) MAP3Kδ4, an Arabidopsis Raf-like MAP3K, regulates plant growth and shoot branching. Plant Biotechnol 28: 463–470 [Google Scholar]
- Shitamichi N, Matsuoka D, Sasayama D, Furuya T, Nanmori T (2013) Over-expression of MAP3Kδ4, an ABA-inducible Raf-like MAP3K that confers salt tolerance in Arabidopsis. Plant Biotechnol 30: 111–118 [Google Scholar]
- Takahashi Y, Soyano T, Sasabe M, Machida Y (2004) A MAP kinase cascade that controls plant cytokinesis. J Biochem 136: 127–132 [DOI] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Eulgem T, Dóczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15: 141–152 [DOI] [PubMed] [Google Scholar]
- Torada A, Koike M, Ogawa T, Takenouchi Y, Tadamura K, Wu J, Matsumoto T, Kawaura K, Ogihara Y (2016) A causal gene for seed dormancy on wheat chromosome 4A encodes a MAP kinase kinase. Curr Biol 26: 782–787 [DOI] [PubMed] [Google Scholar]
- Wang C, Lu W, He X, Wang F, Zhou Y, Guo X, Guo X (2016) The cotton mitogen-activated protein kinase kinase 3 functions in drought tolerance by regulating stomatal responses and root growth. Plant Cell Physiol 57: 1629–1642 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li L, Ye T, Lu Y, Chen X, Wu Y (2013) The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J Exp Bot 64: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Ullah H, Jones AM, Assmann SM (2001) G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292: 2070–2072 [DOI] [PubMed] [Google Scholar]
- Wise AA, Liu Z, Binns AN (2006) Three methods for the introduction of foreign DNA into Agrobacterium. Methods Mol Biol 343: 43–53 [DOI] [PubMed] [Google Scholar]
- Xing Y, Jia W, Zhang J (2008) AtMKK1 mediates ABA‐induced CAT1 expression and H2O2 production via AtMPK6‐coupled signaling in Arabidopsis. Plant J 54: 440–451 [DOI] [PubMed] [Google Scholar]