Abstract
The repertoire of redox-active enzymes produced by the marine fungus Peniophora sp. CBMAI 1063, a laccase hyper-producer strain, was characterized by omics analyses. The genome revealed 309 Carbohydrate-Active Enzymes (CAZymes) genes, including 48 predicted genes related to the modification and degradation of lignin, whith 303 being transcribed under cultivation in optimized saline conditions for laccase production. The secretome confirmed that the fungus can produce a versatile ligninolytic enzyme cocktail. It secretes 56 CAZymes, including 11 oxidative enzymes classified as members of auxiliary activity families (AAs), comprising two laccases, Pnh_Lac1 and Pnh_Lac2, the first is the major secretory protein of the fungi. The Pnh_Lac1-mediator system was able to promote the depolymerization of lignin fragments and polymeric lignin removal from pretreated sugarcane bagasse, confirming viability of this fungus enzymatic system for lignocellulose-based bioproducts applications.
Subject terms: Enzymes, Industrial microbiology
Introduction
Marine-derived fungal species have been considered attractive producers of ligninolytic, hemicellulolytic and other industrial enzymes, presenting different properties compared to terrestrial enzymes, such as high salt tolerance and thermostability1. The white-rot basidiomycete Peniophora sp. CBMAI 1063, isolated from the seawater sponge Amphimedon viridis, has been reported as a producer of oxidative enzymes under saline and non-saline conditions, and recently a transcriptome analysis revealed several sequences encoding for putative laccases2,3.
Fungi from the Basidiomycota phylum are considered the most efficient lignin degraders, and according to their lifestyle and ability to degrade polymeric constituents, they can be classified as white-rot or brown-rot fungi4. White-rot fungi simultaneously attack lignin, hemicellulose and cellulose, the main components of the plant cell wall, and their genomes generally have more genes encoding for oxidative enzymes when compared to brown-rot and other groups of basidiomycetes5,6. Lignin degradation/modification in white-rot fungi is generally performed via the action of enzymes such as laccases and peroxidases while in brown-rot fungi it is driven by Fenton reactions (Fe2+ + H2O2 → Fe3+ + HO• + OH− and Fe3+ + H2O2 → Fe2+ + HOO• + H+). Laccase and peroxidases genes are abundant in white-rot fungi genomes and reduced or absent in brown-rot fungi5,7.
The degradation and modification of aromatic compounds by white-rot fungi involves several enzymes classified as Auxiliary Activity (AA) families in the CAZy database7. The AA1 family contains laccases, which can be classified in three subfamilies AA1_1, AA1_2 and AA1_3. The subfamily AA1_1 has members of the ‘true’ laccases EC 1.10.3.2 or the blue copper oxidases, which are able to oxidize a wide range of aromatic compounds, including lignin, with an oxygen molecule as the final electron acceptor8. The AA2 family includes lignin peroxidases, manganese peroxidases and versatile peroxidases, which all use hydrogen peroxide as a cofactor for lignin degradation7. Moreover, enzymes from families AA4 (vanillyl-alcohol oxidases), AA5 (glyoxal oxidases/alcohol oxidases) and AA6 (1,4-benzoquinone reductases) are also correlated with the oxidation or reduction of phenolic compounds derived from lignin9,10. Other AA families are also reported to potentially drive lignin degradation and modification through Fenton reactions, such as AA3, AA7 and AA8, since the majority of their members can generate H2O2 as a by-product7,11.
In this report, we unveiled the repertoire of genes encoding for ligninolytic enzymes from the genome of Peniophora sp. CBMAI 1063, allied with the transcriptome and secretome analyses of the fungus growth in an optimized cultivation media for laccase production, which was formulated without any complex lignocellulosic component. Although the omics analyses were focused on the Peniophora sp. CBMAI 1063 oxidative enzyme system, cellulose and hemicellulose-degrading enzymes were identified from the secretome analysis as well. Furthermore, the major laccase secreted by Peniophora sp. CBMAI 1063 was identified by mass spectrometry. Structural characteristics of the protein and its potential application to promote lignin modification and degradation were explored.
Material and Methods
Microorganism
The marine-derived fungus Peniophora sp. was isolated from the Brazilian sponge A. viridis, collected from a coral reef in São Sebastião, São Paulo State, Brazil12. The strain was deposited at the Brazilian Collection of Microorganisms from the Environment and Industry—CBMAI, under accession number CBMAI 1063.
Fungal growth
Solid cultivations of Peniophora sp. CBMAI 1063 were carried out in Petri dishes containing malt extract (20 g L−1) and agar-agar (15 g L−1) at 28 °C. After 7 days of growth, six agar-plugs measuring approximately 0.9 cm in diameter were inoculated in non-baffled 125 mL Erlenmeyer flasks containing 50 mL of an optimized medium which includes yeast extract (2.0 g L−1), peptone extract from casein (2.7 g L−1), malt extract powder (1.4 g L−1) and glucose (2.7 g L−1). The reagents were dissolved in distilled water with 65% (v/v) of adapted artificial seawater, containing: MgCl2 (10.83 g L−1), CaCl2 (1.51 g L−1), SrCl2 (0.02 g L−1), NaCl (23.9 g L−1), Na2SO4 (4,0 g L−1), KCl (0.68 g L−1), NaHCO3 (0.2 g L−1), KBr (0.1 g L−1) and H3BO3 (0.03 g L−1)2. The culture medium was supplemented with aqueous CuSO4 to a final copper concentration of 2 mM. The flasks were maintained at 28 °C under constant agitation (140 rpm) for 7 days. Contents of the inoculated flasks was used as inoculum in a Fermac320 bioreactor (Electrolab Limited, UK) with 6.4 L total volume at an equivalent proportion of 8.5% (v/v) for 5 days. The bioreactor was placed in a temperature-controlled room at 20 °C and air filtered through two 0.2 µm filters (Sartorius-GER) was injected to the system via an “L” type sparger with 5 bottom holes2.
DNA extraction and sequencing
Genomic DNA was extracted from the bioreactor culture using the cetyltrimethyl ammonium bromide method adapted from the literature13,14, and purified using a Power Clean® DNA Clean-Up Kit (Mo Bio Laboratories). A sequencing library was built from high-quality genomic DNA using the Nextera DNA library preparation Kit (Illumina) with fragment size around 300 pb. Next, the library was submitted to sequencing on an Illumina Hiseq 2500 instrument with the HiSeq Rapid kit v2 chemistry in paired-end mode (2 × 100 bp), at the NGS sequencing facility of the Brazilian Biorenewables National Laboratory (LNBR/CNPEM).
Genome assembly
The genome of Peniophora sp. CBMAI 1063 was sequenced and generated 55 million paired-end reads (2 × 100 bp). Reads were processed with Trimmomatic 0.3215 to remove low quality reads and adapter sequences, resulting in 39 million quality-filtered reads. The genome size was estimated based on k-mer count statistics (Kmergenie). Before assembly, reads were normalized based on k-mer abundance with Khmer16,17 to a coverage of 20X and subjected to de novo assembly using Velvet version 1.2.1018 with k = 55. The complete subset of quality filtered reads was used to improve the assembly and for scaffolding using Pilon version 1.1619 and SSPACE version 3.020, respectively.
Gene prediction and annotation
A transcriptome analysis of Peniophora sp. CBMAI 1063 cultivated for 7 days at 28 °C in Erlenmeyer flasks was previously reported by Otero et al.3 and the raw sequence data, available in the Short Read Archives (SRA) GenBank database (accession number SRR5799684), was used in the present study as evidence for gene prediction and to determine the expression profile of the CAZymes. In order to use RNA-seq data for gene prediction, QC reads were aligned to the scaffolds produced by SSPACE using Hisat221 and subjected to the BRAKER1 v1.922 an automated genome annotation pipeline, which uses RNA-seq spliced alignments as evidence for GeneMark-ET and Augustus gene structure predictions22. “Non-coding rRNA genes were identified using both ITSx23 version 1.0.11 and RNAmmer24 version 1.2, and tRNA genes were annotated using tRNAscan25 version 1.3.1. Completeness of the genome gene set was estimated using the Benchmarking Universal Single-Copy Orthologs (BUSCO)26 version 1.1. Functional annotation was performed by comparison of the predicted protein sequences in the SwissProt database27 UniRef90 database28, PFAM29, Hidden Markov models (HMM) available at dbCAN 2.0 (automated carbohydrate-active enzyme annotation)30 and EggNOG (evolutionary genealogy of genes with enhanced non-supervised orthologous groups) databases31.
Phylogenetic analysis
Phylogenetic analysis was conducted using 36 organisms from the Basidiomycota phylum with public available genome sequences at JGI and the genome sequence of Peniophora sp. CBMAI 1063. Among the genomes analyzed, 35 are from the Agaricomycetes class and 2 from Ustilaginomycotina, which was used as the out-group. BUSCO was used to search for conserved genes among the 37 genomes analyzed. A set of 92 single copy gene markers present in all species was used to perform the phylogenetic analysis. The protein sequences of each marker gene were aligned using MAFFT version v7.299b32 and concatenated into a super matrix using FASconCAT-G version 1.2. The phylogenetic inference was performed using RAxML version 8.2.033 with PROTGAMMAWAG model.
Secretome analysis
The crude fungal extract from one growth culture batch in the bioreactor was centrifuged at 5,000 g for 30 minutes and submitted to electrophoresis analysis in polyacrylamide gel containing 0.1% (w/v) sodium dodecyl sulfate (SDS-PAGE). The gel-bands were excised from the SDS-PAGE and protein digestion for the mass spectrometry-based analyses was performed in two steps over two days. Firstly, SDS and Coomassie stain were removed with 500 µL of destaining solution (10% glacial acetic acid and 10% ethanol) for 2 h. The bands were dehydrated for 5 min with 200 µL acetonitrile, reduced for 30 min with 30 µL dithiothreitol solution (0.01 mol L−1) and alkylated for 30 min with 30 µL iodoacetamide solution (0.05 mol L−1) and washed with ammonium bicarbonate solution (0.1 mol L−1) for 10 min. A second dehydration step was performed with acetonitrile, followed by rehydration with sodium bicarbonate solution (0.05 mol L−1). Proteins were digested with 30 µL of trypsin (1.0 mg mL−1) in ammonium bicarbonate (0.05 mol L−1) at 37 °C for 12 h. Then, 20 µL of extraction solution composed of 5% (v/v) formic acid was added to each microtube and incubated for 10 min at room temperature. After centrifugation, the supernatant was transferred to another microtube and 30 µL of extraction solution composed of 5% (v/v) formic acid in 50% (v/v) acetonitrile were added. The supernatant was transferred to a tube containing the extract from the previous step after 10 min. The last procedure was repeated, and the samples were evaporated under vacuum to approximately 1 µL final volume. The samples were stored at −20 °C until further analysis by LC–MS/MS. Each sample was mixed with 12 μL of 0.1% (v/v) formic acid, and 4.5 μL of the peptide mixture were injected into the liquid chromatography-tandem mass spectrometry (LC–MS/MS) chromatograph (RP-nanoUPLC, nanoAcquity, Waters, Milford, MA). Peptide separations were performed in a C18 column (100 nm × 100 mm) previously equilibrated with a 0.1% (v/v) formic acid buffer. The elution gradient ranged from 2 to 90% (v/v) acetonitrile in 0.1% (v/v) formic acid at 0.6 μL min−1. Eluted peptides were analyzed in a quadrupole time of flight (Q-TOF) spectrometer (Ultima Mass Spectrometer, Waters Milford, MA) operating in the “top three-MS and MS/MS mode” (Ultima Mass Spectrometer, Waters software). Spectra were acquired using the Mass Lynx v.4.1 software (Waters, Milford, MA, USA), and the raw data were converted to “peak list format (mgf)” using the Mascot Distiller software v.2.3.02, 2009 (Matrix Science Ltd., London, UK). Results were processed using the Mascot v.2.3.02 engine software (Matrix Science Ltd.) against the Peniophora sp. CBMAI 1063 genome sequence database generated in this work. The following parameters were used in this process: carbamidomethylation as a fixed modification, oxidation of methionine as a variable modification, one trypsin cleavage error and a maximum allowable peptide mass error of 0.1 Da. The resulting Mascot data were analyzed for protein identification using Scaffold 3.5.1 (Proteome Software, Portland, OR). The defined parameters were as follows: minimum protein probability of 80%, minimum peptide probability of 90% and uniquely different minimum peptide of 1. Proteins with scores up to 10% false discovery rate (FDR) for a protein and 5% FDR for a peptide were accepted. The presence of a signal peptide on identified proteins was predicted by SignalP v.4.0 (http://www.cbs.dtu.dk/services/SignalP/)34 and the subcellular localization of proteins was predicted by YLoc (abi.inf.unituebingen.de/Services/YLoc/webloc.cgi)35.
Laccase production and purification
Laccase production and purification by ion exchange chromatography followed by size exclusion chromatography were performed according Mainardi et al.2. The fractions corresponding to the two intense peaks signal detected at 260 and 280 nm were concentrated individually and submitted to electrophoresis analysis in polyacrylamide gel containing 0.1% (w/v) SDS-PAGE. The gel-bands were excised from the SDS-PAGE and laccases identities were confirmed in duplicate by mass spectrometry as described above with FDR of 1.35%; and protein concentration was determined by the Bradford method36.
Laccase 3D-structure prediction by homology modeling
The three-dimensional structure of the major secreted laccase was generated by homology modeling on the I-TASSER server37. The laccase amino acid sequence was determined by mass spectrometry analysis; and the final model was generated based on the crystallographic structure of five different laccases from the AA1_1 family (PDBid: 1A65, 1V10, 2QT6, 5DAO and 5E9N) and one unclassified laccase (PDBid: 2HRG).
Laccase activity assay
Laccase activity was determined using syringaldazine (SGD) as the substrate as described in Mainardi et al.2. Oxidation of SGD was monitored at 525 nm for 5 minutes (readings every 20 seconds) at room temperature in a Tecan® Infinite spectrophotometer. One unit of enzyme activity was defined as the amount of enzyme required to oxidize 1 µmol of SGD per minute.
Laccase-Mediator system (LMS) assays for lignin modification
The alkali-lignin used in this study (96% purity) was extracted from steam-exploded sugarcane bagasse according Brenelli et al.38. Initially, a lignin stock solution at 10 mg mL−1 was prepared in NaOH 0.1 mol L−1. In the reaction system, 100 µL of the lignin stock solution was incubated with sodium acetate buffer 0.05 M at pH 5.0, purified laccase at 0.6 U g lignin−1 and 2,2′-Azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS) (Sigma-Aldrich®) as a mediator at a final concentration of 1 mmol L−1. The reaction (1.0 mL final volume) was incubated at 30 °C and 1000 rpm shaking in a Thermomixer (Eppendorf®) for 72 h. Incubations containing inactivated laccase and the ABTS mediator were used as controls. After incubation, the laccase activity was stopped by adding NaN3 at a final concentration of 0.05% (w V−1)39. The reaction mixture was centrifuged (12,000 x g for 10 min at 4 °C) and the supernatant collected for subsequent analysis. Changes in the molecular weight distribution of the soluble lignin and lignin-derived compounds were detected by gel permeation chromatography (GPC) using a Superdex 30 column (65 cm × 1.6 cm) in an automated AKTATM Purifier system (GE Healthcare) equipped with a UV detector (280 nm) and NaOH 0.1 mol L−1 as the eluent. The flow was 0.1 mL min−1 at room temperature with 500 µL sample injection volume. Tannic acid, ferulic acid, coumaric acid, cinnamic acid, hydroquinone and vanillin from Sigma Aldrich® and lignins with known molecular weight were used as internal standards. A UV-vis spectroscopy analysis was performed in a Tecan® Infinite spectrophotometer. The samples were diluted 20-fold in NaOH 0.1 mol L−1 and the UV-vis absorption spectrum (220 to 500 nm) was recorded using a 1 cm quartz cuvette.
LMS-based pretreatment
The pretreated sugarcane bagasse (SCB) used in this study was obtained by steam explosion (190 °C, 1.3 MPa, 15 min) as described previously40. The chemical composition of SCB was determined using the NREL method41 for lignocellulosic biomasses and was reported to be 52% glucan, 6% xylan, 24% lignin, 0.2% galactan and 0.5% arabinan. The commercial cellulolytic cocktails employed in this study were Accellerase® (DuPont) and Cellic® Ctec2 (Novozymes A/S, Bagsvaerd, Denmark). Cellulase activities of the enzymatic preparations measured via the filter paper assay were 168 and 250 filter paper units (FPU) mL−1, respectively. Both preparations were stored at 4 °C until performing the enzymatic hydrolysis assay. First, LMS assays were performed in 2.0 mL tubes with 2.5% (w V−1) dry matter (DM) in sodium acetate buffer pH 5.0; the reaction volume was 1 mL and it was maintained at 30 °C and 1000 rpm shaking in a Thermomixer® (Eppendorf). Purified laccase enzymes were added at a dosage of 0.02 U g biomass−1 as well as the ABTS mediator to a final concentration of 1 mmol L−1. Assays containing laccase inactivated by boiling and the ABTS mediator were used as controls. After 72 h the laccase activity was stopped by adding sodium azide (NaN3) at a final concentration of 0.05% (w V−1), then the commercial cocktails Accellerase® or Cellic® CTec2 were added in a low dosage of 2.5 FPU g cellulose−1 and incubated for 72 h (50 °C, 1000 rpm) in a ThermoMixer® (Eppendorf). The supernatant was separated by centrifugation and filtered (0.45 µM, Millipore®). Reducing sugars were measured by reacting 100 µL of the supernatant with 100 µL of 3,5-dinitrosalicylic acid for 5 min at 99 °C42. The cooled solution was analyzed at 540 nm in an Infinite M200® spectrophotometer (Tecan-Switzerland) and compared with an internal calibration curve using glucose. All enzymatic hydrolysis assays were carried out in triplicate and the average and standard deviation values were determined. The supernatant from non-hydrolyzed LMS-treated SCB was analyzed by GPC as described previously38.
Statistical analysis
A statistical analysis was performed by analysis of variance (One-way ANOVA) with a probability level (P) less than 5% (P < 0.05), using the program STATISTICA 5.5 from StatSoft Inc. (Tulsa, OK, USA).
Results and Discussion
The Peniophora sp. CBMAI 1063 genome content is distinct to other related species
The genome of the marine-derived Peniophora sp. CBMAI 1063 was sequenced using Illumina sequencing, with 165X coverage and reaching 93% genome completeness. The resulting draft genome assembly of Peniophora sp. CBMAI 1063, access number PRJEB28379, is 47.9 Mb in length with an N50 of 155.8 Kb, and average G + C content of 55% (Table S1). The genome size is similar to the other two Peniophora genomes available on the JGI Mycocosm portal: 48.4 Mb for Peniophora sp. CONTA (Lopni1) and 46.0 Mb for Peniophora aff. cinerea (Ricme1), both of which are plant pathogens.
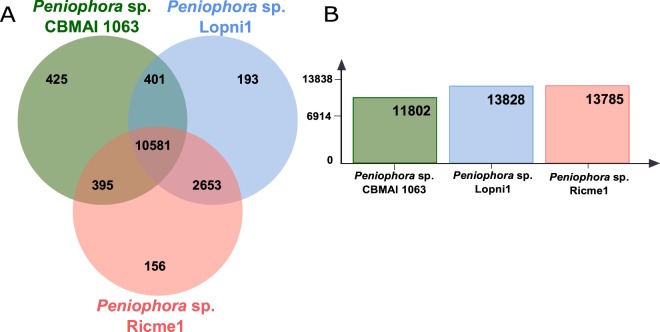
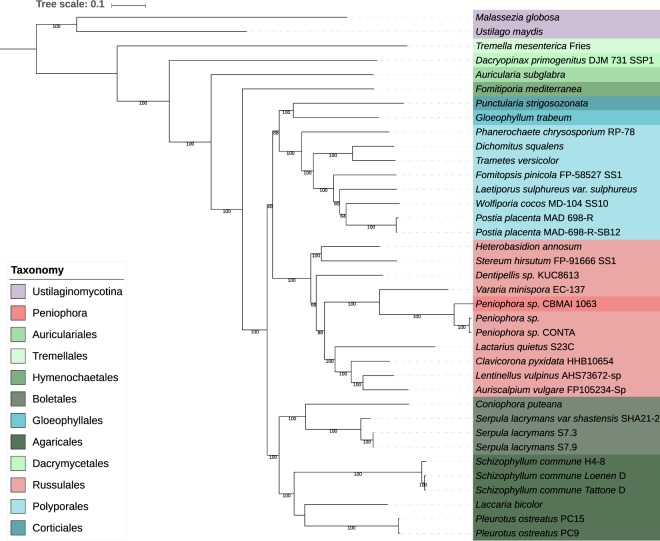
The majority of the predicted genes are common among the three strains analyzed, however a significant number of genes were found to be specific to each strain (Fig. 1). Among these genes, 457 were found to be specific to CBMAI 1063, 187 to Lopni1, and 164 to Ricme1 strains. The two plant-pathogen fungi strains, Lopni1 and Ricme1, shared 2791 orthologous genes (Fig. 1). The number of orthologous genes shared between Peniophora sp. CBMAI 1063 with Lopni1 or Ricme1 is significantly lower, only 370 and 381, respectively. The phylogenetic analysis using a set of 92 single-copy gene markers and 37 genomes from Basidiomycetes phylum (Fig. 2) indicated that Peniophora sp. CBMAI 1063 clustered together with the other two Peniophora species previously sequenced by JGI, forming a monophyletic clade in the Russulales order.
Figure 1.
A Venn diagram showing the distribution of orthologous gene clusters across the marine-derived Peniophora sp. CBMAI 1063, Peniophora sp. CONTA (Lopni1) and Peniophora aff. cinerea (Ricme1) (A) and the total number of orthologous gene clusters of each organism (B).
Figure 2.
Phylogenetic tree of the Agaricomycetes class based on 34 fungi genomes distributed among the Basidiomycota phylum. A set of 92 single copy gene markers was used to perform the phylogenetic analysis. Bootstrap values for 1000 replicates are show in the branches.
The Peniophora sp. CBMAI 1063 ligninolytic enzyme content
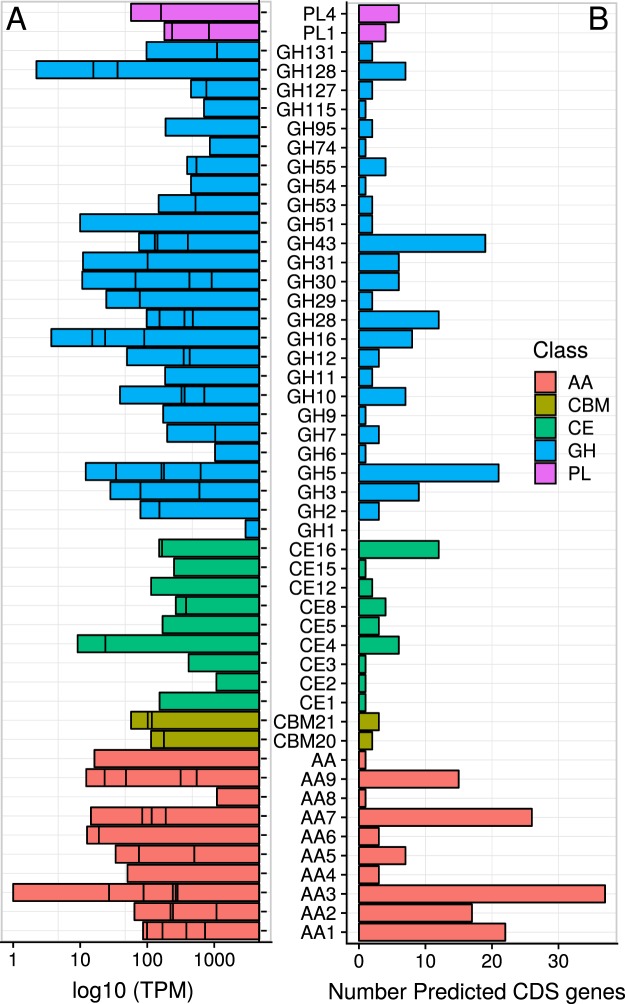
The genome of Peniophora sp. CBMAI 1063 encodes 17,714 predicted/putative genes, including 11,827 clusters of orthologous genes. A comparison of the predicted genes against the Carbohydrate-Active Enzymes database7 (CAZy) identified 310 predicted coding genes related to ligninolytic and carbohydrate-active enzymes (Fig. 3). Among the predicted genes encoding for the degradation/modification of lignin and the derived phenolic compounds, Peniophora sp. CBMAI 1063 exhibited 18 genes from family AA1 (laccases), 17 genes from AA2 (peroxidases), 3 genes from AA4 (vanillyl-alcohol oxidases), 7 genes from AA5 (glyoxal and alchool oxidases) and 3 from AA6 (1,4-benzoquinone reductase). Among those genes, 17 laccases AA1, 16 peroxidases AA2 and 7 glyoxal/alcohol oxidases AA5 are predicted to possess a secretion signal while all genes of AA4 and AA6 lack this signal.
Figure 3.
Transcriptome and genome profile of the marine-derived Peniophora sp. CBMAI 1063. (A) Transcriptome profile represented as log10 of TPM (Transcripts Per Million). (B) Genome profile represented as the number of predicted genes encoding CAZymes among the different classes of enzymes according to the CAZy database (http://www.cazy.org/).
A total of 41 coding genes of protein family AA3 (glucose oxidase, glucose dehydrogenases and cellobiose dehydrogenases) along with 16 genes of AA9 (LPMOs), 27 genes of AA7 (glucooligosaccharide oxidases) and 1 gene of AA8 (cellobiose dehydrogenases) were present in the genome of Peniophora sp. CBMAI 1063 (Fig. 3). The LPMOs from family AA9 have been found only in fungi and they are often co-expressed with sugar oxidases, such as those from the families AA3, AA7 and AA843. All AA9 genes exhibit secretion signals while for the AA3-coding genes the signal peptide is absent.
Furthermore, several genes encoding glycoside hydrolases (GH) were identified, including cellobiohydrolases from families GH6 and GH7, xylanases from GH10 and GH11, and pectinases from family GH28. Protein-coding genes for carbohydrate esterases (CE) families CE4, CE8 and CE16, and CBM families 20 and 21 (Fig. 3) were also identified. A high number of putative genes encoding for family GH5 (21 genes) and GH43 (19 genes), whose characterized members are typically endoglucanases and xylosidases, respectively, were found in the Peniophora sp. CBMAI 1063 repertoire of CAZymes. Interestingly, the fungus contains putative/predicted GH9-encoding genes, a family of endoglucanases found in anaerobic bacteria producing cellulosomes, plants and termites44.
The lignocellulolytic capabilities of marine fungi associated with algae, sponges, and mangrove habitats have previously been highlighted in other studies1,45, although the number of available sequenced genomes is still restricted. The high content of lignocellulosic materials from terrestrial sources that enter the ocean, as well as the symbiotic relationships with other organisms, justify the presence of genes encoding putative CAZymes in marine fungi, which may be found in high number compared to their terrestrial plant-degrading counterparts46.
Furthermore, the oceans are the largest source of biogenic organohalogens containing chlorine or bromine, which are biosynthesized by myriad seaweeds, sponges, corals, tunicates, bacteria, and other marine life47. In particular, the function of organohalogens in sponges is presumably to prevent feeding by fish and fouling by barnacles, bacteria, and fungi48. Since pyrroles, indoles, phenols, and tyrosines are commonly found to be halogenated in sponges, it is not a surprise that associated bacteria, microalgae or fungi are adapted to biosynthesize specific metabolites49. This work does not intend to elucidate fungal-sponge relationships by genomic comparisons, however this wide repertoire of CAZymes, in particular oxidoreductive enzymes, may be important to enable Peniophora sp. CBMAI 1063 to live in close association with its sponge host (Amphimedon viridis) in a marine environment.
Transcript encoding extracellular AA family members were expressed by Peniophora sp. CBMAI 1063 during growth in optimized conditions
Otero et al.3 described a preliminary global transcriptome analysis of Peniophora sp. CBMAI 1063 cultivated in a medium optimized for laccase production for 7 days in Erlenmeyer flask. Herein, the former RNA-Seq data set was used to validate the genome analysis, and also to depict the set of other partner genes encoding for lignin degradation and modifications enzymes. Additionally, the transcriptome data assisted in understanding not only the ligninolytic system, but also the cellulolytic genes co-expressed in media, which was formulated without a complex carbon source, i.e. lignocellulosic biomass.
From the 310 predicted genes related to lignin modification and degradation and CAZymes, 303 were expressed in the optimized media (Fig. 3). The genes encoding ligninolytic enzymes from families AA1, AA2, AA4, AA5 and AA6 were abundant in the transcriptome (Fig. 3). Peniophora sp. CBMAI 1063 exhibited high laccase activity in the optimized cultivation media1,2, corroborating with the present analyses, which identified the complete set of 18 genes expressed as predicted for the family AA1 (Fig. 3).
The AA1 family includes multicopper oxidases, including laccases, ferroxidases and laccase-like multicopper oxidases. Among all genes predicted as AA1 in the fungus, 15721.t1, g1591.t1, and g714.t1 were the most abundant transcripts found, considering the TPM of 53.2, 46.2 and 26.7, respectively. Concerning the family AA2, the genes encoding for peroxidases g10529.t1, g14863.t1, and g8820.t1 were the most abundant (Fig. 3). The gene of highest TPM among all CAZymes was the AA3 gene (g15979.t1). This transcript encodes a cytoplasmatic protein containing a glucose-methanol-choline (GMC) oxidoreductase domain, which is related to hydrogen peroxide generation and may act as a co-factor for peroxidases50. LPMOs from family AA9 and their electron donor protein partners, as well as the genes coding for AA3, AA7, and AA8, were also present in the transcriptome. Among the GH families involved in cellulose, hemicellulose and pectin degradation, genes coding for the families GH1, GH3, GH5, GH7, GH10, GH11, GH43 and GH51 were identified along with the transcription of CE genes from several families (CE4, CE12, CE5, CE16) (Fig. 3).
Secretomic analysis
A wide variety of oxidoreductases and CAZymes related to the degradation of plant cell wall polymers are present in Basidiomycota species51. However, depending on the species and lifestyles, the repertoire of enzymes and their gene numbers can differ significantly52.
In this work the Peniophora sp. CBMAI 1063 secretome was obtained when cultivated under optimized conditions for laccase production in bioreactor2. Additionally, the FDR based on use of a randomized decoy was 0.03%, indicating that the database employed was of high quality for mass spectrometry-based proteomics analysis.
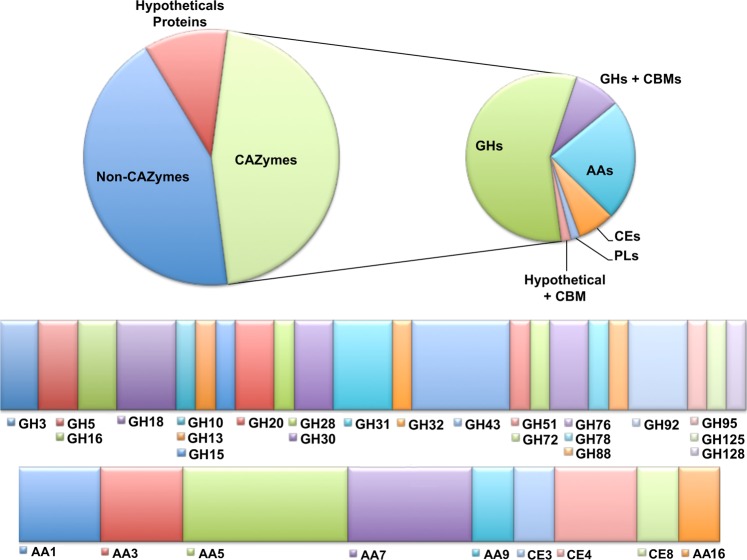
Collectively, 126 proteins were identified in the secretome, of which 56 were classified as CAZymes, 57 were non-CAZymes and 13 were hypothetical proteins (Fig. 4, Table S2). As predicted by SignalP v.4.034 and Yloc35, the majority of the enzymes identified (67%) in the secretome exhibited signal peptides. Among the ligninolytic enzymes, two laccases were identified which both presented predicted signal peptides. Although g15721.t1 presented the highest TPM value among all laccases in the transcriptome, the laccase g1591.t1 exhibited high spectrum counts (255 peptides) in comparison with all CAZymes identified in the secretome (Table 1). The laccase g1591.t1, herein named Pnh_Lac1, was identified with 12 unique peptides matches, covering 41% of the protein sequence. Four enzymes from family AA5 (related to glyoxal oxidases) were also identified in the secretome, all of which presented signal peptides. Peptides for peroxidases from family AA2, vanillyl-alcohol oxidases from AA4 and 1,4-benzoquinone reductase from AA6 were not found in the secretome, despite their relative abundance according to the genome and transcriptome data. However, this result should be considered expected since they lack peptide signals.
Figure 4.
Distribution of proteins identified in the Peniophora sp. CBMAI 1063 secretome obtained after cultivation under saline conditions by mass spectrometry analysis (LC-MS/MS). Non-hypothetical proteins were classified into non-CAZymes and CAZymes groups.
Table 1.
Predicted lignin-active enzymes identified in the secretome of Peniophora sp. CBMAI 1063 cultivated in a bioreactor under saline conditions.
| Accession Number | Molecular Weighta | Amino acid lengtha | dbCANb | PFAMc | Description | Signal Peptided | Locatione | Unique peptides | Spectrum counts |
|---|---|---|---|---|---|---|---|---|---|
| Lignin-Active Enzymes identified in the Secretome | |||||||||
| g1591.t1 | 58 kDa | 546 | AA1 |
PF00394 PF07731 PF07732 |
Multicopper oxidase | YES | SP | 12 | 255 |
| g17194.t1 | 58 kDa | 540 | AA1 |
PF00394 PF07732 PF07731 |
Multicopper oxidase | YES | SP | 3 | 2 |
| g5706.t1 | 71 kDa | 671 | AA5 |
PF07250 PF09118 |
Glyoxal oxidase N- terminus |
YES | SP | 2 | 1 |
| g5707.t1 | 81 kDa | 769 | AA5 |
PF07250 PF09118 |
Glyoxal oxidase N-terminus |
YES | SP | 3 | 1 |
| g5709.t1 | 84 kDa | 799 | AA5 |
PF07250 PF09118 |
Glyoxal oxidase N-terminus |
YES | SP | 2 | 1 |
| g9556.t1 | 59 kDa | 554 | AA5 |
PF07250 PF09118 |
Glyoxal oxidase N-terminus |
YES | SP | 4 | 5 |
| g13672.t1 | 60 kDa | 567 | AA7 |
PF01565 PF08031 |
FAD binding domain berberine |
YES | SP | 5 | 6 |
| g17067.t1 | 51 kDa | 485 | AA7 | PF01565 | FAD binding domain | YES | SP | 3 | 5 |
| g7475.t1 | 59 kDa | 548 | AA7 |
PF01565 PF08031 |
FAD binding domain berberine |
YES | SP | 3 | 1 |
aMolecular Weight and aAmino acid length determined by LC-MS/MS. The results were processed by Mascot v.2.3.01 engine (Matrix Science Ltd.) software against the genome sequencing database of Peniophora sp. CBMAI and Scaffold – Proteome Software (version Scaffold_4.3.2 20140225). bWeb server and database for automated carbohydrate-active enzyme annotation generated based on the family classification from CAZy database: AA –Auxiliary Activity. cProtein Family Domain analysis. dThe presence of a signal peptide of secreted proteins predicted by SignalP v.4.0. eThe subcellular localization of proteins predicted by YLoc (Interpretable Subcellular Localization Prediction): SP – secreted pathway; C – cytoplasm; M – mitochondrial location.
Among the cellulolytic and hemicellulolytic-active enzymes, the secretome contained 38 proteins classified in different families of GHs, which are involved in cellulose and hemicellulose breakdown (Tables 2–4). Enzymes from families GH6 and GH7 were absent in the secretome. Two GH5 were identified, where this family contains cellulase and hemicellulose activities. Five GHs exhibited CBMs: GH18 with CBM5, GH72 with CBM43, GH43 with CBM35, GH72 with CBM43 and GH15 with CBM20. Carbohydrate esterases (CE) from family 4 and 8, involved in deacetylation of hemicelluloses, and a polysaccharide lyase (PL) from family 22 (oligogalacturonate lyase) were also identified (Tables 3 and 5). Interestingly, a GH78 family whose characterized members are typically α-l-Rhamnosidases [E.C. 3.2.1.40] was identified along with a pectin-active enzymes group. These enzymes specifically cleave terminal α-l-rhamnose from a wide range of natural products and have important biotechnological applications in the food and pharmaceutical industries53.
Table 2.
Predicted cellulose-active enzymes identified in the secretome of Peniophora sp. CBMAI 1063 cultivated in a bioreactor under saline conditions.
| Accession Number | Molecular Weighta | Amino acid lengtha | dbCANb | PFAMc | Description | Signal Peptided | Locatione | Unique peptides | Spectrum counts |
|---|---|---|---|---|---|---|---|---|---|
| Cellulose-Active Enzymes | |||||||||
| g13682.t1 | 71 kDa | 640 | AA3 |
PF05199 PF00732 |
Glucose-methanol-choline oxidoreductase | NO | C | 18 | 19 |
| g16244.t1 | 62 kDa | 585 | AA3 |
PF00732 PF05199 |
Glucose-methanol-choline oxidoreductase | NO | SP | 6 | 7 |
| g6504.t1 | 26 kDa | 242 | AA9 | PF03443 | Glycoside hydrolase family 61 | YES | SP | 3 | 71 |
| g6425.t1 | 35 kDa | 348 | AA16 | PF03067 |
Lytic polysaccharide monooxygenase |
YES | SP | 2 | 1 |
| g11705.t1 | 81 kDa | 760 | GH3 |
PF01915 PF00933 PF14310 |
Glycoside hydrolase family 3 | YES | SP | 9 | 18 |
| g15376.t1 | 93 kDa | 879 | GH3 |
PF01915 PF00933 PF14310 |
Glycoside hydrolase family 3 | YES | SP | 2 | 1 |
| g1658.t1 | 37 kDa | 353 | GH5 | PF00150 | Glycoside hydrolase family 5 | YES | SP | 2 | 2 |
| g5589.t1 | 49 kDa | 452 | GH5 | PF00150 | Glycoside hydrolase family 5 | YES | SP | 4 | 7 |
aMolecular Weight and aAmino acid length determined by LC-MS/MS. The results were processed by Mascot v.2.3.01 engine (Matrix Science Ltd.) software against the genome sequencing database of Peniophora sp. CBMAI and Scaffold – Proteome Software (version Scaffold_4.3.2 20140225). bWeb server and database for automated carbohydrate-active enzyme annotation generated based on the family classification from CAZy database: GH- Glycoside Hydrolases; AA- Auxiliary Activities. cProtein Family Domain analysis. dThe presence of a signal peptide of secreted proteins predicted by SignalP v.4.0. eThe subcellular localization of proteins predicted by YLoc (Interpretable Subcellular Localization Prediction): SP – secreted pathway; C – cytoplasm; M – mitochondrial location.
Table 4.
Predicted chitin, starch & other carbohydrate-active enzymes identified in the secretome of Peniophora sp. CBMAI 1063 cultivated in a bioreactor under saline conditions.
| Accession Number | Molecular Weighta | Amino acid lengtha | dbCANb | PFAMc | Description | Signal Peptided | Locatione | Unique peptides | Spectrum counts |
|---|---|---|---|---|---|---|---|---|---|
| Chitin, Starch & Others Carbohydrate-Active Enzymes | |||||||||
| g13668.t1 | 61 kDa | 580 |
CBM20 GH15 |
PF00723 PF00686 |
Glycoside hydrolases family 15 | YES | SP | 20 | 197 |
| g7315.t1 | 82 kDa | 769 | CBM5 |
PF17168 PF16335 PF08760 PF02839 |
Carbohydrate binding domain | YES | SP | 10 | 15 |
| g7861.t1 | 50 kDa | 474 | GH13 |
PF00128 PF02806 |
Alpha amylase | YES | SP | 5 | 18 |
| g11641.t1 | 97 kDa | 890 | GH31 |
PF01055 PF16863 PF13802 |
Glycoside hydrolases family 31 | YES | SP | 3 | 4 |
| g15819.t1 | 109 kDa | 997 | GH31 |
PF01055 PF16863 |
Glycoside hydrolases family 31 | NO | SP | 12 | 9 |
| g15820.t1 | 104 kDa | 944 | GH31 |
PF01055 PF16863 |
Glycoside hydrolases family 31 | YES | SP | 6 | 6 |
| g11828.t1 | 60 kDa | 547 | GH32 | PF00251 | Glycoside hydrolases family 32 | NO | SP | 8 | 35 |
| g11153.t1 | 49 kDa | 460 | GH88 | PF07470 | Glycoside hydrolase Family 88 | YES | SP | 3 | 3 |
| g11942.t1 | 45 kDa | 418 | GH18 | PF00704 | Glycoside hydrolases family 18 | YES | SP | 2 | 12 |
| g11425.t1 | 49 kDa | 472 |
GH18; CBM5 |
PF02839 | Carbohydrate binding domain | YES | SP | 2 | 5 |
| g6083.t1 | 136 kDa | 1254 |
GH18; CBM5 |
PF00009 PF03764 PF14492 PF00679 PF00704 PF02839 PF03144 |
Elongation factor Tu GTP binding domain18 | YES | M | 7 | 23 |
| g10368.t1 | 60 kDa | 560 | GH20 |
PF00728 PF14845 |
Glycoside hydrolase family 20 | YES | SP | 5 | 2 |
| g15989.t1 | 134 kDa | 1230 | GH20 |
PF00728 PF14845 PF02838 |
Glycoside hydrolase family 20 | YES | SP | 7 | 9 |
aMolecular Weight and aAmino acid length determined by LC-MS/MS. The results were processed by Mascot v.2.3.01 engine (Matrix Science Ltd.) software against the genome sequencing database of Peniophora sp. CBMAI and Scaffold – Proteome Software (version Scaffold_4.3.2 20140225). bWeb server and database for automated carbohydrate-active enzyme annotation generated based on the family classification from CAZy database: GH - Glycoside Hydrolases; CBM- Carbohydrate-binding module. cProtein Family Domain analysis. dThe presence of a signal peptide of secreted proteins predicted by SignalP v.4.0. eThe subcellular localization of proteins predicted by YLoc (Interpretable Subcellular Localization Prediction): SP – secreted pathway; C – cytoplasm; M – mitochondrial location.
Table 3.
Predicted hemicellulose-active enzymes identified in the secretome of Peniophora sp. CBMAI 1063 cultivated in a bioreactor under saline conditions.
| Accession Number | Molecular Weighta | Amino acid lengtha | dbCANb | PFAMc | Description | Signal Peptided | Locatione | Unique peptides | Spectrum counts |
|---|---|---|---|---|---|---|---|---|---|
| Hemicellulose-Active Enzymes | |||||||||
| g14451.t1 | 46 kDa | 429 | CE3 |
PF13472 PF00657 |
GDSL-like lipase/acyl hydrolase | YES | SP | 7 | 44 |
| g10047.t1 | 55 kDa | 513 | CE4 | Carbohydrate esterase | YES | SP | 3 | 5 | |
| g4642.t1 | 38 kDa | 354 | CE4 | PF01522 | Polysaccharide deacetylase | YES | SP | 6 | 14 |
| g11401.t1 | 37 kDa | 345 | GH10 | PF00331 | Glycoside hydrolase family 10 | YES | SP | 4 | 8 |
| g11177.t1 | 59 kDa | 540 | GH125 | PF06824 | Protein of unknown function | NO | SP | 5 | 4 |
| g14331.t1 | 30 kDa | 277 | GH128 | PF11790 | Glycoside hydrolase catalytic core | YES | SP | 2 | 6 |
| g7518.t1 | 34 kDa | 318 | GH16 | Glycoside hydrolases family 16 | YES | SP | 4 | 18 | |
| g7519.t1 | 33 kDa | 313 | GH16 | Glycoside hydrolases family 16 | YES | SP | 6 | 66 | |
| g16729.t1 | 57 kDa | 524 | GH30 | PF02055 | Glycoside hydrolase family 30 | YES | SP | 3 | 3 |
| g3624.t1 | 53 kDa | 499 | GH30 | PF14587 | O-Glycoside hydrolase family 30 | YES | SP | 10 | 57 |
| g14525.t1 | 36 kDa | 341 | GH43 | PF04616 | Glycoside hydrolases family 43 | YES | SP | 2 | 2 |
| g4370.t1 | 33 kDa | 321 | GH43 | PF04616 | Glycoside hydrolases family 43 | YES | SP | 4 | 54 |
| g5765.t1 | 33 kDa | 310 | GH43 | PF04616 | Glycoside hydrolases family 43 | YES | SP | 5 | 55 |
| g9420.t1 | 35 kDa | 329 | GH43 | PF04616 | Glycoside hydrolases family 43 | YES | SP | 2 | 13 |
| g15433.t1 | 47 kDa | 444 |
GH43 CBM35 |
PF04616 | Glycoside hydrolases family 43 | YES | SP | 2 | 2 |
| g3571.t1 | 69 kDa | 638 | GH51 | PF06964 | α-L-arabinofuranosidase | YES | SP | 10 | 33 |
| g14175.t1 | 58 kDa | 564 |
GH72 CBM43 |
PF03198 PF07983 |
Glucanosyltransferase X8 domain |
YES | SP | 4 | 3 |
| g11973.t1 | 40 kDa | 375 | GH76 | PF03663 | Glycoside hydrolase family 76 | YES | SP | 5 | 8 |
| g4124.t1 | 40 kDa | 370 | GH76 | PF03663 | Glycoside hydrolase family 76 | YES | SP | 4 | 5 |
| g11336.t1 | 91 kDa | 827 | GH92 | PF07971 | Glycoside hydrolase family 92 | YES | SP | 13 | 14 |
| g1967.t1 | 67 kDa | 606 | GH92 | PF07971 | Glycoside hydrolase family 92 | NO | SP | 10 | 10 |
| g5807.t1 | 88 kDa | 812 | GH92 | PF07971 | Glycoside hydrolase family 92 | YES | SP | 13 | 15 |
| g5441.t1 | 186 kDa | 1691 | GH95 | PF14498 | Glycoside hydrolase family 95 | NO | SP | 2 | 1 |
aMolecular Weight and aAmino acid length determined by LC-MS/MS. The results were processed by Mascot v.2.3.01 engine (Matrix Science Ltd.) software against the genome sequencing database of Peniophora sp. CBMAI and Scaffold – Proteome Software (version Scaffold_4.3.2 20140225). bWeb server and database for automated carbohydrate-active enzyme annotation generated based on the family classification from CAZy database: GH - Glycoside Hydrolases; CE - Carbohydrate Esterases; CBM: Carbohydrate-binding module. cProtein Family Domain analysis. dThe presence of a signal peptide of secreted proteins predicted by SignalP v.4.0. eThe subcellular localization of proteins predicted by YLoc (Interpretable Subcellular Localization Prediction): SP – secreted pathway; C – cytoplasm; M – mitochondrial location.
Table 5.
Predicted pectin-active enzymes identified in the secretome of Peniophora sp. CBMAI 1063 cultivated in a bioreactor under saline conditions.
| Accession Number | Molecular Weighta | Amino acid lengtha | dbCANb | PFAMc | Description | Signal Peptided | Locatione | Unique peptides | Spectrum counts |
|---|---|---|---|---|---|---|---|---|---|
| Pectin-Active Enzymes | |||||||||
| g8265.t1 | 35 kDa | 330 | CE8 | PF01095 | Pectin esterase | YES | SP | 2 | 2 |
| g8087.t1 | 70 kDa | 665 | GH78 | PF05592 | Bacterial alpha-L-rhamnosidase | YES | SP | 7 | 30 |
| g10469.t1 | 74 kDa | 673 | PL22 | PF07676 | WD40-like beta propeller repeat | YES | SP | 2 | 1 |
| g1538.t1 | 45 kDa | 413 | GH28 | PF00295 | Glycoside hydrolase family 28 | YES | SP | 2 | 1 |
aMolecular Weight and aAmino acid length determined by LC-MS/MS. The results were processed by Mascot v.2.3.01 engine (Matrix Science Ltd.) software against the genome sequencing database of Peniophora sp. CBMAI and Scaffold – Proteome Software (version Scaffold_4.3.2 20140225). bWeb server and database for automated carbohydrate-active enzyme annotation generated based on the family classification from CAZy database: GH- Glycoside Hydrolases; PL- Polysaccharide Lyases; CE- Carbohydrate Esterases. cProtein Family Domain analysis. dThe presence of a signal peptide of secreted proteins predicted by SignalP v.4.0. eThe subcellular localization of proteins predicted by YLoc (Interpretable Subcellular Localization Prediction): SP – secreted pathway; C – cytoplasm; M – mitochondrial location.
Omics data integration
Omics approaches can provide hypotheses regarding function for the large number of genes predicted from genome sequences. In this study, an integrative analysis of genomic, transcriptomic and proteomic data was performed for Peniophora sp. CBMAI 1063 Although the cultivation medium had the same composition, an important point to take in account is that the transcriptomic analysis was performed at the seventh day of small-scale cultivation (200 mL) while the secretome analysis was at the fifth day of cultivation in bioreactor scale (5 L). Moreover, it is well documented in the literature the poor correlation between transcriptomic and proteomic data due to several factors, including pre and pos translational processes54,55. Even though, omic data integration was performed in this work in order to explore the biotechnological potential of Peniophora sp. CBMAI 1063 under its optimized cultivation medium.
Thus, according to our omics data, of 18 genes encoding for laccases from family AA1 were found in the genome, only 2 proteins were identified as secreted in the condition analyzed (Figs. 3 and 4 and S1). Post-transcriptional, translational and degradation regulation events regarding the gene Pnh_Lac1 may be involved to explain the differences between gene expression and protein secretion under the conditions evaluated. Conversely, the fungus preferentially secrets Pnh_Lac1 in the optimized media (Fig. S1).
Family members from AA4 were not identified in the proteomic data, although 3 genes were identified in the genome and in the transcriptome (Figs. 3 and S2). This was similar to the observations for AA2, AA6 and AA8 family members, where the transcripts were expressed but the proteins were not identified in the secretome (Figs. 3 and 4). Regarding the family AA5, 4 proteins were found to be secreted from the 7 encoding genes identified in the genome (Figs. 3 and S2). Assigned to a glyoxal oxidase family domain according to PFAM, AA5 members are copper-containing enzymes that mainly oxidize aldehydes generated during lignin and carbohydrates degradation56. Although genes encoding glyoxal oxidases enzyme are widely distributed among white-rot fungi and symbiotic fungi, the number of characterized enzymes is still restricted10. The presence of these proteins in the secretome suggests that the fungus possesses a strong ligninolytic capacity. In this context, further studies of the AA5 enzymes and their role in Peniophora sp. CBMAI 1063 are extremely relevant. Furthermore, among the 27 genes encoding AA7 proteins, only 3 were found to be secreted.
According to the secretome data, in the case of the carbohydrate-binding module (CBM), predicted genes for CBM5, CBM20, CBM 21 and CBM 42 were found; while the transcriptomic data exhibited 4 transcripts for CBM21 and one for CBM20 (Figs. 3 and 4). Among all GH-coding genes identified, GH5 and GH43 exhibited the highest number of predicted genes (Fig. 3), and 2 and 5 proteins were identified in the secretome, respectively (Fig. 3).
Homology and structural insights of the major laccase (Pnh_Lac1) secreted by Peniophora sp. CBMAI 1063
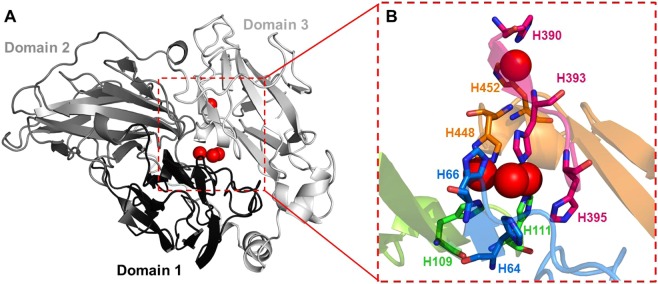
According to a sequence search using BLASTp, the most similar proteins to Pnh_Lac1 are multicopper oxidases from Peniophora sp. (87% identity to KZV66389.1 and 66% identity to KZV69698.1) and laccases from P. lycii (AWC08468.1) and Meripilus giganteus (CBV46340.1), both showing 61% identity. The laccase signature composed of four ungapped sequence segments L1–L457 was in Pnh_Lac1 (Fig. S3). Segments L1-L4 contain the amino acids that bind to the copper centers: H64 and H66 from L1; H109 and H111 from L2; H390, H393 and H395 from L3; and H448 and H452 from L4 (Fig. 5B).
Figure 5.
3D structure of Pnh_Lac1 generated by homology modeling. General structure of laccases, consisting of three cupredoxin domains (A) Zoom on the copper ion centers surrounded by the fungal laccase signature segments L1 (blue), L2 (green), L3 (magenta) and L4 (orange). The ion coordination is performed by the histidines presented in the L1-L4 regions (B).
Performing a sequence search in the Protein Data Bank (PDB), the Pnh_Lac1 shows between 55–60% identity with different laccases from subfamily AA1_1. Due to the highly conserved structure of this family, the three-dimensional model of Pnh_Lac1 could be generated by homology modelling (Fig. 5), which was considered reliable according to the C-score value of 0.7 (range from −5 to 2)37. The enzyme presents typical folding of laccases, consisting of three cupredoxin domains with a mononuclear copper center located in domain 3, and a trinuclear center between domains 1 and 358 (Fig. 5A). Collectively, these results, including the sequence alignment of the conserved regions, as well as the structural alignment of Pnh_Lac1 with other members of family AA1_1, are important to support the gene prediction data (Figs. 3 and S4) described in the present work.
Pnh_Lac1 effects on lignin depolymerization and enzymatic digestibility of steam-exploded sugarcane bagasse
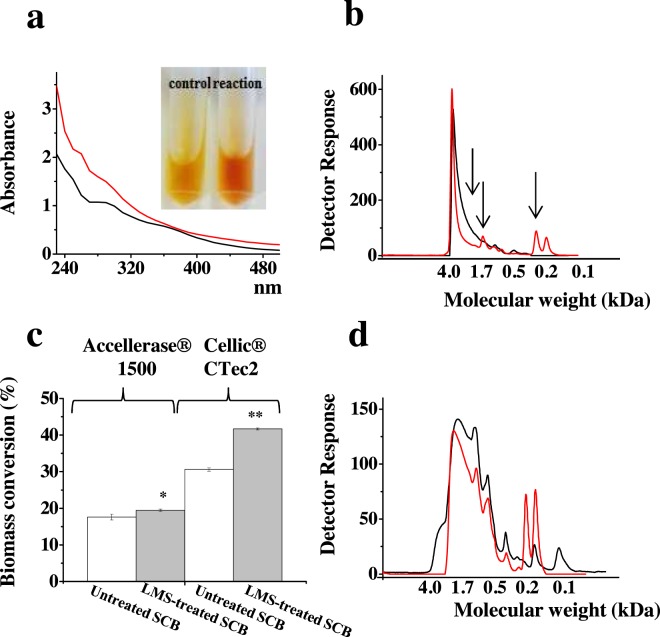
The biochemical properties of purified Pnh_Lac1 were previously characterized by Mainardi et al.2: presenting 986.0 and 30.8 U mg−1, using ABTS and SGD as substrates, respectively, and optimum activity at pH 5.0 and 30 °C. To illustrate a potential biotechnological application, the purified Pnh_Lac1 was employed in combination with a synthetic mediator (ABTS) to promote the depolymerization of a lignin isolated from sugarcane bagasse by alkaline pretreatment39. The soluble fraction after incubation with LMS was analyzed by UV-light absorbance and GPC. An increase of 50% in UV-light absorbance at 280 nm was observed compared to the control experiment without Pnh_Lac1 (Fig. 6a). According to Arzola et al.59, spectra changes detected between 260 and 270 nm (in LMS-treated samples) indicate the introduction of new functional groups in the phenylpropane unit or aromatic rings of lignin fragments (Fig. 6a), resulting in auxochrome groups, new nonconjugated hydroxyl groups in the side-chain phenylpropane unit or new Cα–Cβ double bonds.
Figure 6.
UV-light absorbance (a) and GPC chromatograms (b) showing the spectroscopic profile and molecular weight distribution of the lignin derived products obtained after the incubation of lignin extracted from SCB with purified Pnh_Lac1 and ABTS as the mediator (LMS); Enzymatic hydrolysis of LMS-treated SCB (grey bars) or non-treated SCB (white bars) performed with commercial cocktails at low dosage. The y-axis shows the cellulose conversion in percentage of the maximum theoretical cellulose conversion after 72 h at 50 °C. Error bars represent the standard errors of the means of triplicate experiments (c); GPC chromatograms showing the molecular weight distribution of the solubilized lignin-derived products and polyphenolics from LMS-treated SCB. Red lines (in figures a,b,d) represent experiments with LMS and black lines refer to control experiments without Pnh_Lac1.
Spectral changes in the visible region were also detected, especially at 480 nm. Chromogen groups are commonly introduced after laccase activity, which is related to the reddish color appearance, as can be observed in Fig. 6a. The GPC analysis shows that soluble low molecular weight lignin derived compounds (<500 Da) were generated after LMS treatment (Fig. 6b), indicated by the arrows. The intense peak corresponding to high molecular weight (HMW) lignin (~4 kDa) appeared thinner when compared to the untreated sample, which is indicative of polymer depolymerization. The results indicate that Pnh_Lac1, when combined with a mediator, was able to promote lignin solubilization and depolymerization. This enzyme is therefore should be of biotechnological interest especially for the bioconversion of industrial lignins streams, including those generated from lignocellulose processing biorefineries where utilization is hampered by its chemically heterogeneous nature, low solubility and reactivity38.
Previously works have shown that LMS treatments can improve the enzymatic hydrolysis of lignocellulose materials, based on lignin solubilization, depolymerization and modification mechanisms60. However, there is also evidence that LMS can have detrimental effects on enzymatic hydrolysis yields, as a result of enzymes inhibition, oxygen competition and grafting of phenoxy radicals39,61. We are therefore led to believe that positive or negative effects can depend on singularities of the laccase employed and assay conditions.
In this sense, we evaluated the ability of the LMS based on Pnh_Lac1 and ABTS to remove lignin from pretreated SCB, which could improve the subsequent saccharification of lignocellulosic material. The SCB was first treated with LMS and then with two different commercial cocktails. Increases of 1.9% and 11.0% were observed in saccharification yield using Accellerase® 1500 and CellicTM Ctec2, respectively, compared to the untreated samples (Fig. 6c). The GPC analysis of the soluble fraction after LMS treatment confirmed that the lignin fragments were partially degraded, leading to its solubilization into the supernatant (Fig. 6d).
The new peaks detected in the LMS-treated sample correspond to soluble low molecular weight phenolic compounds, between 400–200 Da (indicated by arrows) (Fig. 6). The HMW lignin fragments from 4–1.7 kDa were found in lower intensity in the chromatogram. Interesting, as can be observed in the LMS-treated supernatant, the peak corresponding to low molecular weight phenolic compounds (around 100 Da) is absent (Fig. 6).
The LPMO-containing cellulose cocktail Cellic™ Ctec2 has better performance than traditional enzyme cocktails mainly consisted of hydrolytic cellulases for biomass saccharification62. In addition, it has been demonstrated that soluble lignin derived compounds act as electron donors to LPMOs, boosting its catalytic efficiency39,63. Thus, the higher saccharification yield using Cellic™ Ctec2 for pretreated SCB may indicate that the soluble low molecular weight phenolic compounds generated by the LMS based on Pnh_Lac1 and ABTS enhanced the LPMO activity.
Collectively, our results demonstrate that Pnh_Lac1 could be applied be applied in bioconversion technologies, based on promoting lignin depolymerization and solubilization from the lignocellulosic substrate, thus facilitating the action of cellulolytic enzymes. However, further studies are necessary to combine the laccase with different mediators and higher enzyme loadings, which could improve the effectiveness of the LMS pre-treatment.
Conclusions
For the first time, the genomic and secretomic analyses of marine-derived Peniophora sp. CBMAI 1063 revealed an important repertoire of valuable extracellular CAZymes, especially lignin and polyphenols-degrading enzymes. Interestingly, this marine fungus presents a higher number of unique orthologous gene clusters compared to the other two genomes from Peniophora species, demonstrating its singularity. In addition, our findings revealed that Peniophora sp. CBMAI 1063 has the ability to secrete a powerful set of oxidative enzymes of biotechnological interest, mainly the laccase Pnh_Lac1 which could be of interest in lignin modification and depolymerization strategies, bioconversion in industries and bioremediation.
Supplementary information
Acknowledgements
This work was supported by São Paulo Research Foundation (FAPESP) research grants: 2015/50590-4 to FMS and 2016/0795-7 to LDS and; fellowships: 2013/03061-0 and 2015/07008-2 to LBB, 2013/19486-0 and 2016/0795-7 to LDS, 2015/06971-3 and 2016/09950-0 to JPLFC, 2014/04105-4, 14/04105-4 to MVL and 17/16089-1 for TAG. This work was supported by the National Council for Scientific and Technological Development (CNPq) grants: 305748/2017-3 and 428527/2018-3 to FMS, 304103/2013-6 to LDS and; fellowships: 159488/2014-1 to IVRO. We would like to thank Brazilian Biorenewables National Laboratory (LNBR) (LNBR/CNPEM) NGS Sequencing Facility for generating the sequencing data described here, Brazilian Biosciences National Laboratory (LNBIO/CNPEM) for Mass spectrometry-based proteomics analysis.
Author contributions
L.B.B., G.F.P. and F.M.S. conceived the study and drafted the manuscript. L.B.B. and F.M.S. designed the experiments. L.B.B., I.V.R.O., P.H.M., J.P.L.F.C. and M.V.L. contributed to the experimental work. G.F.P. contributed to genome assembly, gene prediction and annotation and phylogenetic analysis. L.B.B., G.F.P., I.V.R.O., T.A.G., J.P.L.F.C. and M.V.L. contributed to interpretation of the results. C.F., L.D.S. and F.M.S. supervised the final version. All authors read and approved the final manuscript.
Data availability
The authors promise the availability of supporting data.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lívia B. Brenelli and Gabriela F. Persinoti.
Supplementary information
is available for this paper at 10.1038/s41598-019-53608-1.
References
- 1.Bonugli-Santos RC, et al. Marine-derived fungi: diversity of enzymes and biotechnological applications. Frontiers Microbiol. 2015;6:269. doi: 10.3389/fmicb.2015.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mainardi PH, et al. Laccase production in bioreactor scale under saline condition by the marine-derived basidiomycete Peniophora sp. CBMAI 1063. Fungal Biol. 2018;122(5):302–309. doi: 10.1016/j.funbio.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Otero IVR, Ferro M, Bacci M, Ferreira H, Sette LD. De novo transcriptome assembly: a new laccase multigene family from the marine-derived basidiomycete Peniophora sp. CBMAI 1063. AMB Express. 2017;7(1):222. doi: 10.1186/s13568-017-0526-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohm RA, et al. Genomics of wood-degrading fungi. Fungal Genet. Biol. 2014;72:82–90. doi: 10.1016/j.fgb.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Riley R, et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. PNAS. 2014;111(27):9923–9928. doi: 10.1073/pnas.1400592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Floudas D, et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336(6089):1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 7.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels. 2013;6(1):41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldrian P. Fungal laccases–occurrence and properties. FEMS Microbiol. Rev. 2006;30(2):215–242. doi: 10.1111/j.1574-4976.2005.00010.x. [DOI] [PubMed] [Google Scholar]
- 9.Gygli G, de Vries RP, van Berkel WJ. On the origin of vanillyl alcohol oxidases. Fungal Gen. Biol. 2018;116:24–32. doi: 10.1016/j.fgb.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Daou M, Faulds CB. Glyoxal oxidases: their nature and properties. World J. Microbiol. Biotechnol. 2017;33(5):87. doi: 10.1007/s11274-017-2254-1. [DOI] [PubMed] [Google Scholar]
- 11.Vuong TV, Foumani M, MacCormick B, Kwan R, Master ER. Direct comparison of gluco-oligosaccharide oxidase variants and glucose oxidase: substrate range and H2O2 stability. Sci. Rep. 2016;6:37356. doi: 10.1038/srep37356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menezes CB, et al. Microbial diversity associated with algae, ascidians and sponges from the north coast of São Paulo state, Brazil. Microbiol. Res. 2010;165(6):466–482. doi: 10.1016/j.micres.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Möller EM, Bahnweg G, Sandermann H, Geiger HH. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 1992;20(22):6115. doi: 10.1093/nar/20.22.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerardo NM, Mueller UG, Price SL, Currie CR. Exploiting a mutualism: parasite specialization on cultivars within the fungus–growing ant symbiosis. Proceedings of the Royal Society of London. Series B: Biol. Sci. 2004;271(1550):1791–1798. doi: 10.1098/rspb.2004.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chikhi R, Medvedev P. Informed and automated k-mer size selection for genome assembly. Bioinformatics. 2013;30(1):31–37. doi: 10.1093/bioinformatics/btt310. [DOI] [PubMed] [Google Scholar]
- 17.Crusoe, M. R. et al. The khmer software package: enabling efficient nucleotide sequence analysis. F1000Research, 4 (2015). [DOI] [PMC free article] [PubMed]
- 18.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS one. 2014;9(11):e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2010;27(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 21.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 2015;12(4):357. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 2015;32(5):767–769. doi: 10.1093/bioinformatics/btv661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bengtsson-Palme J, et al. Improved software detection and extraction of ITS1 and ITS 2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013;4(10):914–919. [Google Scholar]
- 24.Lagesen K, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 27.The UniProt Consortium; UniProt: the universal protein knowledgebase. Nucleic Acids Res, (D1), D158–D169, 10.1093/nar/gkw1099 (2017). [DOI] [PMC free article] [PubMed]
- 28.Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH. UniProt Consortium. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2014;31(6):926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn RD, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2015;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin Y, et al. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40(W1):W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huerta-Cepas J, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2015;44(D1):D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen TN, Brunak S, Von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods. 2011;8(10):785. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 35.Briesemeister S, Rahnenführer J, Kohlbacher O. Going from where to why—interpretable prediction of protein subcellular localization. Bioinformatics. 2010;26(9):1232–1238. doi: 10.1093/bioinformatics/btq115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72(1-2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 37.Yang J, et al. The I-TASSER Suite: protein structure and function prediction. Nature Methods. 2015;12(1):7. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brenelli LB, et al. Acidification treatment of lignin from sugarcane bagasse results in fractions of reduced polydispersity and high free-radical scavenging capacity. Ind. Crops Prod. 2016;83:94–103. doi: 10.1016/j.indcrop.2015.12.013. [DOI] [Google Scholar]
- 39.Brenelli L, Squina FM, Felby C, Cannella D. Laccase-derived lignin compounds boost cellulose oxidative enzymes AA9. Biotechnol. Biofuels. 2018;11(1):10. doi: 10.1186/s13068-017-0985-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rocha GDM, Gonçalves AR, Oliveira BR, Olivares EG, Rossell CEV. Steam explosion pretreatment reproduction and alkaline delignification reactions performed on a pilot scale with sugarcane bagasse for bioethanol production. Ind. Crops Prod. 2012;35(1):274–279. doi: 10.1016/j.indcrop.2011.07.010. [DOI] [Google Scholar]
- 41.Sluiter, A. et al. Determination of structural carbohydrates and lignin in biomass. Laboratory analytical procedure. (TP-510-42618). (2010).
- 42.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31(3):426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 43.Bey M, et al. Cello-oligosaccharide oxidation reveals differences between two lytic polysaccharide monooxygenases (family GH61) from Podospora anserina. Appl. Environ. Microbiol. 2013;79(2):488–496. doi: 10.1128/AEM.02942-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ni J, Takehara M, Watanabe H. Identification of activity related amino acid mutations of a GH9 termite cellulase. Bioresour. Technol. 2010;101(16):6438–6443. doi: 10.1016/j.biortech.2010.03.045. [DOI] [PubMed] [Google Scholar]
- 45.Arfi Y, et al. Characterization of salt-adapted secreted lignocellulolytic enzymes from the mangrove fungus Pestalotiopsis sp. Nature communications. 2013;4:1810. doi: 10.1038/ncomms2850. [DOI] [PubMed] [Google Scholar]
- 46.Zhao Z, Liu H, Wang C, Xu JR. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics. 2013;14(1):274. doi: 10.1186/1471-2164-14-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gribble GW. The diversity of naturally produced organohalogens. Chemosphere. 2003;52(2):289–297. doi: 10.1016/S0045-6535(03)00207-8. [DOI] [PubMed] [Google Scholar]
- 48.Caballero-George C, et al. Protocol to isolate sponge-associated fungi from tropical waters and an examination of their cardioprotective potential. Curr. Trends Biotechnol. Pharm. 2010;4(4):881–899. [Google Scholar]
- 49.Kelman D, et al. Antimicrobial activity of the reef sponge Amphimedon viridis from the Red Sea: evidence for selective toxicity. Aquat. Microb. Ecol. 2001;24(1):9–16. doi: 10.3354/ame024009. [DOI] [Google Scholar]
- 50.Ferreira P, Carro J, Serrano A, Martínez AT. A survey of genes encoding H2O2-producing GMC oxidoreductases in 10 Polyporales genomes. Mycologia. 2015;107(6):1105–1119. doi: 10.3852/15-027. [DOI] [PubMed] [Google Scholar]
- 51.Rytioja J, et al. Plant-polysaccharide-degrading enzymes from basidiomycetes. Microbiol. Mol. Biol. Rev. 2014;78(4):614–649. doi: 10.1128/MMBR.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lundell TK, Mäkelä MR, Hildén K. Lignin-modifying enzymes in filamentous basidiomycetes–ecological, functional and phylogenetic review. J. Basic Microbiol. 2010;50(1):5–20. doi: 10.1002/jobm.200900338. [DOI] [PubMed] [Google Scholar]
- 53.Yadav V, Yadav PK, Yadav S, Yadav KDS. α-L-Rhamnosidase: a review. Process Biochem. 2010;45(8):1226–1235. doi: 10.1016/j.procbio.2010.05.025. [DOI] [Google Scholar]
- 54.Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583(24):3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 55.Haider S, Pal R. Integrated analysis of transcriptomic and proteomic data. Curr Genomics. 2013;14(2):91–110. doi: 10.2174/1389202911314020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kersten P, Cullen D. Copper radical oxidases and related extracellular oxidoreductases of wood-decay Agaricomycetes. Fungal Genet. Biol. 2014;72:124–130. doi: 10.1016/j.fgb.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 57.Kumar SS, Phale PS, Durani S, Wangikar PP. Combined sequence and structure analysis of the fungal laccase family. Biotech. Bioeng. 2003;83(4):386–394. doi: 10.1002/bit.10681. [DOI] [PubMed] [Google Scholar]
- 58.Bento I, et al. Mechanisms underlying dioxygen reduction in laccases. Structural and modelling studies focusing on proton transfer. BMC Struct. Biol. 2010;10(1):28. doi: 10.1186/1472-6807-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arzola KG, et al. Early attack and subsequent changes produced in an industrial lignin by a fungal laccase and a laccase-mediator system: an analytical approach. Appl. Microbiol. Biotechnol. 2006;73(1):141–150. doi: 10.1007/s00253-006-0630-0. [DOI] [PubMed] [Google Scholar]
- 60.Moilanen U, Kellock M, Várnai A, Andberg M, Viikari L. Mechanisms of laccase-mediator treatments improving the enzymatic hydrolysis of pre-treated spruce. Biotechnol. Biofuels. 2014;7(1):177. doi: 10.1186/s13068-014-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oliva-Taravilla A, Tomás-Pejó E, Demuez M, González-Fernández C, Ballesteros M. Phenols and lignin: key players in reducing enzymatic hydrolysis yields of steam-pretreated biomass in presence of laccase. J. Biol. 2016;218:94–101. doi: 10.1016/j.jbiotec.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 62.Müller G, Várnai A, Johansen KS, Eijsink VG, Horn SJ. Harnessing the potential of LPMO-containing cellulase cocktails poses new demands on processing conditions. Biotechnol. Biofuels. 2015;8(1):187. doi: 10.1186/s13068-015-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muraleedharan MN, et al. Effect of lignin fractions isolated from different biomass sources on cellulose oxidation by fungal lytic polysaccharide monooxygenases. Biotechnol. Biofuels. 2018;11(1):296. doi: 10.1186/s13068-018-1294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors promise the availability of supporting data.