Abstract
To what extent multi-omic techniques could reflect in situ microbial process rates remains unclear, especially for highly diverse habitats like soils. Here, we performed microcosm incubations using sandy soil from an agricultural site in Midwest USA. Microcosms amended with isotopically labeled ammonium and urea to simulate a fertilization event showed nitrification (up to 4.1 ± 0.87 µg N-NO3− g−1 dry soil d−1) and accumulation of N2O after 192 hours of incubation. Nitrification activity (NH4+ → NH2OH → NO → NO2- → NO3−) was accompanied by a 6-fold increase in relative expression of the 16S rRNA gene (RNA/DNA) between 10 and 192 hours of incubation for ammonia-oxidizing bacteria Nitrosomonas and Nitrosospira, unlike archaea and comammox bacteria, which showed stable gene expression. A strong relationship between nitrification activity and betaproteobacterial ammonia monooxygenase and nitrite oxidoreductase transcript abundances revealed that mRNA quantitatively reflected measured activity and was generally more sensitive than DNA under these conditions. Although peptides related to housekeeping proteins from nitrite-oxidizing microorganisms were detected, their abundance was not significantly correlated with activity, revealing that meta-proteomics provided only a qualitative assessment of activity. Altogether, these findings underscore the strengths and limitations of multi-omic approaches for assessing diverse microbial communities in soils and provide new insights into nitrification.
Subject terms: Element cycles, Soil microbiology, Metagenomics
Introduction
Even though the central role of microbes in the cycling of nitrogen is recognized, the dynamics and controls of the interrelated microbial nitrogen pathways in agricultural soils are not completely understood. This knowledge gap limits the development of accurate, predictive models of nitrogen flux that encompass the role of microbes in the generation and consumption of nitrogen substrates, as well as the emission of greenhouse gases, including nitrous oxide (N2O)1. In agricultural soils receiving large inputs of nitrogen fertilizer, nitrifiers such as ammonia-oxidizing bacteria (AOB), ammonia-oxidizing archaea (AOA) and nitrite-oxidizing bacteria (NOB) collectively are responsible for the conversion of ammonium to nitrate2,3. In addition, the recent discovery of Nitrospira bacteria capable of complete oxidation of ammonia to nitrate (comammox) has suggested that the process of nitrification in natural environments might be carried out by a single taxon4,5. Under anoxic conditions (e.g., water saturated soils), nitrate (NO3−) can be reduced to gaseous forms such as dinitrogen (N2), nitric oxide (NO) or N2O by denitrifying organisms and consequently be lost to the atmosphere. It has also been reported that nitrification is a major N2O source under low oxygen concentrations3,6, although detailed mechanistic understanding is lacking7. Despite the importance of nitrification in the generation of N2O and NO3−, the relative contributions of AOA, AOB and NOB populations in this process, especially during soil fertilization events, are still subjects of intensive research8, and the relative contribution of the comammox bacteria to the process is not clear9. Thus, understanding the niche specialization and diversity of nitrifiers in terrestrial ecosystems is essential for better prediction of the contributions of these microbial taxa to the nitrogen cycle and the modeling of the corresponding activities and products. High-throughput sequencing and proteomic approaches can characterize the diversity of nitrogen pathways in the environment. However, to what extent these omic approaches could also reflect microbial activity is less clear.
Although DNA, RNA, and protein abundances all reflect microbial potential and responses to environmental changes and thus, could be used to study nitrogen cycling in soils, each measurement generally offers different types of information. For instance, metagenomics (DNA level) offers a comprehensive overview of the functional potential of microbial communities but does not reflect active community members or functions. Short-term microbial responses to external changes (e.g., nitrogen addition) can be tracked by analyzing the actively expressed genes (i.e., metatranscriptomics). For instance, the relationship between measured nitrification processes and the ammonia monooxygenase (amoA) transcripts have revealed differences between archaeal and bacterial activity in acidic soils10. Proteomics provides a third level of molecular information by reflecting synthesized enzymes that catalyze reactions. Although proteomics has been applied to only a limited number of natural microbial communities, the results have provided new insights about metabolic pathways and interdependencies among microbial groups [reviewed before11]. However, most of these advances are hindered by the intrinsic complexity of soils. For instance, soil samples are challenging to analyze not only because of their heterogeneous structure and chemical composition (e.g., low quality and quantity of extracted nucleic acids), but also because of the highly diverse microbial communities and slow growth kinetics. Nonetheless, recent advances in metagenomic and metaproteomic techniques as well as integration with stable isotope probing (SIP) have helped elucidate the role of previously elusive keystone microbial populations12. For instance, the combination of multi-omic datasets provided new insights into diversity and gene potential of microbial communities of permafrost ecosystems, but the datasets were less predictive of measured process rates13.
Toward closing the abovementioned knowledge gaps, we examined nitrogen-amended sandy soils obtained from a site with a history of agricultural management and application of synthetic nitrogen fertilizer. A prior year-round characterization of field samples from the same agricultural site revealed increased abundance of novel Thaumarchaeota and comammox nitrifiers, but the findings were limited to metagenomics14. Here, our goal was to assess the strengths and limitations of multi-omics in detecting microbial activity by correlating measurements of DNA, RNA, and protein abundances with measured rates of nitrate formation and N2O production in soils incubated under controlled conditions in the laboratory. The results revealed that metatranscriptomic data best reflected the measured nitrification rates under the tested experimental conditions and provided novel insights about nitrifier gene expression dynamics after a simulated nitrogen fertilization event.
Results
Nitrification activity in soil microcosms
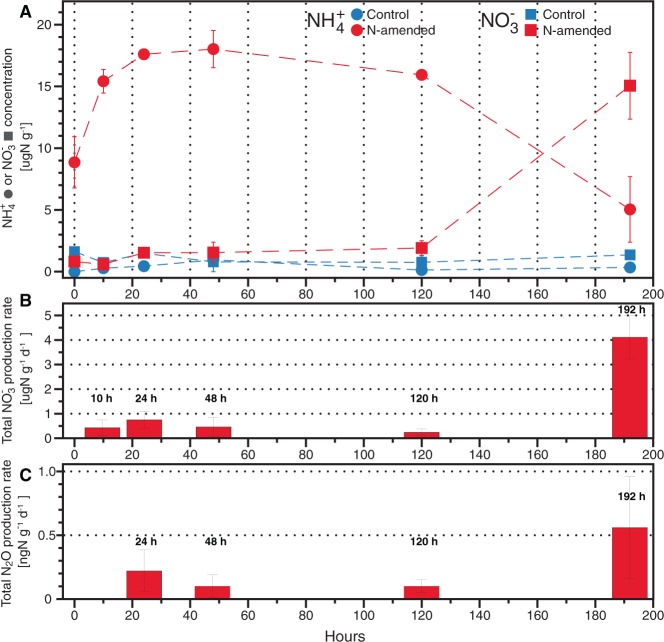
Patterns in nitrification rates were consistent with NO3− formation and NH4+ disappearance during an eight-day period following the amendment of soil microcosms with an equimolar nitrogen mixture of NH4+ and urea, representative of fertilizer application in the field. Nitrate was not supplied as part of the amendment because it could represent both product and substrate for different nitrogen cycle pathways. Based on the NH4+ concentration patterns, urea quickly hydrolyzed to release NH4+ within the first two days of incubation (Fig. 1a). Specifically, the NH4+ concentrations peaked at 48 hours of incubation (avg = 18.02 and SD = ± 1.5 µgN-NH4+ g−1 dry soil) from urea hydrolysis, and decreased to 5.4 ± 2.5 µgN-NH4+ g−1 dry soil by 192 hours of incubation. Consistent with the disappearance of NH4+, the NO3− concentrations increased from an initial value of 0.81 ± 0.28 µgN-NO3- g−1 dry soil to 1.91 ± 0.5 µgN-NO3- g−1 dry soil at 120 hours of incubation, and then increased at a faster rate to 15.06 ± 2.7 µgN-NO3− g−1 dry soil at 192 hours of incubation (Fig. 1a). Measured nitrate accumulation and nitrification rates confirm that nitrification activity was relatively low (<1 µgN-NO3 g−1 dry soil) for the first 120 hours of incubation and increased five to eight days after the addition of NH4+ and urea mixtures, reaching an average nitrification rate of 4.1 ± 0.87 µg N-NO3− g−1 dry soil d−1 (n = 6) after 192 hours of the incubation (Fig. 1b). As a result of nitrification activity, pH values decreased across replicated nitrogen-amended microcosms during the incubation (Table S1). In order to examine the generation of N2O possibly generated as a by-product of oxidation reactions during the incubation, we measured the production of N2O in nitrogen-amended incubations. Net N2O production rates in the incubation headspace increased from 0.22 ± 0.16 ng N-N2O g−1 dry soil d−1 after 24 hours to 0.56 ± 0.40 ng N-N2O g−1 dry soil d−1 at the end of the incubations (Fig. 1c). Control microcosms receiving only irrigation water (i.e., no nitrogen amendment) did not show net NH4+oxidation.
Figure 1.
Nitrogen pools and fluxes in soil incubations amended with NH4+ and urea. Mean NH4+ and NO3− concentrations (A), total NO3− production rate (B), and total N2O production rate (C) for the nitrogen-amended microcosms at each incubation time point. Error bars represent the standard deviation from replicate samples (n = 6 for nitrogen-amended and n = 3 for control).
To evaluate possible differences between the use of NH4+ or urea in nitrifying activity, we examined patterns in 15N-NO3− production rates in microcosms that received 15N-labeled NH4+ versus 15N-labeled urea by tracing the fate of 15 N label in the fertilized treatments. In general, 15NO3− production was similar between 15NH4+ and 15N-urea microcosms, although rates were higher after 10 hours in 15NH4+ microcosms and 48 hours in 15N-urea microcosms (two tailed t-test, P < 0.01), but converged thereafter (Fig. S1). By the end of the incubations, approximately half of the added 15N was converted to 15N-NO3− (49–55% for both labelled solutions/treatments), only a small fraction was converted to 15N-N2O (0.006–0.01%), and a large percentage remained as 15N-NH4+ (19 ± 11%) (Fig. S1). The remaining added nitrogen was presumably rapidly lost as N gas, assimilated into microbial biomass, or adsorbed to soil particles, all well-known nitrogen sinks of 15N tracer studies in soils15,16.
Taxonomy of microbial soil populations based on 16S rRNA gene sequences
The taxonomic composition and abundances of the main microbial groups determined from recovered 16S rRNA gene sequences (DNA level) from nitrogen-amended incubations, were generally stable during incubations (Fig. S2). In agreement with our previous results based on field samples from the same agricultural site14, bacterial and archaeal groups associated with nitrification were comparatively less abundant than (non-nitrifer) abundant bacterial taxa (e.g., Actinobacteria, Flavobacteria, and Acidobacteria) in both DNA and cDNA datasets. For instance, known AOB and NOB genera such as Nitrosomonas and Nitrospira had average relative abundances of 0.01% and 1.6% of the total populations in the metagenomes from incubated soils. Additionally, the relative abundances of the AOA genera related to Nitrososphaera and Nitrosopumilus were 0.9% and 0.3% in the microcosm metagenomes. We note however that due to the high coverage obtained by our datasets, the 16S rRNA gene (and protein-coding genes; see below) of these nitrifiers were adequately sampled. Notably, the 16S rRNA gene transcript abundances for AOB conspicuously increased during the incubation period (Fig. S3). In fact, relative 16 S rRNA gene expression ratios (cDNA/DNA) for AOB and NOB belonging to Nitrosospira, Nitrosomonas and Nitrospira increased 3-,6-, and 14-fold between 10 and 192 hours. In contrast, the 16S rRNA gene expression ratios for the archaeal groups Nitrososphaera and Nitrosopumilus were stable during the same incubation period, although with ~50% increase in relative expression at 48 h of incubation (Fig. S3). A description of detected functions in metatranscriptomes is available in the Supplementary Material. We sought to examine next the dynamics of individual populations/genomes.
Individual populations from microcosm metagenomes
Given that none of the recovered metagenome-assembled genomes (MAGs) represented AOA, AOB, NOB, or comammox populations, we included MAGs obtained from a previous analysis of field samples from the same site (Havana county, Illinois, USA) and depth as the soil used in the soil microcosms in the present study14. MAGs potentially involved in nitrification processes were likely missed in the microcosm metagenomes due to comparatively lower sequencing effort than the field samples and the relatively low abundances of these groups. The MAGs (designated with the letter F at the end of their name for Field metagenomes) consisted of two complete ammonia oxidizer (comammox) Nitrospira MAGs (MAG021F and MAG017F) and five ammonia-oxidizing archaea MAGs representing the Thaumarchaeota lineages I.1b (MAG032F and MAG019F) and I.1a (MAG004F, MAG109F, and MAG001F) (Fig. S4b). The genetic relatedness of the field MAGs compared to the populations found in the microcosm metagenomes based on the identity of reads17 from latter metagenomes recruited against these MAGs was high (ANIr avg = 99.3, SD = 0.27). These results confirmed that the MAGs from the field metagenomes well represented the populations in the microcosms (Fig. S4c). These seven nitrifier MAGs recruited between 0.36% to 0.77% of the microcosm metagenomic reads in each dataset, with an average of 0.55%, and between 0.46% and 0.92% of the metatranscriptomic libraries, which was comparable to the 16S rRNA gene-based abundances mentioned above. This level of relative abundance was adequate for assessing the gene and transcript dynamics of the MAGs since it provided, on average, more than 7.5X coverage of the corresponding gene sequences (i.e., the time each base is covered by transcriptomic reads). Additional statistics for metagenomes and metatranscriptomes datasets, including sequencing effort, coverage obtained, and statistics of all recovered MAGs are available in the Supplementary Material and Tables S2–5.
Relative expression values of MAGs (measured as transcriptomic reads per kilobase million, RPKM) were used as a proxy for comparing the response and metabolic activity among nitrifying bacteria and archaea between samples (incubation time points). Even though expression values for most nitrifying MAGs belonging to Nitrospira and Thaumarchaeota were stable and relatively low, AOA MAG004F, MAG019F and comammox MAG017F had, on average, the highest expression values throughout the incubations (Fig. S4a). For instance, the increase in expression values for AOA belonging to the I.1b clade, MAG004F and MAG032F, were 39% and 50% after 48 hours of incubation (compared to expression levels at 10 hours incubation), respectively. In contrast, gene expression of comammox MAG017F increased by 59% after 120 and 192 hours of incubation (Fig. S4a). Note that AOB and NOB were not included in the RPKM analysis due to lack of recovered MAGs representing these populations (see above). Given the technical limitations in recovering high quality nitrifier MAGs, a gene-based approach was also employed in order to assess changes in gene fragments (DNA) and transcript (cDNA) abundances of genes involved in nitrification activity for AOB and NOB nitrifiers (see below).
Quantification of nitrification genes in microcosms
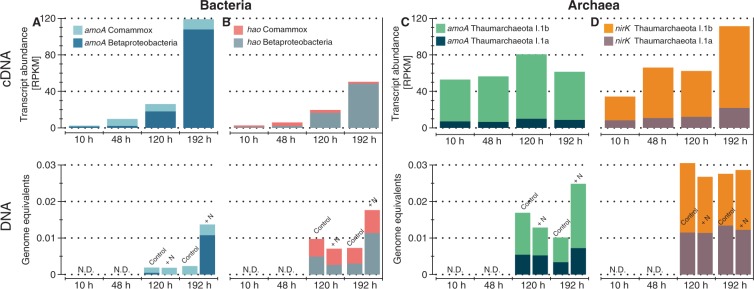
To further explore the microbial nitrification processes in incubated soils at the gene level, we specifically quantified gene fragments and transcripts directly involved in nitrification reactions using ROCker, a tool developed to accurately differentiate between reads encoding different gene families or (distinct) phylogenetic clades within a gene family18 (see Methods for details). Relative expression values belonging to the gene encoding urease subunit c (ureC) were relatively high after 48 and 192 h of incubation but the average abundances were lower compared to other nitrification genes (e.g., amoA; see Fig. S5a). For instance, the relative expression of the bacterial gene encoding ammonia monooxygenase subunit alpha (amoA) was 53.5-fold higher compared to the expression values at 10 h of incubation. Most of the detected amoA transcripts (cDNA) were phylogenetically affiliated with Betaproteobacteria and corresponded to up to 90% of the total detected bacterial amoA transcripts at 192 hours of incubation (Fig. 2a). Notably, regression analysis between amoA transcript abundances and measured nitrate concentrations at 10, 48, 120 and 192 h of incubation had a strong relationship (Coefficient of determination, r2 > 0.97, Fig. S5c). The r2 values determined for transcripts belonging to other nitrification genes during the same incubation points were lower and ranged from ~0 to 0.7. Similarly, low r2 values were observed when using the DNA level (i.e., metagenomic reads) for other nitrification genes. After 192 hours of incubation, betaproteobacterial amoA transcript abundance increased 66-fold, whereas comammox amoA gene transcripts remained stable (Fig. 2a). The latter results indicated that the comammox amoA may be more abundant under field conditions but betaproteobacterial amoA might show a faster response upon ammonia addition, which was also consistent with a previous study14.
Figure 2.
Nitrification genes in incubated soils. Transcript abundance (RPKM, top panel) and gene abundance (genome equivalent, lower panel) for bacterial amoA (A), hao (B), thaumarchaeotal amoA (C), and nirK (D) are shown. Transcript or DNA abundances for specific clades are showed in different colors.
Although the relative expression for the archaeal amoA was more stable throughout the incubation compared to its betaproteobacterial counterparts, maximum expression was reached after 120 hours of incubation, suggesting that archaeal AmoA activity temporarily increased at later time points during the incubation. Archaeal amoA transcripts belonging to the group I.1b were ~7 times more abundant than their I.1a counterpart across the incubations (Fig. 2c). Similar to amoA patterns, the relative expression for the betaproteobacterial hydroxylamine oxidoreductase (haoA; NH2OH → NO2−) steadily increased during the incubations, whereas comammox haoA transcripts were stable throughout the incubations (Fig. 2b). Expression values for the nitrite oxidoreductase subunit alpha (nxrA; NO2− → NO3−) had a 12.4-fold increase compared to the 10-hour time point, consistent with the patterns observed for the previous nitrification genes and NO3− accumulation (Fig. S5a). Unexpectedly, expression values for nirK (NO2− → NO) affiliated to Thaumarchaeota were higher compared to nirK transcripts assigned to the Nitrospira clade (Fig. 2d). In fact, Thaumarchaeota nirK transcripts had a 3.4-fold increase after 192 hours of incubation relative to earlier sampling points, indicating that Thaumarchaeota might have been more active in the reduction of nitrite compared to other steps of nitrification. Specifically, there was a 3.4-fold increase for clade I.1b nirK transcripts during the 10 to 192 hours of incubation period, whereas the abundance of transcripts from clade I.1a were stable throughout the incubations (Fig. 2d). We were also able to assign several of the gene sequences used above to individual MAGs using a phylogenetic approach. Consistent results with those reported above were observed when examining MAG-specific expression patterns. For instance, a high fraction of the total comammox hao and thaumarchaeotal nirk transcripts, ranging from ~62 to 82%, were assigned to MAGs. However, the majority of detected amoA and hao transcripts were derived from soil betaproteobacterial AOB communities (Fig. 2a) not represented by the recovered MAGs (i.e., these genes were part of the un-binned soil predicted sequences). Thus, a MAG-centered approach would have missed the activity of these communities since no betaproteobacterial AOB MAGs were recovered.
In summary, the metatranscriptomic profiles suggested that AOB, but not comammox, responded rapidly to the nitrogen amendment, whereas AOA transcriptome shifts were less pronounced. The response of AOB, and to a lesser extent AOA, was also reflected at the DNA level, albeit with a substantial time delay (Fig. 2, lower panels). For instance, shifts were observed early at the transcript level while at the DNA level changes were mostly evident 192 hours after the start of incubation (Fig. 2a,b). One exception was the urease gene (ureC) that showed high abundance at all incubation time points although its transcription response was lower compared to nitrification genes (Fig. S5a,b). Nonetheless, these results were consistent across the individual nitrification steps and indicated that at least the AOB nitrifiers grew in response to nitrogen addition.
A proteomic perspective in soil microcosms
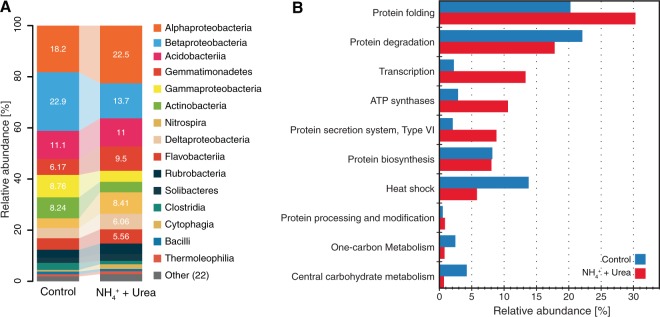
A metaproteomic analysis of the control and nitrogen-amended microcosms at 192 hours of incubation detected a total of 2,892 and 1,629 non-redundant peptides, respectively. A total of 844 peptides were shared among control and nitrogen-amended incubations, whereas 2,048 and 785 were exclusively present in each microcosm, respectively. Most of peptides detected in control and nitrogen-amended incubations matched protein sequences predicted from metagenomic assemblies (89.4% and 88.2%, respectively) and the remaining fraction matched reference proteomes (Table S6). The top 20 most abundant proteins in control and nitrogen-amended treatment microcosms were related to housekeeping and transport proteins whereas in the latter incubation, oxidoreductases for small carbon and alcohol molecules and ATP synthesis were among the most abundant proteins detected (Table S7). These results were also consistent for some of the highly expressed genes in metatranscriptomes related to protein and RNA metabolism at 192 h of incubation (See Supplementary Material). The taxonomic affiliation, at the class level, for the most relatively abundant annotated peptides belonged to Alphaproteobacteria, Betaprotebacteria, and Acidobacteria in control and nitrogen-amended incubations. Although there were major differences in abundances for groups such as Betaproteobacteria (40% decrease) and Gammaproteobacteria (50% decrease) (Fig. 3a), higher abundances were detected for less abundant groups commonly associated with the nitrification process. For instance, close to a 2.2-fold increased abundance for nitrogen-amended incubations were detected for peptides belonging to Nitrospira. Detected peptides related to folding and synthesis were the most abundant and had similar abundances in the control and nitrogen-amended microcosms after 192 hours of incubation. However, the relative abundance was higher for annotated functions related to ATP synthases and transcription categories in the nitrogen-amended samples relative to the control, presumably as a consequence of a higher microbial activity generated after the nitrogen input. On the other hand, heat-shock and degradation proteins were more abundant in the control incubation, probably reflecting a more prevailing dormant state for the microbial communities in these samples (Fig. 3b). However, unlike the metagenomic and metatranscriptomic datasets, only some peptides involved in nitrification were identified using metaproteomics. For instance, the detected peptides directly involved in nitrification pathways corresponded to the nitrite oxidoreductase subunit B (NxrB), which had a 31.3% abundance increase in the nitrogen-amended samples compared to the control.
Figure 3.
Metaproteomic analyses of incubated soils at 192 hours of incubation. Panel (A) shows taxonomic affiliation (class) and abundance (average spectral counts) for peptides detected in control and N-amended incubations. Panel (B) shows summarized functional annotation of detected peptides using SEED functional categories.
Discussion
Using multi-omic approaches for examining process rates
Measuring nitrification rates in incubated soils allowed us to evaluate the explanatory and predictive power of omic approaches in a highly diverse soil system. Although all three omic approaches revealed increased abundance for target genes, transcripts, and proteins related to nitrification pathways, they differed in temporal resolution and detection capabilities. For instance, the strongest agreement to the observed nitrification processes (i.e., ammonia or nitrite oxidation) was for the metatranscriptomic data within the first days of incubations (e.g., Fig. 2a,b,d), whereas metagenomes lagged behind and only reflected the ongoing nitrification process after 192 hours of incubation (e.g., lower panels Fig. 2a,b). These data were presumably attributed to the fact that growth (e.g., at least a few replication cycles) should occur before metagenomics can reveal shifts in relative abundance over time. Note that microbial growth was not explicitly measured by our study to further corroborate the above interpretation. Therefore, metagenomics could also reflect ongoing microbial processes if the processes are ongoing for a period of time and are coupled with the growth of the corresponding organisms, even in soils. In contrast, if the goal is to see immediate responses to a perturbation or if the perturbation is short-lived (e.g., lasting a few hours), metatranscriptomic data will be preferable. We also observed that metatranscriptomes were as good as metagenomics, if not better, at reflecting microbial activity for nitrification processes even at later incubation time points. In contrast, the metaproteomes offered, at most, a qualitative glimpse at nitrification processes and were less definitive in identifying common nitrification markers. The latter was largely attributable to the computational challenges associated with proteomic data such as high peptide redundancy and the requirement of high-quality assemblies which are still challenging for highly complex soil metagenomes. Furthermore, many challenges remain for efficient extraction of membrane proteins from low abundance organisms such as nitrifiers. Ultimately, these technical limitations were reflected in a lower number of detected proteins compared to the number of metagenomic and metatranscriptomic reads recovered that encoded the proteins of interest in our datasets.
While shifts in 16S rRNA gene ratios (cDNA/DNA) were relatively small for AOA, the 16S rRNA and functional gene ratio shifts (e.g., amoA) for AOB/NOB were much more pronounced throughout the incubations (Fig. S3). These results might reflect an active and growing state for AOB/NOB and mostly active AOA communities as observed before for agricultural soil microcosms19. The differences observed between (high) target gene abundances and (low) 16S rRNA gene ratios for AOA could reflect a limitation of the latter approach when used as a proxy for assessing microbial activity (e.g., inconsistent correlation between rRNA and activity)20. Thus, targeting specific functional genes in AOA could offer an alternative approach for tracking their activity in metatranscriptomes. However, more frequent sampling and incubations under different physicochemical conditions will be required for more robust conclusions to emerge on the exact relationship(s) between rRNA marker abundances and process rates.
Future incubation studies could increase the number of samples analyzed and shed light on the intrinsic differences between nitrifier (and denitrifier) communities by testing variables such as oxygen availability (i.e., water saturation) and different agricultural soil types. For instance, the incubation conditions used in our study deliberately promoted nitrification over denitrification processes and as a result, the N2O production was detected due to the former process. Consequently, nitric oxide (e.g., norB) and nitrous oxide reductases (e.g., nosZ) transcripts, which are responsible for N2O production and consumption during denitrification, respectively, were not detected in our metatranscriptomes datasets (i.e., abundance below detection limit). Also, the use of nitrification inhibitors could help to elucidate the origin of the measured N2O whether production was biotic or abiotic, for which our data are limited in predicting.
Previous studies have also found metatranscriptomic approaches to be better predictors of measured microbial activity21 in controlled laboratory systems amended with exogenous organic compounds or natural communities, but have been more limited in providing insights into the whole-microbial community response to the amendment. For instance, a high correlation between environmental conditions and the expression of adaptation mechanisms (transcription level) was observed in acid mine drainage communities22. On the other hand, multi-omic approaches applied to permafrost microbial communities were less predictive of biogeochemical processes such as methane oxidation but provided a higher overview of active (transcription and proteomic levels) microbial members encoding these pathways13. Similar approaches led to the discovery of unexcepted microbial pathways such as the methanogenesis in oxygenated soils23. Here, our metatranscriptomic results were effective in reflecting measured nitrification even though our observations were based on single samples at different time points. Nonetheless, temporal data typically require less replication for statistically robust results and likely offset the experimental noise between sampling time points compared to cross-sectional data. Hence, our main conclusions were likely not affected by the relatively small number of samples analyzed. Taken all together, our analysis showed that metatranscriptomics could reliably and quantitively reflect undergoing microbial processes even in the soil microcosms which represent an intrinsically challenging matrix for RNA work compared to other ecosystems (e.g., aquatic environments24). It should be noted that different soil types may not be as amenable to perform RNA extractions (e.g., not enough high-quality RNA). Hence, metatranscriptomics may continue to be a challenging task for certain soil types and conditions.
New insights into nitrification pathways
In terms of the ecological adaptation of the nitrifiers analyzed here, the Havana agricultural site has had a long history of cyclical seasonal inputs (e.g., fertilizers) that have shaped the structure of microbial communities differently between soil layers. The AOA and AOB communities in the Havana site have legacy establishments at the 20–30 cm soil depth and are under relatively stable environmental conditions compared to the top soil layer that receives most of the nitrogen fertilizer14. Thus, nitrogen amendments tested in our experiment and experimental conditions might not represent closely the conditions usually experienced by the examined, deep-layer AOA and AOB communities (20–30 cm). The rapid response of AOB observed here might be a reflection of physiological adaptations of AOB to thrive under high nitrogen content as reported previously19. In contrast, the low response observed for comammox and some AOA communities might reflect their limited physiological capabilities to respond to high nitrogen concentrations4,5 that were assayed in our experimental setup.
Assessing the individual gene level, as opposed to whole genome transcript level, provided more robust results for relating population response to measured nitrification reactions, presumably due to higher sequence coverage (less noise). Our results showed that even though betaproteobacterial amoA transcripts responded to the addition of ammonium and urea, the relative abundance of comammox amoA transcripts was stable (i.e., not responding to the nitrogen amendment), although comammox populations were relatively more abundant than AOB in the microcosms. This observation is consistent with previous metagenomic results from the same agricultural soil, where comammox amoA genes and the organisms encoding these genes represented the highest fraction of nitrifying bacteria14. The differences between measured genes and transcripts indicated that the incubation conditions favored the activity of Betaproteobacteria over comammox nitrifying bacteria, suggesting ecophysiological differences among these taxa for the incubation conditions or added substrates compared to field conditions.
The sequencing of isolates and environmental AOA genomes has shown that even though they encode an AmoA protein, they lack a canonical hydroxylamine oxidation pathway25. Previous studies have proposed that nitric oxide is essential for hydroxylamine oxidation to nitrite in archaea26. The proposed mechanism involves oxidation of ammonium to hydroxylamine followed by oxidation to nitrite catalyzed by a putative Cu-protein that uses nitric oxide as co-reactant for the oxidation of hydroxylamine. Interestingly, nitric oxide has been proposed to be derived from the activity of the NirK enzyme present in most AOA sequenced genomes. Our results show that unlike AOA amoA or bacterial nirK transcripts, Thaumarchaeota nirK transcripts increased in abundance in the incubated soils, supporting the abovementioned hypothesis. Therefore, even though AOA amoA transcripts did not show clear changes in abundances compared to their bacterial counterparts, these results agree with the previous observations in marine and terrestrial systems26,27, and likely denote an unaccounted role for Thaumarchaeota nirK in nitrification in agricultural soils.
Challenges and opportunities for multi-omic studies
Here we analyzed total RNA extractions from soils where ribosomal RNA transcripts represented 94–98% of the total sample, limiting our study to a small fraction of transcripts related to functional genes. Current experimental approaches offer successful rRNA depletion for environmental samples, when RNA yields are not limiting28, which was not the case in our study. Additionally, all the results represented here provided only relative abundances for measured microbial markers. For instance, approaches such as qPCR or internal standards spiked into the DNA or cDNA library for sequencing28 can strengthen and provide improved quantification compared to those presented here.
The relative low correspondence between detected peptides (metaproteomics) and active microbial processes compared to the DNA or RNA levels was likely due, at least partially, to the low biomass of nitrifiers and extraction biases due to the complexity of soil matrices as well as limited extraction of membrane proteins, such AmoA, as suggested previously29. Alternative proteomic approaches focused on a preselected set of proteins (i.e., selected reaction monitoring or target proteomics) could be used to explore low abundance nitrification proteins. For instance, targeted proteomic approaches have been used to study proteins in low abundance involved in bioremediation pathways in highly-diverse environmental systems30. Therefore, targeted proteomics might offer new opportunities for researchers interested in detecting low-abundance peptides and prediction of process rates in complex samples31. Finally, abundant peptides related to C1 dehydrogenases that were detected in our metaproteomic profiles (Table S7) were likely not related to denitrification as they did not respond to the nitrogen amendment and were detected in both control and N-amended incubations, and our microcosms were kept under aerobic (non-denitrifying) conditions. Additionally, the annotations of these soil predicted protein sequences might be limited due to generally low identity matches to experimentally verified dehydrogenases.
Conclusions
The analyses of different omic levels obtained from the incubations showed a high correspondence between nitrification gene markers/transcripts and nitrification process rates. The gene fragments and transcripts were mostly affiliated to novel nitrifier populations similar to those previously described in field soil metagenomes from the same agricultural site14. Therefore, the gene and genome sequences reported here could facilitate future investigations of nitrogen cycling in agricultural fields; for instance, by applying qPCR assay targeting the key taxa and biomarker genes and transcripts. The combination of metagenomic and metatranscriptomic approaches used in our study provided a promising strategy for examining microbial activity in agricultural soil environments. Therefore, the findings presented here highlighted the potential of omics data to serve as reliable proxies for examining microbial processes in situ, especially in soils, which has been proven to be among the most challenging tasks for environmental studies.
Methods
Soil sampling
Our study was focused on an agricultural plot located in the Havana County, Illinois, USA (lat 40.296, long 89.944; elevation, 150 m). The site is representative of the US Midwest and has a long history of conventionally managed corn and soybean crop rotation. In October 2014, we collected ~2 kg of bulk soil from a 20–30 cm soil depth as previous results have shown significant presence of ammonia-oxidizing microorganisms in this layer14.
Soil incubations, gas and chemical analyses
Soil microcosms were established in triplicate, using ~120 g of soil (~8% moisture content) in 500 ml gas-tight canning jars equipped with gas sampling ports, and were sampled at six time points (0, 10, 24, 48, 120, and 192 hours). To simulate a fertilization event in microcosms, 6 ml of 40 mM NH4Cl and 20 mM urea (80 mM N) in irrigation water was added to two separate batches of 400 g of soil (Final concentration = 1.2 µmoles-N/g or 18.3 µg-N/g dry weight). Gross rates of nitrification stimulated by the NH4+ versus the urea fertilizer were estimated using the 15N tracer approach with two separate stable isotope treatments (n = 6 per treatment): a 50% 15N-NH4Cl and 50% 14N-NH4Cl, 99.7% 14N-NH2CONH2 treatment versus a 50% 15N-NH2CONH2 and 50% 14N- NH2CONH2, 100% 14N-NH4-Cl treatment. After vigorously mixing, 120 g were dispensed into three separate microcosm jars and incubated in a dark growth chamber with diurnal temperature fluctuation of 22–24 °C as observed in Havana field soil at 20–30 cm during the spring fertilization period (early June). Triplicate microcosms each receiving 6 ml of filtered irrigation water (no nitrogen amendment) served as controls. Individual incubations (whole jar) were sacrificed at each sampling point and all soil (~120 gr) was used for chemical, nucleic acid and protein analyses. After each sampling point, headspace gas was collected from closed jars and the N2O concentration was measured on a Shimadzu GC-2014 gas chromatograph (Columbia, MD) equipped with an electron capture detector. Jars were opened for soil sampling and to reestablish equilibration with atmospheric air before being resealed until the next sampling. Ammonium and nitrate in soil subsamples (20 g) were extracted in 2 M KCL and the concentrations were determined using colorimetric analysis on a flow injection auto-analyzer (Lachat Instruments, Milwaukee, WI)32. Soil pH (1:1 in water) and gravimetric water content were measured at each time point (Table S1). 15N isotopic composition of N2O in collected jar headspace samples was determined using an IsoPrime 100 isotope ratio mass spectrometer interfaced with an IsoPrime trace gas analyzer (Cheadle Hulme, UK) at the University of Illinois at Urbana-Champaign. The 15N atom % enrichment of the NO3− pool was determined using acid trap diffusion33 and analysis of the diffusion disks on a Vario Micro Cube elemental analyzer (Elementar, Hanau, Germany) interfaced to an IsoPrime 100 continuous flow isotope ratio mass spectrometer (Cheadle Hulme, UK). 15NO3− and 15N2O production rates were calculated from the change in 15NO3− and 15N2O concentrations, respectively, from one time point to the following sampling time point. NO3− and N2O production rates were estimated from the 15NO3− and 15N2O production rates based on the mean 15N excess atom % of the NH4+ source pool as described before for N2O production rates34. No inhibitors of nitrogen cycle pathways were used in the incubations.
Nucleic acid extractions
DNA and RNA were extracted independently but from the same soil of each incubation at each time point. DNA was extracted from ~0.5 g of soil from individual incubations (i.e., replicates were not mixed) using a modified phenol-chloroform and purification protocol as previously described35. For RNA extraction, 2 gr of soil was preserved in LifeGuard (MoBio) and stored at − 80 °C. A modified protocol derived from the PowerMax Soil DNA kit for extracting RNA was used for total RNA extractions (MoBio). TURBO DNAse (Ambion) was used to remove DNA according to the recommendations of the manufacturer. Nucleic acid extracts were quantified using Quant-it ds DNA HS and HS RNA assays (Invitrogen) according to the instructions of the manufacturer. RNA quality was assessed using Agilent RNA 6000 pico kit (Agilent Technologies) and samples having RNA integrity number (RIN) above 7 were used. Additional details about nucleic acid sequencing are available in Supplementary Material.
Short-read analyses
Metagenomic and metatranscriptomic raw reads (FASTQ) for sequenced samples were trimmed using SolexaQA36 using a Phred score cutoff of 20 and minimum fragment length of 50 bp. Short-reads derived from metatranscriptomes were merged using PEAR using default parameters37. Average coverage for each sequenced metagenome was determined by Nonpareil38 using default settings except that 2,000 reads were used as query (-X option). Short-read sequences encoding 16S rRNA gene fragments were extracted from each metagenome and metatranscriptome by SortMeRNA39 and their taxonomy was assigned using RDP classifier (cutoff 50)40.
To identify and quantify reads encoding specific protein sequences of interest, we used the previously published protein sequences as references14,41. Independent ROCker18 models (length = 125 bp) were subsequently built based on these reference protein sequences with the exception of NarG and NxrA, where the sequences were combined into a single model. Trimmed short-reads from soil metagenomes were used as query for BLASTx searches (e-value 0.01) against the latter protein databases and outputs were filtered using the previously generated ROCker models. For metagenomes, target gene abundance in metagenomes was determined as genome equivalents by calculating the ratio between normalized target reads (number of reads matching divided by median protein length) and normalized RpoB reads (number of reads matching divided by median RpoB protein length), a universal single-copy gene. For metatranscriptomes, target transcripts abundance was calculated as reads per kilobase of transcript per million mapped reads (RPKM). Protein databases and ROCker models are available through http://enve-omics.ce.gatech.edu/.
Assembly and binning of metagenomic populations
Short-read metagenomes from control and treatments (t = 0,120 and 192 hours) were co-assembled using IDBA_UD v1.1.142 and binning was performed as previously described14. Taxonomic classification and degree of novelty (novel species, genus, etc) of the MAGs were obtained from the Microbial Genomes Atlas (MiGA) webserver43. MAG abundance was determined as the total length of all matching metagenomic or metatranscriptomic reads to the binned contigs from BLASTn searches (identity >= 98% and fraction of read aligned >= 50%) divided by the metagenomic or metatranscriptomic sample sizes (in millions of reads) and the length of the bin genomes in Kbp (Kilo base pairs). Reads encoding rRNA sequences (such as 5S, 5.8S, 16S, and 23S) were identified by SortMeRNA, and removed for non-rRNA analyses in order to avoid overestimating abundances.
N cycle protein sequences in the co-assembly and MAGs were detected using hidden Markov models obtained from FUNGENE44, using HMMer45. Detected target N cycle proteins were manually curated, when necessary, by assessing the presence of characteristic amino acid and phylogenetic congruency.
Phylogenetic trees and placement of short-reads
To assess the phylogenetic affiliation of metagenomic or metatranscriptomic reads, reference and fully assembled protein sequences were aligned using ClustalΩ46 with default parameters. Resulting alignments were used to build phylogenetic trees in RAxML v8.0.1947. Short-reads encoding the protein of interest were extracted from metagenomes or metatranscriptomes using ROCker (BLASTx) and placed in their corresponding phylogenetic tree using the methodology previously described14. Quantification of the number of reads assigned to a specific clade (e.g., to distinguish between nxrA or narG reads) was done using the “JPlace.distances.rb” script, also available in the enveomics collection. To quantify nirK gene fragments assigned to specific clades, the same process as described above was repeated except that all reads detected by multiple ROCker models to previously described clades48 (clades I + II, III and Thaumarchaeotea) were used. Abundances of target genes were determined as reads per kilobase million (RPKM) for metatranscriptomics. Equivalent results were obtained when using transcripts per kilobase million (TPM) but RPKM values were preferred for our targeted approach that focused on a reduced set of gene transcripts (ureC, amoA, haoA, nxrA, and nirK) and nitrifier populations.
Shotgun metaproteomics
Approximately 10 g of soil were collected from the 192 hours control and 15N-NH4+ amended microcosms and stored at − 80 °C. Frozen soil (5 g) was used for protein extractions. A detailed procedure is available in the Supplementary Material.
NanoLC-MS/MS analysis
Peptides (75 ug) were loaded onto in-house prepared biphasic resin packed column [SCX (Luna, Phenomenex, Torrance, CA) and C18 (Aqua, Phenomenex, Torrance, CA)] as described earlier49,50 and subjected to an offline wash for 15 min as previously described51. The sample column was aligned with an in-house C18 packed nanospray tip (New Objective, Woburn, MA) connected to a Proxeon (Odense, Denmark) nanospray source as previously detailed51. Peptides were eluted and subjected to chromatographic separation and measurements via 24-hr Multi-Dimensional Protein Identification Technology (MuDPIT) approach as described earlier49–51. Measurements were carried out using LTQ mass spectrometer (Thermo Fisher Scientific, Germany) coupled to the Ultimate 3000 HPLC system (Dionex, USA) and operated in data dependent mode, via Thermo Xcalibur software V2.1.0 as described earlier49.
For protein identification, the raw spectra from each run were searched against a custom database and was constructed using protein sequences predicted from metagenome assemblies obtained from the same soil and 20–30 cm depth14, metagenome assemblies from incubations (Table S2), and reference proteomes for 47 common soil organisms (Table S6). Detected proteins predicted from metagenomic assemblies were annotated using BLASTp52 and UniProt database as reference53 (downloaded in May of 2017). Additional details are available in the Supplementary Material.
Supplementary information
Acknowledgements
We thank Joel Kostka, Alissa Hooker and three anonymous reviewers for helpful discussions related to the manuscript. This work was supported in part by U.S. Department of Energy, Office of Biological and Environmental Research, Genomic Science Program [award DE-SC0006662], US National Science Foundation [Awards 1831582, 1831842, and 1831599], and the Chilean Fulbright-Conicyt doctoral scholarship [L.H.O.].
Author contributions
L.H.O., W.H.Y., J.C.C.-S., R.A.S., and K.T.K. conceived and designed the experiment. L.H.O., W.H.Y., J.C.C.-S., R.A.S. conducted the experiment. W.H.Y., J.C.C.-S., and R.A.S. conducted chemical analyses. J.K.H. and L.H.O. conducted sequencing and bioinformatic analyses. R.I., K.C., R.L.H. conducted proteomic analyses. L.H.O., J.K.H., R.I., K.C., J.C.S., W.H.Y., J.C.C.-S., R.A.S., F.E.L., and K.T.K. interpreted the data. L.H.O and K.T.K. wrote the manuscript. All authors reviewed the manuscript.
Data availability
Raw metagenomic and metatranscriptomic soil datasets and MAGs are deposited in the European Nucleotide archive (ENA) under study number PRJEB27434. MAGs previously recovered from the same agricultural site14 are deposited in ENA under study number PRJEB20068 and are also available at http://enve-omics.ce.gatech.edu/data/.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-53679-0.
References
- 1.Ravishankara AR, Daniel JS, Portmann RW. Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science. 2009;326:123–125. doi: 10.1126/science.1176985. [DOI] [PubMed] [Google Scholar]
- 2.van Groenigen JW, et al. The soil N cycle: new insights and key challenges. Soil. 2015;1:235–256. doi: 10.5194/soil-1-235-2015. [DOI] [Google Scholar]
- 3.Kuypers MMM, Marchant HK, Kartal B. The microbial nitrogen-cycling network. Nature Rev Microbiol. 2018;16:263–276. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 4.Daims H, et al. Complete nitrification by Nitrospira bacteria. Nature. 2015;528:504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Kessel MAHJ, et al. Complete nitrification by a single microorganism. Nature. 2015;528:555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kool DM, Dolfing J, Wrage N, Van Groenigen JW. Nitrifier denitrification as a distinct and significant source of nitrous oxide from soil. Soil Biol Biochem. 2011;43:174–178. doi: 10.1016/j.soilbio.2010.09.030. [DOI] [Google Scholar]
- 7.Zhu X, Burger M, Doane TA, Horwath WR. Ammonia oxidation pathways and nitrifier denitrification are significant sources of N2O and NO under low oxygen availability. Proc. Natl. Acad. Sci. USA. 2013;110:6328–6333. doi: 10.1073/pnas.1219993110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hink L, Gubry-Rangin C, Nicol GW, Prosser JI. The consequences of niche and physiological differentiation of archaeal and bacterial ammonia oxidisers for nitrous oxide emissions. ISME J. 2018;12:1084–1093. doi: 10.1038/s41396-017-0025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu H-W, He J-Z. Comammox—a newly discovered nitrification process in the terrestrial nitrogen cycle. J Soils Sediments. 2017;17:2709–2717. doi: 10.1007/s11368-017-1851-9. [DOI] [Google Scholar]
- 10.Gubry-Rangin C, Nicol GW, Prosser JI. Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol. 2010;74:566–574. doi: 10.1111/j.1574-6941.2010.00971.x. [DOI] [PubMed] [Google Scholar]
- 11.Hettich RL, Sharma R, Chourey K, Giannone RJ. Microbial metaproteomics: identifying the repertoire of proteins that microorganisms use to compete and cooperate in complex environmental communities. Curr Opin Microbiol. 2012;15:373–380. doi: 10.1016/j.mib.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Jiang X, et al. pH regulates key players of nitrification in paddy soils. Soil Biol Biochem. 2015;81:9–16. doi: 10.1016/j.soilbio.2014.10.025. [DOI] [Google Scholar]
- 13.Hultman J, et al. Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature. 2015;521:208–212. doi: 10.1038/nature14238. [DOI] [PubMed] [Google Scholar]
- 14.Orellana LH, Chee-Sanford JC, Sanford RA, Löffler FE, Konstantinidis KT. Year-Round Shotgun Metagenomes Reveal Stable Microbial Communities in Agricultural Soils and Novel Ammonia Oxidizers Responding to Fertilization. Appl. Environ. Microbiol. 2018;84:e01646–17. doi: 10.1128/AEM.01646-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davidson EA, Hart SC, Shanks CA, Firestone MK. Measuring gross nitrogen mineralization, and nitrification by 15 N isotopic pool dilution in intact soil cores. Journal of Soil Science. 1991;42:335–349. doi: 10.1111/j.1365-2389.1991.tb00413.x. [DOI] [Google Scholar]
- 16.Corre MD, Veldkamp E, Arnold J, Wright SJ. Impact of elevated N input on soil N cycling and losses in old-growth lowland and montane forests in Panama. Ecology. 2010;91:1715–1729. doi: 10.1890/09-0274.1. [DOI] [PubMed] [Google Scholar]
- 17.Meziti A, et al. Quantifying the changes in genetic diversity within sequence-discrete bacterial populations across a spatial and temporal riverine gradient. ISME J. 2019;13:767–779. doi: 10.1038/s41396-018-0307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orellana LH, Rodriguez-R LM, Konstantinidis KT. ROCker: accurate detection and quantification of target genes in short-read metagenomic data sets by modeling sliding-window bitscores. Nucleic Acids Res. 2017;45:e14. doi: 10.1093/nar/gkw900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia Z, Conrad R. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol. 2009;11:1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- 20.Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 2013;7:2061–2068. doi: 10.1038/ismej.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helbling DE, Ackermann M, Fenner K, Kohler H-PE, Johnson DR. The activity level of a microbial community function can be predicted from its metatranscriptome. ISME J. 2012;6:902–904. doi: 10.1038/ismej.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L-X, et al. Comparative metagenomic and metatranscriptomic analyses of microbial communities in acid mine drainage. ISME J. 2015;9:1579–1592. doi: 10.1038/ismej.2014.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angle JC, et al. Methanogenesis in oxygenated soils is a substantial fraction of wetland methane emissions. Nature. Communications 2016 7. 2017;8:1567. doi: 10.1038/s41467-017-01753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart FJ, Ulloa O, DeLong EF. Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol. 2012;14:23–40. doi: 10.1111/j.1462-2920.2010.02400.x. [DOI] [PubMed] [Google Scholar]
- 25.Stahl DA, de la Torre JR. Physiology and diversity of ammonia-oxidizing archaea. Annu Rev Microbiol. 2012;66:83–101. doi: 10.1146/annurev-micro-092611-150128. [DOI] [PubMed] [Google Scholar]
- 26.Kozlowski JA, Stieglmeier M, Schleper C, Klotz MG, Stein LY. Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota. ISME J. 2016;10:1836–1845. doi: 10.1038/ismej.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollibaugh JT, Gifford S, Sharma S, Bano N, Moran MA. Metatranscriptomic analysis of ammonia-oxidizing organisms in an estuarine bacterioplankton assemblage. ISME J. 2011;5:866–878. doi: 10.1038/ismej.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsementzi D, Poretsky R, Rodriguez-R LM, Luo C, Konstantinidis KT. Evaluation of metatranscriptomic protocols and application to the study of freshwater microbial communities. Environ Microbiol Rep. 2014;6:640–655. doi: 10.1111/1758-2229.12180. [DOI] [PubMed] [Google Scholar]
- 29.VerBerkmoes NC, Denef VJ, Hettich RL, Banfield JF. Systems Biology: Functional analysis of natural microbial consortia using community proteomics. Nat Rev Microbiol. 2009;7:196–205. doi: 10.1038/nrmicro2080. [DOI] [PubMed] [Google Scholar]
- 30.Werner JJ, Ptak AC, Rahm BG, Zhang S, Richardson RE. Absolute quantification of Dehalococcoides proteins: enzyme bioindicators of chlorinated ethene dehalorespiration. Environ Microbiol. 2009;11:2687–2697. doi: 10.1111/j.1462-2920.2009.01996.x. [DOI] [PubMed] [Google Scholar]
- 31.Hood LE, et al. New and improved proteomics technologies for understanding complex biological systems: Addressing a grand challenge in the life sciences. Proteomics. 2012;12:2773–2783. doi: 10.1002/pmic.201270086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang WH, Traut BH, Silver WL. Microbially mediated nitrogen retention and loss in a salt marsh soil. Ecosphere. 2015;6:7. doi: 10.1890/ES14-00179.1. [DOI] [Google Scholar]
- 33.Herman DJ, Brooks PD, Ashraf M, Azam F, Mulvaney RL. Evaluation of methods for nitrogen-15 analysis of inorganic nitrogen in soil extracts. II. Diffusion methods. Commun Soil Sci Plant Anal. 1995;26:1675–1685. doi: 10.1080/00103629509369400. [DOI] [Google Scholar]
- 34.Templer PH, Silver WL, Pett-Ridge J, DeAngelis KM, Firestone MK. Plant and microbial controls on nitrogen retention and loss in a humid tropical forest. Ecology. 2008;89:3030–3040. doi: 10.1890/07-1631.1. [DOI] [PubMed] [Google Scholar]
- 35.Welsh A, Chee-Sanford JC, Connor LM, Löffler FE, Sanford RA. Refined NrfA Phylogeny Improves PCR-Based nrfAGene Detection. Appl. Environ. Microbiol. 2014;80:2110–2119. doi: 10.1128/AEM.03443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox MP, Peterson DA, Biggs PJ. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez-R LM, Konstantinidis KT. Nonpareil: a redundancy-based approach to assess the level of coverage in metagenomic datasets. Bioinformatics. 2014;30:629–635. doi: 10.1093/bioinformatics/btt584. [DOI] [PubMed] [Google Scholar]
- 39.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 40.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orellana LH, et al. Detecting nitrous oxide reductase (NosZ) genes in soil metagenomes: method development and implications for the nitrogen cycle. mBio. 2014;5:e01193–14. doi: 10.1128/mBio.01193-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez-R LM, et al. The Microbial Genomes Atlas (MiGA) webserver: taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Res. 2018;84:e00014. doi: 10.1093/nar/gky467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fish JA, et al. FunGene: the functional gene pipeline and repository. Frontiers in Microbiology. 2013;4:291. doi: 10.3389/fmicb.2013.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539–539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 48.Wei W, et al. Higher diversity and abundance of denitrifying microorganisms in environments than considered previously. ISME J. 2015;9:1–12. doi: 10.1038/ismej.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown SD, et al. Molecular dynamics of the Shewanella oneidensis response to chromate stress. Mol. Cell Proteomics. 2006;5:1054–1071. doi: 10.1074/mcp.M500394-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Thompson MR, et al. Dosage-dependent proteome response of Shewanella oneidensis MR-1 to acute chromate challenge. J. Proteome Res. 2007;6:1745–1757. doi: 10.1021/pr060502x. [DOI] [PubMed] [Google Scholar]
- 51.Sharma R, et al. Coupling a detergent lysis/cleanup methodology with intact protein fractionation for enhanced proteome characterization. J. Proteome Res. 2012;11:6008–6018. doi: 10.1021/pr300709k. [DOI] [PubMed] [Google Scholar]
- 52.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–12. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw metagenomic and metatranscriptomic soil datasets and MAGs are deposited in the European Nucleotide archive (ENA) under study number PRJEB27434. MAGs previously recovered from the same agricultural site14 are deposited in ENA under study number PRJEB20068 and are also available at http://enve-omics.ce.gatech.edu/data/.