Short abstract
Primary liver cancer is a major public health challenge that ranks as the third most common cause of cancer worldwide despite therapeutic improvement. Reovirus has been emerging as a potential anti-cancer agent and is undergoing multiple clinical trials, and it is reported that reovirus can preferentially cause the cell death of a variety of cancers in a manner of apoptosis. As few studies have reported the efficacy of oncolytic activity and safety profile of avian reovirus, in our study, LDH assay, MTT assay, DAPI staining, and flow cytometry assay were performed to demonstrate the oncolytic effects of avian reovirus against the HepG2 cells, and quantitative real-time PCR (qRT-PCR) and animal experiments were conducted to investigate the dynamic distribution of avian reovirus in infected mice and then illustrate the safety and tissue tropism of avian reovirus. LDH assay, DAPI staining, and flow cytometry assay confirmed the efficacy of the oncotherapeutic effects of avian reovirus, and MTT assay has indicated that avian reovirus suppressed the proliferation of HepG2 cells and decreased their viability significantly. qRT-PCR revealed the dynamic distribution of avian reovirus in infected mice that avian reovirus might replicate better and have more powerful oncolytic activity in liver, kidney, and spleen tissues. Furthermore, histopathological examination clearly supported that avian reovirus appeared non-pathogenic to the normal host, so our study may provide the new insights and rationale for the new strategy of removing liver cancer.
Impact statement
We demonstrated the efficacy of oncolytic activity of avian reovirus (ARV) by LDH assay, MTT assay, DAPI staining, and flow cytometry assay, and also investigated the dynamic distribution of ARV in infected mice and then illustrated the safety and tissue tropism of ARV by quantitative real-time PCR (qRT-PCR) and animal experiments. Collectively, our study may provide the new insights and rationale for the new strategy of removing liver cancer.
Keywords: Hepatocellular carcinoma, avian reovirus, oncolytic virus, safety, dynamic distribution, real-time PCR
Introduction
Primary liver cancer (PLC) ranks as the third leading cause of cancer-associated death in developing countries, and is characterized with high aggression and poor prognosis.1 Hepatocellular carcinoma (HCC) is one of the pathological subtypes of PLC, and is responsible for the majority of PLC, accounting for about 80% of all patients, with approximately 800,000 new cases every year.2 The high rate is especially evident in Asia, due to the infection of hepatitis virus, mainly hepatitis B virus and hepatitis C virus,3,4 for which the five-year-survival rates rank from 5% to 19% during 1995–2004,2,5,6 and therapeutic options for patients with liver cancer are confusing and limited, with surgical resection being the main treatment for patients who are in the early stage of the disease, such as by laparoscopic liver resection,7 however, most patients have missed the suitable time for operation. Vascular intervention may cause incomplete embolization and postoperative collateral circulation, chemotherapy therapy applied to advanced liver cancer has a response rate less than 20%,8 and patients with liver cancer often experience liver dysfunction, thus limiting the application of conventional therapies.
Oncolytic viruses (OVs) have been emerging as effective anti-cancer agents and have made great breakthroughs in cancer treatment,9 especially reovirus, which is a natural oncolytic virus selectively infecting and killing various tumor cells without harming normal tissues,10 as more than 80% of tumor cells are sensitive to reovirus infection. Mammalian reovirus (MRV) is one of the reovirus genera, and Reolysin based on MRV has been tested in clinical trials with phase I to III in head and neck-related carcinomas.11 Avian reovirus (ARV), which can lead to some diseases in poultry, shares similarities with MRV in oncolytic activity. Apoptosis is one of the mechanisms through which the reovirus treats various cancer cells,12 but few studies have researched the apoptosis of liver cancer cells induced by ARV.
Reovirus is reported to specifically infect human tumor cells and spare the normal tissues, and its name is derived from its initial isolation from the respiratory and intestinal tract. Park and Kim13 reported that wild-type reovirus cannot affect the biological activity of human adipose-derived stem cells, such as proliferation, and although many safety studies and clinical trials of MRV have been carried out, no safety studies of ARV have been identified.
The purpose of this report is to detect apoptosis of liver cancer cells induced by ARV by LDH assay, DAPI staining, and flow cytometry assay, to demonstrate the oncotherapy of ARV. Meanwhile, a combination of quantitative real-time PCR and histopathological examination is conducted to investigate the dynamic distribution of ARV in infected mice and confirm the clinical safety and tissue tropism of ARV.
Materials and methods
Ethics statement
The animal experiment with SPF Kunming mice was approved by the Animal Care and Use Committee of Shandong Agriculture University (permit number: 2018085, August, 2018), and performed with the guidelines of the Ethical and Animal Welfare Committee of Shandong Province, China.
Cell culture
Human liver hepatocellular cells (HepG2) were obtained from the American Type Cell Collection(HB-8065,ATCC, Manassas, VA, USA) and maintained in DMEM/F12 (Dulbecco's modified Eagle's medium: Nutrient Mixture F-12) (Gibco, Gaithersburg, MD, USA) with 15% fetal bovine serum (FBS) (Gibco, Gaithersburg, MD, USA) and 1% Penicillin (100 IU/mL)/streptomycin (100 µg/mL) and 2 mM L-glutamine (Solarbio, Beijing Solarbio Science & Technology Co., Ltd, Beijing, China), cells were cultured at 37°C in a humidified 5% CO2 incubator.
Viruses and titer assay
ARV strain S1133(GeneBank accession number:AF330703) was obtained from the Avian Disease Laboratory of Shandong Agricultural University(Taian, China), and propagated in the chicken hepatoma cells (LMH). LMH cells were cultured with ARV S1133 at a MOI of 1 for 60 min in the 37°C, 5% CO2 incubator; then the virus inoculum was removed, and overlaid with DMEM/F12 supplemented with 2% FBS, and the infected cells suspension was subjected to three freeze–thaw cycles at 24 hours post infection (hpi), followed by centrifugation at 12,000 r/min for 10 min at 4°C. Then the cultured supernatant was sub-packed and stored at −80°C. LMH cells were plated for 12 h before infection in the 96-well plates for the TCID50 assay. The virus solution was serially diluted after thawing, the dilution and cell culture medium being DMEM/F12 supplemented with 2% FBS, and the result of virus titer calculated by the previously described method.14
Animal experiment
Sixty five-week-old female SPF Kunming mice in the study were purchased from the Shandong Laboratory Animal Center(Jinan, China), and kept in the SPF animal laboratory with isolators at Shandong Agriculture University (Taian, China), and fed a proper diet. They were separated randomly into four groups, oral group (n = 15), intramuscular injection group (n = 15) and two control groups (n = 30). The mice in the oral group and the intramuscular injection group were inculcated orally and intramuscularly with 3 × 106TCID50/0.2 mL of ARV S1133, respectively, while the control group was challenged with 0.2 mL phosphate-buffered saline (PBS) orally and intramuscularly, respectively.
Three mice selected randomly in all groups were sacrificed by cervical dislocation at 1, 3, 5, 7, 14 days post infection (dpi) for sample collection, in which organ samples of heart, liver, spleen, lung, and kidney were collected for virus detection by qPCR and hematoxylin and eosin(HE) staining. In this study, the weight of the four groups was measured daily by the weighing scales, and other clinical signs including behavior, appetite, breathing, and death were observed daily for 14 days during the study.
RNA extraction and detection of virus RNA
The ARV S1133 RNA of organ samples was extracted from homogenates using the RNAprep pure Tissue Kit (Tiangen, Beijing, China) according to the manufacturer's instructions. Relative real-time polymerase chain reaction (rRT-PCR) was performed with a 7300 real-time PCR system (Applied Biosystems, Foster city, CA, USA) using a one-step SYBR primescript RT-PCR Kit (Takara, Dalian, China) in a 20 µL reaction mixture containing 10 µL one-step SYBR RT-PCR Buffer III, 0.4 µL for each primer, ROX reference dye, Ex Taq HS and PrimeScript RT Enzyme Mix II, 2 µL sample RNA and 6 µL RNase-free dH2O. The specific primers of S1133 and GAPDH are shown in Table 1. The reaction was conducted using the thermal cycling procedure with a reverse transcription reaction at 42°C for 5 min, 95°C for 10 s and PCR reaction followed by 40 cycles of denaturation at 95°C for 5 s, and elongation at 61°C for 31 s, then the fluorescent signal of each sample was collected at the elongation step, and the results of relative expression level of ARV S1133 were determined by the fold changes(2−ΔΔCt).
Table 1.
Primer sequences of ARV S1133 and GAPDH.
| Genes | Primers |
|---|---|
| ARV S1133 | F:5′-TGCCTACTCTGTTATCTCAACCT-3′R:5′-AGTAGAGCACAATCGTACTCACA-3′ |
| GAPDH | F:5′-ACATGGCCTCCAAGGAGTAAGA-3′R:5′-GATCGAGTTGGGGCTGTGACT-3′ |
Histopathology
Organ samples of heart, liver, spleen, lung, and kidney collected at 1, 3, 5, 7, 14 dpi from Kunming mice of four groups were used for histopathological analysis. Samples were maintained in 10% formalin at room temperature (RT) (25°C) until use. The pathological section for all the samples of four groups was completed in the laboratory of pathology of Shandong Agriculture University and examined using a light microscope.
DAPI staining
HepG2 cells were seeded in 6-well plates for 12 h before being infected with ARV S1133 (MOI of 1). Plates containing HepG2 cells were washed with cold PBS twice, fixed with 4% paraformaldehyde fix solution (Beyotime, China) for 15 min at 24, 48, 72 hpi and permeabilized by Triton X-100 (Beyotime) for 10 min at RT, then cells were washed with PBS three times followed by 2–(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) staining for 25 min, and then washed with PBS three times again. An inverted fluorescent microscope (Nikon Eclipse Ti2, Japan) was used to analyze the cells according to the manufacturer’s instructions.
Flow cytometry assay
A flow cytometry assay was conducted using the FITC Annexin V apoptosis detection kit (BD Biosciences, SanJose, CA, USA). Cultured cells of the experimental groups (infected at a MOI of 1) and control groups were washed twice with cold PBS and resuspended in binding buffer at RT, then 100 µL of the aforementioned solution (1 × 105 cells) was transferred to each 1.5 mL amicrobic centrifuge tube, adding 5 µL FITC Annexin V and 10 µL PI to each tube, respectively, and the mixture was vortexed gently and incubated for 15 min at RT(25°C) in the dark. Finally, 400 µL binding buffer was added to each tube, and samples of each tube were analyzed by a flow cytometer (BD Biosciences, SanJose, CA, USA) within 1 hour, and the flow cytometry data were analyzed with FlowJo software version X (https://www.flowjo.com/).
Lactate dehydrogenase release assay
The apoptosis of HepG2 cells treated with ARV S1133 (MOI of 1) was accessed by the release of LDH into the culture medium using the LDH cytotoxicity assay kit (Beyotime, China). HepG2 cells were seeded in the 96-well plates for 12 h before ARV S1133 infection, and the control group was treated with the same volume of the DMEM/F-12 medium containing 2% FBS. After 24, 48, 72 hpi, according to the manufacturer’s instructions, the 96-well plates containing all samples were centrifuged at 400 × g for 5 min, then 100 µL of culture medium of all samples was transferred to new 96-well plates, and centrifuged at 12,000 × g for 10 min to remove any cells. The absorbance of all samples at 490 nm was measured by an Elx808 microplate reader (BioTeK Instruments, VT, USA) with 600 nm as the reference wavelength.
MTT assay
To demonstrate the viability of ARV S1133-treated HepG2 cells, cells grown in DMEM/F-12 medium containing 2% FBS were seeded in a 96-well culture plate in a 37°C, 5% CO2 incubator for 12 h, then cells were exposed to ARV S1133 at MOI of 1, and the viability of HepG2 cells was assessed by a MTT cell proliferation and cytotoxicity assay kit (Beyotime, China); 10 µL MTT solution (5 mg/mL) was added without removing the medium at 24, 48, 72 hpi, respectively, then cells were incubated at 37°C. After 4 h, the formazan crystals were dissolved in 100 µL formazan solution(Beyotime, China) for 3 h at 37°C, then the absorbance was measured at 630 nm using a microplate reader as mentioned in the lactate dehydrogenase (LDH) release assay, and compared with control groups.
Quantitation of ARV S1133 RNA in HepG2 cells
A rRT-PCR method was performed to quantitate ARV S1133 in cell samples. HepG2 cells were cultured in the 6-well plates for 12 h before ARV S1133 infection, and the culture medium was removed and medium containing 2% FBS was added to plates after HepG2 cells were incubated with ARV S1133 for 60 min in a 37°C, 5% CO2 incubator, and after 24, 48, 72 hpi, the total RNA was extracted by using RNAprep pure cell kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions, and the rRT-PCR reaction conditions were the same as described above.
Statistical analysis
The results of LDH released assay, apoptosis analysis, and viral load data were analyzed with the two-tailed t test to access the significant difference of data between different groups by GraphPad Prism software(version 7.0, Inc., La Jolla, CA, USA) (https://www.graphpad.com/). Data were represented as mean ± SE, P < 0.05 was considered statistically significant, and results were from at least three repeated experiments.
Results
Clinical observation
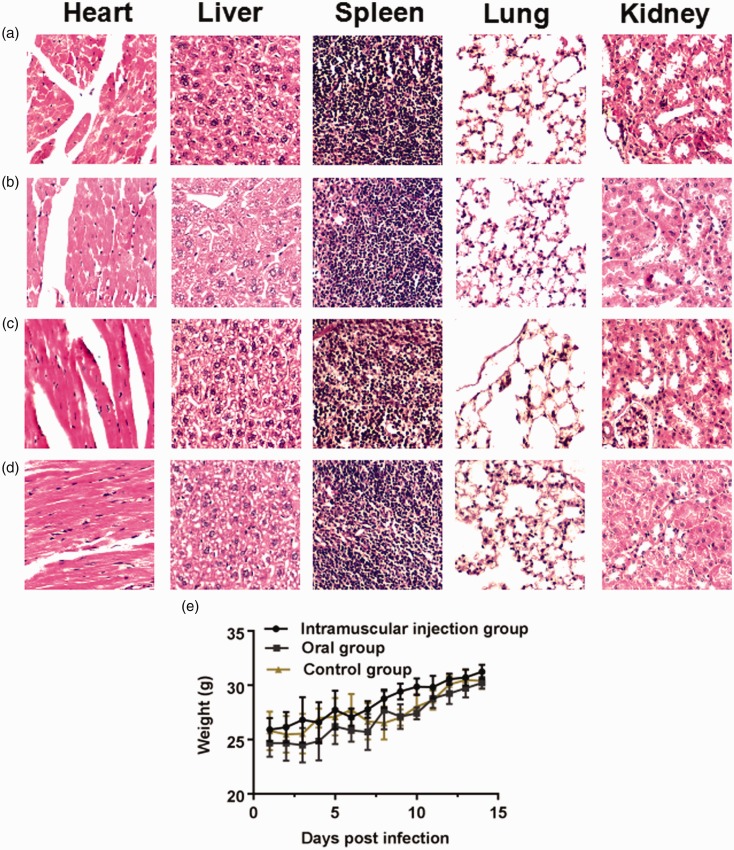
Interestingly, the infected Kunming mice showed no clinical signs, including fever, dyspnoea, anorexia, weight loss and behavioral disorders, and there was no significant difference between the oral group, and intramuscular injection group from 1 dpi to 14 dpi, and there were also no clinical symptoms observed in the control groups. There was no significant difference of body weight among the four groups (Figure 1(e)), and all mice in the four groups remained alive and the mortality rate was zero throughout the study.
Figure 1.
Histopathology in five organs stained with H&E from Kunming mice of three groups. 100×view of five organs from Kunming mice of the oral groups challenged with 3 × 106TCID50/0.2 mL of ARV S1133 (a) and 0.2 mL PBS (b) at 14 dpi, 100×view of five organs from Kunming mice of intramuscular injection groups challenged with 3 × 106TCID50/0.2 mL of ARV S1133 (c) and 0.2 mL PBS (d)at 14 dpi. (e) The body weight of the four groups, the control group in the graph represents the average weight of mice challenged with PBS orally and intramuscularly; error bars here and later indicate the standard deviations. (A color version of this figure is available in the online journal.)
Dynamic distributions of ARV S1133 strain in infected mice
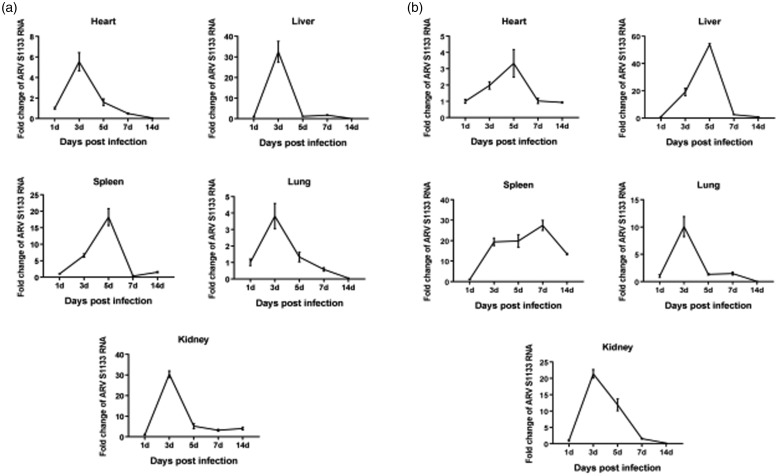
The dynamic distributions of the ARV S1133 strain were detected in the heart, liver, spleen, lung, and kidney by the rRT-PCR, and all the samples of four groups were examined for ARV S1133 using the rRT-PCR foldchange.
In oral groups (Figure 2(a)), the viral load peak point appeared at 3 dpi in all tissues except the spleen tissue with the peak point at 5 dpi, after ARV S1133 infection, and we observed the viral load reached a peak point with 5-fold increase in the heart, 30-fold increase in the liver and kidney, 15-fold increase in the spleen, and 4-fold increase in the lung, approximately, as compared to 1 dpi. The concentration of viral load was stable from 5 dpi to 14 dpi in liver and kidney tissues and decreased from 5 dpi in heart and lung tissues without another peak point in all tissues at the end of the study.
Figure 2.
Dynamic distribution of the ARV S1133 in infected Kunming mice. The foldchange represents the relative expression of ARV S1133 in infected Kunming mice of oral group (a) and intramuscular injection group (b) at 1, 3, 5, 7 and 14 dpi.
In intramuscular injection groups (Figure 2(b)), the viral load peak point appeared at 5 dpi in heart and liver tissues with 3-fold and 50-fold increase and 3 dpi in lung and kidney tissues with 10-fold and 20-fold increase approximately compared to 1 dpi. Interestingly, there were two viral load peak point in spleen tissue; at 3 dpi with 20-fold increase, and 7 dpi with 25-fold increase, respectively, and it was then stable from 7 dpi to 14 dpi in heart, liver, and kidney tissues.
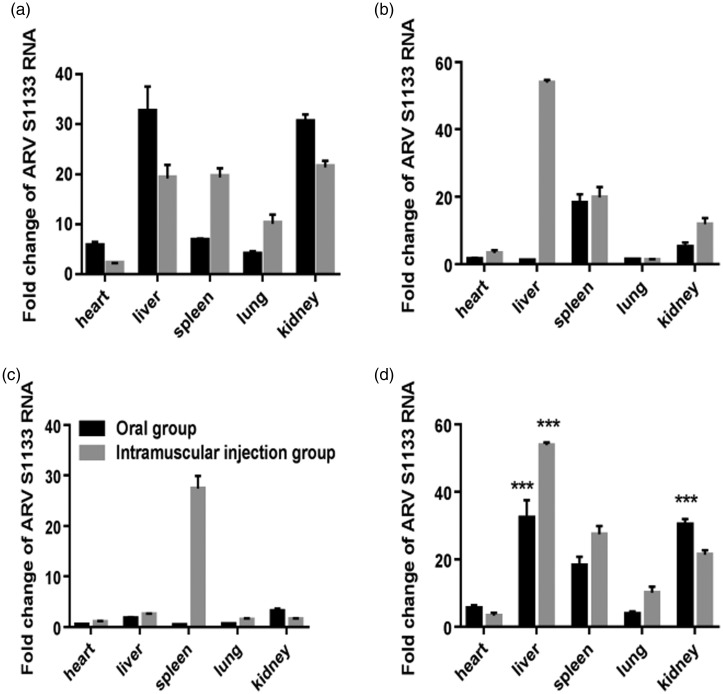
Comparison of foldchange between the oral groups and intramuscular injection groups at 3 dpi (Figure 3(a)), 5 dpi (Figure 3(b)) and 7 dpi (Figure 3(c)) is shown. The foldchange of liver and kidney tissues in oral groups was the highest at 3 dpi, and the liver tissue in intramuscular injection groups obtained the highest foldchange at 5 dpi. However, at 7 dpi, the foldchange of spleen tissue in intramuscular injection groups reached the highest.
Figure 3.
Comparison of relative expression of ARV S1133 of five organs in infected Kunming mice of two experimental groups. Fold change of ARV S1133 between the two groups at 3 dpi (a), 5 dpi (b) and 7 dpi (c). The peak foldchange of virus replication of five organs between the two groups (d) showed the viral load in liver and kidney was significantly higher than that in other tissues in oral groups and the highest viral load occurred in liver tissue of intramuscular injection groups.***P < 0.001.
The peak foldchange of virus replication is shown among the heart, liver, spleen, lung, and kidney tissues between the oral groups and intramuscular injection groups (Figure 3(d)). It can be seen that the peak foldchange of viral load in liver and kidney tissues was significantly higher (P < 0.001) than that in any other tissues in oral groups and the liver tissue in intramuscular injection groups obtained the highest peak foldchange of viral load and it was significantly higher (P < 0.001) than that in the oral groups.
Histopathology in different organs
The results of HE staining of the four groups show that there was no apparent pathological damages observed in the four groups at 14 dpi (Figure 1(a) to (d)) and 1–7 dpi.
DAPI staining of HepG2 cell lesions
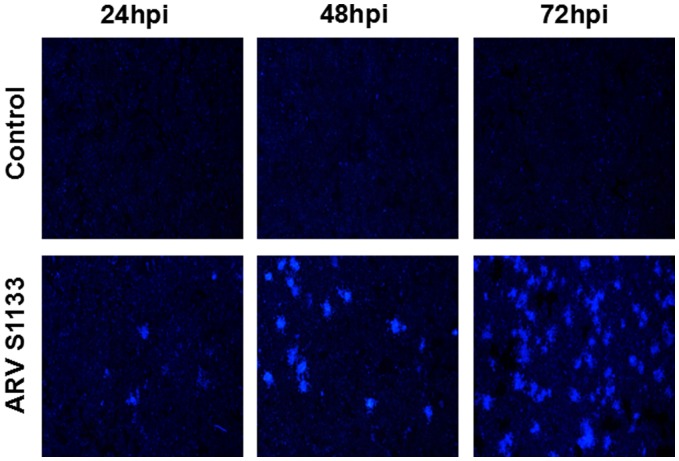
The increase of ARV S1133-treated time caused a trend of increasing numbers of apoptotic HepG2 cells, which demonstrated significant nuclear rounding, shrinkage, and fragmentation compared to the control groups treated with culture medium alone. The syncytia of cell-to-cell fusion was detected at 24, 48 and 72 hpi, and the DAPI stained area became largest after ARV S1133 treatment at 72 hpi compared to 24 and 48 hpi (Figure 4).
Figure 4.
Syncytia formation of HepG2 cells infected with ARV S1133. Experimental groups infected at a MOI of 1 and control groups followed by DAPI staining under the fluorescent microscope (100×magnification). (A color version of this figure is available in the online journal.)
LDH release assay uncovered the cell death
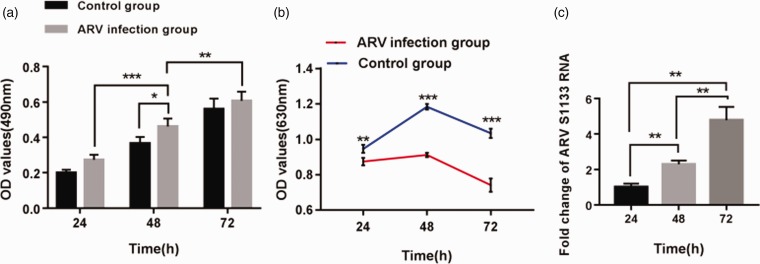
The results obtained from the LDH cytotoxicity assay were an indicator of the apoptosis of HepG2 cells treated with ARV S1133, and it was apparent that the cytotoxicity after treatment with ARV S1133 was in a time-dependent manner. There was a significant difference between 24 hpi and 48 hpi (P < 0.001) or 48 hpi and 72 hpi (P < 0.01), and the occurrence of apoptosis was significant at 48 hpi (P < 0.05) compared to the control groups (Figure 5(a)).
Figure 5.
Cytotoxic effects of ARV S1133 on growth and proliferation of HepG2 cells. Cells were treated with ARV S1133 at a MOI of 1, and the apoptosis was measured by LDH-cytotoxicity assay as the rise of absorbance (a). ARV S1133 on growth and proliferation of HepG2 cells was measured by MTT assay as the decline of absorbance (b). The increase of viral load in HepG2 cells (c).*P < 0.05,**P < 0.01,***P < 0.001. (A color version of this figure is available in the online journal.)
Cell viability analysis
According to the results of MTT assay, the mean OD values of ARV-infected groups and control groups are shown in Table 2, and the OD values of ARV-infected groups were significantly lower than that of control groups at any endpoints (P < 0.001) (Figure 5(b)).Comparing with the cell viability of control groups (100%), the cell viability of ARV-infected groups was 91.2%, 74.2% and 66.5%, respectively, after 24 h-,48 h- and 72 h-incubation of HepG2 cells with ARV S1133. Therefore, it was demonstrated that ARV suppressed the proliferation of HepG2 cells and decreased the viability of target cells significantly compared to the control groups.
Table 2.
Mean OD values of two groups at different endpoint and the corresponding cell viability.
| 24 hpi | 48 hpi | 72 hpi | |
|---|---|---|---|
| ARV infection groups | 0.874 ± 0.017 | 0.911 ± 0.010 | 0.741 ± 0.030 |
| Control groups | 0.946 ± 0.018 | 1.183 ± 0.013 | 1.034 ± 0.021 |
| % cell viability | 91.2 | 74.2 | 66.5 |
Note: The OD values were expressed as mean values ± SE, and cell viability of ARV infection groups at 24, 48, 72 hpi was compared to the control groups (100%).
ARV: avian reovirus.
Flow cytometry analysis
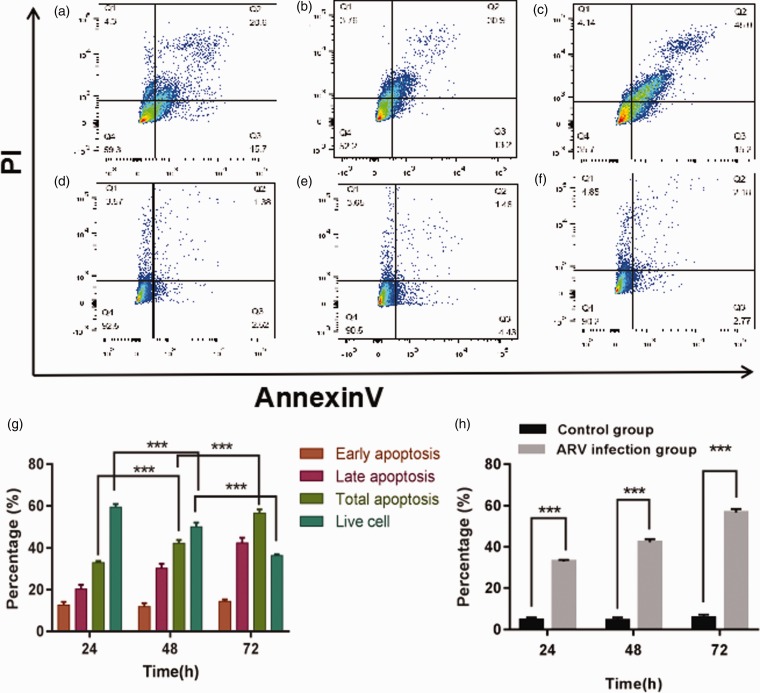
The data analysis was acquired after the HepG2 cells infected with ARV S1133 at 24, 48, 72 hpi, and the early apoptotic rate of the ARV S1133-infected groups varied at the same level, however the rate of HepG2 cells at the late apoptotic stage increased in a time-dependent manner, the late apoptotic rate among 24 hpi, 48 hpi and 72 hpi was 20%, 30% and 45% on average, respectively (Figure 6(a) to (f)). Moreover, the total apoptotic proportion of HepG2 cells detected by flow cytometry was significantly increased at 72 hpi (P < 0.001), and the results also showed that there was a clear trend of decreasing of viable cells of infected groups along with the extension of infected hours (Figure 6(g)), compared to the control groups, and the total apoptosis rate was significantly higher in the ARV S1133-infected groups compared to the control groups (P < 0.001) (Figure 6(h)).
Figure 6.
Apoptosis of HepG2 cells was analyzed by flow cytometry with double staining of Annexin V and PI. The apoptosis was detected after the HepG2 cells infected with ARV S1133 at 24 hpi (a), 48 hpi (b), 72 hpi (c) and the corresponding control groups (d–f). The percentage of early apoptosis, late apoptosis, total apoptosis and live cells was compared among 24 hpi, 48 hpi and 72 hpi (g).The percentage of total apoptosis was compared between ARV S1133 groups and control groups at 24, 48 and 72 hpi (h). ***P < 0.001. (A color version of this figure is available in the online journal.)
The increase of viral load in HepG2 cells
The viral load of ARV S1133 had a significant increase (P < 0.01) at 48 and 72 hpi compared with 24 hpi followed by ARV S1133 infection (Figure 5(c)), and these findings suggested that viral load during treatment of HepG2 cells with ARV S1133 was in a time-dependent manner and revealed that the replication and reproduction of ARV S1133 in HepG2 cells were successful during the study.
Discussion
Reovirus is reported to selectively replicate in the target cells, but not in normal cells, and specifically lyse cancer cells, which has currently been the potential therapeutic option for the cancer treatment. In reviewing the literature, several reports have shown that reovirus can trigger apoptosis in various cancer cells, including solid and hematologic tumors. The Ras pathway is a crucial factor to sensitizing cancer cells to reovirus, and Ras mutations can promote reovirus oncolysis. It is reported that accumulation of Ras induced the apoptosis of Ras-transformed fibroblasts.15 NF-kB also mediates oncolytic action of breast cancer cells induced by reovirus in a manner of apoptosis,16 while reovirus strain T1L can destroy the host restriction by inducing epithelial cell apoptosis with inflammatory signals.17 In addition, reovirus triggers cell death in the apoptotic mode significantly in the gastrointestinal stromal tumor cells by the Fas-FasL pathway.18 However, as the most present studies on the anti-tumor activity of reovirus are based on the MRV, it is unclear whether there is a trend that MRV may induce a toxicity risk to the host when the viral titer is increased. Meanwhile, viral genomes were modified by gene deletions and insertion to improve the oncolytic potential, and gene deletion can make the virus escape from the immune system of host.19 Once the MRV has caused negative effects, it is difficult for the host to remove the viruses to protect itself, and when an oncolytic virus is inserted with other genes, it also gives rise to toxicity and may cause problems of its own. In our study, we implemented a series of assays to detect the apoptosis of HepG2 cells induced by ARV, and we tentatively put forward that the safety profile of ARV is superior to the MRV due to the interspecies difference.
In this study, we have demonstrated the clinical potential of ARV using HepG2 cells in vitro, and in order to identify the oncolytic activity of ARV against HepG2 cells and determine the safety profile of ARV, the DAPI staining, flow cytometry assay, and LDH assay were employed to identify the apoptotic cell death of HepG2 cells. In addition, cell viability was confirmed by the MTT assay, as described previously. Furthermore, we obtained the dynamic tissue distribution of ARV in infected mice with rRT-PCR. The decrease of live target cells along with increasing of ARV exposure time, as measured by MTT assay, resulted from the ARV-induced apoptotic cell death of HepG2 cells, and this is in accordance with a previous similar biological assay.20 The DAPI staining, flow cytometry assay, and LDH assay together indicated the apparent apoptosis of HepG2 cells treated with ARV but not of the uninfected ones, and a comparison of the findings with those of another study confirmed the efficacy of anti-cancer effects of ARV,21 the advantage of our study was that we researched the oncolytic activity of ARV at different endpoints, and both studies demonstrated the oncolytic activity of ARV at 72 hpi when the apoptosis of target cells was significant despite the different MOIs, and although there was only HepG2 cell line in our study. The current study found that ARV caused no pathogenicity to experimental groups and the control groups according to the histopathologic examinations, confirming the safety of ARV, and suggesting the tissue tropism of ARV according to the results of dynamic tissue distribution of ARV S1133. In addition, an important finding was that the trend of tissue tropism of the five organs from all infected mice in our study according to the foldchange (three highest) was: liver, kidney, and spleen, no matter in oral groups or in intramuscular injection groups, and the peak foldchange of liver tissue was exceptionally high, so it can therefore be assumed that the ARV would obtain the greater efficacy in liver cancers, followed by the kidney and spleen cancers.
Oncotherapy of reovirus is a promising option for the treatment of cancer, and it seems that the immune stimulation also plays a very important role in reovirus anti-tumor activity according to a previous study,22 and gene modification has become a new field of the oncotherapy. Further research should be undertaken to improve some aspects of reovirus therapy, such as the safety and delivery methods. As multiple ways of treating cancers have become an exciting prospect, a combination of reovirus with chemotherapeutic drugs has shown significant potential advantages, and it is reported that an agent of treating malignant melanoma, rapamycin which combined with reovirus can improve the anti-tumor efficacy,23and a combination of reovirus and the immune checkpoint blockade also seems to be a powerful immunotherapy of treating breast cancer.24 In our study, the ARV showed the significant viral infectivity and efficacy of its oncolytic activity, along with a clear safety profile. Further work is required to put ARV into clinical trials as the single agent or in a combination with conventional chemotherapy and radiotherapy. A further study with RNA-seq analysis on the apoptosis of HepG2 cells induced by ARV is in progress to seek the potential mechanisms involved.
Collectively, our study has provided significant data for the therapeutic efficacy of ARV and its safety profile. Further work will focus on the potential mechanisms and demonstrate the oncolytic effects of ARV on other cancer types. Our study provides the rationale for the new strategy of removing liver cancers.
ACKNOWLEDGEMENTS
We thank all the experts in Department of Clinical Laboratory, Taian Central Hospital and Shandong Agricultural University.
Authors’ contributions
QF and YL designed research, provided methods and reviewed the paper; YT provided resources; WW validated and the data; RC performed the experiments and wrote the manuscript; GM and SZ supervised the study; YD contributed to the project administration. All authors gave final approval of the article.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
The present study was funded by the Projects of medical and health technology development program of Shandong province (2016WS0060) and Experimental study on reovirus for the treatment of liver cancer (SDBJKT20180023).
References
- 1.Bakiri L, Wagner EF. Mouse models for liver cancer. Mol Oncol 2013; 7:206–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics. CA Cancer J Clin 2015; 65:87–108 [DOI] [PubMed] [Google Scholar]
- 3.Vucenik I, Zhang ZS, Shamsuddin AM. IP6 in treatment of liver cancer II. Intra-tumoral injection of IP6 regresses pre-existing human liver cancer xenotransplanted in nude mice. Anticancer Res 1998; 18:4091–6 [PubMed] [Google Scholar]
- 4.Chuang SC, Lee YC, Hashibe M, Dai M, Zheng T, Boffetta P. Interaction between cigarette smoking and hepatitis B and C virus infection on the risk of liver cancer: a meta-analysis. Cancer Epidemiol Biomark Prevent 2016; 19:1261–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kowdley KV, Hassanein T, Kaur S, Farrell FJ, Van Thiel DH, Keeffe EB, Sorrell MF, Bacon BR, Weber FJ, Tavill AS. Primary liver cancer and survival in patients undergoing liver transplantation for hemochromatosis. Liver Transpl Surg 1995; 1:237–41 [DOI] [PubMed] [Google Scholar]
- 6.Berrino F, De Angelis R, Sant M, Rosso S, Bielska-Lasota M, Coebergh JW, Santaquilani M. Survival for eight major cancers and all cancers combined for European adults diagnosed in 1995-99: results of the EUROCARE-4 study. Lancet Oncol 2007; 8:773–83 [DOI] [PubMed] [Google Scholar]
- 7.Kamiyama T, Tahara M, Nakanishi K, Yokoo H, Kamachi H, Kakisaka T, Tsuruga Y, Matsushita M, Todo S. Long-term outcome of laparoscopic hepatectomy in patients with hepatocellular carcinoma. Hepato Gastroenterol 2014; 61:405–9 [PubMed] [Google Scholar]
- 8.Burroughs A, Hochhauser D, Meyer T. Systemic treatment and liver transplantation for hepatocellular carcinoma: two ends of the therapeutic spectrum. Lancet Oncol 2004; 5:409–18 [DOI] [PubMed] [Google Scholar]
- 9.Meisen WH, Kaur B. How can we trick the immune system into overcoming the detrimental effects of oncolytic viral therapy to treat glioblastoma? Exp Rev Neurother 2013; 13:341–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paola M, Maya S, Lee PW. Connecting reovirus oncolysis and Ras signaling. Cell Cycle 2005; 4:556. [DOI] [PubMed] [Google Scholar]
- 11.Jaime-Ramirez AC, Yu JG, Caserta E, JYY, Zhang J, Lee TJ, Hofmeister C, Lee JH, Kumar B, Pan Q. Reolysin and histone deacetylase inhibition in the treatment of head and neck squamous cell carcinoma. Mol Ther Oncolyt 2017; 5:87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartlett DL, Zuqiang L, Magesh S, Roshni R, Zongbi G, Yukai H, Zong Sheng G. Oncolytic viruses as therapeutic cancer vaccines. Mol Cancer 2013; 12:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park JS, Kim M. Reovirus safety study for proliferation and differentiation of human adipose-derived mesenchymal stem cells. J Microbiol 2017; 55:75–9 [DOI] [PubMed] [Google Scholar]
- 14.Reed LJ., Müench H. A simple method of estimating fifty percent endpoints. Am J Hyg 1938; 27:493–7 [Google Scholar]
- 15.Garant KA, Shmulevitz M, Pan L, Daigle RM, Ahn DG, Gujar SA, Lee P. Oncolytic reovirus induces intracellular redistribution of Ras to promote apoptosis and progeny virus release. Oncogene 2015; 35:771. [DOI] [PubMed] [Google Scholar]
- 16.Thirukkumaran C, Shi ZQ, Thirukkumaran P, Luider J, Kopciuk K, Spurrell J, Elzinga K, Morris D. PUMA and NF-kB are cell signaling predictors of reovirus oncolysis of breast cancer. PLoS One 2017; 12:e0168233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown JJ, Short SP, Stencel-Baerenwald J, Urbanek K, Pruijssers AJ, Mcallister N, Ikizler M, Taylor G, Aravamudhan P, Khomandiak S. Reovirus-induced apoptosis in the intestine limits establishment of enteric infection. J Virol 2018; 92:JVI.02062–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inagaki Y, Kubota E, Mori Y, Aoyama M, Kataoka H, Johnston RN, Joh T. Anti-tumor efficacy of oncolytic reovirus against gastrointestinal stromal tumor cells. Oncotarget 2017; 8:115632–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howells A, Marelli G, Lemoine NR, Wang Y. Oncolytic viruses-interaction of virus and tumor cells in the battle to eliminate cancer. Front Oncol 2017; 7:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang CR, Mahmood J, Zhang QR, Vedadi A, Warrington J, Ou N, Bristow RG, Jaffray DA, Lu QB. In vitro and in vivo studies of a new class of anticancer molecules for targeted radiotherapy of cancer. Mol Cancer Ther 2016; 15:640–50 [DOI] [PubMed] [Google Scholar]
- 21.Kozak RA, Hattin L, Biondi MJ, Corredor JC, Walsh S, Xue-Zhong M, Manuel J, Mcgilvray ID, Morgenstern J, Lusty E. Replication and oncolytic activity of an avian orthoreovirus in human hepatocellular carcinoma cells. Viruses 2017; 9:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gujar SA, Clements D, Dielschneider R, Helson E, Marcato P, Lee P. Gemcitabine enhances the efficacy of reovirus-based oncotherapy through anti-tumour immunological mechanisms. Br J Cancer 2014; 110:83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comins C, Simpson GR, Rogers W, Relph K, Harrington K, Melcher A, Roulstone V, Kyula J, Pandha H. Synergistic antitumour effects of rapamycin and oncolytic reovirus. Cancer Gene Ther 2018; 25:148–160 [DOI] [PubMed] [Google Scholar]
- 24.Mostafa A, Meyers D, Thirukkumaran C, Liu P, Gratton K, Spurrell J, QS, Thakur S, Morris D. Oncolytic reovirus and immune checkpoint inhibition as a novel immunotherapeutic strategy for breast cancer. Cancers 2018; 10:205. [DOI] [PMC free article] [PubMed] [Google Scholar]