Abstract
Purpose:
Reliable monitoring of tissue nicotinamide adenine dinucleotide (NAD+) concentration may provide insights on its roles in normal and pathological aging. In the present study, we report a 1H MRS pulse sequence for the in vivo, localized 1H MRS detection of NAD+ from the human brain.
Methods:
Studies were performed on a 7T Siemens MRI scanner using a 32-channel product volume coil. The pulse sequence consisted of a spectrally selective low bandwidth E-BURP-1 90° pulse. PRESS localization was achieved using optimized Shinnar-Le Roux 180° pulses and overlapping gradients were used to minimize the echo time. The reproducibility of NAD+ quantification was measured in 11 healthy volunteers. The association of cerebral NAD+ with age was assessed in 16 healthy subjects 26–78 years old.
Results:
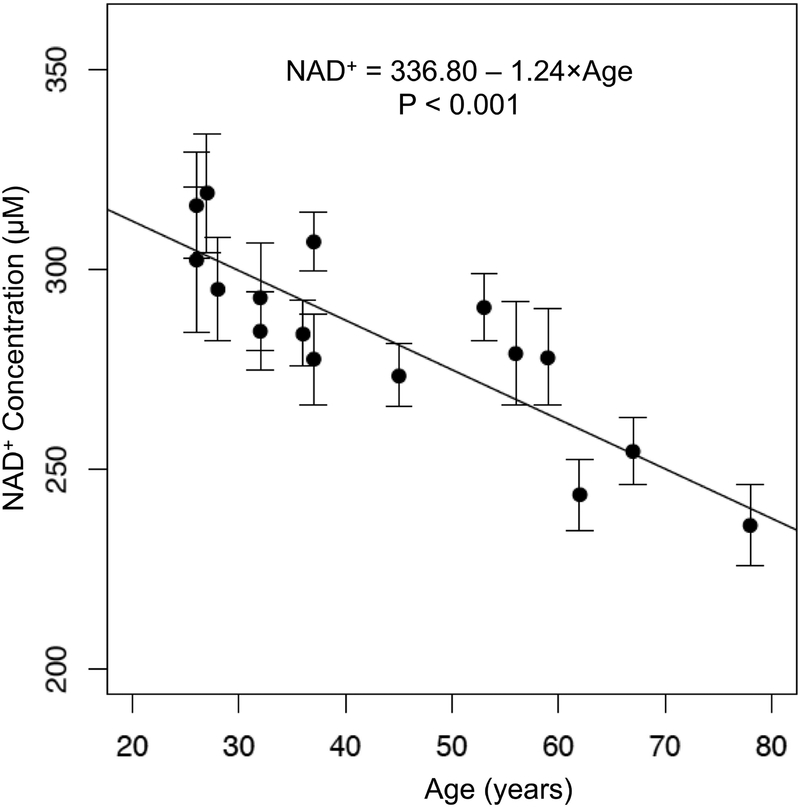
Spectra acquired from a voxel placed in subjects’ occipital lobe consisted of downfield peaks from the H2, H4 and H6 protons of the nicotinamide moiety of NAD+ between 8.9–9.35 ppm. The mean ± SD within-session and between-session coefficients of variation were found to be 6.14 ± 2.03 and 6.09 ± 3.20 % respectively. In healthy volunteers, an age-dependent decline of the NAD+ levels in the brain was also observed (β = −1.24 μM/yr, SE=0.21, p<0.001).
Conclusion:
We demonstrated the feasibility and robustness of a newly developed 1H MRS technique to measure localized cerebral NAD+ at 7T MRI using a commercially available RF head coil. This technique may be further applied to detect and quantify NAD+ from different regions of the brain as well as from other tissues.
Keywords: Aging, Brain, E-BURP-1, 1H MRS, NAD+
Introduction
Nicotinamide adenine dinucleotide (NAD+) is present in all living cells and it plays a vital role in cellular metabolism as a coenzyme for reduction-oxidation (redox) reactions, including those required for mitochondrial energy production (1). It also serves as a cofactor for other enzymes such as sirtuins, Poly (ADP-ribose) polymerases (PARPs) and ADP-ribosyl cyclases that are involved in cell survival and metabolism (2–4). The nicotinamide moiety of NAD+ serves as the site for redox reactions, allowing the reduced form of NAD+ (NADH) to function as an energy-transfer intermediate between various metabolic pathways. The ratio and concentration of NAD+ and NADH influence the rate of reactive oxygen species production in the mitochondria (5,6). An increasing number of studies suggest roles for NAD+ in mediating key cellular processes including gene expression, aging, neurodegeneration and cell death (7–9). The roles of NAD+ dependent enzymes such as sirtuins, PARPs and CD38 are also well documented in many reports (10,11). Reduced NAD+ levels can be restored by treatment with nicotinamide mononucleotide (NMN), an intermediate in the major NAD+ biosynthesis pathway (12,13), or by its precursor, nicotinamide riboside (14). Thus, cellular NAD+ may serve as a potential therapeutic target for metabolic and age-related disorders.
While NAD+ is most commonly quantified using invasive biochemical or auto-fluorescence methods (15,16), non-invasive quantification of tissue NAD+ is highly desirable for in vivo studies, particularly in organs not amenable to biopsy such as brain. Non-invasive in vivo detection and quantification of NAD species (NAD+/NADH) has been performed using 31P magnetic resonance spectroscopy (MRS) with ultra-high magnetic field MR scanners. NAD+ and NADH concentrations have been quantified by modeling the partially overlapping NAD+, NADH, and α-ATP resonances in the 31P MR spectrum (17,18). One study using 31P MRS showed a decline of NAD+ with aging in the human brain (19).
Recently, a non-water suppressed 1H MRS technique with surface coil localization was used to detect protons of the nicotinamide moiety in NAD+ downfield of water in both the rodent and human brain (20,21). Due to cross-relaxation of the NAD+ protons with water, traditional water suppressed 1H MRS methods cannot be used to detect the NAD+ protons (20). This 1H MRS method has an advantage of being free from contamination from other metabolite peaks and offers ~15-fold higher sensitivity compared to 31P MRS.
Here, we demonstrate the development and validation of a spectroscopy (SVS) 1H MRS pulse sequence to detect spatially-localized NAD+ signal from the human brain at 7T with a volume radiofrequency (RF) coil. NAD+ signals were quantified using water signal as an internal concentration reference. In a small cohort of healthy volunteers, we found an age dependent decline in the NAD+ concentration in the occipital cortex.
Methods
Sequence design
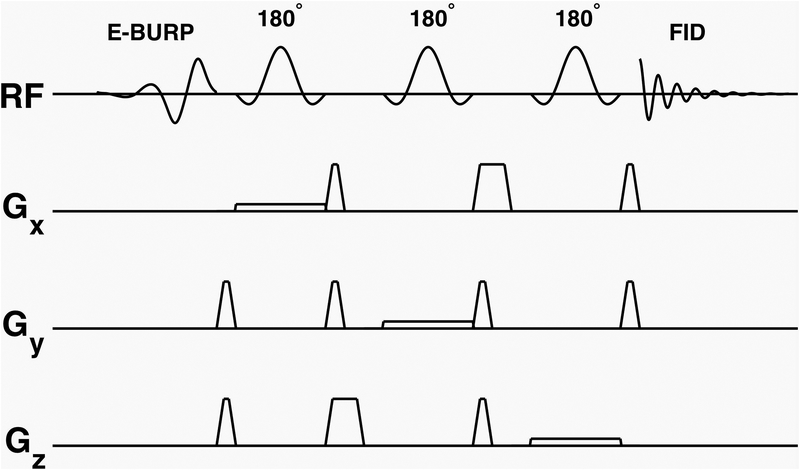
The pulse sequence uses a spectrally selective self-refocused Band-selective Uniform-Response Pure-phase (E-BURP-1) 90° pulse (22,23) and 3 narrow band spatially selective refocusing Shinnar-Le Roux (SLR) 180° pulses (24–27) for localization (Figure 1). The details of the excitation and refocusing pulses are given below.
Figure 1.
Pulse sequence for NAD+ spectroscopy. A spectrally selective self-refocused E-BURP-1 90° pulse (2 ppm bandwidth) followed by three narrow band spatially selective refocusing 180° SLR pulses for localization providing selective excitation and refocusing of NAD+ signal from the MRS voxel
The 90° pulse is 7 ms long to achieve 2 ppm (600 Hz) spectral excitation at 7T. The pulse shape and Bloch simulation MY profile for the E-BURP-1 pulse are shown in Supporting Information Figure S1. The excitation pulse used in the human scans was centered at 9.1 ppm with a range of 8.1 – 10.1 ppm so as to excite all three of the nicotinamide moiety protons of the NAD+. The spatially selective refocusing pulses for localization are based on the SLR pulse design and are 3.2 ms long with a bandwidth of 800 Hz (Fig. S1). The crusher gradient waveform timings were optimized to reduce the overall echo time (TE) to 19.3 ms, which is close to that reported in the previous study using a surface coil (21). The spectrally selective 90° pulse and three 180° pulses are centered at 9.1 ppm for NAD+. For NAD+ quantification and phase estimation, water spectra were acquired using the same SVS localization by moving the center frequency of E-BURP-1 and SLR pulses to 4.7 ppm.
Human studies
All human studies were conducted under an approved Institutional Review Board (IRB) protocol of the University of Pennsylvania on 7T Siemens scanner using a Siemens volume coil transmit/32-channel receive proton phased-array head coil. A total of 16 healthy subjects (12 males and 4 females) with age ranging from 26–78 years (43.8 ± 16.5 yrs) were scanned twice per session in one or two sessions.
MR scanning procedure
Data from 11 subjects with two scanning sessions were used to determine the reproducibility of the acquisition and data processing. After positioning each subject in the scanner, two successive NAD+ MRS scans were performed (first session). Following this, the subject was taken out of the scanner, given a 15–30 minutes break, then re-positioned in the scanner and two more NAD+ MRS scans were performed in succession (second session). For each session, NAD+ levels were measured in a 64 ml cubic voxel in occipital cortex, and the [NAD+] obtained from the two scans were used to estimate within-session coefficient of variation (CV) for each subject. Within-session CVs were averaged within subjects and then across subjects to obtain mean ± SD within-session CV. For each subject, between-session CV was determined separately for the first and second scans, and then these values were averaged first within and then across subjects to obtain mean ± SD between-session CV.
1H MRS acquisition
T1-weighted anatomical three-dimensional magnetization-prepared rapid gradient echo (MPRAGE) images of whole brain were acquired for voxel localization using the following parameters: 176 axial slices, repetition time/echo time/inversion time (TR/TE/TI) = 2800/4.4/1500 ms, Flip angle = 7°, Resolution 0.8×0.8×0.8 mm3, iPAT=2, scan time 7:43 min. A voxel of 40×40×40 mm3 was placed in the occipital lobe of the head as shown in Figure 2.
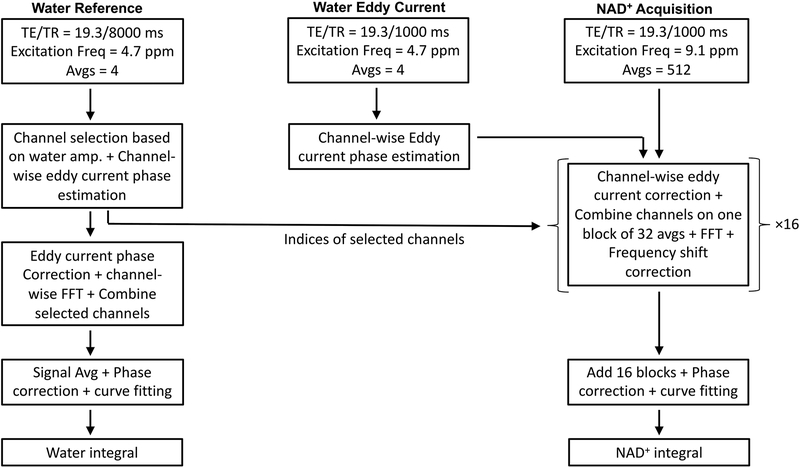
Figure 2.
Acquisition and processing of 1H MRS data. The top row describes the three acquisitions. The left column describes the steps for processing the water reference scan to calculate the final water integral. Channel selection, eddy current phase estimation, signal averaging, phase correction, and curve fitting was performed on the reference water scan. The middle column describes the use of a water eddy current scan for eddy current phase estimation. The right column describes NAD+ spectra processing by channel wise eddy current correction, channel combination, Fourier transform, and frequency alignment of 16 blocks containing 32 averages each. Spectra from all the blocks were then added to obtain the final NAD+ spectrum. The NAD+ spectrum was phase corrected as fitted to determine the integral of NAD+ H2, H4 and H6 peaks
The 1H MRS protocol consisted of 3 scans, (i) Water Reference acquisition (TR = 8 s, excitation and refocusing pulses set at 4.7 ppm, 8 averages), (ii) Water Eddy Current acquisition (TR = 1 s, excitation and refocusing pulses set at 4.7 ppm, 8 averages), and (iii) NAD+ acquisition (TR = 1 s, excitation and refocusing pulses set at 9.1 ppm, 512 averages). The total acquisition time for NAD+ and water 1H MRS was 10 minutes.
Data Processing
A flow chart illustrating the data acquisition and processing steps is depicted in Figure 2. Raw multi-channel time domain data from the scanner were used for post-processing using an in-house MATLAB script. The water reference scan was used to select channels with at least 5% of maximum signal. For the selected channels, channel-wise eddy current phase estimation as a function of time (28,29) was performed using water eddy current estimation scan. The selected channels were combined to obtain the final complex water reference spectrum. The real part of reference water spectrum was phase corrected and curve fitted to derive the water integral. Next, eddy current correction was performed for the NAD+ scan using the eddy current estimation derived for water. To generate the NAD+ spectrum, block averaging of 32 acquisitions was used, and each block spectrum was eddy current corrected and channel combined, Fourier transformed, spectral frequency aligned using the broad NAA amide/unassigned purine nucleotide signal between 7.8 and 8.6 ppm as frequency reference, and phase corrected. The final spectrum was produced as the sum of all the individual blocks. For NAD+ level quantification, the spectrum in the range of 8.8 to 9.6 ppm was fitted with 3 Lorentzian peaks and the integral of the fit at 9.35 ppm was used. NAD+ concentration was calculated using the water integral and estimated tissue water concentration. We applied a T1, T2 correction factor using equation 1:
| Equation 1 |
where INAD and Iwater are the integrals of the NAD-H2 and water peaks, respectively, T1W and T2W are the average water T1 and T2 of human brain at 7T (30,31). T1NAD and T2NAD values were taken from de Graaf et al (20). Further, since de Graaf et al (21) reported a 50% visibility of NAD+ using 1H MRS, an NMR visibility correction factor CFNADvis was included in the calculation of the final NAD+ concentration.
Calculation of contribution of CSF, gray and white matter in the 1H MRS voxel
The tissue composition of the SVS region-of-interest was determined using the following procedure: First, a binary mask of the location of the SVS voxel was created in the T1 structural image space using the header information contained in the dicom image files (both SVS and T1) produced by the scanner. The software used to perform this step is available in the ImScribe package described previously (https://www.med.upenn.edu/cmroi/imscribe.html) (32,33). Next, the T1 structural image was segmented into maps of predominantly gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) voxels using the FAST tool from the FMRIB software library, FSL image analysis package (34). Finally, the SVS spatial mask was multiplied with the tissue segmentation maps to determine the relative contribution from GM, WM and CSF voxels within the SVS region-of-interest. The total water content in the voxel was calculated as a weighted contribution of GM (80% or 44 M water), WM (70% or 38.5 M water) and CSF (100% or 55 M water) (35,36).
Reproducible placement of MRS voxel in brain
In order to enhance overlap of SVS voxel positioning across subjects, we maintained the consistency in voxel placement for multiple acquisitions of 1H MRS from the same voxel using ImScribe. ImScribe uses high resolution T1-weighted images and the voxel information from the spectroscopy file acquired from the first scan as target template for subsequent scans and performs affine co-registration to derive the information needed for a new voxel placement as illustrated in Fig. 2.
Association of NAD+ levels with age
To determine the variability in [NAD+] explained by age, data from all 16 subjects were used in a linear mixed model, including a random subject intercept to account for repeated within-subject measures and age as a fixed-effect covariate. The statistical significance of the age fixed effect was assessed using a Kenward-Roger’s F-test. Additionally, we calculated the Pearson correlation of the within-subject mean [NAD+] with age. Analyses were conducted using R version 3.5.3 (37) with the packages lme4 (38) and pbkrtest (39).
1H MRS acquisition in parietal lobe
To demonstrate the feasibility of this method for detection of NAD+ in brain regions other than the occipital lobe, we performed 1H MRS using a smaller voxel (30×30×30 mm3) positioned in the frontoparietal cortex of two subjects. All other parameters were the same as previously described.
Results
1H MR spectrum of NAD+ from occipital lobe
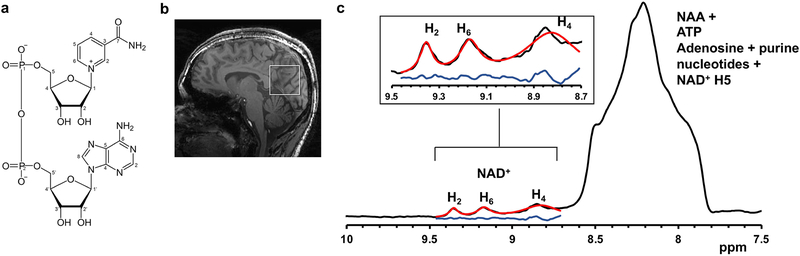
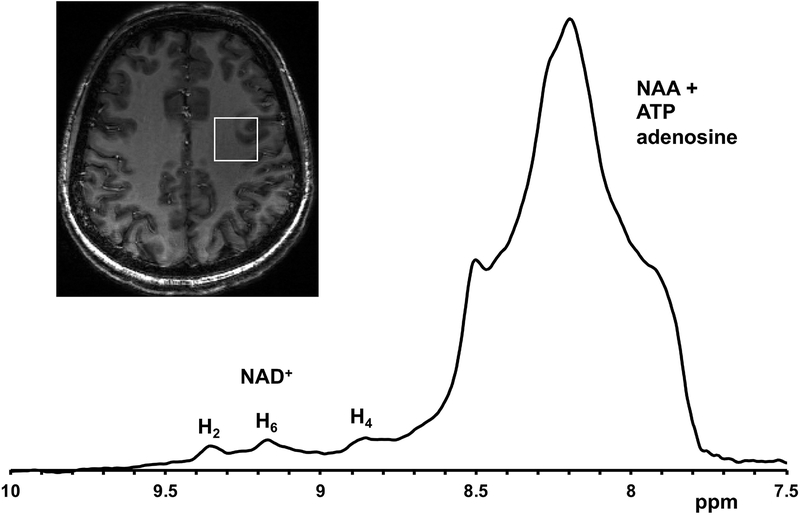
The downfield region of a 1H MR spectrum acquired from a voxel placed in the occipital lobe of the human brain in vivo at 7T is shown in Figure 3. The downfield region displays three peaks of NAD+ at 8.9, 9.15 and 9.35 ppm as well as a broad signal between 7.8–8.6 ppm containing NAA amide-bound proton signal at 7.84 ppm and multiple unassigned signals from ATP and other purine nucleotides between 8.0 and 8.6 ppm. The well-resolved H2 resonance at 9.35 ppm was used for the quantification.
Figure 3.
NAD+ MRS in human brain. Left, Chemical structure of NAD+ showing the H2, H4 and H6 protons present on the nicotinamide moiety which are the target of downfield spectroscopy. Inset, Sagittal section from the anatomical images of the brain showing the position of MR voxel. Right, MR spectra acquired from the healthy human brain showing the three peaks arising from NAD+ at 8.9, 9.1 and 9.35 ppm
Motion related frequency drift during the scans may affect the signal-to-noise ratio of the spectra generated after adding blocks with high number of averages. This was addressed by taking spectra from each average and adding them individually. However, the spectrum from one average may not have enough signal to determine the purine peak between 8–8.5 ppm, which is used as a reference peak to align all the spectra from individual blocks in this study. To find the optimal number of spectra to be added per block, we performed a summation of blocks with 1,8, 32 and 256 averages per block on the spectra acquired from one young volunteer (Supporting Information, Figure S2). We found that using 32 averages per block produced spectra with well resolved peaks of NAD+ H2, H4 and H6 protons with acceptable reproducibility.
Reproducibility of NAD+ 1H MR spectroscopy
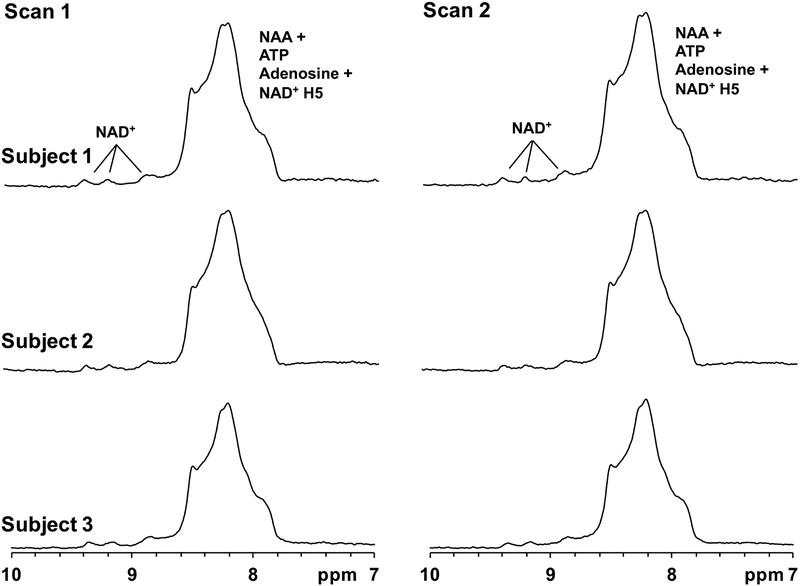
Figure 4 depicts the spectra acquired from two scans in three healthy subjects, which show high reproducibility of the NAD+ peaks downfield of the unassigned NAA/amine peaks between 8–8.6 ppm. The mean ± SD within-session coefficient of variation for 11 subjects was found to be 6.14 ± 2.03 %. Additionally, the mean ± SD between-session coefficient of variation was 6.09 ± 3.20 %, showing satisfactory reproducibility in the MRS scans for the quantification of NAD+.
Figure 4.
Demonstration of 1H MRS reproducibility. NAD+ spectra acquired from three subjects in two scans acquired in the same session showing the intra-scan reproducibility of the pulse sequence
Decline of cerebral NAD+ with normal aging
The linear mixed model for [NAD+] resulted in an estimated coefficient (SE) for age of −1.24 (0.21) (Kenward-Roger’s F1,15.62 = 33.37, p < 0.001), corresponding to a mean decrease in NAD+ concentration of 1.24 μM per year. The standard error was calculated as within-subject standard deviation divided by the square root of the number of measurements for that subject. The negative association between measured [NAD+] and age is plotted in Figure 5. Additionally, the estimated variance of the subject-level random effect was 391.3 in the model without age, and 31.5 in the model with age, indicating that age explains approximately 91.9% of the between-subject variability in these data. Within-subject mean [NAD+] had a Pearson correlation with age of r = −0.86.
Figure 5.
Decline of cerebral NAD+ with aging. Individual NAD+ levels measured from the occipital cortex of healthy volunteers plotted against the age showing a linear decrease of NAD+ level with healthy aging. Filled circles represent within-subject means and error bars depict ± one standard error. Standard error was calculated as the within-subject standard deviation divided by the square root of the number of measurements for that subject
Detection of NAD+ in frontoparietal cortex
Figure 6 shows a spectrum acquired from frontoparietal cortex of a subject. The three distinct peaks of the nicotinamide moiety H2, H4 and H6 protons of NAD+ are well-resolved downfield of the broad NAA+ATP adenosine resonances.
Figure 6.
1H MRS from deeper brain region. 1H MR spectrum from a voxel positioned in the frontoparietal cortex of a 33-year-old volunteer showing well-resolved NAD+ H4, H6 and H2 proton resonances at 8.9, 9.15 and 9.35 ppm, respectively
Discussion
In this study, we demonstrate the non-invasive detection of NAD+ in the brains of healthy individuals using a spectrally- and spatially-selective 1H MRS pulse sequence with a product 32-channel volume coil at 7T. The pulse sequence allows the use of a standard volume coil to acquire spectra from a voxel placed in arbitrary regions of the brain with a reasonable acquisition time. We also tested the reproducibility of NAD+ measured using this method in a sub-group of healthy volunteers by evaluating the within-session and between-session coefficients of variation of the method. Finally, we demonstrate that [NAD+] in the occipital lobe of the brain declines with age. Importantly, this method can be applied obtain the spectrum from a voxel placed anywhere in the brain using a volume coil.
Although 31P MRS provides more comprehensive information regarding phosphorylated biomolecules (40,41), there is a substantial overlap between the NAD+/NADH and ATP peaks necessitating fitting and simulation to quantify NAD+/NADH (17). This limitation was recently addressed by using unlocalized 1H MRS (20,21) wherein the H2, H4 and H6 protons of the nicotinamide moiety in NAD+ were reported to be easily detectable due to the absence of any spectral overlap at the downfield region of water. However, the use of surface coil in these studies limits its application to superficial regions of the brain and complicates the incorporation of NAD+ measurement in multimodal imaging protocols.
The use of a self-refocused E-BURP-1 excitation pulse with a low bandwidth (2 ppm in present study) centered at 9.1 ppm provides a 103-104-fold reduction in the water signal (Supporting Information Figure S3) comparable to the standard water suppression methods (42–45) in addition to producing a pure absorption spectrum in the passband region of the 90° pulse. This is required since traditional water suppression methods eliminate the NAD+ signal due to cross-relaxation between the nicotinamide and water protons (20,21). The sequence additionally used three SLR refocusing pulses centered at 9.1 ppm for voxel localization. However, the phase response of the E-BURP-1 pulse inside the passband (8.1–10.1 ppm) is constant, while outside (>10.1 and <8.1 ppm) it is non-linear (Supporting Information Figure S3). This explains the observed phase distortions of the broad combined peak between 7.8–8.6 ppm and other upfield peaks, making peak fitting for the broad peak with a combination of unspecified singlets hard to achieve. Accordingly, in this study, the fitting range was limited to the H2, H4, and H6 peaks of NAD+. And as the H2 peak at 9.35 ppm is least influenced by the broad resonance, it was selected for NAD+ quantification.
In vivo, NAD+ was shown by de Graaf et al (21) to have 86% and 49% NMR visibility with 31P and 1H MRS, respectively. We used these data to correct for the 1H MRS visibility of NAD+ in the present work. While de Graaf et al (21) used NAA and creatine as a concentration reference to measure [NAD+], we used the water reference signal obtained with the same SVS localization. Due to the use of low bandwidth E-BURP-1 pulse (2 ppm) for the spectrally selective excitation, we did not observe the complete NAA peak resonance at 7.9 ppm, which may also be affected by a large chemical shift displacement contribution. Further, since creatine resonates at close proximity to water at 3 ppm, the partial excitation of water signal can lead to large water signal leading to the necessity for water suppression. The use of low bandwidth pulses positioned at 4.4 ppm downfield to water in the current method allows the suppression of water signal without the requirement of traditional water suppression techniques.
We performed NAD+ SVS in the occipital lobe for comparison with previously published reports employing 31P (19) and 1H MRS (21). In addition, to assess the potential of the technique in other brain regions, 1H MR spectra were also acquired from voxels placed in the parietal lobes of two subjects. The measured cerebral NAD+ concentrations were found to be similar to those previously reported in the literature (4,17,21). Since detection and quantification of NAD+ using this pulse sequence relies on accurate reference voltage settings for the E-BURP-1 excitation pulse, B1 variations currently limit the application of this method to a single voxel.
Cellular NAD+ is reduced and converted into NADH during glycolysis, the Krebs cycle, and a host of other reactions. NADH is then recycled back to NAD+ by donating the electrons to the mitochondrial electron transport chain or to support reactions such as the conversion of pyruvate to lactate by lactate dehydrogenase. All of these processes are directly linked with cerebral glucose metabolism (CMRGlc) and neurotransmitter cycling. It has been shown recently that CMRGlc steadily reduces with healthy aging by 18F-FDG PET (46) and 13C MRS studies (47). A recent study of mice gastrocnemius muscle has shown by biochemical analysis that NAD+ levels of 24 and 30-month old mice were reduced by a factor of 2 compared to 6-month old young adults (13). The authors also showed recovery of reduced NAD+ levels of 22-month old mice to the same level as the 6-month old mice following treatment with NMN (13). Our results also support a reduction in the level of cerebral NAD+ with age, though this should be replicated in a larger sample. Future work can leverage ultra-high field 1H MRS of cerebral NAD+ to study the effects of NMN and other supplements that enhance NAD+ levels in older humans. This technique may further be useful in probing the role of NAD+ changes in neurodegenerative diseases by studying subjects with disease and age matched controls.
Conclusions
We demonstrated a new pulse sequence capable of spatially localized measurement of brain NAD+ concentrations using 1H MRS at 7T with a volume head coil and showed a decline in the cerebral NAD+ in with healthy aging. This non-invasive technique provides a biomarker of brain redox state and enables the evaluation of potential therapies targeting the enhancement of NAD+ concentration in humans.
Supplementary Material
Supporting Information Figure S1. Pulse shapes and the Bloch simulation MY profile of 90° and 180° pulses. a,b) pulse shape and simulation profile of E-BURP-1 90° pulse. c,d) pulse shape and simulation profile of SLR 180° pulse
Supporting Information Figure S2. Effect of block averaging of the NAD+ spectra. Number of blocks used was a) 256, b) 32, c) 8, and d) 1, on a dataset acquired from one volunteer
Supporting Information Figure S3. Water and NAD+ 1H MR spectra. The full spectra from water and NAD+ scans are depicted in a and b respectively showing the selective excitation of water and downfield region using the pulse sequence. The spectral bandwidth for all scans was 4 kHz centered at 4.7 ppm
Acknowledgements
We thank Prof. Robin de Graaf for valuable discussions. We also thank Kevin D’Aquilla and Dr. Ravi Prakash Reddy Nanga for their help in MRI scans. We thank Dr. Craig Jones for publicly sharing his MR pulse sequence diagram code. This project was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health through Grant Number P41-EB015893, the National Institute of Neurological Disorders and Stroke through Award Number R01NS087516, and the National Institute of Diabetes and Digestive and Kidney Diseases through award number DK098656.
References
- 1.Stein LR, Imai S. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol Metab 2012;23(9):420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol 2014;24(8):464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houtkooper RH, Cantó C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev 2010;31(2):194–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonkowski MS, Sinclair DA. Slowing ageing by design: the rise of NAD(+) and sirtuin-activating compounds. Nat Rev Mol Cell Biol 2016;17(11):679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pryde KR, Hirst J. Superoxide is produced by the reduced flavin in mitochondrial complex I: a single, unified mechanism that applies during both forward and reverse electron transfer. J Biol Chem 2011;286(20):18056–18065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kussmaul L, Hirst J. The mechanism of superoxide production by NADH:ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proc Natl Acad Sci U S A 2006;103(20):7607–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang EF, Lautrup S, Hou Y, Demarest TG, Croteau DL, Mattson MP, Bohr VA. NAD+ in Aging: Molecular Mechanisms and Translational Implications. Trends Mol Med 2017;23(10):899–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrido A, Djouder N. NAD+ Deficits in Age-Related Diseases and Cancer. Trends Cancer 2017;3(8):593–610. [DOI] [PubMed] [Google Scholar]
- 9.Satoh A, Imai SI, Guarente L. The brain, sirtuins, and ageing. Nat Rev Neurosci 2017;18(6):362–374. [DOI] [PubMed] [Google Scholar]
- 10.Verdin E NAD⁺ in aging, metabolism, and neurodegeneration. Science 2015;350(6265):1208–1213. [DOI] [PubMed] [Google Scholar]
- 11.Pehar M, Harlan BA, Killoy KM, Vargas MR. Nicotinamide Adenine Dinucleotide Metabolism and Neurodegeneration. Antioxid Redox Signal 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Bonkowski MS, Moniot S, Zhang D, Hubbard BP, Ling AJ, Rajman LA, Qin B, Lou Z, Gorbunova V, Aravind L, Steegborn C, Sinclair DA. A conserved NAD(+) binding pocket that regulates protein-protein interactions during aging. Science 2017;355(6331):1312–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, de Cabo R, Rolo AP, Turner N, Bell EL, Sinclair DA. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell 2013;155(7):1624–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bieganowski P, Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 2004;117(4):495–502. [DOI] [PubMed] [Google Scholar]
- 15.Yoshino J, Imai S. Accurate measurement of nicotinamide adenine dinucleotide (NAD⁺) with high-performance liquid chromatography. Methods Mol Biol 2013;1077:203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang X, Yang L, Qin AR, Ly J, Liederer BM, Messick K, Ma S, Zak M, Dragovich PS, Dean BJ, Hop CE, Deng Y. Measuring NAD(+) levels in mouse blood and tissue samples via a surrogate matrix approach using LC-MS/MS. Bioanalysis 2014;6(11):1445–1457. [DOI] [PubMed] [Google Scholar]
- 17.Lu M, Zhu XH, Zhang Y, Chen W. Intracellular redox state revealed by in vivo (31) P MRS measurement of NAD(+) and NADH contents in brains. Magn Reson Med 2014;71(6):1959–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu M, Zhu XH, Chen W. In vivo (31) P MRS assessment of intracellular NAD metabolites and NAD(+)/NADH redox state in human brain at 4 T. NMR Biomed 2016;29(7):1010–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu XH, Lu M, Lee BY, Ugurbil K, Chen W. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc Natl Acad Sci U S A 2015;112(9):2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Graaf RA, Behar KL. Detection of cerebral NAD(+) by in vivo (1)H NMR spectroscopy. NMR Biomed 2014;27(7):802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Graaf RA, De Feyter HM, Brown PB, Nixon TW, Rothman DL, Behar KL. Detection of cerebral NAD(+) in humans at 7T. Magn Reson Med 2017;78(3):828–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geen HaF Ray. Band-selective radiofrequency pulses. Volume 93 Journal of Magnetic Resonance: Elsevier; 1991. p 93–141. [Google Scholar]
- 23.Bendel P, Margalit R, Salomon Y. Optimized 1H MRS and MRSI methods for the in vivo detection of boronophenylalanine. Magn Reson Med 2005;53(5):1166–1171. [DOI] [PubMed] [Google Scholar]
- 24.Pauly J, Le Roux P, Nishimura D, Macovski A. Parameter relations for the Shinnar-Le Roux selective excitation pulse design algorithm [NMR imaging]. IEEE Trans Med Imaging 1991;10(1):53–65. [DOI] [PubMed] [Google Scholar]
- 25.Shinnar M, Leigh JS. The application of spinors to pulse synthesis and analysis. Magn Reson Med 1989;12(1):93–98. [DOI] [PubMed] [Google Scholar]
- 26.Shinnar M, Eleff S, Subramanian H, Leigh JS. The synthesis of pulse sequences yielding arbitrary magnetization vectors. Magn Reson Med 1989;12(1):74–80. [DOI] [PubMed] [Google Scholar]
- 27.Shinnar M, Bolinger L, Leigh JS. The synthesis of soft pulses with a specified frequency response. Magn Reson Med 1989;12(1):88–92. [DOI] [PubMed] [Google Scholar]
- 28.O RJ, C ID. The correction of transient B 0 field shifts following the application of pulsed gradients by phase correction in the time domain. Journal of Magnetic Resonance 1986;69:151–155. [Google Scholar]
- 29.Klose U In vivo proton spectroscopy in presence of eddy currents. Magn Reson Med 1990;14(1):26–30. [DOI] [PubMed] [Google Scholar]
- 30.Michaeli S, Garwood M, Zhu XH, DelaBarre L, Andersen P, Adriany G, Merkle H, Ugurbil K, Chen W. Proton T2 relaxation study of water, N-acetylaspartate, and creatine in human brain using Hahn and Carr-Purcell spin echoes at 4T and 7T. Magn Reson Med 2002;47(4):629–633. [DOI] [PubMed] [Google Scholar]
- 31.Wright PJ, Mougin OE, Totman JJ, Peters AM, Brookes MJ, Coxon R, Morris PE, Clemence M, Francis ST, Bowtell RW, Gowland PA. Water proton T1 measurements in brain tissue at 7, 3, and 1.5 T using IR-EPI, IR-TSE, and MPRAGE: results and optimization. MAGMA 2008;21(1–2):121–130. [DOI] [PubMed] [Google Scholar]
- 32.Wolf DH, Satterthwaite TD, Loughead J, Pinkham A, Overton E, Elliott MA, Dent GW, Smith MA, Gur RC, Gur RE. Amygdala abnormalities in first-degree relatives of individuals with schizophrenia unmasked by benzodiazepine challenge. Psychopharmacology (Berl) 2011;218(3):503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roalf DR, Nanga RP, Rupert PE, Hariharan H, Quarmley M, Calkins ME, Dress E, Prabhakaran K, Elliott MA, Moberg PJ, Gur RC, Gur RE, Reddy R, Turetsky BI. Glutamate imaging (GluCEST) reveals lower brain GluCEST contrast in patients on the psychosis spectrum. Mol Psychiatry 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging 2001;20(1):45–57. [DOI] [PubMed] [Google Scholar]
- 35.Ernst T, Kreis R, Ross BD. Absolute Quantitation of Water and Metabolites in the Human Brain. I. Compartments and Water. Journal of Magnetic Resonance, Series B 1993:1–8. [Google Scholar]
- 36.Christiansen P, Toft PB, Gideon P, Danielsen ER, Ring P, Henriksen O. MR-visible water content in human brain: a proton MRS study. Magn Reson Imaging 1994;12(8):1237–1244. [DOI] [PubMed] [Google Scholar]
- 37.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2019. [Google Scholar]
- 38.Bates D, Mächler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. 2015 2015;67(1):48. [Google Scholar]
- 39.Halekoh U, Højsgaard S. A Kenward-Roger Approximation and Parametric Bootstrap Methods for Tests in Linear Mixed Models – The R Package pbkrtest. 2014 2014;59(9):32. [Google Scholar]
- 40.Arias-Mendoza F, Brown TR. In vivo measurement of phosphorous markers of disease. Dis Markers 2003;19(2–3):49–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kemp GJ, Ahmad RE, Nicolay K, Prompers JJ. Quantification of skeletal muscle mitochondrial function by 31P magnetic resonance spectroscopy techniques: a quantitative review. Acta Physiol (Oxf) 2015;213(1):107–144. [DOI] [PubMed] [Google Scholar]
- 42.de Graaf RA, Nicolay K. Adiabatic water suppression using frequency selective excitation. Magn Reson Med 1998;40(5):690–696. [DOI] [PubMed] [Google Scholar]
- 43.Ogg RJ, Kingsley PB, Taylor JS. WET, a T1- and B1-insensitive water-suppression method for in vivo localized 1H NMR spectroscopy. J Magn Reson B 1994;104(1):1–10. [DOI] [PubMed] [Google Scholar]
- 44.Tkác I, Starcuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med 1999;41(4):649–656. [DOI] [PubMed] [Google Scholar]
- 45.Haase A, Frahm J, Hänicke W, Matthaei D. 1H NMR chemical shift selective (CHESS) imaging. Phys Med Biol 1985;30(4):341–344. [DOI] [PubMed] [Google Scholar]
- 46.Kalpouzos G, Chételat G, Baron JC, Landeau B, Mevel K, Godeau C, Barré L, Constans JM, Viader F, Eustache F, Desgranges B. Voxel-based mapping of brain gray matter volume and glucose metabolism profiles in normal aging. Neurobiol Aging 2009;30(1):112–124. [DOI] [PubMed] [Google Scholar]
- 47.Boumezbeur F, Mason GF, de Graaf RA, Behar KL, Cline GW, Shulman GI, Rothman DL, Petersen KF. Altered brain mitochondrial metabolism in healthy aging as assessed by in vivo magnetic resonance spectroscopy. J Cereb Blood Flow Metab 2010;30(1):211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1. Pulse shapes and the Bloch simulation MY profile of 90° and 180° pulses. a,b) pulse shape and simulation profile of E-BURP-1 90° pulse. c,d) pulse shape and simulation profile of SLR 180° pulse
Supporting Information Figure S2. Effect of block averaging of the NAD+ spectra. Number of blocks used was a) 256, b) 32, c) 8, and d) 1, on a dataset acquired from one volunteer
Supporting Information Figure S3. Water and NAD+ 1H MR spectra. The full spectra from water and NAD+ scans are depicted in a and b respectively showing the selective excitation of water and downfield region using the pulse sequence. The spectral bandwidth for all scans was 4 kHz centered at 4.7 ppm