Abstract
The predominance of A(H3N2) virus in recent influenza seasons has resulted in rigorous investigation on hemagglutinin, but little attention has been paid to the potential role of neuraminidase (NA). Here we show that since 2016, the S245N/S247T (introducing an N-linked glycosylation site at residue 245) and P468H mutations contributed to antigenic drift of the NA of circulating A(H3N2) viruses, compared with earlier viruses represented by the A/Hong Kong/4801/2014 vaccine strain. As a result, some human monoclonal antibodies, including those that have broad-reactivity with NA of the reference 1957 A(H2N2) and 1968 A(H3N2) pandemic viruses as well as contemporary seasonal A(H3N2) strains, lost binding to NA. This antigenic drift also reduced NA antibody-based protection against in vivo virus challenge. X-ray crystallography showed that the NA245 glycosylation site is within a conserved epitope that overlaps the NA active site, explaining why it impacts antibody binding. Our findings suggest that NA antigenic drift may impact protection against influenza virus infection, highlighting the importance of including NA antigenicity for consideration in optimizing influenza vaccines.
Introduction
Despite tremendous efforts to match the hemagglutinin (HA) antigenicity of vaccine strains to that of circulating influenza viruses, vaccine effectiveness for one or more of the vaccine components can be low during some influenza seasons. This has been the case for A(H3N2) virus in recent years. During recent influenza seasons, A(H3N2) virus predominated, causing significant number of hospitalizations in many countries, with excess mortality in elderly people1–4. In the 2017 Southern Hemisphere influenza season, the number of influenza notifications in Australia, mostly due to A(H3N2) virus infection, reached more than 210,000 by mid-October, far exceeding the cases reported during the 2009 A(H1N1) influenza pandemic3. The vaccine effectiveness estimates for A(H3N2) virus in the United States during the 2014–15, 2016–17 and 2017–18 seasons were approximately 6%, 32% and 25%, respectively5,6. The estimates were even lower in other areas of the world: 17% during the 2017–18 season in Canada and 10% during the 2017 season in Australia2,3,7.
While the poor vaccine performance against A(H3N2) virus is probably multifactorial3, it has mostly been attributed to HA, including antigenic mismatch between HAs of vaccine strains and the circulating A(H3N2) viruses in the 2014–15 season8,9, and the egg-adaptive amino acid mutations in HA that occurred during the egg-based virus isolation and vaccine manufacturing process, e.g., L194P and T160K10,11. However, solid data for the superiority of vaccines without these mutations (e.g., cell-based, inactivated vaccines and recombinant HA-based vaccines) are lacking. Neuraminidase (NA)-specific immunity also provides protection against influenza, and antibodies against HA and NA contribute independently to the protection12–14; however, the importance of NA-specific immunity has long been neglected. Like HA, NA acquires point mutations during the circulation of influenza viruses in the host, resulting in change in antigenicity, the so-called antigenic drift15,16. In this study, we identified an N-linked glycosylation site at residue 245 of the NA (NA245) of circulating A(H3N2) viruses that significantly reduced the binding of NA by antibodies, including broadly-reactive human monoclonal antibodies (mAbs), to a previously conserved NA epitope; we demonstrated that the glycosylation at NA245 and a P468H mutation has become fixed in the circulating A(H3N2) viruses since 2016 and contributed to antigenic drift of the NA of these viruses, which is consistent with reduced protection against virus challenge in the mouse model.
Results
Acquisition of an N-linked glycosylation site at NA amino acid position 245 of circulating A(H3N2) viruses.
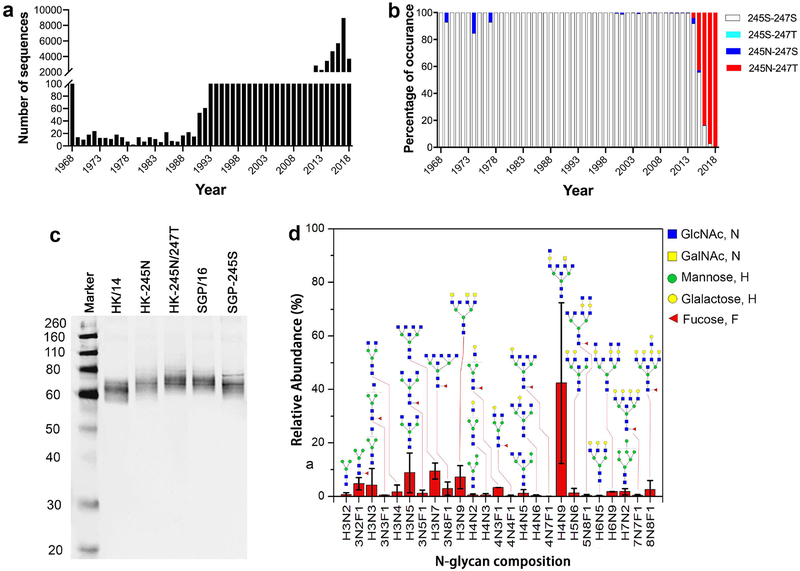
By analyzing ~43,000 NA sequences of human A(H3N2) viruses that are available in public databases (Fig. 1a), we found that amino acid mutations at residues 245 and 247 are of special interest. In 1969, 1974, and 1977, following the outbreak of pandemic A(H3N2) virus in 1968, a single amino acid mutation, S245N, was present in the NA of some A(H3N2) isolates (Fig. 1b), but the mutation was not detected again until >20 years later, when it was found among a small number of A(H3N2) isolates. Then, in 2014, the S245N mutation was present in more A(H3N2) viruses, and the double mutation S245N/S247T also emerged. This double mutation rapidly surged to be in ~50% of A(H3N2) isolates in 2015, and by 2016 and 2017, it was present in nearly all A(H3N2) viruses circulating in humans (Fig. 1b). Interestingly, this double mutation occurred in the NA of A(H3N2) strains of 3C.2a HA clade and various subclades, but not 3C.3a and 3C.3b clades (Supplementary Table 1). Both S245N and S245N/S247T mutations are predicted to introduce an N-linked glycosylation site at NA245, prompting us to investigate the effect of these mutations on NA antigenicity and the in vivo protection against virus infection provided by the NA-specific immunity.
Fig. 1. An N-linked glycosylation site acquired at NA residue 245 of circulating A(H3N2) viruses.
a, b, The presence of S245N and/or S247T mutations in the NA of seasonal A(H3N2) viruses from 1968 to 2018. Shown are the number of NA sequences analyzed by years (a) and the percentage of the indicated mutations (b). Note that only 2 NAs in 2016 and 1 in 2017 possessed 245S-247T, the percentage is too low to be discernable in panel (b). c, Western blot image of NAs with or without the NA245 glycosylation site. HK/14: A/Hong Kong/4801/2014 NA; HK-245N: mutant HK/14 NA with S245N mutation; HK-245N/247T NA: mutant HK/14 NA with S245N/S247T mutation; SGP/16: A/Singapore/INFIMH-16–0019/2016 NA; SGP-245S: mutant SGP/16 NA with N245S mutation. d, The profile of N-linked glycosylation on wt SGP/16 NA. Analysis was performed by mass-spectrometry with A(H6N2) virus containing wt SGP/16 NA (H6N2SGP/16).
We performed Western blot analysis with the wild-type (wt) NAs of A/Hong Kong/4801/2014 (HK/14) and A/Singapore/INFIMH-16–0019/2016 (SGP/16), as well as mutant HK/14 and SGP/16 NAs that bear mutations to introduce or remove the NA245 glycosylation site (Fig. 1c). HK/14, the A(H3N2) vaccine virus for the 2016–17 and 2017–18 Northern Hemisphere and the 2016 and 2017 Southern Hemisphere influenza seasons, is representative of earlier A(H3N2) viruses that lacked the NA245 glycosylation site (encoded S245/A246/S247, Supplementary Fig. 1). SGP/16, the A(H3N2) vaccine virus for the 2018–19 Northern Hemisphere and 2018 Southern Hemisphere influenza seasons, is typical of recently circulating A(H3N2) viruses that possessed the potential NA245 glycosylation site motif (N245/A246/T247). We used reassortant A(H6N2) viruses in Western blot and other assays in the present study to be consistent with using such viruses in NA inhibition (NI) assays17,18 and in our mouse challenge experiments. As shown in Fig. 1c, HK-245N and HK-245N/247T, i.e., HK/14 NAs with S245N and S245N/S247T mutations, respectively, migrated at a higher molecular weight than wt HK/14 NA, while mutant SGP/16 NA with N245S mutation (SGP-245S) migrated at a lower molecular weight than wt SGP/16 NA, indicating that the S245N and S245N/S247T mutations indeed result in glycosylation at NA245.
The glycosylation at NA245 was also verified by glycoproteomics-mass spectrometry, which showed that glycans are present at residue 245 in wt SGP/16 NA but not mutant SGP-245S NA (Supplementary Fig. 2). The glycans added to residue 245 are mainly complex N-linked glycans (Fig. 1d), which can be expected to provide a large hydrodynamic radius covering protein backbone near it. Based on these results, we conclude that A(H3N2) viruses with the NA245 glycosylation site emerged in 2014 and have rapidly become dominant in A(H3N2) strains circulating in humans since 2016.
Glycosylation at NA245 and a mutation at residue 468 result in significant NA antigenic drift.
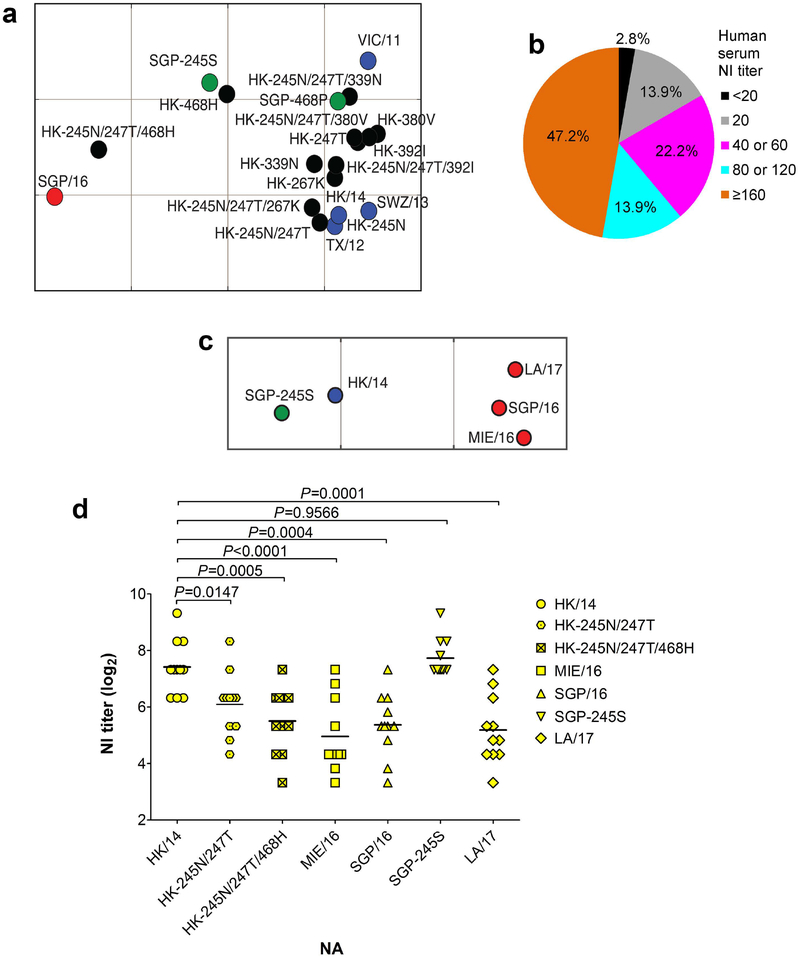
The glycosylation at NA245 decreases the enzymatic activity of NA, as indicated by enzyme-linked lectin assay (ELLA)17, MU-NANA assay18,19, and plaque assay (Supplementary Fig. 3). The 250-loop area is an important antigenic domain on NA18,20,21; therefore, we proceeded to investigate whether the glycosylation at NA245 also affects NA antigenicity. Ferret antisera raised against A(H3N2) vaccine viruses [e.g., A/Victoria/361/2011 (VIC/11), A/Texas/50/2012, A/Switzerland/9715293/2013 (SWZ/13), and HK/14], which lack the NA245 glycosylation site (Supplementary Fig. 1), were tested for inhibition of SGP/16 NA. As measured by ELLA, VIC/11, TX/12, SWZ/13 and HK/14 ferret antisera exhibited significantly (16- to 32-fold) reduced inhibition of SGP/16 NA, while SGP/16 ferret antisera had ≥ 4-fold lower NI titers with these vaccine virus NAs (except VIC/11 NA) than the homologous titers (Fig. 2a and Supplementary Table 2). These results suggest significant NA antigenic drift between the circulating A(H3N2) viruses and vaccine strains. When N245S mutation was introduced in SGP/16 NA to remove the NA245 glycosylation site, the inhibition by ferret antisera against vaccine strains (e.g., VIC/11 and HK/14) significantly recovered (Supplementary Table 2), highlighting the critical role of the NA245 glycosylation site in the observed NA antigenic drift.
Fig. 2. Antigenic drift of the NA of circulating A(H3N2) viruses demonstrated by ferret and human antisera.
a, Antigenic difference between wt NAs of A/Hong Kong/4801/2014 (HK/14) and A/Singapore/INFIMH-16–0019/2016 (SGP/16) and mutant HK/14 and SGP/16 NAs with various amino acid substitutions, shown by antigenic map generated with NI titers (Supplementary Table 2) of ferret antisera against A(H3N2) vaccine viruses A/Victoria/361/2011 (VIC/11), A/Texas/50/2012 (TX/12), A/Switzerland/9715293/2013 (SWZ/13), HK/14 or SGP/16. Blue dots: wt NAs of VIC/11, TX/12, SWZ/13, and HK/14; black dots: mutant HK/14 NAs; red dot, wt SGP/16 NA; green dots: mutant SGP/16 NAs. The antigenic cartography was constructed using AntigenMap (http://sysbio.cvm.msstate.edu/AntigenMap). Each gridline (horizontal and vertical) in the map represents 1 antigenic unit distance corresponding to a 2-fold difference in NI titers. b, The percentage of the indicated NI antibody titers (against HK/14 NA) of 36 human serum samples from persons who had been administered a 2017‒18 season quadrivalent influenza vaccine, which contained HK/14 as the A(H3N2) vaccine component. c, Antigenic difference between the NA of vaccine strain HK/14 and NAs of circulating A(H3N2) viruses A/Mie/26/2016 (MIE/16), SGP/16, A/Louisiana/32/2017 (LA/17), and mutant SGP/16 NA with N245S mutation (SGP-245S). The antigenic map was generated with NI titers of 22 human serum samples that had NI titers ≥ 80 against HK/14 NA (Supplementary Table 3). Blue dot: wt HK/14 NA; red dots: wt NAs of MIE/16, SGP/16, and LA/17; green dot: SGP-245S NA. d, Eleven of the 22 human serum samples described in (c) have ≥4-fold lower NI titers against at least one of the tested NAs of circulating A(H3N2) viruses (MIE/16, SGP/16, and LA/17) than with HK/14 NA, the titers against HK-245N/247T, HK-245N/247T/468H, and SGP-245S NAs were also included for comparison.
Of interest, the inhibition of HK/14 NA by HK/14 ferret antisera was not significantly affected by introducing the NA245 glycosylation site alone (S245N or S245N/S247T) (Fig. 2a and Supplementary Table 2), implying that other amino acid substitutions also contributed to the change in NA antigenicity. Besides amino acid difference at residues 245 and 247, there are additional sequence variations between HK/14 and SGP/16 NAs at residues 212, 231, 267, 339, 380, 392, and 468 (Supplementary Fig. 1). Of these, residue 212 is on the bottom surface of the NA head and residue 231 is not on the NA surface, such that neither is likely to affect NA antigenicity; furthermore, introduction of the residues found in SGP/16 NA into the HK/14 NA individually at positions 247, 267, 339, 380, 392, or 468 resulted in minimal loss of NI titers of HK/14 ferret antisera (≤ 2-fold) (Supplementary Table 2).
We then introduced each of the residues at positions 267, 339, 380, 392, and 468 of SGP/16 NA to HK-245N/247T NA. Mutations at residues 267, 339, 380, and 392 generally increased the inhibition of HK-245N/247T NA by HK/14 ferret antisera; however, the addition of mutation P468H to HK-245N/247T NA (i.e., from a P in HK-245N/247T NA to H in SGP/16 NA) resulted in 8-fold lower NI titers of HK/14 ferret antisera (Supplementary Table 2). The P468H mutation surged in 2015, and by 2016 the majority of circulating A(H3N2) viruses possessed H468 in NA, with a small proportion of viruses bearing L468 (Supplementary Fig. 4). This mutation alone had little effect on the inhibition of HK/14 NA by the homologous HK/14 ferret antisera, but it enhanced the inhibition (6-fold) by SGP/16 ferret antisera. Similarly, the H468P substitution in SGP/16 NA resulted in efficient inhibition by HK/14 ferret antisera without significant loss of inhibition by SGP/16 ferret antisera. Test with cell-grown viruses bearing HK-245N/247T or HK-245N/247T/468H NA revealed inhibition similar to that with egg-grown viruses (Supplementary Table 2). We conclude from these analyses that the glycosylation at NA245 (due to the S245N/S247T substitutions) and a P468H mutation, which had been dominant in the circulating A(H3N2) viruses since 2016 (Fig. 1b and Supplementary Fig. 4), contributed to NA antigenic drift in relation to earlier strains represented by the vaccine strain HK/14.
NA antigenic drift of circulating A(H3N2) viruses demonstrated by human sera.
The NA antigenic drift was also analyzed with a panel of 36 human serum samples, which were collected following the administration of a 2017–18 season influenza vaccine containing HK/14-like A(H3N2) component. All but one of these samples had detectable NI antibody titers (i.e., ≥ 20, the starting dilution of serum samples in our assays) against HK/14 NA (Fig. 2b). Because a ≥ 4-fold change in antibody titer is generally thought to be antigenically significant, only 22 samples exhibiting NI titers ≥ 80 against HK/14 NA (Supplementary Table 3) were used for further analysis. Most of these 22 samples had lower (≥2-fold) NI titers against the NAs of circulating A(H3N2) viruses A/Mie/26/2016 (MIE/16), SGP/16, and A/Louisiana/32/2017 (LA/17) than with HK/14 NA (Supplementary Table 3), reflecting NA antigenic difference between the circulating A(H3N2) viruses and the vaccine strain (Fig. 2c). Furthermore, 11 of the 22 samples had ≥ 4-fold reduced NI titers against at least one of these three recent virus NAs (MIE/16, SGP/16, and LA/17). Thus, the antigenic difference between HK/14 and recent virus NAs was even greater if we compared only these 11 samples (Fig. 2d). In agreement with the findings with ferret antisera (Supplementary Table 2), NA245 glycosylation site (245N/247T) and P468H mutation together resulted in substantially reduced inhibition of NA, with 8 of the 11 samples exhibiting ≥4-fold lower NI titers than with wt HK/14 NA, while the introduction of NA245 glycosylation site alone has less impact (Fig. 2d and Supplementary Table 3). Removal of the NA245 glycosylation site from SGP/16 NA restored the inhibition by human sera, mostly with NI titers equal to or even higher than those generated with HK/14 NA (Supplementary Table 3). Taken together, with human serum samples we confirmed the NA antigenic drift of circulating A(H3N2) viruses in relation to the vaccine strain HK/14 and the critical role of the NA245 glycosylation site and amino acid mutation at residue 468.
NA antigenic drift is consistent with reduced binding by mAbs.
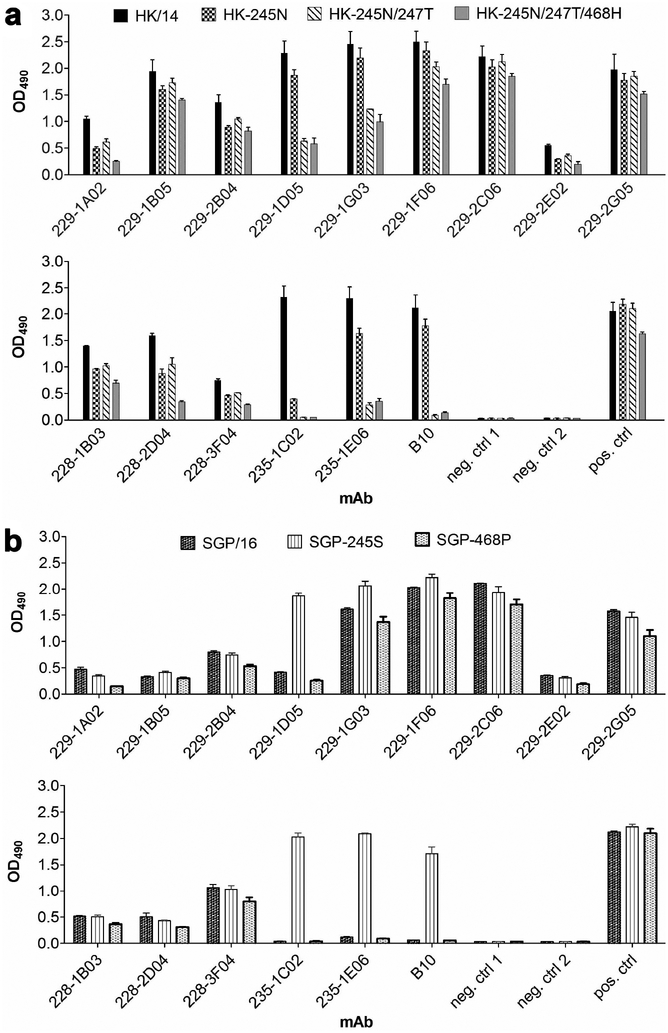
The NA antigenic drift was further investigated by testing the reactivity of a panel of mAbs. These antibodies comprised 14 human mAbs that were isolated from three influenza patients (228, 229, and 235)22 and a mouse mAb B10. Antibody B10 was raised against the NA of an A(H3N2)v swine-origin variant virus, A/Minnesota/11/2010 (MN/10) (Supplementary Fig. 5, and Supplementary Tables 4, 5), and was effective in inhibiting MN/10 NA in ELLA and MU-NANA assay. As measured by ELISA (Fig. 3a), these mAbs efficiently reacted with wt HK/14 NA. However, most of these antibodies exhibited reduced binding to HK-245N and HK-245N/247T. Some antibodies, i.e., 229–1D05, 229–1G03, 235–1C02, 235–1E06, B10, had substantially reduced or no binding. The S245N/S247T/P468H triple mutation, which resulted in significantly reduced inhibition of NA by HK/14 ferret antisera (Fig. 2a and Supplementary Table 2), caused further reduced binding of NA by mAbs 228–2D04 and 229–1A02. The reduced or abolished binding by mAbs was also observed with wt SGP/16 NA; however, with the introduction of N245S mutation, these antibodies, especially 229–1D05, 235–1C02, 235–1E06, and B10, largely recovered binding (Fig. 3b). Interestingly, the H468P mutation did not enhance the binding of SGP/16 NA by the tested mAbs. These data demonstrate that the glycosylation at NA245 interfered with the binding of NA by human mAbs, which is consistent with the NA antigenic drift observed with human sera.
Fig. 3. Glycosylation at NA245 impacts the binding of NA by human and mouse mAbs.
a, Binding of mAbs to the NA of A/Hong Kong/4801/2014 (HK/14) and mutant HK/14 NAs: HK-245N (HK/14 NA with S245N mutation), HK-245N/247T (HK/14 NA with S245N/S247T mutation), and HK-245N/247T/468H (HK/14 NA with S245N/S247T/P468H mutation). The tested mAbs included one mouse mAb, B10, and fourteen human mAbs isolated from patients #228, 229, and 235. b, Binding of these mAbs to the NA of A/Singapore/INFIMH-16–0019/2016 (SGP/16) and mutant SGP/16 NAs: SGP-245S (SGP/16 NA with N245S mutation) and SGP-468P (SGP/16 NA with H468P mutation). The binding was measured by ELISA using A(H6N2) viruses carrying each of the wt and mutant NAs. mAbs were applied at 1 μg ml−1. Mouse antisera against A(H3N2) virus were used as positive control. neg. ctrl 1: negative control 1, virus was detected by an N1-specific mAb 3A221 and Peroxidase-conjugated goat-anti-mouse IgG; neg. ctrl 2: negative control 2, virus was detected by mAb 3A2 and Peroxidase-conjugated goat-anti-human IgG. Shown are mean OD490 values ± SD from two independent assays run in duplicates.
Structural basis for the observed NA antigenic drift.
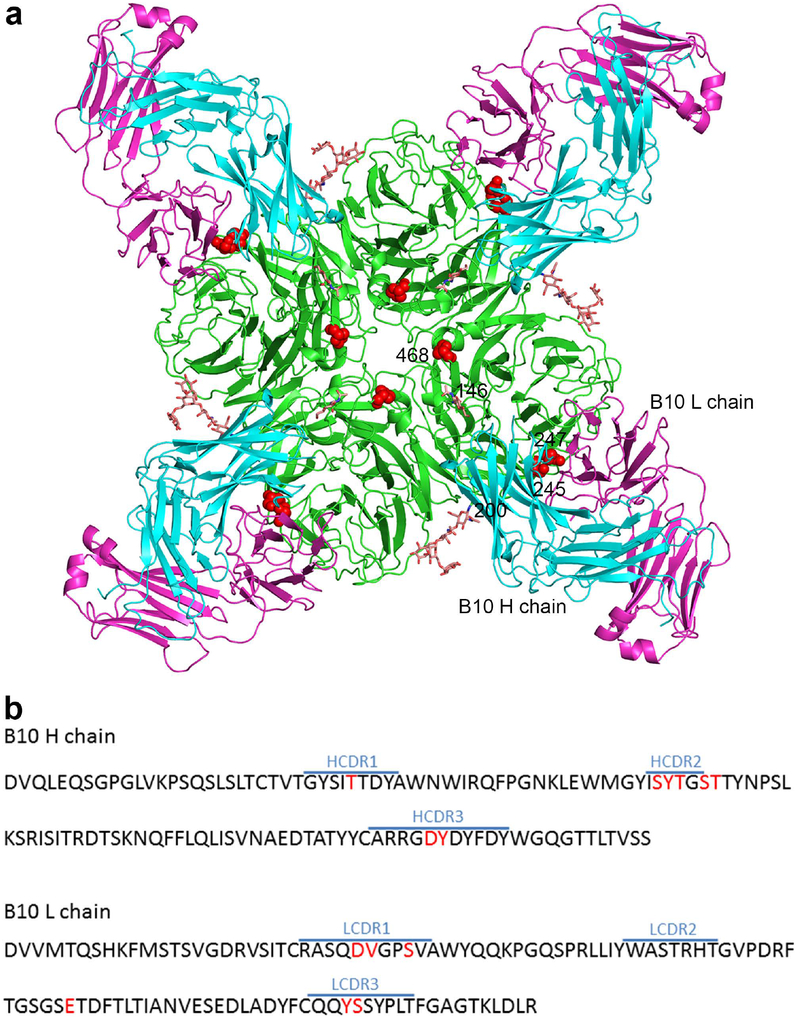
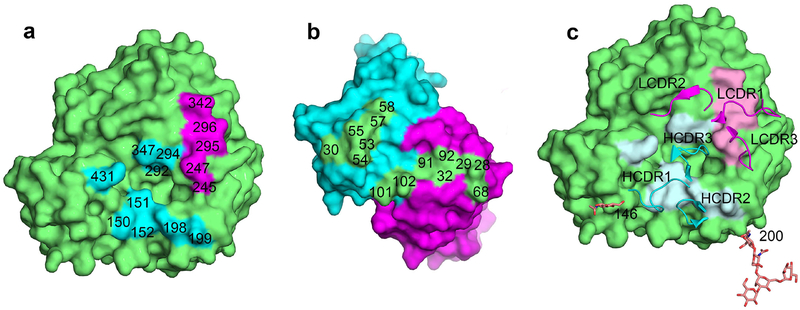
We next wanted to explore the structural basis for the observed NA antigenic change. To do this, we focused on the NA245 glycosylation site because it has a profound impact on the binding of NA by human and mouse mAbs (Fig. 3). The crystal structure of the Fab of mAb B10 in complex with MN/10 NA was solved at 2.3 Å. The statistics for data processing and refinement are presented in Supplementary Table 6. Like all previously reported NA–Fab complexes20,23–25, each B10 Fab binds to one NA monomer (Fig. 4a), and the heavy (H) and light (L) chains both contribute to the antibody footprint (Fig. 4b). As calculated by the Proteins, Interfaces, Structure and Assemblies (PISA) service26, the areas on the NA surface buried by B10 H and L chains are 457 Å2 and 399 Å2, respectively. The NA residues contacted by the H chain are H150, D151, R152, D198, N199, R292, N294, H347, and K431, and those contacted by the L chain are S245, S247, R292, N294, and N342 (Fig. 5a). Fourteen amino acids, eight from the B10 H chain and six from the L chain, are involved in the binding of NA (Fig. 5b and Supplementary Table 7). The complementary determining region 3 (CDR3) of the B10 H chain inserts into the NA active site, with Y102 in this domain forming hydrogen bond with NA residues 292 and 294 (Fig. 5c and Supplementary Table 7). The B10 L chain residue S92, within CDR3, forms a hydrogen bond with NA residue S245, and L chain residues S32 and Y91, located in CDR1 and CDR3, respectively, form hydrogen bonds with NA residue 247.
Fig. 4. Crystal structure of the Fab of mAb B10 in complex with the NA of A(H3N2) Virus.
Antibody B10 was raised against the NA of A/Minnesota/11/2010 (MN/10), and because it reacted with the NA of A(H3N2) virus until the acquisition of NA245 glycosylation site, it was selected in our study to explore the structural basis underlying the impact of this glycosylation site on antibody binding to NA. a, Overall structure of the antibody–antigen complex, showing four B10 Fabs (H chain in cyan, L chain in magenta) bind to an NA tetramer; residues 245, 247, and 468 are highlighted in red, and glycans at residues 146 and 200 are shown as sticks. b, Amino acid sequence of the B10 Fab variable regions. CDR regions of the H and L chains were defined using IMGT/V-QUEST50, and residues that are in contact with NA in the complex structure were highlighted in red.
Fig. 5. NA245 glycosylation site is in an NA epitope that overlaps the enzymatic active site of NA.
a, Footprint of mAb B10 on NA. The NA monomer was colored green, and NA residues interacting with the H chain were colored in cyan and the L chain in magenta. The interacting residues of NA were labelled. Note that residue 245 is contacted by the B10 L chain, and another five residues, 151, 152, 198, 292 and 294 are functionally and structurally critical for the NA enzymatic active site. b, Footprint of NA on B10 (color theme is the same as in a). The interacting residues of B10 were labelled. c, Positions of the B10 CDRs on NA, note that CDR3 of H chain inserts into the NA active site, glycans at residues 146 and 200 are also shown.
The NA active site includes eight highly conserved residues (R118, D151, R152, R224, E276, R292, R371, Y406) that are key for the catalytic function27; furthermore, the active site is structurally stabilized though a network of hydrogen bonds and salt bridges by a constellation of largely conserved framework residues (E119, R156, W178, S179, D198, I222, E227, H274, E277, N294, E425). Among these residues, D151, R152, D198, R292, and N294 are within the B10 epitope and have interactions with the B10 H chain (Supplementary Table 7). Thus, the B10 epitope overlaps the NA active site and involves five residues functionally or structurally critical for the enzymatic activity.
NA antigenic drift results in reduced in vivo protection.
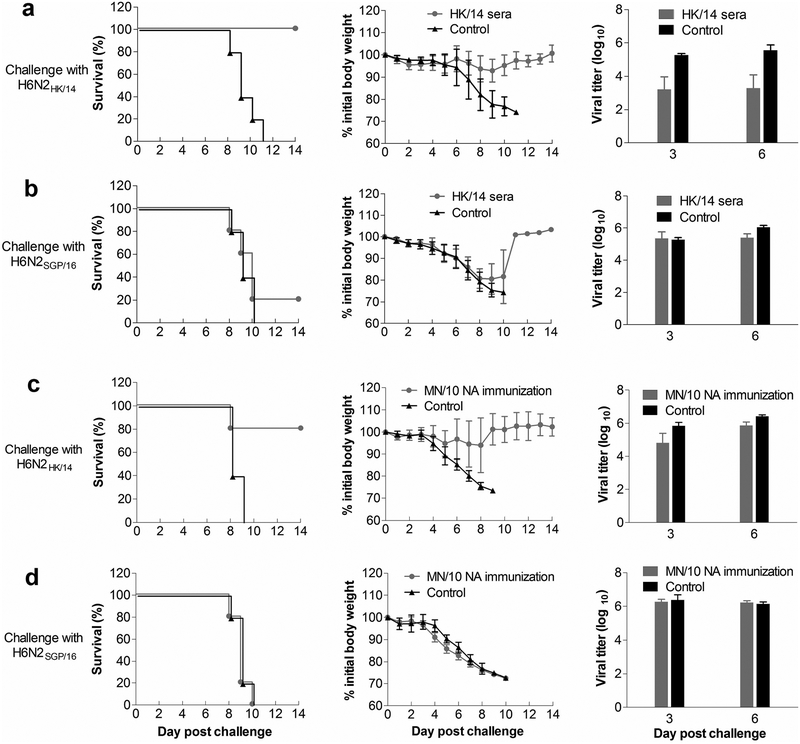
Given the NA antigenic drift demonstrated by in vitro assays, we conducted mouse experiments to assess whether it is associated with reduced protection against virus infection in vivo. The first experiment was a passive serum transfer study. HK/14 ferret antisera [raised against reassortant virus bearing HA and NA genes of HK/14 and the internal genes of an A(H1N1) virus A/Puerto Rico/8/1934] were diluted to an NI titer of 320 and transferred intraperitoneally (i.p.) to mice (200 μl/mouse). Animals were challenged intranasally (i.n.) with 10 mouse median lethal doses (LD50) of H6N2HK/14 or H6N2SGP/16, at ~16 h after serum transfer. This passive serum transfer strategy ensured that the difference in protection was likely due to NA-specific antibodies. HK/14 ferret antisera protected all mice against challenge with H6N2HK/14, but protected only one of five mice from death caused by H6N2SGP/16 (Fig. 6a, b). Compared with the control mice that received sera from naive ferrets, mice that received HK/14 ferret antisera had a substantially reduced (~100-fold) lung virus load of H6N2HK/14 but not H6N2SGP/16. These data suggest that NA antigenic drift resulted in reduced in vivo protection against virus infection provided by NA-specific serum antibodies.
Fig. 6. NA antigenic drift reduces in vivo protection against influenza virus challenge.
a, b, Mice were injected i.p. with ferret antisera against A/Hong Kong/4801/2014 (HK/14) or naïve ferret sera, followed by challenge i.n. with 10 LD50 of reassortant A(H6N2) viruses carrying the NA of HK14 (H6N2HK/14) or A/Singapore/INFIMH-16–0019/2016 (H6N2SGP/16) 16 h later. c, d, Mice were primed and boosted (at a 3-week interval) i.m. with the NA of a swine-origin A(H3N2)v virus A/Minnesota/11/2010 (MN/10) (MN/10 NA shares the B10 epitope with HK/14 NA but not SGP/16 NA) using ploy (I:C) as adjuvant or only with ploy (I:C) as negative control. The immunized mice were challenged i.n. with 10 LD50 of H6N2HK/14 or H6N2SGP/16 virus at 3 weeks after the boost. Survival and body weight loss of 5 mice in each group were monitored, and virus in lungs collected on days 3 and 6 post challenge was titrated in MDCK cells. The titers were expressed as the log10 values of plaque-formation unit (PFU) ml−1.
In the second mouse experiment, we immunized mice with recombinant MN/10 NA prior to virus challenge. MN/10 NA was selected for the experiment because it shares the B10 epitope with HK/14 NA but not SGP/16 NA (Supplementary Fig. 5 and Supplementary Tables 4, 5). After being primed and boosted intramuscularly (i.m.) with MN/10 NA, with poly (I:C) as adjuvant, mice were challenged i.n. with 10 LD50 of H6N2HK/14 or H6N2SGP/16. Animals challenged with H6N2HK/14 experienced slight weight loss and 4 out of 5 mice survived. By contrast, animals challenged with H6N2SGP/16 experienced significant weight loss and all died (Fig. 6c, d), similar to the control group, in which animals were primed and boosted only with poly (I:C) prior to virus challenge. We cannot exclude the possibility that immunogenicity to other unknown antigenic domains common between MN/10 and HK/14 NAs may have played a role in protecting mice against H6N2HK/14 challenge. However, the finding from this second mouse experiment adds evidence that NA antigenic drift of A(H3N2) viruses contributed to the reduced protection against virus infection.
Discussion
Our study demonstrates that during the recent two influenza seasons (the 2016–17 and 2017–18 Northern Hemisphere and 2016 and 2017 Southern Hemisphere influenza seasons), the NA of circulating A(H3N2) viruses was antigenically distinct from the HK/14-like A(H3N2) vaccine strains. Based on the findings from our in vitro assays and in vivo experiments, we propose that this NA antigenic drift, mainly caused by the acquisition of the NA245 glycosylation site (due to the S245N/S247T substitutions) and an amino acid mutation at NA residue 468, might have impacted NA antibody-based protection against A(H3N2) virus infection.
The observed NA antigenic drift did not result solely from the NA245 glycosylation site; however, this glycosylation site is now present in nearly all circulating A(H3N2) strains (Fig. 1b), and we provide structural evidence that it is within a unique epitope. This epitope, defined by mouse mAb B10, overlaps the NA active site (Fig. 5). Amino acid mutations introduced in this epitope may directly impact the antibody–antigen interaction, and more drastic changes, e.g., glycans added to residue 245, sterically hinder antibody binding (Supplementary Fig. 6). The S245N single mutation impacted the binding of HK/4 NA by antibodies such as 229–1A02 and 235–1C02, and the S245N/S247T double mutation further reduced the binding by some of the tested antibodies (Fig. 3a). This was likely due to that the double mutation resulted in more complete glycosylation at residue 245 (Fig.1c) to facilitate the escape from antibody inhibition, explaining why the double mutation, rather than S245N alone, has become predominant in the NA of the circulating A(H3N2) viruses (Fig. 1b). Human mAbs 235–1C02 and 235–1E06 reacted with the NAs of contemporary seasonal A(H3N2) viruses (e.g., SWZ/13 and HK/14), pandemic A(H3N2) virus A/Hong Kong/1/1968 and A(H2N2) virus A/Singapore/1/195722, and likely also target the B10 epitope (Supplementary Fig. 7). Thus, the NA245 glycosylation site disrupted a highly conserved epitope that overlaps the NA active site, likely contributed to the observed NA antigenic drift. The P468H mutation in NA became prevalent in circulating A(H3N2) viruses in 2016 (Supplementary Fig. 4) and has also contributed to the NA antigenic change (Fig. 2 and Supplementary Tables 2,3). The H468P back mutation in SGP/16 NA did not result in enhanced binding as observed with the N245S mutation, however with the presence of S245N/S247T substitutions, P468H further reduced the binding of HK/14 NA by some of the tested mAbs, e.g., 229–1A02 and 228–2D04 (Fig. 3). Unfortunately, residue 468 is not critical for the binding of NA by any of the mAbs available to us, and more efforts are needed to elucidate the role of mutation at residue 468 in determining the distinct NA antigenicity.
Although NA content is not standardized in seasonal influenza vaccines, and may vary with different vaccines, humans do develop NA-specific antibodies, likely due to NA contained in the vaccines administered28–31 or natural virus infection22. NA-specific antibodies have been recognized as a potential independent correlate of protection against influenza virus infection13,32. Most of the human serum samples tested in our assays exhibited high NI antibody titers against HK/14 (Fig. 2b and Supplementary Table 3). However, for ~50% of the tested human serum samples, NI antibody titers against the tested NAs of circulating viruses (MIE/16, SGP/16, and LA/17) were ≥ 4-fold lower than those for HK/14 NA. The reduced inhibition by NA-specific antibodies might have resulted in less effective in vivo protection. This is supported by our mouse experiments, which indicated that ferret antisera against HK/14 provided protection against challenge with A(H6N2) virus carrying HK/14 NA but not the virus possessing the antigenically-drifted SGP/16 NA.
A(H3N2) viruses undergo more frequent genetic and antigenic changes than A(H1N1) and influenza B viruses, necessitating frequent update of the A(H3N2) vaccine strains33. The World Health Organization (WHO) currently selects influenza candidate vaccine viruses based on the HA antigenicity of the circulating viruses but not that of NA. Our findings support continued surveillance of the NA of circulating influenza viruses and the inclusion of NA antigenicity for consideration in optimizing influenza vaccines.
Methods
Viruses.
Viruses used in this study included reassortant H6N2 viruses that contain the HA gene of A/turkey/Massachusetts/3740/1965 (H6N2) and the NA gene of seasonal A(H3N2) viruses A/Wuhan/359/1995 (WH/95), A/Panama/2007/1999 (PA/99), A/Wisconsin/67/2005 (WI/05), A/Victoria/361/2011 (VIC/11), A/Texas/50/2012 (TX/12), A/Switzerland/9715293/2013 (SWZ/13), A/Hong Kong/4801/2014 (HK/14), A/Singapore/INFIMH-16–0019/2016 (SGP/16), A/Mie/26/2016 (MIE/16), A/Louisiana/32/2017 (LA/17), or A(H3N2)v virus A/Minnesota/11/2010 (MN/10). These viruses were rescued with reverse genetics34 and amino acid mutations were introduced into HK/14 and SGP/16 NAs with a QuikChange multisite-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). Viruses were propagated in 9- to 11-day-old specific pathogen–free embryonated chicken eggs and titrated in Madin-Darby canine kidney (MDCK) cells, and those for ELISA were inactivated with β-propiolactone (Sigma-Aldrich, St. Louis, MO, USA) and purified by sucrose gradient centrifugation.
mAbs.
Mouse mAbs were prepared according to routine hybridoma procedures21. In brief, 6-week-old BALB/c mice (The Jackson Laboratory, Bar Harbor, ME, USA) were immunized i.m. with purified MN/10 virus and boosted intravenously on day 3 before the fusion of splenocytes with Sp2/0 cells. Hybridomas secreting N2-specific antibodies were subcloned by limiting dilution. mAbs were purified using protein G columns (GE Healthcare, Uppsala, Sweden). The variable regions of H and L chains of mAb were sequenced by ProMab Biotechnologies, Inc. (Richmond, CA, USA) and GenScript Inc. (Piscataway, NJ, USA). Human mAbs specific to N2 were generated from human peripheral lymph cells as recently reported22.
Human serum samples.
Blood samples were collected from healthy adult volunteers at three weeks after administration of a 2017‒18 quadrivalent seasonal influenza vaccine (containing the following antigens: A(H1N1)pdm09, A/Michigan/45/2015; A(H3N2), A/Hong Kong/4801/2014; B/Brisbane/60/2008; B/Phuket/3073/2013). The samples were allowed to clot at room temperature for 6 h and serum collected after centrifugation. The serum was heat inactivated at 56°C for 30 min then stored at −20°C. Serum collection was approved by the Institutional Review Board, National Institute of Biological Standards and Control, UK.
NA sequence analysis.
A total of 43,733 NA protein sequences of A(H3N2) viruses were downloaded from public databases including GISAID (the Global Initiative on Sharing All Influenza Data; https://www.gisaid.org), the Influenza Research Database (https://www.fludb.org), and the Influenza Virus Resource (https://www.ncbi.nlm.nih.gov/genomes/FLU/). Multiple sequence alignments were generated using Muscle v3.8.3135. The sequence patterns at positions 245, 247, and 468 were analyzed using a Perl program, and the number of each patterns (245S–247S, 245S–247T, 245N–247S, 245N–247T, 468P, 468H, and 468L) was stratified by year.
Western blot.
Purified virus corresponding to 1 μg of viral protein was denatured, separated by SDS-PAGE in 10% Bis-Tris precast gel with MOPS running buffer in the presence of reducing agent (Invitrogen, Calsbad, CA, USA), and transferred onto a nitrocellulose membrane using an iBlot Gel Transfer Device (Invitrogen, Calsbad, CA, USA). The NA was detected with N2-specific rabbit antiserum (Sino Biological, Inc., Beijing, China) and Odyssey IRDye 680RD secondary goat-anti-rabbit antibody (LI-COR, Lincoln, NE, USA), and the target bands were visualized by using the Odyssey Imaging System (LI-COR, Lincoln, NE, USA).
Glycosylation analysis.
Proteins were extracted from viruses by using 8 M urea in 1 M NH4HCO336, reduced by using 12 mM dithiothreitol at 37°C for 1 h, followed by alkylation using 16 mM iodoacetamide at room temperature for 1 h. Samples were diluted with water, and trypsin (Promega, Madison, WI, USA) was added at a trypsin-to-protein ratio of 1:50 and incubated overnight at 37°C. The digest was dried in a Speed-Vac vacuum (Thermo Fisher, Waltham, MA, USA) and re-suspended in 80% acetonitrile in the presence of 0.1% trifluoroacetic acid. Glycopeptides were enriched by use of a hydrophilic interaction liquid chromatography-solid phase extraction column37. The eluate was dried in Speed-Vac vacuum and re-suspended in 0.2% formic acid for liquid chromatography with tandem mass spectrometry analysis. Mass spectrometry was performed using an UltiMate LC and a Fusion Orbitrap Mass Spectrometer (Thermo Fisher, Waltham, MA, USA). Glycopeptides (1 μg) were loaded onto a C18 trap cartridge (Waters, Milford, MA, USA) and further separated by using an Easy-Spray C18 column (Waters, Milford, MA, USA). The collision energy was set as 27 and automatic gain control targets were 1.0×104; other parameters were the same as described in previous studies38. Prediction of N-linked glycosylation for wt and mutant was performed by NetNGlyc39, with scores > 0.5 representing high glycosylation probability. Glycan structures were inferred based on experimental data, presence in GlyTouCAn (https://glytoucan.org) and CFG (www.functionalglycomics.org) glycan databases and consistence with those structures found in Gallus gallus or their close relatives40,41.
ELISA.
The binding of mAbs to NA was measured by ELISA. Immulon® 2HB flat bottom microtiter plates (Thermo Scientific, Rochester, NY, USA) were coated with purified viruses at 10 μg ml−1 (100 μl/well), blocked with 15 % fetal bovine serum (Atlanta Biologics, Flowery Branch, GA) in PBS, and then incubated with mAb (1 μg ml−1) and peroxidase-conjugated goat-anti-mouse IgG (Sigma-Aldrich, St. Louis, MO, USA) or goat-anti-human IgG (SouthernBiotech, Birmingham, AL, USA). To examine the competition for binding of the B10 epitope by different mAbs, plates were coated with virus and blocked, followed by incubation with peroxidase-conjugated mAb B10 dilution [prepared in our laboratory using a peroxidase labeling kit (Roche, Indianapolis, IN, USA)] supplemented with various concentrations of the tested mAbs. After the signals were developed using o-Phenylenediamine dihydrochloride (OPD; Sigma-Aldrich, St. Louis, MO, USA) as substrate, the reaction was stopped with 1 N H2SO4, and OD490 values were read.
MU-NANA assay.
NA activity of virus was measured by MU-NANA assay18,19. Virus was 2-fold serially diluted with PBS (pH 7.4) with MgCl2 and CaCl2 (KD Medical, Columbia, MD, USA) and mixed with an equal volume (50 μl) of 100 μM MU-NANA (Sigma-Aldrich, St. Louis, MO, USA) in Microfluor® 1 Black flat bottom microtiter® plate (Thermo Scientific, Rochester, NY, USA), followed by incubation at 37°C for 1 h. The reaction was stopped with 0.1 M glycine in 25% ethanol (pH 10.7), and the signals were read using an excitation wavelength of 355 nm and an emission wavelength of 460 nm.
NA inhibition (NI) assay.
The inhibition in NA activity was measured with enzyme-linked lection (ELLA)17 and MU-NANA assay18,19 as described previously. To perform ELLA, serial dilutions of antisera or mAbs were mixed with a predetermined amount of virus diluted in MES buffered saline (pH 6.5) (KD Medical, Columbia, MD, USA) containing 1% bovine serum albumin (Sigma-Aldrich, St. Louis, MO, USA) and 0.5% Tween 20 (Sigma-Aldrich, St. Louis, MO, USA). The mixture was added to 96-well plates (Thermo Scientific, Rochester, NY, USA) coated with fetuin (Sigma-Aldrich, St. Louis, MO, USA) and incubated overnight at 37°C. Plates were washed with phosphate buffered saline with Tween-20 (PBST), followed by adding peanut agglutinin conjugated to peroxidase (Sigma-Aldrich, St. Louis, MO, USA). Plates were incubated at room temperature for 2 h in the dark and washed with PBST before the addition of OPD substrate. The reaction was stopped by adding 1 N H2SO4 and OD490 values were read. To measure inhibition of NA activity with the MU-NANA assay, a predetermined amount of virus was mixed with serial dilutions of antibody and incubated in the Microfluor® 1 Black flat bottom microtiter® plates (Thermo Scientific, Rochester, NY) at 37°C for 1 h, followed by the addition of 100 μM MU-NANA (Sigma-Aldrich, St. Louis, MO, USA) and incubation at 37°C for 1 h. The reaction was stopped by the addition of 0.1 M glycine in 25% ethanol (pH 10.7), and the signals were read (excitation, 355 nm; emission, 460 nm). In both assays, The NI titer was expressed as the reciprocal of the highest dilution that exhibited ≥50% inhibition of NA activity.
Construction of the antigenic cartography.
Antigenic cartography was constructed on the basis of NI data by using AntigenMap (http://sysbio.cvm.msstate.edu/AntigenMap)42,43. Each entry in the NI table was normalized by dividing the maximum NI value for the reference antiserum. Missing NI titers and those below the cutoff value for low reactors were analyzed by low-rank matrix completion. A titer of 10 was used as the low-reactor cutoff in the NI assay. Antigenic distance between two viruses was defined as the Euclidean distance between the NI values of the two viruses against all the antisera. Each unit of the antigenic distance corresponded to a 2-fold change in NI titer. Multidimensional scaling was used to project viruses to a 2-dimensional map by minimizing the sum-squared error between map distance and antigenic distance.
Plaque assay.
MDCK cells growing in 6-well plates were infected with diluted virus for 1 h at 37°C. After the virus inoculum was removed and washed with PBS, cells were overlaid with agar supplemented with 1 μg ml−1 of trypsin (Sigma-Aldrich, St. Louis, MO, USA). For identifying mAb escape mutant, the agar was supplemented with trypsin (1 μg ml−1) and mAb (1–2 μg -ml−1). Cells were then incubated at 37°C in 5% CO2 for 3 days and then fixed with methanol and stained with crystal violet solution to visualize plaques.
Selection of antibody escape mutant.
In brief, 0.5 ml of ascitic fluid containing mAbs or purified mAbs (1–2 mg ml−1) was incubated with 106 PFU of virus and inoculated into eggs. The potential escape variants were identified by hemagglutination assay and further cloned in a plaque assay in the presence of mAb. Three plaques with sizes larger than those formed by the parental virus were picked for each variant and propagated in eggs. Allantoic fluid was collected, and the NA gene was sequenced. Sequences were analyzed using DNASTAR Lasergene 13 software (DNASTAR, Inc., Madison, WI, USA) to identify nucleotide and amino acid changes.
NA expression and purification.
The recombinant MN/10 NA, which contains an N-terminal His-tag, tetramerization domain from the human vasodilator-stimulated phosphoprotein and a thrombin cleavage site, was expressed with a baculovirus system. Secreted NA in the culture supernatant was purified by metal affinity chromatography and size-exclusion chromatography (SEC). The protein was further subjected to trypsin cleavage and SEC for structural analyses.
Formation and purification of the Fab—NA complex.
The mAb B10 Fab, prepared by SouthernBiotech (Birmingham, AL, USA) from purified mAb B10, was mixed with purified, His-tag–depleted, recombinant MN/10 NA monomer at a 1:1 molar ratio of Fab to NA. The B10 Fab–MN/10 NA complex was purified from unbound substrates by SEC in a buffer comprising 50 mM Tris-HCl (pH 8.0) and 150 mM NaCl, eluted as a single peak between the 158 and 670-kDa molecular weight markers, and then concentrated to 14 mg ml−1.
Crystallization and data collection of the Fab–NA complex.
Initial sparse-matrix crystallization screening was carried out using Formulatrix NT8 (Formulatrix, Bedford, MA, USA). Following optimization, diffraction quality crystals were obtained at 23°C, using the sitting drop vapor diffusion method against a reservoir containing 0.1 M Tris-HCl (pH 8.5) and 2 M ammonium sulfate (MCSG-1, condition 70; Anatrace, Maumee, OH, USA). The complex dataset was collected from a single crystal to 2.3 Å resolution at the Advanced Photon Source (https://www.aps.anl.gov/) SER-CAT 22-ID beamline (wavelength 1.0 Å), and data were processed with the DENZO-SCALEPACK suite44.
Structure determination and refinement.
The B10 Fab–MN/10 NA complex structure was determined by the molecular replacement method using the program Phaser45. A strong solution was obtained for the NA component of the complex by using a single monomer of NA (PDB: 4GZS). An initial search for B10 Fab determined the position of the variable (Fv) domains using the Fv component of a mouse IgG1 Fab (PDB: 1WEJ) as a search model. A subsequent search using the constant (Fc) domains of a mouse IgG2a Fab (PDB: 3FO9) located the Fc region.
Rigid-body and restrained refinement of the molecular replacement solution was carried out using REFMAC546, and model building was performed in Coot47. 2Fo-Fc electron density was well defined throughout the model. Additional positive electron density was observed in the region of three potential N-linked glycosylation sites. Nine sugar residues were manually fitted into this density at these positions. Restrained refinement of the structure was completed in REFMAC5 using TLS refinement48. The final model was assessed using MolProbity49. All the statistics for data processing and refinement are presented in Supplementary Table 6. In the final model, 95% of the residues were in favored regions of a Ramachandran plot, and the other 5% were in additional allowed regions.
Animal experiment.
Two mouse experiments were conducted to examine the in vivo impact of NA antigenic drift; federal guidelines and protocols approved by the Food and Drug Administration Institutional Animal Care and Use Committee, were followed. The first experiment was a passive serum transfer study, in which groups of 11 DBA/2 mice (12-week old; The Jackson Laboratory, Bar Harbor, ME) were injected i.p. with HK/14 ferret antisera (0.2 ml/animal) that were diluted with PBS to an NI titer of 320, or pooled naive ferret sera (NI titer <10) diluted to the same fold as for HK/14 ferret sera; 16 h later, mice were challenged i.n. with 10 LD50 of H6N2HK/14 or H6N2SGP/16 in 50 μl of PBS. In the second experiment, groups of 11 DBA/2 mice (6-week old; The Jackson Laboratory, Bar Harbor, ME, USA) were immunized i.m. with recombinant MN/10 NA (5 μg/mouse) administered together with poly (I:C) adjuvant (Sigma-Aldrich, St. Louis, MO, USA) and then boosted with the same dose of NA with adjuvant at 3 weeks after the priming. Three weeks after the boost, animals were challenged i.n. with 10 LD50 of H6N2HK/14 or H6N2SGP/16 in 50 μl of PBS. In both experiments, 6 mice from each group were euthanized on days 3 and 6 after challenge, and lungs were sampled for virus titration in MDCK cells; the remaining 5 mice from each group were monitored for weight loss and mortality for up to 14 days; mice that lost 25% weight were euthanized.
Statistical analysis.
NI titers were compared by one-way analysis of variance (ANOVA) using GraphPad Prism version 7.0 (GraphPad Software, Inc., San Diego, CA), and probability (P) values of ≤ 0.05 indicate a statistical significance difference.
Data availability.
The data that support the findings of this study are available from the corresponding author upon request. Atomic coordinates and structure factors for the B10 Fab–MN/10 NA complex have been deposited to the protein Data Bank (www.pdb.org) under the accession code 6N6B.
Supplementary Material
Acknowledgements
This work was supported by intramural funds from the Food and Drug Administration and the Centers for Disease Control and Prevention. Lianlian Jiang were supported by training funds administered by the Oak Ridge Institute for Science and Education. The work was also supported by National Institute of Allergy and Infectious Disease (NIAID), National Institutes of Health grant numbers U19AI082724 (P.C.W.), U19AI109946 (P.C.W.), U19AI057266 (P.C.W.), R01AI116744 (X.F.W.), and the NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS, HSN272201400008C to F.K. and HHSN272201400005C to P.C.W.).
We thank St. Jude Children’s Research Hospital for providing plasmids that were used to rescue viruses. We thank Dr. Vladimir V. Lugovtsev for providing ferret antisera, Dr. Lei Li for help in sequence analysis, and Dr. Li Qi for help in statistical analysis. We thank Dr. Yanming An and Anissa Cheung for helpful discussion in the study. We are indebted to staff of the Division of Veterinary Services, CBER, FDA, for excellent animal care and the Facility for Biotechnology Resources, CBER, FDA, for sequencing work.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Food and Drug Administration and the Centers for Disease Control and Prevention.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Kissling E, Rondy M & team, I. M. I. M. s. Early 2016/17 vaccine effectiveness estimates against influenza A(H3N2): I-MOVE multicentre case control studies at primary care and hospital levels in Europe. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin 22, doi: 10.2807/1560-7917.ES.2017.22.7.30464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan SG et al. Low interim influenza vaccine effectiveness, Australia, 1 May to 24 September 2017. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin 22, doi: 10.2807/1560-7917.ES.2017.22.43.17-00707 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paules CI, Sullivan SG, Subbarao K & Fauci AS Chasing Seasonal Influenza - The Need for a Universal Influenza Vaccine. The New England journal of medicine 378, 7–9, doi: 10.1056/NEJMp1714916 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Appiah GD et al. Influenza activity - United States, 2014–15 season and composition of the 2015–16 influenza vaccine. MMWR Morb Mortal Wkly Rep 64, 583–590 (2015). [PMC free article] [PubMed] [Google Scholar]
- 5.Flannery B et al. Interim Estimates of 2017–18 Seasonal Influenza Vaccine Effectiveness - United States, February 2018. MMWR Morb Mortal Wkly Rep 67, 180–185, doi: 10.15585/mmwr.mm6706a2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmerman RK et al. 2014–2015 Influenza Vaccine Effectiveness in the United States by Vaccine Type. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 63, 1564–1573, doi: 10.1093/cid/ciw635 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skowronski DM et al. Early season co-circulation of influenza A(H3N2) and B(Yamagata): interim estimates of 2017/18 vaccine effectiveness, Canada, January 2018. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin 23, doi: 10.2807/1560-7917.ES.2018.23.5.18-00035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Mello T et al. Update: Influenza activity--United States, September 28, 2014-February 21, 2015. MMWR Morb Mortal Wkly Rep 64, 206–212 (2015). [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers BS, Parkhouse K, Ross TM, Alby K & Hensley SE Identification of Hemagglutinin Residues Responsible for H3N2 Antigenic Drift during the 2014–2015 Influenza Season. Cell Rep 12, 1–6, doi: 10.1016/j.celrep.2015.06.005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zost SJ et al. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proceedings of the National Academy of Sciences of the United States of America 114, 12578–12583, doi: 10.1073/pnas.1712377114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu NC et al. A structural explanation for the low effectiveness of the seasonal influenza H3N2 vaccine. PLoS pathogens 13, e1006682, doi: 10.1371/journal.ppat.1006682 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy BR, Kasel JA & Chanock RM Association of serum anti-neuraminidase antibody with resistance to influenza in man. The New England journal of medicine 286, 1329–1332, doi: 10.1056/NEJM197206222862502 (1972). [DOI] [PubMed] [Google Scholar]
- 13.Monto AS et al. Antibody to Influenza Virus Neuraminidase: An Independent Correlate of Protection. The Journal of infectious diseases 212, 1191–1199, doi: 10.1093/infdis/jiv195 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Monto AS & Kendal AP Effect of neuraminidase antibody on Hong Kong influenza. Lancet 1, 623–625 (1973). [DOI] [PubMed] [Google Scholar]
- 15.Treanor J Influenza vaccine--outmaneuvering antigenic shift and drift. The New England journal of medicine 350, 218–220, doi: 10.1056/NEJMp038238 (2004). [DOI] [PubMed] [Google Scholar]
- 16.De Jong JC, Rimmelzwaan GF, Fouchier RA & Osterhaus AD Influenza virus: a master of metamorphosis. J Infect 40, 218–228, doi: 10.1053/jinf.2000.0652 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Gao J, Couzens L & Eichelberger MC Measuring Influenza Neuraminidase Inhibition Antibody Titers by Enzyme-linked Lectin Assay. J Vis Exp, doi: 10.3791/54573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan H et al. Comparison of the Efficacy of N9 Neuraminidase-Specific Monoclonal Antibodies against Influenza A(H7N9) Virus Infection. Journal of virology 92, doi: 10.1128/JVI.01588-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichelberger MC, Hassantoufighi A, Wu M & Li M Neuraminidase activity provides a practical read-out for a high throughput influenza antiviral screening assay. Virology journal 5, 109, doi: 10.1186/1743-422X-5-109 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venkatramani L et al. An epidemiologically significant epitope of a 1998 human influenza virus neuraminidase forms a highly hydrated interface in the NA-antibody complex. Journal of molecular biology 356, 651–663, doi: 10.1016/j.jmb.2005.11.061 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Wan H et al. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. Journal of virology 87, 9290–9300, doi: 10.1128/JVI.01203-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen YQ et al. Influenza Infection in Humans Induces Broadly Cross-Reactive and Protective Neuraminidase-Reactive Antibodies. Cell 173, 417–429 e410, doi: 10.1016/j.cell.2018.03.030 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan H et al. Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers. Nat Commun 6, 6114, doi: 10.1038/ncomms7114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JT & Air GM Contacts between influenza virus N9 neuraminidase and monoclonal antibody NC10. Virology 300, 255–268 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Colman PM et al. Three-dimensional structure of a complex of antibody with influenza virus neuraminidase. Nature 326, 358–363, doi: 10.1038/326358a0 (1987). [DOI] [PubMed] [Google Scholar]
- 26.Krissinel E & Henrick K Inference of macromolecular assemblies from crystalline state. Journal of molecular biology 372, 774–797, doi: 10.1016/j.jmb.2007.05.022 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Jagadesh A, Salam AA, Mudgal PP & Arunkumar G Influenza virus neuraminidase (NA): a target for antivirals and vaccines. Archives of virology 161, 2087–2094, doi: 10.1007/s00705-016-2907-7 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Cate TR et al. A high dosage influenza vaccine induced significantly more neuraminidase antibody than standard vaccine among elderly subjects. Vaccine 28, 2076–2079, doi: 10.1016/j.vaccine.2009.12.041 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassantoufighi A et al. A practical influenza neutralization assay to simultaneously quantify hemagglutinin and neuraminidase-inhibiting antibody responses. Vaccine 28, 790–797, doi: 10.1016/j.vaccine.2009.10.066 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Eichelberger MC, Morens DM & Taubenberger JK Neuraminidase as an influenza vaccine antigen: a low hanging fruit, ready for picking to improve vaccine effectiveness. Curr Opin Immunol 53, 38–44, doi: 10.1016/j.coi.2018.03.025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Couch RB et al. Randomized comparative study of the serum antihemagglutinin and antineuraminidase antibody responses to six licensed trivalent influenza vaccines. Vaccine 31, 190–195, doi: 10.1016/j.vaccine.2012.10.065 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Memoli MJ et al. Evaluation of Antihemagglutinin and Antineuraminidase Antibodies as Correlates of Protection in an Influenza A/H1N1 Virus Healthy Human Challenge Model. mBio 7, e00417–00416, doi: 10.1128/mBio.00417-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flannery B et al. Interim Estimates of 2016–17 Seasonal Influenza Vaccine Effectiveness - United States, February 2017. MMWR Morb Mortal Wkly Rep 66, 167–171, doi: 10.15585/mmwr.mm6606a3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann G et al. Generation of influenza A viruses entirely from cloned cDNAs. Proceedings of the National Academy of Sciences of the United States of America 96, 9345–9350 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113, doi: 10.1186/1471-2105-5-113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang S et al. Integrated glycoprotein immobilization method for glycopeptide and glycan analysis of cardiac hypertrophy. Analytical chemistry 87, 9671–9678, doi: 10.1021/acs.analchem.5b01663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.An Y et al. Comparative glycomics analysis of influenza Hemagglutinin (H5N1) produced in vaccine relevant cell platforms. J Proteome Res 12, 3707–3720, doi: 10.1021/pr400329k (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang S, Wu WW, Shen RF, Bern M & Cipollo J Identification of Sialic Acid Linkages on Intact Glycopeptides via Differential Chemical Modification Using IntactGIG-HILIC. J Am Soc Mass Spectrom 29, 1273–1283, doi: 10.1007/s13361-018-1931-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S & Brunak S Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649, doi: 10.1002/pmic.200300771 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Paz-Parente J et al. Primary structure of a novel N-glycosidic carbohydrate unit, derived from hen ovomucoid. A 500-MHz 1H-NMR study. FEBS letters 152, 145–152 (1983). [DOI] [PubMed] [Google Scholar]
- 41.Parente JP et al. A novel type of carbohydrate structure present in hen ovomucoid. The Journal of biological chemistry 257, 13173–13176 (1982). [PubMed] [Google Scholar]
- 42.Barnett JL, Yang J, Cai Z, Zhang T & Wan XF AntigenMap 3D: an online antigenic cartography resource. Bioinformatics 28, 1292–1293, doi: 10.1093/bioinformatics/bts105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai Z, Zhang T & Wan XF A computational framework for influenza antigenic cartography. PLoS Comput Biol 6, e1000949, doi: 10.1371/journal.pcbi.1000949 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otwinowski Z & Minor W Processing of X-ray diffraction data collected in oscillation mode. Methods in enzymology 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 45.McCoy AJ, Grosse-Kunstleve RW, Storoni LC & Read RJ Likelihood-enhanced fast translation functions. Acta crystallographica. Section D, Biological crystallography 61, 458–464, doi: 10.1107/S0907444905001617 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Collaborative Computational Project N The CCP4 suite: programs for protein crystallography. Acta crystallographica. Section D, Biological crystallography 50, 760–763, doi: 10.1107/S0907444994003112 (1994). [DOI] [PubMed] [Google Scholar]
- 47.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography 60, 2126–2132, doi: 10.1107/S0907444904019158 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Winn MD, Isupov MN & Murshudov GN Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta crystallographica. Section D, Biological crystallography 57, 122–133 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Davis IW et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35, W375–383, doi: 10.1093/nar/gkm216 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brochet X, Lefranc MP & Giudicelli V IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36, W503–508, doi: 10.1093/nar/gkn316 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. Atomic coordinates and structure factors for the B10 Fab–MN/10 NA complex have been deposited to the protein Data Bank (www.pdb.org) under the accession code 6N6B.