Abstract
Nanomaterials are a relatively new class of materials that acquire novel properties based on their reduced size. While these materials have widespread use in consumer products and industrial applications, the potential health risks associated with exposure to them remain to be fully characterized. Carbon nanotubes are among the most widely used nanomaterials and have high potential for human exposure by inhalation. These nanomaterials are known to penetrate the cell membrane and interact with intracellular molecules, resulting in a multitude of documented effects, including oxidative stress, genotoxicity, impaired metabolism, and apoptosis. While the capacity for carbon nanotubes to damage nuclear DNA has been established, the effect of exposure on mitochondrial DNA (mtDNA) is relatively unexplored. In this study, we investigated the potential of multi-walled carbon nanotubes (MWCNTs) to impair mitochondrial gene expression and function in human bronchial epithelial cells (BECs). Primary BECs were exposed to sub-cytotoxic doses (up to 3 μg/ml) of MWCNTs for 5 days and assessed for changes in expression of all mitochondrial protein-coding genes, heteroplasmies, and insertion/deletion mutations (indels). Exposed cells were also measured for cytotoxicity, metabolic function, mitochondrial abundance, and mitophagy. We found that MWCNTs upregulated mitochondrial gene expression, while significantly decreasing oxygen consumption rate and mitochondrial abundance. Confocal microscopy revealed induction of mitophagy by 2 hours of exposure. Mitochondrial DNA heteroplasmy and insertion/deletion mutations were not significantly affected by any treatment. We conclude that carbon nanotubes cause mitochondrial dysfunction that leads to mitophagy in exposed BECs via a mechanism unrelated to its reported genotoxicity.
Keywords: carbon nanotubes, mitochondria, genotoxicity, epithelial, in vitro
Introduction
Over the last 20 years, nanomaterials have emerged as one of the most promising advances in material science. The unique properties particles acquire at the nano-scale (having at least 1 dimension less than 100 nm) have resulted in an explosion of new applications and products, including construction materials1, medical advances2–3, industrial applications4–5, hydrophobic or absorbent textiles6, superblack or reflective surfaces7, and many others8–10. Carbon nanotubes are already ubiquitous in consumer products, second only to antimicrobial nanosilver in its prevalence11. Multi-walled carbon nanotubes (MWCNTs), which consist of multiple layers of graphene rolled into a tube, possess the highest tensile-strength of any other man-made material12, as well as semiconductivity and hydrophobicity13–14, properties which have made them valuable for use in concrete reinforcement, sporting goods, space travel, electronics/sensors, stain-free fabrics, and even cancer treatments15–16. Consequently, the public’s exposure to MWCNTs and risks of inhalation, both by consumers and as an occupational hazard, are of great concern.
Despite the incorporation of these materials into the public domain, early studies into the health risks associated with exposure to carbon nanotubes have discovered toxic effects including oxidative stress17–18, cytotoxicity/apoptosis19–21, genotoxicity22–23, and impaired cellular function and differentiation24–26. Clearance of nanotubes following inhalation exposure is typically very slow, persisting in the lungs for several months afterwards27. As a fibrous material with a high aspect ratio, carbon nanotubes can cause frustrated phagocytosis20 by resident macrophages, which can lead to chronic inflammation28–29, granulomas30, and pulmonary fibrosis17, 31. Nanoparticles are small enough to penetrate the cell membrane32–33, and can enter the cytosol either passively33–34 or following phagocytosis35–36 by the cell. The ability of carbon nanotubes to passively penetrate the plasma membrane, though potentially therapeutically useful as a drug-delivery vehicle34, 37, may also elevate the threat these particles pose to intracellular organelles, particularly at shorter (<200nm) fiber lengths38. Genotoxicity from nanotube exposure has also been described22–23, indicating that carbon nanotubes are capable of damaging DNA within the nucleus following uptake by the cell.
In this study, we hypothesized that MWCNTs can also exert genotoxic effects on mitochondrial (mt)DNA, which is less protected than nuclear DNA39–40, thereby impairing mitochondrial function in exposed cells. To test this hypothesis, primary bronchial epithelial cells were obtained from healthy human volunteers via bronchoscopy and grown to confluence for five (5) days in submerged culture containing non-cytotoxic doses of MWCNTs. Cellular DNA and RNA were harvested for mitogenomic endpoints including changes in mtDNA heteroplasmy, insertion/deletion (indel) mutations, and expression of mitochondrially-encoded genes by QPCR. Exposed cells were also assessed for oxygen consumption rate using the Seahorse XF extracellular flux assay, as well as verification of cytotoxicity by lactate dehydrogenase (LDH) release and ultrastructural imaging of nanoparticles within the cells by transmission electron microscopy (TEM).
Materials and Methods
Nanomaterial Characterization
The MWCNTs we used were sourced from Helix Nanomaterial Solutions, and were characterized by an independent laboratory (Millennium Research Laboratories, Woburn, MA), in a previous publication41 to be >95% purity as measured by thermogravimetric analysis (TGA) with <2% amorphous carbon. Inductively-coupled plasma Auger electron spectroscopy (ICP-AES) performed by MRL determined the bulk sample to consist of 99% carbon, 0.63% oxygen, 0.34% nickel, and 0.03% lanthanum. Particle agglomeration in the dispersion medium used in these studies was measured and reported previously25 using dynamic light scattering (DLS) analysis. Agglomerates have a Z-average of ~200 nm in hydrodynamic diameter, with most of the mass of the agglomerates forming between 10–100 nm. Individual fibers were present and abundant in TEM imaging during this study, and were usually found between 30–50 nm in diameter, and 100–1000 nm in fiber length. Qualitatively, fiber morphology varied in apparent stiffness, with agglomerates of longer fibers being tangled/bent and individual or shorter fibers appearing more rigid.
Cell Culture and Treatment
Bronchial epithelial primary cells (BECs) were obtained from bronchoscopy brushings of 8 “healthy” (non-smokers without current lung disease) human donors, aged 23–32 years (donor demographics can be found in Table 1). Bronchoscopy of human donors and the use of cells derived from this procedure was performed after review and approval by the National Institute of Environmental Health Sciences Intramural Review Board (NIEHS-IRB, clinicaltrials.gov identifier: ). Bronchoscopies were performed with the donors’ written consent by a trained pulmonologist (S. Garantziotis) at the NIEHS Clinical Research Unit (CRU) per the methods described in NIH protocol 11-E-0006. Following a saline lavage, 3 to 5 cytology brushings were conducted per donor to obtain 4 to 6 million surface mucosal epithelial cells. After each brushing, the brush was removed and the recovered cells were dislodged from the brush by stirring in a 15 mL conical vial containing Bronchial Epithelial Growth Medium (BEGM, Lonza) supplemented with 2× amphotericin B. Cells from brushes were seeded onto T-25 vented flasks in serum-free Lonza BEGM medium and frozen at passage 1. For each experiment, cells were thawed in BEGM for expansion in T-75 flasks, further subpassaged onto either 6-well or 96-well plates, and were exposed to nanomaterials at passage 3 while the cultures reached confluence. Cultures received a media refresh every 2 days containing freshly-dispersed nanotubes, and were exposed again at days 1, 3, and 5 following seeding.
Table 1.
Demographical information for each donor (Bronchial Epithelial Cells BEC1–8). The study included a narrow age range (between 23 and 32 years) of donors to control for age-related mitochondrial heteroplasmy. Donors were a mix of races (black, non-hispanic white, and Asian) and included 5 females and 3 males.
| Age | Gender | Race | |
|---|---|---|---|
| BEC1 | 27 | F | Black |
| BEC2 | 27 | F | Black |
| BEC3 | 32 | M | Black |
| BEC4 | 23 | F | Black |
| BEC5 | 27 | F | Asian |
| BEC6 | 28 | M | White |
| BEC7 | 27 | F | White |
| BEC8 | 31 | M | Asian |
MWCNTs were dispersed in BEGM culture medium supplemented with sterile 600 μg/ml bovine serum albumin (BSA) and 10 μg/ml 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) surfactant. Pulmonary surfactant, while not secreted by bronchial epithelia in the adult lung, does protect non-ciliated BECs in vivo in the bronchioles near alveoli42, and its use is consistent with recommendations by the Nano GO Consortium for optimal dispersion of MWCNTs43. Control vehicle (CoV) exposures used the supplemented medium without nanomaterials. Prior to exposure, stock solutions of MWCNT (2 mg/ml) were sonicated in a cup horn for 15 minutes (5 pulses of 3 minutes each) at 100 amplitude to break up agglomerated materials, then diluted to treatment concentrations in dispersion vehicle at 0.7–12 μg/ml MWCNT. Some sedimentation of the MWCNTs did occur between treatments upon visual examination, though precisely what percentage of the MWCNT mass sedimented could not be conclusively determined. The 0.7, 3, and 12 μg/ml MWCNT doses would equate to approximately 0.25, 1, and 4 ug/cm2 if the nanomaterial sedimented out completely between treatments, and approximately 0.75, 3, and 12 ug/cm2 if the MWCNTs were completely absorbed by the cells before each wash, placing these as upper bounds on the potential MWCNT dose. While 3 μg/ml was the highest non-cytotoxic dose we determined for these cells previously25, we also verified cytotoxicity for this experiment by lactate dehydrogenase (LDH) release assay and trypan blue exclusion.
LDH Release Cytotoxicity Assay
Supernatant from BECs exposed to nanomaterials or dispersion vehicle (24 hours following the most recent media refresh at 24 hours and 5 days) was centrifuged and transferred to a clear-bottom 96-well plate. Acellular treated wells were also included to account for interference from the particles. LDH accumulated over 24 hours was assessed using the CytoTox colormetric assay (Promega) and compared to dispersion vehicle controls to verify that the nanomaterial exposures used in this study were not causing significant cytotoxicity.
Trypan Blue Exclusion Assay
BECs treated with MWCNTs for 24 hours and for 5 days were assessed for membrane permeability by staining with trypan blue dye (Sigma Aldrich). Cell cultures were washed 3× with phosphate buffered saline (PBS) and lifted into suspension using 0.25% trypsin-EDTA solution (Sigma Aldrich) at 37°C for 4 minutes. The reaction was terminated with 2 mg/ml soybean trypsin-inhibitor (SBTI) and cell suspensions were pelleted in a centrifuge at 1000 rcf for 5 minutes, then resuspended in a 1:1 solution of BEGM and trypan blue dye. Counts of live and dead cells were performed manually using a hemocytometer at 20× magnification.
Quantitative PCR
Following 5-day nanomaterial exposures in a 6-well plate, BECs were harvested for total RNA using the RNeasy columns/reagents and QiaCube automated isolation/purification device (Qiagen), according to manufacturer’s protocol. Total RNA was converted to cDNA libraries using iScript reverse transcription kits (BioRad) containing oligo dT’s and randomized hexamers. PCR Prime arrays (BioRad) pre-loaded with primer pairs (see Supplement for amplicon context sequences) for all 13 mitochondrially-encoded protein-coding genes, plus sequences for ubiquitin C (UBC), 18S, and 16S endogenous controls, were used for gene expression analysis of the cDNA libraries by quantitative PCR (QPCR). SSO Advanced SYBR Green master mix was used as a reporter dye in a StepOne real-time sequencer (ABI Technologies). Assays were measured over 40 cycles, denaturing at 95°C for 5 seconds and annealing at 60°C for 30 seconds.
Amplification of mtDNA
Total DNA was harvested from BECs exposed to nanomaterials for 5 days in 6-well plates using a cell scraper in 4°C phosphate buffered saline (PBS). Cell suspensions were lysed with protease K and DNA extracted using DNeasy Blood and Tissue columns and QiaCube automated isolation/purification device (Qiagen). Mitochondrial DNA (mtDNA) was amplified from these samples using the SequalPrep Long PCR kits with dNTPs and the following primer sequences, resulting in “short” (7626 bp) and “long” (9286 bp) mtDNA fragments44:
Long Fragment:
forward-AAC CAA ACC CCA AAG ACA CC
reverse-GCC AAT AAT GAC GTG AAG TCC
Short Fragment:
forward-TCC CAC TCC TAA ACA CAT CC
reverse-TTT ATG GGG TGA TGT GAG CC
Agarose (0.8%, SeaKemp) gel electrophoresis was used to verify the presence and size of mtDNA fragments (see Supplemental Information). Removal of nuclear DNA, dNTPs, polymerases, and other contaminants was performed using DNA Clean and Concentrator columns (Zymo) according to manufacturer instructions to prepare the mtDNA for sequencing.
Ultra-Deep Sequencing
Purified mtDNA from MWCNT-exposed BECs were sequenced for the appearance of heteroplasmies and indel mutations compared to the sequences of vehicle-treated control cultures. Tagmentation of mtDNA (combined short and long fragments) was carried out using a Nextera DNA Library Prep kit (Illumina) according to the manufacturer’s instructions. DNA samples were added to a 96-well thermal cycler plate with Amplicon Tagment Mix and Tagment DNA buffer from the kit and heated at 55°C for 5 minutes. Libraries were then amplified with indexed adapters and Nextera PCR Master Mix, first denaturing at 72°C and then running 15 cycles of 95°C for 10 seconds, 55°C for 30 seconds, and 72°C for 30 seconds. AMPure XP magnetic beads (Illumina) were used to purify the resulting mtDNA libraries, according to manufacturer instructions. AMPure beads were added to each sample and allowed to incubate at room temperature for 7 minutes on a magnetic plate to pellet the DNA/beads. Supernatants were discarded and the bead pellets were washed twice with 80% ethanol, then allowed to air dry completely (approximately 10 minutes). Purified DNA was resuspended in AMPure Resuspension Buffer and the DNA concentration/quality was verified by QuBit High-Sensitivity Analysis (Thermo-Fisher) and the Agilent Bioanalyzer.
Alignment and Variant Calling
Read pairs were aligned using bowtie2, version 2.0.0-beta7, to an index composed of the human mitochondrial genome only, acquired from GenBank May 2015 (accession NC_012920). The alignments were performed in “--local” mode using the “--sensitive-local” preset options to allow insertions and deletions relative to the reference, as well as clipping of ends extending beyond the edges of the artificially linearized reference sequence. Fragment lengths of up to 10kb were allowed, as well as a single mismatch per seed alignment (−X 10000, −N 1).
Variants were identified with a custom script, utilizing a method adapted from Hodgkinson, et al.45. For each sample, depth per allele, per strand was determined, allowing a minimum base and alignment quality score of 20. Indels relative to the reference were counted only in the case that both flanking bases’, and any inserted bases’ quality scores met or exceeded this threshold. Additionally, the depth calculation for sites adjacent to or within homopolymer runs considered only reads traversing the entire repeat. Sites with less than 2000× coverage were not considered, and variants were required to be observed at a frequency of 1% or greater, with a plus to minus strand coverage ratio greater than 0.1 and less than 0.9. The probability of observing each alternate allele by chance was calculated using a Poisson distribution, with an expected error rate of 0.01 for SNPs (derived from the quality score threshold), and 0.02 for indels, based on observations reported by Minoche, et al46. These p-values were adjusted for multiple testing using the Benjamini and Hochberg FDR method, and a significance threshold of 0.05 was applied. Alternate amino acids were identified for all SNPs based on annotations and protein sequences downloaded from Ensembl, June 2015.
Extracellular Flux Assay (Seahorse XF)
An extracellular flux assay was used to determine changes in oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in BECs exposed to nanomaterials. Primary BECs were seeded onto a 96-well Seahorse XF96 microplate in BEGM. Cells were allowed to reach confluence, with fresh changes of medium containing MWCNTs or vehicle every 2 days, as described above (Cell Culture and Treatment). On the day of the assay, cells were washed 3 times with PBS and fed 180 μl/well of pre-warmed (37°C) Seahorse XF medium, consisting of XF Base medium with additional supplements of 1mM sodium pyruvate, 11 mM D-glucose, and 2 mM L-glutamine, with the pH adjusted to 7.4 using NaOH and HCl. The sensor cartridge (following an overnight incubation in XF calibrant solution at 37°C and 0% carbon dioxide) was loaded with reagents from the Seahorse Mito Stress Test: 2 uM oligomycin (20 ul of 20 uM per well), 2uM Carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP, at 22 ul of 20 uM per well), and 0.5 uM rotenone/antimycin A (25 ul of 5 uM per well). The Seahorse XFe96 extracellular flux instrument was programmed to take 3 measurements before and after the injection of each of the 3 compounds, with each measurement having taken approximately 8.7 seconds. All reagents used for Seahorse XF analysis were obtained from Agilent.
MitoTracker and Annexin V Staining
Live BECs from 4 “responding” donors (donors which exhibited a gene expression response to the MWCNTs) were treated with MWCNTs at 0, 0.7, and 3 μg/ml for 5 days, washed with BEGM, then incubated in 6-well culture plates with 100uM MitoTracker Deep Red FM dye (Thermo Fisher) for 45 minutes at 37°C. Wells were then washed with BEGM, passaged into 2 ml microcentrifuge tubes (using 0.25% trypsin/EDTA and 2 mg/ml SBTI), and resuspended in 1× calcium binding buffer for annexin V staining (TACS Apoptosis Detection kit, Trevigen). Cells were incubated in suspension with 15 ul of propidium iodide and 1ul of FITC-conjugated annexin V per 100 ul of binding buffer for 15 minutes at room temperature. Cells were then resuspended in 500 ul of ice-cold binding buffer for flow cytometry. Flow cytometry was performed using a BD LSRFortessa instrument, with 10,000 events counted per sample.
Succinate Dehydrogenase Immunostaining
In addition to MitoTracker, mitochondrial abundance per cell was also determined by immunostaining fixed cells for succinate dehydrogenase A (SDHA), a nuclear-encoded subunit of mitochondrial complex II. BECs from 4 donors were exposed to MWCNTs 0.7, 3, and 12 μg/ml for 5 days, lifted from the 6-well plate with trypsin, fixed with 4% paraformaldehyde, washed 3× with PBS, then resuspended in 1:1000 anti-SDHA FITC-labeled primary antibody (Goat-derived, Abcam) and a DAPI counterstain. A FACSAria II flow cytometer was used to quantify FITC-labeled cells, with 2000 events counted per sample.
Transmission Electron Microscopy (TEM)
For ultrastructural imaging of nanomaterials within the cell, BECs exposed to nanomaterials for 24 hours were washed with PBS, removed from the 6-well plate with cell lifters, centrifuged at 800 rcf, and resuspended in fixative (4 parts formaldehyde and 1 part glutaraldehyde). Cells were then re-pelleted at 1000 rcf and embedded in 3% water agar. Embedded samples were then processed using a Leica EM TP processor. Samples were post-fixed in 1% osmium tetroxide, rinsed in water, then dehydrated in an ethanol series culminating in acetone. Following dehydration, samples were infiltrated with Poly/Bed 812 epoxide resin. Blocks were trimmed and semi-thin (~ 0.5 μm) sections were cut, mounted on glass slides, and stained with 1% toluidine blue O in 1% sodium borate to visualize areas of interest via light microscopy. Ultra-thin sections (80–90 nm thick) were cut from selected blocks and placed onto 200 mesh copper grids. Digital images were captured with a Gatan Orius SC1000/SC600 attached to a FEICO Tecnai T12 transmission electron microscope.
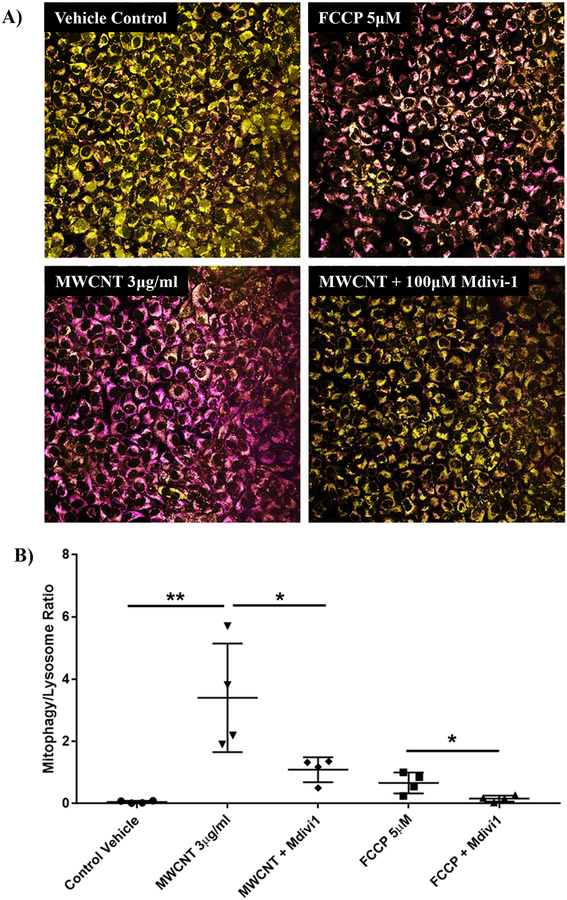
Confocal Imaging of Mitophagy
BECs from responding donors (BECs 2, 4, 6, and 8) were treated with 3 μg/ml MWCNTs for 2 hours to observe their early mitophagy response. Confluent cultures in 4-well glass chamber slides were pre-treated with “Mtphagy” Mitophagy Detection Dye (Dojindo Molecular Technologies, Inc., Washington, DC) at 1/1000 dilution in BEGM for 30 minutes at 37°C. This dye locates to live mitochondria in vitro and becomes fluorescent on exposure to low pH, indicating interaction with lysosomes during mitophagy. Vehicle medium containing 3 μg/ml MWCNTs was applied to BECs following 3× wash with PBS. FCCP was used at 5 μM as a positive control for mitophagy. Mitochondrial division inhibitor 1 (Mdivi-1) inhibits dynamin related proteinase 1 (drp1), thereby inhibiting mitophagy, and was used to verify that MWCNTs were triggering mitophagy in the cells and not interacting with the dye or otherwise confounding the assay. In these wells, 100 μM Mdivi-1 was added to MWCNT-containing treatment medium. Following a 2-hour nanotube treatment, wells were again washed 3× with PBS and counterstained with the lysosome dye included in the kit, at 1/1000 dilution in BEGM, for 30 minutes. Following a final wash step, the cultures were mounted with coverslips using Prolong Gold antifade reagent (Thermo Fisher) and imaged using a Zeiss 780 confocal microscope. Z-stacks of 10 μm of cell monolayer were collapsed into a summation image, with a threshold intensity of 40 applied to generate a binary image of dye distribution. Pixel density analysis was performed using Image J software and the ratio of mitophagic vesicles to lysosomes was calculated for each image. Parameters such as gain, pinhole, laser intensity, z-planes per stack, and image post-processing were kept consistent between treatments.
Statistics
Unless noted otherwise, the statistical significance of differences found between treatments was determined by one-way analysis of variance (ANOVA) on the means averaged from all the donors tested, with Tukey’s post hoc test for multiple comparison correction. An adjusted p-value of 0.05 or less was considered statistically significant. Analysis of mitochondrial gene expression was performed using a 2-way ANOVA for each treatment, making comparisons between donor reactions to nanomaterials for each of the 13 mitochondrial genes measured.
Results
Nanomaterial doses were non-cytotoxic
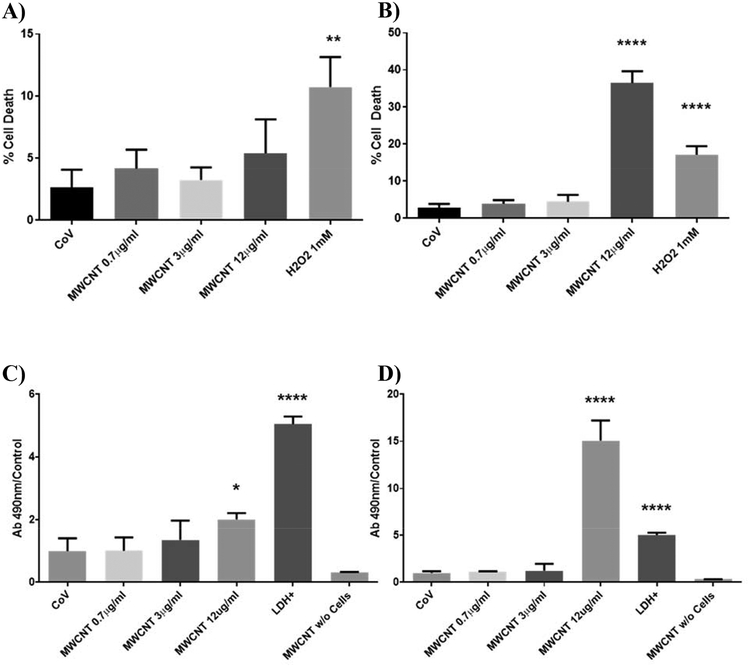
No statistically-significant difference in trypan blue exclusion (Figures 1A and 1B) or extracellular LDH (Figures 1C and 1D) was found at MWCNT doses of 3 μg/ml or less compared to vehicle-treated controls. Results were similar for a 24-hour exposure and a 5-day repeated exposure, though 12 μg/ml MWCNT doses were found to be cytotoxic by day 5. The 3 μg/ml treatments we used were therefore determined to be non-cytotoxic over the time course and conditions used in this study, and the observed cellular and metabolic effects of MWCNTs at this dose were therefore not a consequence of cell death.
Figure 1.
Cytotoxicity in BECs at days 1 and 5 of MWCNT exposure, measured by trypan blue exclusion (A-B) and extracellular LDH (C-D). Exposure to MWCNTs did not affect the percentage of cells permeable to trypan blue at any study dose by day 1 (A), but sharply increased cell death at the 12 μg/ml dose by day 5 (B). Extracellular LDH was similarly unaffected by MWCNT treatments of 3 μg/ml or less, with a slight increase in LDH at day 1 with 12 μg/ml MWCNTs (C) and a large increase at day 5 (D). MWCNT doses of 3 μg/ml or less were determined to be non-cytotoxic for the purposes of this study. (N = 8, ANOVA, Tukey post-hoc, *p < 0.05, **p < 0.01, ****p < 0.0001).
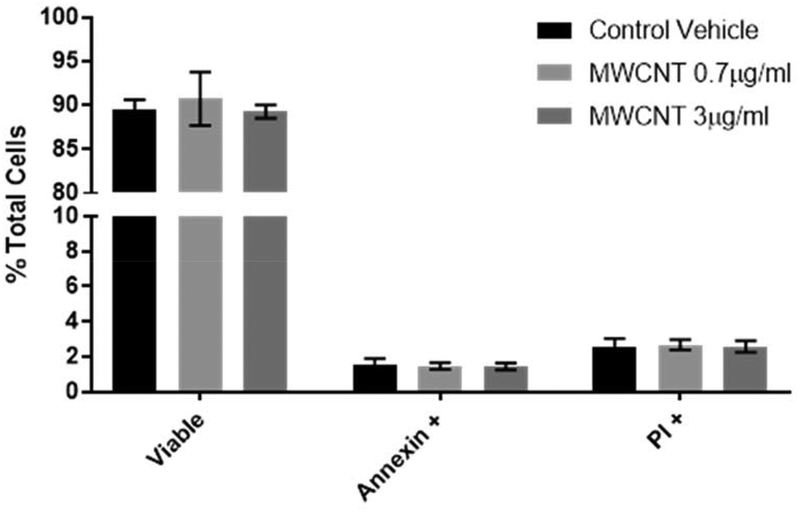
Likewise, annexin/PI staining of 5-day treated BECs revealed no significant increase in apoptosis or necrotic cell death in MWCNT doses up to 3 μg/ml (Figure 2). This provides further evidence that the mitochondrial effects observed in this study occur independently of apoptosis and do not result in apoptosis within the 5-day time course of this study.
Figure 2.
Apoptosis in MWCNT-exposed BECs was measured at day 5 by flow cytometry using annexin V-FITC staining with a propidium iodide counterstain. MWCNT treatments of 3 μg/ml or less were not found to elevate annexin-positive cells or PI-permeable cells, indicating that apoptosis was not significantly induced by these treatments by day 5. (N = 4, ANOVA, Tukey post-hoc)
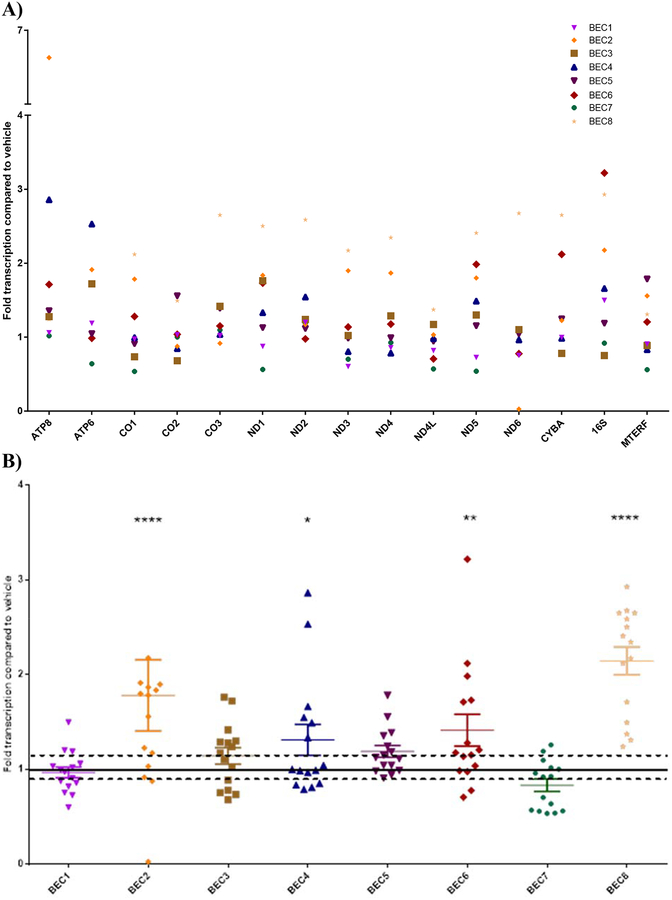
MWCNT exposure upregulated mitochondrial gene expression
Expression of all mitochondrially-encoded genes was measured by QPCR (Figure 3). Cycles to reach threshold (Ct) values were normalized to expression of 18S ribosomal RNA using ddCt analysis. BEC responses to 3 μg/ml MWCNT exposure varied between donors, with upregulated expression of mitochondrially-encoded genes in some donors (BECs 2, 4, 6, and 8) by up to 2–3-fold vehicle-treated values (with a combined average of 1.4-fold overall upregulation, p<0.0001, 2-way ANOVA, Tukey comparison correction). These donors were considered “responders” and metabolic function assays performed later focused on these 4 donors. Expression of these genes in donor BECs 1, 3, 5, and 7 treated with MWCNTs were not statistically different from vehicle-treated controls.
Figure 3.
Mitochondrial gene expression in BECs exposed to 3 μg/ml MWCNTs, as measured by QPCR. Values are expressed as fold control (dispersion vehicle-treated) BECs, with each symbol representing the mean of 3 experimental replicates of a different donor (A). Inter-individual variation was found across the mitochondrial genes in response to MWCNT exposure, while MTERF (a nuclear-encoded mitochondrial regulatory gene) transcription was not significantly altered by treatment. Some donors were “responders” (such as BECs 2, 4, 6, and 8, identified by p < 0.05 on 2-way ANOVA) that consistently upregulated all or most mitochondrial genes in response to MWCNTs, while others were not significantly changed from controls. This donor variation is illustrated in Figure 3B, showing donor-specific patterns of up/downregulation when all genes from a given donor are combined. Dotted lines in Figure 3B indicate the standard deviation around the mean value for vehicle-treated controls (*p < 0.05, **p < 0.02, ****p < 0.0001).
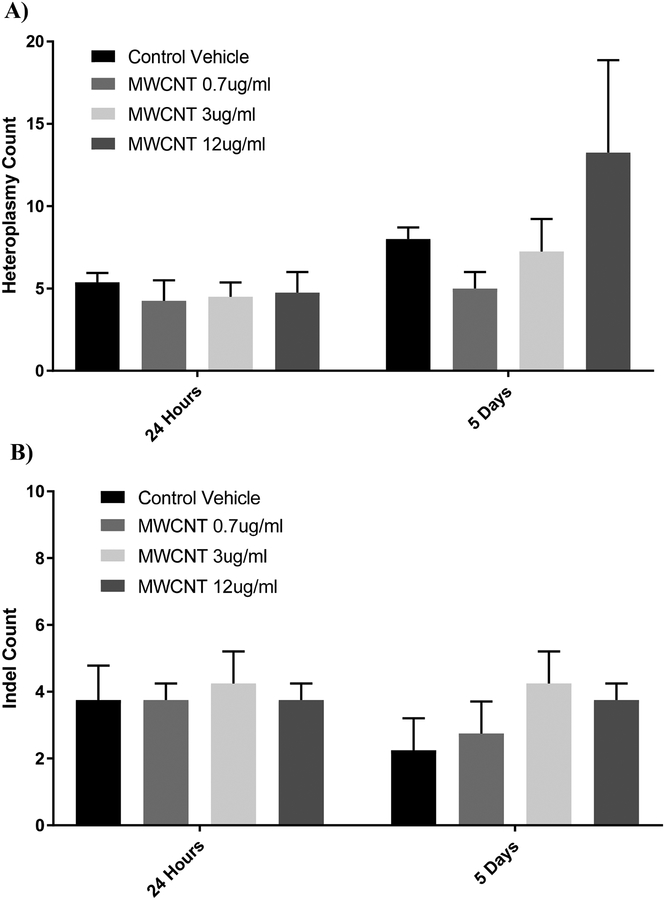
No effect on heteroplasmy or mitochondrial mutations
The mitochondrial genomes of BECs treated with 0.7–12 μg/ml MWCNTs were sequenced and examined for heteroplasmy/SNP variants (Figure 4A) and insertion/deletion mutations (Figure 4B) compared to the hg19 human reference sequence, both at 24 hours following their first treatment and after 5 days. Across all doses, we found no differences in major sequence variants compared to respective control groups. We found an average of approximately 3 ± 1 indels (with sizes ranging from 1–3 base pairs) in mtDNA compared to hg19 (N = 8), and no treatment was statistically different from vehicle treatments. Heteroplasmy of single-nucleotide polymorphisms (SNPs) in which 10% or more of the mtDNA sequences contained the SNP compared to hg19 were averaged among the 8 donor BECs. Vehicle-treated control BECs (CoV) averaged 8.58 ± 2.18 heteroplasmies compared to the hg19 reference sequence, and nanomaterial treatment did not cause heteroplasmy counts that were statistically different from vehicle controls.
Figure 4.
Frequency of heteroplasmy and insertion/deletion mutations (indels), as measured by ultra-deep sequencing. Heteroplasmies with greater than 10% alternate allele frequency were counted (A) for each donor following treatment with dispersion vehicle (CoV) and MWCNTs at 0.7, 3, or 12 μg/ml. Indels were also counted for each donor/treatment (B). No statistically-significant differences were observed between vehicle controls and any treatment, at either day 1 or day 5, including the cytotoxic 12 μg/ml dose. (N = 8, ANOVA, Tukey post-hoc)
MWCNTs decreased intracellular mitochondrial abundance
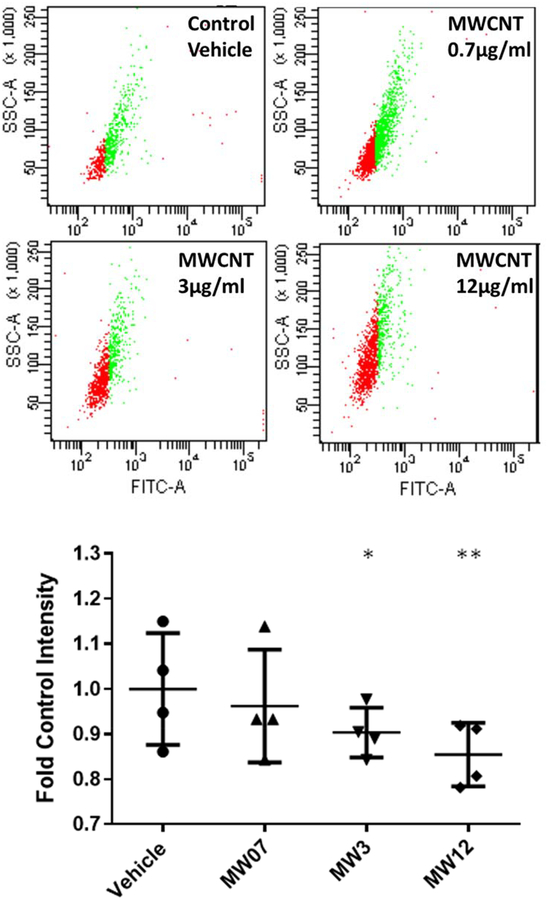
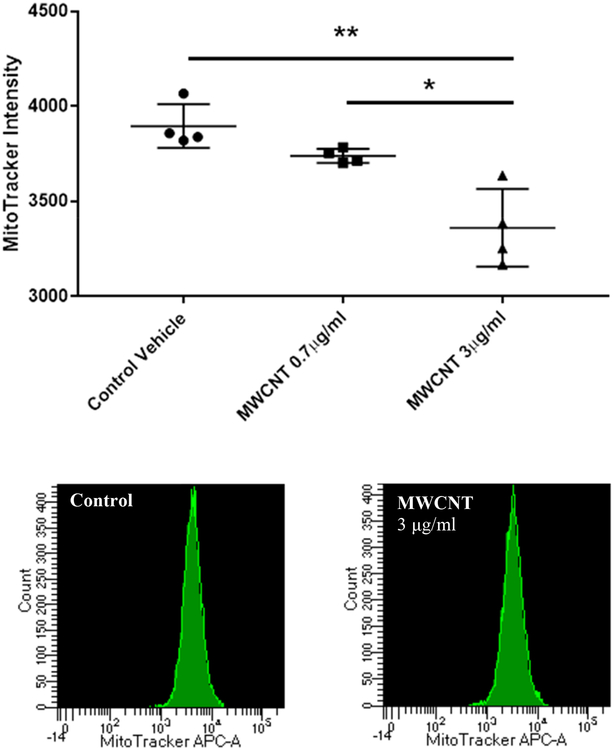
Mitochondrial abundance per cell was determined by flow cytometry, using 1) a FITC-labeled primary antibody against succinate dehydrogenase (SDHA), a nuclear-encoded mitochondrial membrane protein, and 2) in a separate experiment, MitoTracker Deep Red FM dye in live cells. Fluorescent intensity of FITC staining per cell/event was averaged for 4 donors (the highest gene expression responders, BEC2, 4, 6, and 8) and compared between vehicle controls and 3 doses of MWCNTs (0.7, 3, and 12 μg/ml) to determine whether the changes in mitochondrial gene expression observed with MWCNT exposure correlated with a change in mitochondrial abundance (Figure 5). A statistically significant decrease in SDHA staining was found with increasing doses of MWCNTs (N = 4, paired t-test, p < 0.05). A similar pattern was found with MitoTracker (Figure 6), and a significant and dose-dependent decrease in MitoTracker staining intensity per cell was found compared to vehicle-treated controls (N = 4, paired t-test, p < 0.001).
Figure 5.
Succinate Dehydrogenase A (SDHA) staining per cell, as measured by flow cytometry. Average intensity of cellular FITC-labeled SDHA was used to determine mitochondrial abundance per cell, independent of metabolic activity. Values are expressed as fold control FITC-staining intensity and show a dose-dependent decrease in mitochondrial abundance with increasing concentrations of MWCNTs from 0.7–12 μg/ml (MW07, MW3, MW12). Statistical significance determined by paired t-tests (N = 4, *p < 0.1, **p < 0.05)
Figure 6.
MitoTracker staining per cell, as measured by flow cytometry. Similar to SDHA immunostaining, the average fluorescent intensity of MitoTracker Deep Red FM dye taken up by the cell was used to measure mitochondrial abundance. Results are expressed as mean raw intensity for each donor and show a dose-dependent decrease in intracellular mitochondrial mass in BECs treated with MWCNTs for 5 days. (N = 4, t-test, *p < 0.05, **p < 0.01)
Decreased oxygen consumption rate
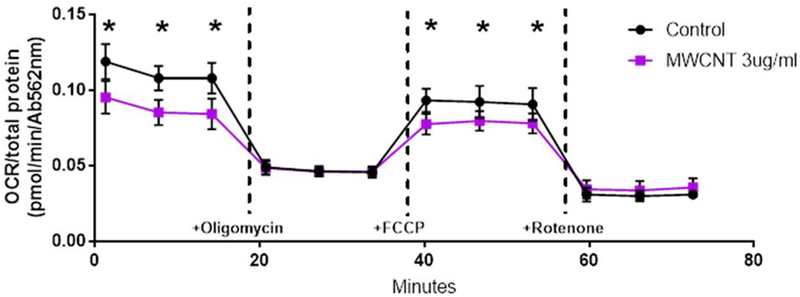
Oxygen consumption rate (OCR) before and after addition of oligomycin, FCCP uncoupler, and rotenone/antimycin was measured in a real-time extracellular flux assay in a 96-well plate. OCR values were then normalized to the total protein content in each well, determined by BCA assay, and averaged over 6 duplicate wells (Figure 7) of cells from responder BECs 2, 4, 6, and 8. OCRs for MWCNT-treated cells were consistently and significantly reduced compared to vehicle-treated control cultures (p < 0.025) at baseline and following FCCP uncoupling.
Figure 7.
Oxygen consumption rate (OCR) in donor primary BECs following overnight exposure to MWCNTs, measured by Seahorse Extracellular Flux assay. OCR measurements at all time points were normalized to total well protein measured after the assay. Baseline OCR was significantly reduced in MWCNT-treated cells compared to controls. ATP-coupled respiration measured following the addition of oligomycin was not significantly altered by MWCNT treatment, however, total reserve capacity of mitochondria uncoupled by FCCP was reduced. No differences in non-mitochondrial respiration (following the addition of rotenone) were found. (N = 4, ANOVA, Tukey post-hoc, *p < 0.05).
MWCNTs found in cytoplasm and intracellular organelles
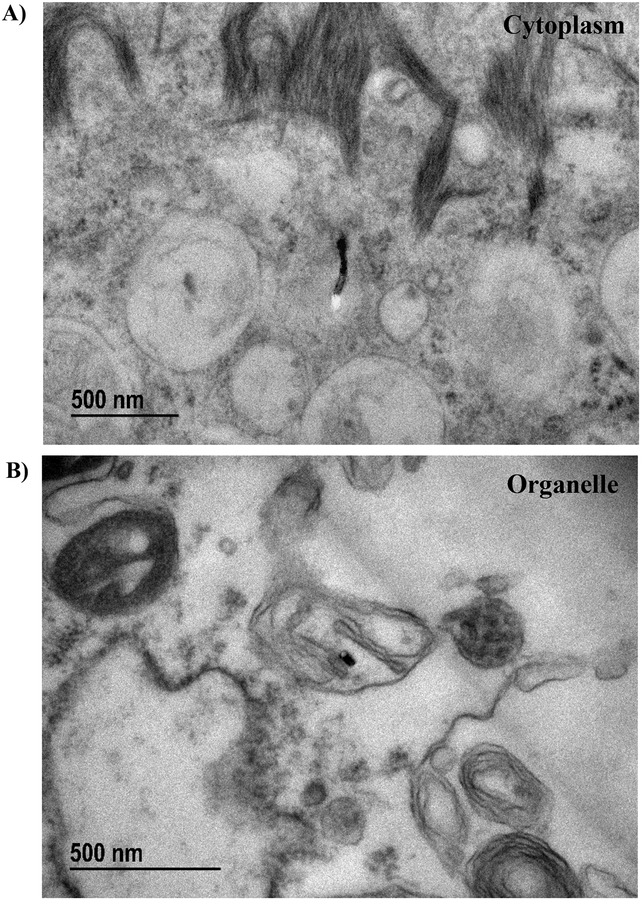
Transmission electron micrographs of BECs treated for 24 hours with 3 μg/ml MWCNTs enabled qualitative analysis of the intracellular distribution of these materials (Figure 8). MWCNT fibers were found outside of endosomes/phagolysosomes within the cytoplasm (Figure 8A), which suggested that these fibers were either capable of penetrating the plasma membrane (as seen in previous studies33–34) or were capable of escaping phagosomes following endocytosis by the cell35. Particles were also found within double-membrane bound structures that were suggestive of mitochondria (Figure 8B). While these structures were too damaged for positive identification as mitochondria, the presence of individual MWCNT fibers within them provide non-quantitative evidence of MWCNT’s ability to penetrate intracellular membranes.
Figure 8.
Sample images of transmission electron microscopy (TEM) of BECs treated with 3 μg/ml MWCNTs for 24 hours. A) MWCNT fiber found within the cytoplasm, unbound by any endosomal structure. B) MWCNT fiber found within a double-membrane bound organelle structurally consistent with a depolarized/damaged mitochondrion.
Mitophagy response to MWCNT exposure
BECs exposed to 3 μg/ml MWCNTs for 2 hours were assessed for mitophagy using confocal microscopy (Figure 9). An uncoupling agent, FCCP, was used as a positive control of mitophagy. Activation of the mitophagy dye by mitochondria/lysosome mergers was significant and widespread following MWCNT exposure compared to vehicle-treated controls (p < 0.001, N = 4, ANOVA, Tukey post-hoc). This response was attenuated in BECs that were co-treated with Mdivi1, a drp1 and mitophagy inhibitor (p < 0.01).
Figure 9.
Confocal microscopy of mitophagic vesicles in BECs following a 2-hour exposure to MWCNTs. FCCP (5 μM), a mitochondrial uncoupling agent, was used as a positive control. A) MWCNTs induced a significant increase in mitophagic vesicles which was attenuated by co-exposure with Mdivi-1, a mitophagy inhibitor. B) Pixel density analysis was used to quantify staining of mitophagic vesicles and normalize it to lysosomal staining for each of the 4 responding donors (BECs 2, 4, 6, and 8). (N = 4, ANOVA, Tukey post-hoc, *p < 0.1, **p < 0.05)
Discussion
While genotoxicity resulting from exposure to nanomaterials has been previously documented22, specific effects on the mitochondrial genome, and function of mitochondrial organelles, have been relatively unexplored. In this study, we tested whether one of the most widely-used nanomaterials, carbon nanotubes, could cause mitochondrial genotoxicity and impaired oxidative phosphorylation in human BECs. BECs obtained from eight healthy human donors (via bronchoscopy) were exposed in submerged culture to MWCNTs or a dispersion vehicle control for five days, until they reached confluence. We found that while MWCNT exposure did not induce significant mutations or heteroplasmy in primary BECs, they did cause significant upregulation of mitochondrial genes with inter-individual variation between donors. MWCNTs also decreased intracellular mitochondrial abundance and oxygen consumption, and induced mitophagy in exposed cells.
We used a primary bronchial epithelial cell model for these studies to better replicate the mitogenomic diversity found in the human population47 than would a cell line. Epithelial cells were an ideal model for this study as they would be expected to encounter the highest concentrations of inhaled nanomaterials in vivo and can be grown in medium without serum supplementation. MWCNTs bind to serum proteins via hydrophobic interactions and could potentially affect concentrations of serum components in the medium48. We used an exposure period of five days for this study to provide enough time for nanomaterials to cause detectable mitogenomic effects. MWCNTs persist in the lungs of exposed mice for several months27, therefore, this exposure duration is realistic without being so long as to introduce additional variables such as cellular differentiation and over-passaging. The doses used in this study were intentionally high (short of cytotoxicity) in comparison to a typical occupational exposure49. One reason for this is because the purpose of this study was to determine whether MWCNT exposure could have effects on the expression and function of mitochondrial genes, and this question could best be answered initially with the highest dosage that would not cause necrosis or apoptosis. The results we have reported with an MWCNT exposure of 3 μg/ml over five days indicate a modest effect on mitogenomic endpoints even in susceptible responder donors, while the more pronounced mitochondrial function and mitophagy findings suggest that mitochondrial injury might be present at lower doses. Secondly, the use of bronchoscopy-derived BECs from volunteer donors inherently limited the scope of experiments which could be performed, due to the small numbers of P0 cells (approximately 1–4 million) which could be obtained from each donor. Consequently, performing this study with lower doses over longer timeframes was not possible. Finally, comparing the dosimetry of inhaled MWCNT deposition into the human lung with that of a submerged BEC culture is inherently imprecise. It would not be plausible to conclude that our in vitro nanotube exposure approximates a 6 month accumulated inhaled dose, for example, as the kinetics of mucociliary clearance, macrophage uptake, agglomeration in airway mucus, and many other factors would confound such an approximation. Therefore, the doses and timepoints used in this study should be taken only as evidence that MWCNTs can cause mitochondrial dysfunction in human bronchial epithelial cells in isolation. Whether or not inhaled MWCNTs cause similar dysfunction in the lungs of those who interact with or manufacture these materials over a typical chronic exposure is beyond the scope of this study.
Carbon nanotubes are used in commercial applications in a variety of fiber lengths, structures, purities, added functional groups, and other variables, and so the choice of which nanotubes to use in this study was based primarily on whether they were found in products which could realistically result in inhalation exposure by consumers. Unfunctionalized MWCNTs, such as the samples we sourced from Helix Material Solutions25, are commonly used in construction applications and textiles where a risk of inhalation exposure is present to consumers and manufacturers50. While it is possible that greater or lesser mitogenomic effects could be obtained from testing nanotubes with other structural/chemical characteristics, the MWCNTs chosen for the current investigation are representative of the types affecting human health through inhalation exposure. We also chose to use a dispersion medium supplemented with BSA during each MWCNT treatment. This results in a protein corona around agglomerates in suspension which is known to increase active uptake of nanotubes in cell culture and reduce cytotoxicity51–52. This method, which is recommended for optimum dispersion of MWCNTs in cell culture medium in accordance with the findings of the Nano GO consortium43, is justified for use in our exposure model as inhalation of MWCNTs would result in contact with extracellular mucus in vivo, therefore, it is highly likely that these nanoparticle aggregates would be surrounded in protein before encountering the epithelium. MWCNTs absent a protein corona may conceivably have resulted in a lesser effect (from increased agglomeration leading to decreased cellular uptake) or greater effect (from a decreased proportion of MWCNT mass undergoing active transport leading to greater passive entry), but we believe the addition of the BSA to our dispersion vehicle more closely replicates in vivo exposure.
Heteroplasmy, defined as the presence of intracellular variability in mitochondrial genome sequences, is only recently gaining recognition53–55 as a toxicological endpoint. Mitochondrial DNA sequences are usually well-conserved, even between species56, and so sequence variants resulting from exogenous insult/mutation often elicit functional detriments. As detection of heteroplasmy requires thousands of repeated sequencing reads at each position of the mitochondrial genome57, it is only due to the recent advancements in ultradeep DNA sequencing that a study measuring the effect of nanomaterials on shifting heteroplasmy of allele variants could be conducted. Additionally, an increase in heteroplasmy, even at low levels, is linked to altered mitochondrial function58, a parameter which itself required recent advances in instrumentation (particularly the extracellular flux assay) to measure. Following the 5-day exposure, heteroplasmy frequency was not changed (above 10% variant allele frequency) for any of the MWCNT doses used, compared to the same cells treated with dispersion vehicle (CoV). Further, no change in the occurrence of indels was observed with MWCNT exposure. The lack of changes in heteroplasmy and indels were unexpected findings, as genotoxicity in similar nanomaterials has been reported in previous publications22–23, but they indicate that either cellular defense mechanisms protecting mtDNA from damage (primarily mitophagy) were more robust than anticipated, or that the nanomaterials were not used in doses/timeframes sufficient to induce such damage. Longer-term in vitro exposures would be possible through repeated treatments over multiple passages, though we have found donor-to-donor variability in the number of passages that primary BECs tolerate, and so the effect of chronic, low-dose MWCNTs on mtDNA may require use of a fully-differentiated air-liquid interface model or in vivo experiments. The value of increasing the dosage to obtain genotoxic effects in mitochondria would be questionable, however, as the doses higher than those implemented in our study were cytotoxic and would be an improbable acute human exposure even in an occupational setting49. Our results thus suggest that heteroplasmy and mutation in mtDNA are not important adverse effects of acute MWCNT inhalation in humans.
However, MWCNTs did significantly affect mitochondrial gene transcription. We found inter-individual variation in the responses between donors, with four of the donors having a 2-fold or higher upregulation of one or more mitochondrial genes in response to MWCNT exposure, and the remaining four were relatively non-responsive. With no demographic similarities or shared SNP/heteroplasmies to separate responders from non-responders, the source of this susceptibility is not currently known, though the variation may be due to differences in nuclear-encoded mitochondrial genes or increased susceptibility to redox stress or particulate matter in general. The upregulation of mitochondrial genes in response to MWCNT exposure contradicted our original expectations that transcription would be impaired by induction of heteroplasmy. We hypothesize that this upregulation indicates compensation by the cells to generate new mitochondrial subunits to replace those lost through mitophagy59–60 or direct damage from physical intrusion36 by the nanotubes. This hypothesis would predict that MWCNT exposure depletes mitochondrial abundance per cell, which we measured by immunostaining for SDHA (a nuclear-encoded mitochondrial Complex II subunit) and MitoTracker dye in flow cytometry experiments. We found a statistically-significant and dose-dependent decrease in mitochondrial abundance with MWCNT exposure. We also used extracellular flux assay to measure the changes in metabolic function resulting from this loss of mitochondria and found that oxygen consumption rate (per unit of total protein), both basally and following uncoupling by FCCP, was significantly decreased by MWCNT exposure. These results suggest an overall impairment of mitochondrial abundance and function in human BECs by MWCNT exposure, despite the elevated transcription of mtDNA.
Another outcome predicted by our hypothesis is that mitochondria damaged by MWCNT exposure would be recycled through mitophagy, thereby preventing the accumulation of damaged mtDNA and consistent with the absence of heteroplasmy. We performed a fluorescent assay to measure formation of mitophagic vesicles in MWCNT-exposed BECs and found that mitophagy was highly induced after 2 hours of exposure. This induction was attenuated by Mdivi-1, which inhibits mitophagy through drp1. Mitophagy in MWCNT-exposed BECs was notably more pronounced than in the FCCP-treated positive controls. This may be due to kinetic effects, wherein the chemical induction of mitophagy by FCCP peaks earlier and is more resolved by 2 hours, while the MWCNTs induce mitophagy later due to their having to penetrate the cells. Electron micrographs following 5-day MWCNT treatments demonstrate the capability of the MWCNTs used in this study to enter the cytoplasm of exposed BECs, whether by escaping endosomes following active uptake35–36 or by passively penetrating the plasma membrane33–34, though whether the physical presence of cytoplasmic MWCNTs is necessary for the induction of mitophagy is not yet known. Future experiments to evaluate the kinetics of initiation and resolution of mitophagy in MWCNT-exposed BECs are planned, but the current results present a plausible mechanism that reconciles the upregulation of mtDNA genes with reduced mitochondrial abundance and function.
Our study has a few inherent limitations related to its translation to human toxicology. The study was performed in vitro to investigate whether, under the most ideal conditions, nanomaterial exposure could cause alterations in the mitochondrial genome. While this eliminates many potential variables, factors that could attenuate or exacerbate any direct mitogenomic effects from the MWCNTs in an intact organism are beyond the scope of this study. These factors include deposition61 of material in the lungs, other cells/tissues62 (e.g., resident macrophages), inflammatory responses63 to the nanomaterials, and co-exposures with other pollutants/chemicals/pathogens64. Another inherent limitation is the use of a submerged epithelial culture model instead of an (arguably more physiologically-relevant65) air-liquid interface model. This was done to eliminate donor-to-donor variation in mucociliary differentiation as a confounding factor in the study, as well as to allow comparison of mitochondrial genomics data with our extracellular flux data (an assay that, at present, cannot be performed using an air-liquid interface).
In summary, our findings suggest that mitochondrial turnover may be important for protection of the lung epithelium’s mtDNA from nanoparticle exposure. Future studies are necessary to determine whether inhibition of mitophagy would result in increased mitogenomic effects from nanomaterials or whether the cell has other, undescribed mechanisms for protecting its mtDNA. Additionally, the investigation of other forms of genotoxic stress for mitogenomic endpoints could result in new therapeutic measures for this relatively unexplored form of cellular damage.
Supplementary Material
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (NIEHS). We would like to acknowledge Greg Solomon, Jason Malphurs, and Nicole Reeves for their assistance with ultradeep sequencing, Jianying Li for statistical analysis, and Dr. Michael Dykstra for ultrastructural imaging. We would also like to acknowledge Annette Rice and the NIEHS Clinical Research Unit for the recruitment and bronchoscopy of healthy donors to obtain bronchial cell samples, as well as Wesley Gladwell and Jacqui Marzec for their technical assistance with this study. We also acknowledge the assistance of Dr. James Samet at the US Environmental Protection Agency (EPA) in acquiring the darkfield microscopy images used in the supplemental information. Finally, we wish to acknowledge the assistance and expertise of Dr. Scott Randell and his laboratory in the Cystic Fibrosis Center the University of North Carolina Chapel Hill.
Footnotes
Disclosure of Interest
The authors report no conflict of interest.
References
- 1.Wang X; Rhee I; Wang Y; Xi Y, Compressive strength, chloride permeability, and freeze-thaw resistance of MWNT concretes under different chemical treatments. TheScientificWorldJournal 2014, 2014, 572102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bianco A; Kostarelos K; Prato M, Applications of carbon nanotubes in drug delivery. Current opinion in chemical biology 2005, 9 (6), 674–9. [DOI] [PubMed] [Google Scholar]
- 3.Liang XJ; Chen C; Zhao Y; Jia L; Wang PC, Biopharmaceutics and therapeutic potential of engineered nanomaterials. Curr Drug Metab 2008, 9 (8), 697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun YF; Liu SB; Meng FL; Liu JY; Jin Z; Kong LT; Liu JH, Metal oxide nanostructures and their gas sensing properties: a review. Sensors (Basel) 2012, 12 (3), 2610–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller K; Bugnicourt E; Latorre M; Jorda M; Echegoyen Sanz Y; Lagaron JM; Miesbauer O; Bianchin A; Hankin S; Bolz U; Perez G; Jesdinszki M; Lindner M; Scheuerer Z; Castello S; Schmid M, Review on the Processing and Properties of Polymer Nanocomposites and Nanocoatings and Their Applications in the Packaging, Automotive and Solar Energy Fields. Nanomaterials (Basel) 2017, 7 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivero PJ; Urrutia A; Goicoechea J; Arregui FJ, Nanomaterials for Functional Textiles and Fibers. Nanoscale Res Lett 2015, 10 (1), 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hecht DS; Hu L; Irvin G, Emerging transparent electrodes based on thin films of carbon nanotubes, graphene, and metallic nanostructures. Adv Mater 2011, 23 (13), 1482–513. [DOI] [PubMed] [Google Scholar]
- 8.De Volder MF; Tawfick SH; Baughman RH; Hart AJ, Carbon nanotubes: present and future commercial applications. Science 2013, 339 (6119), 535–9. [DOI] [PubMed] [Google Scholar]
- 9.Wanekaya AK, Applications of nanoscale carbon-based materials in heavy metal sensing and detection. The Analyst 2011, 136 (21), 4383–91. [DOI] [PubMed] [Google Scholar]
- 10.Du Choi S; Kim D; Kang YP; Jang DH, Effect of spray process conditions on uniformity of carbon nanotube thin films. J Nanosci Nanotechnol 2012, 12 (7), 5290–6. [DOI] [PubMed] [Google Scholar]
- 11.Nanotechnology Consumer Product Inventory. http://www.nanotechproject.org/cpi/about/analysis/ (accessed May 6).
- 12.Bai Y; Zhang R; Ye X; Zhu Z; Xie H; Shen B; Cai D; Liu B; Zhang C; Jia Z; Zhang S; Li X; Wei F, Carbon nanotube bundles with tensile strength over 80 GPa. Nat Nanotechnol 2018. [DOI] [PubMed] [Google Scholar]
- 13.Baughman RH; Zakhidov AA; de Heer WA, Carbon nanotubes - the route toward applications. Science 2002, 297 (5582), 787–792. [DOI] [PubMed] [Google Scholar]
- 14.Law M; Goldberger J; Yang PD, Semiconductor nanowires and nanotubes. Annu Rev Mater Res 2004, 34, 83–122. [Google Scholar]
- 15.Garcia-Hevia L; Fernandez F; Gravalos C; Garcia A; Villegas JC; Fanarraga ML, Nanotube interactions with microtubules: implications for cancer medicine. Nanomedicine (Lond) 2014, 9 (10), 1581–8. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Hevia L; Villegas JC; Fernandez F; Casafont I; Gonzalez J; Valiente R; Fanarraga ML, Multiwalled Carbon Nanotubes Inhibit Tumor Progression in a Mouse Model. Advanced healthcare materials 2016, 5 (9), 1080–7. [DOI] [PubMed] [Google Scholar]
- 17.He XQ; Young SH; Schwegler-Berry D; Chisholm WP; Fernback JE; Ma Q, Multiwalled Carbon Nanotubes Induce a Fibrogenic Response by Stimulating Reactive Oxygen Species Production, Activating NF-kappa B Signaling, and Promoting Fibroblast-to-Myofibroblast Transformation. Chem Res Toxicol 2011, 24 (12), 2237–2248. [DOI] [PubMed] [Google Scholar]
- 18.Manke A; Wang L; Rojanasakul Y, Mechanisms of nanoparticle-induced oxidative stress and toxicity. Biomed Res Int 2013, 2013, 942916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ali A; Suhail M; Mathew S; Shah MA; Harakeh SM; Ahmad S; Kazmi Z; Alhamdan MA; Chaudhary A; Damanhouri GA; Qadri I, Nanomaterial Induced Immune Responses and Cytotoxicity. J Nanosci Nanotechnol 2016, 16 (1), 40–57. [DOI] [PubMed] [Google Scholar]
- 20.Boyles MS; Young L; Brown DM; MacCalman L; Cowie H; Moisala A; Smail F; Smith PJ; Proudfoot L; Windle AH; Stone V, Multi-walled carbon nanotube induced frustrated phagocytosis, cytotoxicity and pro-inflammatory conditions in macrophages are length dependent and greater than that of asbestos. Toxicology in vitro : an international journal published in association with BIBRA 2015, 29 (7), 1513–28. [DOI] [PubMed] [Google Scholar]
- 21.Braydich-Stolle L; Hussain S; Schlager JJ; Hofmann MC, In vitro cytotoxicity of nanoparticles in mammalian germline stem cells. Toxicological sciences : an official journal of the Society of Toxicology 2005, 88 (2), 412–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegrist KJ; Reynolds SH; Kashon ML; Lowry DT; Dong C; Hubbs AF; Young SH; Salisbury JL; Porter DW; Benkovic SA; McCawley M; Keane MJ; Mastovich JT; Bunker KL; Cena LG; Sparrow MC; Sturgeon JL; Dinu CZ; Sargent LM, Genotoxicity of multi-walled carbon nanotubes at occupationally relevant doses. Part Fibre Toxicol 2014, 11, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan J; Gou N; Gao C; He M; Gu AZ, Comparative and mechanistic genotoxicity assessment of nanomaterials via a quantitative toxicogenomics approach across multiple species. Environmental science & technology 2014, 48 (21), 12937–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snyder RJ; Hussain S; Tucker CJ; Randell SH; Garantziotis S, Impaired Ciliogenesis in differentiating human bronchial epithelia exposed to non-Cytotoxic doses of multi-walled carbon Nanotubes. Part Fibre Toxicol 2017, 14 (1), 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snyder RJ; Hussain S; Rice AB; Garantziotis S, Multiwalled carbon nanotubes induce altered morphology and loss of barrier function in human bronchial epithelium at noncytotoxic doses. International journal of nanomedicine 2014, 9, 4093–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rotoli BM; Bussolati O; Barilli A; Zanello PP; Bianchi MG; Magrini A; Pietroiusti A; Bergamaschi A; Bergamaschi E, Airway barrier dysfunction induced by exposure to carbon nanotubes in vitro: which role for fiber length? Human & experimental toxicology 2009, 28 (6–7), 361–8. [DOI] [PubMed] [Google Scholar]
- 27.Lam CW; James JT; McCluskey R; Hunter RL, Pulmonary toxicity of single-wall carbon nanotubes in mice 7 and 90 days after intratracheal instillation. Toxicological sciences : an official journal of the Society of Toxicology 2004, 77 (1), 126–34. [DOI] [PubMed] [Google Scholar]
- 28.Shvedova AA; Kisin ER; Mercer R; Murray AR; Johnson VJ; Potapovich AI; Tyurina YY; Gorelik O; Arepalli S; Schwegler-Berry D; Hubbs AF; Antonini J; Evans DE; Ku BK; Ramsey D; Maynard A; Kagan VE; Castranova V; Baron P, Unusual inflammatory and fibrogenic pulmonary responses to single-walled carbon nanotubes in mice. American journal of physiology. Lung cellular and molecular physiology 2005, 289 (5), L698–708. [DOI] [PubMed] [Google Scholar]
- 29.Donaldson K; Poland CA; Murphy FA; MacFarlane M; Chernova T; Schinwald A, Pulmonary toxicity of carbon nanotubes and asbestos - similarities and differences. Advanced drug delivery reviews 2013, 65 (15), 2078–86. [DOI] [PubMed] [Google Scholar]
- 30.Wang X; Katwa P; Podila R; Chen P; Ke PC; Rao AM; Walters DM; Wingard CJ; Brown JM, Multi-walled carbon nanotube instillation impairs pulmonary function in C57BL/6 mice. Part Fibre Toxicol 2011, 8, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park EJ; Roh J; Kim SN; Kang MS; Han YA; Kim Y; Hong JT; Choi K, A single intratracheal instillation of single-walled carbon nanotubes induced early lung fibrosis and subchronic tissue damage in mice. Archives of toxicology 2011, 85 (9), 1121–31. [DOI] [PubMed] [Google Scholar]
- 32.Bossi E; Zanella D; Gornati R; Bernardini G, Cobalt oxide nanoparticles can enter inside the cells by crossing plasma membranes. Sci Rep 2016, 6, 22254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nerl HC; Cheng C; Goode AE; Bergin SD; Lich B; Gass M; Porter AE, Imaging methods for determining uptake and toxicity of carbon nanotubes in vitro and in vivo. Nanomedicine (Lond) 2011, 6 (5), 849–65. [DOI] [PubMed] [Google Scholar]
- 34.Skandani AA; Al-Haik M, Reciprocal effects of the chirality and the surface functionalization on the drug delivery permissibility of carbon nanotubes. Soft Matter 2013, 9 (48), 11645–9. [DOI] [PubMed] [Google Scholar]
- 35.Mu Q; Broughton DL; Yan B, Endosomal leakage and nuclear translocation of multiwalled carbon nanotubes: developing a model for cell uptake. Nano letters 2009, 9 (12), 4370–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu S; Zhu B; Huang A; Hu Y; Wang G; Ling F, Toxicological effects of multi-walled carbon nanotubes on Saccharomyces cerevisiae: The uptake kinetics and mechanisms and the toxic responses. Journal of hazardous materials 2016, 318, 650–662. [DOI] [PubMed] [Google Scholar]
- 37.Costa PM; Bourgognon M; Wang JT; Al-Jamal KT, Functionalised carbon nanotubes: From intracellular uptake and cell-related toxicity to systemic brain delivery. J Control Release 2016, 241, 200–219. [DOI] [PubMed] [Google Scholar]
- 38.Kang B; Chang S; Dai Y; Yu D; Chen D, Cell response to carbon nanotubes: size-dependent intracellular uptake mechanism and subcellular fate. Small 2010, 6 (21), 2362–6. [DOI] [PubMed] [Google Scholar]
- 39.Liu X; Chen Z, The pathophysiological role of mitochondrial oxidative stress in lung diseases. J Transl Med 2017, 15 (1), 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shukla A; Jung M; Stern M; Fukagawa NK; Taatjes DJ; Sawyer D; Van Houten B; Mossman BT, Asbestos induces mitochondrial DNA damage and dysfunction linked to the development of apoptosis. American journal of physiology. Lung cellular and molecular physiology 2003, 285 (5), L1018–25. [DOI] [PubMed] [Google Scholar]
- 41.Ryman-Rasmussen JP; Tewksbury EW; Moss OR; Cesta MF; Wong BA; Bonner JC , Inhaled Multiwalled Carbon Nanotubes Potentiate Airway Fibrosis in Murine Allergic Asthma. Am J Resp Cell Mol 2009, 40 (3), 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khoor A; Stahlman MT; Gray ME; Whitsett JA, Temporal-spatial distribution of SP-B and SP-C proteins and mRNAs in developing respiratory epithelium of human lung. J Histochem Cytochem 1994, 42 (9), 1187–99. [DOI] [PubMed] [Google Scholar]
- 43.Xia T; Hamilton RF Jr; Bonner JC; Crandall ED; Elder A; Fazlollahi F; Girtsman TA; Kim K; Mitra S; Ntim SA; Orr G; Tagmount M; Taylor AJ; Telesca D; Tolic A; Vulpe CD; Walker AJ; Wang X; Witzmann FA; Wu N; Xie Y; Zink JI; Nel A; Holian A, Interlaboratory Evaluation of Cytotoxicity and Inflammatory Responses to Engineered Nanomaterials: The NIEHS Nano Go Consortium. Environmental health perspectives 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dames S; Chou LS; Xiao Y; Wayman T; Stocks J; Singleton M; Eilbeck K; Mao R, The development of next-generation sequencing assays for the mitochondrial genome and 108 nuclear genes associated with mitochondrial disorders. J Mol Diagn 2013, 15 (4), 526–34. [DOI] [PubMed] [Google Scholar]
- 45.Hodgkinson A; Idaghdour Y; Gbeha E; Grenier JC; Hip-Ki E; Bruat V; Goulet JP; de Malliard T; Awadalla P, High-resolution genomic analysis of human mitochondrial RNA sequence variation. Science 2014, 344 (6182), 413–5. [DOI] [PubMed] [Google Scholar]
- 46.Minoche AE; Dohm JC; Himmelbauer H, Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol 2011, 12 (11), R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hauswirth WW; Clayton DA, Length heterogeneity of a conserved displacement-loop sequence in human mitochondrial DNA. Nucleic Acids Res 1985, 13 (22), 8093–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du J; Ge C; Liu Y; Bai R; Li D; Yang Y; Liao L; Chen C, The interaction of serum proteins with carbon nanotubes depend on the physicochemical properties of nanotubes. J Nanosci Nanotechnol 2011, 11 (11), 10102–10. [DOI] [PubMed] [Google Scholar]
- 49.Birch ME, Exposure and emissions monitoring during carbon nanofiber production--Part II: polycyclic aromatic hydrocarbons. Ann Occup Hyg 2011, 55 (9), 1037–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Donaldson K; Aitken R; Tran L; Stone V; Duffin R; Forrest G; Alexander A, Carbon nanotubes: A review of their properties in relation to pulmonary toxicology and workplace safety. Toxicological Sciences 2006, 92 (1), 5–22. [DOI] [PubMed] [Google Scholar]
- 51.Zhang T; Tang M; Yao Y; Ma Y; Pu Y, MWCNT interactions with protein: surface-induced changes in protein adsorption and the impact of protein corona on cellular uptake and cytotoxicity. International journal of nanomedicine 2019, 14, 993–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bai W; Wu Z; Mitra S; Brown JM, Effects of multiwalled carbon nanotube surface modification and purification on bovine serum albumin binding and biological responses. J Nanomater 2016, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickliffe JK; Rodgers BE; Chesser RK; Phillips CJ; Gaschak SP; Baker RJ, Mitochondrial DNA heteroplasmy in laboratory mice experimentally enclosed in the radioactive Chernobyl environment. Radiat Res 2003, 159 (4), 458–64. [DOI] [PubMed] [Google Scholar]
- 54.Tan D; Goerlitz DS; Dumitrescu RG; Han D; Seillier-Moiseiwitsch F; Spernak SM; Orden RA; Chen J; Goldman R; Shields PG, Associations between cigarette smoking and mitochondrial DNA abnormalities in buccal cells. Carcinogenesis 2008, 29 (6), 1170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matson CW; Lambert MM; McDonald TJ; Autenrieth RL; Donnelly KC; Islamzadeh A; Politov DI; Bickham JW, Evolutionary toxicology: population-level effects of chronic contaminant exposure on the marsh frogs (Rana ridibunda) of Azerbaijan. Environmental health perspectives 2006, 114 (4), 547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bibb MJ; Van Etten RA; Wright CT; Walberg MW; Clayton DA, Sequence and gene organization of mouse mitochondrial DNA. Cell 1981, 26 (2 Pt 2), 167–80. [DOI] [PubMed] [Google Scholar]
- 57.Skonieczna K; Malyarchuk B; Jawien A; Marszalek A; Banaszkiewicz Z; Jarmocik P; Borcz M; Bala P; Grzybowski T, Heteroplasmic substitutions in the entire mitochondrial genomes of human colon cells detected by ultra-deep 454 sequencing. Forensic Sci Int Genet 2015, 15, 16–20. [DOI] [PubMed] [Google Scholar]
- 58.Hirose M; Schilf P; Gupta Y; Zarse K; Kunstner A; Fahnrich A; Busch H; Yin J; Wright MN; Ziegler A; Vallier M; Belheouane M; Baines JF; Tautz D; Johann K; Oelkrug R; Mittag J; Lehnert H; Othman A; Johren O; Schwaninger M; Prehn C; Adamski J; Shima K; Rupp J; Hasler R; Fuellen G; Kohling R; Ristow M; Ibrahim SM, Low-level mitochondrial heteroplasmy modulates DNA replication, glucose metabolism and lifespan in mice. Sci Rep 2018, 8 (1), 5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim I; Lemasters JJ, Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxid Redox Signal 2011, 14 (10), 1919–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kagan VE; Jiang J; Huang Z; Tyurina YY; Desbourdes C; Cottet-Rousselle C; Dar HH; Verma M; Tyurin VA; Kapralov AA; Cheikhi A; Mao G; Stolz D; St Croix CM; Watkins S; Shen Z; Li Y; Greenberg ML; Tokarska-Schlattner M; Boissan M; Lacombe ML; Epand RM; Chu CT; Mallampalli RK; Bayir H; Schlattner U, NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell Death Differ 2016, 23 (7), 1140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sturm R, A stochastic model of carbon nanotube deposition in the airways and alveoli of the human respiratory tract. Inhalation toxicology 2016, 28 (2), 49–60. [DOI] [PubMed] [Google Scholar]
- 62.Cao Y; Roursgaard M; Jacobsen NR; Moller P; Loft S, Monocyte adhesion induced by multi-walled carbon nanotubes and palmitic acid in endothelial cells and alveolar-endothelial co-cultures. Nanotoxicology 2016, 10 (2), 235–44. [DOI] [PubMed] [Google Scholar]
- 63.Hussain S; Sangtian S; Anderson SM; Snyder RJ; Marshburn JD; Rice AB; Bonner JC; Garantziotis S, Inflammasome activation in airway epithelial cells after multi-walled carbon nanotube exposure mediates a profibrotic response in lung fibroblasts. Part Fibre Toxicol 2014, 11, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morozesk M; Franqui LS; Mansano AS; Martinez DST; Fernandes MN, Interactions of oxidized multiwalled carbon nanotube with cadmium on zebrafish cell line: The influence of two co-exposure protocols on in vitro toxicity tests. Aquat Toxicol 2018, 200, 136–147. [DOI] [PubMed] [Google Scholar]
- 65.Panas A; Comouth A; Saathoff H; Leisner T; Al-Rawi M; Simon M; Seemann G; Dossel O; Mulhopt S; Paur HR; Fritsch-Decker S; Weiss C; Diabate S, Silica nanoparticles are less toxic to human lung cells when deposited at the air-liquid interface compared to conventional submerged exposure. Beilstein J Nanotechnol 2014, 5, 1590–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.