Abstract
Objective
To determine the proportion of infant deaths occurring in the setting of a confirmed genetic disorder.
Study Design
A retrospective analysis of the electronic medical records of infants born from January 1, 2011 to June 1, 2017 who died prior to one year of age.
Results
573 deceased infants were identified. 117 were confirmed to have a molecular or cytogenetic diagnosis in a clinical diagnostic laboratory and an additional 7 were diagnosed by research testing for a total of 124/573 (22%) diagnosed infants. 67/124 (54%) had chromosomal disorders and 58/124 (47%) had single gene disorders (one infant had both). The proportion of diagnoses made by sequencing technologies, such as exome sequencing, increased over the years.
Conclusions
The prevalence of confirmed genetic disorders within our cohort of infant deaths is higher than previously reported. Increased efforts are needed to further understand the mortality burden of genetic disorders in infancy.
INTRODUCTION
Infants with genetic disorders, including chromosomal abnormalities (aneuploidy syndromes or chromosomal deletion or duplication disorders) and monogenic Mendelian disorders (caused by variants in single genes) contribute considerably to mortality in neonatal intensive care units (NICUs)1, 2, 3, 4, 5, 6. In the United States, major congenital malformations, affecting approximately 2% of births7, are also reported to be the leading cause of infant mortality8, 9, though the underlying etiology of these malformations may or may not be genetic. The overlapping contributions of genetic disorders and congenital malformations to neonatal and infant mortality has previously been estimated at 20–50% depending upon the population examined, although these figures are in many instances calculated in the absence of comprehensive genetic testing and have relied upon clinical diagnoses instead, therefore likely underestimating the proportion of infants with a pathogenic molecular or cytogenetic aberration1, 2, 5, 6, 8, 9, 10, 11, 12, 13, 14. As such, the true contribution of genetic disorders to neonatal and infant mortality is not fully understood.
As increasing use of massively parallel sequencing techniques, particularly exome sequencing (ES), has resulted in heightened awareness of monogenic conditions presenting in infancy15, 16, 17, 18, it is likely that the known contribution of genetic disorders to infant mortality will continue to increase. Without fully recognizing the contribution of genetic disorders, particularly Mendelian disorders, to infant mortality, adequate efforts towards reducing infant mortality cannot be made. We therefore sought to characterize the genetic evaluation and diagnostic yield of genetic testing for infant mortality cases identified within our institution.
MATERIALS AND METHODS
We identified and reviewed all patients in the electronic medical records (EMR) at our institution with a date of birth from January 1, 2011 to June 1, 2017 and a status of “deceased” in our EMR, indicating that either the patient died at our institution, or died at an outside location with subsequent notification sent to our institution. The date of death was used to select only infants who died at less than 365 days (one year) of age. Patients with the exact date of death missing from the EMR were included only if documentation was clear enough to indicate an age at death of less than 365 days. Stillbirths or products of conception after pregnancy termination were excluded. We performed a review of the EMR for these infants, focusing on the genetic evaluation and genetic diagnosis. Data were collected in a REDCap (Research Electronic Data Capture) database hosted at Boston Children’s Hospital19. Infants were considered to have had a genetic evaluation if a postnatal genetics consultation was performed or a genetic test was sent. They were considered to have had a genetic test sent if a molecular or cytogenetic test was sent on a germline sample (genotyping and karyotype analysis of tumor tissue, for example, were not included). The criteria used to determine whether a genetic diagnosis was made via laboratory testing and via clinical ascertainment have been described previously4. Redirection of care was defined as having necessary medical treatments withheld, if a decision was made to not escalate care or to redirect the goals of care towards comfort measures, or if a do not resuscitate (DNR) or partial DNR order was in place. Withdrawal of life support was defined as death occurring following the planned removal of life-sustaining therapies such as mechanical ventilation, oxygen, or vasoactive medications (infants who were terminally extubated in the setting of an unsuccessful code event were not considered to have had life support withdrawn). Infants were considered to have a major congenital anomaly if documentation indicated the presence of a congenital structural abnormality with “significant medical, social or cosmetic consequences for the affected individual“ that “typically require medical intervention” per the definition of the Centers for Disease Control and Prevention20.
The retrospective portion of our study was approved by the Boston Children’s Hospital Institutional Review Board (BCH IRB) with a waiver of informed consent due to the nature of the study. A subset of these patients who were admitted to the NICU at our institution has been published previously4.
Several of the deceased probands and their parents had been enrolled in a multi-disciplinary BCH IRB-approved protocol within the Gene Discovery Core of The Manton Center for Orphan Disease Research (ClinicalTrials.gov Identifier: NCT02743845), enrolling patients with rare diseases for genetic diagnosis, novel disease gene discovery, and therapeutic investigation (written informed consent or assent, where appropriate, was obtained). Via this mechanism and collaboration with the Center for Mendelian Genomics at the Broad Institute of MIT and Harvard, ES and data processing for 8 patients and genome sequencing (GS) for 2 additional patients was performed by the Genomics Platform at the Broad Institute of MIT and Harvard. ES was performed using an Illumina (San Diego, CA) exome capture (38 Mb target) and sequenced (150 bp paired reads) to cover greater than 90% of targets at 20x and a mean target coverage of greater than 100x. ES data was processed through a pipeline based on Picard and mapping was done using the BWA aligner to the human genome build 37 (hg19). Variants were called using Genome Analysis Toolkit (GATK) HaplotypeCaller package version 3.4. For GS, PCR-free preparation of sample DNA (350 ng input at >2 ng/ul) was accomplished using Illumina HiSeq X Ten v2 chemistry (San Diego, CA). Libraries were sequenced to a mean target coverage of more than 30x. GS data were processed through a pipeline based on Picard, using base quality score recalibration and local realignment at known indels. The BWA aligner was used for mapping reads to the human genome build 37 (hg19). Single Nucleotide Polymorphism (SNPs) and insertions/deletions (indels) were jointly called across all samples using GATK HaplotypeCaller package version 3.4. Default filters were applied to SNP and indel calls using the GATK Variant Quality Score Recalibration (VQSR) approach. Annotation was performed using Variant Effect Predictor (VEP).
Research ES for 4 patients was performed by methods described previously21, two had ES performed by a commercial lab (Ambry Genetics, Aliso Viejo, CA) on a research basis (this was the second ES test for one of these patients), and an additional patient had research ES performed by Beijing Genomics Institute (Cambridge, MA). Sanger sequencing by standard methods was used for diagnosis in one patient and to confirm all research ES findings. Two infants had clinical ES performed as part of the BabySeq study22 and five had clinical ES performed by GeneDx (Gaithersburg, MD) as part of a BCH IRB-approved rapid ES study.
Statistical analysis was performed using SPSS (Version 23.0, IBM Corp, Armonk, NY), using descriptive and Chi-square analyses and a 2-sided Fisher’s exact test to compare variables when appropriate.
RESULTS
Characteristics of the study population
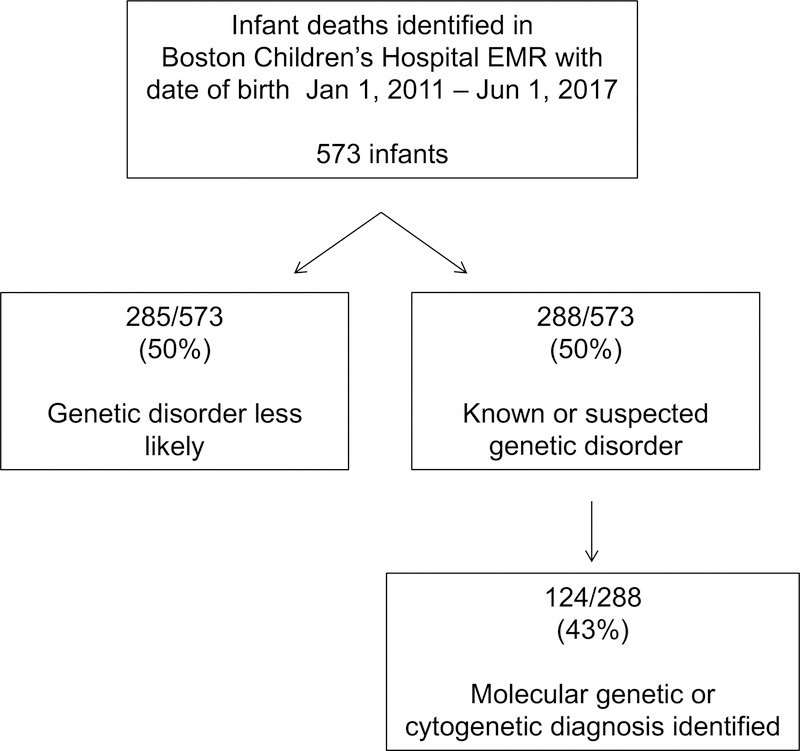
573 deceased infants were identified (Figure 1). 330/573 (58%) were male and 243/573 (42%) were female. 526 infants were evaluated at our hospital (clinic visit or admission) or by one of our physicians consulting at another hospital; 28 infants had records available at our hospital but were never seen, and 19 were registered at our hospital but had no medical records. Additional patient characteristics are presented in Table 1.
Figure 1. Cohort overview.
“Known or suspected disorder” includes infants who had either a prenatal diagnosis made, a clinical genetics consultation, or a molecular or cytogenetic test sent (testing sent at our hospital or elsewhere).
Table 1.
Characteristics of the study population
| Age at Death, days1 | Median (Q1-Q3) | Minimum, Maximum | |
|---|---|---|---|
| 55 (13–161) | 0, 361 | ||
| Neonatal death (<28 days) 1 | N (%) | ||
| 195/573 (34%) | |||
| Gestational age (GA) at birth, weeks | GA Category, weeks | N (%) | |
| 37 0/7 – 42 0/7 | 296/573 (52%) | ||
| < 37 0/7 | 239/573 (42%) | ||
| 32 0/7 to 36 6/7 | 131/239 (55%) | ||
| 28 0/7 to 31 6/7 | 38/239 (16%) | ||
| < 28 0/7 | 66/239 (28%) | ||
| Unknown preterm | 4/239 (2%) | ||
| Unknown GA | 38/573 (7%) | ||
| Location of death | N (%) | ||
| Home | 21/573 (4%) | ||
| Intensive care unit | 384/573 (67%) | ||
| Inpatient2 | 15/573 (3%) | ||
| Other hospital | 31/573 (5%) | ||
| Unknown/no records | 122/573 (21%) | ||
| Circumstances of death | N (%) | ||
| Redirection of care3 | 390/470 (83%) | ||
| Life support withdrawn4 | 313/432 (72%) | ||
Exact date of death unknown for 12 infants.
Includes deaths in the emergency department.
Data not available for 103 infants
Data not available for 141 infants
Genetic diagnosis
A genetics consultation was performed for 211/573 (37%) infants, either at our institution (195/573, 34%) or at an outside institution (16/573, 3%). 236/573 (41%) had a postnatal molecular or cytogenetic test sent either at our institution or elsewhere (results of testing sent at our institution are presented in Table 2). Overall, 288/573 (50%) patients had either a prenatal diagnosis made or had a postnatal genetics evaluation consisting of either a consultation, clinic visit, or molecular or cytogenetic test sent.
Table 2.
Genetic testing performed at our institution.
| Genetic Test | Number of patients | Number of tests | Positive 1 N (%) | Negative 1 N (%) | VUS N (%) | No result N (%) |
|---|---|---|---|---|---|---|
| FISH | 7 | 8 | 1 (13%) | 7 (88%) | 0 (0%) | 0 (0%) |
| Karyotype | 42 | 44 | 7 (16%) | 36 (82%) | 0 (0%) | 1 (2%) |
| Chromosomal microarray2 | 82 | 84 | 14 (17%) | 46 (55%) | 24 (29%) | 1 (1%) |
| Chromosomal deletion/duplication test | 14 | 14 | 3 (21%) | 11 (79%) | 0 (0%) | 0 (0%) |
| Single gene sequencing2,3 | 70 | 122 | 18 (15%) | 97 (80%) | 11 (9%) | 0 (0%) |
| Single gene or gene panel deletion/duplication | 29 | 36 | 3 (8%) | 28 (78%) | 2 (6%) | 3 (8%) |
| Methylation analysis | 4 | 4 | 0 (0%) | 3 (75%) | 0 (0%) | 1 (25%) |
| Gene panel2,4 | 45 | 49 | 7 (14%) | 29 (59%) | 14 (29%) | 1 (2%) |
|
Mitochondrial genetic testing2 (nuclear gene panel or mitochondrial genome) |
10 | 12 | 1 (8%) | 6 (50%) | 6 (50%) | 0 (0%) |
| Clinical exome sequencing | 14 | 14 | 6 (43%) | 4 (29%) | 4 (29%) | 0 (0%) |
| Research exome/genome sequencing5 | 16 | 17 | 6 (35%) | 8 (47%) | 36 (18%) | 0 (0%) |
Testing performed elsewhere included FISH for 20 infants, karyotype for 45, chromosomal microarray for 45, single gene tests for 12, gene panels for 13, deletion/duplication tests for 9, methylation studies for 5, mitochondrial genetic tests for 3, and ES for 6.
“Positive” includes pathogenic or likely pathogenic variants (one infant had a positive result indicating a diagnosis via two different testing modalities at our institution); “Negative” includes benign/likely benign variants and results indicating carrier status or that are nondiagnostic (e.g. Robertsonian translocation without aneuploidy, secondary findings from ES, carrier of one pathogenic variant for a recessive condition).
Test could have more than one category of result (e.g. one pathogenic variant and one VUS).
Includes targeted mutation analysis and sequencing of select exons.
Includes sequencing and deletion/duplication panels, which are often combined as a single test. Gene tests ordered as a panel but that resulted individually were counted as separate single gene tests.
Additionally, one patient had research single gene sequencing with a positive result.
Candidate novel disease genes.
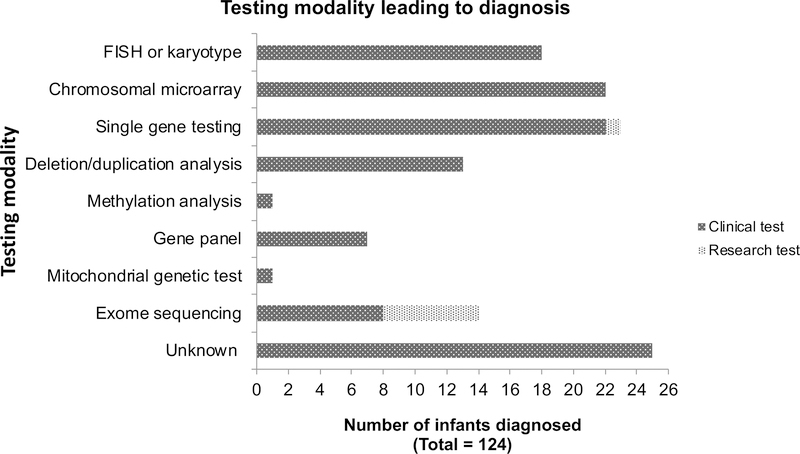
In total, 124/573 infants (22%) had laboratory-confirmed genetic diagnoses through either a clinical or a research laboratory (Table 3a–d, online, with diagnoses made or confirmed at our institution highlighted in grey). 27/124 (22%) diagnoses were made prenatally, 64/124 (52%) in the postnatal period prior to demise, and 24/124 (19%) postmortem, with an additional 7/124 (6%) postmortem diagnoses made via research testing (the timing was unclear from the medical record for 2 pre-mortem diagnoses). The exact date of postnatal / pre-mortem diagnosis was available for 44 infants with a median age at diagnosis of 47.5 days (interquartile range [IQR] 19–72 days). Definitive clinical diagnoses were made prior to laboratory confirmation for 39/65 (60%) of postnatal or postmortem diagnoses for whom sufficient information was available in the medical record to make this determination. The testing modality leading to diagnosis is shown in Figure 2. Notably, only 14 infants had postnatal clinical ES at our institution and 6 at outside institutions, 8/20 (40%) of which led to a diagnosis; two infants had prenatal ES and both were non-diagnostic. Most of these ES were performed recently, with 18/20 (90%) returning results in 2016 or later. Additionally, 16 infants had research ES (14) or GS (2), with 6/16 infants diagnosed (38%). Five of these diagnoses were caused by known disease-associated genes and one was a novel syndrome that had not previously been described23. Three additional candidate novel disease genes are currently under investigation and these results are considered variants of uncertain significance (VUS) at this time (Table 2). The total yield of ES/GS, including clinical and research tests performed at both our and at outside institutions, was therefore 14/36 (39%) infants diagnosed. The majority of postnatal ES/GS were performed as a trio: 25/36 had trio ES (69%), 5/36 (14%) had singleton ES performed, 3/36 had quad ES/GS (8%; one quad GS was performed with 2 affected sibs), and 1/36 had duo ES (3%; proband and mother); type of ES (proband vs trio) was not known for 2/36 (8%) infants. The yield of ES/GS performed with at least one parent was not significantly higher than proband-only ES/GS (12/29, 41% vs 2/5, 40%, p = 1.0).
Figure 2. Testing modality leading to diagnosis.
Of the 25 “unknown” modalities, 18 were chromosomal aneuploidy syndromes, three were diagnoses of chromosome 22q11 deletion syndrome, two were single gene disorders, one was a tumor variant confirmed in a germline tissue, and one was a chromosomal translocation. Three of the deletion/duplication diagnoses were chromosomal, and the remainder represented deletions in single genes. FISH, fluorescence in situ hybridization.
A major congenital anomaly was present in 333/548 (61%) infants (this information was unknown for 25 infants). Of these, 191/333 (57%) had a genetics evaluation and 93/333 (28%) had a laboratory-confirmed diagnosis. Of the infants without a major congenital anomaly, 31/215 (14%) had a genetic diagnosis (p < 0.001). Infants with a major congenital anomaly represented 93/124 (75%) of the confirmed genetic diagnoses.
Types of genetic disorders
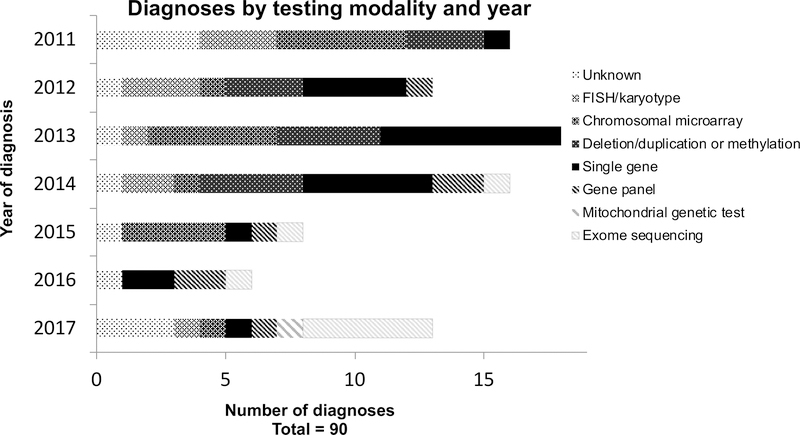
Genetic diagnoses are presented in Table 3a–d (online), with diagnoses made or confirmed at our institution highlighted in grey. 67/124 (54%) infants had chromosomal disorders, including aneuploidy syndromes: most commonly Patau Syndrome/Trisomy 13 (5/67, 7%), Edward Syndrome/Trisomy 18 (8/67, 12%), and Down Syndrome/Trisomy 21 (20/67, 30%); and microdeletion disorders: most commonly Jacobsen syndrome (chromosome 11q23 deletion) in 4/67 (6%) and 22q11 deletion syndrome in 8/67 (12%). Additionally, 58/124 (47%) had monogenic conditions: most commonly inborn errors of metabolism (13/58, 22%), spinal muscular atrophy (9/58, 16%), autosomal recessive polycystic kidney disease (ARPKD) (4/58, 7%), or CHARGE (coloboma, heart defect, atresia choanae, restricted growth and development, genital and ear abnormalities) syndrome (3/58, 5%). Two infants had two syndromes diagnosed (one with Trisomy 21 and galactosemia, and one with both Cat Eye and Klinefelter syndromes) and 9 had two chromosomal diagnoses (such as that resulting from an unbalanced translocation). Diagnoses by testing modality and year are presented in Figure 3, illustrating the increasing use of massively-parallel sequencing-based tests such as gene panels and ES in more recent years.
Figure 3. Diagnoses by testing modality and year.
This includes only postnatal diagnoses made by a clinical laboratory. For infants for whom the exact date of diagnosis was unknown, the date of birth was used.
Infants with diagnoses presented in Table 3a–d (online) were classified as having “positive” results in Table 2 (reflecting the results of testing performed at our institution), with the exception of 24 infants with prenatal diagnoses that were not confirmed post-natally at our institution, 34 infants diagnosed at an outside institution without a positive test at our institution, and one infant with single gene sequencing performed on a research basis that returned positive. Three infants had results reported by the clinical diagnostic laboratory as VUS that we considered disease-causing: one infant with a congenital heart defect had a diagnosis of chromosome 22q11 duplication with evidence of possible pathogenicity in the literature24; a second infant with a clinical diagnosis of tuberous sclerosis had a variant in TSC2 that was reported as a VUS, but the infant’s clinician had determined it to be likely pathogenic; a third infant had a reported VUS in IKBKG that was later found to be pathogenic25. An additional five infants had “positive” results found on molecular testing but were not considered to be “diagnosed” as these variants did not explain the predominant clinical presentation: one with a de novo likely pathogenic variant in EP300 who lacked clinical features of Rubinstein-Taybi syndrome as ascertained by her clinical geneticist, one with an inherited variant in FLG that did not explain the phenotype, one with a chromosome 16p12.2 deletion that did not explain the phenotype, one with a Factor V Leiden variant, and one with a prothrombin (c.20210G>A / c.*97G>A) variant; these were counted as VUS in Table 2 as their relevance to the underlying presentation is unclear. These intricacies highlight the complexity of diagnostic genetic testing and the challenges in variant interpretation, particularly in a population of deceased infants that may not be fully manifesting features of the genetic condition and for whom follow-up testing is difficult post-demise.
Reporting of genetic disorders
Death certificates were available in the EMR for 67 infants with clinical laboratory-confirmed genetic diagnoses. 41/67 (61%) indicated an underlying genetic disorder, including 8/12 (67%) infants with prenatal diagnoses, 25/37 (68%) infants with postnatal diagnoses, and 8/18 (44%) infants with postmortem diagnoses (Table 3a–d, online).
DISCUSSION
We present a description of the prevalence of laboratory-confirmed genetic disorders within a large cohort of deceased infants using data from our institution, which, at 22%, is higher than prior estimates. While prior studies have suggested that this prevalence is in the range of 5–32%1, 2, 5, 6, 11, information regarding confirmed molecular or cytogenetic diagnoses in these patients is often not provided. For example, a report of 32% of infant deaths (16 deaths total) in a NICU attributable to a “recognized syndrome or genetic or metabolic disorder” in the year 2002 did not provide further diagnostic detail1. These diagnoses may therefore include associations such as VACTERL (Vertebral anomalies, Anal atresia, Cardiac anomalies, Tracheo-Esophageal fistula, Renal anomalies, Limb anomalies) for which the genetic etiology, if any, remains unknown, in addition to laboratory-confirmed genetic disorders such as Down Syndrome, thus offering limited insight into this issue. Amongst the studies that do provide details on presumed laboratory-confirmed diagnoses, the prevalence is much lower, around 5–15% 2, 5, 11, likely because the majority of patients in those studies have not had a comprehensive genetic evaluation. Additionally, studies published prior to the widespread use of massively-parallel sequencing generally focused on findings detectable by fluorescence in situ hybridization (FISH), karyotype, or chromosomal microarray, and do not include testing for many single gene disorders (since these often require exome sequencing or the use of gene panels to make a diagnosis in the neonatal period), thus likely underestimating the prevalence of monogenic disorders in particular. Indeed, we have demonstrated that within our cohort, diagnoses made by sequencing-based tests have risen in the past several years owing to the increasing use of massively-parallel sequencing technology in clinical diagnostic laboratories. This number will likely continue to increase, as both we and others have shown that deceased infants represent a high-yield cohort for genetic diagnosis by ES, with diagnostic yields of 48%15 and 55%18 in recent studies. Though we found relatively few infants in our study that had ES as the sole, or first-tier, molecular genetic testing modality, this practice is becoming more commonplace both at our institution and others. Therefore, it is important to bear in mind the limitations of ES, particularly the lower sensitivity for exon-level deletions and duplications and triplet repeat disorders and the inability to detect disorders of methylation. With the exception of spinal muscular atrophy (SMA), we did not find many disorders of this type within our cohort; it is unclear whether this reflects a lack of targeted testing (e.g. for imprinting or triplet repeat disorders) or whether infants with these disorders were absent from our cohort. However, our results do demonstrate that SMA is an important contributor to infant mortality that may be missed if ES were relied upon as the sole testing modality.
As massively-parallel sequencing technologies for diagnosis, particularly ES and GS, are increasingly applied in the neonatal or infant period and have demonstrated great diagnostic utility15, 16, 17, 18, 26, 27, it is likely that the understanding of the phenotypic spectra for many Mendelian disorders will continue to broaden and that additional Mendelian disorders will be recognized as contributors to infant mortality. The increasing use of ES and GS is therefore likely to continue to influence the landscape of known genetic contributions to infant mortality. Furthermore, it has been argued that increasing usage of ES/GS in the NICU will increase neonatal (less than 28 days of age) mortality by aiding parents in the difficult decision to transition to palliative care approaches that may shorten life or to withdraw life-sustaining treatments, even if infant (less than one year of age) mortality is decreased overall via the identification of treatable conditions28. Infants diagnosed with genetic disorders (by rapid ES / GS or standard techniques) have been shown to have a higher 120-day mortality rate than those without a diagnosis, which may reflect either the high diagnostic yield of particularly severe phenotypes or an acceleration of the time to redirection of the goals of care or withdrawal of life support18. In our study, most infants died after redirection of the goals of care or withdrawal of life-sustaining technology, as has been shown in the NICU6 and, more recently, in deaths in a pediatric hospital29, which also suggests that the information gained from a comprehensive diagnostic genetic evaluation could be helpful for parents faced with these difficult decisions.
Prior studies regarding the genetic diagnostic evaluation of infants have generally focused on living infants for whom a diagnosis will ideally be identified prior to death and hence impact management15, 18, 26, 27. We report on our efforts at diagnosis not only during the infant’s life, but also postmortem. In an era of declining rates of traditional autopsy30, 31, pursuing a “molecular autopsy”, particularly via ES, may be a clinically useful surrogate32. Clinical genetic testing, including ES, is not routinely offered in the postmortem setting, due to lack of insurance coverage or the belief that the result may not be impactful. However, for the parents of infants who have died, the diagnostic odyssey may still be ongoing, and the establishment of a confirmed diagnosis may provide a sense of closure. It may also provide information important for future childbearing and can provide the opportunity for use of pre-implantation genetic diagnosis or prenatal genetic testing for parents interested in preventing the birth of future affected children. Finally, establishing a confirmed diagnosis, even after death, affords the opportunity to learn more about disorders contributing to infant mortality. As an example, one infant (Study ID 171) for whom research ES was performed post-mortem, was ultimately diagnosed with early-onset encephalopathy due to biallelic variants in TBCD. This is a recently described disorder33, 34 for which mortality data are scarce, thus the clinical suspicion for this diagnosis in a deceased infant may be low. Indeed, this diagnosis was not suspected in this infant prior to death, and without the use of ES, the parents would be left without a definitive diagnosis. Correctly identifying this recessive disorder, with a 25% recurrence risk, allowed for reproductive counseling to be provided. A second example of the utility of postmortem ES is an infant (Study ID 104) clinically diagnosed with CHARGE syndrome prior to death; however, postmortem ES identified a de novo variant in KMT2D, consistent with a diagnosis of Kabuki syndrome instead. While the phenotypic overlap between these disorders has been described previously35, and both are generally de novo, Kabuki syndrome was not suspected prior to the infant’s death. Finding this explanation is instructive for the evaluation of future infants with multiple congenital anomalies, even for those who are clinically diagnosed with a particular disorder, and it strengthens the argument for expanded sequencing such as ES in such cases.
In this era of emerging gene therapies and fetal interventions, understanding of the mortality burden of genetic disorders is crucial in order to advocate for the allocation of resources for research and therapeutics. For example, we report a substantial proportion of infants with spinal muscular atrophy, for which antisense oligonucleotide therapy is now available and could potential prevent infant deaths associated with this condition. As other new therapies are developed, it is important to understand the potential public health impact of these therapies and data such as ours offer an opportunity to do so.
Finally, we have also demonstrated in our study that the category of deaths due to congenital anomalies may not be a suitable proxy for deaths due to genetic disorders. While a significantly larger proportion of infants with a major congenital anomaly had a confirmed genetic diagnosis (28%) compared to those without a major congenital anomaly (14%), 25% of diagnosed infants overall did not have a major congenital anomaly. As the reproductive implications may be drastically different between cases with an “isolated” congenital anomaly versus those attributed to genetic disorders, there is an urgent need for an increased understanding of both the distinctions and the overlap between these two categories of infant deaths.
Limitations of our study include the retrospective nature of our approach and our access only to information available in our EMR. Therefore, diagnoses may have been made or deaths may have occurred of which we were not aware. Our data regarding both the diagnostic genetic evaluation and the number of known diagnoses may therefore be an underestimate owing to the incomplete data that we have on these infants. Our cohort also appears to be relatively enriched for major congenital anomalies, reflecting our hospital as a quaternary care referral center, which may have led to an overestimate of the contribution of genetic disorders to infant mortality. As such, it is unclear whether our results could be extrapolated to infant mortality at the state or national level. While these are limitations of our study design, they reflect the difficulties in linking detailed phenotypic information to mortality statistics in the absence of a nationalized healthcare system. By capturing all registered cases of infant mortality at our hospital, we have provided the most diverse and phenotypically-detailed cohort possible, though future efforts are certainly needed to incorporate genotype and phenotype information into mortality statistics.
While improving, infant mortality rates in the United States remain relatively high for a developed country36. Our data suggest a greater contribution of genetic disorders than has been previously appreciated. Our data also support the need for the expanded application of genetic testing, particularly ES, not only for critically ill infants but in the postmortem period as well, towards a greater understanding of the mortality burden of genetic disorders in infancy.
Supplementary Material
Acknowledgments
We would like to thank the patients and families whom we care for at our hospital who continue to teach and enrich us all. Dr. Wojcik is supported by NIH T32 GM007748. Dr. Agrawal is supported by NICHD/NHGRI/NIH U19HD077671. Dr. Gubbels was supported by a Harvard University Milton Fund award. Dr. Yu is supported by NIH/NIMH R01MH113761 and NICHD/NHGRI/NIH U19HD077671. Ms. Thiele received research support from the Summer Student Research Program of the Division of Newborn Medicine at Boston Children’s Hospital and Harvard Medical School and from The Brody School of Medicine at East Carolina University.
The Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung and Blood Institute grant UM1HG008900 to Daniel MacArthur and Heidi Rehm.
We appreciate the assistance of Anthony Capraro, MD (Department of Emergency Medicine, Boston Children’s Hospital) in identifying our cohort of deceased infants.
Funding Sources:
Supported by the National Institutes of Health (NIH T32 GM007748 [to MHW], and NICHD/NHGRI/NIH U19HD077671 [to PBA], NIH/NIMH R01MH113761 and NICHD/NHGRI/NIH U19HD077671 [to TWY]). Additional support provided by Harvard University (to CSG) and Boston Children’s Hospital/Harvard Medical School and from The Brody School of Medicine at East Carolina University (to KET). The Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung and Blood Institute grant UM1HG008900 to Daniel MacArthur and Heidi Rehm.
Footnotes
Potential Conflicts of Interest:
The authors have no conflicts of interest relevant to this article to disclose (Dr. Mullen is currently an employee of Quest Diagnostics who performed the work pertaining to this article while at the Broad Institute of MIT and Harvard).
Supplementary information is available at JPER’s website.
References:
- 1.Simpson CD, Ye XY, Hellmann J, Tomlinson C. Trends in cause-specific mortality at a Canadian outborn NICU. Pediatrics 2010, 126(6): 1538. [DOI] [PubMed] [Google Scholar]
- 2.Jacob J, Kamitsuka M, Clark RH, Kelleher AS, Spitzer AR. Etiologies of NICU deaths. Pediatrics 2015, 135(1): e59–65. [DOI] [PubMed] [Google Scholar]
- 3.Meng M, Zhang YP. Impact of inborn errors of metabolism on admission in a neonatal intensive care unit: a 4-year report. J Pediatr Endocrinol Metab 2013, 26(7–8): 689–693. [DOI] [PubMed] [Google Scholar]
- 4.Wojcik MH, Schwartz TS, Yamin I, Edward HL, Genetti CA, Towne MC, et al. Genetic disorders and mortality in infancy and early childhood: delayed diagnoses and missed opportunities. Genet Med 2018, 20: 1396–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudome SM, Kirby RS, Senner JW, Cunniff C. Contribution of genetic disorders to neonatal mortality in a regional intensive care setting. Am J Perinatol 1994, 11(2): 100–103. [DOI] [PubMed] [Google Scholar]
- 6.Weiner J, Sharma J, Lantos J, Kilbride H. How infants die in the neonatal intensive care unit: trends from 1999 through 2008. Arch Pediatr Adolesc Med 2011, 165(7): 630–634. [DOI] [PubMed] [Google Scholar]
- 7.Nelson K, Holmes LB. Malformations due to presumed spontaneous mutations in newborn infants. N Engl J Med 1989, 320(1): 19–23. [DOI] [PubMed] [Google Scholar]
- 8.Mathews TJ, Driscoll AK. Trends in Infant Mortality in the United States, 2005–2014. NCHS data brief, no 279. Hyattsville, MD: National Center for Health Statistics; 2017. [Google Scholar]
- 9.Murphy SL, Mathews TJ, Martin JA, Minkovitz CS, Strobino DM. Annual Summary of Vital Statistics: 2013–2014. Pediatrics 2017, 139(6): 3239. [DOI] [PubMed] [Google Scholar]
- 10.Sankaran K, Chien LY, Walker R, Seshia M, Ohlsson A, Canadian Neonatal N. Variations in mortality rates among Canadian neonatal intensive care units. CMAJ 2002, 166(2): 173–178. [PMC free article] [PubMed] [Google Scholar]
- 11.Wen SW, Liu S, Joseph KS, Rouleau J, Allen A. Patterns of infant mortality caused by major congenital anomalies. Teratology 2000, 61(5): 342–346. [DOI] [PubMed] [Google Scholar]
- 12.Costa S, Rodrigues M, Centeno MJ, Martins A, Vilan A, Brandao O, et al. Diagnosis and cause of death in a neonatal intensive care unit--how important is autopsy? J Matern Fetal Neonatal Med 2011, 24(5): 760–763. [DOI] [PubMed] [Google Scholar]
- 13.Michel MC, Colaizy TT, Klein JM, Segar JL, Bell EF. Causes and circumstances of death in a neonatal unit over 20 years. Pediatr Res 2018, 83(4):829–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seske LM, Muglia LJ, Hall ES, Bove KE, Greenberg JM. Infant Mortality, Cause of Death, and Vital Records Reporting in Ohio, United States. Matern Child Health J 2017, 21(4): 727–733. [DOI] [PubMed] [Google Scholar]
- 15.Meng L, Pammi M, Saronwala A, Magoulas P, Ghazi AR, Vetrini F, et al. Use of Exome Sequencing for Infants in Intensive Care Units: Ascertainment of Severe Single-Gene Disorders and Effect on Medical Management. JAMA Pediatr 2017: e173438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med 2012, 4(154): 154ra135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith LD, Willig LK, Kingsmore SF. Whole-Exome Sequencing and Whole-Genome Sequencing in Critically Ill Neonates Suspected to Have Single-Gene Disorders. Cold Spring Harb Perspect Med 2015, 6(2): a023168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willig LK, Petrikin JE, Smith LD, Saunders CJ, Thiffault I, Miller NA, et al. Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respir Med 2015, 3(5): 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap) - A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009, 42(2): 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Centers for Disease Control and Prevention. 1.4 Congenital Anomalies - Definitions. https://www.cdc.gov/ncbddd/birthdefects/surveillancemanual/chapters/chapter-1/chapter1-4.html. Accessed October 4, 2018.
- 21.Joshi M, Anselm I, Shi J, Bale TA, Towne M, Schmitz-Abe K, et al. Mutations in the substrate binding glycine-rich loop of the mitochondrial processing peptidase-alpha protein (PMPCA) cause a severe mitochondrial disease. Cold Spring Harb Mol Case Stud 2016, 2(3): a000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm IA, Agrawal PB, Ceyhan-Birsoy O, Christensen KD, Fayer S, Frankel LA, et al. The BabySeq project: implementing genomic sequencing in newborns. BMC Pediatr 2018, 18(1): 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wojcik MH, Okada K, Prabhu SP, Nowakowski DW, Ramsey K, Balak C, et al. De novo variant in KIF26B is associated with pontocerebellar hypoplasia with infantile spinal muscular atrophy. Am J Med Genet A 2018, 176(12):2623–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Portnoï MF, Lebas F, Gruchy N, Ardalan A, Biran-Mucignat V, Malan V, et al. 22q11.2 duplication syndrome: two new familial cases with some overlapping features with DiGeorge/velocardiofacial syndromes. Am J Med Genet A 2005, 137(1): 47–51. [DOI] [PubMed] [Google Scholar]
- 25.Johnston AM, Niemela J, Rosenzweig SD, Fried AJ, Delmonte OM, Fleisher TA, et al. A Novel Mutation in IKBKG/NEMO Leads to Ectodermal Dysplasia with Severe Immunodeficiency (EDA-ID). J Clin Immunol 2016, 36(6): 541–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Diemen CC, Kerstjens-Frederikse WS, Bergman KA, de Koning TJ, Sikkema-Raddatz B, van der Velde JK, et al. Rapid Targeted Genomics in Critically Ill Newborns. Pediatrics 2017, 140(4): 2854. [DOI] [PubMed] [Google Scholar]
- 27.Stark Z, Tan TY, Chong B, Brett GR, Yap P, Walsh M, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med 2016, 18(11): 1090–1096. [DOI] [PubMed] [Google Scholar]
- 28.Petrikin JE, Willig LK, Smith LD, Kingsmore SF. Rapid whole genome sequencing and precision neonatology. Semin Perinatol 2015, 39(8): 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trowbridge A, Walter JK, McConathey E, Morrison W, Feudtner C. Modes of Death Within a Children’s Hospital. Pediatrics 2018, 142:e20174182. [DOI] [PubMed] [Google Scholar]
- 30.McPherson E, Nestoridi E, Heinke D, Roberts DJ, Fretts R, Yazdy MM, et al. Alternatives to Autopsy for Fetal and Early Neonatal (Perinatal) Deaths: Insights from the Wisconsin Stillbirth Service Program. Birth Defects Res 2017, 109(18):1430–1441. [DOI] [PubMed] [Google Scholar]
- 31.Auger N, Bilodeau-Bertrand M, Poissant J, Shah PS. Decreasing use of autopsy for stillbirths and infant deaths: missed opportunity. J Perinatol 2018, 38(10):1414–1419. [DOI] [PubMed] [Google Scholar]
- 32.Wojcik MH, Brodsky D, Stewart JE, Picker J. Peri-mortem evaluation of infants who die without a diagnosis: focus on advances in genomic technology. J Perinatol 2018, 38(9): 1125–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flex E, Niceta M, Cecchetti S, Thiffault I, Au MG, Capuano A, et al. Biallelic Mutations in TBCD, Encoding the Tubulin Folding Cofactor D, Perturb Microtubule Dynamics and Cause Early-Onset Encephalopathy. Am J Hum Genet 2016, 99(4): 962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyake N, Fukai R, Ohba C, Chihara T, Miura M, Shimizu H, et al. Biallelic TBCD Mutations Cause Early-Onset Neurodegenerative Encephalopathy. Am J Hum Genet 2016, 99(4): 950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butcher DT, Cytrynbaum C, Turinsky AL, Siu MT, Inbar-Feigenberg M, Mendoza-Londono R, et al. CHARGE and Kabuki Syndromes: Gene-Specific DNA Methylation Signatures Identify Epigenetic Mechanisms Linking These Clinically Overlapping Conditions. Am J Hum Genet 2017, 100(5): 773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan SQ, Berrington de Gonzalez A, Best AF, Chen Y, Haozous EA, Rodriquez EJ, et al. Infant and Youth Mortality Trends by Race/Ethnicity and Cause of Death in the United States. JAMA Pediatr 2018: e183317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.