Abstract
The gut microbiota regulates levels of serotonin (5-hydroxytryptamine, 5-HT) in the intestinal epithelium and lumen1–5. However, whether 5-HT plays a functional role in bacteria from the gut microbiota remains unknown. We demonstrate that elevating levels of intestinal lumenal 5-HT by oral supplementation or by genetic deficiency in the host 5-HT transporter (SERT) increases the relative abundance of spore-forming members of the gut microbiota, which were previously reported to promote host 5-HT biosynthesis. Within this microbial community, we identify Turicibacter sanguinis as a gut bacterium that expresses a neurotransmitter sodium symporter (NSS)-related protein with sequence and structural homology to mammalian SERT. T. sanguinis imports 5-HT through a mechanism that is inhibited by the selective 5-HT reuptake inhibitor, fluoxetine. 5-HT reduces expression of sporulation factors and membrane transporters in T. sanguinis, which is reversed by fluoxetine exposure. Treating T. sanguinis with 5-HT or fluoxetine modulates its competitive colonization in the gastrointestinal tract of antibiotic-treated mice. In addition, fluoxetine reduces the membership of T. sanguinis in the gut microbiota of conventionally-colonized mice. Host association with T. sanguinis alters intestinal expression of multiple gene pathways, including those important for lipid and steroid metabolism, with corresponding reductions in host systemic triglyceride levels and inguinal adipocyte size. Altogether, these findings support the notion that select bacteria indigenous to the gut microbiota signal bidirectionally with the host serotonergic system to promote their fitness in the intestine.
The gut microbiota regulates several host biochemicals with known neuromodulatory properties, including endocannabinoids, neuropeptides and biogenic amines6. Of these, the hormone and neurotransmitter serotonin (5-hydroxytryptamine, 5-HT) is expressed highly in the gastrointestinal tract, synthesized by enterochromaffin cells (ECs) to levels that account for over 90% of the body’s 5-HT content7. Approximately 50% of gut-derived 5-HT is regulated by the gut microbiota, particularly spore-forming bacteria dominated by the families Clostridiaceae and Turicibacteraceae, with downstream consequences on host intestinal motility, hemostasis and ossification1–5. However, whether microbial regulation of host 5-HT occurs as a side effect of bacterial metabolism, or whether there is a functional role for microbial regulation of host 5-HT on bacterial physiology remains unclear. Interestingly, while the majority of gut 5-HT is secreted basolaterally by ECs into surrounding intestinal tissues, some EC-contained 5-HT is secreted apically into the intestinal lumen3,5,8, suggesting that gut microbes are exposed to host-derived 5- HT. Indeed, microbial influences on host ECs regulate 5-HT levels not only in intestinal tissue and blood1–4, but also in the intestinal lumen and feces3,5,8, raising the question of whether there are direct effects of host-derived 5-HT on gut bacteria.
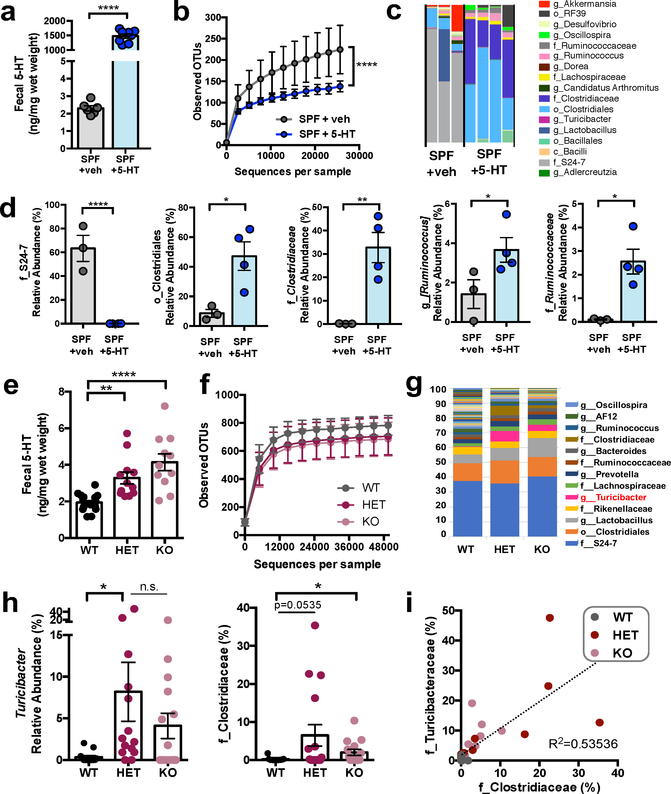
To determine whether the microbiota responds to intestinal 5-HT, we used three approaches to elevate lumenal 5-HT levels and measure their effects on microbiota composition. As a first proof- of-concept, we supplemented specific pathogen-free (SPF) mice orally with 5-HT to increase fecal 5-HT levels relative to vehicle-treated controls (Fig. 1a). 5-HT substantially enriched species of Clostridia from a combined relative abundance of 10.3 ± 1.9% (mean ± S.E.M.) in SPF mice to 86.2 ± 5.8% in 5-HT-supplemented mice (Fig. 1b–d and Supplementary Table 1). This is consistent with prior research demonstrating that spore-forming bacteria dominated by Clostridiaceae and Turicibacteraceae sufficiently promote gut 5-HT3. To determine whether physiological increases in host-derived 5-HT similarly alter the gut microbiota, we utilized serotonin transporter (SERT)- deficient mice which exhibit modest elevations in 5-HT along the intestine9. SERT deficiency increased fecal 5-HT by 1.7-fold in SERT+/− mice (Fig. 1e) and enriched Clostridiaceae and Turicibacter from a combined 0.9 ± 0.3% in wildtype mice to 13.7 ± 5.5% (Fig. 1f–i and Supplementary Table 2). Notably, these changes were more pronounced in SERT+/− mice, which lack confounding abnormalities in gastrointestinal motility and behavior, as compared to SERT−/− mice10. To further determine whether genetic predisposition to elevated intestinal 5-HT shapes the gut microbiota, we re-derived the SERT mouse line as germ-free (GF) and inoculated SERT+/− GF mice and wildtype GF controls with the same SPF fecal microbiota (Supplementary Data Fig. 1a). At 1 day after inoculation, we observed no overt taxonomic differences; however, at 35 days, SERT+/− mice exhibited significantly increased levels of Clostridia, including Clostridiales, Lachnospiraceae and Ruminococcaceae (Supplementary Data Fig. 1b–e and Supplementary Table 3). Overall, these findings reveal that increasing intestinal 5-HT enriches for spore-forming bacteria. This supports the notion that induction of host 5-HT by spore-forming bacteria3 promotes their own community membership in the gut microbiota.
Figure 1: Elevating intestinal 5-HT enriches for spore-forming bacteria in the gut.
a, Fecal 5-HT levels from SPF mice orally supplemented with 5-HT or vehicle in drinking water (two- tailed, unpaired Student’s t-test; n = 6, 8 cages). b, Alpha-diversity of OTUs derived from 16S rDNA sequencing of feces following vehicle vs. 5-HT treatment (two-way ANOVA with Bonferroni, n=3, 4 cages). c, Taxonomic diversity of the fecal microbiota after vehicle vs. 5-HT treatment (n=3, 4 cages). d, Relative abundance of bacterial taxa in fecal microbiota after vehicle vs. 5-HT treatment (two-way ANOVA with Kruskal-Wallis, n=3, 4 cages). e, Fecal 5-HT levels from SPF wild-type (WT), SERT+/− (HET) and SERT−/− (KO) mice. (one-way ANOVA with Bonferroni, n=13–15 cages). f, Alpha-diversity of OTUs derived from 16S rDNA sequencing of feces from SERT-deficient mice relative to WT controls (n=10–12 cages). g, Taxonomic diversity of the fecal microbiota after vehicle vs. 5-HT treatment (n=10–12 cages). h, Relative abundance of bacterial taxa in fecal microbiota of SERT-deficient mice relative to WT controls (one-way ANOVA with Kruskal Wallis, n=10, 11, 12 cages; n.s.: p=0.2134). i, Linear correlation between relative abundance of Turicibacteraceae vs. Clostridiaceae in fecal microbiota from SERT- deficient mice vs. WT controls (n=13, 15, 14 cages). (Mean ± SEM, * p < 0.05, ** p < 0.01, **** p < 0.0001, n.s. = not statistically significant; refer to Supplementary Table 11 for detailed statistical information)
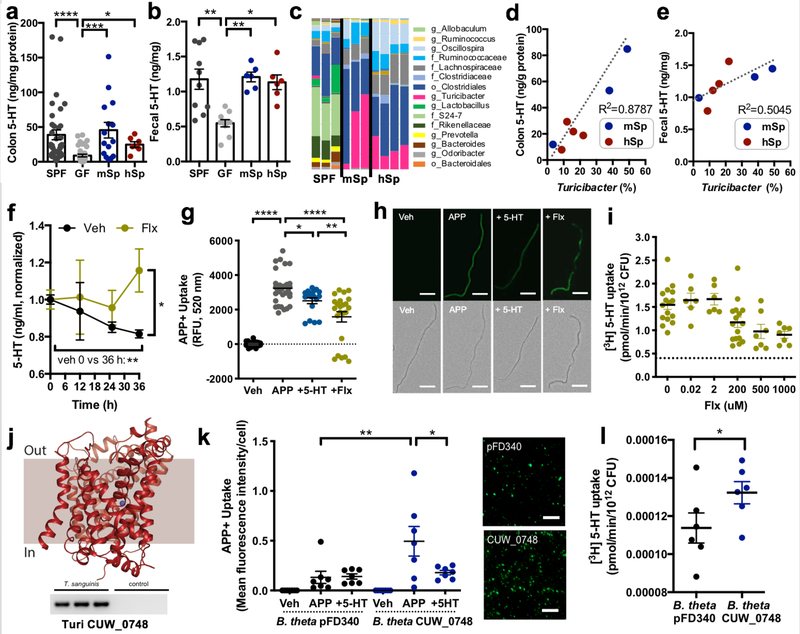
The ability of the microbiota to respond to elevations in host 5-HT suggests that select gut bacteria may encode elements for sensing 5-HT. To evaluate this possibility, we mined bacterial genomes for orthologs of mammalian (human or mouse) SERT. We identified a top candidate with 34% amino acid sequence identity (Supplementary Data Fig. 2a–b) and predicted structural homology to mammalian SERT (Fig. 2j), which is encoded by all four sequenced strains of the filamentous gut bacterium Turicibacter sanguinis and Turicibacter spp. (Supplementary Table 4). The gene is annotated for T. sanguinis HGF1 as a “putative sodium-dependent 5-HT transporter,” and has greater homology to SERT than to LeuT, a promiscuous bacterial amino acid transporter used to model eukaryotic neurotransmitter sodium symporter (NSS) proteins (Supplementary Data Fig. 2a). Interestingly, Turicibacter was enriched particularly in SERT+/− mice, which exhibited increased lumenal bioavailability of 5-HT (Fig. 1g–h). Moreover, Turicibacter was the most abundant member of the spore-forming bacterial consortium previously shown to induce host 5- HT biosynthesis (Fig. 2a–c, Supplementary Table 5 and 3), and correlated with intestinal 5-HT levels (Fig. 2d–e).
Figure 2: Turicibacter sanguinis uptakes 5-HT, which is inhibited by the selective serotonin reuptake inhibitor, fluoxetine.
a, Colon and fecal levels of 5-HT in mice that are conventionally colonized (SPF), germ-free (GF) or colonized with mouse-derived (m) or human-derived (h) spore-forming bacteria (Sp) (one-way ANOVA with Bonferroni, n=36, 37, 15, 7 mice). b, Fecal 5-HT in SPF, GF or mSp- or hSp-colonized mice (one-way ANOVA with Bonferroni, n=10, 10, 6, 6 mice). c, Taxonomic diversity of the fecal microbiota of SPF, mSp- and hSp-colonized mice (n=3, 4 cages). d, Linear correlation between fecal Turicibacter and colon 5-HT (n=3, 4 cages). e, Linear correlation between fecal Turicibacter and fecal 5-HT (n=3, 4 cages). f, 5-HT in supernatant after 0, 12, 25 and 36 hours of T. sanguinis MOL361 growth in Schaedler broth supplemented with vehicle or 200 uM fluoxetine (two-way ANOVA with Bonferroni, n=3, 4 cultures). g, Uptake by T. sanguinis exposed to vehicle (Veh) or APP+, either alone (APP) or with 200 uM 5-HT (+5-HT) or fluoxetine (+Flx) pre-treatment for 30 min (one-way ANOVA with Bonferroni, n=33, 33, 18, 23 cultures). h, Representative images of T. sanguinis exposed to Veh, APP, +5-HT or +Flx for 30 min (representing n=33, 33, 18, 23 cultures). Scale bar: 20 um. i, [3H] 5-HT uptake by T. sanguinis at 20 min after pre-treatment with Veh or +Flx. Dotted line: average background signal in negative controls. (one-way ANOVA with Bonferroni, n=7–16 cultures). j, Top: Predicted structural model for the putative SERT ortholog CUW_0748 in T. sanguinis MOL361. Bottom: qPCR product for CUW_0748 cDNA generated from T. sanguinis RNA (n=3 cultures). k, APP+ uptake in B. theta expressing CUW_0748 or vector control (pFD340) at 20 min after exposure to Veh, APP or +5-HT (one-way ANOVA with Bonferroni, n=7 cultures). Right: Representative images of APP+ uptake in B. theta expressing CUW_0748 or pFD340 control, with brightness at 90% higher than that in panel H. Scale bar: 20 um. l, Uptake of 1 uM [3H] 5-HT by B. theta expressing CUW_0748 or pFD340 after 20 min (two-tailed unpaired Student’s t-test, n=6 cultures). (Mean ± SEM, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, n.s. = not statistically significant; refer to Supplementary Table 11 for detailed statistical information)
We generated a model of the Turicibacter protein based on available structures of human SERT. Given the level of amino acid sequence identity (>30%) between the template and the target sequence, the expected accuracy of this model is 0.7–2.0 Å for the backbone of the transmembrane regions11. Indeed, the model scores very highly, consistent with the Turicibacter protein being globally similar to hSERT (Supplementary Data Fig. 3a–e). Residues contributing to the 5-HT and selective 5-HT reuptake inhibitor (SSRI) binding sites in mammalian SERT12 are strongly conserved (~50% identical and ~70% similar), and the residues required to bind a Na+ ion at the so-called Na2 site are identical. However, there were differences in key residues that mediate Cl−-dependence (N368, replaced by D262) and neutralization of charged amines (D98, replaced by G22) in mammalian SERT13,14. The latter in particular raises the question of whether the bacterial ortholog is capable of transporting 5-HT. However, the concurrent substitution of N368 in hSERT by an acidic side chain (D262) just 7 Å away raises the possibility that D262 can play a similar role to D98, allowing for a slightly altered arrangement for 5-HT or inhibitors in a similar binding region.
To determine whether Turicibacter is able to detect, import or metabolize 5-HT, we examined the ability of T. sanguinis MOL361 to i) consume 5-HT in culture, ii) import 4-(4- (dimethylamino)phenyl)-1-methylpyridinium (APP+), a molecular substrate for SERT15, iii) uptake radiolabeled 5-HT, and iv) respond transcriptionally to 5-HT. Culturing T. sanguinis in nutrient-rich broth decreased 5-HT levels in the media over time (Fig. 2f). This was prevented by supplementation with the SSRI fluoxetine, a reversible inhibitor of SERT (Fig. 2f), which exhibited cytostatic effects over long-term culture (Supplementary Data Fig. 4) but not short- term exposure (Fig. 3h, Supplementary Discussion). Exposing T. sanguinis to APP+ resulted in import, which was partially decreased by bacterial pre-treatment with unlabeled 5-HT or fluoxetine (Fig. 2g–h). There was also a modest, but not statistically significant, effect of norepinephrine, but not dopamine, on decreasing APP+ uptake in the assay (Supplementary Data Fig. 5a). Moreover, T. sanguinis imported tritiated 5-HT ([3H]5-HT), which was inhibited by fluoxetine (Fig. 2i). Notably, fluoxetine had no effect on bacterial uptake of norepinephrine (Supplementary Data Fig. 5b), suggesting some selectivity to inhibition of 5-HT import.
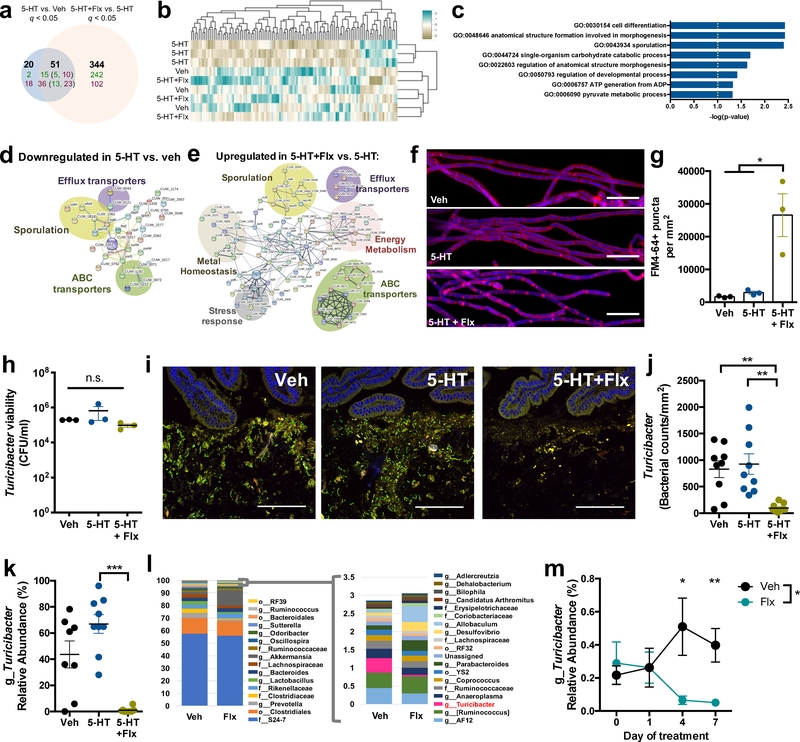
Figure 3: 5-HT and the selective serotonin reuptake inhibitor, fluoxetine, regulate gene expression and intestinal colonization of Turicibacter sanguinis.
a, Differentially expressed genes (q < 0.05) in T. sanguinis MOL361 after 4 hr exposure to vehicle, 200 uM 5-HT, or 5-HT with fluoxetine (Flx). Bold:total genes. Green:upregulated genes; red: downregulated genes. Numbers in parentheses: 5-HT-regulated genes that were further differentially expressed by Flx (Benjamini-Hochberg, n=3 cultures). b, Heatmap of genes with coefficient of variation < 3 (n=3 cultures). c, GO term enrichment analysis of genes differentially expressed in 5-HT vs. vehicle and 5-HT + Flx vs. 5-HT. (Fisher exact, n=3 cultures) d, Protein network analysis of 54 genes downregulated in T. sanguinis treated with 5-HT vs. vehicle. (n=3 cultures) e, Protein network analysis of 257 genes upregulated in T. sanguinis treated with 5-HT + Flx vs. 5-HT. (n=3 cultures) f, Representative images of T. sanguinis treated for 4 hr with vehicle, 200 uM 5-HT, or 5-HT with Flx, and stained with FM 4–64 membrane dye (pink) and DAPI (blue). (n=3 cultures). Scale bar: 10 um. g, Number of FM 4–64 puncta per area of T. sanguinis (one-way ANOVA with Bonferroni, n=3 cultures). h, Viability of T. sanguinis after 4 hr exposure to vehicle, 200 uM 5- HT, or 5-HT with Flx. (one-way ANOVA with Bonferroni, n=3 cultures; n.s.: p=0.9891). i, Representative FISH images of T. sanguinis (green) and small intestinal epithelial cells (DAPI, blue) from antibiotic-treated mice at 3 days after gavage with T. sanguinis pre-treated for 4 hr with vehicle, 200 uM 5-HT, or 5-HT with Flx (n=9 cages). Scale bar: 100 um. j, T. sanguinis cell counts from FISH images (one-way ANOVA with Bonferroni, n=9 cages). k, Relative abundance of Turicibacter by 16S rDNA sequencing of small intestinal lumenal contents (one- way ANOVA with Bonferroni, n=9 cages). l, Taxonomic diversity based on 16S rDNA sequencing of fecal microbiota on day 7 of Flx treatment. m, Relative abundance of Turicibacter in feces from SPF mice at 0, 1, 4, and 7 days after gavage with 10 mg/kg Flx (two-way ANOVA with Bonferroni, n=8, 18 cages). (Mean ± SEM, * p < 0.05, ** p < 0.01, *** p < 0.001, n.s. = not statistically significant; refer to Supplementary Table 11 for detailed statistical information).
Inhibition of 5-HT uptake was not observed by treatment with reserpine or tetrabenazine, inhibitors of mammalian vesicular monoamine transporter (VMAT) (Supplementary Data Fig. 5c), suggesting that import of 5-HT by T. sanguinis is mediated by an NSS-like protein such as the SERT ortholog (Fig. 2j) rather than a protein in the major facilitator superfamily (MFS), such as VMAT.
To gain insight into whether 5-HT uptake by T. sanguinis is mediated by the candidate protein, we cloned the gene from T. sanguinis MOL361 (“CUW_0748”, as annotated for T. sanguinis PC909) into the intestinal bacterium Bacteroides thetaiotaomicron, which shows no evidence of endogenous APP+ import (Supplementary Data Fig. 6a). Heterologous expression of CUW_0748 in B. thetaiotaomicron (Supplementary Data Fig. 6b–c) conferred uptake of APP+ and 1 uM [3H]5- HT, which was inhibited by unlabeled 5-HT (Fig. 2k–l). CUW_0748 in B. thetaiotaomicron exhibited time-dependent saturation of [3H]5-HT import (Supplementary Data Fig. 6d), requiring longer durations than those reported for SERT expressed in mammalian systems16,17. Notably, however, CUW_0748 in B. thetaiotaomicron failed to show dose-dependent saturation of [3H]5- HT import; biological variation increased substantially with concentrations of [3H]5-HT higher than physiological range (>100 uM), which may be due to stress to B. thetaiotaomicron. This indicates that additional studies are needed to validate active transporter-mediated 5-HT import. Notably, there was no effect of CUW_0748 expression on uptake of tryptophan (Supplementary Data Fig. 6f), suggesting that it does not function as a tryptophan transporter or broadly disrupt bacterial membrane integrity. These findings suggest that CUW_0748 contributes to 5-HT uptake in T. sanguinis, but also leave open the possibility that additional mechanisms for 5-HT import exist. Moreover, it is unclear whether the SERT ortholog, or any unknown 5-HT response elements in Turicibacter, are specific to 5-HT as opposed to other biogenic amines, indole derivatives or amino acids. While more research is required to detail the structural and biochemical properties of CUW_0748, these findings demonstrate that T. sanguinis imports 5-HT at physiological levels, which is inhibited by fluoxetine, suggesting that transport through an NSS-like protein mediates bacterial 5-HT uptake.
To identify effects of 5-HT and fluoxetine on bacterial physiology, we profiled transcriptomes of T. sanguinis after acute exposure to vehicle, 5-HT, fluoxetine or 5-HT with fluoxetine. 5-HT significantly altered bacterial expression of 71 genes (Fig. 3a–b), suggesting that T. sanguinis responds functionally to 5-HT. Co-treatment of 5-HT with fluoxetine significantly altered 51 of the 71 genes that were differentially expressed in response to 5-HT alone, with additional changes in 344 genes, revealing substantial effects of fluoxetine on bacterial gene expression (Fig. 3a–b and Supplementary Table 6). In particular, 5-HT downregulated expression of genes that clustered into pathways for cell differentiation, morphogenesis and sporulation (Fig. 3c) and encoded proteins related to efflux transporters, ABC transporters and sporulation (Fig. 3d). Compared to 5- HT-treated controls, fluoxetine upregulated expression of genes encoding proteins relevant to efflux transporters, ABC transporters and sporulation (Fig. 3e), as well as energy metabolism, metal homeostasis and stress response, suggesting that fluoxetine prevents and/or reverses a subset of 5-HT-mediated changes in T. sanguinis gene expression. Consistent with fluoxetine-associated increases in expression of sporulation-related genes, imaging of T. sanguinis exposed to 5-HT with fluoxetine revealed punctate staining of the membrane dye FM 4–64 (Fig. 3f, g), a phenotype observed during spore formation18. Notably, fluoxetine treatment in the absence of 5-HT yielded little change in T. sanguinis gene expression (Supplementary Tables 6 and 7), indicating that 5- HT is required for fluoxetine-induced alterations in gene expression. In addition, T. sanguinis survival was not affected by acute treatment with 5-HT and/or fluoxetine (Fig. 3h), suggesting that gene expression changes were not due to altered bacterial viability. Overall, these data demonstrate that 5-HT directly alters T. sanguinis physiology to reduce the expression of genes related to sporulation and membrane transport, which is abrogated by fluoxetine.
Sporulation is an important determinant of microbial colonization and transmission19. Given that 5-HT and fluoxetine regulate the expression of sporulation-related genes in T. sanguinis, we tested whether 5-HT and fluoxetine modulate T. sanguinis fitness in the intestine. T. sanguinis pre-treated with vehicle, 5-HT or 5-HT with fluoxetine was orally-gavaged into antibiotic-treated SPF mice supplemented with the same chemical treatments in drinking water. Both vehicle and 5-HT treatment resulted in detectable T. sanguinis colonization (Fig. 3i–j). However, treatment with 5-HT and fluoxetine significantly reduced T. sanguinis levels in the small intestine (Fig. 3i– j, Supplementary Discussion) and colon (Supplementary Data Fig. 7a–b), suggesting that fluoxetine-mediated inhibition of 5-HT uptake impairs T. sanguinis colonization. Reductions in T. sanguinis were similarly seen without additional chemical supplementation in the drinking water, where pre-treatment of T. sanguinis with 5-HT alone promoted its competitive colonization in antibiotic-treated mice (Supplementary Data Fig. 8a–d). In contrast, this effect was not seen with host supplementation alone (Supplementary Data Fig. 9a–b), suggesting that 5- HT and fluoxetine act directly on T. sanguinis to modulate intestinal colonization. Moreover, there was no effect of 5-HT and fluoxetine on T. sanguinis in monocolonized mice (Supplementary Data Fig. 10a–b) suggesting that 5-HT and fluoxetine do not affect the stability of T. sanguinis that has already colonized the intestine. In addition, there was no effect of 5-HT and fluoxetine on bacterial colonization of GF mice (Supplementary Data Fig. 11a–b), suggesting that 5-HT sensing by T. sanguinis is important for its competitive colonization within complex microbial communities and that fluoxetine is not broadly antimicrobial to T. sanguinis under these conditions. Consistent with this, SPF mice gavaged with fluoxetine exhibited decreased Turicibacter (Fig. 3k–m) and Clostridiaceae, with no significant differences in bacterial alpha diversity (Supplementary Data Fig. 12a–c). Collectively, these results suggest that inhibiting bacterial uptake of 5-HT and altering bacterial gene expression by fluoxetine limits the ability of Turicibacter to competitively colonize the intestine.
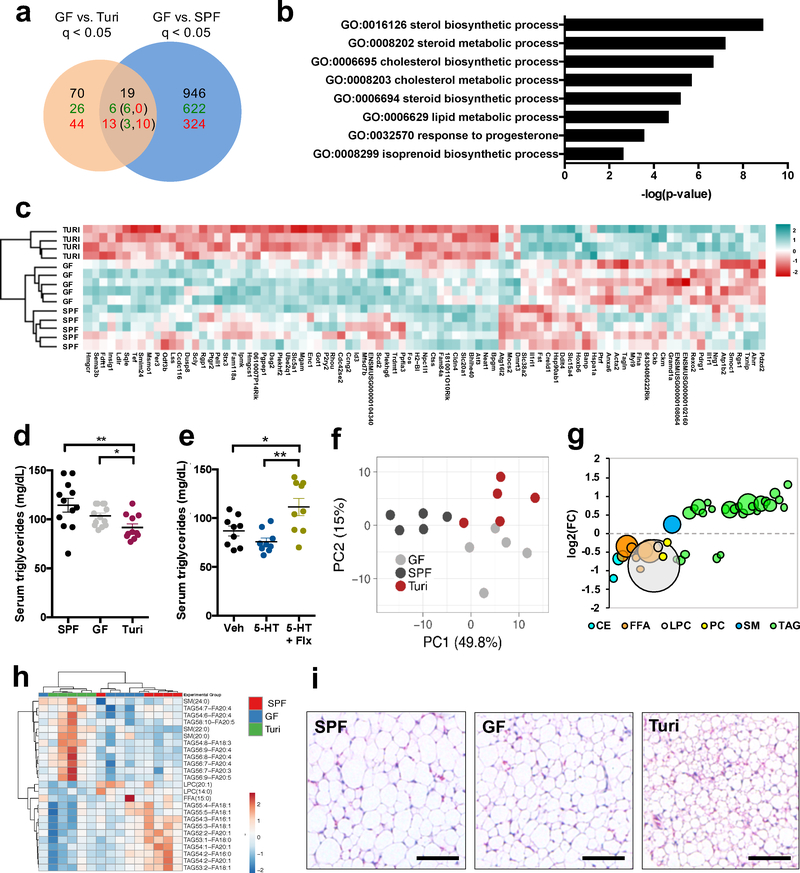
Changes in Turicibacter levels have been correlated with particular disease states20; however, causal effects of Turicibacter on the host remain unknown. To determine how Turicibacter colonization impacts host physiology, we first examined intestinal transcriptomes from T. sanguinis monocolonized mice, relative to GF and SPF controls. T. sanguinis differentially regulated 89 genes in the small intestine (Fig. 4a and Supplementary Table 8) and 87 genes in the colon (Supplementary Data Fig. 13a and Supplementary Table 9), which were enriched in pathways for steroid and lipid metabolism (e.g. Hmgcs1, Insig1, Fift1, Msmo1, Hmgcr, Npc1l1) (Fig. 4b–c and Supplementary Data Fig. 13b–c). This is likely not due to bacterial colonization in general, as there was little overlap between the effects of T. sanguinis versus SPF colonization on intestinal gene expression (Fig. 4a–b, Supplementary Data Fig. 13a–b and 14a–b). Moreover, T. sanguinis-induced transcriptomic changes were distinct from those reported in mice monocolonized with other species of gut bacteria21. In addition to altering intestinal expression of genes related to lipid metabolism, T. sanguinis also reduced host serum triglyceride levels (Fig. 4d). Consistent with this, fluoxetine-induced decreases in T. sanguinis were associated with increased serum triglyceride levels compared to mice colonized with vehicle-treated T. sanguinis (Fig. 4e). There was no effect of T. sanguinis on total serum cholesterol, free fatty acid, low-density lipoprotein or high-density lipoprotein levels (Supplementary Data Fig. 15). To determine whether T. sanguinis modulates concentrations of specific lipid species, we profiled sera for 1100 lipids spanning cholesterol esters, ceramides, diacylglycerides, free fatty acids, hexosyl ceramides, lysophosphatidylcholines, lysophosphatidylethanolamines, phosphatidylcholines, sphingomyelins and triacylglycerides (Fig. 4f–h, Supplementary Data Fig. 16 and Supplementary Table 10). Principal components analysis yielded clustering of lipidomic profiles from T. sanguinis- monocolonized mice distinctly from SPF and GF controls (Supplementary Data Fig. 16a and Fig. 4f). 184 lipids were significantly altered by T. sanguinis (Supplementary Data Fig. 16b and Supplementary Table 10), including several triacylglycerides (Fig. 4g and Supplementary Data Fig. 16c). In particular, T. sanguinis reduced levels of long-chain triglycerides containing 18:1 and 20:1 fatty acids such as oleate and gadoleate, while increasing levels of long-chain triglycerides containing 20:4 and 20:5 polyunsaturated fatty acids such as arachidonate and eicosapentaenoate (Fig. 4h). The reductions in monoenoic triacylglycerides were consistent with transcriptomic results wherein T. sanguinis downregulated expression of stearoyl-coenzyme A desaturase 2 (Scd2) (Fig. 4c), a rate-limiting enzyme for oleate synthesis22. Consistent with reductions in triglyceride levels (Fig. 4d, e), T. sanguinis-colonized mice exhibited small multilocular adipocytes in inguinal white adipose tissue (Fig. 4i, Supplementary Data Fig. 17) and reduced mass of inguinal white adipose tissue in female, but not male, mice (Supplementary Data Fig. 18), suggesting that T. sanguinis regulates fat mass in a sex-specific manner. These results reveal that T. sanguinis modulates host lipid metabolism, and further suggest that regulation of T. sanguinis colonization by 5-HT and fluoxetine could have downstream consequences on host physiology (Supplementary Data Fig. 19).
Figure 4: Turicibacter sanguinis colonization regulates host lipid metabolism.
a, Differentially expressed genes (q < 0.05) in small intestines from T. sanguinis- monocolonized, germ-free (GF) or SPF mice. Bolded numbers represent total differentially expressed genes. Numbers in green denote upregulated genes; numbers in red denote downregulated genes. Numbers in parentheses denote subsets of T. sanguinis regulated genes that were further differentially expressed by SPF (Wald test, n=5, 5, 4 mice). b, GO term enrichment analysis of genes differentially expressed in small intestine in response to T. sanguinis relative to GF controls (Fisher exact, n=4, 5, 5 mice). c, Heatmap of the 89 differentially expressed genes in the small intestine in response to T. sanguinis colonization (n=5, 5, 4 mice). d, Total serum triglyceride levels in GF, SPF and T. sanguinis- monocolonized mice (one-way ANOVA with Bonferroni, n=12, 13, 11 mice). e, Total serum triglyceride levels from antibiotic-treated mice gavaged with T. sanguinis pre-treated for 4 hr with vehicle, 200 uM 5-HT, or 200 uM 5-HT with Flx (one-way ANOVA with Bonferroni, n=9 mice). f, Principal components analysis of serum lipidomic data for lipid species with p < 0.05 for T. sanguinis vs. GF and T. sanguinis vs. SPF mice (two-way ANOVA + Bonferroni, n=5 mice). g, Average fold change (FC) of serum lipid species (p < 0.05) differentially regulated by T. sanguinis compared to GF controls. Largest circle = p < 0.001, smallest circle = p<0.05. CE=cholesterol esters, FFA=free fatty acids, LPC=lysophosphatidylcholines, PC=phosphatidylcholines, SM=sphingomyelins, TAG=triacylglycerides. (two-way ANOVA + Bonferroni, n=5 mice). h, Heatmap of 25 serum lipid species similarly increased (red) or decreased (blue) by T. sanguinis colonization relative to both GF and SPF controls (n=5 mice). i, Representative image of inguinal white adipose tissue from GF, SPF and T. sanguinis-monocolonized mice. Scale bar = 100 um. (representing n=5 mice) (Mean ± SEM, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, n.s. = not statistically significant; refer to Supplementary Table 11 for detailed statistical information)
Considerable progress has been made in understanding host-microbial interactions that control intestinal colonization of select microbial species such as Bacteroides23 but little is known about the molecular determinants of microbial colonization and community membership for other groups of bacteria, including those belonging to the dominant phylum Firmicutes. In addition, previous studies on bacterial responses to norepinephrine fueled the concept of “microbial endocrinology”24, but whether host-microbial interactions occur via other canonical neurotransmitters remains poorly understood. Here we identify a role for host-derived 5-HT and fluoxetine treatment in regulating the gut microbiota, including intestinal colonization of the gut bacterium, T. sanguinis. Based on our findings, we propose that particular bacteria indigenous to the gut microbiota, including Turicibacter, have co-evolved to induce host 5-HT1,3,5,8 and further sense host-derived 5-HT in order to promote their competitive colonization in the intestine. We hypothesize that additional members of the gut microbiota, including particular Clostridia, may respond to 5-HT directly through as-yet uncharacterized 5-HT response elements or indirectly through other bacteria, such as Turicibacter. Findings from our study support the notion that bidirectional host-microbial signaling via the serotonergic system shapes bacterial communities within the gastrointestinal tract. Notably, a recent human study of >2700 twins concluded that SSRI use is associated particularly with reduced Turicibacteraceae in humans, which is consistent with findings from our in silico, in vitro and in vivo animal experiments25. These findings, along with increasing associations of Turicibacter spp. with altered immune and metabolic conditions26–28, raise the question of whether Turicibacter impacts host health and disease. Supporting this, we find that monocolonization with T. sanguinis alters intestinal lipid metabolism, systemic triglyceride profiles and white adipose tissue physiology in mice, which could be relevant to reported links between SSRI use and symptoms of metabolic syndrome29,30. Future studies are needed to investigate the cellular mechanisms by which 5-HT and fluoxetine regulate T. sanguinis colonization in the gut and whether fluoxetine-induced changes in the gut microbiota mediate any effects of the SSRI on host intestinal function, neurophysiology and behavior.
Methods
Mice
C57Bl/6 and SERT−/− mice were purchased from Jackson Laboratories, reared as SPF or C-section rederived as GF and bred in flexible film isolators at the UCLA Center for Health Sciences barrier facility. Breeding animals were fed “breeder” chow (Lab Diets 5K52). Experimental animals were fed standard chow (Lab Diets 5010). Colonization status was monitored weekly by aerobic and anaerobic bacterial cfu plating and by 16S rDNA qPCR from fecal DNA extracted and amplified using the MoBio PowerSoil kit, SYBR green master mix (ThermoFisher) and QuantStudio 5 thermocycler (ThermoFisher). 6–12 week old mice were randomly assigned to experimental groups, which included age- and sex-matched cohorts of males and females. Sample sizes for animal experiments were determined based on prior experience with the experimental paradigm, existing literature utilizing the experimental paradigm, or by power calculation. Samples derived from animals were blinded to the experimenter for imaging- and sequencing-based analyses. All experiments were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals using protocols approved by the Institutional Animal Care and Use Committee at UCLA.
5-HT supplementation in mice for microbiota profiling
Water was supplemented with 5-HT (Sigma Aldrich) at 1.5 mg/ml based on calculations from 31 and provided ad libitum to mice for 2 weeks. Amount of water consumed and mouse weight were measured on days 3, 7, 10 and 14 of treatment. Mice were sacrificed one day after treatment for 5- HT assays and fecal 16S rDNA profiling was conducted as described below.
5-HT measurements
Blood samples were collected by cardiac puncture and spun through SST vacutainers (Becton Dickinson) for serum separation. The entire length of the colon or 1 cm regions of the distal, medial and proximal small intestine was washed in PBS to remove luminal contents, and sonicated on ice in 10 s intervals at 20 mV in ELISA standard buffer supplemented with ascorbic acid (Eagle Biosciences). Fecal pellets and intestinal contents were weighed and homogenized at 50 mg/ml in ELISA standard buffer supplemented with ascorbic acid. Serotonin levels were detected by ELISA according to the manufacturer’s instructions (Eagle Biosciences). Readings from tissue samples were normalized to total protein content as detected by BCA assay or the 660nm Protein Assay (Thermo Pierce).
16S rDNA sequencing
Bacterial genomic DNA was extracted from mouse fecal samples using the MoBio PowerSoil Kit. The library was generated according to methods adapted from 32. The V4 regions of the 16S rDNA gene were PCR amplified using individually barcoded universal primers and 30 ng of the extracted genomic DNA. The PCR reaction was set up in triplicate, and the PCR product was purified using the Qiaquick PCR purification kit (Qiagen). 250 ng of purified PCR product from each sample were pooled and sequenced by Laragen, Inc. using the Illumina MiSeq platform and 2 × 250bp reagent kit for paired-end sequencing. Operational taxonomic units (OTUs) were chosen by open reference OTU picking based on 97% sequence similarity to the Greengenes 13_5 database. Taxonomy assignment and rarefaction were performed using QIIME1.8.033. Metagenomes were inferred from closed reference OTU tables using PICRUSt34.
Sequence alignment and structural modeling
The Joint Genome Institute Integrated Microbial Genomes and Microbiomes and NCBI microbial genomes databases were searched for proteins with sequence similarity to human SERT. Phylogenetic trees were generated for the 15 bacterial candidates with highest alignment scores using MAFFT35 and Phylo.io36. Primer sets were generated against the SERT homolog ZP_06621923.1 from T. sanguinis PC909 and used to sequence the homologous gene encoding CUW_0748 in T. sanguinis MOL361. Protein structural models were created in Phyre237 and with MODELLER 9v1538, based on the highest-resolution available structure of human SERT, namely protein data bank, PDB39 entry 5i6x chain A, an outward-open conformation bound to paroxetine at 3.14 Å resolution. After initially aligning their sequences using AlignMe v1.1 in PST mode40, and inspection of the Ramachandran plot of the initial model, the alignment was refined at positions with strained backbone dihedral angles (see Supplementary Data Fig. 3a for final alignment), and the model was rebuilt. Based on analysis of the binding site conservation in the initial model, a Na+ ion was included at the Na2 site, using the position observed in a related hSERT template structure (PDB entry 5i71 chain A). 500 models were built using different seeds, and a single model was selected, specifically that with the highest ProQM41 score and fewest Ramachandran outliers, according to PROCHECK42. To remove local clashes and optimize geometry, this model was energy-minimized using with the CHARMM36 force field43 with NAMD v2.944, while constraining the backbone atoms and ion at Na2. Further quality assessment was carried out using MolProbity45, and ConSurf46 (Supplementary Data Fig. 3b, 3e). Structural models were visualized in Chimera47 and PyMol v1.7 [Schrödinger, LLC]. Residues involved in 5-HT, Cl− and Na+ binding in mammalian SERT were assessed based on existing literature13,14,48. The model is provided in the supplementary materials (Database S1).
Bacterial isolation and cultivation
Turicibacter sanguinis MOL361 (DSM-14220, DSMZ) was grown in Schaedler’s Broth (BD Biosciences) in an anaerobic chamber of 5% hydrogen, 10% carbon dioxide and balanced nitrogen. Bacteroides thetaiotaomicron (ATCC 29148) was grown in Brain Heart Infusion media (BD Biosciences) supplemented with 5 ug/ml hemin (Frontier Scientific) and 0.5 ug/ml vitamin K1 (Sigma Aldrich) under the same anaerobic conditions. Mouse- and human-derived spore-forming bacterial consortia were isolated by chloroform treatment and propagated in mice as described in3.
Heterologous expression in B. theta
CUW_0748 from T. sanguinis MOL361was PCR amplified and cloned into the expression vector pFD340 using one-step sequence- and ligation-independent cloning (SLIC) protocols described in 49. The vector was transformed into E. coli with selection using 100 ug/ml ampicillin and conjugated into B. theta using the E. coli helper plasmid RK231 with 50 ug/ml kanamycin selection. E. coli was killed using 200 ug/ml gentamicin and transconjugant strains were selected using 5 ug/ml erythromycin. Successful conjugation and gene expression were confirmed by PCR and qPCR for CUW_0748 in DNA and RNA extracted from B. theta clones.
Turicibacter colonization of germ-free mice
T. sanguinis MOL361 was anaerobically cultured as described above, washed, pelleted and re- suspended at 106 cfu/ml in pre-reduced PBS. GF mice were gavaged with 200 ul bacterial suspension and maintained under sterile conditions for at least 2 weeks.
APP+ uptake assay
T. sanguinis and B. theta were cultured anaerobically as described above and subcultured for 15– 17 hours to reach stationary phase. Bacterial APP+ transport activity was measured according to the manufacturer’s instructions using the Neurotransmitter Transporter Uptake Assay Kit (Molecular Devices). Briefly, cells were pre-treated with vehicle, 20 nM to 200 uM unlabeled 5- HT (Sigma Aldrich), fluoxetine (Tocris), dopamine (Sigma Aldrich) or norepinephrine (Sigma Aldrich) for 30 min at 37°C. Dosages were determined based on physiologically- and pharmacologically-relevant concentrations of ~170 uM 5-HT in mouse colon3 and ~130 uM fluoxetine scaled from human clinical treatments50, respectively. Then cells were treated with APP+ for 4 hours at 37°C. Intracellular fluorescence signal was measured at 520 nm using a multimodal plate reader (Biotek Synergy H1). Bacterial suspensions were mounted on slides and imaged using an epifluorescence microscope (EVOS). Images of B. theta were brightened by 90% to resolve fluorescence signal.
[3H]5-HT, [3H]NE and [3H]Trp uptake assays
T. sanguinis or B. theta was cultured anaerobically as described above and subcultured for 15–17 hours to reach stationary phase. Cells were washed and resuspended in Kreb-Ringer’s buffer, then left untreated or pre-treated with vehicle, unlabeled 5-HT (Sigma Aldrich), fluoxetine (Santa Cruz), reserpine (Sigma Aldrich) or tetrabenazine (Sigma Aldrich) for 30 min at 37°C. Cells were then incubated with 1 uM tritiated 5-HT (plus 0–500 uM unlabeled 5-HT for dose-dependent assays), NE or Trp (Perkin Elmer). For T. sanguinis, all uptake reactions were performed at room temperature. For B. theta, uptake reactions were performed at either room temperature or 37°C.
Uptake reactions were terminated by washing in ice cold Kreb-Ringer’s buffer. Washed T. sanguinis was trapped through 0.45 um PVDF filters on MultiScreenHTS plates connected to a vacuum manifold (EMD Millipore). For B. theta, washed cells were either trapped through 0.45 um PVDF filters or lysed by incubating in TE buffer plus triton and lysozyme. Extracted filters or lysed cells were incubated in 4 ml Filter Count scintillation fluid (Perkin Elmer) for 1 hour at room temp and radioactivity was measured using a liquid scintillation counter (Beckman LS6500).
Bacterial transcriptomic analysis
T. sanguinis was cultured anaerobically as described above and subcultured to mid-log-phase. Cells were incubated in culture media supplemented with vehicle, 200 uM 5-HT (Sigma Aldrich), fluoxetine (Tocris) or 5-HT with fluoxetine for 4 hours at 37°C. Bacteria were washed with PBS and lysed in TE buffer containing 1.2% Triton X100 and 15 mg/ml lysozyme. RNA was extracted using the RNAeasy Mini kit with on-column genomic DNA-digest (Qiagen). cDNA synthesis was performed using the qScript cDNA synthesis kit (Quantabio). RNA quality of RIN > 8.0 was confirmed using the 4200 Tapestation system (Agilent). rRNA were removed with the Ribo-Zero Bacterial kit (Illumina), RNA was prepared using the TruSeq Stranded RNA kit (Illumina) and 2 × 75 bp paired end reads were sequenced using the Illumina HiSeq platform by the UCLA Neuroscience Genomics Core. The bacterial RNAseq analysis package Rockhopper51 was used for quality filtering, mapping against the T. sanguinis PC909 genome and differential expression analysis. Over 23.79 million reads were obtained for each sample, with 86–88% aligning to protein- coding genes. Heatmaps containing differentially expressed genes with adjusted p < 0.05 and coefficient of variation (standard deviation / mean) < 3 were generated using the pheatmap package for R. GO term enrichment analysis of differentially expressed genes with adjusted p < 0.05 was conducted using DAVID52. Functional protein networks were generated with differentially expressed genes with adjusted p < 0.05 using STRING53.
Bacterial FM 4–64 staining and imaging
Overnight cultures of T. sanguinis MOL361 were pooled and treated with vehicle, 5-HT (200 uM), fluoxetine (200 uM) or 5-HT and fluoxetine (both 200 uM) for 24 hours at 37°C. Cells were washed with PBS, fixed in 4% PFA, and stained with FM4–64 (5 ug/ml) and DAPI (0.2 ug/ml) according to manufacturer’s instructions. Cells were wet-mounted on glass slides and cover slips. Images were acquired at 63X using the Zeiss LSM 780 confocal microscope. Images were analyzed using Fiji software54 by a researcher blinded to experimental group. FM 4–64 puncta were counted manually based on circularity and fluorescence intensity. Counts were normalized to cell area as determined by thresholding of DAPI staining.
Turicibacter colonization of antibiotic-treated mice
SPF mice were treated with ampicillin, gentamicin, neomycin and vancomycin (AGNV) in their drinking water at 0.5 g/L for 4 days. Fecal samples were collected and plated anaerobically to confirm bacterial clearance. After an additional day on sterile water, antibiotic-treated mice were orally gavaged once daily for 5 days with 106 cfu/mouse T. sanguinis MOL361 that was pre-treated with 200 uM 5-HT, fluoxetine or vehicle for 4 hours at 37°C. Mice were also maintained on regular water, 5-HT (24 μg/ml) or 5-HT (24 μg/ml) with fluoxetine (40 μg/ml) in the drinking water over the same 5 days of Turicibacter oral gavage. Fluoxetine drinking water concentration was determined so each mouse would receive an effective dose of 10 mg/kg/day. 5-HT drinking water concentration was determined by calculating the same molar concentration as Flx in drinking water. This dose of 5-HT elevated fecal levels by approximately 6-fold over vehicle. Three days after the final gavage, mice were euthanized by CO2 and the small and large intestines were collected for FISH as described below and lumenal contents from the small and large intestines were harvested for 16S rDNA sequencing. In the “drinking water only” experimental approach, mice were gavaged with untreated T. sanguinis once daily for 5 days and maintained on regular water, 5-HT or 5-HT with fluoxetine in the drinking water. Tissues were collected and analyzed 3 days after the final gavage. In the “bacterial pre-treatment only” experimental approach, mice were gavaged with one or three doses of vehicle, 5-HT, or 5-HT with fluoxetine-pretreated T. sanguinis once daily and sacrificed 12–24 hours after the final gavage.
Bacterial Fluorescence In Situ Hybridization (FISH)
Mouse intestines were fixed in Carnoys fixative overnight at 4°C, washed and transferred to 70% ethanol. Intestinal samples were then paraffin-embedded and cut into 5 μm longitudinal sections by IDEXX Bioresearch. Slides were deparaffinized with xylene, rehydrated in ethanol and incubated in 1 μM FISH probe (Sigma Aldrich) in hybridization buffer (0.9 M NaCl, 20 mM Tris HCl, pH 7.2, 0.1% SDS) for 4 hours at 50°C in a humidified chamber. Turicibacter probe TUR176 was used: 5’ [6FAM]-GCAYCTTTAAACTTTCGTCCTATCCG. Slides were then washed 3 times in pre-heated wash buffer (0.9 M NaCl, 20 mM Tris-HCl, pH 7.2) and mounted with ProLong Gold Antifade Mountant with DAPI (Invitrogen). Images were acquired on a Zeiss LSM 780 confocal microscope at 20X magnification. Image analyses were performed using ImageJ.
Fluoxetine treatment for 16S rDNA sequencing
SPF mice were orally gavaged daily for 7 days with 10 mg/kg fluoxetine (Santa Cruz) or treated with fluoxetine (40 μg/ml) in the drinking water for 14 days as indicated in figure legends. Fecal samples were harvested on day 0, 1, 4, 7 or 14 of treatment as indicated in figure legends and processed for 16S rDNA sequencing as described above.
Intestinal transcriptomic analysis
1 cm sections of the terminal ileum and distal colon were harvested from SPF, GF or Turicibacter monocolonized mice, washed with PBS to remove lumenal contents and homogenized using 5 mm stainless steel beads (Qiagen) for 1 min in a Mini-Beadbeater-16 (Biospec Products). RNA was extracted using the RNAeasy Mini kit with on-column genomic DNA-digest (Qiagen), and cDNA synthesis was performed using the qScript cDNA synthesis kit (Quantabio). RNA quality of RIN> 8.9 was confirmed using the 4200 Tapestation system (Agilent). RNA was prepared using the QuantSeq mRNA-Seq Library Prep kit (Lexogen) and 1 × 65 bp 3’ reads were sequenced using the Illumina HiSeq platform by the UCLA Neuroscience Genomics Core. The Bluebee analysis platform (Lexogen) was used for quality filtering and mapping. Differential expression analysis was conducted using DESeq255. At least 8.47 million aligned reads were obtained for each sample. Heatmaps containing differentially expressed genes with q < 0.05 were generated using the pheatmap package for R. GO term enrichment analysis of differentially expressed genes with q <0.05 was conducted using DAVID52.
Lipid clinical chemistry analysis
Serum samples were collected from SPF, GF, Turicibacter-monocolonized or Turicibacter- enriched mice and submitted to Charles River Clinical Pathology for analysis of lipid species including total cholesterol, free fatty acids, high-density lipoprotein, low-density lipoprotein and total triglycerides.
Serum lipidomic analysis
Lipids from 25 ul of plasma were extracted using a modified Bligh and Dyer extraction method by the UCLA lipidomics core facility56. Prior to biphasic extraction, a 13-lipid class Lipidyzer Internal Standard Mix is added to each sample (AB Sciex, 5040156). Following two successive extractions, pooled organic layers were dried down in a Genevac EZ-2 Elite evaporator. Lipid samples were resuspended in 1:1 methanol/dichloromethane with 10mM ammonium acetate and transferred to robovials (Thermo 10800107) for analysis. Samples were analyzed on the Sciex Lipidyzer Platform for targeted quantitative measurement of 1100 lipid species across 13 classes. Differential Mobility Device on Lipidyzer was tuned with SelexION tuning kit (Sciex 5040141). Instrument settings, tuning settings, and MRM list are available upon request.
Adipocyte histology and imaging
Epididymal, gonadal and inguinal white adipose tissues were collected from SPF, GF and Turicibacter-monocolonized mice. Tissues were weighed, fixed in 4% PFA for 48 hours, transferred to 70% ethanol, and submitted to IDEXX Pathology Services for paraffin sectioning and embedding. 5 um sections were stained with H&E and imaged at 20X magnification.
Adipocyte cell morphology and number were quantified using ImageJ.
Statistical Methods
All statistical data are included in Supplementary Table 11. Statistical analysis was performed using Prism software (GraphPad). Data were plotted in the figures as mean ± SEM. For each figure, n = the number of independent biological replicates. Differences between two treatment groups were assessed using two-tailed, unpaired Student t test with Welch’s correction. Differences among > 2 groups with only one variable were assessed using one-way ANOVA with Bonferroni post hoc test. Taxonomic comparisons from 16S rDNA sequencing analysis were analyzed by Kruskal-Wallis test with Bonferroni post hoc test. Two-way ANOVA with Bonferroni post-hoc test was used for ≥ 2 groups with two variables. Significant differences emerging from the above tests are indicated in the figures by *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 Notable near-significant differences (0.5 < p < 0.1) are indicated in the figures. Notable non-significant (and non-near significant) differences are indicated in the figures by “n.s.”.
Data Availability
Data generated or analyzed during this study are included in this published article and its supplementary information files. Structural modeling files that support the findings of this study are available from Zenodo with the identifier doi: 10.5281/zenodo.3266444. The 16S rDNA sequencing data that support the findings of this study are available from the Qiita database with study IDs 12585, 12596, and 12597 and are also available in the Supplementary Tables.
Bacterial transcriptomic data that support the findings of this study are available in Gene Expression Omnibus (GEO) repository with the accession number GSE133810 and are also available in the Supplementary Tables. Intestinal transcriptomic data that support the findings of this study are available in GEO repository with the accession number GSE133809 and are also available in the Supplementary Tables.
Supplementary Material
Acknowledgments
We thank members of the Hsiao lab and Gregory Donaldson for critical review of the manuscript; Ron Kaback (UCLA), Dongxue Yang and Eric Gouaux (Vollum Institute, OHSU), KC Huang (Stanford), Matthias Quick and Jonathan Javitch (Columbia University) for helpful advice; John Murowski, David Nusbaum and Jessica Yano (UCLA) for assistance with initial pilot experiments; Yanling Wang and Jeffrey F. Miller (UCLA) for providing Bacteroides strains and expression constructs; Kevin Williams (UCLA lipidomic core facility) for performing lipidomic measurements; Peter Bradley and Robert Gunsalus (UCLA) for facilitating radioisotope experiments; Gerard Karsenty (Columbia), Yin Tintut (UCLA), and Fredrik Bäckhed (University of Gothernburg) for providing Tph1 mice and mouse samples. Support for this research was provided by the NIH Director’s Early Independence Award (5DP5OD017924) to E.Y.H., Klingenstein-Simons Award to E.Y.H., Packard Fellowship in Science and Engineering to E.Y.H., UPLIFT: UCLA Postdocs’ Longitudinal Investment in Faculty (Award # K12 GM106996) to H.E.V., Ruth L. Kirschstein National Research Service (Award # AI007323) to G.N.P. and the Division of Intramural Research of the NIH, National Institute of Neurological Disorders and Stroke to L.R.F. All data and materials to understand and assess the conclusions of this research are available in the main text and supplementary materials.
Footnotes
Competing Interests: The authors declare no competing interests. Findings regarding the host effect of T. sanguinis reported in the manuscript are the subject of provisional patent application US 62/815,760, owned by UCLA.
References
- 1.Vuong HE, Yano JM, Fung TC & Hsiao EY The Microbiome and Host Behavior. Annu Rev Neurosci 40, 21–49, doi: 10.1146/annurev-neuro-072116-031347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gershon MD & Tack J The serotonin signaling system: from basic understanding to drug development for functional GI disorders. Gastroenterology 132, 397–414, doi: 10.1053/j.gastro.2006.11.002 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Reigstad CS et al. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J 29, 1395–1403, doi: 10.1096/fj.14-259598 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wikoff WR et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. PloS one 106, 3698–3703, doi: 10.1371/journal.pone.018074510.1073/pnas.0812874106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yano JM et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276, doi: 10.1016/j.cell.2015.02.047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sjogren K et al. The gut microbiota regulates bone mass in mice. J Bone Miner Res 27, 1357–1367, doi: 10.1002/jbmr.1588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hata T et al. Regulation of gut luminal serotonin by commensal microbiota in mice. PloS one 12, e0180745, doi: 10.1371/journal.pone.0180745 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujimiya M, Okumiya K & Kuwahara A Immunoelectron microscopic study of the luminal release of serotonin from rat enterochromaffin cells induced by high intraluminal pressure. Histochem Cell Biol 108, 105–113 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Mawe GM & Hoffman JM Serotonin signalling in the gut--functions, dysfunctions and therapeutic targets. Nat Rev Gastroenterol Hepatol 10, 473–486, doi: 10.1038/nrgastro.2013.105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen JJ et al. Maintenance of serotonin in the intestinal mucosa and ganglia of mice that lack the high-affinity serotonin transporter: Abnormal intestinal motility and the expression of cation transporters. The Journal of neuroscience : the official journal of the Society for Neuroscience 21, 6348–6361 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olivella M, Gonzalez A, Pardo L & Deupi X Relation between sequence and structure in membrane proteins. Bioinformatics 29, 1589–1592, doi: 10.1093/bioinformatics/btt249 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Coleman JA & Gouaux E Structural basis for recognition of diverse antidepressants by the human serotonin transporter. Nat Struct Mol Biol 25, 170–175, doi: 10.1038/s41594-018-0026-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forrest LR, Tavoulari S, Zhang YW, Rudnick G & Honig B Identification of a chloride ion binding site in Na+/Cl -dependent transporters. Proceedings of the National Academy of Sciences of the United States of America 104, 12761–12766, doi: 10.1073/pnas.0705600104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barker EL, Moore KR, Rakhshan F & Blakely RD Transmembrane domain I contributes to the permeation pathway for serotonin and ions in the serotonin transporter. J Neurosci 19, 4705–4717 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solis E Jr. et al. 4-(4-(dimethylamino)phenyl)-1-methylpyridinium (APP+) is a fluorescent substrate for the human serotonin transporter. J Biol Chem 287, 8852–8863, doi: 10.1074/jbc.M111.267757 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chanrion B et al. Physical interaction between the serotonin transporter and neuronal nitric oxide synthase underlies reciprocal modulation of their activity. Proceedings of the National Academy of Sciences of the United States of America 104, 8119–8124, doi: 10.1073/pnas.0610964104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seimandi M et al. Calcineurin interacts with the serotonin transporter C-terminus to modulate its plasma membrane expression and serotonin uptake. J Neurosci 33, 16189–16199, doi: 10.1523/JNEUROSCI.0076-13.2013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramijan K et al. Stress-induced formation of cell wall-deficient cells in filamentous actinomycetes. Nat Commun 9, 5164, doi: 10.1038/s41467-018-07560-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Browne HP et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546, doi: 10.1038/nature17645 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodrich JK, Davenport ER, Waters JL, Clark AG & Ley RE Cross-species comparisons of host genetic associations with the microbiome. Science 352, 532–535, doi: 10.1126/science.aad9379 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geva-Zatorsky N et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 168, 928–943 e911, doi: 10.1016/j.cell.2017.01.022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyazaki M, Dobrzyn A, Elias PM & Ntambi JM Stearoyl-CoA desaturase-2 gene expression is required for lipid synthesis during early skin and liver development. Proceedings of the National Academy of Sciences of the United States of America 102, 12501–12506, doi: 10.1073/pnas.0503132102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porter NT, Canales P, Peterson DA & Martens EC A Subset of Polysaccharide Capsules in the Human Symbiont Bacteroides thetaiotaomicron Promote Increased Competitive Fitness in the Mouse Gut. Cell host & microbe 22, 494–506 e498, doi: 10.1016/j.chom.2017.08.020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyte M, Villageliu DN, Crooker BA & Brown DR Symposium review: Microbial endocrinology-Why the integration of microbes, epithelial cells, and neurochemical signals in the digestive tract matters to ruminant health. J Dairy Sci 101, 5619–5628, doi: 10.3168/jds.2017-13589 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Jackson MA et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun 9, 2655, doi: 10.1038/s41467-018-05184-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodrich JK et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 19, 731–743, doi: 10.1016/j.chom.2016.04.017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein CN & Forbes JD Gut Microbiome in Inflammatory Bowel Disease and Other Chronic Immune-Mediated Inflammatory Diseases. Inflamm Intest Dis 2, 116–123, doi: 10.1159/000481401 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X et al. High Fat Diet Alters Gut Microbiota and the Expression of Paneth Cell- Antimicrobial Peptides Preceding Changes of Circulating Inflammatory Cytokines. Mediators Inflamm 2017, 9474896, doi: 10.1155/2017/9474896 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SH, Paz-Filho G, Mastronardi C, Licinio J & Wong ML Is increased antidepressant exposure a contributory factor to the obesity pandemic? Transl Psychiatry 6, e759, doi: 10.1038/tp.2016.25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beyazyuz M, Albayrak Y, Egilmez OB, Albayrak N & Beyazyuz E Relationship between SSRIs and Metabolic Syndrome Abnormalities in Patients with Generalized Anxiety Disorder: A Prospective Study. Psychiatry Investig 10, 148–154, doi: 10.4306/pi.2013.10.2.148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 31.Abdala-Valencia H et al. Inhibition of allergic inflammation by supplementation with 5- hydroxytryptophan. Am J Physiol Lung Cell Mol Physiol 303, L642–660, doi: 10.1152/ajplung.00406.2011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America 108 Suppl 1, 4516–4522, doi: 10.1073/pnas.1000080107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods 7, 335–336, doi: 10.1038/nmeth.f.303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langille MG et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31, 814–821, doi: 10.1038/nbt.2676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh K, Rozewicki J & Yamada KD MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform, doi: 10.1093/bib/bbx108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson O, Dylus D & Dessimoz C Phylo.io: Interactive Viewing and Comparison of Large Phylogenetic Trees on the Web. Mol Biol Evol 33, 2163–2166, doi: 10.1093/molbev/msw080 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelley LA, Mezulis S, Yates CM, Wass MN & Sternberg MJ The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10, 845–858, doi: 10.1038/nprot.2015.053 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webb B & Sali A Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Protein Sci 86, 2 9 1–2 9 37, doi: 10.1002/cpps.20 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Berman HM et al. The Protein Data Bank. Nucleic Acids Res 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamm M, Staritzbichler R, Khafizov K & Forrest LR AlignMe--a membrane protein sequence alignment web server. Nucleic Acids Res 42, W246–251, doi: 10.1093/nar/gku291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallner B ProQM-resample: improved model quality assessment for membrane proteins by limited conformational sampling. Bioinformatics 30, 2221–2223, doi: 10.1093/bioinformatics/btu187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R & Thornton JM AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8, 477–486 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Best RB et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J Chem Theory Comput 8, 3257–3273, doi: 10.1021/ct300400x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips JC et al. Scalable molecular dynamics with NAMD. J Comput Chem 26, 1781–1802, doi: 10.1002/jcc.20289 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams CJ et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci 27, 293–315, doi: 10.1002/pro.3330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashkenazy H et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44, W344–350, doi: 10.1093/nar/gkw408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612, doi: 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Coleman JA, Green EM & Gouaux E X-ray structures and mechanism of the human serotonin transporter. Nature 532, 334–339, doi: 10.1038/nature17629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Forrest LR, Tavoulari S, Zhang YW, Rudnick G & Honig B Identification of a chloride ion binding site in Na+/Cl -dependent transporters. Proceedings of the National Academy of Sciences of the United States of America 104, 12761–12766, doi: 10.1073/pnas.0705600104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barker EL, Moore KR, Rakhshan F & Blakely RD Transmembrane domain I contributes to the permeation pathway for serotonin and ions in the serotonin transporter. J Neurosci 19, 4705–4717 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yano JM et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276, doi: 10.1016/j.cell.2015.02.047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeong JY et al. One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl Environ Microbiol 78, 5440–5443, doi: 10.1128/AEM.00844-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hiemke C & Hartter S Pharmacokinetics of selective serotonin reuptake inhibitors. Pharmacol Ther 85, 11–28 (2000). [DOI] [PubMed] [Google Scholar]
- 54.McClure R et al. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41, e140, doi: 10.1093/nar/gkt444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57, doi: 10.1038/nprot.2008.211 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Szklarczyk D et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45, D362–D368, doi: 10.1093/nar/gkw937 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682, doi: 10.1038/nmeth.2019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bligh EG & Dyer WJ A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37, 911–917, doi: 10.1139/o59-099 (1959). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated or analyzed during this study are included in this published article and its supplementary information files. Structural modeling files that support the findings of this study are available from Zenodo with the identifier doi: 10.5281/zenodo.3266444. The 16S rDNA sequencing data that support the findings of this study are available from the Qiita database with study IDs 12585, 12596, and 12597 and are also available in the Supplementary Tables.
Bacterial transcriptomic data that support the findings of this study are available in Gene Expression Omnibus (GEO) repository with the accession number GSE133810 and are also available in the Supplementary Tables. Intestinal transcriptomic data that support the findings of this study are available in GEO repository with the accession number GSE133809 and are also available in the Supplementary Tables.