Abstract
Agonists for PPARα are used clinically to reduce triglycerides and improve high-density lipoprotein (HDL) cholesterol levels in patients with hyperlipidemia. Whether the mechanism of PPARα activation to lower serum lipids occurs in the liver or other tissues is unknown. To determine the function of hepatic PPARα on lipid profiles in diet-induced obese mice, we placed hepatocyte-specific peroxisome proliferator-activated receptor-α (PPARα) knockout (PparaHepKO) and wild-type (Pparafl/fl) mice on high-fat diet (HFD) or normal fat diet (NFD) for 12 wk. There was no significant difference in weight gain, percent body fat mass, or percent body lean mass between the groups of mice in response to HFD or NFD. Interestingly, the PparaHepKO mice on HFD had worsened hepatic inflammation and a significant shift in the proinflammatory M1 macrophage population. These changes were associated with higher hepatic fat mass and decreased hepatic lean mass in the PparαHepKO on HFD but not in NFD as measured by Oil Red O and noninvasive EchoMRI analysis (31.1 ± 2.8 vs. 20.2 ± 1.5, 66.6 ± 2.5 vs. 76.4 ± 1.5%, P < 0.05). We did find that this was related to significantly reduced peroxisomal gene function and lower plasma β-hydroxybutyrate in the PparaHepKO on HFD, indicative of reduced metabolism of fats in the liver. Together, these provoked higher plasma triglyceride and apolipoprotein B100 levels in the PparaHepKO mice compared with Pparafl/fl on HFD. These data indicate that hepatic PPARα functions to control inflammation and liver triglyceride accumulation that prevent hyperlipidemia.
Keywords: apolipoprotein, cholesterol, obesity, nonalcoholic fatty liver disease, peroxisomes
INTRODUCTION
Because of the epidemic of obesity, hyperlipidemia has significantly increased over the past several decades and has led to the expansion of other related disorders, such as nonalcoholic fatty liver disease (NAFLD) and insulin-resistant type II diabetes (4, 69). NAFLD has been shown to worsen with hepatic inflammation (4, 56, 69), which also causes hyperlipidemia and glucose intolerance (4, 56). Hyperlipidemia is characterized by abnormally high levels of lipids (triglycerides and cholesterol) in the blood and can lead to complications like cardiovascular disease and stroke, reducing life expectancy (44). Current research for new therapeutic options is focused on targeting fat burning and inflammatory pathways (4, 56). Drugs such as statins and fibrates are commonly prescribed clinically for the treatment of hyperlipidemia. Fibrates activate the nuclear receptor peroxisome proliferator-activated receptor-α (PPARα), which is a transcription factor that regulates numerous genes involved in fatty acid metabolism. Fibrates are effective at reducing plasma triglyceride levels (61), possibly by lowering plasma apolipoprotein B100 (ApoB100) (35, 57). ApoB100 is essential for very-low-density lipoprotein (VLDL) excretion from the liver (8) that carries triglycerides and cholesterol out of the liver to the blood. In weight gain and obesity, plasma ApoB100 and triglyceride levels are increased (14), which indicates hypersecretion of VLDL and possibly the development of hyperlipidemia (8, 66). Reducing either NAFLD or fat mass lowers hyperlipidemia, but whether PPARα mediates these effects via the liver, adipose, muscle, or unknown tissue is yet to be determined.
PPARα is highly expressed in the liver where it plays an essential function in lipid metabolism, especially in response to fasting (1, 6, 21, 24, 25, 29, 62). However, it is also present in other tissues, such as brown adipose tissue (BAT) and white adipose tissue (WAT), where it mediates mitochondrial function and the browning of WAT for the burning of fat (50–52, 67). However, others have shown, using global PPARα knockout (PPARα ΚΟ) mice that PPARα is dispensable for cold-induced browning and BAT function, which was reported to occur by another isoform, PPARγ (11, 32). While the critical role of PPARα in the metabolic response in the liver, WAT, and BAT has been well studied, tissue-specific knockout of PPARα has not been well established.
Whole body PPARα knockout mice develop obesity in a sexually dimorphic fashion while additional studies on whole body knockout mice on Sv/129 or C57BL/6N genetic backgrounds observed no effect of global PPARα deficiency on the development of obesity (2, 9). Further studies by Kim et al. (30) demonstrated that mice deficient for PPARα on a mixed Sv/129/C57BL/6N background gained more weight in response to high-fat feeding as compared with wild-type control mice. The role of PPARα in the development of insulin resistance has been contentious. One study in global PPARα KO mice reported they are resistant to the development of high-fat-induced insulin resistance and hyperglycemia, while another study showed rodents with a PPARα deletion developed insulin resistance (18, 19). One possible explanation for the discrepancy in the observed phenotypes between these studies is the differences in methods used to determine insulin sensitivity with one study measuring in the fasted state and the other in the fed state (18, 19). Regardless, studies in tissue-specific PPARα KO mice are needed to reveal which tissue the nuclear receptor is responsible for improving hyperlipidemia. The effects may be tissue specific, or factors such as its role in insulin sensitivity may be compounding or from another set of tissues.
To elucidate the role of PPARα in tissues, floxed PPARα mice have been recently generated (43). We utilized hepatocyte-specific PPARα KO (PparaHepKO) mice to determine the capacity of hepatic PPARα in mitigating hyperlipidemia from diet-induced obesity. We examined the effect of hepatocyte PPARα on the plasma lipid and metabolite profile in response to high-fat (HFD) and normal fat (NFD) diets. We found that the PparaHepKO mice had significantly more inflammation, plasma triglycerides, and ApoB100 on HFD, which was associated with exacerbated hepatic steatosis in these mice. Our results demonstrate a principle role for PPARα in the protection against hepatic inflammation that leads to hyperlipidemia and fatty liver disease.
METHODS
Animals.
The experimental procedures and protocols of this study conform to the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of the University of Mississippi Medical Center. All mice had free access to food and water ad libitum. Animals were housed in a temperature-controlled environment with 12/12-h dark/light cycle. PparaHepKO and Pparafl/fl mice were obtained from Dr. Walter Wahli, Lee Kong Chian School of Medicine, Nanyang Technological University the Academia, Singapore as originally described (43). Albumin-Cre mice were used for PparaHepKO and obtained from Dr. Wahli. The mice were housed under standard temperatures between 24 and 25°C. The control NFD consisted of 17% fat diet (Teklad 22/5 rodent diet, cat. no. 860; Harland Laboratories, Indianapolis, IN). Studies were performed on 8-wk-old male mice initially housed under standard conditions with full access to standard mouse chow described above and water. After this time, mice were switched with full access to a 60% HFD (cat. no. D12492; Research Diets, New Brunswick, NJ) for 12 wk and allowed access to water as previously described (1, 24, 40, 62). Mice were fasted 8 h before euthanasia via isoflurane anesthesia and blood and tissues were extracted for further analysis.
Body and liver composition.
Body composition changes were assessed at 4-wk intervals throughout the study using magnetic resonance imaging (EchoMRI-900TM; Echo Medical System, Houston, TX). MRI measurements were performed in conscious mice placed in a thin-walled plastic cylinder with a cylindrical plastic insert added to limit movement of the mice. Mice were briefly submitted to a low-intensity electromagnetic field where fat mass, lean mass, free water, and total water were measured. Liver composition was measured in excised tissues in the same EchoMRI system at the end of 12 wk of HFD, immediately after euthanasia.
Liver triglyceride measurement.
Triglycerides were measured from 100 mg of liver tissue homogenized in 1 ml of 5% NP-40 in water. Homogenized tissues were then heated to 95°C for 5 min and then centrifuged (13,000 g) for 2 min. Tissue triglyceride levels were measured using a fluorometric assay kit according to the manufacturer’s guidelines (PicoProbe Trigylceride Fluorometric Assay Kit; BioVision, Milpitas, CA). Samples from individual mice were run in duplicate and averaged. The averages of individual mice were then used to obtain group averages.
Liver staining.
To determine the effects of treatment on hepatic lipid accumulation, livers were mounted and frozen in Tissue-Tek OCT and sectioned at 10 µm. Frozen sections were air dried and fixed in 10% neutral buffered formalin. Sections were briefly rinsed in tap water, followed by 60% isopropanol, and stained for 15 min in Oil Red O solution. Sections were further rinsed in 60% isopropanol and nuclei stained with hematoxylin followed by aqueous mounting and coverslipping. The degree of Oil Red O staining was determined at ×40 magnification using a color Axiocam 105 camera with Zen 2 software attached to a Zeiss microscope. Images were analyzed using ImageJ software, and to ensure the accuracy of measurement, six images of each animal were analyzed and averaged into a single measure. Measurements were obtained from three individual animals per group. Data are presented as the average mean ± SE of the percent Oil Red O staining for each group. Hemotoxylin-eosin (H&E) and Periodic Acid–Schiff staining were performed as previously described (23–25).
Fasting glucose and insulin.
After an 8-h fast, a blood sample was obtained via the orbital sinus under isoflurane anesthesia. Blood glucose was measured using an Accu-Chek Advantage glucometer (Roche, Mannheim, Germany). Fasting plasma insulin concentrations were determined by a LINCO ELISA kit).
Analysis of plasma lipids and metabolites.
After an 8-h fast, a blood sample was obtained via the orbital sinus under isoflurane anesthesia for plasma lipids and metabolites. Nuclear magnetic resonance (NMR) spectroscopy experiments were acquired using a 14.0-T Bruker magnet equipped with a Bruker AV-III console operating at 600.13 MHz. All spectra were acquired in 3-mm NMR tubes using a Bruker 5-mm QCI cryogenically cooled NMR probe. Plasma samples were prepared and analyzed according to the Bruker In Vitro Diagnostics research (IVDr) protocol. Sample preparation consisted of combining 50 μl plasma with 150 μl buffer supplied by Bruker Biospin specifically for the IVDr protocol. For 1D 1H NMR, data were acquired using the 1D-NOE experiment that filters NMR signals associated with broad line widths arising from proteins that might be present in plasma samples that adversely affect spectral quality. Experiment conditions included: sample temperature of 310 K, 96 k data points, 20 ppm sweep width, a recycle delay of 4 s, a mixing time of 150 ms, and 32 scans. Lipoprotein subclass analysis was performed using regression analysis of the NMR data, which is done automatically as part of the IVDr platform as previously described (46).
Quantitative real-time PCR analysis.
Total RNA was harvested from Pparafl/fl and PparaHepKO mice by lysing livers using a Tissue Lyser LT (Qiagen) and then extraction by 5-Prime PerfectPure RNA Tissue Kit (Fisher Scientific). Total RNA was read on a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) and cDNA was synthesized using high capacity cDNA reverse transcription kit (Applied Biosystems). PCR amplification of the cDNA was performed by quantitative real-time PCR using TrueAmp SYBR Green qPCR SuperMix (Alkali Scientific) for gene-specific primers as previously described (23–26, 40, 62). The thermocycling protocol consisted of 5 min at 95°C, 40 cycles of 15 s at 95°C, and 30 sFF at 60°C, finished with a melting curve ranging from 60 to 95°C to allow distinction of specific products. Normalization was performed in separate reactions with primers to GAPDH.
Gel electrophoresis and Western blot analysis.
Mouse tissues were flash frozen in liquid nitrogen during harvesting and stored at −80°C. For gel electrophoresis, 50–100 mg of cut tissue was then resuspended in three volumes of CelLytic Buffer (Sigma C3228) plus 10% protease inhibitor cocktail (Sigma P2714–1BTL) and Halt phosphatase inhibitor cocktail (Fisher PI78420), and then incubated on ice for 30 min. The livers were lysed using a Qiagen Tissue Lyser LT and then centrifuged at 100,000 g at 4°C. Protein samples were resolved by SDS PAGE and electrophoretically transferred to Immobilon-FL membranes. Membranes were blocked at room temperature for 2 h in TBS (10 mM Tris·HCl (pH 7.4) and 150 mM NaCl) containing 3% BSA. Subsequently, the membranes were incubated overnight at 4°C with the following antibodies: PPARα (cat. no.sc-9000; Santa Cruz Biotechnology, Santa Cruz, CA) or heat shock protein 90 (HSP90) (cat. no. 13119; Santa Cruz). After three washes in TBS + 0.1% Tween 20, the membrane was incubated with an infrared anti-rabbit (IRDye 800, green) or anti-mouse (IRDye 680, red) secondary antibody labeled with IRDye infrared dye (LI-COR Biosciences) (1:10,000 dilution in TBS) for 2 h at 4°C. Immunoreactivity was visualized and quantified by infrared scanning in the Odyssey system (LI-COR Biosciences).
Statistical analysis.
Data were analyzed with Prism 7 (GraphPad Software, San Diego, CA) using ANOVA combined with Tukey’s posttest to compare pairs of group means or unpaired Student’s t tests. Results are expressed as means ± SE. Additionally, one-way ANOVA with a least significant difference post hoc test was used to compare mean values between multiple groups, and a Student’s two-tailed, and a two-way ANOVA was utilized in multiple comparisons, followed by the Bonferroni post hoc analysis to identify interactions. P values of 0.05 or smaller were considered statistically significant.
RESULTS
Body weights, fat, and lean mass of mice with a PPARα hepatocyte-specific knockout are comparable to floxed control mice on HFD or NFD.
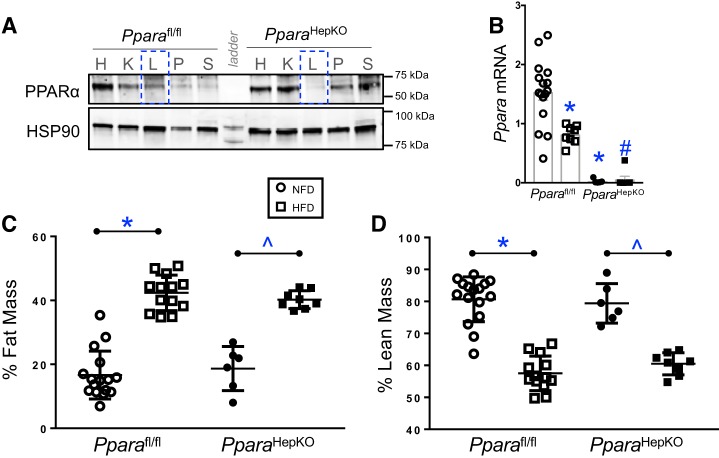
The global PPARα null mice compared with their littermate controls have significant differences in body weight and in their metabolic phenotypes (43). Previously, Montagner et al. (43) showed that mice with a hepatocyte-specific PPARα KO had no change in body weight on a standard chow diet, but significantly lower body weight in response to a methionine-deficient and choline-deficient diet (MCD). Here, we used Pparafl/fl and PparaHepKO mice, confirmed by Western blot analysis in Fig. 1A, which showed the specific loss of PPARα in the liver (dotted blue line) but not in other organs in PparaHepKO mice. To determine how a diet high in fat may impact hyperlipidemia in mice with PPARα not expressed in hepatocytes, we placed the Pparafl/fl and PparaHepKO on HFD or NFD for 12 wk. We found that Pparafl/fl on HFD had significantly (P < 0.05) lower PPARα mRNA in the liver compared with NFD (Fig. 1B). The PparaHepKO had almost no detectable expression on NFD or HFD. The Pparafl/fl and PparaHepKO mice exhibited no difference with NFD between the groups and similar increases in body weight on HFD (Table 1). Also, body weight, body length, and heart and fat pad masses had no differences between the groups on NFD or HFD, but liver weights were significantly (P = 0.0353) higher for the PparaHepKO on HFD. There was no difference in percent body fat mass or lean mass on HFD or NFD (Fig. 1, C and D). These data show that body weights of mice with a hepatocyte-specific PPARα KO on NFD or HFD are not different and that the loss of PPARα in the liver does not significantly change body weight during diet-induced obesity. However, PPARα in the liver does regulate liver mass, which impacts hyperlipidemia and hepatic insulin resistance that typically occur in obesity.
Fig. 1.
Percent body fat and lean mass in peroxisome proliferator-activated receptor-α (PPARα) wild-type (Pparafl/fl) and PPARα hepatocyte-specific knockout (PparaHepKO) mice fed a high-fat (HFD) or normal fat diet (NFD). A: Western blot analysis for PPARα and heat shock protein 90 (HSP90) as loading control in various tissues from Pparafl/fl and PparaHepKO mice. H, heart, K, kidney, L, liver, P, pancreas, S, spleen. Dotted blue line, specific loss of PPARα in the liver. B: Ppara mRNA levels in NFD and HFD in Pparafl/fl and PparaHepKO mice. C: fat mass was directly measured at the end of the experimental protocol by noninvasive EchoMRI. D: lean mass was determined by noninvasive EchoMRI. Values are means ± SE; *P < 0.05 vs. NFD; #P < 0.05 vs. HFD Pparafl/fl; ^P < 0.05 vs. NFD PparaHepKO; n = 16 and13, Pparafl/fl NFD, HFD; n = 6 and 8, PparaHepKO NFD, HFD. n, number of mice. All circles, NFD; all squares, HFD.
Table 1.
Body and organ weights in Pparafl/fl and PparaHepKO mice
| Parameter | Pparafl/fl NFD | Pparafl/fl HFD | PparaHepKO NFD | PparaHepKO HFD | Diet P value | Genotype P value | Interaction P value |
|---|---|---|---|---|---|---|---|
| n | 16 | 13 | 6 | 8 | |||
| BW, g | 34.9 ± 1.4 | 55.2 ± 1.5 | 37.1 ± 1.5 | 51.1 ± 1.1 | 0.0115* | 0.3629 | 0.6625 |
| BL, cm | 9.9 ± 0.09 | 10.5 ± 0.08 | 9.9 ± 0.13 | 10.5 ± 0.04 | <0.0001* | 0.3389 | 0.6402 |
| HW, mg | 159.3 ± 5.2 | 172.2 ± 10.8 | 176.2 ± 9.5 | 171.6 ± 4.3 | 0.4659 | 0.1258 | 0.6675 |
| HW:BW, mg/g | 4.6 ± 0.19 | 3.1 ± 0.15 | 4.7 ± 0.37 | 3.3 ± 0.08 | <0.0001* | 0.3215 | 0.8271 |
| HW:BL, mg/cm | 16.1 ± 0.55 | 16.5 ± 1 | 17.7 ± 1.1 | 16.2 ± 0.4 | 0.5029 | 0.4054 | 0.2561 |
| LW, mg | 1.3 ± 0.4 | 2.5 ± 0.25 | 1.3 ± 0.12 | 3 ± 0.4 | <0.0001* | 0.2465 | 0.2833 |
| LW/BW, mg/g | 0.038 ± 0.001 | 0.044 ± 0.003 | 0.035 ± 0.002 | 0.058 ± 0.002 | 0.0005* | 0.1249 | 0.0353* |
| LW/BL, mg/cm | 0.13 ± 0.004 | 0.23 ± 0.04 | 0.13 ± 0.01 | 0.28 ± 0.03 | <0.0001* | 0.1045 | 0.1139 |
| Epidydmal FW, g | 0.91 ± 0.13 | 1.8 ± 0.13 | 1.2 ± 0.2 | 1.57 ± 0.13 | 0.0167* | 0.4358 | 0.6399 |
| Visceral FW g | 0.75 ± 0.1 | 2.9 ± 0.08 | 0.87 ± 0.16 | 2.8 ± 0.14 | <0.0001* | 0.3189 | 0.5906 |
| Total fat, g | 1.66 ± 0.24 | 4.77 ± 0.17 | 2.03 ± 0.37 | 4.37 ± 0.21 | <0.0001* | 0.3345 | 0.9053 |
Values are means ± SE: n = no. of mice. Pparafl/fl, peroxisome proliferator-activated receptor-α (PPARα) wild-type mice; PparaHepKO, knockout mice; NFD, normal fat diet; HFD, high-fat diet; BW, body weight; BL, body length; HW, heart weight; LW, liver weight; FW, fat weight.
P < 0.05.
PparaHepKO mice have HFD-induced increased plasma insulin and reduced hepatic glycogen levels.
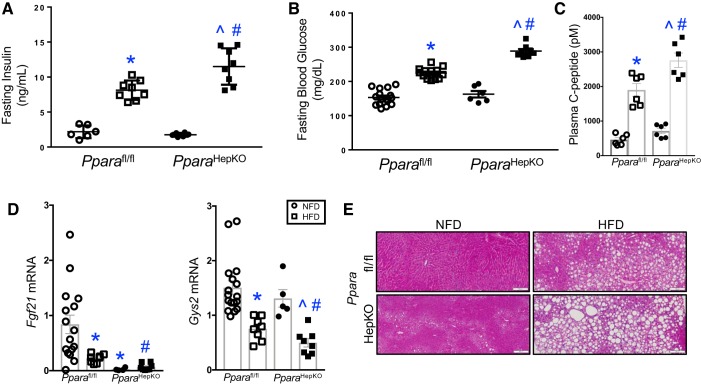
PPARα global null mice have been shown to have altered glucose and insulin plasma levels (43, 62). The role of hepatic PPARα in HFD-induced insulin and glucose intolerance has yet to be elucidated. Both groups of mice on NFD had normal fasting insulin and glucose levels (Fig. 2, A and B). However, HFD significantly increased fasting plasma insulin (Fig. 2A) and glucose (Fig. 2B) levels in both groups as compared with NFD. The insulin and glucose were significantly (P < 0.05) higher in PparaHepKO as compared with Pparafl/fl mice in HFD groups. The proinsulin C-peptide serum levels indicate insulin turnover and secretion, where increased levels suggest less insulin clearance by the liver (10). The C-peptide plasma levels were not changed in NFD between PparaHepKO and Pparafl/fl mice but were significantly (P < 0.05) higher with HFD between the groups). Diet-induced obesity causes less insulin clearance and higher circulating C-peptide levels (20, 55), and the PparaHepKO mice (Fig. 2C e had significantly higher C-peptide plasma levels compared with Pparafl/fl. These data imply that hepatic PPARα regulates insulin clearance, which with reduced levels increases circulating insulin, leading to peripheral insulin resistance.
Fig. 2.
Altered glucose and insulin levels in peroxisome proliferator-activated receptor-α (PPARα) hepatocyte-specific knockout (PparaHepKO) mice fed a high-fat diet (HFD). A: fasting blood insulin levels. B: fasting blood glucose levels. C: serum C-peptide levels. D: hepatic Fgf21 and Gys2 mRNA levels in normal fat diet (NFD) and HFD in PPAR wild-type (Pparafl/fl) and PparaHepKO mice. E: representative Periodic-Acid-Schiff staining in livers from Pparafl/fl and PparaHepKO mice. Scale bar = 100 nm. Values are means + SE. *P < 0.05 vs. NFD Pparafl/fl, #P < 0.05 vs. HFD Pparafl/fl, ^P < 0.05 vs. NFD PparaHepKO; A: n = 6 and 8 mice, Pparfl/fl NFD, HFD; n = 6 and 8 mice, PparaHepKO NFD, HFD; B: n = 16 mice/group NFD, n = 13 mice/group HFD Pparafl/fl, n =6 mice/group NFD, n = 8 mice/group HFD, PparaHEPKO; C: n = 6 mice/group; D: n = 16 mice/group NFD, 13 mice/group HFD, Pparafl/fl, n = 6 mice/group NFD, n = 8 mice/group HDF, PparaHEPKO. All circles, NFD; all squares, HFD.
Two PPARα target genes in the liver that are known to mediate glucose levels in mice are Fgf21 (27, 36) and Gys2 (38). The Fgf21 produces the FGF21 hormone that is known to regulate lipid accumulation and glucose intolerance (5, 7). The Gys2 gene produces glycogen synthase 2 that regulates hepatic glycogen storage for glucose storage (23, 25). Both genes were significantly (P < 0.05) lower in HFD Pparafl/fl mice compared with NFD (Fig. 2D). In the PparaHepKO mice, Fgf21 levels were abolished in NFD and HFD compared with Pparafl/fl. The Gys2 mRNA levels were significantly (P < 0.05) lower in the PparaHepKO mice on HFD compared with Pparafl/fl. In comparison with the lower glycogen synthase 2 mRNA, Periodic acid-Schiff (PAS) staining, an indicator of glycogen levels (25), was also reduced in the livers of HFD for PparaHepKO and Pparafl/fl mice, with PparaHepKO mice lower compared with Pparafl/fl (Fig. 2E). Reduced PPARα expression in the obese lessens β-oxidation and fat burning leading to hepatic steatosis that corresponds to reduced glycogen levels. These data show that hepatic PPARα regulates insulin resistance and glycogen storage, which may alter circulating insulin levels leading to peripheral insulin resistance.
PparaHepKO mice have higher fat accumulation and reduced fat burning in the liver that is worsened on HFD.
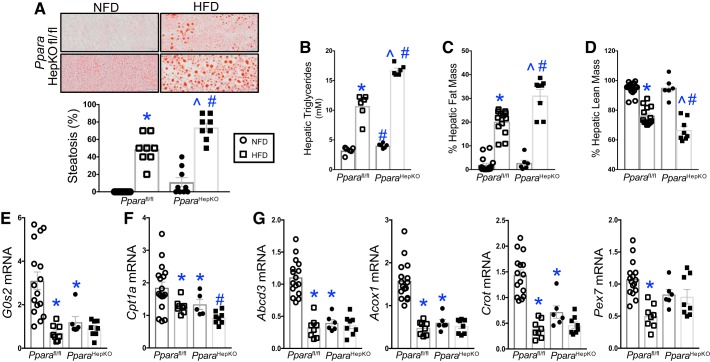
The higher liver weights in the PparaHepKO mice indicate that the loss of hepatic PPARα impairs liver function. PPARα has a known role in increasing fatty acid β-oxidation in the liver, WAT, and BAT. Obese mice have lower liver PPARα expression and hepatic steatosis and hyperlipidemia (25, 40). Oil Red O staining showed higher fat accumulation in the PparaHepKO as compared with Pparafl/fl mice on NFD (Fig. 3A), and these mice have normal body weights and are not obese. The presence of lipid droplets in the PparaHepKO indicates the beginning stages of NAFLD, commonly referred to as steatosis. Diet-induced obesity by HFD in the Pparafl/fl and PparaHepKO mice showed significantly more steatosis in the liver in both groups compared with NFD. However, the PparaHepKO had more micro- and macrosteatosis on HFD and NFD compared with Pparafl/fl. These findings were confirmed biochemically by measurement of hepatic triglycerides that were significantly higher in the PparaHepKO in NFD and HFD as compared with Pparafl/fl mice (Fig. 3B). Hepatic fat mass was also measured by EchoMRI and was significantly (P < 0.05) increased in both groups in response to HFD (Fig. 3C), and similar findings showed that hepatic fat mass was higher in PparaHepKO compared with Pparafl/fl mice. Hepatic lean mass was decreased in both groups in response to HFD but was further suppressed in the PparaHepKO in comparison with Pparafl/fl mice (Fig. 3D). PPARα transcriptionally regulates several genes involved in metabolic processes, such as the G0/G1 switch gene 2 (G0s2) gene, which has been shown to mediate metabolism and the cell cycle (73). Previously, G0s2 was shown to be significantly reduced in global PPARα KO animals (73). Here, we found similar results that G0s2 was substantially lower in the PparaHepKO compared with Pparafl/fl mice (Fig. 3E). These indicate that hepatic fat accumulation may be higher in the PparaHepKO by reduced hepatic metabolic gene actions.
Fig. 3.
Peroxisome proliferator-activated receptor-α (PPARα) hepatocyte-specific knockout (PparaHepKO) mice have enhanced high-fat diet (HFD)-induced hepatic steatosis. A: Oil red O staining in livers from PPARα wild-type (Pparafl/fl) and PparaHepKO mice on normal fat diet (NFD) and HFD. B: biochemical measurement of hepatic triglyceride levels. C: hepatic fat mass as measured by noninvasive EchoMRI at the end of the study. D: hepatic lean mass as measured by noninvasive EchoMRI at the end of the study. Measurement of hepatic metabolic gene expression in NFD and HFD in Pparafl/fl and PparaHepKO mice for G0s2 (E), Ctp1a (F), and peroxisomal gene expression for Abcd3, Acox1, Crot, and Pex7 (G). Values are means ± SE; *P < 0.05 vs. NFD Pparafl/fl; #P < 0.05 vs. HFD Pparafl/fl; ^P < 0.05 vs. NFD PparaHepKO. A: n = 9 and 8 Pparafl/fl and PparaHEPKO NFD and HFD mice. B: n = 6 mice. C and D: n = 16 and 13 Pparafl/fl NFD and HFD mice and n = 6 and 8 PparaHepko NFD and HFD mice. E–G, n = 16 and 8 Pparafl/fl NFD and HFD mice and n = 6 and 8 PparaHepKO NFD and HFD mice. All circles, NFD; all squares HFD.
PPARα mediation of fat-burning β-oxidation is targeted by several genes in the pathway. One major contributor is the carnitine palmitoyltransferase 1a (Cpt1a), which brings fatty acids into the mitochondria for the burning of fat. The Pparafl/fl mice had significantly (P < 0.05) lower Cpt1a mRNA expression on HFD compared with NFD (Fig. 3F). The PparaHepKO mice had significantly (P < 0.01) reduced Cpt1a mRNA expression on HFD compared with Pparafl/fl. Another contributor is peroxisomes that are critical in aiding the mitochondria for the oxidation of very-long-chain-fatty acids and preventing mitochondrial and cellular DNA damage by reactive oxygen species (ROS) generated by β-oxidation of fatty acids (16). Peroxisomal gene expressions for Abcd3, Acox1, Crot, and Pex7 were significantly (P < 0.05) reduced in HFD Pparafl/fl mice compared with NFD (Fig. 3G). PparaHepKO mice showed a significant (P < 0.05) reduction in peroxisomal gene expression in NFD and HFD compared with Pparafl/fl mice. These genes represent peroxisomal biogenesis, fatty acid import, fatty acid degradation, and fatty acid export. These data indicate that mitochondrial dysfunction (Cpt1a) is mirrored by reduced peroxisomal activity (peroxisomal genes), which supports the generally linked activity of these protective organelles against NAFLD.
The major metabolite that PPARα increases during β-oxidation is the ketone β-hydroxybutyrate, which is released from the liver into blood (29). Plasma lipids as well as other metabolites such as amino, carboxylic, and keto acids were analyzed using a novel NMR spectroscopy approach. The PparaHepKO mice on HFD had significantly (P < 0.05) reduced plasma levels of β-hydroxybutyrate (measured as 3-hydroxybutyric acid) compared with control (Table 2). Interestingly, there was no difference between the groups for plasma pyruvic acid, the simplest of the α-keto acids. Other metabolites measured were carboxylic and amino acids. There was no difference between PparaHepKO compared with Pparafl/fl for plasma levels of carboxylic acid groups: acetic acid, citric acid, formic acid, or lactic acid (Table 2). Plasma amino acids between the mice groups showed significantly (P < 0.05) higher levels of alanine and significantly (P < 0.05) lower levels of glutamic acid in the HFD PparaHepKO group (Table 2). No change was observed in the other amino acids measured. These results show that PparaHepKO mice have reduced fat burning via the β-oxidation pathway, have microsteatosis on NFD, and early-stage NAFLD, which is worsened on HFD.
Table 2.
Metabolite analysis in Pparafl/fl and PparaHepKO mice
| Plasma metabolite | Pparafl/fl NFD | Pparafl/fl HFD | PparaHepKO NFD | PparaHepKO HFD | Diet P Value | Genotype P Value | Interaction P Value |
|---|---|---|---|---|---|---|---|
| n | 6 | 12 | 6 | 8 | |||
| Keto acids | |||||||
| 3-Hydroxybutyric acid, mmol/l | 0.228 ± 0.05 | 0.294 ± 0.01 | 0.171 ± 0.01 | 0.21 ± 0.02 | 0.2045 | 0.0366* | 0.9781 |
| Pyruvic acid, mmol/l | 0.048 ± 0.006 | 0.055 ± 0.005 | 0.05 ± 0.005 | 0.046 ± 0.003 | 0.6424 | 0.9641 | 0.7987 |
| Carboxylic acids | |||||||
| Acetic acid, mmol/l | 0.12 ± 0.01 | 0.046 ± 0.004 | 0.11 ± 0.02 | 0.056 ± 0.005 | 0.001* | 0.4856 | 0.9938 |
| Citric acid, mmol/l | 0.188 ± 0.04 | 0.096 ± 0.006 | 0.188 ± 0.04 | 0.088 ± 0.004 | 0.002* | 0.8615 | 0.8615 |
| Formic acid, mmol/l | 0.026 ± 0.006 | 0.028 ± 0.004 | 0.035 ± 0.007 | 0.026 ± 0.001 | 0.1275 | 0.1529 | 0.5817 |
| Lactic acid, mmol/l | 5.55 ± 1.8 | 1.56 ± 0.12 | 4.42 ± 1.9 | 1.72 ± 0.16 | 0.012* | 0.6062 | 0.4943 |
| Amino acids | |||||||
| Alanine, nmol/l | 0.193 ± 0.012 | 0.143 ± 0.005 | 0.156 ± 0.021 | 0.167 ± 0.005 | 0.1816 | 0.3625 | 0.0271* |
| Glutamine, nmol/l | 0.03 ± 0.033 | 0.353 ± 0.0008 | 0.098 ± 0.069 | 0.306 ± 0.0008 | 0.0262* | 0.0312* | 0.0495* |
| Glycine, nmol/l | 0.181 ± 0.005 | 0.105 ± 0.011 | 0.171 ± 0.014 | 0.125 ± 0.004 | 0.001* | 0.374 | 0.1202 |
| Histidine, nmol/l | 0.031 ± 0.004 | 0.04 ± 0.001 | 0.04 ± 0.007 | 0.042 ± 0.005 | 0.5002 | 0.1356 | 0.8656 |
| Isoleucine, nmol/l | 0.035 ± 0.002 | 0.035 ± 0.003 | 0.035 ± 0.002 | 0.033 ± 0.002 | 0.3506 | 0.3506 | 0.1656 |
| Leucine, nmol/l | 0.046 ± 0.006 | 0.033 ± 0.002 | 0.038 ± 0.005 | 0.036 ± 0.002 | 0.0553 | 0.488 | 0.1555 |
| Tyrosine, nmol/l | 0.023 ± 0.008 | 0.026 ± 0.003 | 0.01 ± 0.007 | 0.031 ± 0.005 | 0.6644 | 0.5774 | 0.1608 |
| Valine, nmol/l | 0.088 ± 0.005 | 0.074 ± 0.003 | 0.068 ± 0.01 | 0.077 ± 0.004 | 0.6517 | 0.1395 | 0.0422* |
Values are means ± SE: n = no. of mice. NFD, normal fat diet; HFD, high-fat diet; Pparafl/fl, peroxisome proliferator-activated receptor-α (PPARα) wild-type mice; PparaHepKO, knockout mice.
P < 0.05.
PparaHepKO mice on HFD exhibit inflammation and higher proinflammatory cytokines expression.
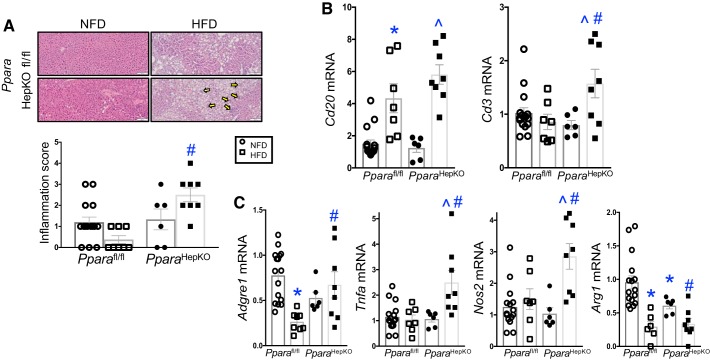
Hepatic inflammation induces liver fat accumulation (40), which most likely occurs from oxidative stress and suppression of PPARα (25). To ascertain if there were inflammation in the PparaHepKO and Pparafl/fl mice in response to diet, we performed H&E staining and determined visible inflammatory foci that assembled in the livers. The Pparafl/fl mice on NFD showed no significant visible inflammatory foci (Fig. 4A). The Pparafl/fl on HFD displayed a variable, low level of inflammatory foci, but no signficant inflammation was observed. The PparaHepKO on NFD had obvious inflammatory foci present, but no significant difference was obeserved from Pparafl/fl mice on NFD for the inflammation score. However, the PparaHepKO on HFD had focal-to-mild lobular inflammation in 100% of the mice tested (Fig. 4A, arrows). These data suggest that PPARα protects against HFD-induced inflammation in the liver.
Fig. 4.
The peroxisome proliferator-activated receptor-α (PPARα) hepatocyte-specific knockout (PparaHepKO) mice develop hepatic inflammation on high-fat diet (HFD). A: H&E staining in livers from PPAR wild-type (Pparafl/fl) and PparaHepKO mice on normal fat diet (NFD) and HFD and inflammation score. Yellow arrows, PparaHepKO had focal-to-mild lobular inflammation on 100% of the HFD mice tested. B and C: biochemical measurement of hepatic triglyceride levels. Measurement of hepatic immune gene expression in NFD and HFD in Pparafl/fl and PparaHepKO mice for Cd20 and Cd3 (B) and macrophage markers Adgre1, Tnfa, Nos2, and Arg1 (C). Values are means ± SE; *P < 0.05 vs. NFD Pparafl/fl; #P < 0.05 vs. HFD Pparafl/fl; ^P < 0.05 vs. NFD PparaHepKO. A: n = 13 and 8, Pparafl/fl NFD, HFD; n = 6 and 8, PparaHepKO NFD, HFD. B and C: n = 13 and 7, Pparafl/fl NFD, HFD; n = 6 and 8, PparaHepKO NFD, HFD. n, number of mice. All circles, NFD; all squares, HFD.
Next, to ascertain the type of immune response that was elicited in the PparaHepKO and Pparafl/fl mice in response to diet, we measured established markers of immune cells, Cd20 and Cd3, which are markers for B-lymphocytes and thymocytes, respectively. In both groups of mice, hepatic Cd20 expression signficantly (P < 0.05) increased (Fig. 4B). However, there was no change in Cd20 expression between Pparafl/fl and PparaHepKO on HFD. The Cd3 expression, however, was significantly higher only in the PparaHepKO on HFD. We have previously shown that hepatic steatosis can occur by inducing a shift in macrophage population from anti-inflammatory M2 to proinflammatory M1 macrophages (40). We found that the Adgre1 mRNA expression that encodes for the general macrophage marker F480 had no significant difference between Pparafl/fl and PparaHepKO on NFD or HFD (Fig. 4C). This finding was not surprising as we have reported no change in total macrophage numbers, but that the change occurred in the M2 and M1 populations (40). In Fig. 4, D and E, the PparaHepKO on HFD had significantly higher expression of proinflammatory M1 macrophage markers TNFα (Tnfa) and inducible nitric oxide synthases iNOS (Nos2), and in Fig. 4F reduced anti-inflammatory M2 macrophage markers arginase 1 (Arg1). These data imply that hepatic PPARα reduces HFD-induced inflammation by mediating the M2 to M1 macrophage population shift.
PparaHepKO mice exhibited worsened HFD-induced hyperlipidemia.
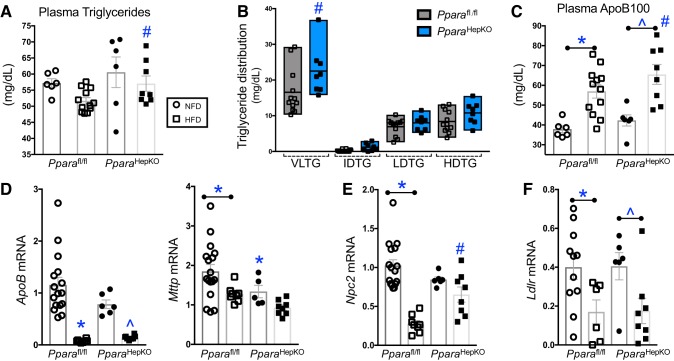
A clinically known function of the activation of PPARα is to reduce plasma triglycerides for patients with hyperlipidemia. However, whether this is mediated from PPARα expressed in liver, WAT, BAT, muscle, or another tissue is unknown. The PparaHepKO on HFD had significantly (P < 0.05) higher plasma triglycerides as compared with HFD Pparafl/fl mice (Fig. 5A). Measurement of the triglyceride distribution showed that the triglyceride increase in the PparaHepKO was mostly in the VLDL lipoprotein and not intermediate-density lipoprotein (IDL, LDL, or HDL (Fig. 5B). The VLDL is assembled in the liver with triglycerides, cholesterol, and apolipoproteins, especially ApoB100 (8). Plasma ApoB100 was significantly (P < 0.05) higher in the HFD PparaHepKO compared with HFD Pparafl/fl mice (Fig. 5C). ApoB100 and the microsomal triglyceride transfer protein are critical for excretion of the VLDL molecule from liver (8). Despite the alteration in plasma apolipoprotein levels, the hepatic apoB mRNA expression was not changed between the HFD groups (Fig. 5D). However, hepatic Mttp mRNA was significantly (P < 0.05) lower in the HFD PparaHepKO compared with HFD Pparafl/fl mice, which could be due to the higher circulating levels of ApoB100 found in PparaHepKO mice.
Fig. 5.
Mice with a loss of hepatic peroxisome proliferator-activated receptor-α (PPARα) on a high-fat diet (HFD) have higher triglycerides and apolipoprotein B100 levels. A: plasma triglycerides as measured by NMR spectroscopy. Pparafl/fl, PPAR wild-type; PparaHepKO, PPARα hepatocyte-specific knockout mice. B: plasma triglyceride distribution as measured by NMR spectroscopy. Plasma ApoB100 as measured by NMR spectroscopy. C: plasma ApoB100 as measured by NMR spectroscopy. mRNA levels in livers from Pparafl/fl and PparaHepKO mice on normal fat diet (NFD) and HFD: Apob and Mttp (D), Npc2 (E), and Ldlr (F). Values are means ± SE; *P < 0.05 vs. NFD Pparafl/fl; #P < 0.05 vs. HFD Pparafl/fl; ^P < 0.05 vs. NFD PparaHepKO; n = 6 and 12, Pparafl/fl NFD, HFD; n = 6 and 8, PparaHepKO NFD, HFD; A and C, n = 8 and 12; B, n = 16 and 8 Pparafl/fl NFD, HFD; D, E, and F, n = 6 and 8, PparaHepKO NFD, HFD. n, number of mice. All circles, NFD; all squares, HFD.
The Npc2 gene encodes for the Niemann-Pick C2 (NPC2) protein that mediates cholesterol egress from lysosomes (28) and regulates sterol transport between membranes (72). The Npc2 expression was significantly reduced in Pparafl/fl mice on HFD compared with NFD (Fig. 5E). However, PparaHepKO had no signficant change in Npc2 expression in NFD or HFD. The Pparafl/fl mice on HFD had significantly lower Npc2 compared with PparaHepKO HFD. The movement of cholesterol aids homeostatic responses and the cellular cholesterol pool, including LDL receptor expression and de novo cholesterol synthesis (41). The hepatic LDL receptor mRNA (Ldlr) expression was signficantly (P < 0.05) lower in PparaHepKO and Pparafl/fl mice on HFD compared with their NFD controls. However, comparison between the groups showed no significant difference.
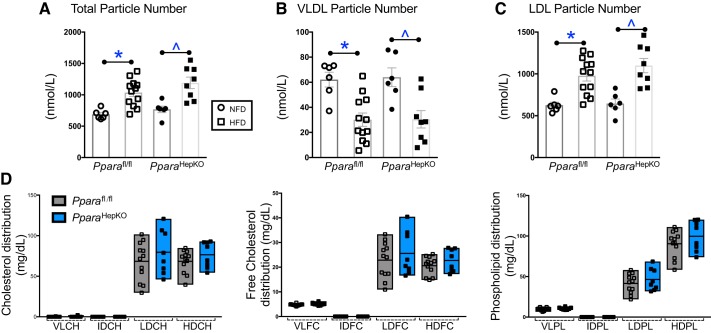
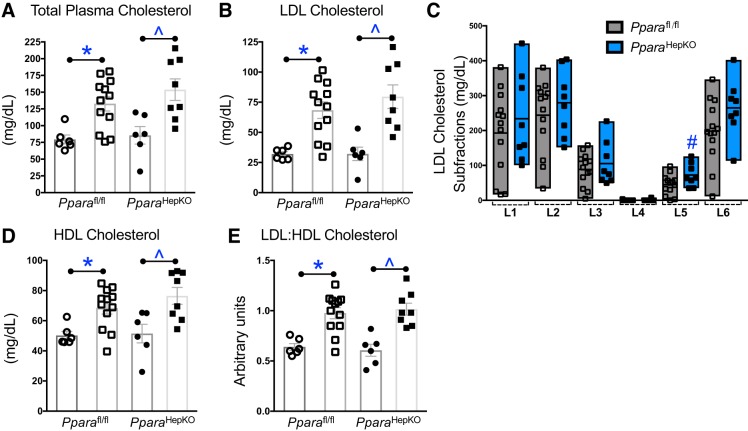
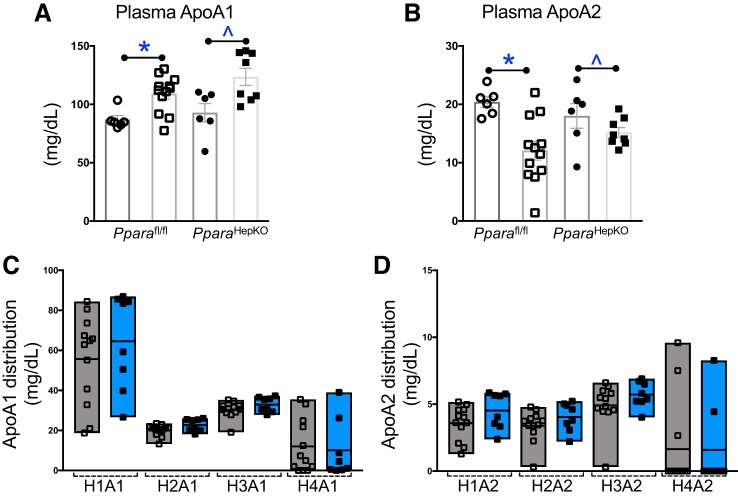
Next, to determine how these signaling mechanisms impact plasma cholesterol levels, we used Bruker NMR spectroscopy with the IVDr) protocol. The total and LDL particle numbers were significantly increased with HFD in both groups (Fig. 6, A and C.). The VLDL particle number was significantly lower with HFD in both groups (Fig. 6B). Interestingly, the total, VLDL, and LDL particle numbers were not changed between the HFD or NFD between the two groups (Fig. 6, A–C). The lipoprotein subclass analysis was performed by Bruker NMR spectroscopy using regression analysis of the NMR data, which is done automatically as part of the IVDr platform as previously described (46). The distribution of cholesterol, free cholesterol, and phospholipid was not changed for VLDL, IDL, LDL, or HDL (Fig. 6D). These data were also supported with no change in total plasma cholesterol, LDL cholesterol, HDL cholesterol, or LDL:HDL ratio (Fig. 7, A–E). However, the L5PN subfraction of LDL was significantly increased in PparaHepKO as compared with Pparafl/fl mice (Fig. 7C). Apolipoproteins ApoA1 and ApoA2 are scavengers that help remove cholesterol from peripheral tissues. There was no significant difference between the HFD groups for plasma ApoA1 (P = 0.0977) or ApoA2 (P = 0.2605) (Fig. 8, A and B). This was also reflected in the ApoA1 and ApoA2 distribution profiles for each fraction (Fig. 8, C and D).
Fig. 6.
Plasma total, very-low-density lipoprotein (VLDL), and low-density lipoprotein (LDL) particle numbers and cholesterol distributions in PPAR wild-type (Pparafl/fl) and PPARα hepatocyte-specific knockout (PparaHepKO) mice are not different between the groups. Total particle number (A), plasma VLDL particle number (B), and plasma LDL particle number (C) as measured by NMR spectroscopy. D: distribution profiles of cholesterol, free cholesterol, and phospholipid in plasma as for VLDL, intermediate-density lipoproteins (IDL), LDL, and high-density lipoprotein (HDL). Values are means ± SE; *P < 0.05 vs. NFD Pparafl/fl; ^P < 0.05 vs. normal fat diet (NFD) PparaHepKO; n = 6 and 12, Pparafl/fl NFD, high-fat diet (HFD); n = 6 and 8, PparaHepKO NFD, HFD. n, number of mice. All circles, NFD; all squares, HFD.
Fig. 7.
Plasma cholesterol in peroxisome proliferator-activated receptor-α (PPARα) wild-type (Pparafl/fl) and PPARα hepatocyte-specific knockout (PparaHepKO) mice fed a high-fat diet (HFD). Plasma total cholesterol (A), low-density lipoprotein (LDL0 cholesterol (B), LDL cholesterol subfractions (C), high-density lipoprotein (HDL) cholesterol (D), and LDL:HDL ratio (E) as measured by NMR spectroscopy. Values are means ± SE; *P < 0.05 vs. normal fat diet (NFD) Pparafl/fl; #P < 0.05 (vs. Pparafl/fl mice); ^P < 0.05 (vs. NFD PparaHepKO); n = 6 and12, Pparafl/fl NFD, HFD; n = 6 and 8, PparaHepKO NFD, HFD). n, number of mice. All circles, NFD; all squares, HFD.
Fig. 8.
Plasma apolipoproteins A1 (ApoA1) and ApoA2 levels in peroxisome proliferator-activated receptor-α (PPARα) wild-type (Pparafl/fl) and PPARα hepatocyte-specific knockout (PparaHepKO) mice fed a high-fat diet (HFD). Plasma ApoA1 (A) and Apo-A2 (B) as measured by NMR spectroscopy. ApoA1 (C) and ApoA2 (D) distribution profiles for very-low-density lipoprotein (VLDL), intermediate-density lipoproteins (IDL), low-density lipoprotein (LDL), and high-density lipoprotein (HDL). Values are means ± SE; *P < 0.05 vs. normal fed diet (NFD) Pparafl/fl; ^P < 0.05 vs. NFD PparaHepKO; n = 6 and12, Pparafl/fl NFD, HFD; n = 6 and 8, PparaHepKO NFD, HFD. n, number of mice. All circles, NFD; all squares, HFD.
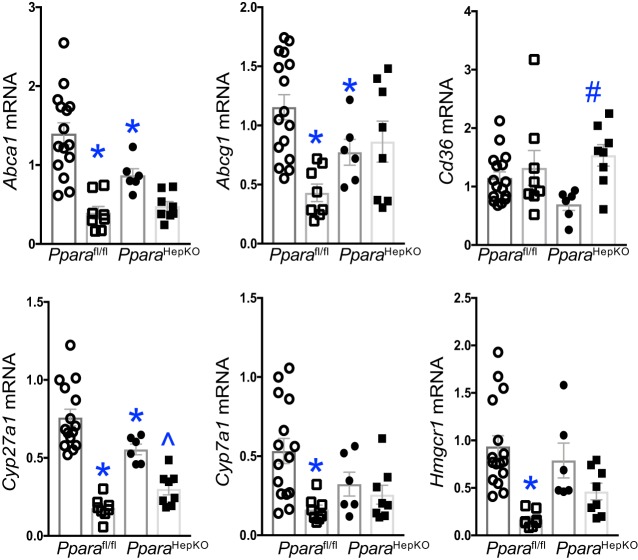
Hyperlipidemia is regulated by the production of cholesterol in the liver (41). To determine changes in cholesterol conversion or transport gene changes, we measured the major players Cyp7a1, Cyp27a1, Hmgcr1, Cd36, Abca1, and Abcg1 (41). The hepatic mRNA expression for Cyp7a1, Cyp27a1, Hmgcr1, Abca1, and Abcg1 was significantly (P < 0.05) reduced in Pparafl/fl mice on HFD compared with NFD, but scavenger receptor Cd36 was unchanged (Fig. 9). The PparaHepKO mice had reduced Cyp27a1 and increased Cd36 expression on NFD compared with HFD. The other genes for PparaHepKO compared with the Pparafl/fl had no significant differences.
Fig. 9.
Cholesterol conversion or transport gene expression in peroxisome proliferator-activated receptor-α (PPARα) wild-type (Pparafl/fl) and PPARα hepatocyte-specific knockout (PparaHepKO) mice. Hepatic Abca1, Abcg1, Cd36, Cyp27a1, Cyp7a1, and Hmgcr1 mRNA levels in NFD (all circles) and HFD (all squares) in Pparafl/fl and PparaHepKO mice. Values are means ± SE; *P < 0.05 vs. NFD Pparafl/fl; ^P < 0.05 vs. NFD PparaHepKO; n = 16 and 13, Pparafl/fl NFD, HFD; n = 6 and 8, PparaHepKO NFD, HFD. n, number of mice.
Overall, these data indicate that hepatic PPARα mediates immune cell foci inclusion in the liver during high-fat feeding and that this causes a shift in the macrophage population from M2 to M1. These events lead to hepatic steatosis, which also increases plasma triglyceride and ApoB100 levels during high-fat diet-induced hyperlipidemia.
DISCUSSION
To determine the specific purpose of hepatic PPARα, here we used mice lacking PPARα specifically in hepatocytes as previously described (43). The effect of the loss of hepatocyte PPARα on inflammation and lipid accumulation in response to fasting and MCD had been studied in these mice (43). However, the role of hepatic PPARα on lipid accumulation in the liver as well as regulation of plasma triglycerides, apolipoproteins, cholesterol, and other metabolites in response to high-fat feeding had not been previously examined. Here, we found no difference in body weight between the Pparafl/fl and PparaHepKO on a high-fat or normal-fat diet. However, there were significant differences observed with the MCD diet in the PparaHepKO but not the global Ppara KO (43). These differences are likely due to the type of calorie load on the liver.
Small, dense LDL is a type of LDL cholesterol that is an emerging risk factor for cardiovascular disease (31, 58, 64). It is smaller and heavier than typical LDL cholesterol and can increase the risk of developing atherosclerosis and other cardiovascular diseases (58, 64). The PparaHepKO mice showed significantly increased levels of LDL subfraction (L5) in response to high-fat feeding. Several studies have demonstrated that alterations in small, dense LDL may mediate the increased risk for atherosclerosis and cardiovascular disease in NAFLD (60, 63). These are the first data to demonstrate a relationship between hepatic PPARα and small, dense LDL particles in the plasma. Further studies are needed to fully elucidate the role of alterations in hepatic PPARα and small, dense LDL particles in the development of cardiovascular disease in NAFLD.
Diet-induced obesity may result in elevated levels of glucose and fatty acids resulting in enhanced production leading to increased formation of ROS. Oxidative stress magnifies the adverse effects of obesity by inducing inflammation in the liver leading to NAFLD and, on occasion, worsens to nonalcoholic steatohepatitis (4). The onset of NAFLD has been linked to cardiovascular events by increasing levels of the proinflammatory markers TNFα and C-reactive protein (CRP) (42). CRP is a liver-specific protein that has been extensively studied as a biomarker of inflammation in cardiovascular disease. Exacerbated ROS, due to obesity-induced NAFLD, increases CRP expression and levels in blood increasing risk of diabetes, hypertension, and cardiovascular disease (48). Both statin and fibrate drugs have been shown to decrease CRP, but if this occurs by their role in regulating hepatic inflammation is unknown. Here, we found that proinflammatory M1 macrophage markers TNFα and iNOS were significantly increased and that intralobular inflammatory foci were increased in the liver of the PparaHepKO on HFD compared with Pparafl/fl control.
Ketone body metabolism is central for fine-tuning metabolic roles that optimize organ and organism performance in varying nutrient states and protect from inflammation and injury in multiple organ systems (49). PparaHepKO mice on a high-fat diet showed significantly lower levels of plasma 3-hydroxybutyric acid, which is converted to β-hydroxybutyrate (βOHB). These are the major metabolites released from the liver during the burning of fat. Interestingly, there was no change in the α-keto acid, pyruvic acid. Ketogenesis is a series of reactions that lead to the formation of ketone bodies, including βOHB. PPARα regulates ketogenesis via a transcriptional network that includes AMP-activated protein kinase (AMPK), PPARγ coactivator 1α (PGC-1α), mammalian target of rapamycin, and FGF21 (17). Lower levels of βOHB production have been associated with NAFLD and NASH (39), which may be reduced from inflammation in the liver, possibly from lipid peroxidation that worsens NAFLD (4). Obesity increases oxidative stress (ROS) and simultaneously decreases expression and activity of key cytoprotective systems including heme oxygenase (HO) and bilirubin as well as PPARα, while increasing inflammatory cytokines and insulin resistance (13, 34, 47, 59, 68). Increasing HO-1 activity produces the antioxidant bilirubin that has been shown to reduce adiposity (53, 65), and recently found to function as a hormone by activating PPARα by direct binding (62). In obese mice, increasing HO-1production of bilirubin reduces oxidative stress and fatty liver by increasing PPARα and FGF21 levels (25, 54). Here, the observed increased hepatic steatosis was associated with lower levels of hepatic Ffg21 and Cpt1a, which are genes involved in fatty acid β-oxidation (22, 45, 69). FGF21 is regulated via PPARα and has been previously reported to reverse hepatic steatosis and glucose intolerance (3, 12, 25, 33). Others have shown that the PparaHepKO mice have significantly lower fasting plasma glucose levels on normal chow diet compared with littermate controls (6, 43). However, here, we found that the PparaHepKO mice in response to high-fat feeding exhibited significantly higher hyperglycemia and hyperinsulinemia compared with Pparafl/fl mice. This alteration in blood glucose regulation in response to high-fat feeding may be due in part to reduced hepatic Fgf21 in PparaHepKO mice, as FGF21 has been shown to function in glucose homeostasis (5, 70, 71).
Other factors such as branched-chain amino acids (BCAAs) are involved in the metabolism of glucose, and oxidation of BCAAs may increase fatty acid oxidation and play a role in obesity (37). Although the loss of hepatic PPARα did not have any significant effect on the levels of plasma BCAAs in response to high-fat diet feeding compared with control, it did have an impact on the levels of alanine and glutamic acid in the present study. The elevations in serum alanine could be reflective of the altered glucose metabolism in the PparaHepKO mice that would interfere with the glucose-alanine cycle (15).
In conclusion, to develop novel therapeutics for the treatment of NAFLD, a better understanding of the ability of the liver to respond to inflammation and how these induce alterations in apolipoprotein and triglyceride metabolism are needed. These may be mediated from the early onset of reduced PPARα, causing inflammation and hepatic steatosis, which also occurs in obesity. PPARα is the major isoform present in the liver where it helps to orchestrate the hepatic responses to fasting (6, 29), and here we show that it is also quintessential for high fat-induced hepatic inflammation. PPARα is also expressed in a variety of extrahepatic tissues, such as muscle, adipose, pancreas, kidney, and brain, which may also play influential roles in the clearance of apolipoproteins for triglycerides and cholesterol homeostasis. This may aid in the protection of the cardiovascular system from lipid peroxidation or oxidized cholesterol and plaque development that can lead to heart attack or stroke. Increasing HDL cholesterol by either exercise or fibrate treatment is beneficial for hyperlipidemia. However, we did not find a significant difference in HDL levels for the hepatic PPARα KO compared with control mice. The PPARα regulation of HDL levels may be mediated by other tissues, such as adipose or muscle. Further studies using tissue-specific PPARα KO in extrahepatic tissues such as adipose and muscle, which has not been performed, will reveal the actions of the nuclear receptor responses to apolipoprotein signaling.
Perspectives and Significance
The results of the present study establish an essential role for hepatic PPARα in protection against high-fat feeding that induced inflammation-induced hyperlipidemia, hepatic steatosis, and insulin resistance. Moreover, our data show a role for hepatic PPARα in the regulation of plasma triglycerides, apolipoproteins, amino acids, and keto acids in response to high-fat feeding. Our results support the development of drugs that target induction of hepatic PPARα as potential novel therapies for dietary obesity-induced inflammation that is known to cause hypertriglyceridemia, hepatic steatosis, and alterations in ketone and fatty acid metabolism.
GRANTS
This work was supported by National Institutes of Health Grant L32MD009154 (to T. D. Hinds, Jr.), National Heart, Lung and Blood Institute Grants K01HL-125445 (to T. D. Hinds, Jr.) and P01 HL05197-11 (to D. E. Stec), and the National Institute of General Medical Sciences Grant P20GM104357-02 (to D. E. Stec). Nuclear magnetic resonance instrumentation was supported in part by National Science Foundation Grant 0922862, National Institutes of Health Grant S10 RR025677, and Vanderbilt University matching funds. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
D.E.S., C.D.A., D.F.S., and T.D.H. conceived and designed research; D.E.S., D.M.G., J.A.H., S.H., Z.L.M., N.R.F., J.W.R., D.F.S., and T.D.H. performed experiments; D.E.S., D.M.G., J.A.H., S.H., Z.L.M., N.R.F., J.W.R., D.F.S., and T.D.H. analyzed data; D.E.S., D.M.G., J.A.H., Z.L.M., N.R.F., J.W.R., C.D.A., D.F.S., and T.D.H. interpreted results of experiments; D.E.S. and T.D.H. prepared figures; D.E.S. and T.D.H. drafted manuscript; D.E.S., D.M.G., J.A.H., S.H., Z.L.M., N.R.F., J.W.R., C.D.A., D.F.S., and T.D.H. edited and revised manuscript; D.E.S., D.M.G., J.A.H., S.H., Z.L.M., N.R.F., J.W.R., C.D.A., D.F.S., and T.D.H. approved final version of manuscript.
REFERENCES
- 1.Adeosun SO, Gordon DM, Weeks MF, Moore KH, Hall JE, Hinds TD Jr, Stec DE. Loss of biliverdin reductase-A promotes lipid accumulation and lipotoxicity in mouse proximal tubule cells. Am J Physiol Renal Physiol 315: F323–F331, 2018. doi: 10.1152/ajprenal.00495.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akiyama TE, Nicol CJ, Fievet C, Staels B, Ward JM, Auwerx J, Lee SS, Gonzalez FJ, Peters JM. Peroxisome proliferator-activated receptor-alpha regulates lipid homeostasis, but is not associated with obesity: studies with congenic mouse lines. J Biol Chem 276: 39088–39093, 2001. doi: 10.1074/jbc.M107073200. [DOI] [PubMed] [Google Scholar]
- 3.Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab 5: 426–437, 2007. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Benedict M, Zhang X. Non-alcoholic fatty liver disease: an expanded review. World J Hepatol 9: 715–732, 2017. doi: 10.4254/wjh.v9.i16.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berglund ED, Li CY, Bina HA, Lynes SE, Michael MD, Shanafelt AB, Kharitonenkov A, Wasserman DH. Fibroblast growth factor 21 controls glycemia via regulation of hepatic glucose flux and insulin sensitivity. Endocrinology 150: 4084–4093, 2009. doi: 10.1210/en.2009-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brocker CN, Patel DP, Velenosi TJ, Kim D, Yan T, Yue J, Li G, Krausz KW, Gonzalez FJ. Extrahepatic PPARα modulates fatty acid oxidation and attenuates fasting-induced hepatosteatosis in mice. J Lipid Res 59: 2140–2152, 2018. doi: 10.1194/jlr.M088419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chau MD, Gao J, Yang Q, Wu Z, Gromada J. Fibroblast growth factor 21 regulates energy metabolism by activating the AMPK-SIRT1-PGC-1alpha pathway. Proc Natl Acad Sci USA 107: 12553–12558, 2010. doi: 10.1073/pnas.1006962107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z, Newberry EP, Norris JY, Xie Y, Luo J, Kennedy SM, Davidson NO. ApoB100 is required for increased VLDL-triglyceride secretion by microsomal triglyceride transfer protein in ob/ob mice. J Lipid Res 49: 2013–2022, 2008. doi: 10.1194/jlr.M800240-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costet P, Legendre C, Moré J, Edgar A, Galtier P, Pineau T. Peroxisome proliferator-activated receptor alpha-isoform deficiency leads to progressive dyslipidemia with sexually dimorphic obesity and steatosis. J Biol Chem 273: 29577–29585, 1998. doi: 10.1074/jbc.273.45.29577. [DOI] [PubMed] [Google Scholar]
- 10.DeAngelis AM, Heinrich G, Dai T, Bowman TA, Patel PR, Lee SJ, Hong EG, Jung DY, Assmann A, Kulkarni RN, Kim JK, Najjar SM. Carcinoembryonic antigen-related cell adhesion molecule 1: a link between insulin and lipid metabolism. Diabetes 57: 2296–2303, 2008. doi: 10.2337/db08-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Defour M, Dijk W, Ruppert P, Nascimento EBM, Schrauwen P, Kersten S. The peroxisome proliferator-activated receptor α is dispensable for cold-induced adipose tissue browning in mice. Mol Metab 10: 39–54, 2018. doi: 10.1016/j.molmet.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dushay J, Chui PC, Gopalakrishnan GS, Varela-Rey M, Crawley M, Fisher FM, Badman MK, Martinez-Chantar ML, Maratos-Flier E. Increased fibroblast growth factor 21 in obesity and nonalcoholic fatty liver disease. Gastroenterology 139: 456–463, 2010. doi: 10.1053/j.gastro.2010.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elmarakby AA, Imig JD. Obesity is the major contributor to vascular dysfunction and inflammation in high-fat diet hypertensive rats. Clin Sci (Lond) 118: 291–301, 2010. doi: 10.1042/CS20090395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabbrini E, Tiemann Luecking C, Love-Gregory L, Okunade AL, Yoshino M, Fraterrigo G, Patterson BW, Klein S. Physiological mechanisms of weight gain-induced steatosis in people with obesity. Gastroenterology 150: 79–81.e2, 2016. doi: 10.1053/j.gastro.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felig P. The glucose-alanine cycle. Metabolism 22: 179–207, 1973. doi: 10.1016/0026-0495(73)90269-2. [DOI] [PubMed] [Google Scholar]
- 16.Gabaldón T. Peroxisome diversity and evolution. Philos Trans R Soc Lond B Biol Sci 365: 765–773, 2010. doi: 10.1098/rstb.2009.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grabacka M, Reiss K. Anticancer properties of PPARα-effects on cellular metabolism and inflammation. PPAR Res 2008: 1–9, 2008. doi: 10.1155/2008/930705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerre-Millo M, Rouault C, Poulain P, André J, Poitout V, Peters JM, Gonzalez FJ, Fruchart JC, Reach G, Staels B. PPAR-alpha-null mice are protected from high-fat diet-induced insulin resistance. Diabetes 50: 2809–2814, 2001. doi: 10.2337/diabetes.50.12.2809. [DOI] [PubMed] [Google Scholar]
- 19.Haluzik M, Gavrilova O, LeRoith D. Peroxisome proliferator-activated receptor-alpha deficiency does not alter insulin sensitivity in mice maintained on regular or high-fat diet: hyperinsulinemic-euglycemic clamp studies. Endocrinology 145: 1662–1667, 2004. doi: 10.1210/en.2003-1015. [DOI] [PubMed] [Google Scholar]
- 20.Heinrich G, Ghadieh HE, Ghanem SS, Muturi HT, Rezaei K, Al-Share QY, Bowman TA, Zhang D, Garofalo RS, Yin L, Najjar SM. Loss of hepatic CEACAM1: a unifying mechanism linking insulin resistance to obesity and non-alcoholic fatty liver disease. Front Endocrinol (Lausanne) 8: 8, 2017. doi: 10.3389/fendo.2017.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinds TD Jr, Adeosun SO, Alamodi AA, Stec DE. Does bilirubin prevent hepatic steatosis through activation of the PPARα nuclear receptor? Med Hypotheses 95: 54–57, 2016. doi: 10.1016/j.mehy.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinds TD Jr, Stec DE. Bilirubin, a cardiometabolics molecule. Hypertension 72: 788–795, 2018. doi: 10.1161/HYPERTENSIONAHA.118.11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinds TD Jr, Burns KA, Hosick PA, McBeth L, Nestor-Kalinoski A, Drummond HA, AlAmodi AA, Hankins MW, Vanden Heuvel JP, Stec DE. Biliverdin reductase A attenuates hepatic steatosis by inhibition of glycogen synthase kinase (GSK) 3β phosphorylation of serine 73 of peroxisome proliferator-activated receptor (PPAR) α. J Biol Chem 291: 25179–25191, 2016. doi: 10.1074/jbc.M116.731703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinds TD Jr, Hosick PA, Chen S, Tukey RH, Hankins MW, Nestor-Kalinoski A, Stec DE. Mice with hyperbilirubinemia due to Gilbert’s syndrome polymorphism are resistant to hepatic steatosis by decreased serine 73 phosphorylation of PPARα. Am J Physiol Endocrinol Metab 312: E244–E252, 2017. doi: 10.1152/ajpendo.00396.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinds TD Jr, Sodhi K, Meadows C, Fedorova L, Puri N, Kim DH, Peterson SJ, Shapiro J, Abraham NG, Kappas A. Increased HO-1 levels ameliorate fatty liver development through a reduction of heme and recruitment of FGF21. Obesity (Silver Spring) 22: 705–712, 2014. doi: 10.1002/oby.20559. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Hinds TD Jr, Stechschulte LA, Cash HA, Whisler D, Banerjee A, Yong W, Khuder SS, Kaw MK, Shou W, Najjar SM, Sanchez ER. Protein phosphatase 5 mediates lipid metabolism through reciprocal control of glucocorticoid receptor and peroxisome proliferator-activated receptor-γ (PPARγ). J Biol Chem 286: 42911–42922, 2011. doi: 10.1074/jbc.M111.311662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V, Elmquist JK, Gerard RD, Burgess SC, Hammer RE, Mangelsdorf DJ, Kliewer SA. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab 5: 415–425, 2007. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, Goldstein JL. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci USA 105: 15287–15292, 2008. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest 103: 1489–1498, 1999. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim BH, Won YS, Kim EY, Yoon M, Nam KT, Oh GT, Kim DY. Phenotype of peroxisome proliferator-activated receptor-alpha (PPARalpha) deficient mice on mixed background fed high fat diet. J Vet Sci 4: 239–244, 2003. doi: 10.4142/jvs.2003.4.3.239. [DOI] [PubMed] [Google Scholar]
- 31.Lamarche B, Tchernof A, Moorjani S, Cantin B, Dagenais GR, Lupien PJ, Després JP. Small, dense low-density lipoprotein particles as a predictor of the risk of ischemic heart disease in men. Prospective results from the Québec Cardiovascular Study. Circulation 95: 69–75, 1997. doi: 10.1161/01.CIR.95.1.69. [DOI] [PubMed] [Google Scholar]
- 32.Lasar D, Rosenwald M, Kiehlmann E, Balaz M, Tall B, Opitz L, Lidell ME, Zamboni N, Krznar P, Sun W, Varga L, Stefanicka P, Ukropec J, Nuutila P, Virtanen K, Amri EZ, Enerbäck S, Wahli W, Wolfrum C. Peroxisome proliferator activated receptor gamma controls mature brown adipocyte inducibility through glycerol kinase. Cell Reports 22: 760–773, 2018. doi: 10.1016/j.celrep.2017.12.067. [DOI] [PubMed] [Google Scholar]
- 33.Li L, Chao QG, Ping LZ, Xue C, Xia ZY, Qian D, Shi-ang H. The prevalence of FOXP3+ regulatory T-cells in peripheral blood of patients with NSCLC. Cancer Biother Radiopharm 24: 357–367, 2009. doi: 10.1089/cbr.2008.0612. [DOI] [PubMed] [Google Scholar]
- 34.Li M, Kim DH, Tsenovoy PL, Peterson SJ, Rezzani R, Rodella LF, Aronow WS, Ikehara S, Abraham NG. Treatment of obese diabetic mice with a heme oxygenase inducer reduces visceral and subcutaneous adiposity, increases adiponectin levels, and improves insulin sensitivity and glucose tolerance. Diabetes 57: 1526–1535, 2008. doi: 10.2337/db07-1764. [DOI] [PubMed] [Google Scholar]
- 35.Lindén D, Lindberg K, Oscarsson J, Claesson C, Asp L, Li L, Gustafsson M, Borén J, Olofsson SO. Influence of peroxisome proliferator-activated receptor alpha agonists on the intracellular turnover and secretion of apolipoprotein (Apo) B-100 and ApoB-48. J Biol Chem 277: 23044–23053, 2002. doi: 10.1074/jbc.M110416200. [DOI] [PubMed] [Google Scholar]
- 36.Lundåsen T, Hunt MC, Nilsson LM, Sanyal S, Angelin B, Alexson SE, Rudling M. PPARalpha is a key regulator of hepatic FGF21. Biochem Biophys Res Commun 360: 437–440, 2007. doi: 10.1016/j.bbrc.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 37.Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol 10: 723–736, 2014. doi: 10.1038/nrendo.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandard S, Stienstra R, Escher P, Tan NS, Kim I, Gonzalez FJ, Wahli W, Desvergne B, Müller M, Kersten S. Glycogen synthase 2 is a novel target gene of peroxisome proliferator-activated receptors. Cell Mol Life Sci 64: 1145–1157, 2007. doi: 10.1007/s00018-007-7006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Männistö VT, Simonen M, Hyysalo J, Soininen P, Kangas AJ, Kaminska D, Matte AK, Venesmaa S, Käkelä P, Kärjä V, Arola J, Gylling H, Cederberg H, Kuusisto J, Laakso M, Yki-Järvinen H, Ala-Korpela M, Pihlajamäki J. Ketone body production is differentially altered in steatosis and non-alcoholic steatohepatitis in obese humans. Liver Int 35: 1853–1861, 2015. doi: 10.1111/liv.12769. [DOI] [PubMed] [Google Scholar]
- 40.Marino JS, Stechschulte LA, Stec DE, Nestor-Kalinoski A, Coleman S, Hinds TD Jr. Glucocorticoid receptor β induces hepatic steatosis by augmenting inflammation and inhibition of the peroxisome proliferator-activated receptor (PPAR) α. J Biol Chem 291: 25776–25788, 2016. doi: 10.1074/jbc.M116.752311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marques LR, Diniz TA, Antunes BM, Rossi FE, Caperuto EC, Lira FS, Gonçalves DC. Reverse cholesterol transport: molecular mechanisms and the non-medical approach to enhance HDL cholesterol. Front Physiol 9: 526, 2018. doi: 10.3389/fphys.2018.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Misra VL, Khashab M, Chalasani N. Nonalcoholic fatty liver disease and cardiovascular risk. Curr Gastroenterol Rep 11: 50–55, 2009. doi: 10.1007/s11894-009-0008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montagner A, Polizzi A, Fouché E, Ducheix S, Lippi Y, Lasserre F, Barquissau V, Régnier M, Lukowicz C, Benhamed F, Iroz A, Bertrand-Michel J, Al Saati T, Cano P, Mselli-Lakhal L, Mithieux G, Rajas F, Lagarrigue S, Pineau T, Loiseau N, Postic C, Langin D, Wahli W, Guillou H. Liver PPARα is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut 65: 1202–1214, 2016. doi: 10.1136/gutjnl-2015-310798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson RH. Hyperlipidemia as a risk factor for cardiovascular disease. Prim Care 40: 195–211, 2013. doi: 10.1016/j.pop.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Brien L, Hosick PA, John K, Stec DE, Hinds TD Jr. Biliverdin reductase isozymes in metabolism. Trends Endocrinol Metab 26: 212–220, 2015. doi: 10.1016/j.tem.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petersen M, Dyrby M, Toubro S, Engelsen SB, Nørgaard L, Pedersen HT, Dyerberg J. Quantification of lipoprotein subclasses by proton nuclear magnetic resonance-based partial least-squares regression models. Clin Chem 51: 1457–1461, 2005. doi: 10.1373/clinchem.2004.046748. [DOI] [PubMed] [Google Scholar]
- 47.Peterson SJ, Drummond G, Kim DH, Li M, Kruger AL, Ikehara S, Abraham NG. L-4F treatment reduces adiposity, increases adiponectin levels, and improves insulin sensitivity in obese mice. J Lipid Res 49: 1658–1669, 2008. doi: 10.1194/jlr.M800046-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pravenec M, Kajiya T, Zídek V, Landa V, Mlejnek P, Simáková M, Silhavý J, Malínská H, Oliyarnyk O, Kazdová L, Fan J, Wang J, Kurtz TW. Effects of human C-reactive protein on pathogenesis of features of the metabolic syndrome. Hypertension 57: 731–737, 2011. doi: 10.1161/HYPERTENSIONAHA.110.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puchalska P, Crawford PA. Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metab 25: 262–284, 2017. doi: 10.1016/j.cmet.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rachid TL, Penna-de-Carvalho A, Bringhenti I, Aguila MB, Mandarim-de-Lacerda CA, Souza-Mello V. Fenofibrate (PPARalpha agonist) induces beige cell formation in subcutaneous white adipose tissue from diet-induced male obese mice. Mol Cell Endocrinol 402: 86–94, 2015. [Erratum in: Mol Cell Encocrinol 413: 249, 2015.] doi: 10.1016/j.mce.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 51.Rachid TL, Penna-de-Carvalho A, Bringhenti I, Aguila MB, Mandarim-de-Lacerda CA, Souza-Mello V. PPAR-α agonist elicits metabolically active brown adipocytes and weight loss in diet-induced obese mice. Cell Biochem Funct 33: 249–256, 2015. doi: 10.1002/cbf.3111. [DOI] [PubMed] [Google Scholar]
- 52.Rachid TL, Silva-Veiga FM, Graus-Nunes F, Bringhenti I, Mandarim-de-Lacerda CA, Souza-Mello V. Differential actions of PPAR-α and PPAR-β/δ on beige adipocyte formation: a study in the subcutaneous white adipose tissue of obese male mice. PLoS One 13: e0191365, 2018. doi: 10.1371/journal.pone.0191365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raffaele M, Barbagallo I, Licari M, Carota G, Sferrazzo G, Spampinato M, Sorrenti V, Vanella L. N-acetylcysteine (NAC) ameliorates lipid-related metabolic dysfunction in bone marrow stromal cells-derived adipocytes. Evid Based Complement Alternat Med 2018: 1–9, 2018. doi: 10.1155/2018/5310961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raffaele M, Bellner L, Singh SP, Favero G, Rezzani R, Rodella LF, Falck JR, Abraham NG, Vanella L. Epoxyeicosatrienoic intervention improves NAFLD in leptin receptor deficient mice by an increase in PGC1α-HO-1-PGC1α-mitochondrial signaling. Exp Cell Res 380: 180–187, 2019. doi: 10.1016/j.yexcr.2019.04.029. [DOI] [PubMed] [Google Scholar]
- 55.Ramakrishnan SK, Russo L, Ghanem SS, Patel PR, Oyarce AM, Heinrich G, Najjar SM. Fenofibrate decreases insulin clearance and insulin secretion to maintain insulin sensitivity. J Biol Chem 291: 23915–23924, 2016. doi: 10.1074/jbc.M116.745778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarwar R, Pierce N, Koppe S. Obesity and nonalcoholic fatty liver disease: current perspectives. Diabetes Metab Syndr Obes 11: 533–542, 2018. doi: 10.2147/DMSO.S146339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shah A, Rader DJ, Millar JS. The effect of PPAR-alpha agonism on apolipoprotein metabolism in humans. Atherosclerosis 210: 35–40, 2010. doi: 10.1016/j.atherosclerosis.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 58.Shiffman D, Louie JZ, Caulfield MP, Nilsson PM, Devlin JJ, Melander O. LDL subfractions are associated with incident cardiovascular disease in the Malmö Prevention Project Study. Atherosclerosis 263: 287–292, 2017. doi: 10.1016/j.atherosclerosis.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 59.Sodhi K, Inoue K, Gotlinger KH, Canestraro M, Vanella L, Kim DH, Manthati VL, Koduru SR, Falck JR, Schwartzman ML, Abraham NG. Epoxyeicosatrienoic acid agonist rescues the metabolic syndrome phenotype of HO-2-null mice. J Pharmacol Exp Ther 331: 906–916, 2009. doi: 10.1124/jpet.109.157545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sonmez A, Nikolic D, Dogru T, Ercin CN, Genc H, Cesur M, Tapan S, Karslioğlu Y, Montalto G, Banach M, Toth PP, Bagci S, Rizzo M. Low- and high-density lipoprotein subclasses in subjects with nonalcoholic fatty liver disease. J Clin Lipidol 9: 576–582, 2015. doi: 10.1016/j.jacl.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 61.Srivastava RA, Jahagirdar R, Azhar S, Sharma S, Bisgaier CL. Peroxisome proliferator-activated receptor-alpha selective ligand reduces adiposity, improves insulin sensitivity and inhibits atherosclerosis in LDL receptor-deficient mice. Mol Cell Biochem 285: 35–50, 2006. doi: 10.1007/s11010-005-9053-y. [DOI] [PubMed] [Google Scholar]
- 62.Stec DE, John K, Trabbic CJ, Luniwal A, Hankins MW, Baum J, Hinds TD Jr. Bilirubin binding to PPARα inhibits lipid accumulation. PLoS One 11: e0153427, 2016. doi: 10.1371/journal.pone.0153427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sugino I, Kuboki K, Matsumoto T, Murakami E, Nishimura C, Yoshino G. Influence of fatty liver on plasma small, dense LDL-cholesterol in subjects with and without metabolic syndrome. J Atheroscler Thromb 18: 1–7, 2011. doi: 10.5551/jat.5447. [DOI] [PubMed] [Google Scholar]
- 64.Toft-Petersen AP, Tilsted HH, Aarøe J, Rasmussen K, Christensen T, Griffin BA, Aardestrup IV, Andreasen A, Schmidt EB. Small dense LDL particles—a predictor of coronary artery disease evaluated by invasive and CT-based techniques: a case-control study. Lipids Health Dis 10: 21, 2011. doi: 10.1186/1476-511X-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vanella L, Sodhi K, Kim DH, Puri N, Maheshwari M, Hinds TD Jr, Bellner L, Goldstein D, Peterson SJ, Shapiro JI, Abraham NG. Increased heme-oxygenase 1 expression in mesenchymal stem cell-derived adipocytes decreases differentiation and lipid accumulation via upregulation of the canonical Wnt signaling cascade. Stem Cell Res Ther 4: 28, 2013. doi: 10.1186/scrt176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vine DF, Wang Y, Jetha MM, Ball GD, Proctor SD. Impaired apoB-lipoprotein and triglyceride metabolism in obese adolescents with polycystic ovary syndrome. J Clin Endocrinol Metab 102: 970–982, 2017. doi: 10.1210/jc.2016-2854. [DOI] [PubMed] [Google Scholar]
- 67.Wang L, Teng R, Di L, Rogers H, Wu H, Kopp JB, Noguchi CT. PPARα and Sirt1 mediate erythropoietin action in increasing metabolic activity and browning of white adipocytes to protect against obesity and metabolic disorders. Diabetes 62: 4122–4131, 2013. doi: 10.2337/db13-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang MH, Smith A, Zhou Y, Chang HH, Lin S, Zhao X, Imig JD, Dorrance AM. Downregulation of renal CYP-derived eicosanoid synthesis in rats with diet-induced hypertension. Hypertension 42: 594–599, 2003. doi: 10.1161/01.HYP.0000090123.55365.BA. [DOI] [PubMed] [Google Scholar]
- 69.Weaver L, Hamoud AR, Stec DE, Hinds TD Jr. Biliverdin reductase and bilirubin in hepatic disease. Am J Physiol Gastrointest Liver Physiol 314: G668–G676, 2018. doi: 10.1152/ajpgi.00026.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu J, Lloyd DJ, Hale C, Stanislaus S, Chen M, Sivits G, Vonderfecht S, Hecht R, Li YS, Lindberg RA, Chen JL, Jung DY, Zhang Z, Ko HJ, Kim JK, Véniant MM. Fibroblast growth factor 21 reverses hepatic steatosis, increases energy expenditure, and improves insulin sensitivity in diet-induced obese mice. Diabetes 58: 250–259, 2009. doi: 10.2337/db08-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu J, Stanislaus S, Chinookoswong N, Lau YY, Hager T, Patel J, Ge H, Weiszmann J, Lu SC, Graham M, Busby J, Hecht R, Li YS, Li Y, Lindberg R, Véniant MM. Acute glucose-lowering and insulin-sensitizing action of FGF21 in insulin-resistant mouse models–association with liver and adipose tissue effects. Am J Physiol Endocrinol Metab 297: E1105–E1114, 2009. doi: 10.1152/ajpendo.00348.2009. [DOI] [PubMed] [Google Scholar]
- 72.Xu Z, Farver W, Kodukula S, Storch J. Regulation of sterol transport between membranes and NPC2. Biochemistry 47: 11134–11143, 2008. doi: 10.1021/bi801328u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zandbergen F, Mandard S, Escher P, Tan NS, Patsouris D, Jatkoe T, Rojas-Caro S, Madore S, Wahli W, Tafuri S, Müller M, Kersten S. The G0/G1 switch gene 2 is a novel PPAR target gene. Biochem J 392: 313–324, 2005. doi: 10.1042/BJ20050636. [DOI] [PMC free article] [PubMed] [Google Scholar]