Keywords: bovine milk, exosomes, microbiome, mouse
Abstract
Exosomes and exosome-like vesicles participate in cell-to-cell communication in animals, plant, and bacteria. Dietary exosomes in bovine milk are bioavailable in nonbovine species, but a fraction of milk exosomes reaches the large intestine. We hypothesized that milk exosomes alter the composition of the gut microbiome in mice. C57BL/6 mice were fed AIN-93G diets, defined by their content of bovine milk exosomes and RNA cargos: exosome/RNA-depleted (ERD) versus exosome/RNA-sufficient (ERS) diets. Feeding was initiated at age 3 wk, and cecum content was collected at ages 7, 15, and 47 wk. Microbial communities were identified by 16S rRNA gene sequencing. Milk exosomes altered bacterial communities in the murine cecum. The abundance of three phyla, seven families, and 52 operational taxonomic units (OTUs) was different in the ceca from mice fed ERD and ERS (P < 0.05). For example, at the phylum level, Tenericutes had more than threefold abundance in ERS mice at ages 15 and 47 wk compared with ERD mice (P < 0.05). At the family level, Verrucomicrobiaceae were much less abundant in ERS mice compared with ERD mice age 47 wk (P < 0.05). At the OTU level, four OTUs from the family of Lachnospiraceae were more than two times more abundant in ERS mice compared with ERD at age 7 and 47 wk (P < 0.05). We conclude that exosomes in bovine milk alter microbial communities in nonbovine species, suggesting that exosomes and their cargos participate in the crosstalk between bacterial and animal kingdoms.
NEW & NOTEWORTHY This is the first report that exosomes from bovine milk alter microbial communities in mice. This report suggests that the gut microbiome facilitates cell-to-cell communication by milk exosomes across species boundaries, and milk exosomes facilitate communication across animal and bacteria kingdoms.
INTRODUCTION
Exosomes are nanoparticles that play essential roles in cell-to-cell communication (1, 54). Exosomes transfer diverse cargos from donor cells to adjacent or distant recipient cells. Cargos include various species of RNA, proteins, and lipids. In recipient cells, exosome cargos alter gene expression and metabolism. For example, miR-30d secreted by the endometrium is taken up by the preimplantation embryo and modifies the transcriptome in human embryos (46). Recently, we made the paradigm-shifting discovery that exosomes and their RNA cargos are not exclusively derived from endogenous synthesis but can also be obtained from dietary sources (3, 26, 52). We demonstrated that human and rat intestinal cells transport exosomes from bovine milk by endocytosis and secrete microRNA cargos across the basolateral membrane (52). A similar transport mechanism operates in human vascular endothelial cells (21). Exosomes accumulate in immune cells if transferred across species boundaries (3, 17, 18). We further demonstrated that bovine milk exosomes contribute to the body pool of microRNAs in human milk feeding and murine exosome depletion studies (3, 41, 48). MicroRNAs in dietary exosomes alter gene expression across species boundaries (3). These discoveries are consistent with observations by other investigators. Ongoing phenotyping studies in our laboratory suggest that dietary depletion of bovine milk exosomes alters plasma cytokine profiles in humans and mice and causes an aberrant flux of purine metabolites in mice (2).
Despite the compelling evidence in support of the theory that milk exosomes are bioavailable, concerns have been raised that the amount of cargos, particularly microRNAs, delivered by bovine exosomes to host organisms is exceedingly low in transgenic mouse models [(22, 45), reviewed in Ref. 56]. In a recent opinion article, we suggested that, while studies of dietary microRNAs are important and warrant investigation, the controversy surrounding the field of dietary microRNAs must not impede the rate of discovery in areas such as dietary exosomes, exosome cargos other than microRNAs, and noncanonical RNA signaling pathways (55). Here, we tested the hypothesis that bovine milk exosomes alter bacterial communities in the murine cecum. This hypothesis was based on the following rationale. First, prokaryotic and eukaryotic microbes communicate with their environment through exosome-like vesicles (51). This observation includes gram-positive bacteria, which use vesicles for communication despite the cell wall posing a barrier for vesicle transport (23, 37). Viruses may participate in exosome signaling through hijacking and modifying exosomes (29). Second, up to 20% and 40% of RNA sequence reads in plasma from healthy adults map to bacterial and fungal genomes, respectively (47). Third, evidence suggests that orally administered, fluorophore-labeled exosomes from bovine milk are delivered to peripheral tissues (33). While that study lacked important controls (unlabeled exosomes, free fluorophore), its findings are largely consistent with our ongoing studies, suggesting that endogenously and exogenously labeled milk exosomes accumulate in liver and spleen, but that a considerable fraction of orally administered exosomes escapes absorption and reaches the large intestine (26).
MATERIALS AND METHODS
Mouse diets.
Exosome and RNA-depleted (ERD) and exosome and RNA-sufficient (ERS) diets are based on the AIN-93G formulation (3, 36). The milk added to the diets provided the equivalent of 0.5 liters milk consumed by a human adult per day, adjusted by body weight in mice. The milk used to prepare the powder for the ERD diet was ultrasonicated, which led to a >98% depletion of microRNA cargos in exosomes, 20% decreased in exosome count (9.1 × 1012 ± 7.1 × 1011 exosomes/mL in ERS milk vs. 7.3 × 1012 ± 3.5 × 1011 exosomes/mL in ERD milk] and a >60% decrease in intestinal exosome transport rates [(3, 24), S. Sukreet and J. Zempleni, unpublished data]. Diet ingredients other than milk were not ultrasonicated, i.e., nutrients other than exosomes and their RNA cargos were the same in ERD and ERS diets. Diets were dried and fed in pelleted form. Diets did not affect food and water consumption, feeding frequency, physical activity, and variables of liver and kidney disease (24).
Mouse feeding studies.
C57BL/6 mice (stock number 000664, Jackson Laboratories) were obtained at age 3 wk when ERS and ERD feeding was initiated. Mice were housed in groups of four mice per cage in eight cages, separated by diet and sex. Males and females were studied. True randomization of group assignment was achieved by labeling mice with numbers and randomly assigning numbers to groups. At timed intervals (ages 7, 15, and 47 wk), mice were sampled from different cages to avoid cage effects (n = 8 for each sex and age), and euthanized for sample collection. Body weight and body fat were measured by using a Scout Pro digital scale (Ohaus, Melrose, MA) and a PIXImus mouse densitometer (LUNAR, Madison, WI), respectively. Cecum content was collected, flash frozen in liquid nitrogen, and stored at −80°C. The study was approved by the Institutional Animal Care and Use Committee at the University of Nebraska-Lincoln (protocol 1229).
Bacterial growth ex vivo.
Cecum content was collected from three male and three female C57BL/6 mice under anaerobic conditions. Aliquots were suspended in medium containing M9 minimum salts (Gibco BRL, Gaithersburg, MD), 100 µM CaCl2, and 2 mM MgSO4 in a sterile, anaerobic work bench. Bovine milk exosomes were isolated from milk and authenticated by transmission electron microscopy, nanosight size analysis, and immunoblot analysis, as previously described (21, 26, 52). Half of the cultures were supplemented with sterile-filtered (0.22 µm) bovine milk exosomes (1.2 × 1012 exosomes/mL) on a daily basis, whereas the other cultures received solvent (PBS). The number of exosomes added was the equivalent of 0.5 liters milk in the total gastrointestinal water in a human adult, adjusted by the body weight of mice. Contamination controls were prepared by culturing exosomes in the same media without cecum content. All samples were incubated in an anaerobic chamber at 37°C for 7 days (n = 3 per treatment). Bacterial growth was measured using the absorbance at 600 nm in a Synergy H1 Hybrid multimode microplate reader (Bio-Tek Instruments, Winooski, VT).
Analysis of bacterial communities.
DNA was extracted from cecum content using the PowerSoil DNA isolation kit following the manufacturer’s instructions (Mo Bio Laboratories, Carlsbad, CA). DNA purity and integrity were confirmed by using the 260-to-280 nm ratio (Nanodrop ND-1000, Nanodrop Technologies, Wilmington, DE) and agarose gel (0.8%) electrophoresis. The V4 region in the 16S rRNA gene was amplified and sequenced, as described previously (19). The sequencing reads were quality filtered and analyzed, as described previously (38). Contigs were generated from paired-end reads and were screened using MOTHUR v.1.38.1 (38) to exclude low-quality sequences and reads containing ambiguous bases or homopolymers longer than eight base pairs (bp). The resulting reads were trimmed to retain only reads between 245 bp and 275 bp. The UPARSE pipeline (USEARCH, v7.0.1090) (11) was used to cluster quality-filtered sequences into operational taxonomic units (OTUs) by using a 97% identity cut-off after removal of chimeras using UCHIME (12). ChimeraSlayer gold.fa was used as the reference database for chimera detection. Sequence alignment was performed using the SILVA v123 reference and was used to build a phylogenetic tree using Clearcut (40). Taxonomy assignment (Greengenes database: gg_13_8_otus) (28) was performed using assign_taxonomy script of QIIME v.1.9.1 (5) and a cutoff value set to 97%. Genera were not annotated due to short sequencing reads; therefore, OTUs (proxy for species) were used for the lower taxonomic rank analysis. We removed 10 samples that had sequencing read counts lower than 4,000. Eighty-two samples with an average read count of 40,468 reads and a range of 4,112–144,788 reads were used for downstream analysis. Reads were normalized by using the cumulative sum of counts (34). Alpha diversity metrics were used to evaluate richness (Chao1), diversity (Shannon-Weiner index), and coverage (Good’s coverage) (5). Rarefaction curves were constructed using Chao1 values. A core measurable microbiome was identified on the basis of the factors diet, sex, and age to reduce mouse-to-mouse variation. The core measurable microbiome was defined as the group of OTUs that are present in at least 80% of the samples within each factor. Differences in bacterial communities were assessed using permutational multivariate analysis of variance (PERMANOVA), utilizing the weighted UniFrac distance matrix. Additionally, the weighted UniFrac distance matrix was used for principal coordinate analysis. The linear discriminant analysis (LDA) of effect size (LEfSe) algorithm with default parameters (α = 0.05 for Kruskal-Wallis and Wilcoxon tests) was used to identify the differences in abundance between ERS and ERD feeding groups at different ages, phyla, family, and OTU levels (39). Differences were considered statistically significant if P < 0.05 and LDA score was >2.0. Multivariate analysis by linear models (MaAsLin) with default parameters was performed to test for associations between microbial community abundance and experimental factors, such as age, sex, and diet (31). Association was considered statistically significant at an FDR corrected P < 0.05. Sequence data were deposited in the NCBI-BioProject database under accession number PRJNA413623.
Statistical analysis.
Student’s unpaired t-test was used to compare body weight and fat in mice on ERS and ERD diets and growth of murine gut bacteria in exosome-defined media ex vivo. Differences were considered statistically significant if P < 0.05.
RESULTS
Effects of ERS and ERD diets on body weight and fat.
Body weight and body fat increased transiently in ERS males compared with ERD males at age 15 wk, which was no longer observed at age 47 wk (Supplemental Table S1 on https://doi.org/10.6084/m9.figshare.9639287.v4; all supplemental material can be found on this site). Diets did not affect body weight and body fat in young males, age 7 wk, and in females.
Growth of murine gut bacteria ex vivo.
Supplementation of culture media with milk exosomes conferred a growth advantage to bacteria from male mice ex vivo compared with control cultures (Supplemental Fig. S1, P < 0.01). The apparent growth advantage of exosome-supplemented bacteria from female mice compared with solvent control was not statistically significant. Bacterial communities were not assessed in ex vivo studies, because the sole purpose of this experiment was to assess whether milk exosomes confer a growth advantage to some bacteria.
Bacterial communities.
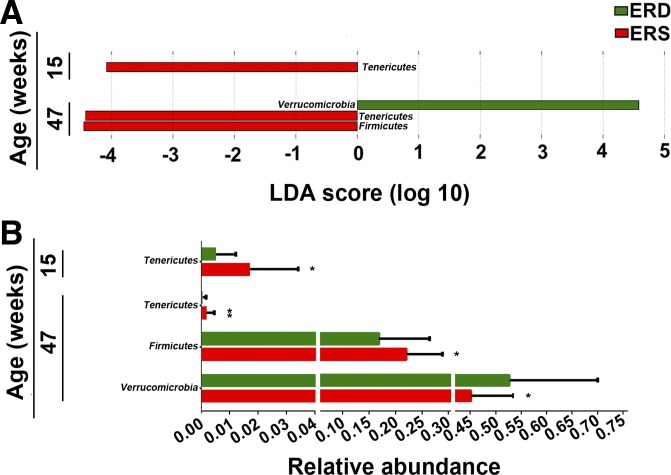
Bacterial communities varied by age with a significant diet × age × sex interaction (P = 0.039; Table 1). Sex alone had no effect on microbial communities, and therefore, sexes were combined in subsequent analyses (P = 0.215; Table 1). At the phylum level, exosome-defined diets significantly affected bacterial communities in mice at ages 15 and 47 wk (Fig. 1A). The abundance of Tenericutes was more than three times greater in ERS mice at ages 15 and 47 wk compared with ERD mice, whereas the abundance decreased in both ERS and ERD mice at age 47 wk compared with age 15 wk (P < 0.05, Fig. 1B). Also, Firmicutes were less abundant in ERD mice than ERS mice at age 47 wk (P = 0.025). The abundance of Verrucomicrobia was greater in ERD mice compared with ERS mice at age 47 wk (P = 0.025). No significant differences at the phylum level were detected at age 7 wk (Supplemental Table S2).
Table 1.
Effect of diet, age, and sex on bacterial communities in the cecum of mice
| DF | SumsOfSqs | MeanSqs | F Model | P | |
|---|---|---|---|---|---|
| Diet × age × sex | 2 | 0.004 | 0.002 | 2.376 | 0.039 |
| Diet × age | 2 | 0.003 | 0.002 | 1.950 | 0.064 |
| Diet × sex | 1 | 0.001 | 0.001 | 1.047 | 0.326 |
| Age × sex | 2 | 0.002 | 0.001 | 1.345 | 0.215 |
| Diet | 1 | 0.002 | 0.002 | 2.342 | 0.075 |
| Age | 2 | 0.065 | 0.032 | 40.297 | 0.001 |
| Sex | 1 | 0.001 | 0.001 | 1.341 | 0.215 |
| Residuals | 70 | 0.056 | 0.001 | ||
| Total | 81 | 0.134 |
DF, degree of freedom; MeanSqs = SumsOfSqs/df, mean squares; F Model, F-tests; SumsOfSqs, sum of squares; n = 8/diet group/sex/time point.
Fig. 1.
Bacterial phyla in ceca from mice fed exosome/RNA-sufficient (ERS) or exosome/RNA-depleted (ERD) diets beginning at age 3 wk. A: linear discriminant analysis (LDA) of effect size (LEfSe) analysis of phyla in ERS and ERD mice. Bars represent the LDA score. Diet effects are statistically significant for LDA scores greater than 2. B: relative abundance of the phyla in A. Values are expressed as means and SD. MaAsLin, multivariate analysis by linear models; n = 16 per diet group and time point. *P < 0.05, **P < 0.01.
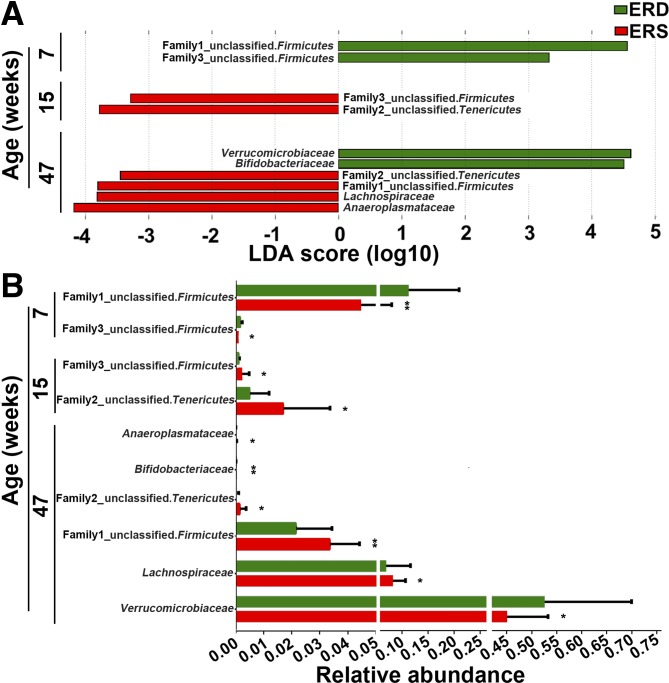
A total of 19 families were identified by 16S rRNA sequencing. Lachnospiraceae, Ruminococcaceae, and an unclassified family in the order Clostridiales (phylum Firmicutes), were the three most abundant families (Supplemental Fig. S2). Exosome-defined diets altered bacterial communities at the family level (Fig. 2). For example, two unclassified families from phylum Firmicutes were more abundant in ERD mice than in ERS mice at age 7 wk (P < 0.05). At age 15 wk, one of the two unclassified families from the phylum of Firmicutes and another unclassified family from phylum of Tenericutes were more abundant in ERS mice compared with ERD mice (P < 0.05). At age 47 wk, the family of Verrucomicrobiaceae was more abundant in ERD mice compared with ERS mice, whereas Lachnospiraceae and two unclassified families from phyla Firmicutes and Tenericutes were more abundant in ERS mice than in ERD mice (P < 0.05).
Fig. 2.
Bacterial families in ceca from mice fed exosome/RNA-sufficient (ERS) or exosome/RNA-depleted (ERD) diets beginning at age 3 wk. A: linear discriminant analysis (LDA) of effect size (LEfSe) analysis of families in ERS and ERD mice. Bars represent the LDA score. Diet effects are statistically significant for LDA scores greater than 2. B: relative abundance of the families in A. Values are expressed as means and SDs. MaAsLin, multivariate analysis by linear models; n = 16 per diet group and time point; *P < 0.05, **P < 0.01.
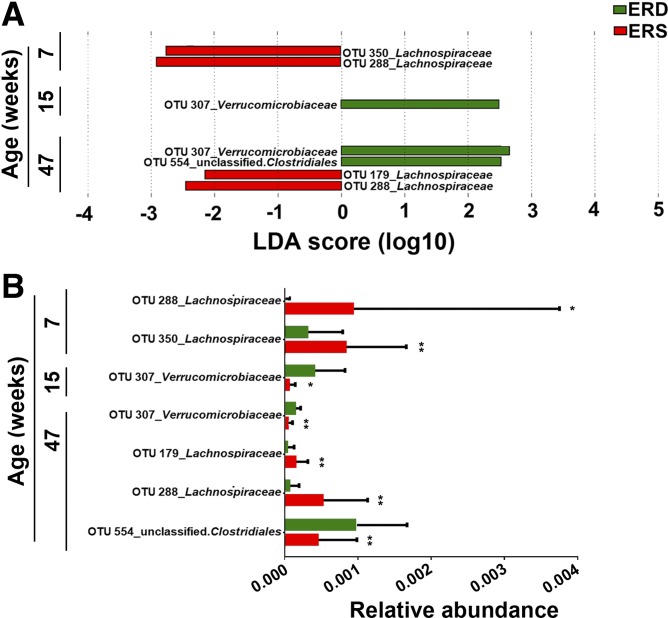
Fifty-two OTUs were different in the ceca from mice fed ERD and ERS as per LEfSe analysis (P < 0.05, Supplemental Table S3). Five of these were confirmed by MaAsLin analysis (Table 2). At age 7 wk, two OTUs from the family of Lachnospiraceae (phylum Firmicutes) were twofold higher in abundance in ERS mice compared with ERD mice (P = 0.009 and P = 0.044, respectively; Fig. 3). In contrast, the abundance of OTUs from the family Verrucomicrobiaceae (phylum Verrucomicrobia) was lower in ERS mice compared with ERD at both age 15 and 47 wk (P < 0.05). Another two OTUs from family Lachnospiraceae were significantly more abundant in ERS mice compared with ERD mice at age 47 wk. Analysis by MaAsLin suggested that 8 and 199 OTUs were associated with sex and age, respectively (Supplemental Table S4).
Table 2.
Associations between diet and OTU abundance in murine ceca
| OTU | Family | Diets | Coefficient | P |
|---|---|---|---|---|
| 307 | Verrucomicrobiaceae | ERD vs. ERS | 0.005 | 0.003 |
| 288 | Lachnospiraceae | ERD vs. ERS | −0.005 | 0.012 |
| 179 | Lachnospiraceae | ERD vs. ERS | −0.002 | 0.023 |
| 554 | o Clostridiales | ERD vs. ERS | 0.006 | 0.035 |
| 350 | Lachnospiraceae | ERD vs. ERS | −0.007 | 0.038 |
A positive coefficient indicates higher relative abundance in exosome and RNA-depleted (ERD) diet and a negative coefficient indicates a lower relative abundance in ERD. ERS, exosome and RNA-sufficient; o, order; OTU, operational taxonomic unit; P, FDR adjusted P value; n = 16/diet group/time point. Multivariate analysis was performed using linear models (MaAsLin).
Fig. 3.
Bacterial operational taxonomic units (OTUs) in ceca from mice fed exosome/RNA-sufficient (ERS) or exosome/RNA-depleted (ERD) diets beginning at age 3 wk. A: linear discriminant analysis (LDA) of effect size (LEfSe) analysis of OTUs associated with diets. The bar column represents the LDA score. Diet effects are statistically significant for LDA scores greater than 2. B: relative abundance of OTUs in A. Values are means and SDs. MaAsLin, multivariate analysis by linear models; n = 16 per diet group and time point; *P < 0.05, **P < 0.01.
DISCUSSION
This paper provides novel insights in the following areas of research. First, this paper provides strong evidence that dietary exosomes, at least those in milk, might be responsible for some of the effects of diet on the gut microbiome (25, 53). To the best of our knowledge, the only evidence that bovine milk compounds alter the microbiome comes from a study reporting that consumption of casein protein alters bacterial communities in the gut of rats (58). A previous paper suggests that exosome-like nanoparticles from ginger alter microbial communities in mice, but it is safe to assume that the consumption of milk by adults and milk and formulas by infants is quantitatively greater than the consumption of ginger in most societies (8, 44, 50). Also, in this study, we administered a nutritionally relevant number of exosomes to mice, whereas the dose of ginger-derived nanoparticles given to mice was the equivalent of 39 kg of ginger consumed by a human adult (44).
Second, this paper suggests that gut microorganisms might act as transmitters or amplifiers of dietary exosome signals. Following early reports that microRNA cargos in dietary exosomes might be bioavailable, it was proposed that dietary microRNAs achieve tissue concentrations that are too low to elicit biological effects and that the bioavailability of milk exosomes might be low (10, 22, 42, 45). Here we provide evidence that exosome-defined diets alter bacterial communities in the murine cecum, and the effects are particularly strong if studied at the OTU level. Importantly, this article suggests that milk exosomes that escape absorption by mucosal cells may still elicit major biological effects, facilitated by the gut microbiome. It is now widely accepted that prokaryotic and eukaryotic microbes communicate with their environment through exosome-like vesicles (23, 37, 51). It is reasonable to speculate that changes in bacterial communities are paralleled by changes in the production of microbial metabolites, which may transmit and amplify milk exosome signals (49).
Third, this paper provides evidence to suggest that sex influences bacterial growth ex vivo in cultures from male mice, but not in cultures from female mice. The factors mediating sex effects are unknown. We speculate that milk exosomes conferred a growth advantage to a quantitatively minor species of bacteria that escaped detection in 16S rRNA sequencing of gut content in vivo, which then outgrew other bacteria ex vivo. We interpret this observation with due caution and consider the possibility that sex of the donor may not affect microbial growth ex vivo.
This study does not establish a causal relation between RNA cargos in milk exosomes and changes in bacterial communities in the murine gut. Ever since a publication in 2012 suggested that microRNAs in rice are bioavailable and alter gene expression in animals, much of the attention has focused on microRNA cargos in exosomes [(57), reviewed in Ref. 56]. It is tempting to give in to this speculation, because microRNAs regulate more than 60% of human genes, milk exosomes contain up to 700 species of microRNAs with nucleotide sequences identical to those in humans, and microRNAs encapsulated in milk exosomes are bioavailable (3, 16, 20, 26, 43). Crude estimates of the bioavailability of microRNAs in milk exosomes suggest that 25% the microRNAs are absorbed, suggesting that the unabsorbed fraction may interact with gut microbes (26). This being said, there is experimental evidence that both microRNAs and exosomes may affect cell signaling. For example, microRNAs elicit biological effects through binding to Toll-like receptors, whereas mere exosome-cell surface interactions activate signaling by mitogen activated kinases in human myeloma cell lines and human T-cell subsets (6, 14, 32, 35). Future studies will need to determine whether effects of dietary exosomes on bacterial communities are caused by exosomes, microRNA cargos, or cargos other than microRNAs.
The bacterial communities altered by milk exosome-defined diets are related to pathological and physiological conditions, as evidenced by the following examples. A loss of Lachnospiraceae in the ileal mucosa has been implicated in inflammatory bowel disease (4, 15). The proportion of Firmicutes and Tenericutes was lower in cecal content of obese diabetic mice compared with controls (13). Dysbiosis in the gut microbiome may cause liver disease due to microbial metabolites altering the metabolism in hepatic cells (27), e.g., a decreased abundance of Verrucomicrobia has been linked with nonalcoholic fatty liver disease (30). Inflammatory bowel disease, obesity, and nonalcoholic fatty liver disease are major health concerns in the United States (7, 9).
The functional consequences of exosome-induced changes in bacterial communities in the gut, if any, and the mechanisms by which these changes contribute phenotypes of health and disease are unknown. We have recently completed a one-year mouse feeding trial to assess phenotypes associated with feeding milk exosome-defined diets to mice. To date, we have published the following phenotypes. First, hepatic concentrations of purine metabolites were higher in ERD mice compared with ERS mice, and similar increases were observed in the urine from infants fed soy formula compared with human milk or milk formula, and in adult dairy avoiders compared with dairy consumers (2). Note that pathways in purine metabolism were among those identified in the metagenomics studies described above. Second, the muscle grip strength was moderately lower in ERD mice compared with ERS mice (24). It will be interesting to test which of these, and other phenotypes depend on the presence of gut microbiota by comparing germ-free and conventional mice or conducting fecal transplant studies.
GRANTS
This work was supported by the National Institute of Food and Agriculture, US Department of Agriculture (USDA), under award numbers 2015-67017-23181 and 2016-67001-25301; National Institutes of Health Grant 1P20GM104320; the University of Nebraska Agricultural Research Division (Hatch Act) and USDA multistate groups W3002 and W4002 (all to J. Zempleni); a Food for Health grant by the University of Nebraska President’s Office (J. Cui and J. Zempleni); a grant from the University of Nebraska Agricultural Research Division (Hatch Act), USDA Agricultural Research Station, United States Meat Animal Research Center (collaborative funding program); and USDA multistate group Grant W2010 (all to S. Fernando).
DISCLOSURES
J. Zempleni is a consultant for PureTech Health in Boston, MA. S. Fernando is a co-owner of NuGUT LLC. The other authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
J.Z. conceived and designed research; F.Z., H.A.P., and M.S. performed experiments; F.Z., H.A.P., J.C., S.D.K., and S.C.F. analyzed data; J.Z. interpreted results of experiments; J.Z. prepared figures; F.Z. drafted manuscript; S.D.K., S.C.F., and J.Z. edited and revised manuscript; F.Z., H.A.P., M.S., J.C., S.D.K., S.C.F., and J.Z. approved final version of manuscript.
ACKNOWLEDGMENTS
We acknowledge the support by the Biomedical and Obesity Research Core in the Nebraska Center for the Prevention of Obesity Diseases through Dietary Molecules (National Institutes of Health Grant 1P20GM104320) and the Holland Computing Center, both at the University of Nebraska-Lincoln. The authors thank Dr. Jiang Shu (Department of Computer Science and Engineering, University of Nebraska-Lincoln) for help with preliminary data analysis.
REFERENCES
- 1.Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo Selection, content, release, and uptake. Cell Mol Neurobiol 36: 301–312, 2016. doi: 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar-Lozano A, Baier S, Grove R, Shu J, Giraud D, Leiferman A, Mercer KE, Cui J, Badger TM, Adamec J, Andres A, Zempleni J. Concentrations of purine metabolites are elevated in fluids from adults and infants and in livers from mice fed diets depleted of bovine milk exosomes and their RNA cargos. J Nutr 148: 1886–1894, 2018. doi: 10.1093/jn/nxy223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J Nutr 144: 1495–1500, 2014. doi: 10.3945/jn.114.196436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, Orsi RH, Wiedmann M, McDonough P, Kim SG, Berg D, Schukken Y, Scherl E, Simpson KW. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J 1: 403–418, 2007. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 5.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336, 2010. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavalieri D, Rizzetto L, Tocci N, Rivero D, Asquini E, Si-Ammour A, Bonechi E, Ballerini C, Viola R. Plant microRNAs as novel immunomodulatory agents. Sci Rep 6: 25761, 2016. doi: 10.1038/srep25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention Inflammatory bowel disease. https://www.cdc.gov/ibd/, 2011.
- 8.Centers for Disease Control and Prevention Breastfeeding among U.S. children born 2002–2012, CDC national immunization surveys. https://www.cdc.gov/breastfeeding/index.htm/, 2015.
- 9.Centers for Disease Control and Prevention Obesity and overweight. https://www.cdc.gov/nchs/fastats/obesity-overweight.htm/, 2015.
- 10.Dickinson B, Zhang Y, Petrick JS, Heck G, Ivashuta S, Marshall WS. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol 31: 965–967, 2013. doi: 10.1038/nbt.2737. [DOI] [PubMed] [Google Scholar]
- 11.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998, 2013. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 12.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200, 2011. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everard A, Matamoros S, Geurts L, Delzenne NM, Cani PD. Saccharomyces boulardii administration changes gut microbiota and reduces hepatic steatosis, low-grade inflammation, and fat mass in obese and type 2 diabetic db/db mice. MBio 5: e01011–e01014, 2014. doi: 10.1128/mBio.01011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA 109: E2110–E2116, 2012. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frank DN, St. Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104: 13780–13785, 2007. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105, 2009. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imai T, Takahashi Y, Nishikawa M, Kato K, Morishita M, Yamashita T, Matsumoto A, Charoenviriyakul C, Takakura Y. Macrophage-dependent clearance of systemically administered B16BL6-derived exosomes from the blood circulation in mice. J Extracell Vesicles 4: 26238, 2015. doi: 10.3402/jev.v4.26238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izumi H, Tsuda M, Sato Y, Kosaka N, Ochiya T, Iwamoto H, Namba K, Takeda Y. Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. J Dairy Sci 98: 2920–2933, 2015. doi: 10.3168/jds.2014-9076. [DOI] [PubMed] [Google Scholar]
- 19.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79: 5112–5120, 2013. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73, 2014. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kusuma RJ, Manca S, Friemel T, Sukreet S, Nguyen C, Zempleni J. Human vascular endothelial cells transport foreign exosomes from cow’s milk by endocytosis. Am J Physiol Cell Physiol 310: C800–C807, 2016. doi: 10.1152/ajpcell.00169.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laubier J, Castille J, Le Guillou S, Le Provost F. No effect of an elevated miR-30b level in mouse milk on its level in pup tissues. RNA Biol 12: 26–29, 2015. doi: 10.1080/15476286.2015.1017212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J, Lee EY, Kim SH, Kim DK, Park KS, Kim KP, Kim YK, Roh TY, Gho YS. Staphylococcus aureus extracellular vesicles carry biologically active β-lactamase. Antimicrob Agents Chemother 57: 2589–2595, 2013. doi: 10.1128/AAC.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leiferman A, Shu J, Grove R, Cui J, Adamec J, Zempleni J. A diet defined by its content of bovine milk exosomes and their RNA cargos has moderate effects on gene expression, amino acid profiles and grip strength in skeletal muscle in C57BL/6 mice. J Nutr Biochem 59: 123–128, 2018. doi: 10.1016/j.jnutbio.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature 444: 1022–1023, 2006. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 26.Manca S, Upadhyaya B, Mutai E, Desaulniers AT, Cederberg RA, White BR, Zempleni J. Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci Rep 8: 11321, 2018. doi: 10.1038/s41598-018-29780-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, Thomas LV, Zoetendal EG, Hart A. The gut microbiota and host health: a new clinical frontier. Gut 65: 330–339, 2016. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618, 2012. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meckes DG., Jr Exosomal communication goes viral. J Virol 89: 5200–5203, 2015. doi: 10.1128/JVI.02470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michail S, Lin M, Frey MR, Fanter R, Paliy O, Hilbush B, Reo NV. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol Ecol 91: 1–9, 2015. doi: 10.1093/femsec/fiu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13: R79, 2012. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller L, Mitsuhashi M, Simms P, Gooding WE, Whiteside TL. Tumor-derived exosomes regulate expression of immune function-related genes in human T cell subsets. Sci Rep 6: 20254, 2016. doi: 10.1038/srep20254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munagala R, Aqil F, Jeyabalan J, Gupta RC. Bovine milk-derived exosomes for drug delivery. Cancer Lett 371: 48–61, 2016. doi: 10.1016/j.canlet.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods 10: 1200–1202, 2013. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purushothaman A, Bandari SK, Liu J, Mobley JA, Brown EE, Sanderson RD. Fibronectin on the surface of myeloma cell-derived exosomes mediates exosome-cell interactions. J Biol Chem 291: 1652–1663, 2016. doi: 10.1074/jbc.M115.686295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reeves PG, Nielsen FH, Fahey GC Jr. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr 123: 1939–1951, 1993. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- 37.Rivera J, Cordero RJ, Nakouzi AS, Frases S, Nicola A, Casadevall A. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc Natl Acad Sci USA 107: 19002–19007, 2010. doi: 10.1073/pnas.1008843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541, 2009. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol 12: R60, 2011. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheneman L, Evans J, Foster JA. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics 22: 2823–2824, 2006. doi: 10.1093/bioinformatics/btl478. [DOI] [PubMed] [Google Scholar]
- 41.Shu J, Chiang K, Zempleni J, Cui J. Computational characterization of exogenous microRNAs that can be transferred into human circulation. PLoS One 10: e0140587, 2015. doi: 10.1371/journal.pone.0140587. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Snow JW, Hale AE, Isaacs SK, Baggish AL, Chan SY. Ineffective delivery of diet-derived microRNAs to recipient animal organisms. RNA Biol 10: 1107–1116, 2013. doi: 10.4161/rna.24909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun J, Aswath K, Schroeder SG, Lippolis JD, Reinhardt TA, Sonstegard TS. MicroRNA expression profiles of bovine milk exosomes in response to Staphylococcus aureus infection. BMC Genomics 16: 806, 2015. doi: 10.1186/s12864-015-2044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teng Y, Ren Y, Sayed M, Hu X, Lei C, Kumar A, Hutchins E, Mu J, Deng Z, Luo C, Sundaram K, Sriwastva MK, Zhang L, Hsieh M, Reiman R, Haribabu B, Yan J, Jala VR, Miller DM, Van Keuren-Jensen K, Merchant ML, McClain CJ, Park JW, Egilmez NK, Zhang HG. Plant-derived exosomal microRNAs shape the gut microbiota. Cell Host Microbe 24: 637–652.e8, 2018. doi: 10.1016/j.chom.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Title AC, Denzler R, Stoffel M. Uptake and function studies of maternal milk-derived microRNAs. J Biol Chem 290: 23680–23691, 2015. doi: 10.1074/jbc.M115.676734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vilella F, Moreno-Moya JM, Balaguer N, Grasso A, Herrero M, Martínez S, Marcilla A, Simón C. Hsa-miR-30d, secreted by the human endometrium, is taken up by the pre-implantation embryo and might modify its transcriptome. Development 142: 3210–3221, 2015. doi: 10.1242/dev.124289. [DOI] [PubMed] [Google Scholar]
- 47.Wang K, Li H, Yuan Y, Etheridge A, Zhou Y, Huang D, Wilmes P, Galas D. The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PLoS One 7: e51009, 2012. doi: 10.1371/journal.pone.0051009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Sadri M, Giraud D, Zempleni J. RNase H2-dependent polymerase chain reaction and elimination of confounders in sample collection, storage, and analysis strengthen evidence that microRNAs in bovine milk are bioavailable in humans. J Nutr 148: 153–159, 2018. doi: 10.1093/jn/nxx024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA 106: 3698–3703, 2009. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wisconsin Milk Marketing Board, Inc. Dairy Statistics. Madison, WI, 2013, https://media.eatwisconsincheese.com/dairyimpact/statistics/dairystatistics.aspx. [Google Scholar]
- 51.Wolf JM, Casadevall A. Challenges posed by extracellular vesicles from eukaryotic microbes. Curr Opin Microbiol 22: 73–78, 2014. doi: 10.1016/j.mib.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolf T, Baier SR, Zempleni J. The intestinal transport of bovine milk exosomes is mediated by endocytosis in human colon carcinoma caco-2 cells and rat small intestinal IEC-6 cells. J Nutr 145: 2201–2206, 2015. doi: 10.3945/jn.115.218586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science 334: 105–108, 2011. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yáñez-Mó M, Siljander PR, Andreu Z, Bedina Zavec A, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colás E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Krämer-Albers EM, Laitinen S, Lässer C, Lener T, Ligeti E, Linē A, Lipps G, Llorente A, Lötvall J, Manček-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-’t Hoen EN, Nyman TA, O’Driscoll L, Olivan M, Oliveira C, Pállinger É, Del Portillo HA, Reventós J, Rigau M, Rohde E, Sammar M, Sánchez-Madrid F, Santarém N, Schallmoser K, Stampe Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MH, De Wever O. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 4: 27066, 2015. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zempleni J. Milk exosomes: beyond dietary microRNAs. Genes Nutr 12: 12, 2017. doi: 10.1186/s12263-017-0562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zempleni J, Aguilar-Lozano A, Sadri M, Sukreet S, Manca S, Wu D, Zhou F, Mutai E. Biological activities of extracellular vesicles and their cargos from bovine and human milk in humans and implications for infants. J Nutr 147: 3–10, 2017. doi: 10.3945/jn.116.238949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, Li J, Bian Z, Liang X, Cai X, Yin Y, Wang C, Zhang T, Zhu D, Zhang D, Xu J, Chen Q, Ba Y, Liu J, Wang Q, Chen J, Wang J, Wang M, Zhang Q, Zhang J, Zen K, Zhang CY. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res 22: 107–126, 2012. [Erratum in Cell Res 22: 273-274, 2012.] doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu Y, Lin X, Zhao F, Shi X, Li H, Li Y, Zhu W, Xu X, Li C, Zhou G. Meat, dairy and plant proteins alter bacterial composition of rat gut bacteria. Sci Rep 5: 15220, 2015. doi: 10.1038/srep15220. [DOI] [PMC free article] [PubMed] [Google Scholar]