Abstract
Alveolar type I (TI) cells are large squamous cells that cover >95% of the internal surface area of the lung; type II (TII) cells are small cuboidal cells with distinctive intracellular surfactant storage organelles. Based on autoradiographic studies in the 1970s, the long-held paradigm of alveolar epithelial development has been a linear progression from undifferentiated progenitor cells through TII cells to TI cells. Subsequent data support the existence of more complex pathways. Recently, a bipotent TI/TII progenitor cell at embryonic day E18 has been inferred both from marker expression in developing airways and from statistical analyses of gene expression data obtained from single-lung embryonic cells. To study cell lineage directly by fate mapping, we developed new transgenic mouse models in which rtTA is driven either by the rat podoplanin or the mouse Sftpc gene to mark cells irreversibly in development. Using these models, we found two distinct lineage pathways. One pathway, evident as early as E12–15, is devoted almost exclusively to TI cell development; a second pathway gives rise predominantly to TII cells but also a subpopulation of TI cells. We have defined the molecular phenotypes of these distinct progenitor populations and have identified potential regulatory factors in TI and TII cell differentiation. By analyzing gene pathways in mature TI and TII cells, we identified potential novel functions of each cell type. These results provide novel insights into lung development and suggest a basis for testing strategies to promote alveolar differentiation and repair, including potential transplantation of lineage-specific progenitor cells.
Keywords: alveolar epithelium, alveolar functions, lung development
INTRODUCTION
The highly specialized pulmonary alveolar epithelium, composed of TI and TII cells, is critical for normal lung function. Anatomically, the epithelium provides a short diffusion pathway for gases. Functionally, the alveolar epithelium prevents air space flooding and collapse, forms a first-line immunologic defense in the lung, and participates in lung repair after injury. Both TI and TII cells are believed to be essential for normal lung function. TI cells are very large squamous cells with calculated diameters of 50–100 μm and volumes of 3,000 μm3 (30). The thin cytoplasmic extensions of TI cells cover 95–98% of the internal surface area of the lung, providing a narrow anatomic barrier between the air and blood compartments critical for efficient gas exchange. The remainder of the alveolar surface is composed of TII cells, smaller, cuboidal cells (diameter ~10 μm) characterized morphologically by distinctive surfactant secretory granules called lamellar bodies. TI cells play an important role in water (10) transport; TII cells are the source of pulmonary surfactant, synthesizing, secreting, and recycling surfactant components (38). Both cell types transport ions (8, 11, 19), perform immunomodulatory functions (36, 37), and act as progenitor cells following injury (2, 14, 18).
Based on autoradiographic data, a long-held paradigm has been that pulmonary alveolar epithelial development occurred by a linear progression from undifferentiated progenitor cells to alveolar epithelial type II cells, some of which then matured into type I cells (1). However, other data suggest more complex pathways in development. Morphologically undifferentiated epithelial cells co-express markers typical of several different types of mature epithelial cells (25, 39), evidence against a simple linear progression. In the last few years, marker analysis in sacculating airways (9) and statistical analyses of RNA sequencing of single-lung cells at different stages of mouse development (31) have been used to infer developmental and lineage hierarchies of distal lung epithelium. The results of these studies suggested that the TI and TII cell lineages emerge from a common progenitor cell observed at embryonic day (E) 18.5 displaying markers of both cell types, after which the two lineages diverge. It has been postulated that these bipotent progenitors generate most, if not all, TI and TII cells in development (31). However, co-expression of markers is not sufficient to define an exclusive cell lineage. In earlier development, epithelial cells co-express multiple markers of mature differentiated cells, from which one cannot necessarily infer the fates of these cells. Furthermore, it is unclear whether a progenitor cell expressing markers of two distinct mature cell types produces equal numbers of each cell type, or unequal numbers, such as 99% of one type and 1% of the other. It is also possible that some cells co-expressing TI and TII cell markers might mature into cell types other than TI or TII cells.
A more traditional approach to cell lineage analysis is to label irreversibly various progenitor cell populations and to trace these marked cells during development. The human SP-C promoter has been used in lineage analysis of alveolar epithelium (27), although the interpretation of some studies is complicated because expression of the human promoter used for these studies occurs as early as E0–E2.5, before the development of the lung buds. We have previously used bacterial artificial chromosomes (BACs) to introduce large regions of genes (33, 34) that direct expression to TI or TII cells in mice. We have now used this strategy to produce novel transgenic mouse models that are useful for cell lineage analysis and gene targeting of TI and TII cells.
The RT22-rtTA line expresses rtTA under control of the rat podoplanin (Pdpn) gene. Line 114 [Sftpc (surfactant protein C)-rtTA] expresses rtTA under control of the mouse Sftpc gene. By crossing each of these lines to both tet-O-Cre- and to reporter mice, we have irreversibly marked cells during development. We have identified two distinct progenitor populations that are identifiable as early as E12–15. One progenitor population, defined in the RT22 line, develops almost exclusively into TI cells. The other population, defined in the 114 line, differentiates mostly into TII cells, and a subpopulation of TI cells. We isolated these progenitor cell populations. By transcriptome analysis, the molecular phenotypes of the pre-TI cells are very similar, whether in the 114 or the R22 lineages, and both are different from the pre-TII cell phenotype. We identified candidate regulatory pathways in late-alveolar epithelial differentiation for development of the TI cell phenotype. We also analyzed TI and TII cells from postnatal (PN) 4 wk mice to identify functional gene pathways and marker genes for each cell phenotype. These transcriptome analyses point to specific factors regulating TI cell differentiation and suggest previously unrecognized functions of both cell types.
MATERIALS AND METHODS
Description of transgenic mice, construction, and breeding.
The goal was to produce mice expressing rtTA under the control of either the mouse SP-C promoter or the rat podoplanin promoter and to cross these mice with tet-O-Cre and a reporter line so that we could irreversibly label cells in both podoplanin and SP-C lineages for the purposes of fate mapping.
We chose the rat podoplanin gene to direct rtTA because of previous work suggesting that the rat promoter is more specific to TI cells than is the mouse podoplanin promoter (33), and we used the mouse Sftpc gene to direct expression to TII cells, as described previously (34). Transgenic lines RT22 and 114 were produced with BACs modified to express rtTA. The starting BACs (BAC RP23–247J9, BACPAC Resources Center, CHORI, Oakland, CA; and RT1bac Clone 240G15, Genome Systems, St. Louis, MO) and the modification process have been described previously (33, 34). rtTA sequences from pOPRTTA (32) were inserted downstream of the internal ribosome entry site elements in the two BACs, replacing the enhanced green fluorescent protein coding sequences described in the previous modifications. Founder lines identified by PCR of tail clip DNA by using primers for rtTA were crossed to tet-O7-CRE mice (no. 006234, Jackson Laboratory, Bar Harbor, ME) to produce doubly transgenic mice for test crossing with the reporter line Ai9 (24). The Ai9 reporter mouse has a floxed STOP cassette flanking TdTomato knocked in at a modified ROSA 26 locus; after recombination, TdTomato is expressed. Dams in these crosses were either given doxycycline from before conception to weaning or were maintained on normal chow. Offspring were genotyped by PCR. Tail clips were homogenized for 30 s by using a Polytron homogenizer on setting 6 at 4°C (Brinkmann, Fisher Scientific, Pittsburgh, PA), genomic DNA was extracted using a QiaAmp Fast DNA Tissue Kit (Qiagen, Valencia, CA), and 0.1 µg of tail clip DNA was assayed for the presence of Ai9, rtTA, or CRE by PCR using standard conditions (35 cycles annealed at 61°C). Primer sequences were as follows: for rtTA forward: 5′-CATCGCAATGGAGCAAAAGTG-3′ and reverse 5′-GCAGCAGTGGAGGCATACTATCAG-3′, for CRE forward 5′-AGAGACGGAAATCCATCGCTCG-3′ and reverse 5′-CTGCCACGACCAAGTGACAGCAATG-3′, for RT1bac forward 5′-GCAGGCTGTGACTATTGTTTACGC-3′ and reverse 5′-GAAAGCAAGCACAACGCCC-3′, for Ai9 forward 5′-GGCATTAAAGCAGCGTATCC-3′ and reverse 5′-CTGTTCCTGTACGGCATGG-3′.
Mice were bred and treated in accordance with the University of California, San Francisco (UCSF), Animal Policies under Institutional Animal Care and Use Committee (IACUC) approval.
Lines 114 and R22 were backcrossed to BL6 for five generations, and then crossed with tet-O-Cre mice (no. 006234, Jackson Laboratory); 114/tet-O-Cre mice were bred to homozygosity. Although we attempted to breed the R22/tet-O-Cre mice to homozygosity, homozygosity was not achieved with this line.
Ai9 reporter mice (24) contain a floxed STOP cassette flanking TdTomato knocked in at a modified ROSA 26 locus; after recombination, TdTomato is expressed. Ai9 mice were maintained as homozygous on the BL6 background and bred to the 114/tet-O-Cre and R22/tet-O-Cre lines.
Mice were housed in the UCSF animal care facility and treated according to approved procedures in the IACUC protocol. For timed matings, E0 was timed as the morning the vaginal plug was first seen. Doxycycline was administered in chow, Bio Serv (San Diego, CA), product no. S4107, doxycycline 600 mg/kg, 1/2” pellets, stored refrigerated in the dark.
Tissue preparation and imaging.
Mice were anesthetized with ketamine-xylazine as per UCSF IACUC protocol. The lungs were removed and fixed by inflation with a solution of 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4, and then submerged in the same fixative for 20 min, minced into ~2-mm cubes that were fixed for 4 h, cryoprotected in 30% sucrose/PBS, followed by OCT for 30 min, and stored at −80°C. Three-micrometer cryosections (Leica CM1850 cryostat) were stained with antibodies [anti-prosurfactant protein C (cat. no. AB3786, Millipore Antibodies); anti-mouse Pdpn (Developmental Studies Hybridoma Bank 8.1.1, University of Iowa), rabbit anti-mouse aquaporin 5 (AQP5-005, Almone Laboratories, Jerusalem, Israel); goat anti-rabbit Alexa 647λ (A 32344, Life Technology); Goat anti-Hamster 488λ (A21110, Life Technology)], rinsed with phosphate buffered saline containing 1% bovine serum albumin (A503, Sigma) and 0.3% Triton X 100 (no. 22140, Electron Microscopy Sciences, Hatfield, PA) for 15 min, and coverslipped with DAPI mounting media. Control sera (for pro-SPC, Rabbit serum, Sigma R4505; for mouse Pdpn, Syrian hamster IgG, 007-000-002, Jackson ImmunoResearch) were diluted to the same concentration as the primary antibodies and incubated on the sections exactly as the antibodies.
Images were collected with a Leica DM 5000B Orthoplan or Leica Orthoplan microscope as described previously. Leica 506507 HCX PL Fluotar 10×/0.30 PH1, Leica 506506 HCX PL Fluotar 20×/0.50 PH2, and Leica 506145 HCX PL Fluotar 40×/0.75 PH23 objectives were used. Filter blocks used included the following: Y5 ET – Ex: BP 620/60 nm, Dichromatic filter: 660 nm, Em: BP 700/75 nm; TX2 ET – Ex.: BP 560/40 nm, Dichromatic filter: 595 nm, Em. BP645/75 nm; L5 ET – Ex. BP 480/40 nm, Dichromatic filter: 505 nm, Em. BP 527/30 nm DAPI ET – Ex: BP 360/40 nm, Dichromatic filter: 400 nm, Em. BP 470/40 nm. Images were acquired with a LEICA DFC3000 camera and Leica LAS AF v4.0 software (Leica Acquisition Suite). Images are 1,298 × 966 pixels. Images from the ×10 objective were 882.95 microns × 657.95 microns = resolution 1.362 microns; ×20 objective, 441.48 µm × 328.98 µm = resolution 0.817 µm; ×40 objective, 220.75 µm × 164.49 µm = resolution 0.545 µm. Widefield imaging was performed as described previously (34).
Number of replicates.
The number of animals used from each line in each time point window is referred to by “n.” For mice examined at PN 4 wk, “n” refers to different individual mice; for fetal samples, “n” represents the number of different litters used for each condition.
Isolation of TdTomato+/ Pdpn+, TdTomato+/ Pdpn−, TdTomato−/ Pdpn−, and TdTomato−/ Pdpn− cells by FACS.
Tail clips and lungs were collected from E18 fetal pups of both 114 and R22 lines that had received doxycycline from E15–18. Tail clips were immediately frozen and stored at −70°C for subsequent genotype analysis (see below). All fetal lungs were checked for TdTomato expression by observing lung fluorescence using a Nikon TE300 inverted microscope equipped with a low-power objective and fluorescent capability; TdTomato-negative lungs were discarded. All the fetuses examined from line 114 litters had TdTomato+ lungs, as expected because line 114/Cre is homozygous for both genes. Because R22 mice are not homozygous, R22 litters varied between none to all the pups being TdTomato+ (with an average of four TdTomato+ lungs per litter, n = 31 litters).
For each litter, TdTomato+ lungs were pooled, submerged in 3 mL of RPMI1640-Hepes (RH), minced with sharp dissecting scissors until fragments were 1 mm3, and washed three times with 40 mL of media by allowing the fragments to settle in a 50-mL tube containing RH and discarding the wash media. After the final wash, 2 ml of a solution of elastase (20 mg 2× crystallized elastase, NJ/8 ml RH, Worthington, Lakewood) was added, and the fragments were incubated in a 37°C water bath. After 15, 30, and 45 min of incubation, 2 mL of fresh elastase solution was added, and fragments were minced 40 additional times, resulting in a final suspension consisting of single cells and undigested fibrous tissue. After an additional 5-min incubation, 0.1 mL of DNase (2 mg/mL RH; Sigma, St. Louis, MO) and 2 mL of fetal bovine serum (Hyclone FBS; Cell Culture Facility, UCSF) were added, the cell suspension was triturated 10 times with a “large orifice” 1-mL pipet tip (cat. no. 02-707-145, Fisher, Pittsburgh, PA) to liberate single cells from aggregates. Single cells were separated from cell clumps by successive filtration through 70- and 20-µm nylon mesh (Fisher Cell Strainers), centrifuged at 150 g for 12 min at 4°C, and suspended in 0.2 mL RH containing 0.05 mL DNase.
For FACS, cells were sorted for the expression of TdTomato and Pdpn. The purpose of FACS was to isolate cells for subsequent gene expression profiling. We harvested half of each density cloud with the higher fluorescent magnitude in an effort to optimize differences between the cell types we were comparing. The cells varied considerably in size and in intensity of expression of fluorophores. We made the assumption that selecting cells expressing greater amounts of phenotypic specific antigens might maximize the chance of achieving homogeneous populations of cells. Because we were unable to “rerun” the collected cells because of the low numbers collected, this served to increase the purity of each respective sample.
Cells were labeled with anti-Pdpn primary antibody (Hybridoma Bank University Iowa clone 8.1.1; Iowa City, Iowa) (1:500) for 15 min, at 22°C, followed by washing in 10 mL RH, and centrifuging at 150 g, 4°C for 12 min. The cell pellet was then suspended in 0.2 mL RH, anti-hamster Alexa-488 (1:500, Life Science/Invitrogen, Carlsbad, CA) was added, incubated at 22°C for 15 min, and the washing procedure was repeated. Cell pellets were suspended in 2.0 ml RH containing 1% FCS and 50 µg DNase/mL. Cells were sorted using a FACS Aria (BD Biosciences, San Jose, CA), and results were analyzed using BD Diva software.
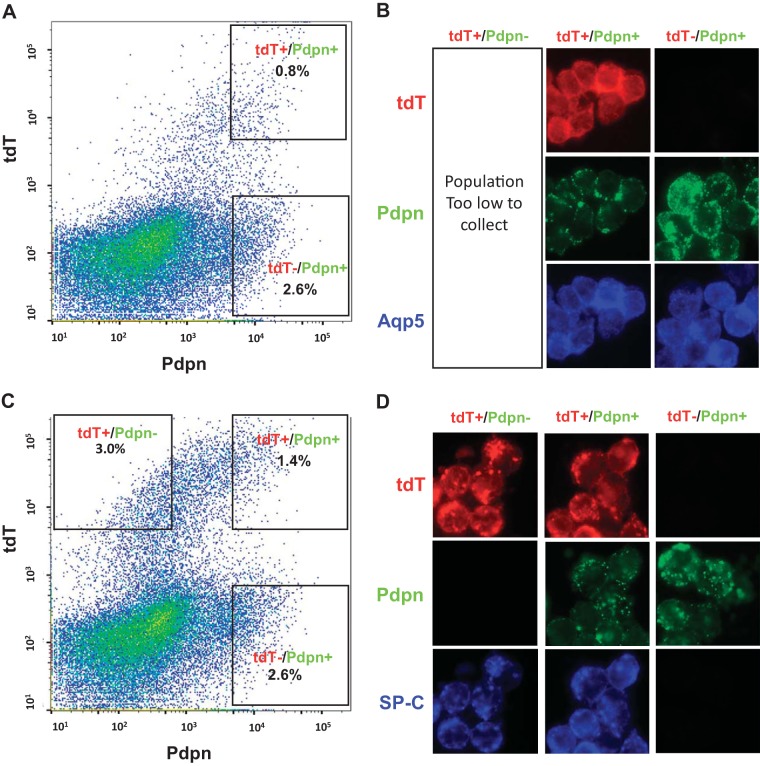
Aliquots from freshly isolated cell populations were deposited onto glass slides by using a Shandon Cytospin (Thermo Fisher Scientific, Waltham, MA). Samples were fixed overnight in freshly prepared 4% paraformaldehyde in PBS at 4°C. Cells were permeabilized by exposure to 0.5% Triton X-100 in PBS for 30 min, and differentially stained with anti-pro-SPC and anti-aquaporin 5 (see Tissue preparation and imaging section). Scattergrams and cytospin results are shown in Fig. 4.
Fig. 4.
Scattergrams and cytocentrifuged preparations of FACS E18 cells in 114 and R22 lineages. In both 114 and R22 lineages, doxycycline (Dox) was administered E15–18, and cells were harvested at E18. A and B: R22E18 cells. A: scattergram showing FACS of R22 E18 cells. There were insufficient numbers of TdTomato+/Pdpn− cells to isolate by these methods. B: cytocentrifuged preparations of each group of R22 E18 cells showing immunofluorescence for TdTomato, Pdpn, and AQP5. C and D: 114 E18 cells. C: scattergram showing FACS of 114 E18 cells. D: cytocentrifuged preparations of each group of 114 E18 isolated cells showing immunofluorescence of the different FACS populations for TdTomato (tdT), Pdpn, and SP-C. The TdTomato+/Pdpn− population is SP-C+. E, embryonic day.
RNA isolation, sequencing, and analyses.
Cells were obtained from E18 mice of each strain by pooling all embryos in a litter as described above. The number of litters in each line used at each time point is referred to by “n.” From E18 mice, the cells isolated from four litters of each strain were used. From 4-wk-old mice, four mice were used for each strain. After RNAseq analysis, we discovered that several samples had been inadvertently contaminated with human keratinocytes, and therefore excluded those samples from further analysis, which reduced the final number of litters studied in each group. For the final analyses, we used three samples from each of the E18 114 TdTomato+/Pdpn−, E18 114 TdTomato+/ Pdpn+, and E18 R22 TdTomato+/ Pdpn+ populations, four samples from PN 4 wk TI cells (R22 TdTomato+/ Pdpn+) and two samples from PN 4 wk TII cells (114 TdTomato+/ Pdpn−).
RNA was isolated from the FACS-isolated cells by using a NucleoSpin RNA XS kit (Macherey-Nagel, Bethlehem, PA). Total RNA samples were precipitated with sodium acetate, suspended in 10 µL of water, quantitated with a Nanodrop 1000 Reader (Thermo Scientific, Wilmington, DE), and RNA integrity was assessed using RNA6000 Pico Chips and an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) before RNAseq.
Sample library preparation was performed according to standard protocols from the SABRE Functional Genomics Core (http://www.arrays.ucsf.edu.). Sequencing libraries were generated using the TruSeq stranded mRNA sample prep kits with multiplexing primers, according to the manufacturer’s protocol (Illumina, San Diego, CA). Fragment size distribution was assessed using the Bioanalyzer 2100 and the DNA high-sensitivity chip. Library concentrations were measured using KAPA Library Quantification Kits (Kapa Biosystems, Woburn, MA), and equal amounts of indexed libraries were pooled and sequenced on the Illumina HiSeq 4000 (Illumina). We obtained a total of 1.3 billion reads across all samples or an average of 45.6 million reads/sample. Reads were demultiplexed, trimmed of any existing adapter sequences, and aligned to Ensembl’s GRCm38.78 reference genome (16) and aggregated on a per gene basis. Sequence alignment and splice junction estimation was performed using STAR (13) (available at https://code.google.com/archive/p/rna-star). Mappings were restricted to those that were uniquely assigned to the mouse genome, as provided by Ensembl (GRCm38.78) and aggregated on a per gene basis. STAR aggregates this subset of mappings on a per gene basis as raw input for the program DESeq2 (23). The data have been uploaded to the Bioproject Database (https://www.ncbi.nlm.nih.gov/bioproject/; the Bioproject number is PRJNA414925). We analyzed these raw data using DESeq2 to assess variance and differential expression in sample groups, filtering differentially expressed genes to have 1) a minimum of a twofold change, 2) an false discovery rate (FDR) < 0.01, and 3) at least one of the groups in the pairwise comparison had a mean read count greater than 100.
We used the Panther Gene Database (http://www.pantherdb.org) to analyze gene ontology and ingenuity pathway analysis (IPA, qiagenbioinformatics.com, QIAGEN, Germantown, MD) to analyze pathways and biological processes in TI and TII cells.
RESULTS
The R22-rtTA/tet-O-Cre/TdTomato cell lineage produces almost exclusively TI cells.
We crossed RT22 to tet-O-Cre and to Ai9 mice to produce R22-rtTA/tet-O-Cre/Ai9+ mice, in which rtTA is driven by the rat podoplanin gene. After doxycycline (Dox) was administered E10–18, mice were analyzed at PN 4 wk. After this time window of Dox administration, the vast majority of Pdpn+ TI cells at PN 4 wk were TdTomato+ (Fig. 1, A and B). When Dox was administered E 0–10 or E 10–12, there was no specific labeling at PN 4 wk (data not shown). With Dox administration from E 12–15, at PN 4 wk there were few TdTomato+/Pdpn+ TI cells (Fig. 1, C and D). Dox E 15–18 analyzed at E18 resulted in TdTomato+/Sftpc−/Pdpn+ cells with a typical TI morphologic pattern (Fig. 1, E and F); these findings are in contrast to the observations in the 114 line when Dox E15–18 analyzed at E18 (Fig. 2, C–E) resulted in most TdTomato+ cells being Sftpc+/ Pdpn−, rather than Sftpc−/Pdpn+. When Dox was administered E15–18 and mice were examined at PN day 7 (d7), most of the TI cells were TdTomato+ (Fig. 1G). Comparing TdTomato+ TI cell expression at PN d7, RT 22 Dox E15–18 had more uniform expression in TI cells (Fig. 1G) than did 114 Dox E15–18 (Fig. 2F). There was widespread TdTomato expression in TI cells across an entire lobe (Fig. 3A). Pleura, which also expresses Pdpn, also contained TdTomato+ cells (Fig. 3, A and B). Dox E15–18 analyzed at 4 wk produced similar results to the images shown in Fig. 1, A and B. Most Sftpc+ cells were TdTomato−; there were rare (~1/1,000) TdTomato+/ Sftpc+ cells present (Fig. 1H). Taken together, these results illustrate that the population identified by Dox E15–18 in R22-rtTA/tet-O-Cre/Ai9 mice is a progenitor population almost exclusively for TI cells. These findings are summarized in Table 1, along with the findings described below in line 114.
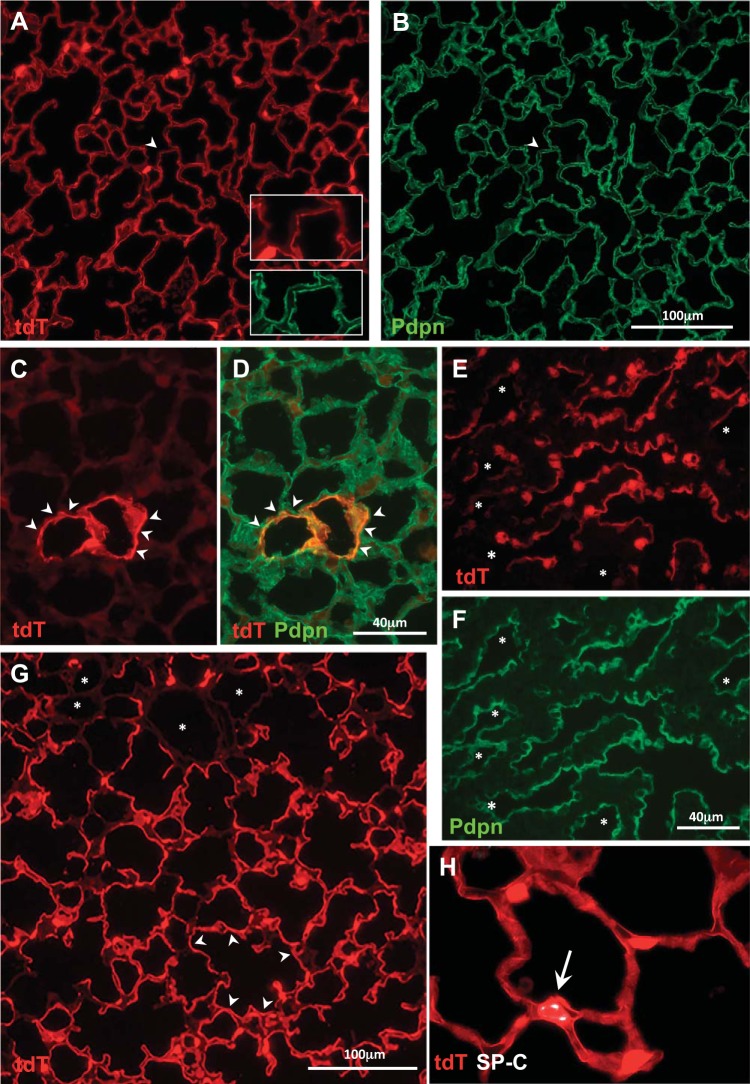
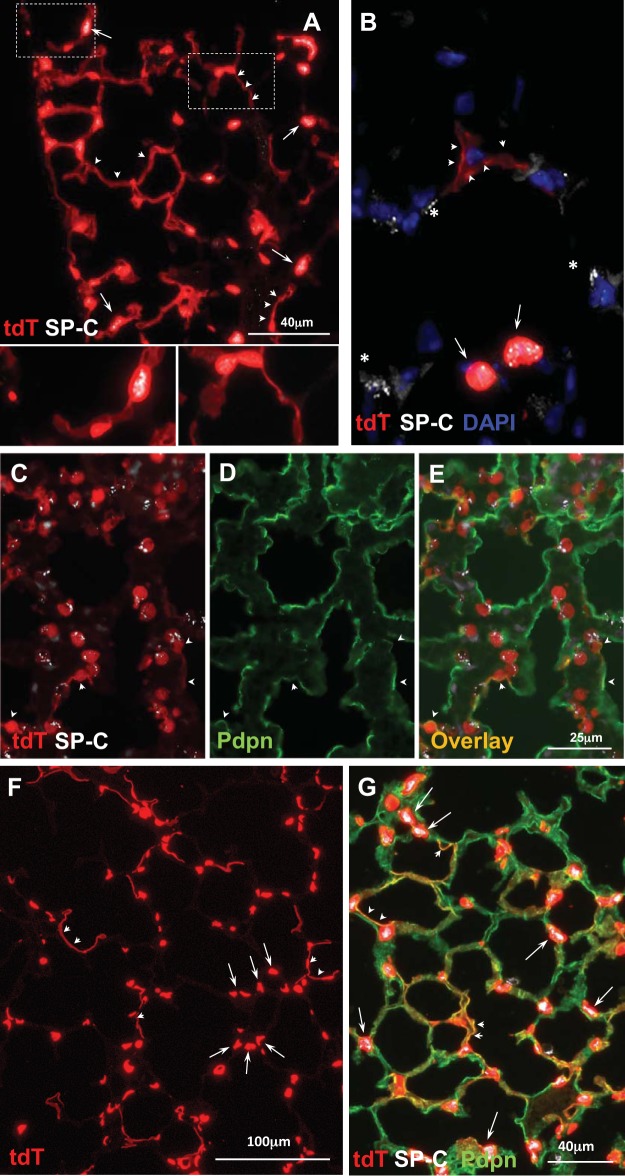
Fig. 1.
RT-22-rtTA/tet-O-Cre/Ai9 line. A and B: doxycycline (Dox) E10–18 analyzed at PN 4 wk (n = 7): widespread TdTomato (tdT) expression in Pdpn+ TI cells. Arrowheads indicates a rare TdTomato−/Pdpn+ area, shown at higher magnification in inset. C and D: Dox E12–15 analyzed at PN 4 wk (n = 4) results in very few TdTomato+/Pdpn+ cells (arrowheads); colocalization of TdTomato (red) and Pdpn (green) results in D orange color. E and F: Dox E15–18 at E18 (n = 3) results in TdTomato+ pre-TI cells that are Pdpn+. Much of the Pdpn+ surface area is TdTomato+; some Pdpn+ areas (asterisks) are TdTomato−. G: Dox E15–18 analyzed at PN day 7 (d7; n = 3): widespread expression of TdTomato in Pdpn+ TI cells (see Fig. 2F for comparison with line 114 at the same time point). Occasional areas (asterisks) are TdTomato−. H: Dox E15–18 analyzed at PN 4 wk (n = 8): rare TdTomato+/ Sftpc+ cell (~0.1% of TdTomato+ cells). E, embryonic day; PN, postnatal.
Fig. 2.
Sftpc-rtTA 114/tet-O-Cre/Ai9 line. A: doxycycline (Dox) E10–18 analyzed at PN 4 wk (n = 7). Extensive TdTomato (tdT) expression, more in TII cells (arrows, Sftpc+) than TI cells (arrowheads). Higher magnification views show TII and TI cells. B: Dox E12–15 analyzed at PN 4 wk (n = 3) results in a few TII cells (arrows, Sftpc+) and rare TI cell-like TdTomato+ extensions (arrowheads). Some, not all, Sftpc+ cells are TdTomato+; arrows denote tdt+/Sftpc+ cells; asterisks indicate tdt−/Sftpc cells. The image was selected to show both TI and TII cells in the same field. C–E: Dox E15–18 analyzed at E18 (n = 3). Most TdTomato+ cells are cuboidal and Sftpc+. A few TdTomato+ cells have Pdpn+ (arrowheads) thin cytoplasmic extensions). F: Dox E15–18 analyzed at PN day 7 (d7; n = 3). Most are TdTomato+ cuboidal cells (arrows), with patchy expression of TdTomato in cells with a TI-like morphology (arrowheads) (see Fig. 1G for comparison with RT22). G: Dox E15–18 analyzed at PN 4 wk (n = 8). Most TdTomato+ TII cells are Sftpc+ (arrows); there is patchy TdTomato expression in Pdpn+ TI cells (arrowheads); merged TdTomato+/Pdpn+ areas are orange (arrowheads). E, embryonic day; PN, postnatal.
Fig. 3.
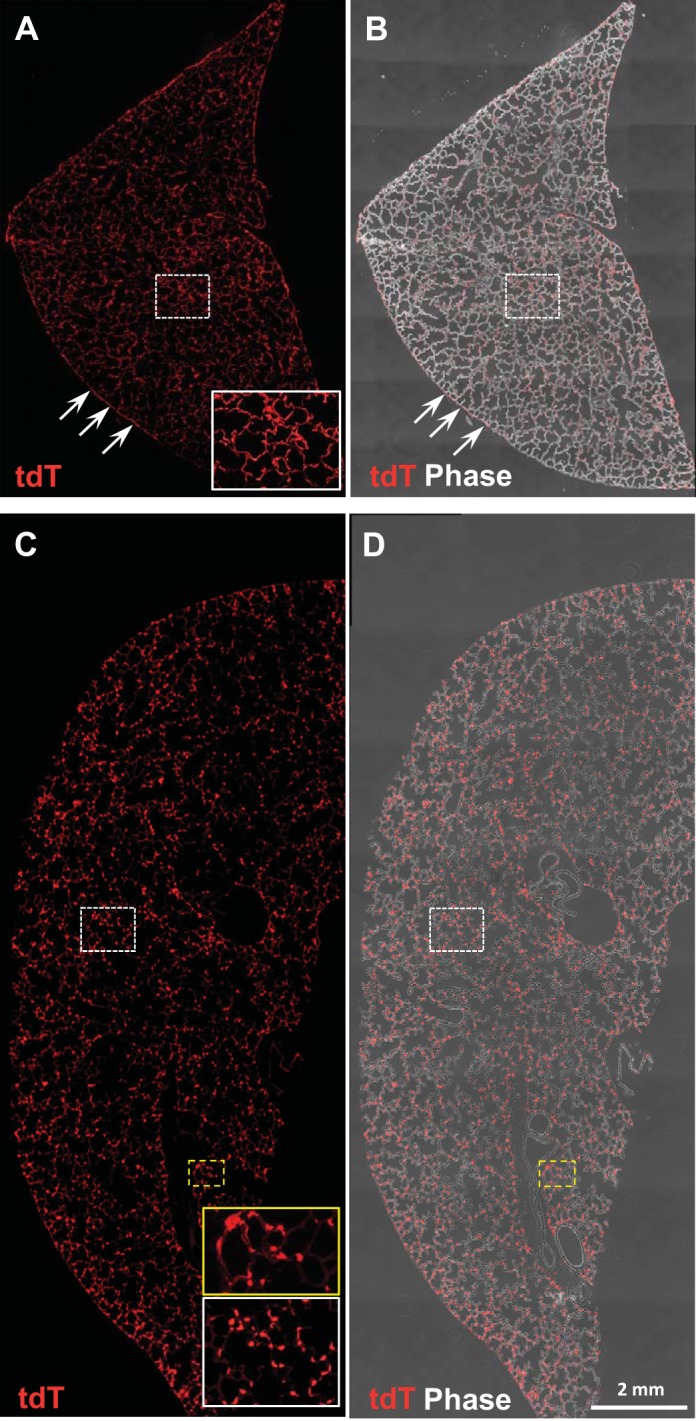
Analysis of intact lung lobes at PN7. A and B: fluorescence and composite fluorescence/phase contrast imaging of a lobe from the R22 line given doxycycline (Dox) 15–18 and examined at PN day 7 (d7). Td/Tomato+ fluorescence is visible almost entirely in thin cytoplasmic extensions (see higher magnification inset), with fairly uniform expression throughout the lobe; pleura (arrows) is also TdTomato+. C and D: in contrast, imaging of a lobe from 114 mice Dox E15–18 analyzed at PN d7. Most of the TdTomato+ fluorescence is in cuboidal type II cells, which at this magnification appear punctuated. Insets showing selected areas at higher magnification illustrate both thin cytpoplasmic extensions of TI cells (yellow box) and cuboidal TII cells (white box). E, embryonic day; PN, postnatal.
Table 1.
Summary of doxycycline time courses and morphologic results
| Reporter Line | Dox Admin | Age When Examined | TI Cell Squamous (Pdpn+, Sftpc−) | TII Cell Cuboidal (Pdpn−, Sftpc+) |
|---|---|---|---|---|
| R22/tet-O-Cre/tdT | E10–18 | PN 4 wk | ++++ | −/+ |
| E12–15 | PN 4 wk | + | ND | |
| E15–18 | E18 | +++++ | ND | |
| E15–18 | PN7 | ++++ | ND | |
| E15–18 | PN 4 wk | ++++ | −/+ | |
| 114/tet-O-Cre/tdT | E10–18 | PN 4 wk | ++ | ++++ |
| E12–15 | PN 4 wk | + | + | |
| E15–18 | E18 | + | +++++ | |
| E15–18 | PN7 | ++ | ++++ | |
| E15–18 | PN 4 wk | ++ | ++++ |
The results of morphologic analyses shown in Figs. 1 and 2 and described in the text are summarized in this table. The amount and pattern of TdTomato cell staining were evaluated qualitatively by inspecting multiple images for each of the time windows of Dox administration in the 114 and R22 lines. “n” is described for each time point in the Figure legends. Note that the patterns fall largely into two groups, with R22 leading to virtually all TI cells and 114 to “mostly” TII cells. Dox, doxycycline; ND, not detected; PN, postnatal: +/−, rare; +, very few/few; ++, patchy; ++++, most; +++++, extensive.
The Line 114 Sftpc-rtTA/tet-O-Cre/TdTomato cell lineage produces predominantly TII cells, but also a subpopulation of TI cells.
We generated Sftpc-rtTA 114/tet-O-Cre/Ai9 reporter mice by crossing Sftpc-rtTA (line 114) mice to tet-O-Cre and to Ai9 reporter mice to produce 114/tet-O-Cre/Ai9 mice, in which rtTA is driven by the mouse SP-C gene. When Dox was given E10–18 (Fig. 2A), there was widespread TdTomato expression at PN 4 wk in TII cells (cuboidal shape, Sftpc+) and in a subpopulation of TI cells (squamous morphology with thin cytoplasmic extensions, Sftpc−). Similar to the results with the R22 line, Dox E0–10 or E10–12 resulted in no specific labeling at PN 4 wk (data not shown). Dox E12–15 at PN 4 wk (Fig. 2B) labeled a few TII cells and very few TI TdTomato+/Pdpn+ cells. After Dox administration E15–18, most epithelial cells were TdTomato+ at E18 and at later time points. At E18 (Fig. 2, C–E), most TdTomato+ cells were cuboidal and Sftpc+/Pdpn−; there were a few TI-like cells that were TdTomato+/ Pdpn+. By both PN d7 (Fig. 2F) and PN 4 wk (Fig. 2G), most TII cells were TdTomato+; there was a subpopulation of TdTomato+ TI cells. Although we did observe some intermouse variation, Dox E15–18 at PN d7 in the 114 line resulted in fewer TdTomato+ TI cells (Fig. 2F), than did Dox E15–18 in the R22 line (Fig. 1G). Imaging of PN d7 entire lobes demonstrated widespread expression throughout the lobe (Fig. 3, C and D). These results show that the progenitor population in the E15–18 period identified by the expressed Sftpc transgene is the source for virtually all TII cells and also gives rise to a subpopulation of TI cells. Unlike some lines of Sftpc-rtTA transgenic mice (27), Cre-mediated recombination can be induced postnatally. Dox administered to PN 4 wk mice yielded ~95% TdTomato+/ Sftpc+ cells (data not shown).
Summary of both lineages.
The results shown in Figs. 1 and 2 are summarized in Table 1 and illustrated in a schematic diagram in Fig. 5A.
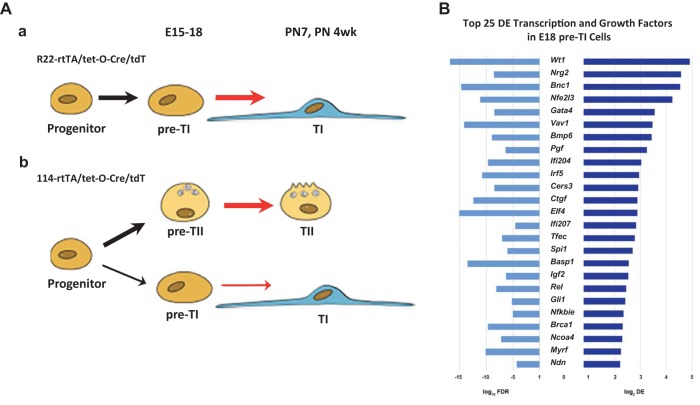
Fig. 5.
Schematic of alveolar epithelial development and potential regulatory factors derived from RNAseq analysis of pre-TI and pre-TII cells. A: schematic of alveolar epithelial development; a: pathway defined in the R22/tet-O-Cre/Ai9 line. At E12–15 and E15–18, a progenitor population gives rise almost completely to TI cells; a very small number of TII cells (~0.1%) is produced by this pathway; b: pathway defined in the 114/tet-O-Cre/Ai9 line. At E12–15 and E15–18, a progenitor population gives rise mostly to TII cells, with a subpopulation of TI cells. B: following RNAseq analysis of pre-TI and pre-TII cells, we analyzed the data for potential regulatory factors important in TI cell development. Bar graphs showing the top 25 (by log2DE) transcription and GFs common to both populations of pre-TI cells, when each is compared with the pre-TII cell population (E18 114 A9+/Pdpn−). For each factor, the log10FDR is shown on the left bar graphs and the log2DE on the right. E, embryonic day.
Control mice/nonspecific labeling.
Control tet-O-Cre/Ai9 mice and no-Dox controls in 114/tet-O-Cre/Ai9 and R22/tet-O-Cre/Ai9 mice showed a few TdTomato+ cells in pleura and in the interstitium (data not shown) at PN d7 and PN 4 wk (data not shown).
Molecular phenotypes of the pre-TI cell populations in the R22 and 114 lineages reveal potential regulatory factors for TI cell differentiation.
We isolated various cell populations for RNAseq analysis at E18 and PN 4 wk following Dox administration E15–18, as described in materials and methods. The FACS data and immunocytochemical characteristics of the various cell populations are shown in Fig. 4. There was variation in both the cell size and fluorescence intensity in each population. The strategy for cell isolation, described in more detail in materials and methods, was to select cells with maximal fluorescence intensity to have distinct cell populations for subsequent RNAseq analyses. The immunocytochemical data (Fig. 4, B and D) confirm the immunologic characteristics of the cell populations isolated by FACS.
In analysis of RNAseq data (see materials and methods), we used criteria for differential expression (DE) of >2-FC (fold change), a FDR <0.01, and a mean of 100 reads (~2.77 reads/million) in at least one of the two comparison groups.
Whether in the 114 or R22 lines, the E18 progenitor cells that subsequently develop into TI cells have almost identical molecular phenotypes. To identify potential transcription (TF) and growth factors (GFs) important in the development of TI cells, we compared the E15–18 pre-TI cells in both 114 (114/TdTomato+/Pdpn+/Sfptc−) and R22 (R22/TdTomato+/Pdpn+/Sftpc−) lineages to the E15–18 pre-TII cell population (114/TdTomato+/Pdpn−/Sfptc+). Among the resultant 30 most highly DE TFs and GFs, 25 were the same in both populations of pre-TI cells (Fig. 5, A and B); the FDRs of these genes are highly significant, between 10−4 and 10−17. Among the mostly highly DE TFs are many with known roles in lung development: these include Wt1 (Wilms tumor-1), Nrg2 (Neuregulin 2), Bnc1 (basonuclin-1), Gata4, and Nfe2l3 (nuclear-factor, erythroid 2 like 3, Nrf3).
Functions of TI and TII cells described by molecular phenotypes of the mature and progenitor cell populations.
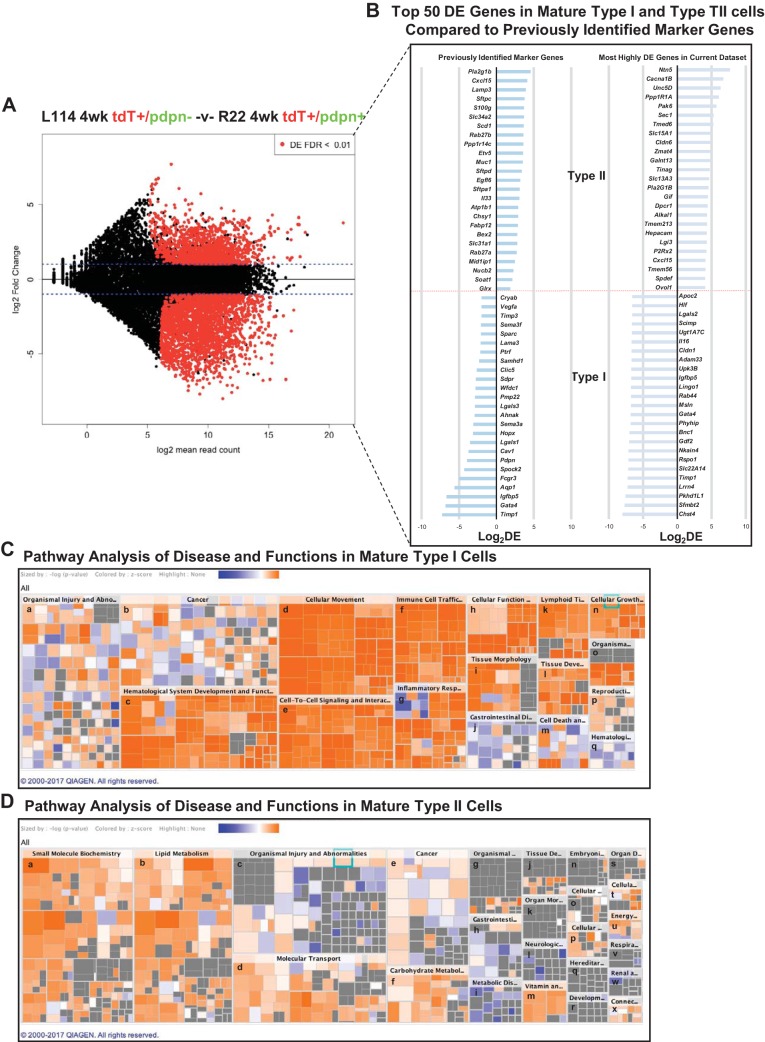
We compared the gene expression profiles of PN 4 wk TI and TII cells (Fig. 6A); as expected, the expression profiles are quite different. After pseudo- and “predicted” genes were removed, there remained 1,983 DE genes in PN 4 wk TI cells and 1,471 DE genes in PN 4 wk TII cells. Although we understand many functions of mature TI and TII cells, potential unknown functions may be elucidated by ingenuity pathway analyses of both canonical pathways and pathways common to various diseases and cellular functions. The results of these analyses are shown in Fig. 6, C and D. For TII cells, it is not surprising that lipid metabolism pathways, used for surfactant synthesis, are predominant. From studies of TI cells in vitro, it is not surprising to find pathways involving cell movement and immune/inflammatory responses in TI cells (see discussion for details).
Fig. 6.
Comparison between doxycycline (Dox) E15–18 4 wk TI and TII cells. A: MA plot comparing Dox E15–18 analyzed at 4 wk 114 TdTomato+/Pdpn− (TII cells) and R22 TdTomato+/Pdpn+ (TI cells) shows a large number of DE genes. B: the right column shows the 50 most highly DE protein-encoding genes between PN 4 wk TI and TII cells in the present data set. The left column shows the level of expression, in the present data set, of various previously identified marker genes (7, 17, 31). C and D: analysis of the DE genes by ingenuity pathway analysis reveals substantial differences between the PN 4 wk TI and TII cells. Depicted is a hierarchical heatmap where larger boxes represent a family of related functions and small squares are individual biological processes. The size of the rectangles is correlated with the overlap of predicted and observed gene sets and serves as a significance score (using FET P value). Color indicates the pathways’ predicted activation state (Z score): increasing likelihood (orange) or decreasing likelihood (blue). In in (6C), Tl cell major boxes are associated with genes suggesting newly ascribed Tl cell functions, such as cellular movement, genes relevant to hematological system development, immune cell trafficking, and inflammatory response. In (6D), major boxes of the Tll heatmap are associated with known functions of Tll cells, such as small molecule biochemistry, lipid metabolism, and molecular transport. The image was generated using Qiagen’s IPA software and cropped to reduce the size to improve readability. The unabbreviated names of the pathway names are as follows; in C, PN 4 wk Tl cells: a, organismal injury and abnormalities; b, cancer; c, hematological system development and function; d, cellular movement; e, cell-to-cell signaling and interactions; f, immune cell trafficking; g, inflammatory response; h, cellular function and maintenance; i, tissue morphology; j, gastrointestinal disease; k, lymphoid tissue structure and development; l, tissue development; m, cell death and survival; n, cellular growth and proliferation; o, organismal development; p, reproductive system; q, hematological disease; in D, PN 4 wk Tll cells: a, small molecule biochemistry; b, lipid metabolism; c, organismal injury and abnormalities; d, molecular transport; e, cancer; f, carbohydrate metabolism; g, organismal injury and abnormalities; h, gastrointestinal disease; i, metabolic disease; j, tissue development; k, organ morphology; l, neurological disease; m, vitamin and mineral metabolism; n, embryonic development; o, cellular function and maintenance; p, cellular assembly and organizational; q, heredity disorder; r, developmental disorder; s, organ development; t, cellular assembly and organization; u, energy production; v, respiratory disease; w, renal and urological disease; x, connective tissue development and function. E, embryonic day; PN, postnatal.
Canonical pathways DE by each cell type are shown in Table 2, and heat maps of disease and functional pathways differentially expressed in TI and TII cells are shown in Fig. 6, C and D. These analyses both confirm previously known functions and point to new functions. When DE canonical functions are compared between pre-TI cells versus PN 4 wk TI cells, there are similar pathways to those found in the pre-TI cell versus 4 wk TI cell comparison (Table 3). In contrast, there are fewer pathways shared between pre-TII and PN 4 wk TII cells (Table 4). Thus, it appears that the functional phenotype of the E18 pre-TI cells is more similar to the PN 4 wk TI cells than is that of the E18 pre-TII cells to PN 4 wk TII cells.
Table 2.
Differentially expressed canonical pathways between PN 4 wk TI and PN 4 wk TII cells
| PN 4 wk TI Cells |
PN 4 wk TII Cells |
||
|---|---|---|---|
| Canonical Pathway | −log(P Value) | Canonical Pathway | −log(P Value) |
| Hepatic fibrosis/hepatic stellate cell activation | 23.1 | Superpathway of cholesterol biosynthesis | 11.7 |
| Axonal guidance signaling | 21.1 | Cholesterol biosynthesis I | 9.43 |
| Role of macrophages, fibroblasts and endothelial cells in rheumatoid arthritis | 17.9 | Cholesterol biosynthesis II (via 24,25- dihydrolanosterol) | 9.43 |
| Leukocyte extravasation signaling | 16 | Cholesterol biosynthesis III (via desmosterol) | 9.43 |
| Phagosome formation | 14 | Fatty acid β-oxidation I | 5.44 |
| Role of osteoblasts, osteoclasts and chondrocytes in rheumatoid arthritis | 13.8 | Zymosterol biosynthesis | 5.14 |
| NF-κB signaling | 13 | Stearate biosynthesis I (animals) | 4.29 |
| Colorectal cancer metastasis signaling | 12.5 | Superpathway of geranylgeranyldiphosphate biosynthesis I (via mevalonate) | 4.23 |
| Molecular mechanisms of cancer | 12.4 | TR/RXR activation | 4.16 |
| Integrin signaling | 12.2 | Ephrin A signaling | 3.91 |
| Granulocyte adhesion and diapedesis | 12 | Glutaryl-CoA degradation | 3.61 |
| PI3K signaling in B lymphocytes | 11.7 | LXR/RXR activation | 3.43 |
| B cell receptor signaling | 11.5 | Glutathione-mediated detoxification | 3.14 |
| Fcγ receptor-mediated phagocytosis in macrophages and monocytes | 10.9 | Mevalonate pathway I | 2.98 |
| Dendritic cell maturation | 10.8 | Arginine degradation I (arginase pathway) | 2.96 |
| Virus entry via endocytic pathways | 10.4 | GDP-glucose biosynthesis | 2.73 |
| IL-8 signaling | 10.3 | p53 signaling | 2.58 |
| Glioblastoma multiforme signaling | 10.3 | Lysine degradation II | 2.58 |
| Thrombin signaling | 10.3 | Triacylglycerol biosynthesis | 2.55 |
| Role of pattern recognition receptors in recognition of bacteria and viruses | 10.2 | Glucose and glucose-1-phosphate degradation | 2.54 |
| PPARα/RXRα activation | 9.62 | Paxillin signaling | 2.5 |
| Phospholipase C signaling | 9.53 | Tryptophan degradation III (eukaryotic) | 2.4 |
| Osteoarthritis pathway | 9.29 | Epoxysqualene biosynthesis | 2.36 |
| Gαq signaling | 9.08 | Spermine biosynthesis | 2.36 |
| TREM1 signaling | 9.06 | Proline degradation | 2.36 |
Differentially expressed (PN 4 wk TI vs. PN 4 wk TII cells) canonical pathways, as analyzed by ingenuity pathways (see materials and methods). PN, postnatal.
Table 3.
Differentially expressed canonical pathways between E18 pre-TI cells and PN 4 wk TII cells
| Canonical Pathway | −log(P Value) |
|---|---|
| Hepatic fibrosis/hepatic stellate cell activation | 16.5 |
| Axonal guidance signaling | 16.3 |
| Molecular mechanisms of cancer | 10.4 |
| Leukocyte extravasation signaling | 9.99 |
| Phagosome formation | 9.29 |
| Role of macrophages, fibroblasts and endothelial cells in rheumatoid arthritis | 9.02 |
| NF-κB signaling | 8.92 |
| B cell receptor signaling | 8.12 |
| Integrin signaling | 7.55 |
| Role of tissue factor in cancer | 7.12 |
| Role of osteoblasts, osteoclasts and chondrocytes in rheumatoid arthritis | 6.95 |
| Glioblastoma multiforme signaling | 6.6 |
| Actin cytoskeleton signaling | 6.59 |
| PTEN signaling | 6.5 |
| PI3K signaling in B lymphocytes | 6.44 |
| ILK signaling | 6.38 |
| NF-κB activation by viruses | 6.21 |
| Virus entry via endocytic pathways | 6.18 |
| PPARα/RXRα activation | 5.91 |
| Dendritic cell maturation | 5.86 |
| Thrombin signaling | 5.83 |
| Osteoarthritis pathway | 5.81 |
| Wnt/Ca+ pathway | 5.74 |
| Colorectal cancer metastasis signaling | 5.65 |
| Sphingosine-1-phosphate signaling | 5.49 |
When E18 pre-TI cells are compared with PN 4 wk TII cells by ingenuity pathway analysis, canonical pathways DE in pre-TI cells are very similar (pathways in common are shown in bold) to the DE canonical pathways in adult TI cells shown in Table 2. DE, differential expression; PN, postnatal.
Table 4.
Differentially expressed canonical pathways between E18 pre-TII cells and PN 4 wk TI cells
| Canonical Pathway | −log(P Value) |
|---|---|
| Ephrin A signaling | 7.27 |
| Growth hormone signaling | 5.96 |
| EIF2 signaling | 5.79 |
| AMPK signaling | 4.34 |
| Fc Epsilon RI signaling | 4.25 |
| TR/RXR activation | 4.19 |
| IGF-1 signaling | 4.14 |
| Mouse embryonic stem cell pluripotency | 4.14 |
| Axonal guidance signaling | 3.9 |
| Insulin receptor signaling | 3.87 |
| p53 signaling | 3.83 |
| EGF signaling | 3.83 |
| Melanocyte development and pigmentation signaling | 3.78 |
| JAK/Stat signaling | 3.75 |
| ErbB signaling | 3.72 |
| Paxillin signaling | 3.71 |
| mTOR signaling | 3.71 |
| CNTF signaling | 3.69 |
| UVA-induced MAPK signaling | 3.68 |
| Breast cancer regulation by Stathmin1 | 3.63 |
| Regulation of the epithelial-mesenchymal transition pathway | 3.52 |
| Superpathway of cholesterol biosynthesis | 3.45 |
| ERK/MAPK signaling | 3.45 |
| NGF signaling | 3.33 |
| PDGF signaling | 3.29 |
When embryonic day E18 pre-TII cells are compared with postnatal (PN) 4 wk TI cells by ingenuity pathway analysis, the canonical pathways differentially expressed in pre-TII cells have fewer similarities (shown in bold) to those in adult TII cells than the comparison between E18 pre-TI and 4 wk TII cells shown in Table 3. TI, TII, alveolar type I and II cells; CNTF, ciliary neurotrophic factor; EIF, eukaryotic translation initiation factor; mTOR, mechanistic target of rapamycin; NGF, nerve growth factor..
Additional marker genes of mature TI and TII cell phenotypes.
The most highly DE genes in PN 4 wk TI and TII cells identify potential marker genes for each cellular phenotype. Within our data set, one can identify most previously identified marker genes (7, 17, 31) (Fig. 6B, left column). In the present data set, the 25 most highly DE protein-encoding genes in each population (Fig. 6B, right column) exhibit from 17-fold (Ovol1) to 256-fold (Chst4) differences in expression between the two cell types.
DISCUSSION
Cell lineage and cell fate analysis.
Studies in the 1970s used autoradiographic techniques at the light and electron microscopic level to develop the hypothesis that TII cells are the progenitor cells in the lung for TI cells, both after lung injury (14) and in development (1). Despite subsequent data demonstrating that embryonic lung cells co-express markers associated with fully differentiated TI and TII cellular phenotypes in adult lung (25, 39), findings that did not support this concept, this hypothesis was largely accepted until the more recent studies of Desai (9) and Treutlein (31).
In these latter studies, lung embryonic cells expressing a subset of TI and TII cell markers progressively extinguished expression of markers of one or the other cell phenotype as they matured. In clonal analysis of cells irreversibly marked by the Shh-Cre transgene at E15, clones of both TI and TII cells were identified, suggesting that TI and TII cells share a common progenitor (9). In a different approach, cell lineage was inferred from statistical analyses of RNAseq data obtained from single cells at different stages of lung development (31). From this analysis, a model of a bipotent TI/TII progenitor cell in development was also proposed. Although these studies both support a bipotent TI/TII progenitor cell model, other pathways for generation of alveolar epithelial cells may exist. For example, in repair following lung injury, several different pathways may play a role in generating both TI and TII cells. TII cells can proliferate and yield either TI or TII cells (15), and mature TI cells can produce TII cells (18). Furthermore, there may be subpopulations within each cell type that have different properties. For example, a subpopulation of Wnt+ TII cells may be important in regenerating alveolar epithelium following injury (26, 40).
To perform cell lineage analysis, we developed two novel transgenic mouse models to label cells irreversibly. One transgenic mouse expresses rtTA under control of the rat podoplanin (a TI cell marker) promoter (33). A second transgenic mouse has rtTA under control of the mouse surfactant protein C (Sftpc) gene, which is expressed uniquely in TII cells in the adult lung (20, 34). The pathway identified as early as E12–15 by the R22 (rat podoplanin) transgene appears dedicated almost completely to TI cell development; very few (~1/1,000) TII cells are irreversibly marked in this pathway. The elucidation of this pathway adds to our knowledge about potential pathways involved in lung development. A second pathway, identified by the 114 line (Sftpc transgene), produces virtually all TII cells and also gives rise to a subpopulation of TI cells, confirming by cell lineage analysis the bipotent TI/TII progenitor pathway previously proposed. Although our data (Figs. 1–3) suggest that this second pathway is more important for TII cell development than for TI cell development, we are not able to determine the relative contributions of the two pathways to TI cell development from the experiments we performed.
Transgenes have been used in many different systems to identify precursor cells and to ascertain their fates, from which cell lineage is deduced. What is important in these types of studies is to identify a population of cells at one time point and then assess what the fates of these cells will be over time. The relationship of transgene expression to expression of endogenous genes is of interest, but not critical to the results. In the system we used, detection of the marker, TdTomato, occurs after a chain of sequential events: doxycycline absorption and placental transfer to the fetus, activation of rtTA, induction of tet-O-Cre expression followed by synthesis of Cre-recombinase, recombination to excise the floxed STOP TdTomato cassette, and finally synthesis and accumulation of TdTomato protein to detectable levels. Some of these steps involve several processes, including synthesis/degradation of mRNAs and proteins. In preliminary experiments (data not shown), we found that it took ~48 h from Dox administration to induce Cre-recombinase mRNA 20-fold over baseline and ~72 h to achieve maximal levels. This time course is consistent with that described for oral administration by doxycycline observed by others (27). The mouse podoplanin gene is expressed from E13.5 onward by RT-PCR, but can be detected by in situ hybridization in neuronal tissue before formation of the lung bud (28). Endogenous mouse SP-C can be detected by ~E10 by in situ hybridization. However, one cannot infer from mRNA expression levels that are detected either by in situ hybridization or by RT-PCR the time course of subsequent events leading to the synthesis of downstream proteins. Although one can detect SP-C mRNA E10–12 by in situ hybridization, it may be that higher levels of SP-C mRNA are required for activation of rtTA than are expressed within this time frame. We do not know the ontogeny of the rat podoplanin and mouse Sftpc transgenes used in our experiments. From the lag time between Dox administration and Cre-recombinase mRNA expression, it is reasonable to postulate that it might take 48–72 h from Dox administration to induction of detectable levels of TdTomato. Our results, showing no significant TdTomato expression in time windows Dox E10–12 or earlier, but some expression resulting from Dox E12–15 are consistent with a hypothesis that transgene expression occurs somewhere in the E11–13 time period.
Rat podoplanin and transgenic mouse containing a BAC containing the rat podoplanin gene appear to have a more restricted expression pattern than does endogenous mouse podoplanin (Ref. 33 and Figs. 1 and 3C). However, as with the endogenous gene, pleural cells express the transgene (Fig. 1, A and B). Because the pleural surface area constitutes <1% of the alveolar surface area (29, 30), the majority of TdTomato+ cells in the lung are TI cells. Therefore, we would expect that pleural cell mRNA would provide a minimal contribution to gene expression profiling data.
Potential regulatory factors important in TI and TII cell maturation.
We compared the E15–18 pre-TI cells in both R22 and 114 lineages to the pre-TII cells to identify potential transcription (TF) and growth factors (GF) important in the development of TI cells. In the 30 most highly DE TFs and GFs resulting from these two comparisons, 25 were the same in both populations of pre-TI cells (Fig. 5B). The FDRs were highly significant, as high as 10−17. Among the most highly DE TFs are Wt1 (Wilms tumor-1), Nrg2 (Neuregulin 2), Bnc1 (basonuclin-1), Gata4, and Nfe2l3 (nuclear-factor, erythroid 2 like 3, Nrf3), factors that are known to be involved either directly or indirectly in lung development or in differentiation in other organs (4–6, 21, 22). Future experiments may elucidate the precise combination of factors that are critical in regulating TI and TII cell differentiation.
TI and TII cell markers.
Not surprisingly, when we compared the gene expression patterns of PN 4 wk TI and TII cells, there were marked differences (Fig. 6A), with almost 2,000 DE genes per each cell type. Many of the most highly DE genes expressed large fold differences between the mature TI and TII cells. Within our data set, one can identify most previously identified marker genes (7, 17, 31) (Fig. 6B, left column). Many of the novel putative marker genes elucidated in our data (Fig. 6B, right column), have larger fold differences than previously identified marker genes. However, the utility of some of these potential new marker genes may be limited because they exhibit much lower levels of expression than the previously identified marker genes (mean expression levels ~25%, median ~5% of the previously identified marker genes). Nevertheless, because they are highly DE, these genes may be of functional interest. For example, Ovol1 regulates growth arrest of embryonic epidermal progenitor cells, Gdf2 and Bnc1 are known growth factors, Netrin 5 and Unc5d are a chemotropic factor/receptor pair involved in axonal guidance, and Slca2a14 is an integral membrane cotransporter of Na+ and the antioxidant ergothioneine.
Comparison of cellular functions of TI and TII cells by ingenuity pathway analysis.
Although many functions of mature TI and TII cells are well understood, potential unknown functions can be elucidated by analyzing downstream effects and inferring from these likely biochemical and biologic pathways. Pathway analysis can point to both canonical pathways, as well as pathways common to various diseases and cellular functions. We used ingenuity pathway analyses to assess our data. Heat maps of disease and functional pathways DE in TI and TII cells are shown in Fig. 6, C and D, and Table 2. Some of these pathways, such as lipid metabolism for surfactant biosynthesis in TII cells (3), and TI cell movement and immune/inflammatory responses are known (12, 35, 36). Canonical pathways DE by each cell type are shown in Table 2. For TII cells, pathways involving cholesterol and fatty acid metabolism feature predominantly. For TI cells, pathways such as “phagosome formation” and “leukocyte extravasation signaling” seem appropriate from known or inferred functions of this cell type. Functions in TI cells parallel to other canonical or disease pathways seem reasonable. Some examples follow: the “hepatic fibrosis” pathway contains multiple genes encoding collagen molecules, metallopeptidase inhibitors, and growth factors; the “axonal guidance” pathway contains various metallopeptidases and G proteins. These pathways suggest previously unexplored potential functions of TI cells, such as matrix regulation and cell migration. These analyses also confirm previously known functions and point to new functions. The predicted upregulated genes (Fig. 6, C and D, those with high activation z-scores, orange color) in TI cells are in functions such as cellular movement, immune cell trafficking, cell-cell signaling, and inflammatory responses, whereas in TII cells, the upregulated genes in several categories were virtually all related to phospholipid metabolism.
When DE canonical functions are compared between pre-TI cells and PN 4 wk TII cells (Table 3), there are similar pathways to those found in the PN 4 wk TI versus PN 4 wk TII comparison. In contrast, there are fewer pathways shared between pre-TII and PN 4 wk TII cells (Table 4). Thus, it appears that the functional phenotype of the E18 pre-TI cells is more similar to the PN 4 wk TI cells than is that of the E18 pre-TII cells to PN 4 wk TII cells. These observations suggest that late embryonic pre-TI cells have similar functions to mature TI cells. In contrast, embryonic pre-TII cells appear to acquire functions of mature TII cells, such as those involved in surfactant biosynthesis, at a later period of time.
Summary.
In summary, we developed novel transgenic mouse models expressing rtTA under control of either the mouse TII cell-specific surfactant protein (Sftpc) gene (line 114) or an equivalent TI transgene, rat podoplanin (line R22). The pathway, detectable as early as E15, defined by the R22 transgene, is almost completely dedicated to TI cell development; very few (~1/1,000) TII cells are labeled in this pathway. A second pathway, identified by the Sftpc transgene, produces virtually all TII cells and also gives rise to a subpopulation of TI cells, confirming by cell lineage analysis the previously proposed bipotent TI/TII progenitor pathway. By comparing the pre-TI cell populations in both the Sftpc and RT22 lineages, there is a common group of transcription and growth factors that should be useful in determining regulatory factors for development of TI cells. Transcriptome analyses of both mature TI and TII cells point to unrecognized functions of both cell types. Finally, specific pre-TI and pre-TII progenitor cells could be used for potential cell transplantation in lung injury and development.
GRANTS
This work was funded by National Heart, Lung, and Blood Institute Grant PPG HL-24075.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
R.G. and L.D. conceived and designed research; R.G., D.L., and A.G. performed experiments; R.G., D.L., C.C., W.E., and L.D. analyzed data; R.G., C.C., W.E., and L.D. interpreted results of experiments; R.G. and C.C. prepared figures; R.G. and L.D. drafted manuscript; R.G., C.C., and L.D. edited and revised manuscript; R.G., D.L., C.C., A.M.G., W.E., and L.D. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank J. Vanderbilt for his role in producing the transgenic mouse strains and for the images shown in Fig. 4, A. Barczak for advice regarding sample preparation for RNAseq, and J. Pollack for his help and advice in statistical analyses.
Present addresses: A. M. Gillespie, Dept. of Radiology and Biomedical Imaging, Byers Hall, 1700 4th St., Box 2532, San Francisco, CA 94158-2330; R. Gonzalez, Terrace Biotech, 241 Upper Terrace, Suite 3, San Francisco, CA, 94117-4550; D. Leaffer, 1861 16th Ave. San Francisco, Ca 94122.
REFERENCES
- 1.Adamson IY, Bowden DH. Derivation of type 1 epithelium from type 2 cells in the developing rat lung. Lab Invest 32: 736–745, 1975. [PubMed] [Google Scholar]
- 2.Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, Hogan BL. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 123: 3025–3036, 2013. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batenburg JJ. Surfactant phospholipids: synthesis and storage. Am J Physiol Lung Cell Mol Physiol 262: L367–L385, 1992. doi: 10.1152/ajplung.1992.262.4.L367. [DOI] [PubMed] [Google Scholar]
- 4.Cano E, Carmona R, Muñoz-Chápuli R. Wt1-expressing progenitors contribute to multiple tissues in the developing lung. Am J Physiol Lung Cell Mol Physiol 305: L322–L332, 2013. doi: 10.1152/ajplung.00424.2012. [DOI] [PubMed] [Google Scholar]
- 5.Chevillard G, Blank V. NFE2L3 (NRF3): the Cinderella of the Cap’n’Collar transcription factors. Cell Mol Life Sci 68: 3337–3348, 2011. doi: 10.1007/s00018-011-0747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chevillard G, Nouhi Z, Anna D, Paquet M, Blank V. Nrf3-deficient mice are not protected against acute lung and adipose tissue damages induced by butylated hydroxytoluene. FEBS Lett 584: 923–928, 2010. doi: 10.1016/j.febslet.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 7.Dahlin K, Mager EM, Allen L, Tigue Z, Goodglick L, Wadehra M, Dobbs L. Identification of genes differentially expressed in rat alveolar type I cells. Am J Respir Cell Mol Biol 31: 309–316, 2004. doi: 10.1165/rcmb.2003-0423OC. [DOI] [PubMed] [Google Scholar]
- 8.Davis IC, Matalon S. Epithelial sodium channels in the adult lung—important modulators of pulmonary health and disease. Adv Exp Med Biol 618: 127–140, 2007. doi: 10.1007/978-0-387-75434-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507: 190–194, 2014. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobbs LG, Gonzalez R, Matthay MA, Carter EP, Allen L, Verkman AS. Highly water-permeable type I alveolar epithelial cells confer high water permeability between the airspace and vasculature in rat lung. Proc Natl Acad Sci USA 95: 2991–2996, 1998. doi: 10.1073/pnas.95.6.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobbs LG, Johnson MD. Alveolar epithelial transport in the adult lung. Respir Physiol Neurobiol 159: 283–300, 2007. doi: 10.1016/j.resp.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Dobbs LG, Johnson MD, Vanderbilt J, Allen L, Gonzalez R. The great big alveolar TI cell: evolving concepts and paradigms. Cell Physiol Biochem 25: 55–62, 2010. doi: 10.1159/000272063. [DOI] [PubMed] [Google Scholar]
- 13.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21, 2013. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans MJ, Cabral LJ, Stephens RJ, Freeman G. Transformation of alveolar type 2 cells to type 1 cells following exposure to NO2. Exp Mol Pathol 22: 142–150, 1975. doi: 10.1016/0014-4800(75)90059-3. [DOI] [PubMed] [Google Scholar]
- 15.Evans MJ, Johnson LV, Stephens RJ, Freeman G. Renewal of the terminal bronchiolar epithelium in the rat following exposure to NO2 or O3. Lab Invest 35: 246–257, 1976. [PubMed] [Google Scholar]
- 16.Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Girón CG, Gordon L, Hourlier T, Hunt S, Johnson N, Juettemann T, Kähäri AK, Keenan S, Kulesha E, Martin FJ, Maurel T, McLaren WM, Murphy DN, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ruffier M, Sheppard D, Taylor K, Thormann A, Trevanion SJ, Vullo A, Wilder SP, Wilson M, Zadissa A, Aken BL, Birney E, Cunningham F, Harrow J, Herrero J, Hubbard TJ, Kinsella R, Muffato M, Parker A, Spudich G, Yates A, Zerbino DR, Searle SM. Ensembl 2014. Nucleic Acids Res 42: D749–D755, 2014. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez R, Yang YH, Griffin C, Allen L, Tigue Z, Dobbs L. Freshly-isolated rat alveolar type I cells, type II cells, and cultured type II cells have distinct molecular phenotypes. Am J Physiol Lung Cell Mol Physiol 288: L179–L189, 2004. [DOI] [PubMed] [Google Scholar]
- 18.Jain R, Barkauskas CE, Takeda N, Bowie EJ, Aghajanian H, Wang Q, Padmanabhan A, Manderfield LJ, Gupta M, Li D, Li L, Trivedi CM, Hogan BLM, Epstein JA. Plasticity of Hopx+ type I alveolar cells to regenerate type II cells in the lung. Nat Commun 6: 6727, 2015. doi: 10.1038/ncomms7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson MD. Ion transport in alveolar type I cells. Mol Biosyst 3: 178–186, 2007. doi: 10.1039/b614348b. [DOI] [PubMed] [Google Scholar]
- 20.Kalina M, Mason RJ, Shannon JM. Surfactant protein C is expressed in alveolar type II cells but not in Clara cells of rat lung. Am J Respir Cell Mol Biol 6: 594–600, 1992. doi: 10.1165/ajrcmb/6.6.594. [DOI] [PubMed] [Google Scholar]
- 21.Lentjes MH, Niessen HE, Akiyama Y, de Bruïne AP, Melotte V, van Engeland M. The emerging role of GATA transcription factors in development and disease. Expert Rev Mol Med 18: e3, 2016. doi: 10.1017/erm.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C, Morrisey EE, Whitsett JA. GATA-6 is required for maturation of the lung in late gestation. Am J Physiol Lung Cell Mol Physiol 283: L468–L475, 2002. doi: 10.1152/ajplung.00044.2002. [DOI] [PubMed] [Google Scholar]
- 23.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550, 2014. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13: 133–140, 2010. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meneghetti A, Cardoso WV, Brody JS, Williams MC. Epithelial marker genes are expressed in cultured embryonic rat lung and in vivo with similar spatial and temporal patterns. J Histochem Cytochem 44: 1173–1182, 1996. doi: 10.1177/44.10.8813083. [DOI] [PubMed] [Google Scholar]
- 26.Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, Desai TJ. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359: 1118–1123, 2018. doi: 10.1126/science.aam6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perl AK, Wert SE, Nagy A, Lobe CG, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc Natl Acad Sci USA 99: 10482–10487, 2002. doi: 10.1073/pnas.152238499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rishi AK, Joyce-Brady M, Fisher J, Dobbs LG, Floros J, VanderSpek J, Brody JS, Williams MC. Cloning, characterization, and development expression of a rat lung alveolar type I cell gene in embryonic endodermal and neural derivatives. Dev Biol 167: 294–306, 1995. doi: 10.1006/dbio.1995.1024. [DOI] [PubMed] [Google Scholar]
- 29.Staub NC, Wiener-Kronish JP, Albertine KH. Transport through the pleura. In: The Pleura in Health and Disease, edited by Chretien J, Bignon J, Hirsch A. New York: Dekker, 1985, p. 169–193. [Google Scholar]
- 30.Stone KC, Mercer RR, Gehr P, Stockstill B, Crapo JD. Allometric relationships of cell numbers and size in the mammalian lung. Am J Respir Cell Mol Biol 6: 235–243, 1992. doi: 10.1165/ajrcmb/6.2.235. [DOI] [PubMed] [Google Scholar]
- 31.Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509: 371–375, 2014. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valencik ML, McDonald JA. Codon optimization markedly improves doxycycline regulated gene expression in the mouse heart. Transgenic Res 10: 269–275, 2001. doi: 10.1023/A:1016601928465. [DOI] [PubMed] [Google Scholar]
- 33.Vanderbilt JN, Allen L, Gonzalez RF, Tigue Z, Edmondson J, Ansaldi D, Gillespie AM, Dobbs LG. Directed expression of transgenes to alveolar type I cells in the mouse. Am J Respir Cell Mol Biol 39: 253–262, 2008. doi: 10.1165/rcmb.2008-0049OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanderbilt JN, Gonzalez RF, Allen L, Gillespie A, Leaffer D, Dean WB, Chapin C, Dobbs LG. High-efficiency type II cell-enhanced green fluorescent protein expression facilitates cellular identification, tracking, and isolation. Am J Respir Cell Mol Biol 53: 14–21, 2015. doi: 10.1165/rcmb.2014-0348MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong MH, Chapin OC, Johnson MD. LPS-stimulated cytokine production in type I cells is modulated by the renin-angiotensin system. Am J Respir Cell Mol Biol 46: 641–650, 2012. doi: 10.1165/rcmb.2011-0289OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong MH, Johnson MD. Differential response of primary alveolar type I and type II cells to LPS stimulation. PLoS One 8: e55545, 2013. doi: 10.1371/journal.pone.0055545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright JR. Immunoregulatory functions of surfactant proteins. Nat Rev Immunol 5: 58–68, 2005. doi: 10.1038/nri1528. [DOI] [PubMed] [Google Scholar]
- 38.Wright JR, Dobbs LG. Regulation of pulmonary surfactant secretion and clearance. Annu Rev Physiol 53: 395–414, 1991. doi: 10.1146/annurev.ph.53.030191.002143. [DOI] [PubMed] [Google Scholar]
- 39.Wuenschell CW, Sunday ME, Singh G, Minoo P, Slavkin HC, Warburton D. Embryonic mouse lung epithelial progenitor cells co-express immunohistochemical markers of diverse mature cell lineages. J Histochem Cytochem 44: 113–123, 1996. doi: 10.1177/44.2.8609367. [DOI] [PubMed] [Google Scholar]
- 40.Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, Zhou S, Cantu E, Morrisey EE. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555: 251–255, 2018. doi: 10.1038/nature25786. [DOI] [PMC free article] [PubMed] [Google Scholar]