Highlights
-
•
Transgene expressions of simultaneously-administered two plasmid DNA in muscle correlated each other.
-
•
Transgene expressions of secretable luciferase in muscle and plasma also correlated each other.
-
•
It was possible to monitor transgene expression in tissues by blood sampling.
Keywords: Naked plasmid DNA, Therapeutic drug monitoring, In vivo gene transfer, Genetic variability, Individual variation of efficacy
Abstract
In this study, we have developed a novel method to monitor transgene expression in tissues by blood sampling. We administered plasmid DNA (pDNA) encoding non-secretory form of firefly luciferase as a reporter gene and pDNA encoding secretable Gaussia princeps luciferase as a monitor gene simultaneously into mice. Good positive correlations were found between log-transgene expression of the reporter gene and the monitor gene in the treated muscle, between the monitor gene in the treated muscle and plasma, and consequently between the reporter gene in the treated muscle and the monitor gene in plasma after naked pDNA transfer into the muscle of mice. Such positive correlations were also found with gastric serosal surface instillation of naked pDNA, intravenous injection of lipoplex, and hydrodynamics-based injection of naked pDNA. We developed monitoring method of transgene expression in tissues by blood sampling, which was named ‘Therapeutic transgene monitoring (TTM)’, after ‘Therapeutic drug monitoring (TDM)’.
1. Introduction
In vivo transgene expression in gene therapy should be regulated so as it stays in a therapeutic range to avoid low responses or severe toxic effects; however, the efficiency of transgene expression in individuals often varies whether using viral or non-viral vectors [[1], [2], [3]]. To regulate transgene expression, use of a drug-inducible promoter, such as tetracycline-responsive promoter is beneficial [4]. Regulation of transgene expression requires the monitoring of transgene expression to assess whether it is in therapeutic range; however, monitoring transgene expression in tissues is difficult, often requiring biopsy, except for secretory proteins. Here, we have developed a method for monitoring transgene expression in tissues using plasmid DNA (pDNA) encoding a monitoring gene, named ‘therapeutic transgene monitoring (TTM)’. When TTM is achieved, it is possible to make a dosing plan similar to therapeutic drug monitoring (TDM).
In this study, we simultaneously administered two pDNA encoding a non-secretable form of a reporter gene assuming a therapeutic gene and a secretable form of a monitor gene in mice. If transgene products of the monitor gene secreted in plasma are positively correlated with the reporter gene expression in a target tissue, we can monitor transgene expression in the tissue by blood sampling. We used firefly luciferase (Fluc) as a reporter gene due to high sensitivity of the assay. As monitor genes, there were several candidates, including secreted embryonic alkaline phosphatase (SEAP) [5] and Gaussia princeps luciferase (Gluc) [6]. Especially, a humanized form of Gluc has 100- to 1000-fold higher bioluminescent signal intensity than humanized forms of firefly and Renilla luciferases [6]. Using a secretable form of Gluc, quantification of tumor growth [6,7], monitoring of cell viability and proliferation [8], monitoring of microbial infections [9], and in vivo tracking of extracellular vesicles in mice [10]. Thus, we selected the secretable form of Gluc as a monitor gene.
2. Materials and methods
2.1. Materials
Glucose and cholesterol were purchased from Nacalai Tesque (Kyoto, Japan). DOTAP methyl sulfate salt was purchased from Avanti Polar Lipids (Alabaster, AL, USA). TO-PRO-3 was purchased from Molecular Probes (Invitrogen, Carlsbad, CA, USA). All chemicals were of the highest purity available.
2.2. pDNA
pcDNA3/FL (originally referred to as pCMV-Luc), pDNA encoding the non-secretory form of Fluc under the control of CMV promoter, was constructed as reported previously [11]. pcDNA3/GL vector, pDNA encoding the secretable form of Gluc under the control of CMV promoter, was purchased from Lux Biotechnology Ltd. (Edinburgh, UK). pDNA was amplified in the Escherichia coli strain DH5α, isolated, and purified using an EndoFree Plasmid Giga Kit (QIAGEN GmbH, Hilden, Germany). pDNA dissolved in 5 % glucose solution or phosphate-buffered saline (PBS) were stored at −20 °C prior to experiments. Fluorescein or tetramethyl-rhodamine labeling of pDNA were performed using the Label IT Nucleic Acid Labeling Kit (Mirus Co., Madison, WI, USA).
2.3. Preparation of cationic liposome/pDNA complex (lipoplex)
DOTAP and cholesterol were dissolved in chloroform at a molar ratio of 1:1, vacuum-desiccated, and resuspended in sterile 5 % dextrose at a concentration of 4 mg total lipids per ml to form liposomes. Liposomes were extruded 10-times through a polycarbonate membrane filter (100 nm pore size) using a commercially available instrument (Mini-Extruder, Avanti Polar Lipids). For lipoplex preparation, pDNA in 5 % dextrose was mixed with an equal volume of cationic liposomes and was incubated for 30 min. Charge ratio, which is the molar ratio of cationic lipids to pDNA phosphate residue, was 3. The charge ratio of unity was 3.52 μg total lipid/μg pDNA for this formulation.
2.4. Animals
Five-week-old male ddY (22.0–36.6 g), ICR (26.5–28.1 g), C57BL6 (15.1–17.2 g), and 10-week-old male ddY mice (41.3–46.5 g) were housed in cages in an air-conditioned room and maintained on a standard laboratory diet (MF; Oriental Yeast Co., Ltd., Tokyo, Japan) and water ad libitum. All animal experiments were carried out in accordance with the Guidelines for Animal Experimentation of Nagasaki University.
2.5. In Vivo gene transfer
For intramuscular injection of naked pDNA, a solution of 25 μg of pcDNA3/FL and pcDNA3/GL in 50 μL PBS was injected into the left quadriceps femoris muscle in different strains of mice under anesthesia (sodium pentobarbital, 40−60 mg/kg, intraperitoneal injection). One day after naked pDNA injection, blood was collected under anesthesia. Then, the mice were sacrificed under anesthesia, and the treated muscle was removed. For gastric serosal surface instillation of naked pDNA, five-week-old male ddY mice were anesthetized. Laparotomy was performed and the stomach was exposed. Naked pDNA (solution of 0.5 μg pcDNA3/FL and pcDNA3/GL in 10 μL PBS) was instilled onto the gastric serosal surface using a micropipette (PIPETMAN; Gilson Inc., Villiers-le-Bel, France). Six hours after naked pDNA instillation, blood was collected under anesthesia. Then, the mice were sacrificed under anesthesia, and the stomach was removed. For intravenous administration of lipoplex, a solution of lipoplexes containing 15 μg pcDNA3/Fluc and pcDNA3/Gluc in 200 μL or 22.5 μg pcDNA3/Fluc and pcDNA3/Gluc in 300 μL was injected via tail vein in 5-week-old male ddY mice. Six hours after administration, blood was collected under anesthesia. Then, the mice were sacrificed under anesthesia, and the lungs was removed. For hydrodynamics-based intravenous injection of naked pDNA, a solution of 1 μg pcDNA3/FL and 0.1 μg pcDNA3/GL in 2 mL saline was injected via tail vein within 5 s in 5-week-old male ddY mice. At appropriate time intervals, blood was collected under anesthesia. Then, the mice were sacrificed under anesthesia, and the liver was removed. Blood and tissue samples were subjected to luciferase assay.
2.6. Luciferase assay
Blood samples were collected and centrifuged at 15,000 × g for 5 min to obtain plasma. Tissue samples were washed twice with saline and homogenized with lysis buffer containing 0.1 M Tris/HCl buffer (pH 7.8), 0.05 % Triton X-100, and 2 mM EDTA. The volume of lysis buffer was 4 μL/mg tissue. Homogenates were centrifuged at 15,000 × g for 5 min. Ten microliters of the plasma or supernatant of tissue homogenates were mixed with 100 μL Fluc assay substrates (PicaGene; Toyo Ink Mfg Co. Ltd., Tokyo, Japan) or renilla luciferase assay substrates containing Gluc substrate coelenterazine (Promega, Madison, WI, USA), and the light produced was immediately measured using a luminometer (Lumat LB 9507; Berthold Technologies, Bad Wildbad, Germany). Luciferase activity is indicated as the relative light units (RLU) per tissue or whole plasma. Total volume of plasma in mice was estimated using the proportion of blood volume to body weight of individual mice (8 %) and hematocrit value of mice (40 %).
2.7. In vitro stability of gluc proteins
Gluc proteins were obtained from plasma of mice that received hydrodynamics-based intravenous injection of naked pDNA (10 μg pcDNA3/GL in 2 mL saline). Gluc proteins (1.26 × 106 RLU) were incubated in 200 μL plasma of male ddY mice at 37 °C for indicated time periods. Gluc activities in samples were determined as mentioned above.
2.8. In vivo disposition of gluc proteins
Gluc proteins were obtained from plasma of mice that received hydrodynamics-based intravenous injection of naked pDNA (10 μg pcDNA3/GL in 2 mL saline). Gluc proteins (2.62 × 109 RLU in 100 μL) were administered to 5-week-old male ddY mice via tail vein. Gluc activities in plasma, tissues, and urine were determined as mentioned above.
2.9. Intracellular Localization of pcDNA3/FL and pcDNA3/GL
Six hours after instillation of 1 μg of both fluorescein-pDNA and rhodamine-pDNA in 10 μL PBS onto the gastric serosal surface of 5-week-old male ddY mice, imprints of mesothelial cells were prepared as reported previously [12,13]. Briefly, the stomach was washed 5 times with saline and dried for 4−5 min at room temperature. Imprints of gastric mesothelial cells were obtained on MAS-coated microslide glasses (SUPERFROST S-9441; Matsunami Glass Ind. Ltd., Osaka, Japan). Imprints were fixed with 4 % paraformaldehyde in PBS for 10 min and permeabilized for 5 min with PBS containing 0.2 % Triton X-100. Nuclei were stained with TO-PRO-3 (dilution 1:2000 in PBS) by incubating in a humidified chamber for 1 h. SlowFade Gold antifade reagent (Invitrogen) was applied to imprints before mounting. Subsequently, imprints were subjected to confocal laser scanning microscopy (LSM 510 META; Carl Zeiss Microimaging Inc., Thornwood, NY, USA).
2.10. Laser scanning microscopy for imprints
Imprints were analyzed by confocal laser scanning microscopy using LSM 510 META (Plan-Apochromat 63×, NA 1.4 oil immersion objective lens; Carl Zeiss Microimaging Inc., Thornwood, NY, USA). Laser lines used were at 488 nm, 543 nm, and 633 nm to excite fluorescein, rhodamine, and TO-PRO-3, respectively. Each dye was scanned in sequential mode to prevent fluorescence crosstalk. Acquisition software used was ZEN 2007 (Carl Zeiss Microimaging Inc.).
2.11. Statistical analysis
Statistical analysis was performed by Pearson's correlation. Differences among regression lines were analyzed by analysis of covariance. Tukey multiple comparison tests were performed in some experiments.
3. Results and discussion
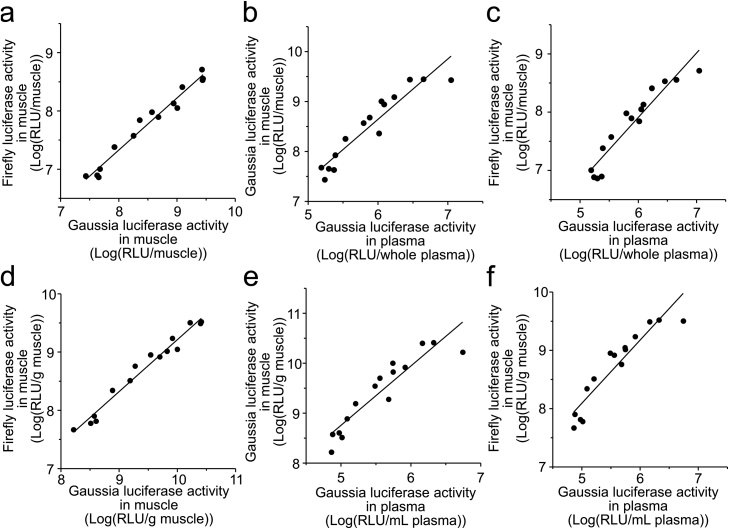
We administered Gluc encoding pcDNA3/GL with pcDNA3/FL (originally referred to as pCMV-Luc), pDNA encoding the non-secretory form of firefly luciferase (Fluc) (same backbone as pcDNA3/GL), into ddY mice via three administration routes. When naked pDNA were administered into muscle, the range of transgene expression was extremely broad 1 day after administration (2 orders of magnitude); therefore, when we assessed the correlation between Fluc and Gluc expression, we took the logarithm of gene expression to uniformly analyze low and high values of gene expression. Log-Fluc expression in the treated muscle was well and positively correlated with log-Gluc expression in the same muscle (Fig. 1a, r2 = 0.970 (P < 0.001)). The slope of regression line was 0.894, while the intercept of regression line was 0.176. Also, log-Gluc expression in the treated muscle was positively correlated with the log-Gluc expression secreted into plasma (Fig. 1b, r2 = 0.889 (P < 0.001), slope = 1.209, intercept = 1.388); as a consequence, log-Fluc expression in the treated muscle was positively correlated with the log-Gluc expression secreted into plasma (Fig. 1c, r2 = 0.899 (P < 0.001), slope = 1.104, intercept = 1.284). When we analyzed the correlation of transgene expression, log-Fluc concentration in the treated muscle positively correlated with log-Gluc concentration in the same muscle (Fig. 1d, r2 = 0.972 (P < 0.001) slope = 0.891, intercept = 0.305). However, correlation between log-Gluc concentration in the treated muscle and the log-Gluc concentration in plasma (Fig. 1e, r2 = 0.855 (P < 0.001), slope = 1.188, intercept = 2.813) was slightly worse than that between log-Gluc in whole treated-muscle and plasma (r2 = 0.889); as a consequence, correlation between log-Fluc concentration in the treated muscle and the log-Gluc concentration in plasma (Fig. 1f, r2 = 0.871 (P < 0.001), slope = 1.084, intercept = 2.670) was also worse than that between log-Fluc in whole treated muscle and log-Gluc in whole plasma. This could be attributed to the inter-individual differences in the ratio between the tissue weight and the plasma volume. These results implied that the expression of the therapeutic gene (Fluc as a reporter gene) in the target tissue could be monitored by the expression of the monitor gene (Gluc) secreted into plasma. Since whole amount of transgene expression produced slightly better correlation than the concentration, we employed RLU/whole tissue and RLU in whole plasma in the following experiments.
Fig. 1.
Correlation of transgene expression levels between reporter gene, as a therapeutic gene, and monitor gene after intramuscular injection of naked pDNA in mice. Symbols and lines express luciferase expression levels in individual mice and regression lines, respectively. (a) Correlation between firefly luciferase (Fluc) and Gaussia princeps luciferase (Gluc) activities in whole treated muscle. (b) Correlation between Gluc activities in whole treated muscle and whole plasma. (c) Correlation between Fluc activity in whole treated muscle and Gluc activity in whole plasma. (d) Correlation between concentrations of Fluc and Gluc activities in the treated muscle (/g muscle). (e) Correlation between concentrations of Gluc activities in the treated muscle (/g muscle) and plasma (/mL plasma). (f) Correlation between concentrations of Fluc activity in the treated muscle (/g muscle) and Gluc activity in plasma (/mL plasma).
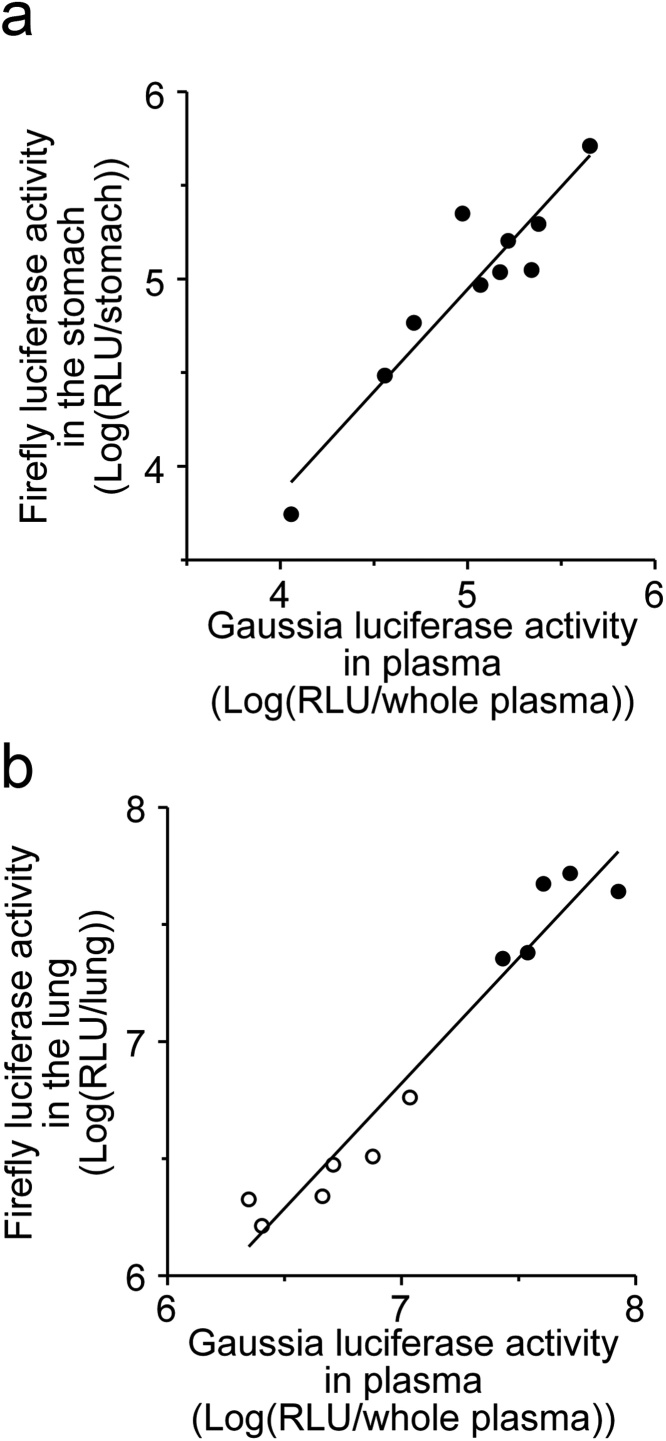
To validate the effectiveness of TTM, it is important to test the correlation between Fluc expression in different target tissue and Gluc expression secreted into plasma. We previously developed a novel gene delivery method targeted to the stomach, in which naked pDNA is simply instilled onto the gastric serosal surface of mice and rats [12,[14], [15], [16]]. We assessed the correlation between Fluc expression in the stomach and Gluc expression secreted into plasma 6 h after gastric serosal instillation of both pcDNA3/FL and pcDNA3/GL into ddY mice. Log-Fluc expression in the stomach was positively correlated with log-Gluc expression secreted into plasma (Fig. 2a, r2 = 0.875 (P < 0.001), slope = 1.094, intercept = -0.524). As other methods, cationic liposomes can deliver pDNA via the systemic circulation. Transgene expression mainly occurred in the lung after intravenous injection of cationic liposomes/pDNA complex (lipoplex). Six hours after intravenous administration of lipoplex, log-Fluc expression in the lung was positively correlated with log-Gluc expression secreted into plasma (Fig. 2b, r2 = 0.948 (P < 0.001), slope = 1.069, intercept = -0.658). Thus, TTM would be valuable for gene carriers as well as naked pDNA.
Fig. 2.
Correlation of transgene expression levels between reporter gene, as a therapeutic gene, and monitor gene after gastric serosal surface instillation of naked pDNA (a) and intravenous injection of Lipoplex (b) in mice. Symbols and lines express luciferase expression in individual mice and regression lines, respectively. (a) Correlation between Fluc activity in the stomach and Gluc activity in plasma 6 h after instillation of naked pDNA onto the gastric serosal surface in mice. (b) Correlation between Fluc activity in the lung and Gluc activity in plasma 6 h after intravenous administration of lipoplex (a solution of 15 μg pcDNA3/Fluc and pcDNA3/Gluc in 200 μL (open circles), and 22.5 μg pcDNA3/Fluc and pcDNA3/Gluc in 300 μL (closed circles)) in mice.
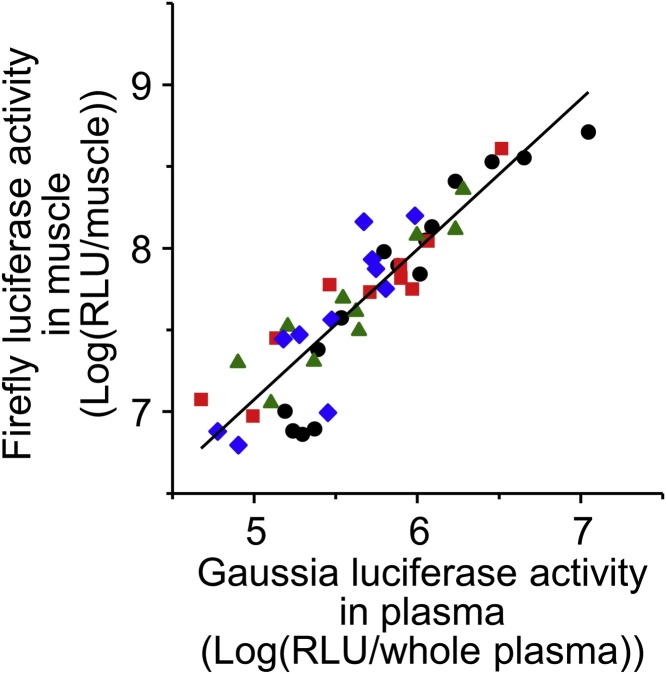
Since ddY mice are an inbred strain, genetic variability is low among individuals. This might contribute to the good correlation between Fluc expression in target tissues and Gluc expression secreted into plasma as genetic variability may affect the ratio between transgene products in tissues and plasma. Also, a narrow body weight range might contribute to a good correlation, because blood volume may affect the concentration of Gluc secreted into plasma. We simultaneously assessed the effects of the strains and body weight of individual mice on the correlation between Fluc expression in the treated muscle and Gluc expression secreted into plasma after intramuscular injection into mice. Again, log-Fluc expression in the treated muscle was positively correlated with log-Gluc expression secreted into plasma (Fig. 3, r2 = 0.823 (P < 0.001), slope = 0.920, intercept = 2.474). For future application to human gene therapy, this monitoring method may be effective in various patients with different genetic backgrounds (such as races) and individual differences (such as body weight). Although the correlation between the monitoring gene and the therapeutic gene needs to be tested in humans, this monitoring system is expected to be useful in human gene therapy in future.
Fig. 3.
Effect of strain of mice on correlation of transgene expression levels between reporter gene, as a therapeutic gene, and monitor gene after intramuscular injection of naked pDNA in mice. Symbols and lines express luciferase expression in individual mice and regression lines, respectively. Symbols: Five-week-old male ddY (circles), ICR (triangles), C57BL6 (diamonds) and 10-week-old male ddY mice (squares).
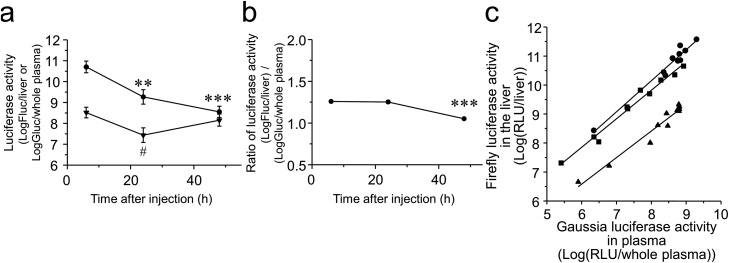
As transgene expression after the administration of non-viral vectors is transient, it is important to test the effectiveness of TTM at various time points. Hydrodynamics-based intravenous injection (high volume injection at a high velocity) of naked pDNA is a potent gene transfer method into the liver [17]. Fig. 4a shows the duration of the expression levels of Fluc in the liver and Gluc in plasma after hydrodynamics injection. Fluc expression in the liver was transient, while duration of Gluc in plasma was relatively longer. Log-Fluc/log-Gluc ratio for 6 and 24 h were almost same, after that the ratio was decreased at 48 h (Fig. 4b). Fig. 4c shows the correlation between log-Fluc expression in the liver and log-Gluc expression secreted into plasma 6, 24 and 48 h after hydrodynamics-based intravenous injection in ddY mice (r2 = 0.964, 0.979, and 0.958 (P < 0.001); slope = 1.070, 0.984, and 0.915; intercept = 1.592, 1.950, and 1.091, at 6, 24 and 48 h, respectively). In this experiment, the dose of pcDNA3/GL was decreased to a tenth of that of pcDNA3/FL; therefore, the amount of the monitor gene could be reduced when transfection activity was sufficiently high. As for the timing of monitoring, regression lines at 6, 24, and 48 h were significantly different (P < 0.001, analysis of covariance), while slopes of these regression lines were similar (not significant). The ratio of Gluc in plasma to Fluc in the liver increased with time, which might indicate the possible accumulation of Gluc proteins in plasma after hydrodynamics-based injection of naked pDNA into mice.
Fig. 4.
(a) Duration of the expression level of Fluc in the liver (circle) and Gluc in plasma (inverted triangle) 6, 24, and 48 h after hydrodynamics-based intravenous injection of naked pDNA in mice. Each point represents the mean ± S.E. at least 10 experiments. Tukey multiple comparison tests were performed (**, P < 0.01; ***, P < 0.001 vs. 6 h group of Fluc; #, P < 0.05 vs. 6 h group of Gluc). (b) Duration of log-Fluc/log-Gluc ratios. Each point represents the mean ± S.E. at least 10 experiments. Tukey multiple comparison test was performed (***, P < 0.001 vs. 6 and 48 h groups). (c) Correlation between Fluc activity in the liver and Gluc activity in plasma 6 (Circles), 24 (Squares) and 48 h (Triangles) after hydrodynamics-based intravenous injection of naked pDNA in mice. Symbols and lines express luciferase expression levels in individual mice and regression lines, respectively.
Ratios of Fluc in a target tissue and Gluc in plasma were different depending on the target tissue and monitoring time point. This may be due to differences in the structure of tissues and stability of Gluc protein. Muscle fibers are outside from the blood vessels; thus, secretion of Gluc to blood circulation may be restricted. On the contrary, in case of intravenous injection of lipoplex, transgene expression occurred mainly in lung capillary blood vessels, which face to the blood stream. In case of hydrodynamics-based injection, target cells are hepatocytes. Hepatocytes are outside of sinusoidal endothelium, but secreted Gluc can easily pass through fenestrae on endothelium. Also, secretion polarity may affect the ratios of Fluc in a target tissue and Gluc in plasma. Hepatocytes may secret Gluc not only to the blood stream, but also to bile. In case of the gastric serosal surface instillation, it is relatively complicated due to secretion to the blood stream and peritoneal cavity. Contrary to the case of hepatocytes, Gluc secreted to peritoneal cavity also can enter the systemic circulation.
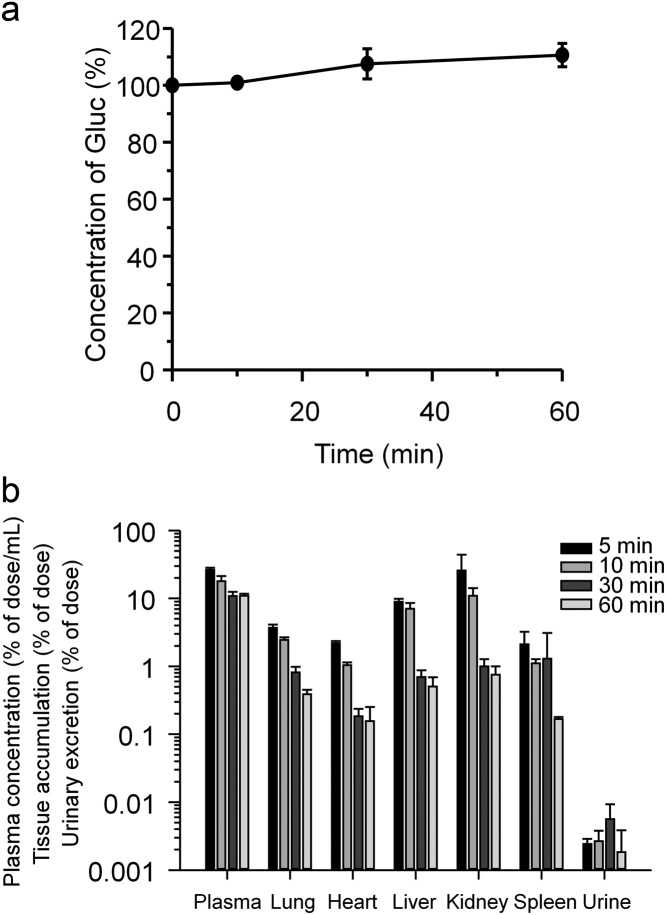
To test the stability of Gluc proteins in vitro, they were incubated in plasma, collected from ddY mice, at 37 °C for 60 min (Fig. 5a). Gluc activity slightly increased with time, probably due to solvent evaporation; therefore, Gluc proteins were stable in plasma. When Gluc proteins were administered via tail vein, Gluc activity decreased in plasma for 30 min, and was maintained thereafter (Fig. 5b). Gluc activity was mainly distributed to the kidney in the early phase. Gluc activity in each tissue also rapidly decreased with time. Urinary excretion of Gluc was detectable, but very low. These results might indicate that Gluc proteins were stable and could remain in plasma for a long time. Fluc is unstable at body temperature [18]; thus, Fluc activity in tissues reflected gene expression at the time, while Gluc activity in plasma might depend on the cumulative amount. The stability of Gluc proteins may be too good to monitor gene expression with time. Since transgene expression by non-viral vectors is generally transient, multiple administrations may be required. However, with multiple administrations, different time profiles of transgene expressions in therapeutic and monitor gene is a problem; i.e., it may be difficult to distinguish transgene expression levels of first and subsequent administrations due to high stability of Gluc. To destabilize Gluc, it may be useful to apply protein-destabilizing elements [19] to Gluc. Such genetic modification to destabilize Gluc activity will be required for future clinical use of TTM using Gluc.
Fig. 5.
In vitro stability of Gluc proteins (a) and in vivo disposition of Gluc proteins after intravenous injection in mice (b). Each value represents the mean ± S.D. of 3 experiments.
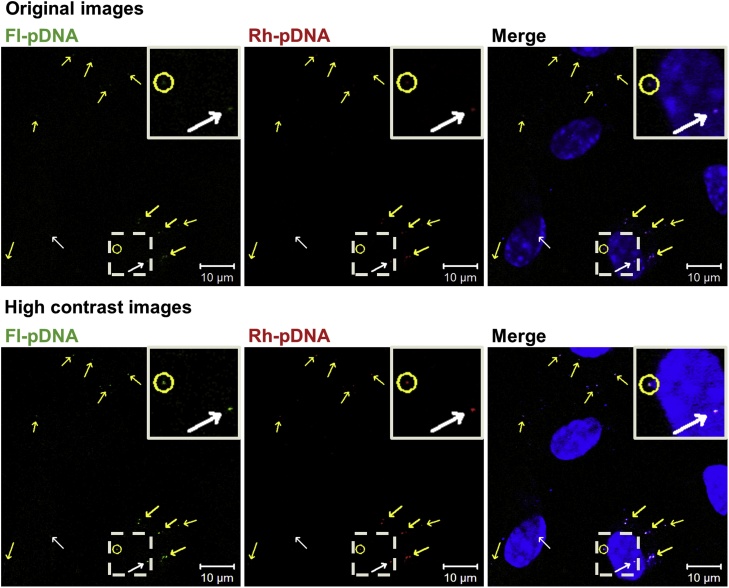
To elucidate the mechanism of the good correlation between Fluc and Gluc expression levels, we observed intracellular localization of both pcDNA3/FL and pcDNA3/GL. After instillation of both fluorescein-pDNA (Fl-pDNA) and rhodamine-pDNA (Rh-pDNA) onto the gastric serosal surface in mice, Fl-pDNA and Rh-pDNA were substantially co-localized outside and inside of nuclei (Fig. 6). Also, we have previously reported similar co-localization of two pDNAs in the liver after hydrodynamics-based injection in mice [20]. These co-localization data can explain the good correlation between Fluc and Gluc expression levels, since it is expected that similar distribution of the two pDNAs in the cells can result in similar gene expression levels. Interestingly, several molecules of pDNA entered the nucleus simultaneously. pDNA was taken up by gastric mesothelial cells via macropinocytosis (unpublished results). Thus, several molecules of pDNA would enter the same macropinosome, and be transferred into the nucleus together. It was reported that the nuclear import of pDNA required both cytoplasmic factors and specific DNA sequences [21]. These cytoplasmic factors are probably transcription factors, which have a nuclear localization signal. This mechanism of the nuclear import of pDNA may not require simultaneous nuclear import and the same nuclear localization of several molecules of pDNA. One possible reason for the simultaneous nuclear import and same nuclear localization was that pDNA might be complexed with cationic proteins, such as a histone, in cytoplasm and subsequently be delivered into the nucleus by transcription factors that bound to pDNA.
Fig. 6.
Co-localization of pcDNA3/FL and pcDNA3/GL in gastric mesothelial cells after instillation of fluorescently labeled pDNA onto the gastric serosal surface of mice. White and yellow arrows express the co-localization of both pDNA inside and outside of the nucleus, respectively. Yellow circle indicates simultaneous nuclear import of both pDNA. Dashed areas are enlarged in the top right corner of each panel (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
In this study, we did not test the antigenicity of Gluc; thus, the neutralization of Gluc by anti-Gluc antibodies may disturb TTM using Gluc with repeated administration. Using the secretory form of other reporter genes, such as firefly and Renilla luciferase, this problem is partially resolved theoretically. Furthermore, cytotoxic T lymphocytes may kill transgene-expressing cells. Utilization of the Tet-on promoter [4] will enable on-demand transgene expression of the monitor gene at the time of monitoring. Administration of immunosuppressive drugs during TTM in combination with the Tet-on system is a useful strategy for repeat monitoring.
Since gene expression is transient after non-viral gene delivery, repeated monitoring may be required; however, it is painful. In humans, it is difficult to examine the correlation between transgene expression of therapeutic gene and monitor gene by frequent biopsies. However, gene expression can be estimated by therapeutic effect as a parameter. For example, if the therapeutic gene is LDL receptor, the therapeutic effect can be evaluated by determining serum LDL cholesterol levels. Also, application of the concept of pharmacokinetic/pharmacodynamic modeling in drug treatment to TTM would be useful to decrease the frequency of biopsies. To decrease the frequency of blood sampling, the concept of population pharmacokinetics [22] is applicable. Regarding the backbones of pDNA, pcDNA3/FL and pcDNA3/GL are identical. This might contribute to the good correlation between Fluc and Gluc expression levels; therefore, it is currently unclear whether there is always a correlation among pDNA with different backbones, such as different promoters. Encoding both the therapeutic gene and monitor gene on the same pDNA molecule, the problem of backbone difference is theoretically negligible; however, co-administration of the therapeutic gene and monitor gene on separate pDNA molecules has several merits. Because the copy number of large pDNA in E. coli is generally low, usage of a separate monitor pDNA with therapeutic pDNA can prevent low yield due to the unnecessary elongation of pDNA base pairs; consequently, industrial manufacturing of the pDNA encoding monitor gene is relatively easy. It was also reported that the increasing size of pDNA reduced the transfection efficiency of lipoplex in vitro [23]; therefore, the problem of backbone difference should be tested in the future study. At present, TTM is useful for monitoring transfection efficiency. The ultimate goal of gene delivery systems is stable (long-term) transgene expression. To monitor inter-individual variability of promoter activity, another technique needs to be established.
Currently, there are possible limitations in the application of TTM. If the target cells is outside from tight blood vessels, the blood vessels may restrict secretion of the monitor gene product from the parenchyma to intravascular space. Blood-brain barrier (BBB) is the tightest barrier in the body. For noninvasive tumor growth monitoring, it was reported that Gluc secreted from Gluc-expressing glioma cells in the orthotropic tumor model can be monitored by blood sampling [24]. Thus, in case of targeting glioma, it may be possible to monitor transgene expression in the tumor by blood sampling. However, BBB leakiness in glioma is dependent on the type of tumor. In Hs683 human oligodendroglioma model, BBB was tighter than that in GL261 mouse glioblastoma model [25]. Recently, Ogawa et al. reported that secretable luciferase was detected in cerebrospinal fluid (CSF) after transfection into the choroid plexus by intracerebroventricular administration of ultrasound-responsive nanobubbles with naked pDNA [26]. So, monitoring transgene expression in the brain by CSF sampling instead of blood sampling might seems feasible.
Applicability of TTM to viral vectors is unclear. For in vivo gene therapy, adeno-associated viral vectors are a good choice due to their long-lasting transgene expression and safety [27]. In contrast, the long-lasting expression of monitor gene would cause a safety concern due to unexpected side effects and the immunogenicity. So, it may be necessary to control the monitor gene expression using a drug-responsive Tet-on system [4]. Also, because the packaging capacity of adeno-associated viral vectors is relatively small compared with that of adenoviral vectors, there is a limitation in packaging the monitor gene in addition to therapeutic gene on the same adeno-associated viral particle. This can theoretically be solved using two viral packages encoding therapeutic and monitor genes, respectively.
4. Conclusion
We have developed a novel method to monitor transgene expression in tissues by simultaneous administration of a monitor gene, secretable Gaussia princeps luciferase. Good correlation between transgene expression levels of the monitor gene and reporter gene, as a therapeutic gene, was evident after the administration of naked pDNA and lipoplex via various administration routes in mice. We named this method, ‘Therapeutic transgene monitoring (TTM)’, after ‘Therapeutic drug monitoring (TDM)’. The basis of analytical methods for TDM, such as population pharmacokinetics, is theoretically applicable for TTM. We believe that TTM will contribute to the safety of gene therapy.
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, a Grant-in-Aid from the Uehara Memorial Foundation, and a Grant-in-Aid for Scientific Research from the President of Nagasaki University. We would like to thank Editage (www.editage.com) for English language editing.
References
- 1.Manthorpe M., Cornefert Jensen F., Hartikka J., Felgner J., Rundell A., Margalith M., Dwarki V. Gene therapy by intramuscular injection of plasmid DNA: studies on firefly luciferase gene expression in mice. Hum. Gene Ther. 1993;4:419–431. doi: 10.1089/hum.1993.4.4-419. [DOI] [PubMed] [Google Scholar]
- 2.Barr D., Tubb J., Ferguson D., Scaria A., Lieber A., Wilson C., Perkins J., Kay M.A. Strain related variations in adenovirally mediated transgene expression from mouse hepatocytes in vivo: comparisons between immunocompetent and immunodeficient inbred strains. Gene Ther. 1995;2:151–155. [PubMed] [Google Scholar]
- 3.Nomura T., Nakajima S., Kawabata K., Yamashita F., Takakura Y., Hashida M. Intratumoral pharmacokinetics and in vivo gene expression of naked plasmid DNA and its cationic liposome complexes after direct gene transfer. Cancer Res. 1997;57:2681–2686. [PubMed] [Google Scholar]
- 4.Baron U., Bujard H. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 2000;327:401–421. doi: 10.1016/s0076-6879(00)27292-3. [DOI] [PubMed] [Google Scholar]
- 5.Wang M., Orsini C., Casanova D., Millán J.L., Mahfoudi A., Thuillier V. MUSEAP, a novel reporter gene for the study of long-term gene expression in immunocompetent mice. Gene. 2001;279:99–108. doi: 10.1016/s0378-1119(01)00754-5. [DOI] [PubMed] [Google Scholar]
- 6.Tannous B.A., Kim D.E., Fernandez J.L., Weissleder R., Breakefield X.O. Codon-optimized gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Chung E., Yamashita H., Au P., Tannous B.A., Fukumura D., Jain R.K. Secreted Gaussia luciferase as a biomarker for monitoring tumor progression and treatment response of systemic metastases. PLoS One. 2009;4:2019. doi: 10.1371/journal.pone.0008316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wurdinger T., Badr C., Pike L., de Kleine R., Weissleder R., Breakefield X.O., Tannous B.A. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat. Methods. 2008;5:171–173. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enjalbert B., Rachini A., Vediyappan G., Pietrella D., Spaccapelo R., Vecchiarelli A., Brown A.J.P., D’Enfert C. A multifunctional, synthetic Gaussia princeps luciferase reporter for live imaging of Candida albicans infections. Infect. Immun. 2009;77:4847–4858. doi: 10.1128/IAI.00223-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi Y., Nishikawa M., Shinotsuka H., Matsui Y., Ohara S., Imai T., Takakura Y. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J. Biotechnol. 2013;165:77–84. doi: 10.1016/j.jbiotec.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Fumoto S., Tsuchimochi M., Nishi J., Ishii H., Kodama Y., Nakashima M., Sasaki H., Nakamura J., Nishida K. Liver- and lobe-specific gene transfer following the continuous microinstillation of plasmid DNA onto the liver surface in mice: Effect of instillation speed. Biol. Pharm. Bull. 2009;32:1298–1302. doi: 10.1248/bpb.32.1298. [DOI] [PubMed] [Google Scholar]
- 12.Nishi J., Fumoto S., Ishii H., Kodama Y., Nakashima M., Sasaki H., Nakamura J., Nishida K. Highly stomach-selective gene transfer following gastric serosal surface instillation of naked plasmid DNA in rats. J. Gastroenterol. 2008;43:912–919. doi: 10.1007/s00535-008-2301-7. [DOI] [PubMed] [Google Scholar]
- 13.Foley-Comer A.J., Herrick S.E., Al-Mishlab T., Prêle C.M., Laurent G.J., Mutsaers S.E. Evidence for incorporation of free-floating mesothelial cells as a mechanism of serosal healing. J. Cell. Sci. 2002;115:1383–1389. doi: 10.1242/jcs.115.7.1383. [DOI] [PubMed] [Google Scholar]
- 14.Fumoto S., Nishi J., Nakamura J., Nishida K. Gene therapy for gastric diseases. Curr. Gene Ther. 2008;8:187–200. doi: 10.2174/156652308784746431. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura J., Fumoto S., Shoji K., Kodama Y., Nishi J., Nakashima M., Sasaki H., Nishida K. Stomach-selective gene transfer following the administration of naked plasmid DNA onto the gastric serosal surface in mice. Biol. Pharm. Bull. 2006;29:2082–2086. doi: 10.1248/bpb.29.2082. [DOI] [PubMed] [Google Scholar]
- 16.Nishi J., Fumoto S., Ishii H., Kodama Y., Nakashima M., Sasaki H., Nakamura J., Nishida K. Improved stomach selectivity of gene expression following microinstillation of plasmid DNA onto the gastric serosal surface in mice. Eur. J. Pharm. Biopharm. 2008;69:633–639. doi: 10.1016/j.ejpb.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 17.Liu F., Song Y.K., Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 18.Chen X., Ren S., Jin Z., Zhu S. Production and purification of firefly luciferase in Escherichia coli. Biotechnol. Tech. 1996;10:89–92. [Google Scholar]
- 19.Voon D.C., Subrata L.S., Baltic S., Leu M.P., Whiteway J.M., Wong A., Knight S.A., Christiansen F.T., Daly J.M. Use of mRNA- and protein-destabilizing elements to develop a highly responsive reporter system. Nucleic Acids Res. 2005;33:e27. doi: 10.1093/nar/gni030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fumoto S., Nishimura K., Nishida K., Kawakami S. Three-dimensional imaging of the intracellular fate of plasmid DNA and transgene expression: ZsGreen1 and tissue clearing method CUBIC are an optimal combination for multicolor deep imaging in murine tissues. PLoS One. 2016;11 doi: 10.1371/journal.pone.0148233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson G.L., Dean B.S., Wang G., Dean D.A. Nuclear import of plasmid DNA in digitonin-permeabilized cells requires both cytoplasmic factors and specific DNA sequences. J. Biol. Chem. 1999;274:22025–22032. doi: 10.1074/jbc.274.31.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ette E.I., Williams P.J. Population pharmacokinetics II: estimation methods. Ann. Pharmacother. 2004;38:1907–1915. doi: 10.1345/aph.1E259. [DOI] [PubMed] [Google Scholar]
- 23.Kreiss P., Cameron B., Rangara R., Mailhe P., Aguerre-Charriol O., Airiau M., Scherman D., Crouzet J., Pitard B. Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic Acids Res. 1999;27:3792–3798. doi: 10.1093/nar/27.19.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alessandrini F., Ceresa D., Appolloni I., Marubbi D., Malatesta P. Noninvasive monitoring of glioma growth in the mouse. J. Cancer. 2016;7:1791–1797. doi: 10.7150/jca.15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leten C., Struys T., Dresselaers T., Himmelreich U. In vivo and ex vivo assessment of the blood brain barrier integrity in different glioblastoma animal models. J. Neurooncol. 2014;119:297–306. doi: 10.1007/s11060-014-1514-2. [DOI] [PubMed] [Google Scholar]
- 26.Ogawa K., Fuchigami Y., Hagimori M., Fumoto S., Maruyama K., Kawakami S. Ultrasound-responsive nanobubble-mediated gene transfection in the cerebroventricular region by intracerebroventricular administration in mice. Eur. J. Pharm. Biopharm. 2019;137:1–8. doi: 10.1016/j.ejpb.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Aponte-Ubillus J.J., Barajas D., Peltier J., Bardliving C., Shamlou P., Gold D. Molecular design for recombinant adeno-associated virus (rAAV) vector production. Appl. Microbiol. Biotechnol. 2018;102:1045–1054. doi: 10.1007/s00253-017-8670-1. [DOI] [PMC free article] [PubMed] [Google Scholar]