V. cholerae is the causative agent of cholera, a life-threatening disease characterized by severe, watery diarrhea. The 7th pandemic started in Indonesia in 1961 and spread globally, currently infecting 1.3 million to 4 million people annually. Here, we applied whole-genome sequencing to analyze a long-term collection of V. cholerae clinical strains to reveal the phylogenetic background and evolutionary dynamics of the 7th pandemic in Shanghai, which had undergone breathtakingly rapid development in the last half-century. All but one of the Shanghai 7th-pandemic strains fell into five “stages” that were dominant in Shanghai and appeared to be closely related to 7th-pandemic strains of South or Southeast Asia. Our findings extended the understanding of the dynamics of the evolution and transmission of the 7th-pandemic clones in East Asia and the relationship between social changes and cholera epidemiology.
KEYWORDS: Vibrio cholerae, Shanghai cholera, 7th pandemic, whole-genome sequencing, comparative genomics
ABSTRACT
The 7th cholera pandemic began in 1961 in Sulawesi, Indonesia, and then spread around the world in at least three waves. However, the lack of genome sequences for Vibrio cholerae strains under long-term surveillance in East Asia, especially in China, has restricted our understanding of the dynamics of the intracountry and intercountry evolution and transmission of the 7th-pandemic clones. In this study, we obtained the genome sequences of 60 V. cholerae strains isolated in Shanghai, the largest port in the world and the largest city in China, from 1961 to 2011. Our whole-genome-based phylogeny of 7th-pandemic strains revealed that all but one fell into five “stages,” most of which are single clades and share independent ancestors. Each stage dominated in succession for a period, with little overlap between them. In addition, two near-identical Shanghai strains belonging to a pre-7th-pandemic precursor and 4 nontoxigenic O1/O139 strains attributed to independent recombination events at the O-antigen loci were present. The major lineages of the 7th pandemic in Shanghai appeared to be closely related to V. cholerae strains isolated from South or Southeast Asia. Stage succession was consistently related to changes in society and human activity, implying that human-caused niche change may play a vital role in the cholera dynamics in Shanghai.
IMPORTANCE V. cholerae is the causative agent of cholera, a life-threatening disease characterized by severe, watery diarrhea. The 7th pandemic started in Indonesia in 1961 and spread globally, currently infecting 1.3 million to 4 million people annually. Here, we applied whole-genome sequencing to analyze a long-term collection of V. cholerae clinical strains to reveal the phylogenetic background and evolutionary dynamics of the 7th pandemic in Shanghai, which had undergone breathtakingly rapid development in the last half-century. All but one of the Shanghai 7th-pandemic strains fell into five “stages” that were dominant in Shanghai and appeared to be closely related to 7th-pandemic strains of South or Southeast Asia. Our findings extended the understanding of the dynamics of the evolution and transmission of the 7th-pandemic clones in East Asia and the relationship between social changes and cholera epidemiology.
INTRODUCTION
Vibrio cholerae is the causative agent of cholera, a life-threatening disease characterized by severe, watery diarrhea (1, 2). There have been seven cholera pandemics since 1817, and all continents except Antarctica have had significant or major incursions by one or more of them (1). The 7th pandemic began in 1961 in Makassar, Sulawesi, Indonesia, and currently causes an estimated 1.3 million to 4 million cases annually (3). An outbreak in Haiti following the 2010 earthquake infected nearly 700,000 people and has caused over 8,500 deaths (4). In a 2008 epidemic in Zimbabwe, more than 90,000 suspected cholera cases were reported, with more than 4,000 patient deaths (5).
The 7th-pandemic El Tor biotype lineage was first observed in the Middle East in 1897 but first caused an outbreak 40 years later (1937) on the Indonesian island of Sulawesi, which was followed by others in 1940, 1944, and 1957 (6, 7). However, after 20 years on Sulawesi, this lineage developed a greater capacity to spread, and a 1960 outbreak in the same location became pandemic in 1961, reaching several countries in Asia, including China, in that year, and then spread around the world in at least three waves. The origins of the 7th pandemic from its appearance in the Middle East to its becoming pandemic in 1961 have been described in a recent paper (8), while the spread of the pandemic strain was covered in an earlier paper (9).
Until very recently, there were very few genome sequences of V. cholerae strains from China. However, Didelot et al. reported a study that included the genome sequences of 71 Chinese isolates and showed that the three major waves in which the 7th pandemic spread around the world all reached China and that one of the strains is related to a prepandemic strain from Indonesia (10). This study provided important insight into the relationship of the strains from the two major centers of cholera, China and South/Southeast Asia.
In this study, we obtained the genome sequences of 60 V. cholerae strains isolated in Shanghai between 1961 and 2011 by the Shanghai Center for Disease Control (CDC). The 60 strains with clear background information, which can be laboratory cultured, were selected as representatives from more than 2,900 isolates in the collection. This is a unique collection of isolates from one city over the entire period of the 7th pandemic. Shanghai is the largest port in the world and the largest city in China; in the past, there have been many outbreaks of cholera, but none of the published genome sequences from China have been reported to be from Shanghai isolates. This study was performed to clarify the evolutionary dynamics of V. cholerae in Shanghai, which had undergone breathtakingly rapid development in the last half-century. Our work may provide novel insights into the relationship between social changes and cholera epidemiology.
In our collection, most of the Shanghai strains were from the 7th pandemic. A phylogenetic analysis was performed for 54 Shanghai 7th-pandemic strain sequences, together with published 7th-pandemic genome data for 146 strains isolated around the world. All but one of the Shanghai 7th-pandemic strains fall into five “stages” that were dominant in Shanghai. Each stage appears to have arrived from South or Southeast Asia. Two Shanghai isolates are related to prepandemic isolates from Indonesia, and four are not related to the major pandemic lineage.
RESULTS AND DISCUSSION
Strain choice and analysis based on genome sequencing.
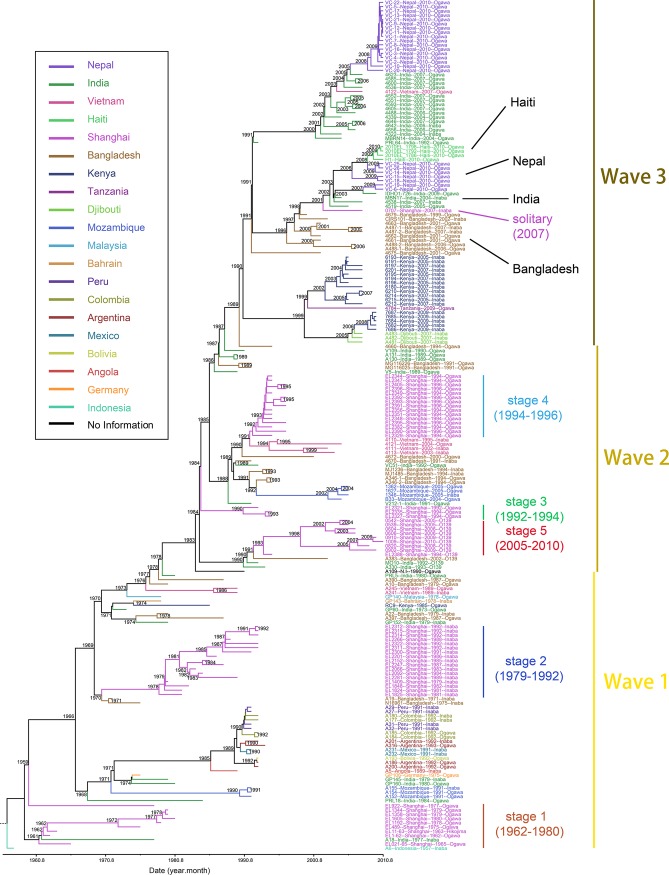
We obtained the genome sequences of 60 V. cholerae strains isolated in Shanghai, which had been retained by the Shanghai CDC and determined to be of the O1 or O139 serogroup and, if O1, the El Tor biotype. We constructed a phylogenetic tree (Fig. 1) based on whole-genome single-nucleotide polymorphisms (SNPs) from our 54 Shanghai 7th-pandemic strains and 146 publicly available 7th-pandemic strains isolated worldwide (see Table S1 in the supplemental material). The SNPs were filtered by a statistical method (11) to identify those introduced by recombination events, and only the recombination event that introduced the O139 O antigen was found. In total, 1,396 of the 1,417 mutational SNPs were congruent with the tree and could be located on specific branches (Fig. S1 and Table S2). Only 21 (1.5%) of the SNPs are not congruent and represent homoplasies involving repeat or reverse events, indicating that the tree structure is very robust. Of the 1,396 congruent SNPs, 1,204 are located in coding regions, of which 846 (70%) are nonsynonymous SNPs (ns SNPs) and 358 are synonymous SNPs. This proportion of nonsynonymous SNPs is very high and indicates positive selection pressure on the 7th-cholera-pandemic strains, but this result could in part be because short-term comparisons are likely to include mutations that will not be retained over longer periods (12).
FIG 1.
Phylogenetic tree of the 7th-pandemic V. cholerae strains. The maximum likelihood (ML) tree of 200 V. cholerae strains was based on mutational SNPs. The branches of Shanghai isolates corresponding to the five stages in the evolution of the 7th pandemic are labeled with epidemic dates. The dates shown are the estimates for the indicated nodes, taken from the results of the BEAST analysis.
Branch identifications and SNP numbers of the 7th-pandemic phylogenetic tree. Download FIG S1, PDF file, 0.6 MB (629.2KB, pdf) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
General characteristics of strains used in this study. Download Table S1, XLS file, 0.06 MB (59.5KB, xls) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SNPs used to build the 7th-pandemic phylogenetic tree and associated information. Download Table S2, XLS file, 0.3 MB (333.5KB, xls) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Additionally, for comparison of our strains with the collection reported by Didelot et al. (10), we incorporated their strains into our tree (Fig. S2). We used this tree to place our discussion of the Shanghai strains in the broader context of the whole of China.
Phylogenetic tree of our 7th-pandemic collection combined with the China strains from the collection of Didelot et al. (10). Download FIG S2, PDF file, 0.8 MB (861.4KB, pdf) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
V. cholerae harbors several key virulence elements, including cholera toxin (CTX), the toxin-coregulated pilus (TCP), Vibrio pathogenicity islands (VPIs), and the Vibrio seventh-pandemic island (VSP). SXT is an integrative conjugative element that harbors multiple antibiotic resistance genes (13). Genomic islands (GIs) also play an important role in the evolution of V. cholerae, and more than 70 have been observed and defined (14). We also investigated the distribution of these elements (Fig. S3).
Virulence-related elements identified among the 216 V. cholerae strains included in this study. The strains are arranged according to the trees shown in Fig. 1 and 3. Download FIG S3, PDF file, 0.7 MB (744.7KB, pdf) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The five major V. cholerae stages in Shanghai between 1961 and 2011.
The phylogenetic tree in Fig. 1 shows that 54 of the 7th-pandemic Shanghai isolates fall into five stages, most of which are single clades that dominated the isolates in succession. Each stage shares an independent ancestor and appears to have arrived independently. The five major stages in Shanghai are shown in Fig. 1. Four of these are in clades reported by Didelot and Wilson (15) (Fig. S3). The features and affiliations of the isolates from the five stages are discussed below. The changes for genetic elements in each stage and the SNPs specific to each stage are also discussed below.
Stage 1 (1962 to 1980).
Stage 1 includes 7 O1 Ogawa strains and one (EL11-63) O1 Hikojima strain (Fig. 1). All have the CTX pathogenicity island and the El Tor RS1, toxin-linked cryptic (TLC), and VPI-1 elements but do not have the self-transmissible, chromosomally integrating genetic (SXT) element drug resistance island (Fig. S3). Most of the stage 1 strains fit within the China 1.A clade described by Didelot et al., and in both collections, the earliest strain was isolated in 1962, and the last strain was isolated in 1978 or 1980. The locations of the isolates described by Didelot et al. were not reported.
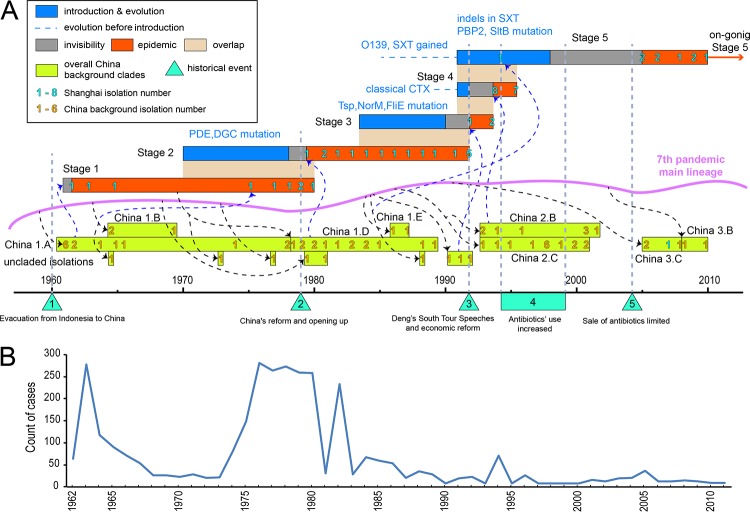
The 7th pandemic began in 1961 and was also first reported in China in 1961 (16). Our first Shanghai isolate is from 1962, and its clade is located at the root of the 7th-pandemic phylogenetic tree. This rapid transfer to China may be related to the migration of many Chinese people from Indonesia to China at that time (16) (Fig. 2A). The last stage 1 strain was isolated in 1980 (Fig. 1 and Fig. 2A), at a time when isolates from South and Southeast Asia were all from later-diverging branches of the tree. According to the case report (Fig. 2B), the outbreaks in the early 1960s and from 1975 to 1980 in Shanghai correspond well to the stage division.
FIG 2.
Details of the 5 stages of the Shanghai cholera outbreak. (A) Date ranges of each evolutionary and transmission process of the five stages, shown as color-coded horizontal bars, with details of the mutational events and sequenced isolate numbers given for each stage. The important historical events in Shanghai and/or China are also labeled. The correspondence of Shanghai isolates to the China background isolates and the introduction of China clades from the 7th-pandemic main lineage are linked by blue and black dotted arrows, respectively. (B) Line chart showing the numbers of cholera cases in Shanghai from 1962 to 2011.
All strains but EL922 share a common ancestor on branch 381 (Fig. S1) and are part of the stage 1 clade, while EL922 arrived independently (Fig. 1). Only one ns SNP on the rho gene (Table S2), which encodes a transcription termination factor (17), is located on branch 381 leading to stage 1. There is no ns SNP on branch 381. That indicates that these strains might already have been able to cause outbreaks in Shanghai as they arrived.
Stage 2 (1979 to 1992).
The stage 2 strains have the same GI patterns as those of the stage 1 strains (Fig. S3) but are O1 Inaba strains with the same rfbT mutation as N16961, which is the root of the stage 2 clade (18). The Shanghai stage 2 strains fall within the China group, China clade 1.D (Fig. S2).
The first isolate is from 1979, just before the last stage 1 isolate from 1980. Transfer occurred on branch 299 (Fig. S1), which diverges very close to the branch to a well-characterized Bangladesh strain, N16961. Two ns SNPs on branch 299 (Table S1) are in pathogenicity-related genes, including genes for a c-di-GMP phosphodiesterase A-related protein (PDE) and a GGDEF family protein (diguanylate cyclases [DGC]) (Fig. S4). The PDE and DGC genes participate in the synthesis and decomposition of c-di-GMP, which is a second messenger generally found in bacteria and can regulate many physiological processes, such as the generation of virulence factors, motility, and the promotion of biomembrane synthesis (19, 20).
Mutations in stages 2, 3, and 5. Download FIG S4, PDF file, 1.1 MB (1.1MB, pdf) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The replacement of stage 1 strains with stage 2 strains occurred between 1978 and 1980. During that time, China started its greatest recent political and economic reform, called “China’s reform and opening up” (Fig. 3). From 1978 to 1980, many special economic zones were established, and private enterprise, especially in the import and export trades, was opened. We suggest that these great changes in the social environment could have driven the succession of cholera in Shanghai during this period, although the connection between the genetic variations and the socioeconomic changes is unclear due to the complexity of China’s reform and opening up, which covered every aspect of China’s society. Future studies are required to confirm the function of those mutations and their correlation with socioeconomic changes.
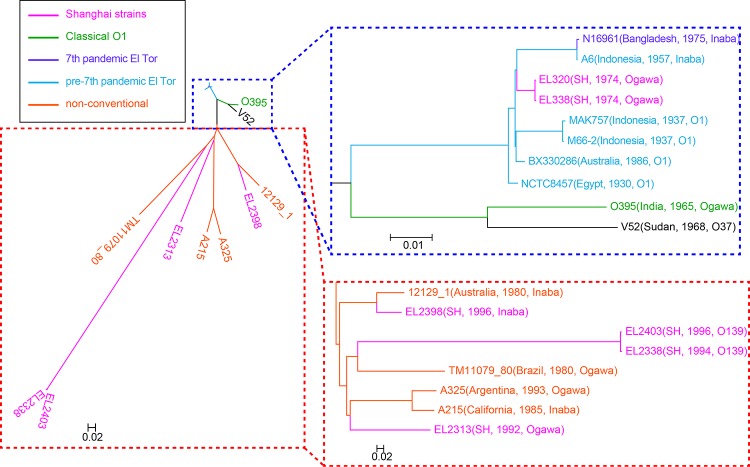
FIG 3.
Phylogenetic tree for 6th-pandemic and nonconventional strains combined with pre-7th- and 7th-pandemic strains. SH, Shanghai.
Stage 3 (1992 to 1994).
There are only 3 Shanghai strains in the stage 3 lineage that are connected to branch 251 (Fig. 1). They are O1 Ogawa strains, as in stage 1, and the GI pattern is the same as that of stages 1 and 2 (Fig. S3). Three strains reported by Didelot et al. are clustered in stage 3, as shown in Fig. S2, but they were not given a clade name. We suggest that this shared group be named China clade 1.F (Fig. S2).
There are 21 mutations on branch 251 (Fig. S1) dating from 1984 to 1990, and some of them are in genes that might be related to bacterial drug resistance, survival, and motility (Fig. S4). They include (i) a gene for a tail-specific protease (TSP), which is involved in the cleavage of a C-terminal peptide of 11 residues from the precursor form of penicillin-binding protein 3 (PBP3) and may be involved in bacterial defense against thermal and osmotic stresses (21); (ii) a gene encoding the multidrug resistance protein NorM, which is a multidrug efflux pump that functions as a Na+/drug antiporter and confers resistance to several drugs, including norfloxacin, ciprofloxacin, ethidium, kanamycin, and streptomycin (21); and (iii) the gene for the flagellar hook-basal body complex protein FliE, which is located in the flagellar basal body and functions as a structural adaptor for flagellin secretion (22, 23). The number and nature of the mutations on branch 251 suggest that clade 3 underwent adaptive evolution before its first isolation in 1992 (Fig. 3).
Similar to the stage 2 lineage, stage 3 strains also underwent a relatively long local evolution process before their first isolation in 1992. Then, a special economic zone was established in Shanghai, which may have caused a second abrupt environmental change. These environmental changes may have caused a lineage with better selective advantages to become the dominant population. The end of the prevalence of stage 3 strains may be related to a change in the use of antibiotics. In approximately 1994, the use of cephalosporin started to increase dramatically (24).
Stage 4 (1994 to 1996).
Stage 4 strains are O1 Ogawa strains, similar to stage 1, but their GI content differs from those of stages 1 to 3 in the replacement of the El Tor CTX island with the classical CTX island, as observed for the parental lineage (Fig. 1 and Fig. 2A). Stage 4 strains fall within China clade 2.B described by Didelot et al. (Fig. S2), when there was an outbreak in Shanghai in 1994 (Fig. 2B).
The introduction to China occurred on branch 194, which has no mutations, but there is one SNP on branch 195, which leads to all isolates but EL2329. The mutation is in the lepA gene, which encodes a GTP-binding protein. Branch 194 diverges close to the branch for a group of Vietnam strains (Fig. 1 and Fig. S1). The classical CTX island of stage 4 strains may have been acquired by their South Asian ancestor in 1988 on branch 190 (Fig. 1, Fig. 2A, and Fig. S1).
Stage 5 (2005 to 2010).
Stage 5 strains are of serogroup O139 and have the same GI pattern as those of the stage 1 to 3 strains, except for the presence of the drug resistance island SXT (Fig. 1 and Fig. S3). The O139 variant of the 7th pandemic was first isolated in India in 1992 and soon came to dominate in South and Southeast Asia. From 1978 to 1984, this branch gained the SXT island (9). However, after 1995, the number of clinical O139 cases decreased (25). In contrast, nearly all of the clinical V. cholerae cases reported in Shanghai after 2005 were of serogroup O139 (Fig. 1 and Fig. 2A). A similar phenomenon has been observed for the prevalence of O139 isolates in Mainland China (26). The O139 lineage reached Shanghai on branch 259 (Fig. S1), which was estimated to span from 1991 to 1993, soon after it was first reported in India, and the first isolate reported in Shanghai was from 1994. However, it did not become dominant until 2005 (Fig. 2A), perhaps due to the increase in antibiotic use between 1994 and 1999 (24). O139 strains were not present in the set described by Didelot et al., but as the latest isolate in that collection was from 2005, this lack does not mean that this serogroup was not widespread in China generally during the period of its presence in Shanghai.
The 3 mutations on branch 259 are all synonymous. However, there are several mutations affecting proteins that may influence virulence on subsequent branches. The mutations on branch 260 represented the genetic changes between EL2388 (in 1994) and other O139 strains (2005 to 2010), which may reveal key evolutionary dynamics during this period. All strains except for EL2388 (Table S1) carry two nonsynonymous mutations in the well-known drug-related genes pbp2, which encodes penicillin-binding protein 2 (PBP2) (27), and sltB, which encodes a lytic murein transglycosylase, with a function in beta-lactam antibiotic resistance (28) (Fig. S4). These mutations are on branch 260, which was estimated to run from 1993 to 1998 (Fig. 1 and Fig. S1), when the use of beta-lactam antibiotics increased dramatically in Shanghai. O139 isolates from other areas do not have mutations in these two genes. These genetic changes may explain how the stage 5 O139 lineage became dominant in Shanghai, and the increment of antibiotic resistance of these stage 5 strains has also been confirmed by antimicrobial susceptibility experiments (Table S3).
Antimicrobial susceptibility of strains isolated in Shanghai. Download Table S3, XLS file, 0.02 MB (23KB, xls) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The SXT islands of strains EL2388 and 0902 are the same as or similar to that of the early O139 strain MO10. However, many stage 5 strains, such as strains 0910, 1009, and 0802, have experienced different deletion events, most of which are related to the loss of trimethoprim resistance (Fig. S5). The lack of trimethoprim resistance in these strains must be a result of convergent evolution. Trimethoprim resistance may have had a weak contribution to the survival of O139 strains in the Shanghai environment at that time (24).
Genetic organization of the SXT regions of stage 5 O139 strains. Download FIG S5, PDF file, 0.2 MB (215.3KB, pdf) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of the 7th pandemic in China and Shanghai.
In general, good concordance exists between the strains in the two collections. The major exception, the absence of clade 2.C isolates in Shanghai, is probably due to a lack of samples from 1997 to 2004 in the Shanghai collection. However, the absence of stage 5 O139 isolates in the rest of China may reflect a real difference in Shanghai. However, the existing data include relatively few strains for such a large country, so it is not possible to say how much the O139 strain actually spread in China.
Solitary strain 0707.
The 2007 0707 O1 Inaba strain is the only wave 3 Shanghai isolate (Fig. 1). It has the “hybrid CTX,” El Tor RS1, TLC, VPI-1, and SXT GI pattern, with some deletion events as well as some other GIs (Fig. S3). It is related to the Bangladesh CIRS1010 strain from a lineage of South Asian strains, which is prevalent in Nepal and was recently transmitted from there to Haiti to cause the Haiti cholera outbreak in 2010 (29).
Two isolates of prepandemic strains related to the Makassar outbreaks.
Two nearly identical 1974 O1 Ogawa clinical isolates (EL320 and EL338) (Fig. 3) diverged from the main pathogenic lineage about midway between the divergence of the 1937 prepandemic outbreak in Indonesia and the divergence of the 1957 prepandemic outbreak in Indonesia from the main lineage leading to the 7th pandemic (8).
These two strains do not have RS1 elements (Fig. S3), and their CTX island is a hybrid type (with the classical ctxB gene and the El Tor rstR gene, while the CTX of wave 3 strains have the El Tor ctxB gene and the classical rstR gene [30]) that has never been found in other V. cholerae strains. The variation in CTX from 6th-pandemic strains to the recent 7th-pandemic strains is displayed in Fig. S6. The CTX types of the two Shanghai strains fill the gap in the evolution of El Tor CTX. From the end of the 6th pandemic to the recent 7th pandemic, the CTX island, which is a key virulence element of V. cholerae, evolved from the classical type to the El Tor type and then back to the classical type, which indicates that V. cholerae can adapt to environmental changes by changing its virulence.
Evolution of the CTX type. Download FIG S6, PDF file, 0.4 MB (402.5KB, pdf) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Four nontoxigenic O1/O139 strains are the result of independent recombination events at the O-antigen loci.
Four of the Shanghai clinical isolates are nontoxigenic and branch from the known pathogenic lineage well before the divergence of the classical and El Tor pandemic lineages (Fig. 3). They do not have the CTX, RS1, TLC, and SXT GIs or even VPI-1, which carries the tcp gene cluster (Fig. S3). Thus, it is concluded that the O1 and O139 antigen phenotypes arose by horizontal gene transfer, respectively, in the evolution of these nontoxigenic strains, in agreement with data from a previous study (9). Except for the wbfA gene, the major region of the O-antigen gene cluster of nonconventional O139 strains shares high identity (near the average identity of housekeeping genes between these two lineages) to that of the typical 7th-pandemic O139 strains, similar to the O-antigen gene cluster of the O22 serogroup, for which the evolution process is thought to be caused mainly by recombination events (Fig. S7). The finding that these strains are from cholera cases can be attributed to independent recombination events at the O-antigen loci, including the O139 antigen.
Alignment of O139 O-antigen gene clusters of Shanghai nonconventional strains with those of related lineages. Download FIG S7, PDF file, 0.7 MB (704.4KB, pdf) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conclusions.
We found that 54 of the 60 7th-pandemic strains isolated in Shanghai between 1962 and 2010 cluster into five clones. Each stage was dominant for a period, with very little overlap between periods of dominance, although the small number of isolates available would not show the presence of a clone at lower numbers outside its period of dominance. This situation is very different from the situation in South and Southeast Asia, where there was much greater diversity during that period, as shown by a previous report (9). It is striking that the South/Southeast Asian lineage was the source of each of the five stages of the samples that we observed. This result was quite unexpected, as historically, South/Southeast Asia and China were the two areas in which cholera had been reported for centuries, but it appears that for the 7th pandemic, at least, South and Southeast Asia were the center of evolution for cholera. Additionally, the transitions between each stage were reliably associated with social changes in China, especially Shanghai, showing the importance of human social activities in the dynamic evolution of the Shanghai 7th pandemic.
MATERIALS AND METHODS
Strains and culture conditions.
All strains used in this are listed in Table S1 in the supplemental material. The 60 V. cholerae strains (listed in Table S1) were kindly provided by the Shanghai Municipal Center for Disease Control and Prevention. All strains were isolated from clinical specimens and stored at −80°C in Luria-Bertani (LB) broth with 20% glycerol. LB broth at 37°C was used as the standard bacterial growth condition.
Genome sequencing.
DNA was prepared from 1 ml of cultures grown overnight with the Wizard genomic DNA purification kit (Promega) according to the manufacturer’s instructions. We obtained draft genome sequences for 60 V. cholerae strains (Table S1) using Solexa pair-end sequencing (Illumina, Little Chesterford, Essex, United Kingdom), with a depth of 90- to 100-fold coverage, and the sequences were de novo assembled using Velvet v1.0 (31). Protein prediction and annotation of the newly sequenced genomes were performed by using Glimmer v3.0 (32) and BLASTp v2.6.0+ (33), using the 1 October 2018 version of the NCBI nr database (34), respectively. The annotation result was verified manually using the annotation of the genome sequence of reference strain N16961.
SNP calling and recombination detection.
The SNPs across the genome of each strain were identified by aligning and mapping against the reference genome sequence of strain N16961 using Mauve v2.3.1 (35) and BWA v0.7.13 (36). We used RecDect v1.0 within SaRTree pipeline v1.1 (11) to analyze and remove SNPs due to recombination.
Phylogenetic analyses and divergence time analysis.
The maximum likelihood (ML) phylogenetic tree was built by using RAxML v8.2.11 (37) with 1,000 bootstraps and the –m GTRGAMMER parameter, including 1,417 mutational SNPs (Table S2). These mutational SNPs were grouped into different patterns according to the strains in which they occur, and these patterns were then allocated to specific branches of the ML tree by using SaRTree pipeline v1.1.
We used BEAST v1.10.4 (38) to infer the divergences dates of the lineages. The BEAST estimations were performed using the evolutionary model GTR+Γ4 (general time reversible model with gamma distributions) with log-normal relaxed molecular clocks, the coalescent constant population size, and 10,000,000 Markov chain Monte Carlo (MCMC) states sampled every 1,000 steps. Different models and parameters were tested by parallel runs of BEAST, and the best model used in this study was selected by comparing the marginal likelihoods, when all the effective sample sizes (ESSs) were >200 for all the parameters, using Tracer v1.5. The first 5% of trees were then discarded as burn-ins, and the posterior probability limit filtering threshold was set to 50%. The divergence dates were estimated using the remaining trees, which were mapped onto the maximum likelihood phylogeny generated in the last step with the mean height of the nodes using TreeAnnotator v1.10.4. The output tree was displayed by using FigTree v1.4.4.
Comparative genomics.
To examine virulence elements and GI-1 to GI-67, we located and screened genes or gene clusters using BLASTp v2.6.0+ (33). The reference sequences of GIs were determined as described in a previous article (14). Protein secondary structure was predicted using PSIPRED v4.0 (39).
Antimicrobial susceptibility.
An antimicrobial susceptibility experiment was performed for the Shanghai strains using the Etest (AB Biodisk, Solna, Sweden), according to the manufacturer’s instructions, on LB agar plates, with incubation at 37°C for 18 to 24 h.
Data availability.
Sequence data are available in the NCBI RefSeq database under project accession no. PRJNA555013. The accession numbers of the 60 strains’ genomes are listed in Table S1 in the supplemental material.
ACKNOWLEDGMENTS
We thank Yamin Sun and Wenchao Lin for technical support of genome analysis.
This work was funded by the National Key Programs for Infectious Diseases of China (no. 2017ZX10303405 and 2017ZX10104002-001-006) and the National Natural Science Foundation of China (no. 31820103002, 31530083, 31770144, 81471904, 81772148, and 81611530714).
REFERENCES
- 1.Barua D. 1992. History of cholera, p 1–36. In Barua D, Greenough WB (ed), Cholera. Current topics in infectious disease. Springer, Boston, MA. [Google Scholar]
- 2.Blake PA. 1994. Historical perspectives on pandemic cholera, p 293–295. In Wachsmuth IK, Blake PA, Olsvik Ø (ed), Vibrio cholerae and cholera: molecular to global perspectives. ASM Press, Washington, DC. [Google Scholar]
- 3.Ali M, Nelson AR, Lopez AL, Sack DA. 2015. Updated global burden of cholera in endemic countries. PLoS Negl Trop Dis 9:e0003832. doi: 10.1371/journal.pntd.0003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan American Health Organization, World Health Organization. 2014. Epidemiological update: cholera—18 February 2014. Pan American Health Organization, World Health Organization, Washington, DC. [Google Scholar]
- 5.Islam MS, Mahmud ZH, Ansaruzzaman M, Faruque SM, Talukder KA, Qadri F, Alam M, Islam S, Bardhan PK, Mazumder RN, Khan AI, Ahmed S, Iqbal A, Chitsatso O, Mudzori J, Patel S, Midzi SM, Charimari L, Endtz HP, Cravioto A. 2011. Phenotypic, genotypic, and antibiotic sensitivity patterns of strains isolated from the cholera epidemic in Zimbabwe. J Clin Microbiol 49:2325–2327. doi: 10.1128/JCM.00432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukhopadhyay AK, Takeda Y, Balakrish Nair G. 2014. Cholera outbreaks in the El Tor biotype era and the impact of the new El Tor variants. Curr Top Microbiol Immunol 379:17–47. doi: 10.1007/82_2014_363. [DOI] [PubMed] [Google Scholar]
- 7.Kaper JB, Morris JG Jr, Levine MM. 1995. Cholera. Clin Microbiol Rev 8:48–86. doi: 10.1128/CMR.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu D, Liu B, Feng L, Ding P, Guo X, Wang M, Cao B, Reeves PR, Wang L. 2016. Origins of the current seventh cholera pandemic. Proc Natl Acad Sci U S A 113:E7730–E7739. doi: 10.1073/pnas.1608732113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, Niyogi SK, Kim EJ, Ramamurthy T, Chun J, Wood JLN, Clemens JD, Czerkinsky C, Nair GB, Holmgren J, Parkhill J, Dougan G. 2011. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Didelot X, Pang B, Zhou Z, McCann A, Ni P, Li D, Achtman M, Kan B. 2015. The role of China in the global spread of the current cholera pandemic. PLoS Genet 11:e1005072. doi: 10.1371/journal.pgen.1005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu D, Liu B, Wang L, Reeves PR. 2019. Living trees: high-quality reproducible and reusable construction of bacterial phylogenetic trees. Mol Biol Evol. doi: 10.1093/molbev/msz241. [DOI] [PubMed] [Google Scholar]
- 12.Ho SYW, Phillips MJ, Cooper A, Drummond AJ. 2005. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol Biol Evol 22:1561–1568. doi: 10.1093/molbev/msi145. [DOI] [PubMed] [Google Scholar]
- 13.Beaber JW, Hochhut B, Waldor MK. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol 184:4259–4269. doi: 10.1128/jb.184.15.4259-4269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, Taviani E, Jeon Y-S, Kim DW, Lee J-H, Brettin TS, Bruce DC, Challacombe JF, Detter JC, Han CS, Munk AC, Chertkov O, Meincke L, Saunders E, Walters RA, Huq A, Nair GB, Colwell RR. 2009. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A 106:15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang X. 2014. The global cholera pandemic reaches Chinese villages: population mobility, political control, and economic incentives in epidemic prevention, 1962–1964. Mod Asian Stud 48:754–790. doi: 10.1017/S0026749X12000807. [DOI] [Google Scholar]
- 17.Banerjee S, Chalissery J, Bandey I, Sen R. 2006. Rho-dependent transcription termination: more questions than answers. J Microbiol 44:11–22. [PMC free article] [PubMed] [Google Scholar]
- 18.Liang W, Wang L, Liang P, Zheng X, Zhou H, Zhang J, Zhang L, Kan B. 2013. Sequence polymorphisms of rfbT among the Vibrio cholerae O1 strains in the Ogawa and Inaba serotype shifts. BMC Microbiol 13:173. doi: 10.1186/1471-2180-13-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rashid MH, Rajanna C, Ali A, Karaolis DKR. 2003. Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol Lett 227:113–119. doi: 10.1016/S0378-1097(03)00657-8. [DOI] [PubMed] [Google Scholar]
- 20.Tamayo R, Schild S, Pratt JT, Camilli A. 2008. Role of cyclic di-GMP during El Tor biotype Vibrio cholerae infection: characterization of the in vivo-induced cyclic di-GMP phosphodiesterase CdpA. Infect Immun 76:1617–1627. doi: 10.1128/IAI.01337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hara H, Yamamoto Y, Higashitani A, Suzuki H, Nishimura Y. 1991. Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J Bacteriol 173:4799–4813. doi: 10.1128/jb.173.15.4799-4813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller V, Jones CJ, Kawagishi I, Aizawa SI, Macnab RM. 1992. Characterization of the fliE genes of Escherichia coli and Salmonella typhimurium and identification of the FliE protein as a component of the flagellar hook-basal body complex. J Bacteriol 174:2298–2304. doi: 10.1128/jb.174.7.2298-2304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reed KA, Hobert ME, Kolenda CE, Sands KA, Rathman M, O’Connor M, Lyons S, Gewirtz AT, Sansonetti PJ, Madara JL. 2002. The Salmonella typhimurium flagellar basal body protein fliE is required for flagellin production and to induce a proinflammatory response in epithelial cells. J Biol Chem 277:13346–13353. doi: 10.1074/jbc.M200149200. [DOI] [PubMed] [Google Scholar]
- 24.Tu L, Chen M, Li W, Zhang X, Chen H-Y, Wang W-J. 2013. Characterization of phenotype and molecular characteristics on Vibrio cholerae strains isolated in Shanghai, 1962-2011. Zhonghua Liu Xing Bing Xue Za Zhi 34:815–818. [PubMed] [Google Scholar]
- 25.Banerjee R, Das B, Balakrish Nair G, Basak S. 2014. Dynamics in genome evolution of Vibrio cholerae. Infect Genet Evol 23:32–41. doi: 10.1016/j.meegid.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Zhang P, Zhou H, Diao B, Li F, Du P, Li J, Kan B, Morris JG, Wang D. 2014. A molecular surveillance reveals the prevalence of Vibrio cholerae O139 isolates in China from 1993 to 2012. J Clin Microbiol 52:1146–1152. doi: 10.1128/JCM.03354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powell AJ, Tomberg J, Deacon AM, Nicholas RA, Davies C. 2009. Crystal structures of penicillin-binding protein 2 from penicillin-susceptible and -resistant strains of Neisseria gonorrhoeae reveal an unexpectedly subtle mechanism for antibiotic resistance. J Biol Chem 284:1202–1212. doi: 10.1074/jbc.M805761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legaree BA, Clarke AJ. 2008. Interaction of penicillin-binding protein 2 with soluble lytic transglycosylase b1 in Pseudomonas aeruginosa. J Bacteriol 190:6922–6926. doi: 10.1128/JB.00934-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, Hine E, Su Q, Tallon LJ, Prosper JB, Furth K, Hoq MM, Li H, Fraser-Liggett CM, Cravioto A, Huq A, Ravel J, Cebula TA, Colwell RR. 2012. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc Natl Acad Sci U S A 109:E2010–E2017. doi: 10.1073/pnas.1207359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim EJ, Lee CH, Nair GB, Kim DW. 2015. Whole-genome sequence comparisons reveal the evolution of Vibrio cholerae O1. Trends Microbiol 23:479–489. doi: 10.1016/j.tim.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 31.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. 2005. GenBank. Nucleic Acids Res 33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darling AE, Treangen TJ, Messeguer X, Perna NT. 2007. Analyzing patterns of microbial evolution using the mauve genome alignment system. Methods Mol Biol 396:135–152. doi: 10.1007/978-1-59745-515-2_10. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 38.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol 4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGuffin LJ, Bryson K, Jones DT. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Branch identifications and SNP numbers of the 7th-pandemic phylogenetic tree. Download FIG S1, PDF file, 0.6 MB (629.2KB, pdf) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
General characteristics of strains used in this study. Download Table S1, XLS file, 0.06 MB (59.5KB, xls) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SNPs used to build the 7th-pandemic phylogenetic tree and associated information. Download Table S2, XLS file, 0.3 MB (333.5KB, xls) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic tree of our 7th-pandemic collection combined with the China strains from the collection of Didelot et al. (10). Download FIG S2, PDF file, 0.8 MB (861.4KB, pdf) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Virulence-related elements identified among the 216 V. cholerae strains included in this study. The strains are arranged according to the trees shown in Fig. 1 and 3. Download FIG S3, PDF file, 0.7 MB (744.7KB, pdf) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutations in stages 2, 3, and 5. Download FIG S4, PDF file, 1.1 MB (1.1MB, pdf) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antimicrobial susceptibility of strains isolated in Shanghai. Download Table S3, XLS file, 0.02 MB (23KB, xls) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genetic organization of the SXT regions of stage 5 O139 strains. Download FIG S5, PDF file, 0.2 MB (215.3KB, pdf) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Evolution of the CTX type. Download FIG S6, PDF file, 0.4 MB (402.5KB, pdf) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Alignment of O139 O-antigen gene clusters of Shanghai nonconventional strains with those of related lineages. Download FIG S7, PDF file, 0.7 MB (704.4KB, pdf) .
Copyright © 2019 Hu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Sequence data are available in the NCBI RefSeq database under project accession no. PRJNA555013. The accession numbers of the 60 strains’ genomes are listed in Table S1 in the supplemental material.