Abstract
Here we present new and original data on the endogenous conversion of tyrosol (Tyr) into hydroxytyrosol (OHTyr) in humans and its effects on the cardiovascular system. A randomized, crossover, controlled clinical trial was performed with individuals at cardiovascular risk (n = 33). They received white wine (WW) (females 1, males 2 standard drinks/day), WW plus Tyr capsules (WW + Tyr) (25mg Tyr capsule, one per WW drink), and water (control) ad libitum. Intervention periods were of 4 weeks preceded by three-week wash-out periods. We assessed the conversion of Tyr to OHTyr, its interaction with a polygenic activity score (PAS) from CYP2A6 and CYP2D6 genotypes, and the effects on cardiovascular risk markers. For further details and experimental findings please refer to the article “Cardiovascular benefits of tyrosol and its endogenous conversion into hydroxytyrosol in humans. A randomized, controlled trial” [1].
Keywords: Tyrosol, Hydroxytyrosol, Endogenous conversion, CYP2A6, CYP2D6, Cardiovascular risk
Specifications Table
| Subject | Nutrition |
| Specific subject area | Nutritional biochemistry and genotype interaction |
| Type of data | Tables, Figures, File text |
| How data were acquired |
HPLC-MS-MS for Tyr and OHTyr Genotyping of allelic variants of CYP2A6 and CYP2D6 .A polygenic score activity was calculated Reactive hyperemia index, RHI for endothelial function Automated methods, HPLC, and ELISA for cardiovascular risk biomarkers Real-time polymerase chain reaction (PCR) for gene expression |
| Data format | Raw data collection and analysis |
| Parameters for data collection | Before and after each one of the three interventions with 1) white wine (WW) (females 1, males 2 standard drinks/day), 2) WW plus Tyr capsules (WW + Tyr) (25mg Tyr capsule, one per WW drink), and 3) water (control) ad libitum. Intervention periods were of 4 weeks preceded by 3-weeks washout periods. |
| Description of data collection | Biological samples were collected and processed in the context of a randomized controlled intervention trial by field investigators |
| Data source location | Barcelona, Spain |
| Data accessibility | With the article |
| Related research article | Boronat A et al. Cardiovascular benefits of tyrosol and its endogenous conversion into hydroxytyrosol in humans. A randomized, controlled trial. Free Radical Biol Med, 2019 Aug 31;143:471-481.https://doi.org/10.1016/j.freeradbiomed.2019.08.032. |
Value of the Data
|
1. Data
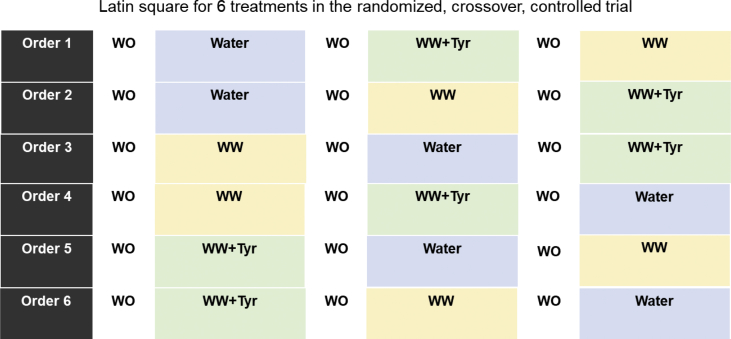
Thirty-three participants (21 men and 12 women) were randomly allocated to participate in a clinical trial, and 32 participants completed the trial. Initially, 192 subjects were assessed for eligibility, 157 were excluded for 1) not meeting the inclusion criteria, 2) refusing to participate, 3) taking medication non compatible with the interventions, 4) suffering from a coronary heart disease, 5) having undergone bariatric surgery, 6) intestinal alterations, 7) mobility problems, 8) chronic inflammatory diseases, 9) dysregulated hypertension, 10) illicit drug consumption, 11) heavy alcohol consumption, and 12) hepatic alterations. The 33 volunteers were randomly allocated to receive the following treatments for 4 weeks: WW, WW + Tyr and control intervention [1] (Fig. 1).
Fig. 1.
Schema of the clinical trial. Intervention periods of 4 weeks. WO: wash-out period (3 weeks) without alcohol and following a low-phenolic content diet. WW (white wine): 2 glasses (270 mL, 27 g of alcohol, 2.8 mg of Tyr and 0.4 mg of OHTyr) for men, and 1 glass (135 mL, 13.5 mg, 1.4 mg of Tyr, and 0.2 mg of OHTyr) for women. WW + Tyr (white wine plus tyrosol): 2 glasses of wine: 270 mL, 27 g of alcohol, 2.8 + 50 mg of Tyr (2 capsules), and 0.4 mg of OHTyr for men, and 1 glass:135 mL, 13.5 g of alcohol, 1.4 mg + 25 mg of Tyr (1 capsule), and 0.2 mg of OHTyr for women.
Baseline characteristics of the participants are shown in Table 1. No dietary differences were observed among interventions (Table 2). Volunteers were genotyped for multiple allelic variants of CYP2A6 and CYP2D6 (Table 3). For each enzyme, an activity score was given to each volunteer according to the alleles identified (Table 4). A pooled polygenic activity score (PAS) was calculated by adding the two activity scores together.
Table 1.
Baseline characteristics of the participants.
| Variable | Values |
|---|---|
| Age, y | 65.3 ± 6.2 |
| Gender, n (%) | |
| Women | 12 (36.4%) |
| Men | 21 (63.6%) |
| BMI, kg/m2 | 32.6 ± 4.2 |
| LDL cholesterol, mg/dL | 118 ± 34.4 |
| HDL cholesterol, mg/dL | 50.2 ± 12.9 |
| Total cholesterol, mg/dL | 192 ± 39.3 |
| Triglycerides, mg/dL | 120 ± 72.2 |
| Cardiovascular Risk factors, n (%) | |
| Current smokers | 6 (18.2%) |
| Family history of premature CHD | 6 (19.4%) |
| Obesity (BMI ≥ 25kg/m2) | 32 (97.0%) |
| Type 2 Diabetes | 13 (39.4%) |
| Hypertension | 28 (84.8%) |
| High LDL cholesterol (>130 mg/dL) | 25 (75.6%) |
| Low HDL cholesterol (<40 mg/dL for men or <50 mg/dL for women) | 8 (24.2%) |
| Medications, n (%) | |
| Alfa blockers | 2 (6.1%) |
| Beta blockers | 6 (18.2%) |
| ACE inhibitors | 14 (42.4%) |
| Angiotensin II receptor antagonists | 11 (33.3%) |
| Diuretics | 13 (39.4%) |
| Statins | 16 (48.5%) |
| Oral hypoglycemic drugs | 12 (36.4%) |
| Acetylsalicylic acid | 10 (30.3%) |
Data presented as mean ± SD or n (%) (n = 33). BMI, body mass index; LDL, low density lipoproteins; HDL, high density lipoproteins; CHD, coronary heart disease.
Table 2.
Energy, nutrients, and fiber at the beginning and at the end of the clinical trial.
| Variable | Treatment |
P* | |||||
|---|---|---|---|---|---|---|---|
| Control | P | WW | P | WW+TYR | P | ||
| Energy, kcal/day | |||||||
| Baseline | 1695 ± 446 | 1663 ± 421 | 1624 ± 370 | NS | |||
| 12-week | 1643 ± 361 | 0.616 | 1650 ± 354 | 0.868 | 1737 ± 450 | 0.082 | |
| HC, % energy | |||||||
| Baseline | 38.2 ± 8.6 | 40.2 ± 6.4 | 38.8 ± 6.3 | NS | |||
| 12-week | 38.3 ± 7.3 | 0.906 | 37.6 ± 7.7 | 0.095 | 37.8 ± 7.5 | 0.360 | |
| HC, grams | |||||||
| Baseline | 159 ± 48 | 165± 43 | 157 ± 38 | NS | |||
| 12-week | 156 ± 41 | 0.811 | 153 ± 40 | 0.209 | 163 ± 45 | 0.532 | |
| Protein, % energy | |||||||
| Baseline | 20.9 ± 4.0 | 19.2 ± 3.0 | 21.5 ± 4.5 | NS | |||
| 12-week | 21.2 ± 4.1 | 0.578 | 19.5 ± 4.8 | 0.028 | 19.0± 3.9 | 0.142 | |
| Protein, grams | |||||||
| Baseline | 88 ± 25 | 79 ± 19 | 86 ± 24 | NS | |||
| 12-week | 87 ± 25 | 0.939 | 81 ± 30 | 0.651 | 82 ± 23 | 0.184 | |
| Total Fat, % energy | |||||||
| Baseline | 40.7 ± 7.5 | 40.3 ± 6.2 | 39.2 ± 6.6 | NS | |||
| 12-week | 40.2 ± 7.2 | 0.625 | 36.7 ± 9.0 | 0.496 | 38.6 ± 6.3 | 0.230 | |
| Total Fat, grams | |||||||
| Baseline | 78 ± 29 | 76 ± 28 | 72 ± 24 | NS | |||
| 12-week | 74 ± 23 | 0.489 | 68 ± 24 | 0.015 | 76± 28 | 0.809 | |
| SFA, % energy | |||||||
| Baseline | 11.4± 3.8 | 10.1 ± 3.0 | 11.4 ± 4.2 | NS | |||
| 12-week | 10.7 ± 3.7 | 0.526 | 10.1 ± 3.9 | 0.953 | 10.4 ± 3.7 | 0.272 | |
| SFA, grams | |||||||
| Baseline | 22 ± 12 | 19 ± 9 | 21 ± 12 | NS | |||
| 12-week | 20 ± 10 | 0.428 | 19 ± 10 | 0.873 | 21 ± 11 | 0.855 | |
| MUFA,% energy | |||||||
| Baseline | 19.8 ± 5.0 | 19.7 ± 4.2 | 18.6 ± 5.0 | NS | |||
| 12-week | 20.1 ± 4.2 | 0.893 | 18.4 ± 5.1 | 0.177 | 19.6 ± 3.5 | 0.172 | |
| MUFA, grams | |||||||
| Baseline | 53.3 ± 21.0 | 53.8 ± 15.5 | 53.0 ± 16.6 | NS | |||
| 12-week | 51.9 ± 21.2 | 0.475 | 46.5 ± 13.3 | 0.274 | 50.0 ± 14.3 | 0.054 | |
| PUFA, % energy | |||||||
| Baseline | 6.1 ± 2.6 | 6.8 ± 2.5 | 6.0 ± 2.1 | NS | |||
| 12-week | 5.9 ± 2.3 | 0.697 | 5.2 ± 2.1 | 0.222 | 5.4 ± 1.9 | 0.179 | |
| PUFA, grams | |||||||
| Baseline | 12 ± 6 | 13 ± 8 | 11 ± 4 | NS | |||
| 12-week | 11 ± 7 | 0.901 | 10 ± 5 | 0.025 | 11 ± 6 | 0.960 | |
| Fiber, g/day | |||||||
| Baseline | 20 ± 7 | 23 ± 11 | 20 ± 8 | NS | |||
| 12-week | 20 ± 8 | 0.894 | 23 ± 11 | 0.848 | 21 ± 9 | 0.408 | |
Dietary data is expressed as mean ± SD (N = 32). HC, carbohydrates; SFA, saturated fatty acids; MUFA, monounsaturated fatty acids; PUFA, polyunsaturated fatty acids. P Intra-treatment comparisons by Student's t-test. *P value for ANOVA repeated measures adjusted by age and sex.
Table 3.
Characteristics of CYP2A6 and CYP2D6 SNPs tested.
| Tested Allelic Variants | Reference Number | Nucleotide Substitution | Amino acid substitution | TaqMan Assay ID |
|---|---|---|---|---|
| CYP2A6 | rs1801272 | 479T > A | Leu160His | C_27861808_60 |
| rs28399433 | - 48T > G | Upstream | C_30634332_10 | |
| CYP2D6 | rs1135840 | 4181G > C | Ser486Thr | C_27102414_10 |
| rs16947 | 2851C > T | Arg296Cys | C_27102425_10 | |
| rs3892097 | 1847G > A | Intron Variant | C_27102431_DO | |
| rs5030656 | 2616_2618delAAG | Lys281del | C_32407229_60 | |
| rs1065852 | 100C > T | Pro34Ser | C_11484460_40 | |
| rs769258 | 31G > A | Val11Met | C_27102444_80 | |
| rs28371725 | 2989G > A | Intron Variant | C_34816116_20 |
Table 4.
Activity score assigned to each tested variant in the PAS model.
| Tested Allelic | Variants | Functional consequence | Activity score | Defining SNP |
|---|---|---|---|---|
| CYP2A6 | *2 | No function | 0 | 479T > A |
| *4 | No expression | 0 | Gene deletion | |
| *9 | Decreased | +0.5 | - 48T > G | |
| *12 | Decreased | +0.5 | Hybrid allele with CYP2A7 | |
| *1xN | Increased | +2 | Multiple copies | |
| CYP2D6 | *2 | Normal | +1 | 2851C > T 4181 G > C |
| *4 | No function | 0 | 1847 G > Aa | |
| *5 | No expression | 0 | Gene deletion | |
| *9 | Decreased | +0.5 | 2616 del AGG | |
| *10 | Decreased | +0.5 | 100C > T 4181 G > C |
|
| *35 | Normal | +1 | 31G > A 2851C > T 4181 G > C |
|
| *41 | Decreased | +0.5 | 2989 G > A 2851C > T 4181 G > C |
|
| *1xN | Increased | +2 | Multiple copies | |
| *2xN | ||||
| *35xN |
*4 sub-alleles can commonly present other SNPs such as 100C > T, 4181 G > C and/or 2851C > T.
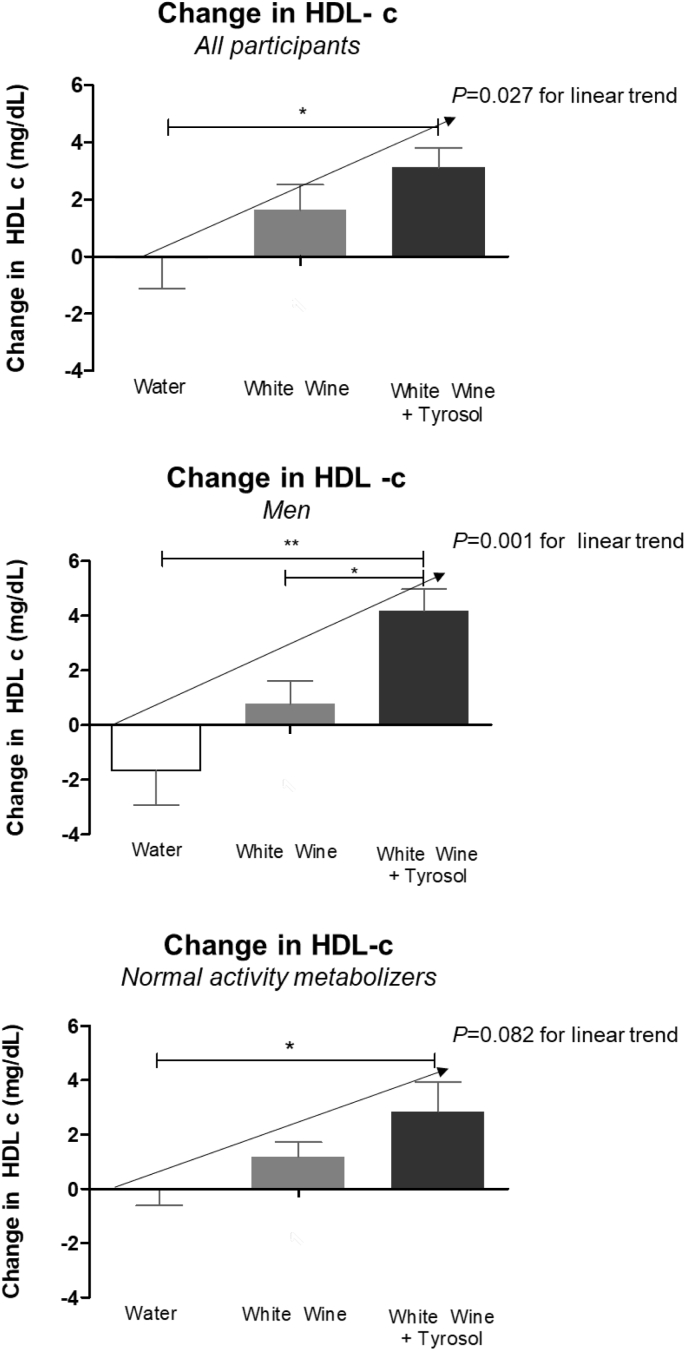
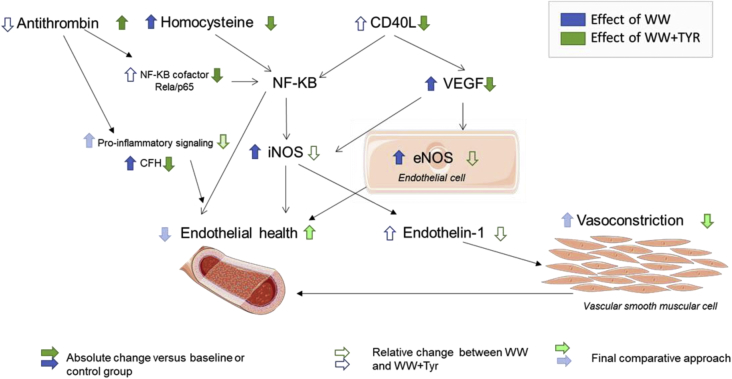
On the basis of PAS, 11 individuals were categorized as low (LA, PAS range: 1–2.5), 19 as normal (NA, PAS range: 3–4), and 2 as rapid (RA, PAS range = 5) activity metabolizers. Due to their low number, RAs were excluded for analyses. Age and gender were equally distributed among the three groups. The conversion of Tyr into OHTyr was assessed by measuring OHTyr and Tyr urinary recovery following each treatment. No changes were observed in lipid biomarkers (Table 5) with exception of HDL-cholesterol (HDLc) which increased after WW and WW + Tyr. HDL-c increased in a dose-dependent manner with the content of alcohol plus Tyr administered in all participants (p = 0.027 for linear trend), among men (p = 0.001 for linear trend), and with a borderline significance in the NA group (p = 0.082) (Fig. 2). Endothelin-1 levels at the end of WW + Tyr were lower than at the end of WW intervention (Table 6). Table 7 outlines the transcriptomic changes observed in genes related with endothelial function. Changes are grouped by sex and PAS. Fig. 3 compares the effects observed in WW and WW + Tyr intervention.
Table 5.
Changes in lipid and inflammatory biomarkers (mg/dL).
| Interventions |
|||||
|---|---|---|---|---|---|
| Control | WW | WW + TYR |
P value for WW + Tyr |
||
| vs Control | vs WW | ||||
| Total Cholesterol | 1.8 ± 12.5 | 2.9 ± 21.8 | 7.7 ± 24.4 | 0.665 | 0.753 |
| LDL cholesterol | −1.0 ± 14.1 | 0.1 ± 17.8 | 4.7 ± 22.3 | 0.511 | 0.761 |
| Triglycerides | 14.7 ± 62.0 | 6.0 ± 25.9 | −0.9 ± 25.7 | 0.290 | 0.788 |
| Glucose | 0.9 ± 13.4 | 2.7 ± 10.3 | 2.6 ± 12.0 | 0.849 | 0.999 |
| hsCRP | −0.2 ± 1.3 | 0.02 ± 0.3 | −0.01 ± 0.2 | 0.443 | 0.516 |
Changes expressed as mean ± SD (N = 32). LDL, low density lipoprotein; hsCRP, high sensitivity C reactive protein. ANOVA adjusted by age, sex and smoking habits, LDL cholesterol at the beginning of the clinical trial, and baseline levels.
Fig. 2.
Changes in HDL cholesterol (HDL-c) after interventions. Change in HDL-c compared to the baseline of the intervention expressed as mean and SD in all participants (A), only men (B) and only normal activity metabolizers (C). ANOVA adjusted by age, sex and smoking habits, LDL cholesterol at the beginning of the clinical trial, and baseline levels * P < 0,05; **P < 0,01.
Table 6.
Endothelin concentrations (ng/dL) after interventions.
| Interventions |
|||||
|---|---|---|---|---|---|
| Control | WW | WW + TYR | P value for WW + Tyr |
||
| vs Control | vs WW | ||||
| All participants | 2.15 ± 0.90 | 2.33 ± 1.07 | 2.03 ± 0.82 | 0.572 | 0.031 |
| Women | 2.38 ± 1.07 | 2.57 ± 1.11 | 2.12 ± 0.81 | 0.479 | 0.108 |
| Men | 2.01 ± 0.80 | 2.16 ± 1.08 | 1.99 ± 0.87 | 0.990 | 0.463 |
| Genotype interaction | |||||
| LA | 1.93 ± 0.69 | 2.25 ± 0.96 | 1.89 ± 0.72 | 0.981 | 0.203 |
| NA | 2.24 ± 0.95 | 2.48 ± 1.32 | 2.13 ± 1.07 | 0.747 | 0.068 |
Endothelin-1 concentrations are expressed as mean ± SD (N = 32). WW, white wine; WW + Tyr, white wine plus tyrosol (Tyr) capsules; LA, low activity group metabolizers; NA, normal activity group metabolizers. ANOVA adjusted by age, sex, smoking, acetylsalicylic acid consumption, and baseline levels. *P < 0.05 versus its baseline; P value, significance for inter-intervention comparisons.
Table 7.
Transcriptomic changes (% change versus baseline) after interventions.
| Intervention |
|||||
|---|---|---|---|---|---|
| Control | WW | WW + Tyr |
P value for WW + Tyr |
||
| vs Control | vs WW | ||||
| CD40L | |||||
| All participants | 8.9 ± 60.6 | 21.7 ± 62.3 | −26.8 ± 34.2† | 0.042 | 0.003 |
| Women | −10.7 ± 75.4 | 28.8 ± 59.5 | −29.9 ± 33.7* | 0.743 | 0.063 |
| Men | 24.6 ± 47.2 | 20.4 ± 66.2 | −25.5 ± 35.8* | 0.016 | 0.024 |
| Genotype interaction | |||||
| LA | −4.8 ± 66.7 | 11.1 ± 59.8 | −17.1 ± 35.6 | 0.874 | 0.514 |
| NA | 20.1 ± 60.0 | 16.7. ± 56.5 | −28.2 ± 33.9† | 0.011 | 0.020 |
| P65/RElA | |||||
| All participants | 1.1 ± 0.5 | 1.18 ± 0.5 | 0.9 ± 0.30 | 0.229 | 0.048 |
| Women | −7.6 ± 54.0 | 26.9 ± 63.3 | −16.9 ± 29.8 | 0.896 | 0.089 |
| Men | 22.9 ± 41.9 | 13.4 ± 48.2 | −2.2 ± 28.6 | 0.157 | 0.484 |
| Genotype interaction | |||||
| LA | 16.7 ± 63.5 | 6.3 ± 45.3 | −3.0 ± 43.4 | 0.584 | 0.886 |
| NA | 6.7 ± 41.1 | 22.5 ± 58.3 | -.12,5 ± 20.9 | 0.414 | 0.054 |
| CFH | |||||
| All participants | 19.8 ± 57.6 | 28.8 ± 56.5* | −9.1 ± 51.5 | 0.115 | 0.025 |
| Women | 9.2 ± 57.5 | 48.4 ± 51.9* | 11.9 ± 63.2 | 0.994 | 0.334 |
| Men | 27.9 ± 59.5 | 16.6 ± 58.9 | −18.1 ± 40.9* | 0.013 | 0.048 |
| Genotype interaction | |||||
| LA | 21.4 ± 77.7 | 35.2 ± 74.8 | −16.7 ± 29.5 | 0.359 | 0.150 |
| NA | 22.4 ± 46.9 | 22.6 ± 45.1 | 0.0 ± 62.8 | 0.438 | 0.433 |
| iNOS | |||||
| All participants | −5.0 ± 38.3 | 36.7 ± 82.6* | −19.7 ± 62.4 | 0.734 | 0.007 |
| Women | −16.9 ± 28.8 | 56.1 ± 109.4 | 2.3 ± 24.1 | 0.897 | 0.303 |
| Men | −2.7 ± 41.2 | 29.9 ± 71.3 | −27.5 ± 70.4 | 0.470 | 0.019 |
| Genotype interaction | |||||
| LA | 6.3 ± 36.2 | 17.0 ± 66.9 | −33.0 ± 52.4 | 0.299 | 0.091 |
| NA | −4.7 ± 38.1 | 46.2 ± 26.7 | −10.4 ± 69.6 | 0.996 | 0.080 |
| eNOS | |||||
| All participants | 11.7 ± 65.6 | 34.9 ± 72.2* | −8.2 ± 50.2 | 0.509 | 0.035 |
| Women | 13.2 ± 71.7 | 42.2 ± 42.1* | 14.7 ± 48.8 | 0.997 | 0.565 |
| Men | 12.9 ± 65.5 | 26.4 ± 83.7 | −20.5 ± 49.4 | 0.351 | 0.115 |
| Genotype interaction | |||||
| LA | −4.8 ± 54.9 | 28.9 ± 77.8 | −16.2 ± 62.6 | 0.910 | 0.344 |
| NA | 28.3 ± 72.9 | 34.8 ± 74.6 | −0.2 ± 44.8 | 0.536 | 0.334 |
| VEGFA | |||||
| All participants | 14.3 ± 58.1 | 32.2 ± 69.7* | −6.2 ± 45.2 | 0.398 | 0.045 |
| Women | 12.6 ± 59.0 | 30.4 ± 68.9 | −3.0 ± 61.6 | 0.533 | 0.112 |
| Men | 9.4 ± 55.4 | 26.2 ± 67.9 | −10.0 ± 36.3* | 0.870 | 0.500 |
| Genotype interaction | |||||
| LA | 26.3 ± 65.9 | 19.4 ± 75.6 | −3.0 ± 40.6 | 0.497 | 0.665 |
| NA | 7,7 ± 57.6 | 27.7 ± 60.1 | −1.1 ± 46.5 | 0.896 | 0.256 |
Changes are expressed as mean ± SD (N = 32). WW, white wine; WW + Tyr, white wine plus tyrosol (Tyr) capsules; LA, low activity group metabolizers; NA, normal activity group metabolizers. CD40L, CD40 ligand; CFH, complement factor H; eNOS, endothelial nitric oxide synthase 3; iNOS, inducible nitric oxide synthase; p65/RELA, transcription factor p65 (RELA); VEGFA, vascular endothelial growth factor. ANOVA adjusted by age and sex. *P < 0.05, †P < 0.001 versus its baseline; P value, significance for inter-intervention comparisons.
Fig. 3.
Comparison of the effects of white wine (WW) (blue) versus those of white wine plus tyrosol (WW + Tyr) (Green), CD40L, CD40 ligand; NF-KB, nuclear factor kappa B; CFH, complement factor H; iNOS, inducible nitric oxide synthase; eNOS, endothelial nitric oxide synthase.
2. Experimental design, materials, and methods
2.1. Study design
A randomized, controlled, clinical trial with 33 individuals at cardiovascular risk (21 men and 12 women) was performed (Fig. 1). Inclusion criteria were to be at high risk for coronary heart disease (CHD) with 3 or more risk factors including: current smoking (>1 cig/day during the last month), hypertension (≥140/90 mmHg or antihypertensive medication), high LDL cholesterol (>130 mg/dl or lipid-lowering therapy), low HDL-cholesterol (≤40 mg/dl in men and ≤50 mg/dl in women), overweight/obesity (body mass index ≥25 kg/m2), a family history of premature CHD, and/or type II diabetes treated with oral hypoglycemic agents; and to have a social or recreational use of ethanol/wine consumption at least once during lifetime. Exclusion criteria were participants with a history of cardiovascular disease or severe chronic illness, chronic inflammatory diseases, BMI > 40 kg/m2, suffered from any severe illness or undergone major surgery in the last three months prior to the clinical trial, an alcohol consumption exceeding 8 units or 80 g per day, a history of alcohol hypersensitivity/intolerance, illicit drug consumption, intake of antioxidant supplement(s), the taking of sedative drugs that could potentially interact with alcohol, multiple allergies or intestinal diseases, being vegetarian of following special diets, a history of food allergies or intolerances, illiteracy, and any condition that limited mobility making trial visits impossible or worsening adherence to treatments.
Participants were asked to follow a controlled diet with a moderate content of antioxidants and to abstain of any alcoholic drinks (except in the framework of treatment allocations) thorough the trial. The consumption of certain food was limited to a maximum of 1) vegetables (including pulses): one serving/day, 2) fruits (or juices): 2 pieces/day, 3) ordinary olive oil: maximum 25 mL/day, 4) drinks containing xanthines (coffee, tea, cola, energy drinks …): maximum 3 cups/day, 5) chocolate: maximum one piece (small 15 gr)/day, 6) nuts: maximum 30 g (a small handful)/week, and 7) fish: maximum 3 times per week (150 g/serving).
During the control intervention participants were allowed to only drink water (no alcohol. wine or supplemented Tyr or OHTyr). The reason for different WW doses for men and women was in order to follow the American Heart Association (AHA) guidelines, which limit alcohol consumption to one drink in women and two in men, preferably taken at meals [2]. The doses of wine administered are within those recommended by the AHA.
A 24 h food recall was performed to assess dietary intake before and after each intervention period. Physical activity was recorded at the beginning and end of the clinical trial and assessed by the Minnesota Leisure Time Physical Activity Questionnaire, validated for the Spanish population [3]. A general physical examination, and routine urine, blood chemical and hematological analyses, were performed at the beginning and end of the trial. Blood and 24h-urine samples were collected at fasting state before and after each intervention period. Blood was collected into 10 mL tubes containing EDTA and centrifuged (1700g, 15 min, 4 °C), and plasma and buffy coat samples were then isolated. Peripheral blood mononuclear cells (PBMC) were isolated using a Vacutainer Cell Preparation Tube (CPT™) and kept for RNA extraction. Plasma, urine, and PBMC samples were frozen at −80 °C until analysis. Genomic DNA isolation from buffy coat was performed with QIAamp DNA Blood Midi Kit (Qiagen, Dusseldorf, Germany).
2.2. Tyr and OHTyr metabolites analysis
The urinary concentrations of Tyr and OHTyr metabolites were determined from samples collected before and after each intervention following a validated methodology [4]. Briefly, 0.5 mL of urine was diluted with 0.5 mL of purified water, spiked with 10 μL of internal standard mixture (containing 10 μg/mL of 3-(4-hydroxyphenyl)-1-propanol, 3-(4-hydroxyphenyl)-1-propanol glucuronide and 10 μg/mL HT-1′-O-sulfate) and stabilized with 1 mL of phosphoric acid 4%. Thereafter, samples went under a solid-phase extraction using Oasis HLB columns 3 mL, 60-mg cartridges from Waters Corporation (Milford, MA USA). First, samples were loaded into cartridges then washed with 2 mL of purified water. Thereafter, the compounds of interest were eluted from the cartridge with 2 mL of pure methanol. The methanol extracts were then evaporated until dryness under a stream of nitrogen (29 °C, 10–15 psi). Finally, the dried extracts were reconstituted with a mixture of mobile phases (95% A and 5% B v/v), transferred into HPLC microvials, and analyzed using LC-MS/MS. To prepare blank samples and calibration curves, urine from volunteers after consuming a diet poor in Tyr and OHTyr- Tyr and OHTyr metabolite concentrations were below quantification limits in these blank samples. Blank urine was spiked with increasing concentrations of the metabolites of interest, and then processed in the same manner as samples (described above). Identification and quantification of the metabolites was performed using an Agilent 1200 series HPLC system coupled to a triple quadrupole (6410 Triple Quad LC/MS) mass spectrometer with an electrospray interface from Agilent Technologies (Santa Clara, CA, USA). For the chromatographic separation, an Acquity UPLC®BEH C18 column (100 mm × 3.0 mm i.d., 1.7 μm particle size) from Waters Corporation (Milford, MA, USA) was used at 40 °C. The composition of mobile phase A was 0.01% (v/v) formic acid in water, and mobile phase B was acetonitrile with 0.01% (v/v) of formic acid. Injection volume was 10 μL and the flow rate was set at 0.25 mL/min. The ion source operated in negative ionization for 27 minutes. Finally, urinary concentrations of each metabolite were standardized with the total urinary excretion volume to obtain the total recovery of each metabolite. Quantified Tyr metabolites included Tyr -4-sulfate and Tyr -4-glucuronide. OHTyr metabolites quantified were OHTyr -3-sulfate, OHTyr-4-sulfate, OHTyr-acetate-3-sulfate, OHTyr-3-glucuronide, OHTyr-4-glucuronide, and homovanillyl alcohol (HVAL)-4-glucuronide. Total Tyr and total OHTyr correspond to the molar sum of their respective quantified metabolites.
2.3. Genotyping
Volunteers were genotyped for multiple allelic variants of CYP2A6 and CYP2D6 using TaqMan genotyping assays (Applied Biosystems, Foster City, CA, USA) SNP genotyping was performed with a TaqMan allelic discrimination system (Applied Biosystems, Foster City, CA, USA). Copy-number variations (CYP2A6 *4, *12, and CYP2D6 *5, and duplications) were analyzed with specific copy number assays. When these allelic variants were not detected, a designation of *1 (e.g. wildtype) was assigned.
2.3.1. SNP genotyping
Table 3 shows the characteristics of CYP2A6 and CYP2D6 tested allelic variants. The following SNPs were analyzed: for CYP2A6 *2 and *9, and for CYP2D6 *2, *4, *9, *10, *35, and *41. TaqMan SNP genotyping assay were used, which included FAM™ and VIC™ dye-labeled TaqMan pre-designed proves specifics for each SNP. PCR was performed in a QuantStudio™ 12K Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Reactions were prepared with 10 ng of DNA, 0.25 μL of TaqMan SNP Genotyping Assay, and 2.5 μL of TaqMan Genotyping Master Mix (Applied Biosystems, Foster City, CA, USA). SNP determination was made using allelic discrimination plots with TaqMan Genotyper Software (Applied Biosystems, Foster City, CA, USA).
2.4. Copy number variation (CNV) detection analysis
TaqMan CNV assays were used to analyze CYP2A6 allelic variants *4, *12 (Hs07545274_cn; Hs07545275_cn), and CYP2D6 allelic variants *5 (deletion),*1xN, *2xN, and *35xN (Hs00010001_cn). Real time qPCR was performed using the specific TaqMan assays. Quantitative PCR was performed in QuantStudio™ 12K Flex Real-Time PCR System (Applied Biosystems, Foster City, USA). Reaction was carried in 384-well plates with a mixture of TaqMan Master Mix (Applied Biosystems, Foster City, CA, USA), CNV assays, 10 ng DNA/well and RNase P as reference (Applied Biosystems, Foster City, CA, USA). Reactions were performed in duplicates. Copy number calls were made with the Expression Suite Software v1.0.3 (Applied Biosystems, Foster City, CA USA). CYP2D6 gene duplications as previously described [5]. First, a specific 6.6 kb long piece of CYP2D6 was amplified. Second, a 3.5 kb fragment was amplified from alleles carrying gene duplications. Every duplication-positive sample was further analyzed using two long-range PCR reactions that allow to discriminate among CYP2D6*1xN, *2xN, and *4xN duplications to determine allele-defining SNPs.
2.5. Polygenic activity score
Tested allelic variants were categorized into: those with no-function (CYP2A6 *2, *4; CYP2D6 *4,*5); decreased function (CYP2A6 *9,*12; CYP2D6 *9,*10,*11); normal function (CYP2A6 *1; CYP2D6 *1,*2,*35); and increased function (CYP2A6 *1xN; CYP2D6 *1xN,*2xN,*35xN). A score of 0, 0.5, 1 or 2 was assigned for the presence of each allele (Table 4). For each enzyme, an activity score was given to each volunteer according to the identified alleles and classified as detailed above based on the method described by Gaedigk et al. [6]. A pooled polygenic activity score (PAS) was calculated by adding together the activity scores of both enzymes. Finally, according to their PAS, individuals were placed into three groups of predicted activity: low (LA), normal (NA), and rapid activity (RA) groups.
2.6. Endothelial function measurement
Endothelial function was assessed before and after interventions by monitoring endothelium-mediated changes (reactive hyperemia index, RHI) in the digital pulse waveform, known as the Peripheral Arterial Tone (PAT) signal (EndoPAT 2000; Itamar Medical Inc., Caesarea, Israel). Specially designed finger probes were placed on the middle finger of each subject's dominant hand. The probes comprised a system of inflatable latex air cuffs connected by pneumatic tubes to an inflating device controlled through a computer algorithm. A constant counter pressure (pre-determined by baseline DBP) was applied through the air cushions. Pulsatile volume changes of the distal digit induced pressure alterations in the finger cuff, which were sensed by pressure transducers and transmitted to and recorded by the EndoPAT 2000 device. EndoPAT 2000 also provides the augmentation index (AI), a measurement of arterial stiffness via pulse-wave analysis, which was normalized to 75 bpm heart rate. Measurements were performed by a trained professional with the participants in resting supine conditions, in a quiet room at a constant temperature after 10 minutes of stabilization. Hyperemic reactivity index measured by EndoPAT 2000 has been shown to predict cardiovascular disease [7].
2.7. Gene expression measurements
On the basis of their relationship with endothelial health and atherosclerosis, and the available data of gene expression response after VOO ingestion several candidate genes were selected. Candidate genes were AKT serine/threonine kinase 2 (AKT2), arachidonate 5-lipoxygenase (ALOX5), CD40 ligand (CD40L), complement factor H (CFH), endothelial nitric oxide synthase 3 (eNOS), endothelial plasminogen activator inhibitor (SERPINE1), inducible nitric oxide synthase (iNOS), interferon gamma (IFNG), interleukins (IL)1B (IL1B) and 6 (IL6), matrix metalloproteinases (MMP) 2 (MMP2) and 9 (MMP9), mitogen-activated protein kinase 14 (MAPK14), monocyte chemoattractant protein 1 (MCP1), nuclear factor (NF) (erythroid-derived 2)-like 2 (NEF2L2), NF-kappa B inhibitor alpha (NFKBIA), platelet-derived growth factor subunit B (PDGFB), peroxisome proliferator-activated receptor alpha (PPARɑ), sirtuins (SIRT) 1 (SIRT1), 2 (SIRT2), and 6 (SIRT6), transcription factor p65 (p65/RELA), tumor necrosis factor alpha (TNF-ɑ), and vascular endothelial growth factor (VEGFA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and B-actin were used as endogenous controls to correct changes in gene expression. Isolation of RNA from PBMC was performed with the RNeasy Mini Kit (Qiagen, Duesseldorf, Germany). DNA complementary conversion was then carried out with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA).
Gene expression was measured, before and after interventions, by a real-time polymerase chain reaction with a QuantStudio™ 12K Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and SYBR Green dye-based analysis. Samples were analyzed in duplicate. Results were obtained with the Expression Suite Software v1.0.3 (Applied Biosystems, Foster City, CA, USA). Changes in gene expression were assessed first by calculating the relative quantification, applying the 2–ΔΔCT of each sample. Thereafter, the fold change of each intervention was extracted by calculating the ratio between values at the end and at baseline of each intervention period.
2.8. Sample size and power analyses
A total sample of 32 participants would allow at least 80% power to detect a statistically significant difference among groups of 0.205 units in the RHI measurement, assuming a dropout rate of 5% and type I error of 0.005 (2-sided). The standard deviation of the measurement was assumed 0.4 [7].
2.9. Statistical analyses
Normality of continuous variables was assessed by normal probability plots and data were log transformed when required. Intra-treatment comparisons were assessed by Student's t-test for paired samples. Comparisons among treatments were made by an ANOVA for repeated measures and adjusted by age, gender, smoking, AAS medication, and baseline concentrations. In the case of lipids an additional adjustment for LDL cholesterol values at the beginning of the clinical trial was performed. A general lineal model was used to assess linear and quadratic trends. For the post-hoc pairwise comparison, the Tuckey test was used. Statistical analyses were performed with R (R Foundation for Statistical Computing, Vienna, Austria). version 3.0.2., and R package multcomp. Significance was defined as p < 0.05.
Acknowledgments
This research was funded by the Instituto de Salud Carlos III (PI14/00072) and by grants from DIUE of Generalitat de Catalunya (2107 SGR 138). AB is recipient of a PFIS predoctoral fellowship from the Instituto Carlos III (PFIS-FI16/00106). JM is recipient of a Rio Hortega fellowship from the Instituto Carlos III (CM17/00024). NS is recipient of fellowship from Centro de Información Cerveza y Salud, Fundación Manuel de Oya. CIBER de Fisiopatología de la Obesidad y Nutrición (CIBEROBN) is an initiative of the ISCIII, Madrid, Spain. RFT is a recipient of a Canada Research Chair in Pharmacogenomics and her work is supported by Canadian Institutes of Health Research (FDN-154294).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2019.104787.
Conflict of Interest
RFT has been consulted by Quinn Emanuel, Ethismos and Apotex on unrelated topics. The other authors declare they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Boronat A., Mateus J., Soldevila-Domenech N., Guerra M., Rodríguez-Morató J., Varón C., Muñoz D., Barbosa F., Morales J.C., Langohr K., Covas M.I., Pérez-Mañá C., Fitó M., Tyndale R.F., de la Torre R. Cardiovascular benefits of tyrosol and its endogenous conversion into hydroxytyrosol in humans, A randomized, controlled trial. Free Radic. Biol. Med. 2019 Aug 31;143:471–481. doi: 10.1016/j.freeradbiomed.2019.08.032. [DOI] [PubMed] [Google Scholar]
- 2.Lichtenstein A.H., Appel L.J., Brands M., Carnethon M., Daniels S., Franch H.A. Summary of American heart association diet and lifestyle recommendations revision 2006. Arterioscler. Thromb. Vasc. Biol. 2006;26:2186–2191. doi: 10.1161/01.ATV.0000238352.25222.5e. [DOI] [PubMed] [Google Scholar]
- 3.Elosua R., Garcia M., Aguilar A., Molina L., Covas M.I., Marrugat J. Validation of the Minnesota leisure time physical activity questionnaire in Spanish women. Med. Sci. Sport. Exerc. 2000;32:1431–1437. doi: 10.1097/00005768-200008000-00011. [DOI] [PubMed] [Google Scholar]
- 4.Kotronoulas A., Pizarro N., Serra A., Robledo P., Joglar J., Rubió L., Hernaéz A., Tormos C., Motilva M.J., Fitó M., Covas M.I., Solà R., Farré M., Saez G., de la Torre R. Dose-dependent metabolic disposition of hydroxytyrosol and formation of mercapturates in rats. Pharmacol. Res. 2013 Nov;77:47–56. doi: 10.1016/j.phrs.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Gaedigk A., Ndjountché L., Divakaran K., Dianne Bradford L., Zineh I., Oberlander T.F., Brousseau D.C., McCarver D.G., Johnson J.A., Alander S.W., Wayne Riggs K., Steven Leeder J. Cytochrome P4502D6 (CYP2D6) gene locus heterogeneity: characterization of gene duplication events. Clin. Pharmacol. Ther. 2007 Feb;81(2):242–251. doi: 10.1038/sj.clpt.6100033. [DOI] [PubMed] [Google Scholar]
- 6.Gaedigk A., Dinh J.C., Jeong H., Prasad B., Leeder J.S. Ten years' experience with the CYP2D6 activity score: a perspective on future investigations to improve clinical predictions for precision therapeutics. J. Personalized Med. 2018;8:15. doi: 10.3390/jpm8020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubinshtein R., Kuvin J.T., Soffler M., Lennon R.J., Lavi S., Nelson R.E. Assessment of endothelial function by non-invasive peripheral arterial tonometry predicts late cardiovascular adverse events. Eur. Heart J. 2010;31:1142–1148. doi: 10.1093/eurheartj/ehq010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.