Abstract
Introduction
The role of TOMM40-APOE 19q13.3 region variants is well documented in Alzheimer's disease (AD) but remains contentious in dementia with Lewy bodies (DLB) and Parkinson's disease dementia (PDD).
Methods
We dissected genetic profiles within the TOMM40-APOE region in 451 individuals from four European brain banks, including DLB and PDD cases with/without neuropathological evidence of AD-related pathology and healthy controls.
Results
TOMM40-L/APOE-ε4 alleles were associated with DLB (ORTOMM40-L = 3.61; P value = 3.23 × 10−9; ORAPOE-ε4 = 3.75; P value = 4.90 × 10−10) and earlier age at onset of DLB (HRTOMM40-L = 1.33, P value = .031; HRAPOE-ε4 = 1.46, P value = .004), but not with PDD. The TOMM40-L/APOE-ε4 effect was most pronounced in DLB individuals with concomitant AD pathology (ORTOMM40-L = 4.40, P value = 1.15 × 10−6; ORAPOE-ε4 = 5.65, P value = 2.97 × 10−8) but was not significant in DLB without AD. Meta-analyses combining all APOE-ε4 data in DLB confirmed our findings (ORDLB = 2.93, P value = 3.78 × 10−99; ORDLB+AD = 5.36, P value = 1.56 × 10−47).
Discussion
APOE-ε4/TOMM40-L alleles increase susceptibility and risk of earlier DLB onset, an effect explained by concomitant AD-related pathology. These findings have important implications in future drug discovery and development efforts in DLB.
Keywords: Parkinson's disease, Alzheimer's disease, Parkinson's disease dementia, Dementia with Lewy bodies, Apolipoprotein E, APOE, TOMM40, Association analysis, Brain banks, Lewy body dementias, Neuropathology
1. Introduction
The two late-onset Lewy body dementia forms, namely dementia with Lewy bodies (DLB) and Parkinson's disease dementia (PDD), represent the third most common type of late-onset dementia after the commonest sporadic late-onset Alzheimer's disease and vascular dementia, representing 7.6%–25% [1], [2] of late-onset dementia cases. DLB is characterized by the occurrence of significant cognitive decline before or within a year of the onset of typical Parkinson's disease (PD) motor signs, whereas the clinical diagnosis of PDD is based on dementia occurring in the subsequent course of PD, as per the third and fourth DLB consortium criteria [3], [4], [5]. It has been estimated that dementia ultimately affects 70–80% of patients with PD [6]. PD, DLB, and PDD share a common neuropathological substrate featuring selective neuronal loss of dopaminergic neurons in the substantia nigra and widespread intraneuronal inclusions of aggregated α-synuclein, known as Lewy bodies (LB) in neuronal somata and Lewy neurites in neuronal processes [5], [7], [8]. The APOE-ε4 allele, the best-validated genetic risk factor for AD [9], has previously been associated with DLB and PDD [10], [11], [12], [13]. As in AD, the APOE-ε2 allele might have a protective effect on these dementia types [14].
Neuropathologically, DLB and PDD cannot be clearly distinguished, and while both may show concomitant typical AD pathology in older individuals, this feature is more commonly observed in DLB. It has even been suggested that cortical and striatal amyloid-β (Aβ) depositions are virtually always present in DLB [15]. We have previously reported a higher neocortical LB burden (mainly in the parietal and temporal cortex) and cortical plaque-load in DLB compared with PDD, underlined by APOE-ε4 carriage, and highlighted that elevated LB and Aβ plaque deposition were correlated with earlier age of onset (AOO) of cognitive decline and faster progression to dementia [16]. Similar observations have also been reported by other investigators [13], [17]. APOE-ε4 has been shown to confer a higher risk for increased Aβ burden [18], [19] as well as faster cognitive decline in cognitively healthy elderly individuals, mainly in female APOE-ε4 carriers [20].
APOE maps to chromosome 19 and several variants within this locus are in tight linkage disequilibrium (LD) with variants in the TOMM40 gene located ~2kb proximal (p-ter). APOE-ε4 and the tightly correlated long (L) poly-thymine [T] repeat polymorphism within the intervening sequence of intron 6 (IVS6, tagged by rs10524523) of TOMM40 have been associated with the risk and AOO of cognitive decline due to AD [21], [22]. Although the associations with susceptibility and AOO in AD could be due to a combination of effects of both APOE and TOMM40, the strong LD between these variants complicates analytical approaches to disentangle possible independent effect(s) [23]. As the APOE-ε3 allele is linked to TOMM40 short (S) and very long (VL) alleles in most individuals, several attempts have been made to disentangle the effects of APOE and TOMM40 variants by selecting AD patients with the APOE ε3/ε3 or ε3/ε4 genotypes. Indeed, an APOE-independent effect of longer poly-T repeats has been proposed on AOO of cognitive decline and dementia [22], risk for pathologically proven AD [23], cognition and grey matter volume in cognitively healthy adults [24], hippocampal thinning [25], cerebrospinal fluid cortisol levels [26], and other AD-related biomarkers [27]. However, other studies have provided contradictory data pertaining to the role of APOE/TOMM40-S/VL alleles on late-onset Alzheimer's disease–related outcomes [28], [29], [30]. A recent study on homozygous ε3/ε3 older adults reported that the S allele of the TOMM40 gene was associated with faster cognitive decline in global cognition, mainly in the domain of episodic and semantic memory, while the VL allele was under-represented in that age group [31]. It was subsequently suggested that VL allele may have a protective effect in the older old, possibly due to a pleiotropic effect of poly-T VL repeats, increasing risk in cohorts with the mean age of 57.8 and 65 years and being protective in older cohorts of a mean age of 78.5 years [32].
Here, we studied the effects of TOMM40 and APOE variants on susceptibility to DLB, PDD, and PD in clinically and neuropathologically well-characterized individuals from one of the largest multicentre data sets assembled to date. Our newly generated data were combined with those from previously published neuropathological data sets using the same phenotypes. To our knowledge, no previous studies have investigated the effects of TOMM40 variants in pathologically confirmed DLB and PDD cases.
2. Methods
2.1. Data set and clinical diagnosis
Our data set includes samples from the Parkinson's UK Brain Bank at Imperial College London (PUK-ICL), the Oxford University Brain Bank (UO), the Newcastle Brain Tissue Resource at Newcastle University, and the Munich Brain Bank at Ludwig-Maximilians-University (Table 1). Overall, these brain banks contributed clinical and neuropathological data and DNA samples from 451 individuals, classified into four clinical phenotypes: normal controls (NC) with no symptoms of PD or forms of dementia (n = 86), PD with no symptoms of dementia (PDnD, n = 84), PDD (n = 102), and DLB (n = 179) (Table 1 and Supplementary Materials). Clinical diagnosis was based on the Parkinson's Society UK Brain Bank criteria for PD [33], and the third DLB consortium criteria for DLB [3]; the diagnosis of PDD was based on the occurrence of significant cognitive and functional decline over a year after the onset of PD motor signs.
Table 1.
Demographic characteristics of clinically and neuropathologically characterized patients and controls by study centre
| Centre | Phenotype Groups |
|||
|---|---|---|---|---|
| NC/PDnD | DLB | PDD | Total N of individuals by centre (males, %) | |
| Newcastle Brain Tissue Resource (NBTR) | ||||
| N (Males, %) | -/31 (67.74) | 86 (59.30) | 38 (63.16) | 155 (60.75) |
| Mean age at onset of PD (SD) | -/64.69 (10.93)∗ | 74.51 (7.13)∗ | 64.92 (8.62) | |
| Mean age at onset of Dementia (SD) | - | 74.03 (8.02) | 71.68 (6.52) | |
| Mean age at death (SD) | -/77.52 (7.4) |
78.98 (7.21) |
76.13 (5.68) |
|
| NC/PDnD |
DLB-AD/DLB+AD |
PDD-AD/PDD+AD |
||
| Parkinson's UK Brain Bank at Imperial College London (PUK-ICL) | ||||
| N (Males, %) | -/42 (69.05) | 8 (87.50)/7 (71.43) | 43 (69.77)/12 (66.67) | 112 (70.54) |
| Mean age at onset of PD (SD) | -/66.50 (±10.43) | 72.38 (±5.34)/69.29 (±8.88) | 62.63 (±8.89)/64.08 (±8.51) | |
| Mean age at onset of Dementia (SD) | - | 72.75 (±5.12)/69.71 (±8.67) | 74.19 (±7.22)/76.08 (±4.98) | |
| Mean age at death (SD) | -/78.24 (±8.12) | 76.62 (±4.34)/79.47 (±7.01) | 77.42 (±6.99)/78.67 (±5.28) | |
| Oxford University Brain Bank (UO) | ||||
| N (Males, %) | 86 (51.16)/- | 29 (67.74)/33 (48.39) | - | 148 (54.05) |
| Mean age at onset of PD (SD) | - | - | - | |
| Mean age at onset of Dementia (SD) | - | 73.07 (±9.12)/72.11 (±7.65) | - | |
| Mean age at death (SD) | 81.81 (±10.90)/81.55 (±9.47) | 81.68 (±7.94)/80.68 (±6.17) | - | |
| Munich Brain Bank at Ludwig-Maximilians-University (LMU) | ||||
| N (males, %) | -/11 (54.55) | 10 (50)/6 (50) | 6 (50)/3 (66.67) | 36 (52.78) |
| Mean age at death (SD) | -/74.92 (±6.4) | 75.30 (±7.72)/75.17 (±8.42) | 73.67 (±3.27)/77.33 (±6.81) | |
| Total N of individuals by phenotype, (Males, %) | 170 (58.82) | 179 (59.78) | 102 (65.68) | 451 (62.57) |
NOTE. Each study contributed data available for each specific phenotype group. Some centres did not have specific phenotypes represented. The ICL centre data contained seven (males, 71.43%) PDnD-AD (Parkinson's disease without dementia with neuropathologically defined Alzheimer's disease), mean age of onset of PD (Parkinson's disease) 66.29 (±11.44) y, mean age at death 80.29 (±9.09) y, which were not used on any analyses presented in this report because of the low number of available subjects. The concomitant AD pathology data were not available for Newcastle cases.
Abbreviations: NC, normal controls; PDnD, Parkinson's disease without dementia; PDD-AD/PDD+AD, Parkinson's disease with dementia with/without neuropathologically defined Alzheimer's disease; DLB-AD/DLB+AD, dementia with Levy bodies with/without neuropathologically defined AD.
Some individuals did not have this information.
In the UO, Ludwig-Maximilians-University, and PUK-ICL samples, pathological assessment allowed further clinicopathological subclassification into PDD with/without AD (PDD+AD/PDD-AD) and DLB with/without AD (DLB+AD/DLB-AD) (Table 1 and Supplementary Materials). The AD pathology was considered to be present if the Braak neurofilament tangle stage was > III and the “Consortium to Establish a Registry for AD” (CERAD) plaque score was C [34]. PDD and DLB cases were subdivided based on the presence of concomitant AD pathology, as follows: PDD-AD/DLB-AD – Braak tau stage I-III and CERAD plaque 0-B; PDD+AD/DLB+AD – Braak tau stage ≥IV and/or CERAD C.
All the controls (UO) were tested for protein deposits or morphological abnormalities. Individuals were only included if, additionally, no clinical information or pathological findings suggested the possibility of an alternative form of a late-onset neurodegenerative disease.
Genotyping was carried out on DNA extracted from cerebellar cortex using standard PCR procedures (Supplementary Materials). The TOMM40 poly-T repeat rs10524523 (hg19 chr19:45403049-45403067 → poly-T) was genotyped directly through PCR, obtaining a direct readout of the poly-T length. We found 16 allelic lengths of the multivariate fragment length of the TOMM40 IVS6 poly-T and applied the previously reported length classification in three groups; short (S<20 T bases), long (20≤L≤30 T bases), and very long (VL≥31 T bases) [35]. For the APOE, we used two tagging SNPs rs429358 (hg19 chr19:4541194T>C) and rs7412 (hg19 chr19:45412079C>T), which uniquely identify haplotypes for the APOE ε2/ε3/ε4 alleles.
2.2. Statistical methods
We used logistic regression (R statistical software, v3.2.0) to test for log-additive genetic effects of individual alleles compared with other alleles grouped together on the study phenotypes. Models based on TOMM40 and APOE variant carrier status were tested and provided very similar results (not shown). All tests were adjusted for sex and age at death of participants. We also tested a model that additionally accounted for the effect of APOE variants on TOMM40 associations and vice versa. We applied Bonferroni correction to adjust for multiple testing (8 tests for each of S/L/VL alleles, total = 24, Table 2, Supplementary Tables 1 and 2); hence, P value <.0021 was used as significance threshold in this study. Furthermore, associations of TOMM40-L/APOE-ε4 alleles with the presence of concomitant AD pathology in DLB and PDD were tested using logistic regression adjusting for sex and age at death. We also conducted a survival analysis and implemented the Cox proportional hazard model to evaluate the effect of TOMM40-L/APOE-ε4 alleles on the age at onset of DLB and PDD among individuals with dementia and visualized the results using Kaplan-Meier plot. Subgroup analysis was further conducted among subjects with APOE ε3/ε3 and ε3/ε4 genotypes to examine additional effects of TOMM40 S/VL alleles.
Table 2.
Effects of TOMM40-L allele in the risk of PD, PDD, and DLB
| Case group, N individuals | Control group, N individuals | Frequency of effect allele (L) |
OR (95% CI) | P value | P value adjusted for APOE-ε4 effect | |
|---|---|---|---|---|---|---|
| Case group, % | Reference group, % | |||||
| PDnD, 84 | NC, 86 | 13.7 | 10.5 | 1.15 (0.59–2.26) | .69 | .58 |
| PDD, 102 | NC and PDnD, 170 | 20.1 | 12.1 | 1.69 (1.05–2.76) | .033 | .73 |
| DLB, 179 | NC and PDnD, 170 | 32.1 | 12.1 | 3.61 (2.39–5.59) | 3.23 × 10−9 | .66 |
| DLB, 179 | PDD, 102 | 32.1 | 20.1 | 2.15 (1.40–3.38) | 6.37 × 10−4 | .64 |
| DLB+AD, 46 | NC and PDnD, 170 | 37.0 | 12.1 | 4.40 (2.42–7.99) | 1.15 × 10−6 | .40 |
| PDD+AD, 15 | NC and PDnD, 170 | 16.7 | 12.1 | 1.38 (0.44–3.57) | .53 | .17 |
| DLB-AD, 47 | NC and PDnD, 170 | 20.2 | 12.1 | 1.71 (0.96–3.03) | .067 | .51 |
| PDD-AD, 49 | NC and PDnD, 170 | 18.4 | 12.1 | 1.46 (0.78–2.64) | .22 | .63 |
NOTE. The results are reported as odds ratios (ORs) with their 95% confidence intervals (CIs). Model tested: TOMM40-L allele log-additive effect on risk compared with combined effect of other alleles, adjusted for age at death and sex. We also adjusted the same model for the additive effect of APOE-ε4 allele.
Results in bold are statistically significant after correction for multiple testing (P value < .0021).
Abbreviations: NC, normal controls; PDnD, Parkinson's disease without dementia; PDD+AD/PDD-AD, Parkinson's disease with dementia with/without neuropathologically defined Alzheimer's disease; DLB+AD/DLB-AD, dementia with Levy bodies with/without neuropathologically defined AD.
2.3. Literature search and meta-analysis methods
To identify previous reports on the association between the TOMM40/APOE locus and DLB, we followed data extraction protocols previously established by our group for meta-analysis of genetic association data [36], [37]. In the absence of previous reports on the effect of TOMM40 poly-T repeat variants, we searched PubMed (www.ncbi.nlm.nih.gov/pubmed/) using the search terms “(“apoe”[All Fields] OR “apolipoprotein e*”[All Fields] OR “Apolipoproteins”[Mesh] OR “AD2” [All Fields] OR “lpg”[All Fields] OR “apo e”([All Fields] OR “19q13*”[All Fields]) AND (lewy*) AND (body*) OR bodies) AND (dementia*) OR (disease*) OR “lewy body disease”[MeSH Terms]”. Among 247 identified publications, we used the following specific criteria to assess for their inclusion eligibility: only studies published in English in peer-reviewed journals and comparing APOE genotypes or allele frequencies in at least 10 DLB cases (with and without concomitant AD pathology) and 10 controls were included in the analyses. This led to the identification of a total of 42 studies published by 15 September, 2017. We extracted information from these studies and assessed them for data overlap. We excluded 16 overlapping data sets, leaving the largest of two overlapping data sets with available genotype data for meta-analysis. We extracted genotype and/or allele distributions from the remaining 26 publications as reported and meta-analyzed in R software environment, assuming an additive genetic model and using a fixed-effects inverse-variance meta-analysis approach [36]. We performed meta-analyses on all available data, and after stratification for diagnosis (clinical and pathological). In addition, we conducted meta-analysis of data sets with preselected individuals that showed presence of concomitant AD pathology. We used Forest plots implemented through a customized R package script to visualize meta-analysis results [36].
3. Results
3.1. Genetic association results in newly ascertained neuropathological data sets
We detected no association between the TOMM40-L or APOE-ε4 alleles and PDnD as compared with NC (P value = .69, P value = .49) (Table 2, Table 3). Thus, the PDnD group was combined with NC to improve power of all subsequent analyses (n = 170). Association of the TOMM40-L or APOE-ε4 with PDD compared with combined NC/PDnD group was only nominally significant (P value = .033, P value = .024) and did not survive the correction for multiple testing. However, DLB showed strong evidence for association with the TOMM40-L allele (OR = 3.61, P value = 3.23 × 10−9) as well as with the APOE-ε4 allele (OR = 3.75, P value = 4.90 × 10−10). There was also a statistically significant difference in the effect of the TOMM40-L allele/APOE-ε4 allele on the risk of DLB as compared with PDD status (ORTOMM40 = 2.15, P value = 6.37 × 10−4; ORAPOE = 2.17; P value = 3.45 × 10−4).
Table 3.
Effects of APOE-ε4 allele on the risk of dementias related to Parkinson's disease and dementia with Lewy bodies
| Case group, N individuals | Control group, N individuals | Frequency of effect allele (ε4) |
OR (95% CI) | P value | P value adjusted for TOMM40-L effect | |
|---|---|---|---|---|---|---|
| Case group, % | Reference group, % | |||||
| PDnD, 84 | NC, 86 | 14.9 | 11.1 | 1.26 (0.66–2.45) | .49 | .43 |
| PDD, 102 | NC and PDnD, 170 | 21.6 | 12.9 | 1.70 (1.07–2.70) | .02 | .40 |
| DLB, 179 | NC and PDnD, 170 | 34.6 | 12.9 | 3.75 (2.50–5.76) | 4.90 × 10−10 | .03 |
| DLB, 179 | PDD, 102 | 34.6 | 21.6 | 2.17 (1.44–3.36) | 3.44 × 10−4 | .22 |
| DLB+AD, 46 | NC and PDnD, 170 | 43.5 | 12.9 | 5.65 (3.06–10.43) | 2.97 × 10−8 | 8.29 × 10−4 |
| PDD+AD, 15 | NC and PDnD, 170 | 13.3 | 12.9 | 1.00 (0.29–2.72) | 1.00 | .24 |
| DLB-AD, 47 | NC and PDnD, 170 | 23.4 | 12.9 | 1.90 (1.09–3.29) | .023 | .13 |
| PDD-AD, 49 | NC and PDnD, 170 | 21.4 | 12.9 | 1.62 (0.92–2.80) | .09 | .20 |
NOTE. The results are reported as odds ratios (ORs) with their 95% confidence intervals (CIs). Model tested: APOE-ε4 allele log-additive effect on risk compared to combined effect of other alleles, adjusted for age at death and sex. We also adjusted the same model for the additive effect of TOMM40-L allele.
Results in bold are statistically significant after correction for multiple testing (P value < .0021).
Abbreviations: NC, normal controls; PDnD, Parkinson's disease without dementia; PDD+AD/PDD-AD, Parkinson's disease with dementia with/without neuropathologically defined Alzheimer's disease; DLB+AD/DLB-AD, dementia with Levy bodies with/without neuropathologically defined AD.
3.2. TOMM40-L/APOE-ε4 effect on AD pathology
Applying the neuropathological criteria for AD, as aforementioned, we observed large number of individuals with concomitant AD pathology in 23% of patients with PDD and 49% of patients with DLB. The TOMM40-L and APOE-ε4 alleles were associated with higher prevalence of AD pathology in DLB (ORTOMM40 = 2.27, P value = .017; ORAPOE = 2.61, P value = .006) but not in PDD patients. In PDD, there was no statistically significant effect of TOMM40-L or APOE-ε4 alleles in either subgroup, PDD+AD (P value = .53, P value = 1.00) or PDD-AD (P value = .22, P value = .09), compared with the combined NC/PDnD control group (Table 2, Table 3). TOMM40-L/APOE-ε4 alleles exerted the largest effect on the DLB+AD group (ORTOMM40 = 4.40, P value = 1.15 × 10−6; ORAPOE = 5.65, P value = 2.97 × 10−8) only; there was no association observed for the similarly sized group of DLB-AD individuals (P value = .067, P value = .023, respectively), after correction for multiple testing. In the sensitivity analyses, the effect estimates of TOMM40-L and APOE-ε4 alleles were similar in magnitude for all phenotypes when compared with NC and the combined NC-PDnD groups (Supplementary Table 1).
3.3. Conditional analyses of the TOMM40/APOE variants
The TOMM40 poly-T repeat length polymorphism and APOE haplotypes show strong LD in our data set (r2 = 0.941), in line with previous reports [21]. We therefore investigated, whether the observed effects at both loci on DLB+AD risk can be disentangled using conditional analyses. As shown in Table 2, the observed effects of the TOMM40-L allele on the DLB+AD neuropathological (NP) phenotype lost significance when adjusted for the APOE-ε4 effect (P value > 0.05). By contrast, the APOE-ε4 association with DLB+AD remained unaffected when adjusted for the TOMM40-L allele carrier status (P value = 8.29 × 10−4), but became insignificant for other NP phenotypes (Table 3). The residual significance exerted by APOE-ε4 in DLB+AD individuals after adjustment for TOMM40-L may be due to the presence of one instance of SL and one of LVL genotypes co-occurring with ε4/ε4 genotype, and 3 SVL and one VLVL co-occurring with ε2/ε4 or ε3/ε4 genotype, in this particular diagnostic group. This proportion of non-L/L genotype carriers is nearly 3 times as high as in the full sample, where we observed 3 instances of SL, one SVL and two LVL genotypes co-occurring with APOE ε4/ε4. Next, we investigated whether any of the other TOMM40 or APOE loci alleles showed additional effects apart from primary L-/ε4-allele associations but found no evidence on any phenotypes to support this hypothesis (Supplementary Tables 2–5). Finally, a subgroup analysis in individuals with APOE ε3/ε3 or ε3/ε4 genotypes showed no effects of the TOMM40 S/VL alleles on dementia risk (Supplementary Tables 6 and 7).
3.4. TOMM40-L/APOE-ε4 effects on age at onset of dementia
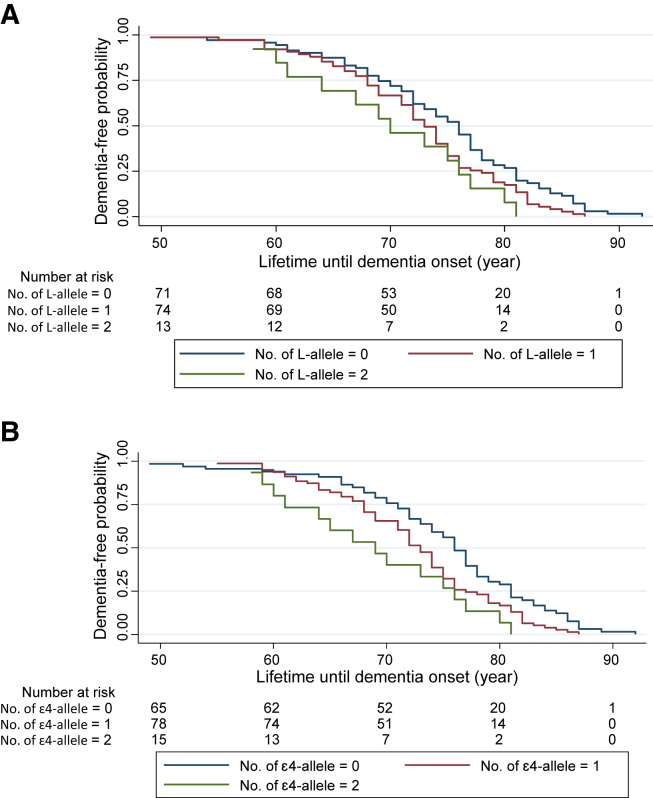
We implemented Cox regression survival analyses to assess whether the TOMM40-L/APOE-ε4 alleles were associated with risk of earlier onset of dementia. This was, indeed, the case in DLB (HRTOMM40 = 1.33, 95% CI [1.03–1.72], P value = .031; HRAPOE = 1.46, 95% CI [1.13–1.89], P value = .004); (Fig. 1). By contrast, no such associations were identified in PDD cases (HRTOMM40 = 1.19, 95% CI [0.82–1.73], P value = .37; HRAPOE = 1.27, 95% CI [0.91–1.78], P value = .17). As shown in Fig. 1, the correlation between APOE-ε4 and TOMM40-L shows marginal differences across the age groups, below the age of 80 years; while in all cases aged 80 years and older, both alleles co-occur in the same individuals. The reason for this is unclear but may be due to the imperfect LD between both variants or, indeed, subtle population differences across age groups and/or statistical fluctuations.
Fig. 1.
Kaplan-Meier survival plot for effects of TOMM40-L and APOE-ε4 alleles on the age at onset of dementia with Lewy bodies (DLB) estimated using Cox proportional hazards model. (A) risks of developing DLB for individuals carrying one or two TOMM40-L alleles compared with noncarriers. (B) risks of developing DLB for individuals carrying one or two APOE-ε4 alleles compared with noncarriers. The Cox proportional hazards model for DLB onset in relation to the number of risk alleles carrier status for either TOMM40 or APOE are adjusted for sex.
3.5. Meta-analysis with previously published data
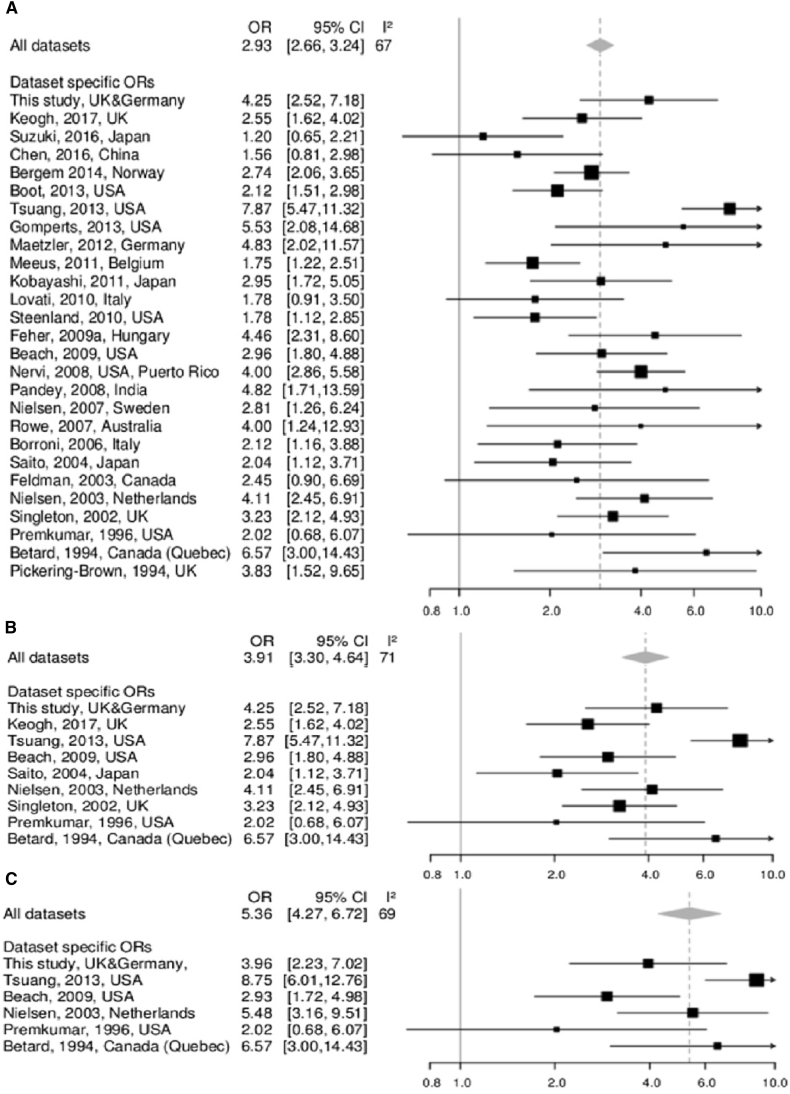
Following our systematic literature screen of studies available through PubMed containing data on the association between APOE-ε4 allele and DLB (with and without concomitant AD pathology), we identified a total of 26 studies that combined information about up to 9400 individuals. Summary statistics limited to these previously published studies alone corroborated our newly generated findings, that is, they showed significant association between APOE-ε4 and DLB (OR = 2.89, 95% CI [2.61–3.20], P value = 3.24 × 10−93) without stratification by AD pathology. The effect became more pronounced when the data were filtered for the presence of NP diagnosis (OR = 3.87, 95% CI [3.24–4.63], P value = 1.92 × 10−49), and even stronger when limited to DLB+AD only (OR = 5.67, 95% CI [4.42–7.26], P value = 5.33 × 10−43). Addition of the associations observed in the present study strengthened the association evidence for DLB (OR = 2.93, 95% CI [2.66–3.24], P value = 3.78 × 10−99) and DLB+AD (OR = 5.36, 95% CI [4.27–6.72], P value = 1.56 × 10−47), (Fig. 2).
Fig. 2.
Meta-analysis of associations between APOE-ε4 allele and risk of dementia with Lewy bodies. (A) Forest plot for unstratified meta-analysis combining results of the present study with those of 26 previous studies. (B and C) Forest plots for analysis of studies with neuropathologically diagnosed DLB cases (B) and DLB cases with concomitant AD pathology (C). Dashed line indicates the pooled OR, with the width of the diamond representing the 95% CI. Abbreviations: AD, Alzheimer's disease; CI, confidence interval; OR, odds ratio; DLB, dementia with Lewy bodies.
4. Discussion
We report one of the largest multicentre studies to date disentangling the TOMM40 and APOE loci genetic variant effects on the risk of pathologically confirmed DLB and PDD. We demonstrated that both the TOMM40-L and APOE-ε4 alleles increase the susceptibility and risk of earlier onset of DLB, while we observed no major effect of these variants on PDD risk. The APOE-ε4 allele effect estimates on DLB obtained in our study are of similar magnitude to those reported in other studies of AD (Table 3) [38]. In this study, we showed that both TOMM40-L and APOE-ε4 alleles are associated with higher prevalence of concomitant AD pathology in DLB but not in PDD. Furthermore, the TOMM40-L/APOE-ε4 effects on DLB are likely driven by their associations with AD-related pathology (DLB+AD) because these alleles do not increase the risk of DLB-AD. These results confirm our previous findings reported using the Parkinson's UK ICL Brain Bank samples [16]; and provide evidence for the TOMM40-L/APOE-ε4 positive associations with DLB risk being explained by their effects on AD pathology. Owing to the strong LD between variants of the two genes at this locus, we were unable to statistically distinguish the effects of TOMM40-L and APOE-ε4 in our series. It has recently been reported that LD structures of genes in the APOE/TOMM40 locus are highly heterogeneous, with significant differences between patients with AD and unaffected controls and that these differing patterns may correspond to heterogenous “molecular signatures” that may denote polygenetically enhanced susceptibility to AD and related diseases [39]. Future work needs to further assess the clinical discriminative value of either variant to distinguish DLB with versus without concomitant AD pathology and to clarify the molecular mechanisms underlying the association between DLB+AD and APOE-ε4 or TOMM40-L (or both). In contrast to the DLB associations, we did not identify significant effects of either TOMM40 or APOE variants on PDD, regardless of concomitant AD pathology. This is in line with a previously published report, which found only weak evidence for a role of APOE in PDD [40]. Further studies are needed in larger, ideally neuropathologically confirmed data sets, to validate the lack of association between APOE-ε4/TOMM40-L and PDD.
The associations between TOMM40/APOE and DLB, observed in our analysis, were corroborated in meta-analyses of the previously published evidence on the topic. Taken together, the statistical support from the meta-analyses in our study exceed those estimated in 2003 using only 61 individuals with DLB [41] by at least five orders of magnitude owing to the much greater power of our analyses. Our results are also in agreement with Bras et al. on AD and PD candidate loci (using the “NeuroX” array) in neuropathologically confirmed DLB cases and controls, although that study did not report association results for APOE-ε4 or DLB+AD specifically [10]. This possibly explains why the OR reported in the study by Bras et al. is much smaller (OR = 2.8) compared with our analyses (OR = 5.65).
Since the completion of our study, a large genome-wide association studies (GWAS) of DLB, has reported the association with DLB for previously established AD and PD loci [11]. Specifically, the study identified the genome-wide significant (P value <5 × 10−8) association between DLB risk and APOE-ε4, and, to a lesser extent, effects of SNCA and GBA locus variants on susceptibility to DLB [11]. Similar to previous GWAS in AD, this GWAS study did not include the poly-T repeat variant rs10524523 (rs523), thus, no data were available with regard to TOMM40 variants.
Our findings, corroborated by others [7], [17], have important implications for drug discovery and development strategies towards novel effective therapies for delaying disease onset and progression for both DLB and AD. First, the current R&D monotherapy model in AD, DLB, and other late-onset dementias targeting individual pathological pathways, such as amyloid, tau or α-synuclein, may not be optimal for treating these diseases due to the co-occurrence of multiple pathological features [5], [7], [8]. Furthermore, DLB and AD are multifactorial conditions, resulting from complex interactions (over time) between gene products, epigenetic and other biological processes related to ageing and a variety of life-long environmental factors and exposures. The gene products encoded by the two genes, namely APOE and the TOMM40 Translocase, have important roles in metabolic and oxidative homeostasis. APOE is a key cholesterol transporter with known effects on lipid metabolism, mitochondrial function, and is possibly involved in immune-modulating mechanisms [42]. TOMM40 Translocase, on the other hand, forms part of the outer mitochondrial membrane apparatus, with a key role in transmembrane transport mechanisms [43]. For AD, DLB, and other age-related neurodegenerative diseases, we still have major gaps in understanding the composition and chronology of the etiological puzzle that leads to synaptic dysfunction and neuronal death. It is also important to note that genetic-based risk prediction algorithms and biomarkers are currently being used in randomized clinical trials involving cognitively healthy elderly individuals for disease stratification and selection of “at–risk for AD” subjects. APOE-ε4 homozygosity or heterozygosity, the latter with biomarker evidence of abnormal amyloid burden, are currently used in randomized clinical trials testing antiamyloid therapies, and, in a trial of pioglitazone (repurposed antidiabetic) a genetics-based risk algorithm uses a combination of age and APOE/TOMM40 poly-T repeat genotypes as a risk selection criterion [44]. In addition, polygenic risk scores, using GWAS-derived common and small-effect variants in addition to APOE have been reported to moderately increase sensitivity and specificity of modulating risk for AD and its AOO [45], [46], [47], thus potentially increasing the sensitivity of current models.
Our study has a number of limitations. First, the data sets used in our association analyses were relatively modest and, thus, potentially would require larger study samples to disentangle the effects of TOMM40-L and APOE-ε4 alleles or the APOE-ε3/TOMM40-S and -VL alleles on DLB. This potential loss in power was somewhat mitigated by the fact that all cases and controls included here were neuropathologically confirmed, thus diminishing the risk of misclassification. We also acknowledge that this study is smaller than currently available GWAS for PD or DLB [10], [11], [48]. However, even recent genome-wide genotyping arrays do not directly genotype all variants investigated in our study (e.g., TOMM40 poly-T repeats) [43], and GWAS are typically based on clinical data, thus diminishing their ability to evaluate genetic effects on accurate DLB and PDD clinicopathological phenotypes. Furthermore, our APOE-ε4 – DLB association findings were corroborated in independent data sets investigating the role of APOE specifically, which we combined in the hitherto largest meta-analysis on the topic. We note that all of the samples analysed here were of European descent. To further assess the contribution of APOE-ε4/TOMM40-L on DLB, further work is required in non-European data sets to generate genotype data in diverse ethnic-descent groups.
Another limitation of our study is the lack of genome-wide genotyping data precluding a detailed assessment of potential population stratification effects on our results. However, we took several measures to minimize the possibility of bias due to admixture in our analyses: First, we have defined all samples in our data set as of European descent based on the clinical information. Second, we adjusted our analyses for study centre, thus taking into account subtle differences in data set composition across sites. Moreover, very similar results were observed in independent data sets identified by our systematic literature screen, suggesting that our findings are not substantially skewed by unadjusted stratification. Finally, our results do not allow any specific insights on the molecular mechanisms underlying the observed associations. This includes an answer to the question as to whether dysfunction of TOMM40 or APOE is responsible for the risk effects on DLB. We did not detect any additional effects of TOMM40-S or VL alleles in APOE ε3/ε3 or ε3/ε4 carriers, possibly because of the small sample size in those genotype subgroups. The ε4-allele is a missense change invoking a cysteine (Cys) to arginine (Arg) exchange to the amino acid sequence at residue 112. There is a plethora of published reports on the potential molecular mechanisms of the effect of APOE-ε4 expression on the amyloid cascade hypothesis, involving enhanced amyloid synthesis, early Aβ fibril formation and significant increase of amyloid load, as well as impaired Aβ clearance [18], [19], [42]. A direct neurotoxic effect of apoE ε4, when expressed in neurons, has also been proposed; this effect may be mediated by mitochondrial dysfunction and cytoskeletal alterations [42], [49]. Notwithstanding, to allow a more detailed assessment of the respective roles of APOE and TOMM40, molecular methods allowing haplotype reconstruction and analysis are needed and should be the focus of future work.
In summary, we report evidence for genome-wide significant association between APOE-ε4/TOMM40-L allele with DLB risk and age of disease onset. This association is explained by the co-occurrence of AD pathology in DLB that is altogether absent in PDD cases. Our findings may have important implications in drug discovery and development strategies targeting late-onset dementias with mixed pathological phenotypes.
Research in context.
-
1.
Systematic review: The presence of Lewy body pathology is a key neuropathological feature in Parkinson's disease (PD) and two forms of late-onset dementias, namely dementia with Lewy bodies (DLB) and Parkinson's disease dementia (PDD). Pathological studies have highlighted the presence of Alzheimer's disease (AD)-related changes in a large proportion of individuals with DLB and PDD. Genome-wide association studies have recently demonstrated the role of the APOE locus on susceptibility to DLB, however, provided contradictory support in the case of PDD. Yet, the respective roles of apolipoprotein E (APOE) ε4 and TOMM40 IVS6 poly-T L alleles in susceptibility to DLB have rarely been addressed. Given their functional impact and potential implications for drug discovery and development, the issue of a distinct biological candidacy between the two genes is an important one, albeit difficult to disentangle due to their tight linkage disequilibrium.
-
2.
Interpretation: Using samples and data from four European Brain banks, along with assessment of co-occurrence of AD pathology, we have attempted to dissect the primary effect of APOE-ε4/TOMM40-L on risk of developing Lewy body dementias. Our findings suggest that presence of APOE-ε4/TOMM40-L has no effect on PDD risk, while their effect on DLB is due to AD co-occurrence.
-
3.
Future directions: Our findings have important implications for drug discovery and development strategies towards novel effective therapies for delaying disease onset and progression of both DLB and AD.
Acknowledgements
The authors are grateful to the patients who have made this study possible by donating brain tissue to the collaborating Brain banks of our study. Tissue material and associated clinical and neuropathological data were obtained from the following brain banks: the Oxford Brain Bank, supported by the Medical Research Council (MRC), Brains for Dementia Research (BDR) (Alzheimer Society and Alzheimer Research UK), Autistica UK and the NIHR Oxford Biomedical Research Centre; the Parkinson's UK Tissue Bank, funded by Parkinson's UK, a charity registered in England and Wales (258197) and Scotland (SCO37554); the Newcastle Brain Tissue Resource, which is funded in part by a grant from the UK Medical Research Council (G0400074) and by Brains for Dementia research, a joint venture between Alzheimer's Society and Alzheimer's Research UK.
Part of this work was performed using the Imperial College High Performance Computing Service, URL: http://www.imperial.ac.uk/admin-services/ict/self-service/research-support/hpc/.
The study was supported by grants from Parkinson's UK (G0909), the Michael J Fox Foundation and UCB Pharmaceuticals to LTM.
I.P. was funded by the European Union's Horizon 2020 research, and innovation programme (LONGITOOLS, H2020- SC1-2019-874739; DYNAhealth, H2020-PHC-2014-633595); and the Wellcome Trust (WT205915). M.A.K. was funded by the European Commission under the Marie Curie Intra-European Fellowship, project MARVEL (PIEF-GA-2013-626461). E.L. was supported by MRC CASE studentship. Further support came from the ERC (as part of the EMIF-AD project to LB, the Possehl foundation, the Renate Maaβ foundation, the German Research Foundation (FOR2488/1, GZ LI 2654/2-1) and the University of Lübeck (medical section, grant number: J21-2016) to C.M.L. L.P. is supported by a grant from Monument Trust Discovery Award (J-1403) from Parkinson's UK.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.trci.2019.08.005.
Supplementary data
References
- 1.Vann Jones S.A., O'Brien J.T. The prevalence and incidence of dementia with Lewy bodies: a systematic review of population and clinical studies. Psychol Med. 2014;44:673–683. doi: 10.1017/S0033291713000494. [DOI] [PubMed] [Google Scholar]
- 2.Tola-Arribas M.A., Yugueros M.I., Garea M.J., Ortega-Valin F., Ceron-Fernandez A., Fernandez-Malvido B. Prevalence of dementia and subtypes in Valladolid, northwestern Spain: the DEMINVALL study. PLoS One. 2013;8:e77688. doi: 10.1371/journal.pone.0077688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emre M., Aarsland D., Brown R., Burn D.J., Duyckaerts C., Mizuno Y. Clinical diagnostic criteria for dementia associated with Parkinson's disease. Mov Disord. 2007;22:1689–1707. doi: 10.1002/mds.21507. quiz 837. [DOI] [PubMed] [Google Scholar]
- 4.McKeith I.G., Boeve B.F., Dickson D.W., Halliday G., Taylor J.P., Weintraub D. Diagnosis and management of dementia with Lewy bodies: fourth consensus report of the DLB Consortium. Neurology. 2017;89:88–100. doi: 10.1212/WNL.0000000000004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKeith I.G., Galasko D., Kosaka K., Perry E.K., Dickson D.W., Hansen L.A. Consensus guidelines for the clinical and pathologic diagnosis of dementia with Lewy bodies (DLB): report of the consortium on DLB international workshop. Neurology. 1996;47:1113–1124. doi: 10.1212/wnl.47.5.1113. [DOI] [PubMed] [Google Scholar]
- 6.Svenningsson P., Westman E., Ballard C., Aarsland D. Cognitive impairment in patients with Parkinson's disease: diagnosis, biomarkers, and treatment. Lancet Neurol. 2012;11:697–707. doi: 10.1016/S1474-4422(12)70152-7. [DOI] [PubMed] [Google Scholar]
- 7.Robinson J.L., Lee E.B., Xie S.X., Rennert L., Suh E., Bredenberg C. Neurodegenerative disease concomitant proteinopathies are prevalent, age-related and APOE4-associated. Brain. 2018;141:2181–2193. doi: 10.1093/brain/awy146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spillantini M.G., Schmidt M.L., Lee V.M., Trojanowski J.Q., Jakes R., Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 9.Bertram L., Tanzi R.E. The genetics of Alzheimer's disease. Prog Mol Biol Transl Sci. 2012;107:79–100. doi: 10.1016/B978-0-12-385883-2.00008-4. [DOI] [PubMed] [Google Scholar]
- 10.Bras J., Guerreiro R., Darwent L., Parkkinen L., Ansorge O., Escott-Price V. Genetic analysis implicates APOE, SNCA and suggests lysosomal dysfunction in the etiology of dementia with Lewy bodies. Hum Mol Genet. 2014;23:6139–6146. doi: 10.1093/hmg/ddu334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerreiro R., Ross O.A., Kun-Rodrigues C., Hernandez D.G., Orme T., Eicher J.D. Investigating the genetic architecture of dementia with Lewy bodies: a two-stage genome-wide association study. Lancet Neurol. 2018;17:64–74. doi: 10.1016/S1474-4422(17)30400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi S., Tateno M., Park T.W., Utsumi K., Sohma H., Ito Y.M. Apolipoprotein E4 frequencies in a Japanese population with Alzheimer's disease and dementia with Lewy bodies. PLoS One. 2011;6:e18569. doi: 10.1371/journal.pone.0018569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuang D., Leverenz J.B., Lopez O.L., Hamilton R.L., Bennett D.A., Schneider J.A. APOE epsilon4 increases risk for dementia in pure synucleinopathies. JAMA Neurol. 2013;70:223–228. doi: 10.1001/jamaneurol.2013.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berge G., Sando S.B., Rongve A., Aarsland D., White L.R. Apolipoprotein E epsilon2 genotype delays onset of dementia with Lewy bodies in a Norwegian cohort. J Neurol Neurosurg Psychiatry. 2014;85:1227–1231. doi: 10.1136/jnnp-2013-307228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jellinger K.A., Attems J. Neuropathological evaluation of mixed dementia. J Neurol Sci. 2007;257:80–87. doi: 10.1016/j.jns.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 16.Ruffmann C., Calboli F.C., Bravi I., Gveric D., Curry L.K., de Smith A. Cortical Lewy bodies and Abeta burden are associated with prevalence and timing of dementia in Lewy body diseases. Neuropathol Appl Neurobiol. 2016;42:436–450. doi: 10.1111/nan.12294. [DOI] [PubMed] [Google Scholar]
- 17.Irwin D.J., Grossman M., Weintraub D., Hurtig H.I., Duda J.E., Xie S.X. Neuropathological and genetic correlates of survival and dementia onset in synucleinopathies: a retrospective analysis. Lancet Neurol. 2017;16:55–65. doi: 10.1016/S1474-4422(16)30291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mormino E.C., Betensky R.A., Hedden T., Schultz A.P., Ward A., Huijbers W. Amyloid and APOE epsilon4 interact to influence short-term decline in preclinical Alzheimer disease. Neurology. 2014;82:1760–1767. doi: 10.1212/WNL.0000000000000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villemagne V.L., Burnham S., Bourgeat P., Brown B., Ellis K.A., Salvado O. Amyloid beta deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer's disease: a prospective cohort study. Lancet Neurol. 2013;12:357–367. doi: 10.1016/S1474-4422(13)70044-9. [DOI] [PubMed] [Google Scholar]
- 20.Buckley R.F., Mormino E.C., Amariglio R.E., Properzi M.J., Rabin J.S., Lim Y.Y. Sex, amyloid, and APOE epsilon4 and risk of cognitive decline in preclinical Alzheimer's disease: findings from three well-characterized cohorts. Alzheimers Dement. 2018;14:1193–1203. doi: 10.1016/j.jalz.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roses A.D., Lutz M.W., Crenshaw D.G., Grossman I., Saunders A.M., Gottschalk W.K. TOMM40 and APOE: requirements for replication studies of association with age of disease onset and enrichment of a clinical trial. Alzheimers Dement. 2013;9:132–136. doi: 10.1016/j.jalz.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Roses A.D., Lutz M.W., Amrine-Madsen H., Saunders A.M., Crenshaw D.G., Sundseth S.S. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer's disease. Pharmacogenomics J. 2010;10:375–384. doi: 10.1038/tpj.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G., Bekris L.M., Leong L., Steinbart E.J., Shofer J.B., Crane P.K. TOMM40 intron 6 poly-T length, age at onset, and neuropathology of AD in individuals with APOE epsilon3/epsilon3. Alzheimers Dement. 2013;9:554–561. doi: 10.1016/j.jalz.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson S.C., La Rue A., Hermann B.P., Xu G., Koscik R.L., Jonaitis E.M. The effect of TOMM40 poly-T length on gray matter volume and cognition in middle-aged persons with APOE epsilon3/epsilon3 genotype. Alzheimers Dement. 2011;7:456–465. doi: 10.1016/j.jalz.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burggren A.C., Mahmood Z., Harrison T.M., Siddarth P., Miller K.J., Small G.W. Hippocampal thinning linked to longer TOMM40 poly-T variant lengths in the absence of the APOE epsilon4 variant. Alzheimers Dement. 2017;13:739–748. doi: 10.1016/j.jalz.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruno D., Nierenberg J.J., Ritchie J.C., Lutz M.W., Pomara N. Cerebrospinal fluid cortisol concentrations in healthy elderly are affected by both APOE and TOMM40 variants. Psychoneuroendocrinology. 2012;37:366–371. doi: 10.1016/j.psyneuen.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruno D., Pomara N., Nierenberg J., Ritchie J.C., Lutz M.W., Zetterberg H. Levels of cerebrospinal fluid neurofilament light protein in healthy elderly vary as a function of TOMM40 variants. Exp Gerontol. 2012;47:347–352. doi: 10.1016/j.exger.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu S.H., Roeder K., Ferrell R.E., Devlin B., DeMichele-Sweet M.A., Kamboh M.I. TOMM40 poly-T repeat lengths, age of onset and psychosis risk in Alzheimer disease. Neurobiol Aging. 2011;32:2328.e1–2328.e9. doi: 10.1016/j.neurobiolaging.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helisalmi S., Hall A., Merilainen E.H., Vaisanen V., Koivisto A.M., Herukka S.K. The effect of TOMM40 poly-T repeat lengths on age of onset and cerebrospinal fluid biomarkers in Finnish Alzheimer's disease patients. Neurodegener Dis. 2014;14:204–208. doi: 10.1159/000367994. [DOI] [PubMed] [Google Scholar]
- 30.Jun G., Vardarajan B.N., Buros J., Yu C.E., Hawk M.V., Dombroski B.A. Comprehensive search for Alzheimer disease susceptibility loci in the APOE region. Arch Neurol. 2012;69:1270–1279. doi: 10.1001/archneurol.2012.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu L., Lutz M.W., Wilson R.S., Burns D.K., Roses A.D., Saunders A.M. TOMM40'523 variant and cognitive decline in older persons with APOE epsilon3/3 genotype. Neurology. 2017;88:661–668. doi: 10.1212/WNL.0000000000003614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiba-Falek O., Gottschalk W.K., Lutz M.W. The effects of the TOMM40 poly-T alleles on Alzheimer's disease phenotypes. Alzheimers Dement. 2018;14:692–698. doi: 10.1016/j.jalz.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daniel S.E., Lees A.J. Parkinson's Disease Society Brain Bank, London: overview and research. J Neural Transm Suppl. 1993;39:165–172. [PubMed] [Google Scholar]
- 34.Hyman B.T., Phelps C.H., Beach T.G., Bigio E.H., Cairns N.J., Carrillo M.C. National Institute on Aging-Alzheimer's Association guidelines for the neuropathologic assessment of Alzheimer's disease. Alzheimers Dement. 2012;8:1–13. doi: 10.1016/j.jalz.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linnertz C., Saunders A.M., Lutz M.W., Crenshaw D.M., Grossman I., Burns D.K. Characterization of the poly-T variant in the TOMM40 gene in diverse populations. PLoS One. 2012;7:e30994. doi: 10.1371/journal.pone.0030994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lill C.M., Roehr J.T., McQueen M.B., Kavvoura F.K., Bagade S., Schjeide B.M. Comprehensive research synopsis and systematic meta-analyses in Parkinson's disease genetics: the PDGene database. PLoS Genet. 2012;8:e1002548. doi: 10.1371/journal.pgen.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertram L., McQueen M.B., Mullin K., Blacker D., Tanzi R.E. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 38.Farrer L.A., Cupples L.A., Haines J.L., Hyman B., Kukull W.A., Mayeux R. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 39.Kulminski A.M., Huang J., Wang J., He L., Loika Y., Culminskaya I. Apolipoprotein E region molecular signatures of Alzheimer's disease. Aging Cell. 2018;17:e12779. doi: 10.1111/acel.12779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams-Gray C.H., Goris A., Saiki M., Foltynie T., Compston D.A., Sawcer S.J. Apolipoprotein E genotype as a risk factor for susceptibility to and dementia in Parkinson's disease. J Neurol. 2009;256:493–498. doi: 10.1007/s00415-009-0119-8. [DOI] [PubMed] [Google Scholar]
- 41.Bang O.Y., Kwak Y.T., Joo I.S., Huh K. Important link between dementia subtype and apolipoprotein E: a meta-analysis. Yonsei Med J. 2003;44:401–413. doi: 10.3349/ymj.2003.44.3.401. [DOI] [PubMed] [Google Scholar]
- 42.Mahley R.W., Huang Y. Apolipoprotein e sets the stage: response to injury triggers neuropathology. Neuron. 2012;76:871–885. doi: 10.1016/j.neuron.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roses A., Sundseth S., Saunders A., Gottschalk W., Burns D., Lutz M. Understanding the genetics of APOE and TOMM40 and role of mitochondrial structure and function in clinical pharmacology of Alzheimer's disease. Alzheimers Dement. 2016;12:687–694. doi: 10.1016/j.jalz.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 44.Chiba-Falek O., Lutz M.W. Towards precision medicine in Alzheimer's disease: deciphering genetic data to establish informative biomarkers. Expert Rev Precis Med Drug Dev. 2017;2:47–55. doi: 10.1080/23808993.2017.1286227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chouraki V., Reitz C., Maury F., Bis J.C., Bellenguez C., Yu L. Evaluation of a genetic risk score to improve risk prediction for Alzheimer's disease. J Alzheimers Dis. 2016;53:921–932. doi: 10.3233/JAD-150749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Escott-Price V., Sims R., Bannister C., Harold D., Vronskaya M., Majounie E. Common polygenic variation enhances risk prediction for Alzheimer's disease. Brain. 2015;138:3673–3684. doi: 10.1093/brain/awv268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Lee S.J., Wolters F.J., Ikram M.K., Hofman A., Ikram M.A., Amin N. The effect of APOE and other common genetic variants on the onset of Alzheimer's disease and dementia: a community-based cohort study. Lancet Neurol. 2018;17:434–444. doi: 10.1016/S1474-4422(18)30053-X. [DOI] [PubMed] [Google Scholar]
- 48.Nalls M.A., Pankratz N., Lill C.M., Do C.B., Hernandez D.G., Saad M. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y., Mahley R.W. Apolipoprotein E: structure and function in lipid metabolism, neurobiology, and Alzheimer's diseases. Neurobiol Dis. 2014;72:3–12. doi: 10.1016/j.nbd.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.