Abstract
Recent findings suggest that dopamine oxidation contributes to the development of Parkinson's disease (PD); however, the mechanistic details remain elusive. Here, we compare 6-hydroxydopamine (6-OHDA), a product of dopamine oxidation that commonly induces dopaminergic neurodegeneration in laboratory animals, with a synthetic alkyne-functionalized 6-OHDA variant. This synthetic molecule provides insights into the reactivity of quinone and neuromelanin formation. Employing Huisgen cycloaddition chemistry (or “click chemistry”) and fluorescence imaging, we found that reactive 6-OHDA p-quinones cause widespread protein modification in isolated proteins, lysates and cells. We identified cysteine thiols as the target site and investigated the impact of proteome modification by quinones on cell viability. Mass spectrometry following cycloaddition chemistry produced a large number of 6-OHDA modified targets including proteins involved in redox regulation. Functional in vitro assays demonstrated that 6-OHDA inactivates protein disulfide isomerase (PDI), which is a central player in protein folding and redox homeostasis. Our study links dopamine oxidation to protein modification and protein folding in dopaminergic neurons and the PD model.
Keywords: Parkinson's disease, Dopamine oxidation, 6-Hydroxydopamine p-quinone, Post-translational protein modification, Neurotoxicity, Protein disulfide isomerase
Graphical abstract
Highlights
-
•
Chemical modification of 6-OHDA increases stability of 6-OHDA p-quinone by preventing neuromelanin formation.
-
•
Modified 6-OHDA enables visualization of thiol-dependent protein modification by p-quinone.
-
•
Wide-spread proteome modification by 6-OHDA p-quinone impairs neuroblastoma viability.
-
•
6-OHDA p-quinone inactivates PDI linking dopamine oxidation to protein unfolding.
1. Introduction
Parkinson's disease (PD), the second most common age-related neurodegenerative disorder, challenges the ageing Western society and its health care sectors [[1], [2], [3]]. Typical clinical symptoms of PD include rigidity, resting tremor, bradykinesia and postural instability [[4], [5], [6]]. These symptoms are due to the loss of dopaminergic neurons in the substantia nigra pars compacta (SNc) yielding low levels of the neurotransmitter dopamine within the brain. Despite the identification of several risk factors and genetic markers, the underlying cause of this slowly progressive disease remains unknown. Research suggests that intracellular accumulation of protein aggregates in abnormal deposits contributes to neuronal cell death [7,8]. These deposits, termed Lewy bodies, contain large amounts of the intrinsically disordered protein alpha-synuclein (SNCA), establishing PD as the most prominent member among the family of synucleinopathies [9,10].
The specific loss of dopaminergic neurons in the SNc is likely due to the presence of dopamine and the associated biological pathways in these cells [11,12]. Dopamine is a highly reactive molecule and is stored in synaptic vesicles, in which the local acidic environment prevents autoxidation. Nonetheless, intracellular dopamine autoxidation can occur because dopamine synthesis and metabolism take place in the cytosol, which provides conditions more conducive for oxidation. Cellular antioxidants, for instance glutathione (GSH), inhibit dopamine oxidation whereas metal ions, in particular iron and copper, accelerate dopamine oxidation [[13], [14], [15], [16]]. Many studies report both reduction of GSH levels as well as iron accumulation in the brain of PD patients implying that dopamine oxidation is a disease-relevant process [17,18]. A recent study showed that dopamine oxidation affects mitochondrial activity, and oxidized dopamine metabolites accumulate in the brain of PD patients [19]. It is widely accepted that oxidation products of dopamine affect several fundamental physiological processes in neuronal cells [20,21]. Perturbation of dopamine metabolism impairs lysosomal activity and protein degradation [19,22], promotes aggregation of SNCA into neurotoxic protofibrils [23,24], and leads to oxidative damage in the brain [25,26].
One-electron or two-electron oxidation of dopamine (DA) produce the dopamine semiquinone (DASQ) or the dopamine o-quinone (DAQ), respectively (Fig. 1A) [20,21]. Dopamine quinones are highly reactive and rapidly undergo intramolecular conversion or react with other molecules and, thus, display only a very short lifetime; in particular, DAQ cyclizes almost instantaneously with a rate constant of 0.15 s–1 to dopaminochrome (DAC) via the unstable intermediate leukodopaminochrome (LDAC) at physiological pH [[27], [28], [29]]. In the brain, DAC polymerizes to form pigments known as neuromelanin [30]. The role of these polymers is controversial as both neuroprotective and neurotoxic properties have been reported [[31], [32], [33]].
Fig. 1.
Characterization of chemically modified 6-hydroxydopamine (6-OHDA). Overview of the dopamine and the 6-OHDA oxidation cascade promoted by oxidizing species indicated as [ox]. DA, dopamine; DAQ, dopamine quinone; DASQ, dopamine semiquinone; LDAC, leukodopaminochrome; DAC, dopaminochrome (A). Structure of the alkyne-functionalized 6-OHDA (6-OHDA-PEG3-yne) used in this study (B). Schematic overview of the applied copper-catalyzed azide-alkyne cycloaddition approach applied in this study (C). Monitoring absorption at 490 nm of 6-OHDA (100 μM, red) and 6-OHDA-PEG3-yne (100 μM, blue) in PBS (pH 7.6; circles) or in water (pH 5.8; triangles) continuously over 4 h (D). Cell viability of SH-SY5Y cells challenged with either 6-OHDA (red) or 6-OHDA-PEG3-yne (blue). IC50 values determined by fitting are presented in the graph (E). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
One notable oxidation product of dopamine is 6-hydroxydopamine (6-OHDA), which forms in the presence of iron via hydroxylation of dopamine [12,[34], [35], [36], [37], [38]]. Furthermore, 6-OHDA has been widely used in preclinical models to investigate PD in the animal model [[39], [40], [41]]. 6-OHDA undergoes a very similar series of reactions like dopamine, giving rise to 6-OHDA p-quinone, and eventually resulting in neuromelanin as well (Fig. 1A) [42,43]. A few studies showed that both DAQ and 6-OHDA p-quinone are capable of modifying proteins or peptides but the biological significance is unclear [20,[43], [44], [45], [46], [47], [48]].
Any attempt to study experimentally the role of dopamine and dopamine oxidation products is challenging due to the high reactivity, the associated series of complex reaction cascades, and the formation of various intermediates and large insoluble polymers, producing opposing effects within the cell. The lack of tools to interrogate specific intermediates of dopamine oxidation, especially within cellular systems, limits our understanding of the role of dopamine oxidation in the development of PD. For instance, the interaction of dopamine metabolites with other biological molecules, such as proteins, remains elusive [20]. In this study, we sought to address this limitation and present the proof-of-concept of a novel chemical biology approach to elucidate modification of proteins by dopamine intermediates. We interrogated a chemically modified 6-OHDA that we presented previously [52]. Here, we found that this modificationprevents intramolecular conversion and limits 6-OHDA's oxidative reaction cascade. Furthermore, this synthetic molecule allows visualization of protein modification by quinones in vitro and in cellulo, and permits interrogation of the associated environmental conditions in order to study the emerging link between dopamine oxidation and protein modification in the development of PD.
2. Material and methods
2.1. Reagents
All reagents, solvents and oligonucleotides were purchased from Sigma Aldrich, unless otherwise noted, and used without further purification. Propargyl-PEG3-acid was purchased from BroadPharm. Alexa Fluor™ 488 azide was obtained from Thermo Fisher Scientific. Rhodamine-azide was synthesized as described previously [[49], [50], [51]].
2.2. Synthesis and characterization of 6-OHDA-PEG3-yne
Initially, 6-OHDA-PEG3-yne was obtained as described previously [52]. Additional 6-OHDA-PEG3-yne was synthesized as described except for the use of nitrogen instead of argon as inert gas. Absorbance in water (pH 5.8) or PBS (pH 7.6) was monitored at 30 °C under repeated shaking using an automated plate reader (Microplate reader Tecan Safire2, Cisbio).
2.3. Animals and cell culture
Primary cortical neurons were derived from E14.5–15.5 wild type (WT) CD1 mice (Charles River Laboratories) of either sex and maintained in Neurobasal media (Invitrogen) supplemented with B27 with antioxidants (Gibco), N2 (Gibco), 0.5 mM l-glutamine, and penicillin-streptomycin (Gibco) as previously reported [53]. Neurons were plated onto poly-d-lysine coated plates as follows: 4 × 106 cells per 10 cm plate and 1 × 106 per well of 6-well plate. Cells were grown in culture for 4 days in the presence of antioxidants. Cultures received new media containing no antioxidants on the fourth day in vitro (DIV4). Treatment and harvest were performed on DIV5.
The dopaminergic human neuroblastoma cell line SH-SY5Y was maintained in Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F-12) and the non-catecholaminergic human liver cell line HepG2 was maintained in DMEM. Media was supplemented with 10% fetal bovine serum (FBS), 100 nmol/L non-essential amino acids, and a penicillin-streptomycin mixture. All cells were cultured at 37 °C in a humidified atmosphere of 95% air and 5% CO2.
2.4. Cell viability assay
Viability of SH-SY5Y cells was determined by measuring the reduction of 3-(4,5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2H-tetrazolium bromide (MTT) to formazan. Cells were seeded in 96-wells (1 × 104 cells per well) and cultured for 24 h or 48 h until confluency reached approximately 70%. Then, culture media was replaced with fresh media supplemented with varying concentrations of 6-OHDA or 6-OHDA-PEG3-yne. After 24 h, cells were washed three times with PBS and incubated for 5 h at 37 °C in culture medium containing 0.5 mg/ml MTT tetrazolium salt. The medium was carefully removed, and the generated formazan was solubilized in DMSO. Absorbance was measured at 562 nm by an automatic plate reader (SpectraMax M2, Molecular Devices or Tecan Safire2, Cisbio). The absorbance at each concentration of 6-OHDA or 6-OHDA-PEG3-yne was expressed as a percentage of the absorbance measured in the absence of any compound. Finally, obtained values were plotted against the log-transformed concentration of the used compound and fit to a four-parameter equation.
2.5. Cloning, expression and purification of proteins
Expression vectors for an N-terminal hexahistidine-tagged human PDIA1 was kindly provided by Dr. Llyod Ruddock [54]. The catalytically active domain of PDI (D18-A137), termed PDIa, was obtained by introducing a stop codon after Alanine 137 as previously described [55,56]. N-terminal hexahistidine-tagged PDI and PDIa were expressed in E. coli Rosetta (DE3) strains (Invitrogen) at 25 °C for 16 h in the presence of 1 mM IPTG. Proteins were purified by Ni-affinity chromatography (HiTrap IMAC HP, GE Healthcare) followed by size-exclusion-chromatography (Superdex75 10/300, GE Healthcare) in 50 mM sodium phosphate, pH 7.7 (25 °C) and 150 mM NaCl.
The expression vector for alpha-synuclein (SNCA) was a gift from Michael J Fox Foundation (Addgene plasmid #51486). SNCA was expressed and purified as described elsewhere [57]. After anion exchange chromatography (HiTrap Q FF column, GE Healthcare), SNCA was dialyzed against 20 mM Tris–HCl, pH 8.0 and 150 mM NaCl.
Protein concentrations were measured by UV spectroscopy using a NanoDrop 1000 (Thermo Scientific). Samples were stored at −80 °C until use.
2.6. Protein disulfide isomerase activity assay
PDI and PDIa were reduced with 20-fold molar excess of DTT at room temperature for 30 min and then applied onto a PD-10 column. Collected fractions were monitored for residual DTT via DTNB and fractions without DTT were pooled and stored at −80 °C as described previously [58]. Reduced PDI or PDIa (75 nM each) were incubated with 150 μM H2O2, 150 μM 6-OHDA or 150 μM PAO in the presence or absence of 1 mM l-cysteine or 1 mM l-methionine. Oxidoreductase activity of PDI or PDIa was determined by reduction of 15 μM pseudo substrate diabzGSSH in the presence of 50 μM DTT in a 384-well plate as described previously [56,58,59]. Fluorescence (excitation filter 312 nm, emission filter 415 nm) was monitored at 5 s intervals for 90 min at 30 °C under repeated shaking using an automated plate reader (Microplate reader Tecan Safire2, Cisbio). Each sample was measured in quadruplicate. Data points are presented as means and error bars represent the standard error of the mean.
2.7. Determination of reduced cysteines with DTNB
First, 70 μM of thiol-containing compound (GSH or cysteine) was incubated in PBS (pH 7.6) without or with 10 μM 6-OHDA or 10 μM 6-OHDA-PEG3-yne at ambient temperature for 1 h. After incubation, 200 μM of 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) was added to the reaction mixtures. Finally, the absorbance of the solution was measured after 25 min at 412 nm using an automated plate reader (Microplate reader Tecan Safire2, Cisbio). Three independent replicates were measured and error bars represent the standard deviation.
2.8. Far-UV circular dichroism spectroscopy
Initially, BSA was reduced as described for PDI above. Far-UV CD spectra of 3 μM reduced BSA were recorded in 40 mM potassium phosphate (pH 7.5) at 30 °C in the presence of buffer, 150 μM H2O2, 150 μM 6-OHDA or 150 μM 6-OHDA-PEG3-yne using a Jasco J-815 spectropolarimeter. Samples were incubated with the corresponding reagents for 12–16 h at 4 °C and transferred to 30 °C 5 min before recording (data interval: 0.5 nm, cell length: 0.1 mm, 8 accumulations). The temperature during the experiment was controlled with a Jasco Peltier device.
2.9. In-gel fluorescence of proteins and cell lysates in vitro
Lysates were prepared by lysing cells in cold 1% Triton X-100 in PBS followed by sonication for 5 s. Samples were centrifuged at 14,000 rpm for 5 min at 4 °C. An aliquot of the supernatant was used for protein quantification via DC Protein Assay (Bio-Rad), whereas the remaining supernatant was used for labelling. Purified proteins (i.e. BSA, SNCA or PDI) were transferred into PBS and incubated with or without freshly prepared 2 mM maleimide for 1 h at ambient temperature. 6-OHDA-PEG3-yne was added to prepared samples and incubated under gentle mixing for 1 h at room temperature (for cell lysates) or 37 °C (for purified proteins) unless otherwise noted. Then, a ‘click mix’ containing 1 mM TCEP, 100 μM TBTA, 1 mM CuSO4 and 50 μM rhodamine-azide in PBS was added for additional 45–60 min. The copper-catalyzed azide-alkyne cycloaddition reaction was terminated, and the proteome was precipitated by addition of cold acetone and incubation at −80 °C for at least 30 min.
Samples were then centrifuged at 14,000 rpm for 15 min at 4 °C. Precipitated protein pellets were air-dried for 10 min, resuspended in 20 μL–35 μL Laemmli buffer, and heated for 10 min at 95 °C. Samples were resolved on SDS Protein gels, and in-gel fluorescence was detected by the ChemiDoc MP system (Bio-Rad). Fluorescence intensity of three experiments was normalized to the corresponding Coomassie bands and quantified using Image lab (Bio-Rad). Statistical significance was evaluated using an unpaired two-tailed Student's t-test.
2.10. Fluorescence microscopy of intracellular protein modification
HepG2 cells or SH-SY5Y cells were seeded (40′000 cells per well) in a 24-well glass bottom plate and cultured for 24 h or 48 h, respectively. Cells were fixed with 4% formaldehyde in PBS for 15 min at ambient temperature and washed three times with PBS. Fixed samples were stored at 4 °C until cell membranes were permeabilized with 0.1% Triton X-100 in PBS for 5 min. After washing with PBS, 6-OHDA-PEG3-yne was added to the samples for 30 min, or samples were treated with solvent only. Alternatively, living cells were treated with 6-OHDA-PEG3-yne, then washed three times with PBS before proceeding with fixation and permeabilization. Cells were incubated for 1 h with a ‘click mix’, containing 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 100 μM tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl] amine (TBTA), 1 mM CuSO4 and Alexa488-azide in PBS. Next, samples were washed with PBS and with cold methanol, and stained with 0.1 μg/ml DAPI (4′,6-diamidino-2-phenylindole). Cells were stored at 4 °C in the dark. Imaging was performed using a Leica SP5-X confocal laser-scanning microscope (Leica Microsystems, Mannheim, Germany) equipped with a standard 20x objective lens (Leica Microsystems, Mannheim, Germany). DAPI and Alexa488 were excited sequentially at 405 nm and at 488 nm. Emission spectra were recorded between 410 nm and 479 nm for DAPI and between 502 nm and 541 nm for Alexa488. Images were analyzed using the Leica Application Suite 2.7.4.10100.
2.11. Biotin-streptavidin pull-down of proteins coupled to 6-OHDA-PEG3-yne
Primary mouse neurons were lysed prior to the application of 6-OHDA-PEG3-yne. Copper-catalyzed azide-alkyne cycloaddition using biotin-azide followed by streptavidin-enrichment, and trypsin digest of modified proteome were performed as previously described [60]. After incubation, an additional acetone precipitation and three washing steps with cold methanol were performed in order to remove the ‘click mix’ from the protein fraction. Following protein enrichment, agarose-streptavidin beads (Pierce) were washed three times with 100 μL PBS in a Biospin column. Using 200 μL of PBS, the beads were transferred to cell lysates modified by 6-OHDA-PEG3-yne and incubated for 90 min. The beads were washed five times with 50 mM ammonium bicarbonate and transferred to an Eppendorf tube. Samples were heated for 15 min at 65 °C with 500 μL of 10 mM DTT in 50 mM ammonium bicarbonate (ABC). After 15 min, 25 μL of 500 mM iodoacetamide was added and samples were incubated in the dark for 30 min. The samples were centrifuged for 2 min at 1400 rpm and 100 μL of ABC was added followed by 2 μL of 0.5 mg/mL of trypsin. After incubation at 37 °C overnight on a rotating wheel, the beads were pelleted. The supernatant was purified using C18 Zip-tip columns and then applied to MS analysis.
2.12. Mass spectrometry for protein characterization
Mass spectra of BSA and SNCA were obtained by electron spray ionization (ESI) liquid chromatography mass spectrometry (LC/MS) on an Agilent 6410 Triple Quadrupole Mass Spectrometer instrument coupled to an Agilent 1200 HPLC system. The system was equipped with C8 (Agilent Zorbax Eclipse XBD-C8, 4.6 × 50 mm) and C18 (Agilent Zorbax Eclipse XBD-C8, 4.6 × 50 mm) reverse phase columns, an autosampler, and a diode-array detector. A linear gradient of the binary solvent system of H2O:acetonitrile:formic acid (from 95:5:0.1 to 5:95:0.1) was used. Data was analysed using MassHunter 01.03 (Agilent Technologies) and the deconvoluted sum spectra is presented.
2.13. Mass spectrometry for protein and peptide identification
Protein identification was performed using an Orbitrap Fusion (Thermo Scientific) coupled to an Ultimate3000 nanoRLSC (Dionex). Peptides were separated via a water/acetonitrile/0.1% formic acid gradient on an in-house packed column (Polymicro Technology), 15 cm × 70 μm ID, Luna C18(2), 3 μm, 100 Å (Phenomenex). Peptides were separated using an initial linear gradient from 2% to 38% of acetonitrile followed by a second gradient from 38% to 98% of acetonitrile. Final separation was achieved with 98% of acetonitrile followed by a reverse gradient from 98% to 2% of acetonitrile and a final wash with 2% of acetonitrile. Eluted peptides were sprayed directly into the mass spectrometer using positive ESI at an ion source temperature of 250 °C and an ionspray voltage of 2.1 kV. The Orbitrap Fusion Tribrid was run in top speed mode. Full-scan MS spectra (m/z 350–2000) were acquired at a resolution of 60,000. Precursor ions were filtered according to monoisotopic precursor selection, charge state (+2 to +7), and dynamic exclusion (30 s with a ±10 ppm window). The automatic gain control settings were 4e5 for full FT-MS scans and 1e4 for MS/MS scans. Fragmentation was performed with collision-induced dissociation (CID) in the linear ion trap. Precursors were isolated using a 2 m/z isolation window and fragmented with a normalized collision energy of 35%.
2.14. Analysis of mass spectrometry data
MaxQuant was used for protein identification and quantification. Search parameters were set to allow for dynamic modification of methionine oxidation, and static modification of cysteine carbamidomethylation. The search database consisted of nonredundant mouse protein sequences in FASTA file format from the UniProt/SwissProt database. The FDR was set to 0.05 for protein identifications. Pathway analysis of selected proteins (Sequest HT Score > 10 and excluding targets in mock pulldown) was performed using the ToppGene Suite [61]. ClustVis was used for Principal component analysis (PCA) [62].
2.15. Statistical analysis and software
Unless otherwise noted, data analysis was performed in GraphPad Prism 7.00 (GraphPad Inc.), Excel 2010 (Microsoft) and R 3.2.3 via R Studio 1.0.136. Chemical structures were drawn with ChemDraw Professional 16.0 (PerkinElmer). Figures were created using Inkscape 0.92 and the auxiliary R packages “gplots” and “ggplot2”.
3. Results
3.1. Prevention of intramolecular cyclization abrogates 6-OHDA pigment formation and extends stability of oxidation product
Previously, we investigated small molecule mediated inhibition of viral replication and, during this process, synthesized 6-OHDA-PEG3-yne by coupling a PEG3-alkyne to the amino group of 6-OHDA [52] (Fig. 1B). In this study, we sought out to characterize the physiochemical properties of 6-OHDA-PEG3-yne, and to apply this molecule to neuronal cell models. Our modification on 6-OHDA abrogates intramolecular ring closure and, hence, limits the subsequent reaction cascades. Moreover, 6-OHDA-PEG3-yne enables us to perform copper-catalyzed azide-alkyne Huisgen cycloaddition (also known as “click chemistry”) in order to study protein modification (Fig. 1C).
First, we monitored the absorption spectra of both molecules in slightly basic conditions (i.e., PBS pH 7.6). In agreement with earlier reports of pH-dependent autoxidation of 6-OHDA, we observed a rapid red coloration for both 6-OHDA and 6-OHDA-PEG3-yne within a few seconds corresponding to the formation of the 6-OHDA p-quinone (Fig. S1A) [43]. There was no significant difference between both absorption spectra in the first 30 min; however, after this time, we noticed a continuous decline in the absorption peak at 490 nm for 6-OHDA whereas 6-OHDA-PEG3-yne remained unchanged for several hours (Fig. 1D). Eventually, the absorption intensity for 6-OHDA-PEG3-yne started to decline over the course of 48 h (Figs. S1A and B). By contrast, both molecules underwent only marginal autoxidation under slightly acid conditions (i.e., de-ionized water, pH 5.8), confirming earlier observations (Figs. S1B and C) [43]. Interestingly, after 48 h in PBS, we observed the formation of black pigment, likely corresponding to neuromelanin, for 6-OHDA but not for 6-OHDA-PEG3-yne (Fig. S1D). We validated the pH-dependent autoxidation of 6-OHDA-PEG3-yne via LC/MS. After 2 h, we found substantially more reduced 6-OHDA-PEG3-yne in water (pH 5.8), which became very acidic (pH 4.0) (Fig. S1E), than slightly basic PBS (Fig. S1F). We also investigated whether the chemical modification on 6-OHDA's amino group affects toxicity on the human neuroblastoma cell line SH-SY5Y, which is widely used in PD research [63]. We conducted an MTT assay to assess cell viability upon treatment with either 6-OHDA or 6-OHDA-PEG3-yne but could not detect a significant difference between both molecules (Fig. 1E). We conclude that our modification of 6-OHDA does not affect autoxidation and formation of 6-OHDA p-quinone but stabilizes the subsequent reaction products.
3.2. Visualization of quinone modification of bovine serum albumin in vitro
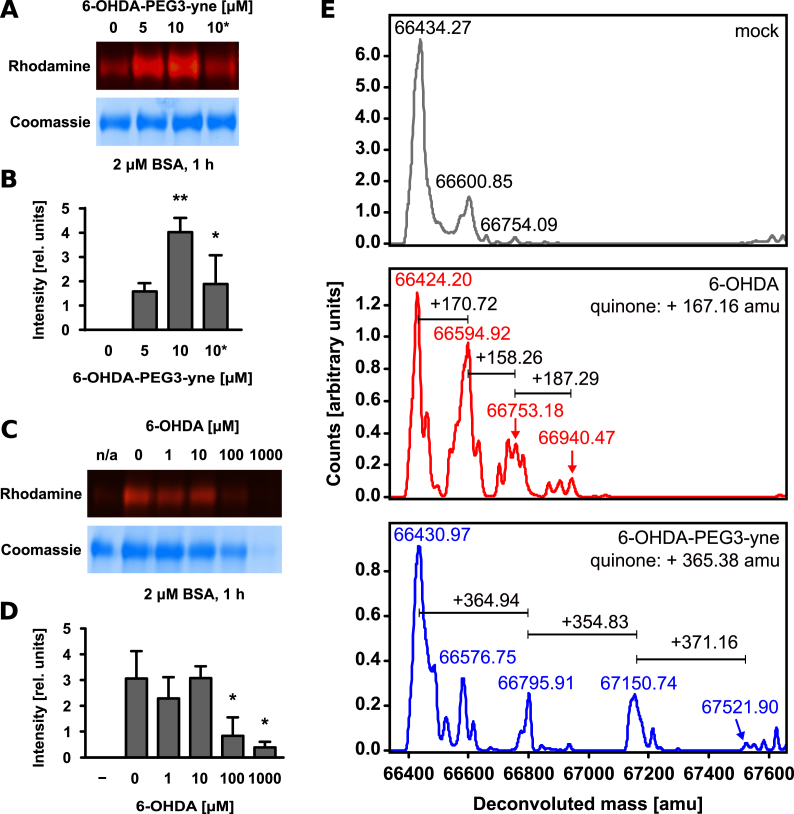
Next, we employed 6-OHDA-PEG3-yne to visualize protein modification by quinones using bovine serum albumin (BSA) as a model protein substrate. We incubated BSA with 6-OHDA-PEG3-yne followed by coupling of rhodamine-azide and in-gel fluorescence imaging. Our analysis demonstrated covalent modification of BSA by 6-OHDA-PEG3-yne (Fig. 2A and B). Previous studies indicated that the electrophilic 6-OHDA p-quinone serves as a Michael acceptor for biological nucleophiles, such as cysteine thiols [43]. Prior to incubation with 6-OHDA-PEG3-yne, we incubated BSA with maleimide, a compound that is commonly used to covalently modify cysteines [64,65]. We found that maleimide pre-treatment prevented alkylation by 6-OHDA-PEG3-yne, confirming cysteines as the main target (Fig. 2A and B). We excluded that our modification on 6-OHDA altered the molecule's reactivity or specificity by incubating BSA with native 6-OHDA prior to applying 6-OHDA-PEG3-yne. Increasing concentration of 6-OHDA correlated with decreasing in-gel fluorescence confirming that both molecules target the same sites in BSA (Fig. 2C and D). At very high concentrations of 6-OHDA, we noticed aggregation of BSA, which could be due to either 6-OHDA itself or due to the release of hydrogen peroxide (H2O2) during autoxidation of 6-OHDA or 6-OHDA-PEG3-yne. We examined this finding by treating BSA with different concentrations of H2O2 prior to incubation with 6-OHDA-PEG3-yne, and observed a similar degree of BSA aggregation (Fig. S2A and B). We are aware that this experiment does not fully clarify whether reduction of in-gel fluorescence is due to the loss of protein or due to cysteine oxidation, eventually rendering the target sites of 6-OHDA inaccessible; however, we concluded that autoxidation of large amounts of 6-OHDA produces large amounts of H2O2 that induce protein aggregation. Further, we applied LC/MS to confirm the observed modification of BSA after the copper-catalyzed azide-alkyne cycloaddition. We were able to detect additional mass on BSA due to the addition of one or more equivalents of 6-OHDA or 6-OHDA-PEG3-yne (Fig. 2E). We would like to point out that the reducing condition during the cycloaddition reaction causes the breakage of a certain amount of BSA's disulfide bonds rendering additional cysteines accessible as described in previous studies [66]; however, we were not able to resolve reliably the spectra for BSA modified with more than three equivalents due to technical limitations. Regardless, we noted that the spectra of BSA modified with 6-OHDA-PEG3-yne displayed more distinct peaks than BSA modified by 6-OHDA, likely reflecting the limited reactivity of 6-OHDA-PEG3-yne. In addition, we tested modification of BSA by 6-OHDA-PEG3-yne at different temperatures but could not detect a significant difference (Figs. S2C and D). Further, we performed this modification reaction at increasing pH, but the observed differences fall within the accuracy of the measurement (Figs. S2E and F). Generally, BSA modification by 6-OHDA-PEG3-yne occurred almost instantaneously (Figs. S2G and H). We also studied the effect of quinone modification of BSA but could not detect changes in overall protein folding (Fig. S2I); however, we observed the formation of black pigment and aggregation of BSA after 24 h for 6-OHDA, whereas BSA treated with 6-OHDA-PEG3-yne did not show any noticeable change (Fig. S2J).
Fig. 2.
Modification of BSA by 6-OHDA quinones. Representative in-gel fluorescence of BSA (2 μM) incubated for 1 h without or with 6-OHDA-PEG3-yne (5 μM or 10 μM). The asterisk denotes incubation of BSA with the cysteine-modifier maleimide (2 mM) for 1 h prior to incubation with 6-OHDA-PEG3-yne. The same gel after staining with Coomassie Brilliant Blue R-250 is shown below (A). Quantification of in-gel fluorescence of three independent experiments (B). Representative in-gel fluorescence and corresponding Coomassie stained gel of BSA (2 μM) incubated for 1 h with 6-OHDA-PEG3-yne (10 μM) or without (denoted by n/a) in the presence of increasing concentration of 6-OHDA (C) and quantitation of in-gel fluorescence of three independent experiments (D). LC/MS analysis of BSA (2 μM) incubated with either mock (top panel), 6-OHDA (10 μM, middle panel) or 6-OHDA-PEG3-yne (10 μM, bottom panel). Expected mass differences are indicated in the individual panels (E). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Protein modification by the 6-OHDA quinone is thiol-dependent
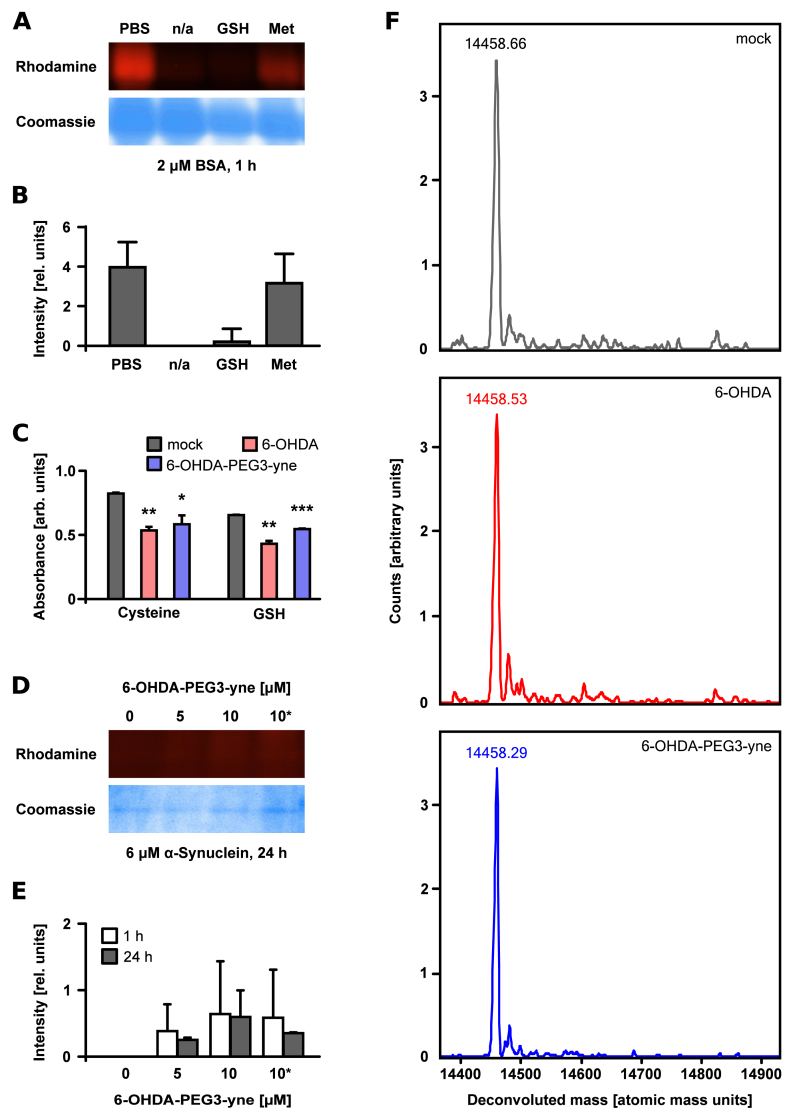
We demonstrated that maleimide treatment abrogated quinone modification of BSA suggesting that cysteines are the predominant target of the 6-OHDA p-quinone. We confirmed this finding by performing the alkylation reaction in the presence of the known antioxidants GSH and methionine. GSH prevented almost all protein modification, whereas methionine did not have a significant effect (Fig. 3A and B). We also performed this experiment in the presence of all 20 natural amino acids in order to exclude 6-OHDA-PEG3-yne reacting with other amino acids besides cysteine in our experiments (Fig. S3). Due to high experimental variability, a sound statistical assessment was not possible; however, we noticed that cysteine affected Coomassie staining or induced protein aggregation, indicating that cysteine plays a crucial role. Subsequently, we applied an indirect method to validate the role of thiols in scavenging 6-OHDA p-quinones. We incubated 6-OHDA or 6-OHDA-PEG3-yne with the physiological relevant GSH before quantifying the free thiols using Ellman's reagent (DTNB). We employed a low concentration of 6-OHDA (and 6-OHDA-PEG3-yne) to keep the interference of the absorbance of the p-quinone with the TNB absorbance at a minimum. We observed that the presence of GSH decreased the detected DTNB absorption, indicating that thiol-containing molecules scavenged 6-OHDA p-quinones (Fig. 3C). We confirmed this by employing cysteine, another thiol-containing compound with relevance for putative protein modification, which yielded the same result (Fig. 3C). In order to confirm that protein modification by 6-OHDA is thiol-dependent, we employed SNCA, which is highly relevant to PD and does not contain any cysteine. Concurring with our data, we were not able to detect any in-gel fluorescence for recombinantly expressed human SNCA (Fig. 3D). Additional mass spectrometry analysis of SNCA incubated with either native 6-OHDA or 6-OHDA-PEG3-yne did not reveal any additional mass, confirming our finding (Fig. 3E). Collectively, 6-OHDA-PEG3-yne is a potent modifier of proteins with a high selectivity for cysteine thiols.
Fig. 3.
Thiol-dependent modification of proteins by 6-OHDA quinones. Representative in-gel fluorescence and corresponding Coomassie stained gel of BSA (2 μM) incubated for 1 h with 6-OHDA-PEG3-yne (10 μM) or without (denoted by n/a) in the absence or presence of methionine (Met, 1 mM) or glutathione (GSH, 1 mM) (A) and quantitation of in-gel fluorescence of three independent experiments (B). Quantitation of free thiols in cysteine (Cys, 70 μM) or glutathione (GSH, 70 μM) via DTNB (200 μM) after incubation with or without 6-OHDA (10 μM, red) or 6-OHDA-PEG3-yne (10 μM, blue) for 1 h. Absorbance was corrected for background of 6-OHDA or 6-OHDA-PEG3-yne. Error bars represent standard deviation of three independent experiments. Statistical significance was evaluated using an unpaired two sample Student's t-test (* p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001). (C). Representative in-gel fluorescence and Coomassie stained gel of alpha-synuclein (SNCA, 6 μM) incubated for 1 h (white) or 24 h (grey) with or without 6-OHDA-PEG3-yne (5 μM or 10 μM) or 6-OHDA-PEG3-yne (10 μM) after pre-incubation for 1 h with maleimide (2 mM, denoted with *) (D) and quantitation of in-gel fluorescence of three independent experiments (E). LC/MS analysis of SNCA (6 μM) incubated with either mock (top panel), 6-OHDA (10 μM, middle panel) or 6-OHDA-PEG3-yne (10 μM, bottom panel) (F). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. 6-OHDA p-quinone modifies the cellular proteome
In the next step, we applied 6-OHDA-PEG3-yne to a complex cellular environment. Besides SH-SY5Y cells, we also used the non-catecholaminergic human liver cell line HepG2 that has been used as a non-neuronal control in previous studies [67]. In addition, we used mouse primary embryonic cortical neurons (herein referred to as ‘mouse cells’) since mice are a pre-clinical model for 6-OHDA-induced dopaminergic neurodegeneration. First, we lysed the cells and treated the cell lysates with 6-OHDA-PEG3-yne followed by copper-catalyzed azide-alkyne cycloaddition but we did not find a specific pattern among the fluorescent bands in SH-SY5Y cells (Fig. S4A). Increasing the concentration of 6-OHDA-PEG3-yne yielded a corresponding increase in in-gel fluorescence (Fig. 4A); however, no specific banding pattern emerged. We obtained the same result for both HepG2 and mouse cell lysates implying that 6-OHDA-PEG3-yne does not target specific proteins (Figs. 4A, S4B,C). Incubation of cell lysates with 6-OHDA prior to treatment with 6-OHDA-PEG3-yne greatly reduced in-gel fluorescence confirming the overlapping reaction sites for both 6-OHDA and 6-OHDA-PEG3-yne (Fig. 4B, S4D-F). Since 6-OHDA is used in mice to generate a PD model [39,41], we sought to identify proteins modified by 6-OHDA-PEG3-yne in mouse cell lysates using affinity purification via biotin-azide and streptavidin-functionalized agarose, followed by mass spectrometry. After correcting for mock pulldown, we identified n = 201 proteins targeted by 6-OHDA-PEG3-yne (Table S1). As expected, we found an overrepresentation of proteins involved in pathways associated with catecholamine-mediated binding and signaling (e.g. adrenergic receptor signaling pathway, 5HT1 type receptor mediated signaling pathway) (Fig. S5). Interestingly, we also found several pathways involved in respiratory electron transport and oxidative phosphorylation indicating that 6-OHDA affects mitochondrial function, which confirms previous reports [19]. It is noteworthy that we found a small number of proteins (n = 12) among the identified targets that do not possess a cysteine. Since we were not able to modify proteins without cysteines (i.e. SNCA) with 6-OHDA as shown before, these identified 12 proteins likely represent indirect binding partners that, given the experimental procedure, probably engage in very strong interactions with other identified proteins. We continued exploring the application of 6-OHDA-PEG3-yne to whole cells but we could not detect any intracellular signal (Fig. S6). This indicates that the alkyne modification of 6-OHDA prevents uptake via the constitutively expressed dopamine transporter (DAT) in SH-SY5Y cells [68]. This supports previous studies interrogating chemical modification of dopamine analogues [69]. Consequently, we permeabilized the cell membrane of fixed SH-SY5Y before applying 6-OHDA-PEG3-yne, and proceeded with an established copper-catalyzed azide-alkyne cycloaddition approach [70]. Our fluorescence microscopy assessment revealed that 6-OHDA-PEG3-yne produced a clearly visible signal in the cytosol indicating substantial intracellular protein modification (Fig. 4C). This demonstrates that 6-OHDA-PEG3-yne permits the visualization of intracellular protein modification by 6-OHDA p-quinones.
Fig. 4.
Visualization of protein modification by 6-OHDA-PEG3-yne in lysates and in cellulo. Quantitation of in-gel fluorescence signal of three independent labelling experiments using SH-SY5Y cells (green), HepG2 cells (light grey) or primary embryonic cortical neurons from mouse (pink) using increasing concentration of 6-OHDA-PEG3-yne (A) or 6-OHDA-PEG3-yne (10 μM) in the presence or absence of 6-OHDA (100 μM) (B). Representative bright-field and fluorescence microscopy image of fixed and permeabilized SH-SY5Y cells incubated for 30 min with or without 6-OHDA-PEG3-yne (10 μM) and stained with DAPI for DNA (blue) and Alexa488-azide for 6-OHDA-PEG3-yne (green). Scale bar denotes 100 μm (C). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Quinone modification of thiols impairs viability of SH-SY5Y cells
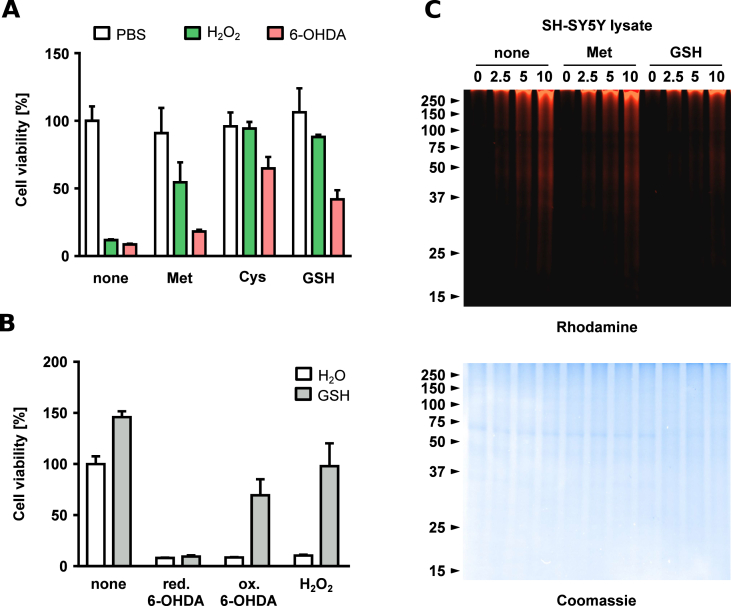
Previous research linked 6-OHDA-induced cell death to the generation of free radicals released during 6-OHDA autoxidation, in particular H2O2 [43,71,72]; however, few studies suggested that the 6-OHDA p-quinone contributes to toxicity as well [73,74]. Intrigued by the large extent of intracellular protein modification, we investigated the impact of protein modification by quinones versus the impact of reactive oxygen species on cell viability. First, we treated SH-SY5Y cells with 6-OHDA or H2O2, and detected a similar impairment in cell viability (Fig. 5A). The presence of methionine significantly reduced the toxicity of H2O2 but not 6-OHDA, whereas the thiol-containing antioxidants cysteine or GSH improved cell viability substantially for both H2O2 and 6-OHDA. This indicates that H2O2 generated by 6-OHDA autoxidation is only one factor in neuronal cell death. After this, we examined whether quinone formation and subsequent protein modification contribute to 6-OHDA induced neuronal cell death. We incubated 6-OHDA in water and immediately added GSH to prevent autoxidation, or we allowed 6-OHDA to undergo autoxidation yielding H2O2 and 6-OHDA p-quinone (indicated by a visible change in the color of the solution). Then, we transferred these mixtures to SH-SY5Y cells rapidly diluting the reaction solutions in the process (Fig. 5B). Cell viability was drastically impaired if reduced 6-OHDA was applied, regardless of the presence of GSH. In contrast, oxidized 6-OHDA impaired cell viability in absence of GSH but showed substantially reduced toxicity in the presence of GSH. This suggests that GSH quenches 6-OHDA p-quinones, and thus, indicates that quinone formation is an important contributor to dopaminergic cell death. To support our conclusion, we assessed whether thiols are able to quench the 6-OHDA p-quinone in SH-SY5Y cell lysates. We employed 6-OHDA-PEG3-yne in the presence of either methionine or GSH and found that the presence of methionine only slightly impaired in-gel fluorescence whereas GSH abrogated fluorescence almost completely (Fig. 5C). Interestingly, application of antioxidants after successful incubation with 6-OHDA-PEG3-yne had no effect implying that protein modification by quinone is irreversible under cellular conditions (Fig. S7).
Fig. 5.
Impact of protein modification by 6-OHDA quinones on cell viability. Cell viability of SH-SY5Y cells challenged with either PBS pH 7.6 (white), H2O2 (60 μM, green) or 6-OHDA (60 μM, red) for 24 h in the absence of antioxidants or presence of either methionine (Met, 1 mM), cysteine (Cys, 1 mM) or glutathione (GSH, 1 mM). Error bars represent standard deviation of three independent experiments. (A). Cell viability of SH-SY5Y cells supplemented with either H2O (white) or GSH (1 mM, grey) were unchallenged or challenged with either reduced 6-OHDA (60 μM), oxidized 6-OHDA (60 μM) or H2O2 (60 μM). Error bars represent standard deviation of three independent experiments. (B). Representative in-gel fluorescence of SH-SY5Y cell lysates incubated for 1 h with different concentrations of 6-OHDA-PEG3-yne (in μM) in the absence of antioxidants or presence of of methionine (Met, 1 mM) or glutathione (GSH, 1 mM). Below is the same gel after staining with Coomassie Brilliant Blue R-250 (C). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. Quinone modification renders protein disulfide isomerase inactive
We sought out to identify functional aspects of protein modification by 6-OHDA p-quinones and impaired cell viability. Our mass spectrometry identification of 6-OHDA-modified proteins revealed a large number of proteins relevant to respiratory electron transport and oxidative phosphorylation, including several peroxiredoxins (Table S1). Several studies demonstrated that peroxiredoxins regulate the cellular redox environment and interact with protein disulfide isomerase (PDI) [[75], [76], [77]]. PDI is also part of the cellular redox network, and catalyzes the formation of disulfide bonds and, hence, protein folding [[78], [79], [80]]. A growing body of research points to a crucial role of PDI in neurodegenerative disorders since PDI is strongly associated with the two fundamental processes underlying neurodegenerative diseases, i.e., protein misfolding and oxidative stress [[81], [82], [83]]. Therefore, we examined the effect of 6-OHDA on the activity of recombinantly expressed human PDI. First, we confirmed modification of PDI's cysteines by 6-OHDA-PEG3-yne (Fig. 6A). We also employed mass spectrometry to validate modification of PDI by native 6-OHDA (Fig. S8). With this in mind, we hypothesized that cysteine modification of the catalytic domain of PDI (PDIa) by 6-OHDA p-quinones inhibits PDIa's oxidoreductase activity similar to the cysteine modification of PDIa by the small molecule phenyl arsenoxide (PAO) [58,59]. We monitored PDIa's oxidoreductase activity and found that H2O2, which is released during autoxidation of 6-OHDA, had no effect, whereas 6-OHDA rendered PDIa inactive (Fig. S9). After this, we continued to study the effect of 6-OHDA on full-length PDI. Similar to the catalytic domain PDIa, 6-OHDA rendered full-length PDI inactive (Fig. 6B). The presence of methionine altered PDI's overall activity to a small degree but did not mitigate 6-OHDA's effect on enzyme activity (Fig. 6C). In contrast, adding cysteine fully restored PDI's oxidoreductase activity (Fig. 6D). This supports our finding that thiols are the predominant targets of 6-OHDA p-quinone. In addition, we demonstrated that quinones impair key players in cellular protein folding and redox homeostasis.
Fig. 6.
Inactivation of protein disulfide isomerase (PDI) by 6-OHDA quinones. Representative in-gel fluorescence and Coomassie stained gel of PDI (2 μM) incubated for 1 h with or without 6-OHDA-PEG3-yne (5 μM or 10 μM) or 6-OHDA-PEG3-yne (10 μM) after pre-incubation for 1 h with maleimide (2 mM, denoted with *) (A). PDI (75 nM) was either mock-treated with H2O (black) or with 6-OHDA (150 μM, red) in the absence of antioxidants (B) or in presence of either methionine (Met, 1 mM) (C) or cysteine (Cys, 1 mM) (D). Oxidoreductase activity was monitored via reduction of diabzGSSG (15 μM) in 5-s intervals. Solid circles represent the average of four measurement. Error bars represent the standard error of the mean. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
A recent study tied dopamine oxidation to mitochondrial and lysosomal dysfunction in PD, indicating a paradigm shift in our perception of the relationship between this neurodegenerative disease and the very complex metabolism of dopamine [19]. Here, we demonstrate a novel tool and associated methods to the research community for studying the vast and complex network of neurochemistry in the brain. Chemical modification of naturally occurring dopamine metabolites permits the acquisition of unparalleled knowledge about the underlying chemistry and its implication for biology. For instance, we were able to prevent intramolecular cyclization and, hence, abrogate neuromelanin formation by the introduction of an alkyne-functionalized PEG at the amino group of 6-OHDA. To our knowledge, this is the first experimental demonstration of cyclization as a prerequisite for neuromelanin formation. At this point, we cannot exclude that other types of polymerization or molecular conversion occur; nevertheless, our approach allows studying the effect of the 6-OHDA p-quinone, which displays an extremely short lifetime otherwise.
Most literature regarding 6-OHDA suggests that the generation of reactive oxygen species (ROS) is the driving force of the molecule's toxicity [34,40,43,71,84,85]. In contrast, only few studies addressed protein modification by dopamine quinones [19,20,[44], [45], [46], [47], [48],86]. We were surprised by the large extent of protein modification, observed for both 6-OHDA and 6-OHDA-PEG3-yne in lysates and in cellulo. Together with the presented viability studies, our observation challenges the view of ROS as major factor in neurotoxicity. Instead, it implies that protein modification by dopamine quinones impairs proteome function and, hence, contributes substantially to dopaminergic cell death. Our work shows that 6-OHDA-PEG3-yne covalently modifies proteins and is specific for cysteine thiols. The ability of 6-OHDA p-quinone to react with thiols in a cellular environment likely depends on additional parameters such as steric accessibility or pKa values. In keeping with this, an earlier study showed that the different pKa values of GSH and cysteine are associated with the protection of neuronal cells towards challenges with dopamine metabolites [87]. Another early study attributed the protective effect of cysteine to very quickly replenishing the GSH/GSSG balance upon challenge with 6-OHDA but did not consider a nucleophilic attack of cysteine's thiol on the 6-OHDA quinone [43]. In addition, a large number of studies investigated the role of different antioxidants and found that compounds containing thiols protect against ROS [43,[88], [89], [90], [91]]. Our finding that thiol modification is a major factor contributing to cell death provides a simple and clear explanation for all these observations. This suggests that cysteine and GSH quench quinones and, thus, prevent damaging protein modification. Consequently, we call for reassessing the role of astrocytes with their very high cytosolic concentrations of GSH and, generally, the role of thiol redox homeostasis in the development of PD [92,93].
We are aware of several limitations of our approach. A synthetic molecule might affect cellular biochemical processes in a different way than the unmodified native molecule. For instance, thiol modification by 6-OHDA-PEG3-yne could prevent protein-protein interaction due to the PEG linker, which would not occur upon application of unmodified 6-OHDA. Furthermore, the administration of synthetic molecules to cell lysates or even cultured cells does not fully mimic physiological conditions. Finally, other dopamine metabolites might display a different physicochemical behavior. Still, we argue that the mechanistic implications and the conclusions of our study do hold true.
Future work could employ strain-promoted alkyne-azide cycloaddition (SPAAC) using cyclooctynes as an alternative to copper-catalyzed azide-alkyne cycloaddition since SPAAC would allow monitoring protein modification in living cells and even animal models [[94], [95], [96]]. First, it is necessary to develop azide-functionalized dopamine metabolites that are able to cross the cell membrane and to reach the intracellular environment. For example, it is worthwhile to explore shorter PEG-linker that might help to overcome this obstacle. Previous research on dopamine modification and dopamine uptake already provides very useful information [69]. SPAAC between an azide-functionalized dopamine metabolite and a cyclooctyne-modified fluorophore provides an alternative method to study the role and function of dopamine uptake without the use of radiolabeled compounds.
In general, our work suggests that 6-OHDA follows a two-step mechanism leading to cell death. The first step is generation of H2O2 by autoxidation causing oxidative stress. The second step is quinone-mediated protein modification interfering with cell physiology and proteostasis. We insinuate that the second step has not been credited accordingly but contributes substantially to the toxicity of dopamine oxidation. Moreover, our study presents a novel strategy to explore role and function of dopamine metabolites and their interaction with biological molecules in physiological relevant conditions.
5. Conclusion
Dopamine oxidation has emerged as an important process in the development of Parkinson's disease (PD), strengthening the link between neurochemistry and the loss of dopaminergic neurons. The molecular mechanism by which dopamine oxidation contributes to dopaminergic cell death is unknown, mainly due to the lack of tools to study (oxidized) dopamine metabolites. This study explores a novel approach to investigate the role of neurochemistry in neurodegenerative diseases by devising a chemically modified dopamine metabolite, which aims to elucidate the chemical reactivity of the products of dopamine oxidation in dopaminergic neurons. This study found that proteins containing thiols form adducts with dopamine metabolites and, by extension, that disrupting cellular protein thiol redox homeostasis contributes to the development of PD. Our findings suggests that there is another poorly explored but important layer of neurochemistry that contributes to dopaminergic cell death. The approach presented in this study is applicable to other neurochemical metabolites and enables the study of a wide range of neurodegenerative and neuropsychiatric disorders.
Author contribution
A.F. performed in vitro experiments and imaging. K.C. performed in vitro experiments. M.S. assisted in cell culture, in vitro experiments and imaging. S.J.H. performed mouse work. D.S.P. and J.P.P. provided resources and general supervision. D.Ö. performed cell culture work, in vitro experiments and imaging. All authors contributed to analysis and interpretation. D.Ö. conceptualized and designed this study, and wrote the manuscript with input from all authors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethics and animal experiments
All animal experiments conformed to the guidelines set forth by the Canadian Council on Animal Care (CCAC) and the Canadian Institutes of Health Research (CIHR), and had approval from the University of Ottawa Animal Care Committee.
Acknowledgments
The authors thank Dr. Mathhew A. Lafreniere for the initial synthesis of 6-OHDA-PEG3-yne. Mass spectrometry services were provided by the University of Ottawa core facility managed by Dr. Maxim Berezovski, Dr. Zoran Minic, and Dr. Gleb Mironov. D.Ö. would like to thank Dr. Eduardo F.A. Fernandes, Dominik J. Essig (University of Copenhagen, Denmark) and Steve Callaghan (University of Ottawa) for technical assistance, and Dr. Kristian Strømgaard for resources and critical reading of the manuscript. Dr. Llyod Ruddock (University of Oulu, Finland) and the Michael J Fox Foundation are acknowledged for the expression vectors for PDI and SNCA, respectively. DÖ was initially supported by a post-doctoral fellowship from CIHR.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101377.
Abbreviations
- 6-OHDA
6-hydroxydopamine
- ABC
ammonium bicarbonate
- ATCC
American Type Culture Collection
- BSA
bovine serum albumin
- CCAC
Canadian Council on Animal Care
- CD
circular dichroism
- CID
collision-induced dissociation
- CIHR
Canadian Institutes of Health Research
- Cys
cysteine
- DAC
dopaminochrome
- DA
dopamine
- DAPI
4′,6-diamidino-2-phenylindole
- DAQ
dopamine o-quinone
- DASQ
dopamine semiquinone
- Diabz
di(o-aminobenzoyl)
- DIV
day in vitro
- DMEM
Dulbecco's modified Eagle medium
- DTNB
5,5′-dithiobis-(2-nitrobenzoic acid)
- DTT
dithiothreitol
- ESI
electron spray ionization
- FBS
fetal bovine serum
- FDR
false discovery rate
- GSH
glutathione
- GSSG
glutathione disulfide
- H2O2
hydrogen peroxide
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- LC/MS
liquid chromatography mass spectrometry
- LDAC
leukodopaminochrome
- Met
methionine
- MTT
3-(4,5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2H-tetrazolium bromide
- PAO
phenyl arsenoxide
- PCA
principal component analysis
- PDI
protein disulfide isomerase
- PDIa
catalytically active domain of protein disulfide isomerase
- PEG
polyethylene glycol
- PBS
phosphate buffered saline
- PD
Parkinson's disease
- ROS
reactive oxygen species
- SDS
sodium dodecyl sulfate
- SNc
substantia nigra pars compacta
- SNCA
alpha-synuclein
- SPAAC
strain-promoted alkyne-azide cycloaddition
- TBTA
tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine
- TCEP
tris(2-carboxyethyl)phosphine hydrochloride
- UV
ultraviolet
- WT
wild type
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Physicochemical characterization of 6-OHDA-PEG3-yne; in-gel fluorescence of BSA modified by 6-OHDA-PEG3-yne under different conditions; reactivity of 6-OHDA quinone towards natural amino acids; in-gel fluorescence of cell lysates modified by 6-OHDA-PEG3-yne; analysis of proteome modified by 6-OHDA; fluorescence microscopy of living cells treated with 6-OHDA-PEG3-yne; reversibility of quinone modification by antioxidants; activity of PDIa upon treatment with 6-OHDA; quinone modification of PDI; list of proteins targeted by 6-OHDA-PEG3-yne.
References
- 1.Habermann B., Davis L.L. Caring for family with Alzheimer's disease and Parkinson's disease: needs, challenges and satisfaction. J. Gerontol. Nurs. 2005;31:49–54. doi: 10.3928/0098-9134-20050601-11. [DOI] [PubMed] [Google Scholar]
- 2.Maresova P., Klimova B., Novotny M., Kuca K. Alzheimer's and Parkinson's diseases: expected economic impact on europe-A call for a uniform European strategy. J. Alzheimer's Dis. 2016;54:1123–1133. doi: 10.3233/JAD-160484. [DOI] [PubMed] [Google Scholar]
- 3.Wu Y.T., Fratiglioni L., Matthews F.E., Lobo A., Breteler M.M., Skoog I., Brayne C. Dementia in western Europe: epidemiological evidence and implications for policy making. Lancet Neurol. 2016;15:116–124. doi: 10.1016/S1474-4422(15)00092-7. [DOI] [PubMed] [Google Scholar]
- 4.Jankovic J. Parkinson's disease: clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry. 2008;79:368–376. doi: 10.1136/jnnp.2007.131045. [DOI] [PubMed] [Google Scholar]
- 5.Kalia L.V., Lang A.E. Parkinson's disease. Lancet. 2015;386:896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- 6.Sveinbjornsdottir S. The clinical symptoms of Parkinson's disease. J. Neurochem. 2016;139(Suppl 1):318–324. doi: 10.1111/jnc.13691. [DOI] [PubMed] [Google Scholar]
- 7.Dhungel N., Eleuteri S., Li L.B., Kramer N.J., Chartron J.W., Spencer B., Kosberg K., Fields J.A., Stafa K., Adame A. Parkinson's disease genes VPS35 and EIF4G1 interact genetically and converge on alpha-synuclein. Neuron. 2015;85:76–87. doi: 10.1016/j.neuron.2014.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesage S., Brice A. Parkinson's disease: from monogenic forms to genetic susceptibility factors. Hum. Mol. Genet. 2009;18:R48–R59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- 9.Henderson M.X., Trojanowski J.Q., Lee V.M. Alpha-synuclein pathology in Parkinson's disease and related alpha-synucleinopathies. Neurosci. Lett. 2019:134316. doi: 10.1016/j.neulet.2019.134316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spillantini M.G., Goedert M. The alpha-synucleinopathies: Parkinson's disease, dementia with Lewy bodies, and multiple system atrophy. Ann. N. Y. Acad. Sci. 2000;920:16–27. doi: 10.1111/j.1749-6632.2000.tb06900.x. [DOI] [PubMed] [Google Scholar]
- 11.Herrera A., Munoz P., Steinbusch H.W.M., Segura-Aguilar J. Are dopamine oxidation metabolites involved in the loss of dopaminergic neurons in the nigrostriatal system in Parkinson's disease? ACS Chem. Neurosci. 2017;8:702–711. doi: 10.1021/acschemneuro.7b00034. [DOI] [PubMed] [Google Scholar]
- 12.Meiser J., Weindl D., Hiller K. Complexity of dopamine metabolism. Cell Commun. Signal. 2013;11:34. doi: 10.1186/1478-811X-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ballatori N., Krance S.M., Notenboom S., Shi S., Tieu K., Hammond C.L. Glutathione dysregulation and the etiology and progression of human diseases. Biol. Chem. 2009;390:191–214. doi: 10.1515/BC.2009.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berndt C., Lillig C.H. Glutathione, glutaredoxins, and iron. Antioxidants Redox Signal. 2017;27:1235–1251. doi: 10.1089/ars.2017.7132. [DOI] [PubMed] [Google Scholar]
- 15.Dichtl S., Haschka D., Nairz M., Seifert M., Volani C., Lutz O., Weiss G. Dopamine promotes cellular iron accumulation and oxidative stress responses in macrophages. Biochem. Pharmacol. 2018;148:193–201. doi: 10.1016/j.bcp.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Hirrlinger J., Schulz J.B., Dringen R. Effects of dopamine on the glutathione metabolism of cultured astroglial cells: implications for Parkinson's disease. J. Neurochem. 2002;82:458–467. doi: 10.1046/j.1471-4159.2002.01013.x. [DOI] [PubMed] [Google Scholar]
- 17.Costa-Mallen P., Gatenby C., Friend S., Maravilla K.R., Hu S.C., Cain K.C., Agarwal P., Anzai Y. Brain iron concentrations in regions of interest and relation with serum iron levels in Parkinson disease. J. Neurol. Sci. 2017;378:38–44. doi: 10.1016/j.jns.2017.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smeyne M., Smeyne R.J. Glutathione metabolism and Parkinson's disease. Free Radic. Biol. Med. 2013;62:13–25. doi: 10.1016/j.freeradbiomed.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burbulla L.F., Song P., Mazzulli J.R., Zampese E., Wong Y.C., Jeon S., Santos D.P., Blanz J., Obermaier C.D., Strojny C. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson's disease. Science. 2017;357:1255–1261. doi: 10.1126/science.aam9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monzani E., Nicolis S., Dell'Acqua S., Capucciati A., Bacchella C., Zucca F.A., Mosharov E.V., Sulzer D., Zecca L., Casella L. Dopamine, oxidative stress and protein-quinone modifications in Parkinson's and other neurodegenerative diseases. Angew Chem. Int. Ed. Engl. 2019;58:6512–6527. doi: 10.1002/anie.201811122. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S., Wang R., Wang G. Impact of dopamine oxidation on dopaminergic neurodegeneration. ACS Chem. Neurosci. 2019;10:945–953. doi: 10.1021/acschemneuro.8b00454. [DOI] [PubMed] [Google Scholar]
- 22.Zucca F.A., Segura-Aguilar J., Ferrari E., Munoz P., Paris I., Sulzer D., Sarna T., Casella L., Zecca L. Interactions of iron, dopamine and neuromelanin pathways in brain aging and Parkinson's disease. Prog. Neurobiol. 2017;155:96–119. doi: 10.1016/j.pneurobio.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cappai R., Leck S.L., Tew D.J., Williamson N.A., Smith D.P., Galatis D., Sharples R.A., Curtain C.C., Ali F.E., Cherny R.A. Dopamine promotes alpha-synuclein aggregation into SDS-resistant soluble oligomers via a distinct folding pathway. FASEB J. 2005;19:1377–1379. doi: 10.1096/fj.04-3437fje. [DOI] [PubMed] [Google Scholar]
- 24.Lee H.J., Baek S.M., Ho D.H., Suk J.E., Cho E.D., Lee S.J. Dopamine promotes formation and secretion of non-fibrillar alpha-synuclein oligomers. Exp. Mol. Med. 2011;43:216–222. doi: 10.3858/emm.2011.43.4.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cobley J.N., Fiorello M.L., Bailey D.M. 13 reasons why the brain is susceptible to oxidative stress. Redox Biol. 2018;15:490–503. doi: 10.1016/j.redox.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puspita L., Chung S.Y., Shim J.W. Oxidative stress and cellular pathologies in Parkinson's disease. Mol. Brain. 2017;10:53. doi: 10.1186/s13041-017-0340-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bisaglia M., Soriano M.E., Arduini I., Mammi S., Bubacco L. Molecular characterization of dopamine-derived quinones reactivity toward NADH and glutathione: implications for mitochondrial dysfunction in Parkinson disease. Biochim. Biophys. Acta. 2010;1802:699–706. doi: 10.1016/j.bbadis.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Segura-Aguilar J., Paris I., Munoz P., Ferrari E., Zecca L., Zucca F.A. Protective and toxic roles of dopamine in Parkinson's disease. J. Neurochem. 2014;129:898–915. doi: 10.1111/jnc.12686. [DOI] [PubMed] [Google Scholar]
- 29.Tse D.C., McCreery R.L., Adams R.N. Potential oxidative pathways of brain catecholamines. J. Med. Chem. 1976;19:37–40. doi: 10.1021/jm00223a008. [DOI] [PubMed] [Google Scholar]
- 30.Sulzer D., Bogulavsky J., Larsen K.E., Behr G., Karatekin E., Kleinman M.H., Turro N., Krantz D., Edwards R.H., Greene L.A. Neuromelanin biosynthesis is driven by excess cytosolic catecholamines not accumulated by synaptic vesicles. Proc. Natl. Acad. Sci. U. S. A. 2000;97:11869–11874. doi: 10.1073/pnas.97.22.11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bustamante J., Bredeston L., Malanga G., Mordoh J. Role of melanin as a scavenger of active oxygen species. Pigment Cell Res. 1993;6:348–353. doi: 10.1111/j.1600-0749.1993.tb00612.x. [DOI] [PubMed] [Google Scholar]
- 32.Hirsch E., Graybiel A.M., Agid Y.A. Melanized dopaminergic neurons are differentially susceptible to degeneration in Parkinson's disease. Nature. 1988;334:345–348. doi: 10.1038/334345a0. [DOI] [PubMed] [Google Scholar]
- 33.Zecca L., Bellei C., Costi P., Albertini A., Monzani E., Casella L., Gallorini M., Bergamaschi L., Moscatelli A., Turro N.J. New melanic pigments in the human brain that accumulate in aging and block environmental toxic metals. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17567–17572. doi: 10.1073/pnas.0808768105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blum D., Torch S., Lambeng N., Nissou M., Benabid A.L., Sadoul R., Verna J.M. Molecular pathways involved in the neurotoxicity of 6-OHDA, dopamine and MPTP: contribution to the apoptotic theory in Parkinson's disease. Prog. Neurobiol. 2001;65:135–172. doi: 10.1016/s0301-0082(01)00003-x. [DOI] [PubMed] [Google Scholar]
- 35.Cohen G., Heikkila R.E. The generation of hydrogen peroxide, superoxide radical, and hydroxyl radical by 6-hydroxydopamine, dialuric acid, and related cytotoxic agents. J. Biol. Chem. 1974;249:2447–2452. [PubMed] [Google Scholar]
- 36.Heikkila R.E., Cohen G. In vivo generation of hydrogen peroxide from 6-hydroxydopamine. Experientia. 1972;28:1197–1198. doi: 10.1007/BF01946168. [DOI] [PubMed] [Google Scholar]
- 37.Napolitano A., Crescenzi O., Pezzella A., Prota G. Generation of the neurotoxin 6-hydroxydopamine by peroxidase/H2O2 oxidation of dopamine. J. Med. Chem. 1995;38:917–922. doi: 10.1021/jm00006a010. [DOI] [PubMed] [Google Scholar]
- 38.Pezzella A., d'Ischia M., Napolitano A., Misuraca G., Prota G. Iron-mediated generation of the neurotoxin 6-hydroxydopamine quinone by reaction of fatty acid hydroperoxides with dopamine: a possible contributory mechanism for neuronal degeneration in Parkinson's disease. J. Med. Chem. 1997;40:2211–2216. doi: 10.1021/jm970099t. [DOI] [PubMed] [Google Scholar]
- 39.Mercanti G., Bazzu G., Giusti P. A 6-hydroxydopamine in vivo model of Parkinson's disease. Methods Mol. Biol. 2012;846:355–364. doi: 10.1007/978-1-61779-536-7_30. [DOI] [PubMed] [Google Scholar]
- 40.Offenburger S.L., Gartner A. 6-hydroxydopamine (6-OHDA) oxidative stress assay for observing dopaminergic neuron loss in Caenorhabditis elegans. Bio Protoc. 2018;8 doi: 10.21769/BioProtoc.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simola N., Morelli M., Carta A.R. The 6-hydroxydopamine model of Parkinson's disease. Neurotox. Res. 2007;11:151–167. doi: 10.1007/BF03033565. [DOI] [PubMed] [Google Scholar]
- 42.Fink J., Pathak H., Smith J., Achat-Mendes C., Haining R.L. Development of a competition-binding assay to determine binding affinity of molecules to neuromelanin via fluorescence spectroscopy. Biomolecules. 2019;9 doi: 10.3390/biom9050175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soto-Otero R., Mendez-Alvarez E., Hermida-Ameijeiras A., Munoz-Patino A.M., Labandeira-Garcia J.L. Autoxidation and neurotoxicity of 6-hydroxydopamine in the presence of some antioxidants: potential implication in relation to the pathogenesis of Parkinson's disease. J. Neurochem. 2000;74:1605–1612. doi: 10.1046/j.1471-4159.2000.0741605.x. [DOI] [PubMed] [Google Scholar]
- 44.Hauser D.N., Dukes A.A., Mortimer A.D., Hastings T.G. Dopamine quinone modifies and decreases the abundance of the mitochondrial selenoprotein glutathione peroxidase 4. Free Radic. Biol. Med. 2013;65:419–427. doi: 10.1016/j.freeradbiomed.2013.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spencer J.P., Jenner P., Daniel S.E., Lees A.J., Marsden D.C., Halliwell B. Conjugates of catecholamines with cysteine and GSH in Parkinson's disease: possible mechanisms of formation involving reactive oxygen species. J. Neurochem. 1998;71:2112–2122. doi: 10.1046/j.1471-4159.1998.71052112.x. [DOI] [PubMed] [Google Scholar]
- 46.Van Laar V.S., Dukes A.A., Cascio M., Hastings T.G. Proteomic analysis of rat brain mitochondria following exposure to dopamine quinone: implications for Parkinson disease. Neurobiol. Dis. 2008;29:477–489. doi: 10.1016/j.nbd.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitehead R.E., Ferrer J.V., Javitch J.A., Justice J.B. Reaction of oxidized dopamine with endogenous cysteine residues in the human dopamine transporter. J. Neurochem. 2001;76:1242–1251. doi: 10.1046/j.1471-4159.2001.00125.x. [DOI] [PubMed] [Google Scholar]
- 48.Xu Y., Stokes A.H., Roskoski R., Jr., Vrana K.E. Dopamine, in the presence of tyrosinase, covalently modifies and inactivates tyrosine hydroxylase. J. Neurosci. Res. 1998;54:691–697. doi: 10.1002/(SICI)1097-4547(19981201)54:5<691::AID-JNR14>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 49.Li K., Chen Y., Li S., Nguyen H.G., Niu Z., You S., Mello C.M., Lu X., Wang Q. Chemical modification of M13 bacteriophage and its application in cancer cell imaging. Bioconjug. Chem. 2010;21:1369–1377. doi: 10.1021/bc900405q. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen T., Francis M.B. Practical synthetic route to functionalized rhodamine dyes. Org. Lett. 2003;5:3245–3248. doi: 10.1021/ol035135z. [DOI] [PubMed] [Google Scholar]
- 51.Yang P.Y., Liu K., Ngai M.H., Lear M.J., Wenk M.R., Yao S.Q. Activity-based proteome profiling of potential cellular targets of Orlistat--an FDA-approved drug with anti-tumor activities. J. Am. Chem. Soc. 2010;132:656–666. doi: 10.1021/ja907716f. [DOI] [PubMed] [Google Scholar]
- 52.Lafreniere M.A., Powdrill M.H., Singaravelu R., Pezacki J.P. 6-Hydroxydopamine inhibits the hepatitis C virus through alkylation of host and viral proteins and the induction of oxidative stress. ACS Infect. Dis. 2016;2:863–871. doi: 10.1021/acsinfecdis.6b00098. [DOI] [PubMed] [Google Scholar]
- 53.Kim R.H., Smith P.D., Aleyasin H., Hayley S., Mount M.P., Pownall S., Wakeham A., You-Ten A.J., Kalia S.K., Horne P. Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5215–5220. doi: 10.1073/pnas.0501282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen V.D., Saaranen M.J., Karala A.R., Lappi A.K., Wang L., Raykhel I.B., Alanen H.I., Salo K.E., Wang C.C., Ruddock L.W. Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. J. Mol. Biol. 2011;406:503–515. doi: 10.1016/j.jmb.2010.12.039. [DOI] [PubMed] [Google Scholar]
- 55.Alanen H.I., Salo K.E., Pekkala M., Siekkinen H.M., Pirneskoski A., Ruddock L.W. Defining the domain boundaries of the human protein disulfide isomerases. Antioxidants Redox Signal. 2003;5:367–374. doi: 10.1089/152308603768295096. [DOI] [PubMed] [Google Scholar]
- 56.Ozcelik D., Pezacki J.P. Small molecule inhibition of protein disulfide isomerase in neuroblastoma cells induces an oxidative stress response and apoptosis pathways. ACS Chem. Neurosci. 2019;10:4068–4075. doi: 10.1021/acschemneuro.9b00301. [DOI] [PubMed] [Google Scholar]
- 57.Huang C., Ren G., Zhou H., Wang C.C. A new method for purification of recombinant human alpha-synuclein in Escherichia coli. Protein Expr. Purif. 2005;42:173–177. doi: 10.1016/j.pep.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 58.Foster C.K., Thorpe C. Challenges in the evaluation of thiol-reactive inhibitors of human protein disulfide Isomerase. Free Radic. Biol. Med. 2017;108:741–749. doi: 10.1016/j.freeradbiomed.2017.04.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raturi A., Vacratsis P.O., Seslija D., Lee L., Mutus B. A direct, continuous, sensitive assay for protein disulphide-isomerase based on fluorescence self-quenching. Biochem. J. 2005;391:351–357. doi: 10.1042/BJ20050770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Speers A.E., Cravatt B.F. Activity-Based protein profiling (ABPP) and click chemistry (CC)-ABPP by MudPIT mass spectrometry. Curr. Protoc. Chem. Biol. 2009;1:29–41. doi: 10.1002/9780470559277.ch090138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Metsalu T., Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xicoy H., Wieringa B., Martens G.J.M. The SH-SY5Y cell line in Parkinson's disease research: a systematic review. Mol. Neurodegener. 2017;12:10. doi: 10.1186/s13024-017-0149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koniev O., Wagner A. Correction: developments and recent advancements in the field of endogenous amino acid selective bond forming reactions for bioconjugation. Chem. Soc. Rev. 2015;44:5743. doi: 10.1039/c5cs90060c. [DOI] [PubMed] [Google Scholar]
- 65.Koniev O., Wagner A. Developments and recent advancements in the field of endogenous amino acid selective bond forming reactions for bioconjugation. Chem. Soc. Rev. 2015;44:5495–5551. doi: 10.1039/c5cs00048c. [DOI] [PubMed] [Google Scholar]
- 66.Bocedi A., Fabrini R., Pedersen J.Z., Federici G., Iavarone F., Martelli C., Castagnola M., Ricci G. The extreme hyper-reactivity of selected cysteines drives hierarchical disulfide bond formation in serum albumin. FEBS J. 2016;283:4113–4127. doi: 10.1111/febs.13909. [DOI] [PubMed] [Google Scholar]
- 67.Kim H.S., Hong S.J., LeDoux M.S., Kim K.S. Regulation of the tyrosine hydroxylase and dopamine beta-hydroxylase genes by the transcription factor AP-2. J. Neurochem. 2001;76:280–294. doi: 10.1046/j.1471-4159.2001.00044.x. [DOI] [PubMed] [Google Scholar]
- 68.Kovalevich J., Langford D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol. Biol. 2013;1078:9–21. doi: 10.1007/978-1-62703-640-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andersen J., Ladefoged L.K., Kristensen T.N., Munro L., Grouleff J., Stuhr-Hansen N., Kristensen A.S., Schiott B., Stromgaard K. Interrogating the molecular basis for substrate recognition in serotonin and dopamine transporters with high-affinity substrate-based bivalent ligands. ACS Chem. Neurosci. 2016;7:1406–1417. doi: 10.1021/acschemneuro.6b00164. [DOI] [PubMed] [Google Scholar]
- 70.Nasheri N., Joyce M., Rouleau Y., Yang P., Yao S., Tyrrell D.L., Pezacki J.P. Modulation of fatty acid synthase enzyme activity and expression during hepatitis C virus replication. Chem. Biol. 2013;20:570–582. doi: 10.1016/j.chembiol.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 71.Bernstein A.I., Garrison S.P., Zambetti G.P., O'Malley K.L. 6-OHDA generated ROS induces DNA damage and p53- and PUMA-dependent cell death. Mol. Neurodegener. 2011;6:2. doi: 10.1186/1750-1326-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumar R., Agarwal A.K., Seth P.K. Free radical-generated neurotoxicity of 6-hydroxydopamine. J. Neurochem. 1995;64:1703–1707. doi: 10.1046/j.1471-4159.1995.64041703.x. [DOI] [PubMed] [Google Scholar]
- 73.Izumi Y., Sawada H., Sakka N., Yamamoto N., Kume T., Katsuki H., Shimohama S., Akaike A. p-Quinone mediates 6-hydroxydopamine-induced dopaminergic neuronal death and ferrous iron accelerates the conversion of p-quinone into melanin extracellularly. J. Neurosci. Res. 2005;79:849–860. doi: 10.1002/jnr.20382. [DOI] [PubMed] [Google Scholar]
- 74.Villa M., Munoz P., Ahumada-Castro U., Paris I., Jimenez A., Martinez I., Sevilla F., Segura-Aguilar J. One-electron reduction of 6-hydroxydopamine quinone is essential in 6-hydroxydopamine neurotoxicity. Neurotox. Res. 2013;24:94–101. doi: 10.1007/s12640-013-9382-7. [DOI] [PubMed] [Google Scholar]
- 75.Kakihana T., Araki K., Vavassori S., Iemura S., Cortini M., Fagioli C., Natsume T., Sitia R., Nagata K. Dynamic regulation of Ero1alpha and peroxiredoxin 4 localization in the secretory pathway. J. Biol. Chem. 2013;288:29586–29594. doi: 10.1074/jbc.M113.467845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pace P.E., Peskin A.V., Han M.H., Hampton M.B., Winterbourn C.C. Hyperoxidized peroxiredoxin 2 interacts with the protein disulfide- isomerase ERp46. Biochem. J. 2013;453:475–485. doi: 10.1042/BJ20130030. [DOI] [PubMed] [Google Scholar]
- 77.Sato Y., Kojima R., Okumura M., Hagiwara M., Masui S., Maegawa K., Saiki M., Horibe T., Suzuki M., Inaba K. Synergistic cooperation of PDI family members in peroxiredoxin 4-driven oxidative protein folding. Sci. Rep. 2013;3:2456. doi: 10.1038/srep02456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ali Khan H., Mutus B. Protein disulfide isomerase a multifunctional protein with multiple physiological roles. Front. Chem. 2014;2:70. doi: 10.3389/fchem.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hatahet F., Ruddock L.W. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxidants Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 80.Soares Moretti A.I., Martins Laurindo F.R. Protein disulfide isomerases: redox connections in and out of the endoplasmic reticulum. Arch. Biochem. Biophys. 2017;617:106–119. doi: 10.1016/j.abb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 81.Barnham K.J., Masters C.L., Bush A.I. Neurodegenerative diseases and oxidative stress. Nat. Rev. Drug Discov. 2004;3:205–214. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- 82.Perri E.R., Thomas C.J., Parakh S., Spencer D.M., Atkin J.D. The unfolded protein response and the role of protein disulfide isomerase in neurodegeneration. Front. Cell Dev. Biol. 2015;3:80. doi: 10.3389/fcell.2015.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Soto C., Estrada L.D. Protein misfolding and neurodegeneration. Arch. Neurol. 2008;65:184–189. doi: 10.1001/archneurol.2007.56. [DOI] [PubMed] [Google Scholar]
- 84.Rodriguez-Pallares J., Parga J.A., Munoz A., Rey P., Guerra M.J., Labandeira-Garcia J.L. Mechanism of 6-hydroxydopamine neurotoxicity: the role of NADPH oxidase and microglial activation in 6-hydroxydopamine-induced degeneration of dopaminergic neurons. J. Neurochem. 2007;103:145–156. doi: 10.1111/j.1471-4159.2007.04699.x. [DOI] [PubMed] [Google Scholar]
- 85.Saito Y., Nishio K., Ogawa Y., Kinumi T., Yoshida Y., Masuo Y., Niki E. Molecular mechanisms of 6-hydroxydopamine-induced cytotoxicity in PC12 cells: involvement of hydrogen peroxide-dependent and -independent action. Free Radic. Biol. Med. 2007;42:675–685. doi: 10.1016/j.freeradbiomed.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 86.LaVoie M.J., Ostaszewski B.L., Weihofen A., Schlossmacher M.G., Selkoe D.J. Dopamine covalently modifies and functionally inactivates parkin. Nat. Med. 2005;11:1214–1221. doi: 10.1038/nm1314. [DOI] [PubMed] [Google Scholar]
- 87.Sabens E.A., Distler A.M., Mieyal J.J. Levodopa deactivates enzymes that regulate thiol-disulfide homeostasis and promotes neuronal cell death: implications for therapy of Parkinson's disease. Biochemistry. 2010;49:2715–2724. doi: 10.1021/bi9018658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Benvenutti R., Marcon M., Reis C.G., Nery L.R., Miguel C., Herrmann A.P., Vianna M.R.M., Piato A. N-acetylcysteine protects against motor, optomotor and morphological deficits induced by 6-OHDA in zebrafish larvae. PeerJ. 2018;6 doi: 10.7717/peerj.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ebadi M., Hiramatsu M., Burke W.J., Folks D.G., el-Sayed M.A. Metallothionein isoforms provide neuroprotection against 6-hydroxydopamine-generated hydroxyl radicals and superoxide anions. Proc. West. Pharmacol. Soc. 1998;41:155–158. [PubMed] [Google Scholar]
- 90.Nouraei N., Zarger L., Weilnau J.N., Han J., Mason D.M., Leak R.K. Investigation of the therapeutic potential of N-acetyl cysteine and the tools used to define nigrostriatal degeneration in vivo. Toxicol. Appl. Pharmacol. 2016;296:19–30. doi: 10.1016/j.taap.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xie L., Tiong C.X., Bian J.S. Hydrogen sulfide protects SH-SY5Y cells against 6-hydroxydopamine-induced endoplasmic reticulum stress. Am. J. Physiol. Cell Physiol. 2012;303:C81–C91. doi: 10.1152/ajpcell.00281.2011. [DOI] [PubMed] [Google Scholar]
- 92.McBean G.J. Cysteine, glutathione, and thiol redox balance in astrocytes. Antioxidants (Basel) 2017;6 doi: 10.3390/antiox6030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McBean G.J., Aslan M., Griffiths H.R., Torrao R.C. Thiol redox homeostasis in neurodegenerative disease. Redox Biol. 2015;5:186–194. doi: 10.1016/j.redox.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baskin J.M., Prescher J.A., Laughlin S.T., Agard N.J., Chang P.V., Miller I.A., Lo A., Codelli J.A., Bertozzi C.R. Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chang P.V., Prescher J.A., Sletten E.M., Baskin J.M., Miller I.A., Agard N.J., Lo A., Bertozzi C.R. Copper-free click chemistry in living animals. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1821–1826. doi: 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Codelli J.A., Baskin J.M., Agard N.J., Bertozzi C.R. Second-generation difluorinated cyclooctynes for copper-free click chemistry. J. Am. Chem. Soc. 2008;130:11486–11493. doi: 10.1021/ja803086r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.