Abstract
Disruption of estrogen receptor beta (ESR2) dysregulates oocyte maturation, which leads to failure of ovulation. We investigated ESR2-regulated genes during gonadotropin-induced oocyte maturation using RNA-sequencing. Through the administration of pregnant mare's serum gonadotropin (PMSG), synchronized follicle development was initiated in four-week-old wildtype and Esr2-null female rats. Forty-eight hours after the PMSG injection, human chorionic gonadotropin (hCG) was used for further maturation. Oocytes were collected from the ovaries 4 h after hCG injection. The total RNA was isolated from the oocytes and the whole oocyte transcriptome was determined by RNA-sequencing on the Illumina HiSeq4000 sequencer. RNA-sequencing data of wildtype and Esr2-null oocytes were analyzed, and differentially expressed genes were identified using the CLC Genomics Workbench. Whole oocyte transcriptome data of wildtype and Esr2-null oocytes were compared to identify the differentially expressed genes. Raw data are deposited to the NCBI Sequence Read Archive (SRA) and analyzed data are presented in this data article. These datasets can be utilized to identify the gonadotropin-induced genes in oocytes that are ESR2-regulated and important to oocyte maturation.
Keywords: Rat models, ESR2 knockout, Gonadotropins, Oocyte transcriptome, RNA-sequencing
Specifications Table
| Subject | Biology |
| Specific subject area | Reproductive biology |
| Type of data | Transcriptomic data, table |
| How data were acquired | High-throughput RNA sequencing using Illumina HiSeq4000 sequencer |
| Data format | Raw (FASTQ) and analyzed (Excel table) |
| Parameters for data collection | RNA sequencing was performed on oocytes collected from gonadotropin-treated wildtype and Esr2-null rats. |
| Description of data collection | Four-week-old wildtype and Esr2-null Holtzman rats were treated with 30IU of pregnant mare's serum gonadotropin (PMSG). Forty-eight hours after the PMSG administration, 30IU of human chorionic gonadotropin (hCG) was injected into the rats. Four hours after the hCG injection, oocytes were collected from the ovaries. RNAs were purified and analyzed by mRNA-sequencing. Differentially expressed gonadotropin-induced genes in Esr2-null oocytes were identified through analyses of the transcriptome data using CLC Genomics Workbench. |
| Data source location | University of Kansas Medical Center, Kansas City, KS 66160, USA |
| Data accessibility | Repository name: Raw data are available in SRA Data identification number: PRJNA562521 Direct URL to data: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA562521 |
Value of the Data
|
1. Data
Transcriptome data generated by RNA-sequencing of wildtype and Esr2-null oocytes were compared to identify the differentially expressed genes. Analyzed data are presented in Excel format and the raw data are deposited to the NCBI SRA (SRR10024657-10024662, included under PRJNA562521). SRR10024660, SRR10024661 and SRR10024662 are the raw data obtained from wildtype oocytes, while SRR10024657, SRR10024658 and SRR10024659 are the raw data obtained from Esr2-null oocytes. The corresponding experiment numbers are SRX6761695, SRX6761696, SRX6761697, SRX6761698, SRX6761699 and SRX6761700. Analyzed data are presented in the following tables: (see Table 1, Table 2)
Table 1.
Top 50 upregulated genes in gonadotropin treated Esr2 null rat oocytes. This data-table shows the top 50 upregulated genes selected from the differentially expressed genes.
| Name | Chr. | Region | Max gr. mean | Log₂ fold change | Fold change | P-value | FDR P-value | Bon-ferroni | ENSEMBL |
|---|---|---|---|---|---|---|---|---|---|
| AABR07001512.1 | 1 | complement(48565160..48565711) | 4.190 | 8.670 | 408.400 | 0.000 | 0.003 | 1.000 | ENSRNOG00000017412 |
| Ms4a6bl | 1 | 227240383..227252250 | 1.200 | 8.250 | 304.330 | 0.001 | 0.007 | 1.000 | ENSRNOG00000050395 |
| Tsen34l1 | 1 | complement(64024240..64030175) | 1.640 | 8.160 | 285.430 | 0.001 | 0.010 | 1.000 | ENSRNOG00000055179 |
| LOC108348062_2 | 6 | 108745895..108756130 | 1.710 | 7.940 | 245.090 | 0.002 | 0.010 | 1.000 | ENSRNOG00000040153 |
| Myh9 | 7 | complement(118741110..118792625) | 3.020 | 6.460 | 88.180 | 0.000 | 0.000 | 0.000 | ENSRNOG00000004860 |
| RGD1561661 | X | 13441558..13443470 | 3.840 | 6.220 | 74.450 | 0.000 | 0.000 | 0.000 | ENSRNOG00000023094 |
| Serpinc1 | 13 | 78805347..78833192 | 3.610 | 5.150 | 35.420 | 0.000 | 0.000 | 0.000 | ENSRNOG00000002783 |
| Cpa2 | 4 | 57855416..57879239 | 53.310 | 4.870 | 29.300 | 0.000 | 0.000 | 0.000 | ENSRNOG00000028092 |
| Cma1 | 15 | complement(34601037..34603819) | 1.070 | 4.830 | 28.480 | 0.000 | 0.000 | 0.040 | ENSRNOG00000020563 |
| Ces1d | 19 | 15033108..15239821 | 8.570 | 4.570 | 23.830 | 0.000 | 0.000 | 0.000 | ENSRNOG00000015519 |
| Ifit1 | 1 | 252944103..252946170 | 1.920 | 4.530 | 23.080 | 0.000 | 0.000 | 0.000 | ENSRNOG00000019050 |
| RT1-Bb | 20 | 4039413..4049711 | 55.030 | 4.170 | 17.980 | 0.000 | 0.000 | 0.000 | ENSRNOG00000032708 |
| AABR07043407.1 | 19 | complement(28142561..28148135) | 2.920 | 4.140 | 17.670 | 0.000 | 0.000 | 0.020 | ENSRNOG00000060719 |
| Cga | 5 | 50381244..50393367 | 201.520 | 4.120 | 17.390 | 0.000 | 0.000 | 0.000 | ENSRNOG00000009269 |
| Mcpt1l1 | 15 | complement(34609776..34612432) | 1.600 | 4.070 | 16.740 | 0.000 | 0.000 | 0.000 | ENSRNOG00000053494 |
| Rgs13 | 13 | complement(60970247..61003744) | 1.120 | 3.940 | 15.400 | 0.000 | 0.000 | 0.000 | ENSRNOG00000003888 |
| Qrfpr | 2 | complement(122891321..122949241) | 1.410 | 3.860 | 14.500 | 0.000 | 0.000 | 0.000 | ENSRNOG00000014414 |
| Aqp11 | 1 | complement(162703442..162713610) | 1.030 | 3.760 | 13.520 | 0.000 | 0.000 | 0.000 | ENSRNOG00000013358 |
| Amtn | 14 | complement(21286510..21299068) | 1.090 | 3.710 | 13.130 | 0.000 | 0.000 | 0.003 | ENSRNOG00000003776 |
| AABR07027872.1 | 17 | 47870611..47878555 | 1.970 | 3.650 | 12.560 | 0.000 | 0.000 | 0.000 | ENSRNOG00000055197 |
| Lrrc17 | 4 | complement(10108192..10138652) | 9.790 | 3.640 | 12.440 | 0.000 | 0.000 | 0.000 | ENSRNOG00000012817 |
| Spag4 | 3 | 151609602..151613942 | 1.200 | 3.640 | 12.430 | 0.000 | 0.000 | 0.000 | ENSRNOG00000048056 |
| Shisal2b | 2 | complement(34946797..34963207) | 9.710 | 3.520 | 11.430 | 0.000 | 0.000 | 0.000 | ENSRNOG00000013372 |
| Cst8 | 3 | 143129248..143156177 | 1.380 | 3.450 | 10.930 | 0.000 | 0.000 | 0.004 | ENSRNOG00000004989 |
| Myorg | 5 | complement(57876498..57881944) | 1.550 | 3.370 | 10.350 | 0.000 | 0.000 | 0.000 | ENSRNOG00000023208 |
| Oit3 | 20 | complement(29009330..29029905) | 13.220 | 3.240 | 9.480 | 0.000 | 0.000 | 0.000 | ENSRNOG00000046365 |
| Lrp2 | 3 | complement(55665145..55822551) | 2.020 | 3.230 | 9.370 | 0.000 | 0.000 | 0.000 | ENSRNOG00000056184 |
| Cd70 | 9 | 9842585..9845728 | 2.030 | 3.120 | 8.700 | 0.000 | 0.000 | 0.000 | ENSRNOG00000051015 |
| LOC100364500 | 20 | complement(2704148..2707120) | 12.940 | 3.110 | 8.640 | 0.000 | 0.000 | 0.000 | ENSRNOG00000048951 |
| Cx3cl1 | 19 | complement(10644244..10653800) | 22.930 | 3.110 | 8.610 | 0.000 | 0.000 | 0.000 | ENSRNOG00000016326 |
| RT1-A1 | 20 | 5351605..5421098 | 169.660 | 3.030 | 8.140 | 0.000 | 0.000 | 0.000 | ENSRNOG00000038999 |
| Higd1b | 10 | 90929423..90931639 | 1.130 | 2.980 | 7.870 | 0.000 | 0.000 | 0.200 | ENSRNOG00000002814 |
| AABR07042609.1 | 19 | 275531..277005 | 1.230 | 2.920 | 7.590 | 0.002 | 0.010 | 1.000 | ENSRNOG00000051666 |
| Ptgds | 3 | complement(2686123..2689084) | 19.660 | 2.920 | 7.570 | 0.000 | 0.000 | 0.000 | ENSRNOG00000015550 |
| Tnfsf13b | 16 | complement(85275678..85306366) | 3.470 | 2.890 | 7.420 | 0.000 | 0.000 | 0.000 | ENSRNOG00000014464 |
| Aard | 7 | 91588458..91593297 | 7.240 | 2.860 | 7.250 | 0.000 | 0.000 | 0.000 | ENSRNOG00000004708 |
| Atp6ap1l | 2 | complement(19781408..19808937) | 12.450 | 2.840 | 7.160 | 0.000 | 0.000 | 0.000 | ENSRNOG00000040201 |
| AABR07046778.1 | 5 | 6373583..6373849 | 1.390 | 2.830 | 7.130 | 0.005 | 0.030 | 1.000 | ENSRNOG00000058589 |
| AABR07058124.4 | 7 | complement(101138549..101138860) | 21.080 | 2.830 | 7.130 | 0.000 | 0.000 | 0.000 | ENSRNOG00000055178 |
| Actc1 | 3 | complement(105507403..105512939) | 1.290 | 2.820 | 7.080 | 0.000 | 0.000 | 0.000 | ENSRNOG00000008536 |
| AABR07043200.1 | 19 | 26416818..26417597 | 1.140 | 2.800 | 6.970 | 0.002 | 0.010 | 1.000 | ENSRNOG00000058276 |
| Fcgr3a | 13 | 89385859..89396051 | 3.080 | 2.800 | 6.940 | 0.000 | 0.000 | 0.000 | ENSRNOG00000024382 |
| Synpo2 | 2 | complement(227255902..227411964) | 2.160 | 2.740 | 6.670 | 0.000 | 0.000 | 0.000 | ENSRNOG00000014867 |
| Calca | 1 | complement(184184020..184188911) | 2.750 | 2.720 | 6.610 | 0.000 | 0.000 | 0.000 | ENSRNOG00000011130 |
| AABR07049499.1 | 5 | 124442293..124542156 | 2.560 | 2.710 | 6.550 | 0.000 | 0.000 | 0.000 | ENSRNOG00000030938 |
| Cntn3 | 4 | complement(134784668..135069970) | 3.130 | 2.690 | 6.460 | 0.000 | 0.000 | 0.000 | ENSRNOG00000006144 |
| Hp | 19 | 42097995..42100804 | 22.850 | 2.690 | 6.440 | 0.000 | 0.000 | 0.000 | ENSRNOG00000014964 |
| Aldob | 5 | complement(64805773..64818824) | 3.070 | 2.680 | 6.400 | 0.000 | 0.000 | 0.000 | ENSRNOG00000006807 |
| Smpx | X | complement(40030248..40086870) | 8.320 | 2.640 | 6.240 | 0.000 | 0.000 | 0.000 | ENSRNOG00000007495 |
| Uchl3_1 | 3 | complement(171092946..171134655) | 3.530 | 2.620 | 6.130 | 0.009 | 0.050 | 1.000 | ENSRNOG00000046120 |
Chr. Chromosome; Max gr. mean, Maximum group mean.
Table 2.
Top 50 downregulated genes in gonadotropin treated Esr2-null rat oocytes. This data-table shows the top 50 downregulated genes selected from the differentially expressed genes.
| Name | Chr. | Region | Max gr. mean | Log₂ fold change | Fold change | P-value | FDR P-value | Bonfe-rroni | ENSEMBL |
|---|---|---|---|---|---|---|---|---|---|
| Impad1_1 | 5 | complement(17633766..17663589) | 1.210 | −8.690 | −413.81 | 0.000 | 0.001 | 1.000 | ENSRNOG00000027079 |
| Ncbp2_2 | 11 | complement(72921282..72929003) | 4.010 | −8.370 | −329.92 | 0.000 | 0.003 | 1.000 | ENSRNOG00000048589 |
| LOC103690018 | 3 | 57286892..57300840 | 1.300 | −8.260 | −307.35 | 0.000 | 0.003 | 1.000 | ENSRNOG00000023386 |
| LOC100361898 | 1 | 248402980..248403399 | 2.890 | −7.740 | −214.50 | 0.001 | 0.006 | 1.000 | ENSRNOG00000033038 |
| Kiss1 | 13 | complement(50529510..50535389) | 27.18 | −7.180 | −144.59 | 0.000 | 0.000 | 0.000 | ENSRNOG00000047481 |
| Spp1 | 14 | complement(6673686..6679901) | 231.1 | −6.450 | −87.560 | 0.000 | 0.000 | 0.000 | ENSRNOG00000043451 |
| Car14 | 2 | complement(198010349..198016898) | 9.110 | −6.380 | −83.380 | 0.000 | 0.000 | 0.000 | ENSRNOG00000023162 |
| Lce1m | 2 | complement(193333800..193335002) | 1.220 | −5.640 | −49.730 | 0.000 | 0.000 | 0.250 | ENSRNOG00000009581 |
| Vstm2l | 3 | 154395187..154424625 | 1.590 | −4.920 | −30.190 | 0.000 | 0.000 | 0.040 | ENSRNOG00000034031 |
| Myh15 | 11 | complement(54204775..54344615) | 28.32 | −4.870 | −29.300 | 0.000 | 0.000 | 0.000 | ENSRNOG00000061038 |
| Npr3 | 2 | complement(61888950..61949926) | 3.600 | −4.550 | −23.370 | 0.000 | 0.000 | 0.000 | ENSRNOG00000019184 |
| Ptgs2 | 13 | 67351087..67359335 | 294.8 | −4.210 | −18.560 | 0.000 | 0.000 | 0.000 | ENSRNOG00000002525 |
| Kcnk12 | 6 | complement(11373917..11494459) | 3.450 | −4.180 | −18.150 | 0.000 | 0.000 | 0.000 | ENSRNOG00000016110 |
| AABR07052431.1 | 3 | 53316026..53316481 | 2.030 | −4.170 | −18.050 | 0.000 | 0.000 | 0.030 | ENSRNOG00000038559 |
| Adcyap1 | 9 | complement(121706979..121725716) | 504.6 | −4.080 | −16.910 | 0.000 | 0.000 | 0.000 | ENSRNOG00000049882 |
| Heatr9 | 10 | complement(70726071..70735742) | 2.570 | −3.980 | −15.810 | 0.000 | 0.000 | 0.000 | ENSRNOG00000037100 |
| Olr154 | 1 | 169575656..169576609 | 2.310 | −3.960 | −15.620 | 0.000 | 0.000 | 0.000 | ENSRNOG00000059092 |
| Wnt16 | 4 | 49369296..49379703 | 2.520 | −3.950 | −15.490 | 0.000 | 0.000 | 0.000 | ENSRNOG00000005781 |
| Lif | 14 | 84482674..84500642 | 10.81 | −3.920 | −15.180 | 0.000 | 0.000 | 0.000 | ENSRNOG00000007002 |
| LOC108348130 | 11 | 33845463..33847793 | 83.93 | −3.910 | −15.010 | 0.000 | 0.003 | 1.000 | ENSRNOG00000049693 |
| Olfr656 | 1 | 169616178..169617571 | 1.570 | −3.890 | −14.780 | 0.000 | 0.000 | 0.000 | ENSRNOG00000017252 |
| Fam25a | 16 | complement(10702263..10706073) | 3.800 | −3.850 | −14.440 | 0.000 | 0.000 | 0.000 | ENSRNOG00000055025 |
| Gal | 1 | complement(218652917..218657925) | 169.4 | −3.810 | −13.990 | 0.000 | 0.000 | 0.000 | ENSRNOG00000015156 |
| Kcnk2 | 13 | complement(107690087..107886476) | 5.400 | −3.680 | −12.800 | 0.000 | 0.000 | 0.000 | ENSRNOG00000002653 |
| Rpl10l | 6 | complement(88231611..88232252) | 9.090 | −3.630 | −12.350 | 0.000 | 0.000 | 0.000 | ENSRNOG00000032720 |
| Olr155 | 1 | 169590308..169591279 | 7.040 | −3.570 | −11.840 | 0.000 | 0.000 | 0.000 | ENSRNOG00000017234 |
| Fndc9 | 10 | 31324512..31325192 | 4.410 | −3.560 | −11.810 | 0.000 | 0.000 | 0.000 | ENSRNOG00000006549 |
| Igsf9 | 13 | complement(90815562..90832469) | 30.57 | −3.540 | −11.660 | 0.000 | 0.000 | 0.000 | ENSRNOG00000008054 |
| Hamp | 1 | complement(89368021..89369960) | 38.27 | −3.510 | −11.380 | 0.000 | 0.000 | 0.000 | ENSRNOG00000021029 |
| Dok6 | 18 | complement(86420361..86878142) | 7.510 | −3.480 | −11.140 | 0.000 | 0.000 | 0.000 | ENSRNOG00000038190 |
| . | 1 | complement(22649081..22661377) | 10.74 | −3.460 | −11.000 | 0.000 | 0.000 | 0.000 | ENSRNOG00000039865 |
| Mt1m | 20 | 3677474..3677847 | 1.010 | −3.410 | −10.640 | 0.001 | 0.009 | 1.000 | ENSRNOG00000028841 |
| LOC102555453 | X | complement(1345684..1346181) | 54.65 | −3.370 | −10.340 | 0.000 | 0.000 | 0.000 | ENSRNOG00000028993 |
| Ms4a4c | 1 | 227640680..227661311 | 3.460 | −3.320 | −9.990 | 0.000 | 0.000 | 0.000 | ENSRNOG00000020997 |
| Snap25 | 3 | 129599353..129788400 | 23.03 | −3.320 | −9.980 | 0.000 | 0.000 | 0.000 | ENSRNOG00000006037 |
| Sult1e1 | 14 | 22072024..22089248 | 71.00 | −3.290 | −9.770 | 0.000 | 0.000 | 0.000 | ENSRNOG00000001957 |
| AABR07044900.1 | 20 | complement(25064702..25826658) | 14.22 | −3.250 | −9.490 | 0.000 | 0.000 | 0.000 | ENSRNOG00000000373 |
| Fam124a | 15 | 45712821..45780405 | 2.12 | −3.210 | −9.240 | 0.000 | 0.000 | 0.000 | ENSRNOG00000009802 |
| Gdf1 | 16 | complement(20845576..20860767) | 2.95 | −3.170 | −9.020 | 0.000 | 0.000 | 0.050 | ENSRNOG00000020142 |
| Adtrp | 17 | 22619891..22688307 | 4.640 | −3.140 | −8.810 | 0.000 | 0.000 | 0.000 | ENSRNOG00000014481 |
| Nup62cl | X | complement(111334252..111365849) | 1.740 | −3.040 | −8.250 | 0.000 | 0.000 | 0.000 | ENSRNOG00000057753 |
| Tdh | 15 | complement(46667926..46681467) | 6.230 | −3.010 | −8.030 | 0.000 | 0.000 | 0.000 | ENSRNOG00000011342 |
| Il13ra2 | X | complement(118443823..118513061) | 11.52 | −2.990 | −7.960 | 0.000 | 0.000 | 0.000 | ENSRNOG00000032973 |
| Adgrf5 | 9 | complement(20091099..20195566) | 7.240 | −2.820 | −7.070 | 0.000 | 0.000 | 0.000 | ENSRNOG00000011154 |
| Sfmbt2 | 17 | complement(71723620..71897972) | 3.820 | −2.790 | −6.900 | 0.000 | 0.000 | 0.000 | ENSRNOG00000029235 |
| Xpnpep2 | X | 134940615..134969996 | 10.73 | −2.760 | −6.790 | 0.000 | 0.000 | 0.000 | ENSRNOG00000004009 |
Chr., Chromosome; Max gr. mean, Maximum group mean.
Transcriptome analyses of gonadotropin-induced genes in Esr2-null rat oocytes were compared to that of wildtype. Of the 933 differentially expressed genes, 535 were ≥2 fold upregulated, whereas the 398 were ≤2 fold downregulated.
2. Experimental design, materials and methods
2.1. Experimental animals
Four-week-old wildtype and Esr2-null female Holtzman Sprague-Dawley (HSD) rats were included in these RNA-sequencing analyses. Holtzman Sprague-Dawley (HSD) Esr2-mutant rat models were generated by the targeted deletion of exon 3 in the Esr2 gene as described previously [3]. Deletion of exon 3 caused a frameshift and null mutation in the ESR2 coding sequence [3].
All animals were screened for mutation by PCR-based genotyping that uses tail-tip DNA samples (RED extract-N-Amp Tissue PCR Kit, Sigma-Aldrich) and primers targeting the flanking intron sequences [3]. All procedures performed and precautions taken were in accordance with the protocols approved by the University of Kansas Medical Center Animal Care and Use Committee. Each sample in the data represents the RNA obtained by pooling oocytes from three different animals of the same genotype. So, a total of 9 wildtype and 9 Esr2-null rats were used in the present study.
2.2. Gonadotropin treatment
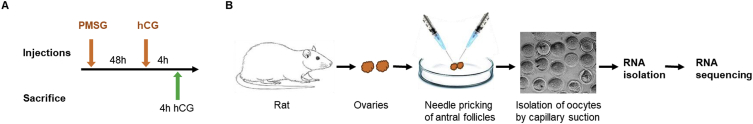
Synchronized follicular growth was initiated through the administration of gonadotropins to four-week-old wildtype and Esr-2 null female rats [[1], [2], [3]]. First, 30 IU of PMSG (Lee Bioscience, MO) was intraperitoneally injected into the rats. Forty-eight hours after this PMSG treatment, 30 IU of hCG (Lee Bioscience, MO) was injected (Fig. 1A).
Fig. 1.
Schematic presentation of the experimental design. A) Four-week-old wildtype or Esr2-null female rats were injected intraperitoneally with 30IU of PMSG, and 48 h after the PMSG injection, with 30IU of hCG. B) Rats were sacrificed 4 h after hCG injection, and the ovaries were collected for oocyte isolation. COCs were isolated from the ovaries by needle puncturing under stereoscope. Cumulus cells were removed by pipetting followed by repeated washings using capillary suction under microscope. The total RNA was extracted, quality assessed, and used for mRNA sequencing.
2.3. Sample collection and processing
Four hours after the hCG injection to PMSG-treated rats, Esr2-null and wildtype rats were sacrificed, and their ovaries were collected (Fig. 1B). Cumulus oocyte complexes (COCs) were isolated from the large antral ovaries by needle puncture under microscopic examination (Fig. 1B) [1,2]. All cumulus cells were removed from the oocytes by pipetting followed by repeated washings into fresh media using capillary suction. The total RNA was extracted from the cumulus-free oocytes using TRI Reagent (Sigma-Aldrich, St. Louis, MO) following the manufacturer's instruction. RNA quality was assessed by a Bioanalyzer and samples with a RIN value over 9 were selected for mRNA-sequencing library preparation. Approximately 500 ng of the total RNA was used for the RNA-sequencing library preparation using a TruSeq Standard mRNA kit (Illumina, San Diego, CA) following the manufacturer's instruction [4]. The cDNA libraries were evaluated for quality and then sequenced on an Illumina HiSeq 4000 sequencer (Novogene Corporation, Sacramento, CA).
2.4. RNA-seq data analyses
RNA-sequencing data were demultiplexed, trimmed, aligned and analyzed using CLC Genomics Workbench 12.2 (Qiagen Bioinformatics, Germantown, MD). Through trimming, low-quality reads were removed, and good-quality reads were aligned with Rattus norvegicus genome (Rnor_6.0) using default guidelines: (a) maximum number of allowable mismatches = 2, (b) minimum length and similarity fraction = 0.8, and (c) minimum number of hits per read = 10. Gene expression values were measured in transcripts per million (TPM). Differentially expressed genes were identified with an absolute fold change of TPM values ≥ 2 showing a false discovery rate (FDR) p-value of ≤0.05.
2.5. Statistical analysis
Each RNA-sequencing library was prepared using pooled RNA samples from three or more individual wildtype or Esr2-null rats. Each group for RNA-sequencing consisted of three libraries. Differentially expressed genes were identified by CLC Genomics Workbench as described previously [4].
Acknowledgements
Generation of these datasets was partially supported by funding from the NIH Clinical and Translational Science Award (Grant UL1TR002366) awarded to University of Kansas Medical Center (KUMC), and the Lied Basic Science Grant Program of the KUMC Research Institute.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2019.104786.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Chakravarthi V.P., Khristi V., Ghosh S., Yerrathota S., Dai E., Roby K.F., Wolfe M.W., Rumi M.A.K. ESR2 is Essential for Gonadotropin-induced Kiss1 expression in granulosa cells. Endocrinology. 2018;159:3860–3873. doi: 10.1210/en.2018-00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khristi V., Chakravarthi V.P., Singh P., Ghosh S., Pramanik A., Ratri A., Borosha S., Roby K.F., Wolfe M.W., Rumi M.A.K. ESR2 regulates granulosa cell genes essential for follicle maturation and ovulation. Mol. Cell. Endocrinol. 2018;474:214–226. doi: 10.1016/j.mce.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Rumi M.A.K., Singh P., Roby K.F., Zhao X., Iqbal K., Ratri A., Lei T., Cui W., Borosha S., Dhakal P., Kubota K., Chakraborty D., Vivian J.L., Wolfe M.W., Soares M.J. Defining the role of estrogen receptor beta in the regulation of female fertility. Endocrinology. 2017;158:2330–2343. doi: 10.1210/en.2016-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khristi V., Chakravarthi V.P., Singh P., Ghosh S., Pramanik A., Ratri A., Borosha S., Roby K.F., Wolfe M.W., Rumi M.A.K. Differentially regulated genes in Esr2-mutant rat granulosa cells. Data Brief. 2018;19:1008–1011. doi: 10.1016/j.dib.2018.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.