Short abstract
Nuclear migration and anchorage, together referred to as nuclear positioning, are central to many cellular and developmental events. Nuclear positioning is mediated by a conserved network of nuclear envelope proteins that interacts with force generators in the cytoskeleton. At the heart of this network are linker of nucleoskeleton and cytoskeleton (LINC) complexes made of Sad1 and UNC-84 (SUN) proteins at the inner nuclear membrane and Klarsicht, ANC-1, and Syne homology (KASH) proteins in the outer nuclear membrane. LINC complexes span the nuclear envelope, maintain nuclear envelope architecture, designate the surface of nuclei distinctly from the contiguous endoplasmic reticulum, and were instrumental in the early evolution of eukaryotes. LINC complexes interact with lamins in the nucleus and with various cytoplasmic KASH effectors from the surface of nuclei. These effectors regulate the cytoskeleton, leading to a variety of cellular outputs including pronuclear migration, nuclear migration through constricted spaces, nuclear anchorage, centrosome attachment to nuclei, meiotic chromosome movements, and DNA damage repair. How LINC complexes are regulated and how they function are reviewed here. The focus is on recent studies elucidating the best-understood network of LINC complexes, those used throughout Caenorhabditis elegans development.
Impact statement
Defects in nuclear positioning disrupt development in many mammalian tissues. In human development, LINC complexes play important cellular functions including nuclear positioning, homolog pairing in meiosis, DNA damage repair, wound healing, and gonadogenesis. The topics reviewed here are relevant to public health because defects in nuclear positioning and mutations in LINC components are associated with a wide variety of human diseases including muscular dystrophies, neurological disorders, progeria, aneurysms, hearing loss, blindness, sterility, and multiple cancers. Although this review focuses on findings in the model nematode Caenorhabditis elegans, the studies are relevant because almost all the findings originally made in C. elegans are conserved to humans. Furthermore, C. elegans remains the best described network for how LINC complexes are regulated and function.
Keywords: Nuclear envelope, linker of nucleoskeleton and cytoskeleton (LINC), nuclear positioning, cellular, nuclear, development
Introduction
Nuclear positioning, which involves moving the nucleus to a specific intracellular location and anchoring it in place, is central to many cellular and developmental processes including pronuclear migration, cellular migration, muscle development, and neuronal differentiation.1
Here I discuss the contributions to our current understanding of nuclear positioning and movements of and within nuclei made using Caenorhabditis elegans as a model. None of the findings presented here work would have been possible without the foundational contributions made by John Sulston and his colleagues. Sulston’s findings include describing the developmental cell lineage and nuclear migration events in the embryo and larva,5,6 isolating the first mutants with nuclear positioning defects,12,13 and spearheading the effort to sequence the C. elegans genome11 (Box 1).
Box 1.
Sir John Sulston (1942–2018), a scientific giant of developmental biology.
| New findings are made by “standing on the shoulders of giants” (Isaac Newton 1610). Our giant is John Sulston (1942–2018).7,8 Sulston, an early member of Sydney Brenner’s group that established C. elegans as a model organism in the 1970s, found that 15 nuclei in the ventral cord of a newly hattched larvae turned into 57 in the young adult. Sulston set out to determine where the cells came from during development. Surprisingly, he found that the original 12 cells did not divide; instead new cells (now known as P cells) “spontaneously appeared within the ventral cord”.9 These P-cell nuclei “were squeezing into the ventral cord from nearby positions out of the plane of focus”.6,9,10 The findings described in this review hailed directly from attempts to determine how P-cell nuclei squeeze into the ventral cord during development. |
| Sulston made at least three other major contributions that accelerated the field. In the seminal paper that led to his Nobel Prize,5,10 Sulston determined the entire cell lineage of the C. elegans embryo. While doing so, he described nuclear migrations in hyp7 precursors on the dorsal surface of the embryo5 that has proven important for understanding nuclear migration in a live animal. Second, Sulston and Horvitz isolated the first alleles of unc-83 and unc-84 in a classic screen for lineage defects.12,13 Cloning unc-83 and unc-84 eventually led to the discovery of LINC complexes as described in this review. Finally, Sulston led teams sequencing the C. elegans genome.14 Part of that effort was to create a physical map of cosmids covering the C. elegans genome,11 which allowed the cloning of genes, including unc-83 and unc-84. This review is dedicated to the memory and honor of John Sulston, a true scientific giant. |
Nuclear positioning requires closely regulated interactions between nuclei and the cytoskeleton. Thus, it is important to understand the nuclear envelope, which lies at the interface between the nucleus and the cytoskeleton.2–4 Connecting nuclei to the cytoskeleton is complicated by two characteristics of the nuclear envelope. First, the cytoplasmic face of the outer nuclear membrane needs to be specifically marked as distinct from the contiguous, and much larger, endoplasmic reticulum (ER) membrane. A failure to designate the nuclear surface could lead to improper connections between the cytoskeleton and the ER. Second, once the outer nuclear membrane is distinguished from the ER, forces generated in the cytoplasm need to be transferred across both membranes and the perinuclear space of the nuclear envelope to structural elements in the nucleoskeleton. Finally, components specific to the inner nuclear membrane must connect the nuclear envelope to structural components in the nucleoplasm, including lamins and chromatin.15,16
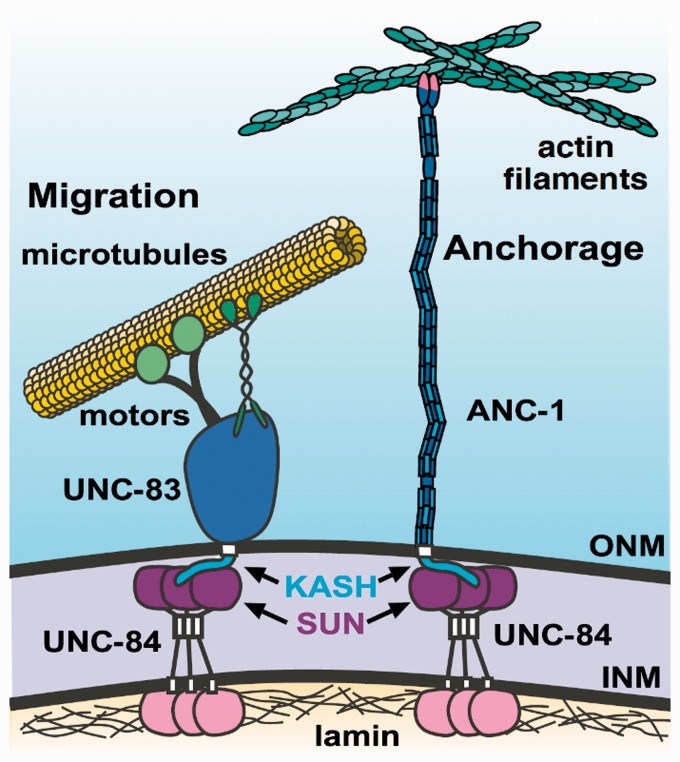
The discovery of Sad1/UNC-84 (SUN) and Klarsicht/ANC-1/Syne homology (KASH) proteins,17–21 led to a model where SUN and KASH proteins form a bridge across the nuclear envelope to transfer mechanical forces generated by the cytoskeleton in the cytoplasm to structural elements inside nuclei.17 This model overcomes the two obstacles presented by the nuclear envelope.22,23 First, KASH proteins localize specifically to the outer nuclear membrane, but not the ER, specifically marking the surface of the nucleus. Second, SUN proteins in the inner nuclear membrane interact with KASH proteins in the outer nuclear membrane to form a physical bridge that transfers mechanical forces across both membranes of the nuclear envelope (Figure 1).
Figure 1.
SUN trimers (UNC-84 in C. elegans) localize to the inner nuclear membrane. SUN domains (purple) recruit KASH proteins (blue) to the outer nuclear membrane. UNC-83 recruits microtubule motors to move nuclei along microtubules. ANC-1 potentially anchors nuclei to actin. See text for more details. (A color version of this figure is available in the online journal.)
SUN-KASH bridges are broadly conserved and are now known as linker of nucleoskeleton and cytoskeleton (LINC) complexes,23–30 LINC complexes play many roles in addition to nuclear positioning. Foremost, they are thought to have been instrumental in the early evolution of eukaryotes.31 Additional examples of some of their many roles include meiotic homolog pairing,32,33 cilliogenesis,34,35 Golgi maintenance,36 DNA damage repair,37,38 wound healing,39 spermatogenesis,40,41 and nuclear pore complex formation.42 Not surprisingly, LINC complex defects are associated with a wide variety of human diseases including cardiomyopathies,43–46 hearing loss,47 blindness,48 neurological disorders,49–52 progeria,53,54 sterility,55 and various cancers.56–60
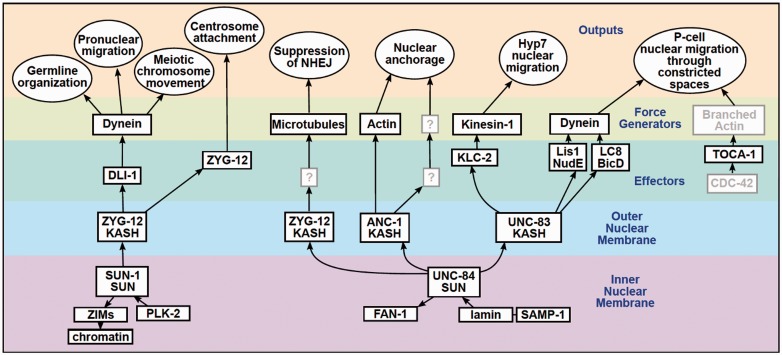
Here, I focus on how LINC complexes are central to a conserved network of nuclear envelope proteins, effectors, and force generators that mediate nuclear positioning in C. elegans (Figure 2). C. elegans has two SUN proteins: the canonical UNC-84 (orthologous to mammalian Sun1 and Sun2) and the more divergent SUN-1. There are also four KASH proteins in C. elegans, ANC-1 (orthologous to Nesprin-1 and -2), UNC-83 (functionally related to mammalian Nesprin-4 and KASH5), ZYG-12, and KDP-1.24 Herein I summarize the current literature on C. elegans LINC complexes starting with what is known about the regulation of different LINC complexes and how LINC complexes maintain the architecture of the nuclear envelope. Next, I describe how SUN proteins interact with lamins and function in DNA damage repair. Turning to the cytoplasmic face of nuclei, I explain how LINC complexes mediate nuclear migration in developing hypodermal tissues, nuclear anchorage in adult hypodermal syncytia, and pronuclear migration in newly fertilized zygotes. I also write about the role of LINC complexes in rapid chromosome movements during meiotic prophase. Finally, I discuss how LINC complexes work with a branched actin network to squeeze nuclei squeeze through constricted spaces.
Figure 2.
A network of LINC proteins and their functions at the nuclear envelope. A large, mostly conserved, network of C. elegans proteins regulates nuclear positioning and other functions throughout development. The purple layer contains SUN proteins and the proteins they interact with in the nuclear lamina. KASH proteins in the outer nuclear membrane are in the blue layer. The green layer contains the effectors that KASH proteins directly interact with. The yellow-green layer lists the cytoskeletal elements regulated by the KASH effectors. The cytoskeletal players then provide the mechanical forces to produce the cellular outputs listed in the top, orange layer. Unknown or hypothesized players are in gray. All black arrows are supported by genetic and/or biochemical evidence as described and referenced throughout the text. (A color version of this figure is available in the online journal.)
Interactions between SUN and KASH proteins to form LINC complexes
The central core of LINC complexes is the physical interaction between conserved domains of SUN and KASH proteins in the perinuclear space between inner and outer nuclear membranes.23 LINC complexes have been proposed to maintain nuclear architecture; disruption of LINC in adherent HeLa cells, which are under considerable mechanical strain to spread across the tissue culture plate,61 results in large blebs between the inner and outer nuclear membranes.62 The role of LINC complexes in maintaining nuclear envelope architecture was tested in C. elegans tissues using electron microscopy in unc-84 null (SUN) mutants. Surprisingly, nuclear envelope architecture was normal in most embryonic and larval tissues, suggesting that LINC is not required to maintain the even spacing of nuclear membranes.63 The exception was in larval body-wall muscles, where the outer nuclear membrane was pulled from about 40 to 500 nm from the inner nuclear membrane.63 Nuclear envelope defects correlate with movement defects in C. elegans L1 larvae,63 suggesting that nuclear envelope morphology might play a role in muscular pathologies. These findings suggest LINC complexes are required to maintain nuclear envelope architecture, but only in tissues subjected to mechanical strain.64
A major breakthrough in our mechanistic understanding of how LINC complexes function occurred when the structure of mammalian SUN–KASH interfaces was determined.65,66 Crystal structures revealed that three KASH peptides interact with a trimer of SUN proteins and that each KASH peptide forms three interfaces with SUN protomers. The four C-terminal residues of KASH interact with a pocket of the first SUN protomer. The next 11 residues fit into a cleft between two SUN protomers. Finally, the next 10 residues interact with the surface of the second protomer. At residue -23 from the C terminus of KASH, a conserved cysteine forms a disulfide bond with a conserved cysteine in SUN. Thus, the interactions between SUN and KASH are predicted to be stable and able to transmit mechanical forces across the nuclear envelope.25,65,67
Genetic approaches in C. elegans are ideal to test functional consequences of predictions raised by the SUN/KASH structure. C. elegans somatic cells have two major LINC complexes—UNC-83 and UNC-84 move nuclei, while ANC-1 and UNC-84 anchor nuclei in place (Figure 1). Yet, little was known about how the UNC-84 SUN protein is regulated to interact with different KASH proteins. One major difference between UNC-83 and ANC-1 KASH domains is that ANC-1 has a full-length KASH domain with all three interaction interfaces and the conserved cysteine at the -23 position,21,68 but UNC-83 has a truncated KASH (only 18 residues long) that lacks the third interaction interface and the conserved cysteine.68,69 In vivo functional analyses of point mutations predicted to disrupt SUN–KASH interactions showed that extending the C terminus of the UNC-83 KASH domain by a single alanine blocks nuclear migration.68 Additionally, mutations of a conserved aromatic amino acid seven residues from the C terminus of KASH cause intermediate nuclear positioning phenotypes. The UNC-83(Y-7A) mutant interacts with SUN well enough to localize to the nuclear envelope but not strongly enough to mediate nuclear migration. In contrast, the ANC-1(F-7A) mutant had no effect on nuclear anchorage.68 Thus, the structure of SUN–KASH interactions led to predictions for how LINC complexes behave that were tested in vivo.
The intermolecular disulfide bond between SUN and KASH identified in the crystal structure hints at a difference in the regulation of how SUN might choose between KASH partners. The conserved cysteine at residue -23 is present in the nuclear anchorage KASH protein, ANC-1, but not in the nuclear migration KASH protein, UNC-83 (Figure 1). Thus, intermolecular cysteine bonds were hypothesized to be important for nuclear anchorage, but dispensable for nuclear migration.68,70 Mutating conserved cysteines in SUN or KASH disrupted ANC-1-dependent nuclear anchorage, but not UNC-83-mediated nuclear migration.68 Click or tap here to enter text.Furthermore, molecular dynamic simulations showed that SUN–KASH pairs lacking intermolecular disulfide bonds could not transmit as much force as wild-type LINC complexes.68 To further test this model, the KASH domains of ANC-1 and UNC-83 were swapped.71 ANC-1 with the short KASH from UNC-83 partially blocked nuclear anchorage, while UNC-83 with the long ANC-1 KASH had a partial nuclear migration defect. Mutation of the cysteine in the UNC-83 with the long ANC-1 KASH construct completely blocked nuclear migration, suggesting that short KASH domains are able to transfer more forces across LINC complexes than long KASH domains missing the intermolecular disulfide bonds.71 Thus, the length of KASH domains and their ability to form intermolecular disulfide bonds with SUN partners contributes to the ability of LINC complexes to transfer forces across the nuclear envelope.
LINC complexes interact with lamins and contribute to DNA damage repair
LINC complexes function like a bolt through the nuclear envelope.72,73 A bolt through a planar surface without a nut or washer on the other side will likely fail when forces are applied to the bolt. The washer and nut serve to dissipate the forces transferred by the bolt, stabilizing the system. In this analogy, the nuclear scaffold, made of lamins, inner nuclear membrane proteins, and associated complexes, serves as the washer to dissipate mechanical forces generated in the cytoplasm. UNC-84 is recruited to the inner nuclear membrane where it is retained by the sole C. elegans lamin, LMN-1.72,74–76 Specifically, the nucleoplasmic domain of UNC-84 interacts with LMN-1 in a two-hybrid assay and this interaction is significantly weaker in the presence of an unc-84(P91S) mutation.72 Interestingly, the unc-84(P91S) allele was originally found as an intermediate disrupter of nuclear migration.20 Live imaging showed that 62% of nuclei in the unc-84(P91S) background migrated normally, 22% started migration normally, but stopped before completion, and 16% failed to initiate migration.72 Consistent with this bolt and nut model, lmn-1(RNAi) animals also have a nuclear migration defect72 and human SUN proteins interact with Lamin A.77 Based on the literature in mammalian cells and fission yeast, the inner nuclear membrane protein NET5/SAMP/Ima1 interacts with LINC complexes and is thought to participate with LINC to help dissipate forces on the nucleoskeleton.73,78 The C. elegans homolog SAMP-1 also localizes to the nuclear membrane and knockdown of samp-1 leads to weak nuclear migration defects.72 Thus, LINC complexes interact with the nucleoskeleton to mediate nuclear migration.
LINC complexes are used to move chromosomes within nuclei and have been recently implicated in the DNA damage response38,79 SUN proteins promote homologous recombination by inhibiting non-homologous end joining (NHEJ).37 Specifically, unc-84 mutants and human cells depleted of Sun1 were both hypersensitive to DNA crosslinking, which was rescued by inactivating NHEJ. UNC-84 also recruited Fanconi anemia nuclease, FAN-1, to the nucleoplasm of germ line nuclei,37 suggesting that UNC-84 alters both the extent of repair by NHEJ and promotes processing of DNA crosslinks. The KASH protein ZYG-12 and microtubules are also required.37 This suggests that LINC complexes play a conserved role in DNA repair through both the inhibition of NHEJ and the promotion of homologous recombination.
UNC-84/UNC-83 LINC complexes recruit microtubule motors to move nuclei in somatic tissues
KASH proteins are positioned on the cytoplasmic surface of the nuclear envelope where they can interact with a variety of different cytoskeletal modules.24,80 The KASH protein UNC-83 interacts with microtubule motors to move nuclei. A yeast two-hybrid screen with the cytoplasmic domain of UNC-83 identified interactions between UNC-83 and four regulators of microtubule motors: the kinesin light chain KLC-2, and the dynein regulators NUD-2 (NudE homolog), BICD-1 (bicaudalD homolog), and DLC-1 (an LC8 dynein light chain).81,82 Knockdown of the kinesin-1 motor caused a severe nuclear migration defect in embryonic hyp7 precursors, but knockdown of dynein components only caused weak nuclear migration defects.81,82 This suggested that kinesin-1 is the major force producer to move nuclei, while dynein plays a regulatory role. Live imaging of the growing plus ends of microtubules showed that embryonic hyp7 nuclei migrate toward plus ends of microtubules, consistent with the model that kinesin-1 is the major force producer.83 Further live imaging showed that dynein was required for backward nuclear movements to move past cellular roadblocks.83,84 Thus, the role of UNC-83 during embryonic hyp7 nuclear migration is to recruit kinesin-1 and dynein to the nuclear envelope, which then move nuclei towards the plus ends of microtubules. Mammalian KASH proteins Nesprin-1, Nesprin-2, Nesprin-4, and KASH5 also interact with microtubule motors.85–88 Although it is not possible to say which one of these KASH proteins is the UNC-83 ortholog, the role of KASH proteins recruiting motors to the surface of nuclei appears conserved.
The UNC-84/UNC-83 LINC complex is also responsible for nuclear migration in larval hypodermal P cells.12,13,17,20,89 P-cell nuclei migrate through a constricted space (discussed below). Surprisingly, P-cell nuclei migrate toward the minus ends of microtubules.90 Furthermore, genetic analyses showed that dynein is the major motor required to move P-cell nuclei and that kinesin-1 plays a regulatory role.90,91 Thus, UNC-83 favors kinesin-1 in embryonic hyp7 precursors but prefers dynein in larval P cells. How UNC-83 dictates the choice between plus and minus-end directed microtubule motors remains an open question.
UNC-84/ANC-1 LINC complexes anchor nuclei in syncytial cells
After nuclei migrate to specific intracellular locations, they must be anchored in place. Mutations in the KASH protein ANC-1 or the SUN protein UNC-84 disrupt nuclear anchorage.20,21,92 ANC-1 is a giant protein of over 8500 amino acids. The C terminus of ANC-1 has a KASH domain, while the N terminus of ANC-1 contains two calponin homology domains that bind actin filaments in vitro.21 In between are extensive repeats predicted to be helical. ANC-1 is orthologous to mammalian Nesprin-1 and -2 and Drosophila MSP-300, all of which contain calponin homology domains and KASH domains at the ends, separated by long repetitive regions.21,23
Nuclear anchorage defects are most easily observed in C. elegans syncytial cells, where normally evenly spaced nuclei are instead clustered together.92 The C. elegans adult hyp7 syncytium contains 139 nuclei that are normally uniformly spread throughout the length of the animal. In anc-1 or unc-84 mutants, syncytial hypodermal nuclei are frequently seen in abnormal clusters.20,21,92 The phenotype and molecular identity of ANC-1 led to the current model, where ANC-1 directly tethers nuclei to the actin cytoskeleton to anchor nuclei in place.21
More recent data suggest that the tethering model for ANC-1 is overly simplistic. This working model predicts that null mutations in anc-1 or unc-84 should have similar phenotypes, as both would equally disrupt the LINC complex. However, a careful quantification of the nuclear anchorage phenotype93 shows that null mutations in anc-1 have much more severe nuclear anchorage defects than null mutations in unc-84.68 This suggests that ANC-1 anchors nuclei through multiple mechanisms. First, the KASH domain of ANC-1 forms a LINC complex with UNC-84 to mediate interactions between nuclei and unknown cytoskeletal components. Second, the large cytoplasmic domain of ANC-1 functions in LINC-independent mechanisms for nuclear anchorage. One possibility is that ANC-1 mediates the formation of a microtubule cage, similar to what has been seen for the mechanism of MSP-300 in Drosophila muscles.94 Further investigations are required to fully understand how ANC-1 functions to anchor nuclei.
SUN-1/ZYG-12 LINC complexes mediate pronuclear migration in the early embryo and organization of the germline
The first nuclear migration event in animals occurs in the newly fertilized zygote when male and female pronuclei migrate toward each other. In most animals, centrosomes are contributed by sperm and remain attached to the male pronucleus, while the microtubule motor dynein mediates female pronuclear migration toward the male pronucleus at the center of the microtubule aster.95 LINC complexes consisting of the SUN protein SUN-1 and the KASH protein ZYG-12 are required for pronuclear migration in the early embryo and for organization of the germline.96,97 Because SUN-1 is divergent from other SUN proteins98 and because the KASH domain of ZYG-12 is short and not well conserved with other KASH peptides,96,99 I refer to SUN-1/ZYG-12 as a non-canonical LINC.
SUN-1 and ZYG-12 were first identified because they cause a severe separation between centrosomes and the male pronucleus in the newly fertilized zygote.96 There are two isoforms of ZYG-12; the KASH-less isoform localizes to centrosomes and the long isoform, which contains a C-terminal KASH domain, localizes to the nuclear envelope in a SUN-1-dependent manner to form a LINC complex.100 KASH-less and full-length ZYG-12 then dimerize to attach centrosomes to the male pronuclear envelope. In zyg-12 or sun-1 mutants, both centrosomes become detached from the male pronucleus, leading to severe pronuclear migration defects.96 ZYG-12 also interacts with the minus-end directed microtubule motor dynein through DLI-1.96 SUN-1/ZYG-12 LINC complexes recruit dynein to the cytoplasmic surface of pronuclei. This allows male pronuclei to capture centrosomes shortly after fertilization.96 Sperm nuclei are extremely compact and failure to expand rapidly after fertilization leads to a phenotype where a single centrosome is detached, suggesting that the surface area of the male pronucleus is important for centrosomal attachment.75 Finally, SUN-1 and ZYG-12 are required to recruit dynein to female pronuclei to mediate pronuclear migration.96
SUN-1 and ZYG-12 also interact with microtubules to organize nuclei in the C. elegans germline. Normally, during meiosis, germline nuclei are localized to the periphery of the syncytial gonad, evenly spaced and surrounding a nuclear-free rachis.101,102 In zyg-12 mutant gonads, microtubules are disorganized, and nuclei fall into the rachis.97 Thus, SUN-1/ZYG-12 LINC complexes are required to position nuclei in the germline and early embryo. In zebrafish and likely human embryos, Lrmp1/KASH5 plays an analogous role to ZYG-12, recruiting dynein to nuclei to mediate pronuclear migration.88,103
LINC complexes move meiotic chromosomes to enable homologous chromosome pairing
Homologous chromosome pairing is one of many important events that occurs during meiotic prophase. Chromosome pairing occurs during a burst of rapid prophase movements driven by telomeres moving along the inner surface of the nuclear envelope. These movements are thought to prevent non-homologous chromosomes from becoming interlocked.104 LINC complexes in fission yeast were found to transfer forces generated by dynein in the cytoplasm, across the nuclear envelope, to meiotic telomeres, leading to rapid chromosome movements.105 Subsequently, LINC complexes have been shown to mediate meiotic chromosome movements in plants and animals.32,33,106
In C. elegans, LINC complexes of SUN-1 and ZYG-12 are required during homologous chromosome pairing.107–109 SUN-1/ZYG-12 LINC complexes transition from an even distribution around the nucleus in interphase to distinct puncta in the nuclear envelope that move throughout early meiotic prophase in conjunction with rapid chromosome movements.110,111 C. elegans LINC complexes interact with pairing centers, subtelomeric repeats, of meiotic chromosomes. The ZIM/HIM proteins mediate interactions between pairing centers and LINC complexes, although how LINC complexes directly interact with meiotic chromosomes is unknown.112,113 Lamins play a significant role in meiotic chromosome pairing. The C. elegans lamin LMN-1 is remodeled during rapid chromosome movements in meiotic prophase. LMN-1 is reduced and more sensitive to detergent, making it more soluble, allowing rapid chromosome movements.114,115 LMN-1 remodeling is regulated by CHK-2 and PLK-2 kinase activity; a phospho-mutant lmn-1 animal, with eight phosphorylation sites mutated, was resistant to detergent and led to significant delays in homolog pairing.115 Additionally, the nucleoplasmic domain of SUN-1 is phosphoregulated by CHK-2 and PLK-2 during rapid chromosome movements. SUN-1 remains phosphorylated until homologous chromosomes are paired. Blocking SUN-1 phosphorylation leads to chromosome entanglements, unpaired chromosomes, and embryonic lethality.114–116 Thus, both LMN-1 and SUN-1 are regulated by kinases to facilitate rapid chromosome movements and homolog pairing in meiosis.
Migration of nuclei through constricted spaces
Cellular migrations through constricted spaces are a critical aspect of many developmental and disease processes including hematopoiesis, inflammation, and metastasis.3,117 Multiple labs have developed in vitro assays to observe cancer or dendritic cells migrating through narrow constrictions and have found that the stiffness of the nucleus is the rate-limiting step for such cell migrations.118,119 An in vivo model where nuclei can be observed moving through constricted spaces is needed to fully understand nuclear squeezing through constricted spaces. Thus, C. elegans larval P cells were developed as a model because their nuclei undergo extreme morphological changes to squeeze through a space about 5% their resting width as a part of normal development.90
P-cell nuclear migration requires unc-83 and unc-84. In fact, screens for mutants with P-cell nuclear migration defects led to the isolation of the first unc-83 and unc-84 alleles (see Box 1).12,13 Because UNC-83 recruits both kinesin-1 and dynein to the surface of nuclei in other cells,81–83 it was proposed that LINC complexes made of UNC-83 and UNC-84 would also mediate P-cell nuclear migration by serving as a cargo adaptor for microtubule motors. P-cell nuclei move toward the minus ends of microtubules.90 This is in contrast to embryonic hyp7 precursors, described above, where nuclei migrate toward the plus ends of growing microtubules.83 In support of P-cell nuclei moving toward the minus end of microtubules, genetic analyses showed that dynein is the major force producer and kinesin-1 plays an unknown regulatory role.90,91 Thus, LINC works through dynein to move nuclei through constricted spaces. How UNC-83 favors kinesin-1 in embryonic hyp7 nuclear migration but dynein in P-cell nuclear migration remains an open question.
Using LINC complexes is not the only mechanism mediating P-cell nuclear migration through constricted spaces. Null alleles of unc-83 or unc-84 are temperature sensitive.13 At restrictive temperatures, about 50% of P-cell nuclei fail to migrate, leading to P-cell death. Mutant animals are egg-laying deficient (Egl) because of missing vulva cells and uncoordinated (Unc) due to missing neurons normally derived from P-cell lineages. However, at permissive temperatures, almost all P-cell nuclei migrate in the absence of LINC complexes.17,20,89 This suggested that other pathways are sufficient to move P-cell nuclei through constricted spaces in the absence of LINC.
To identify proteins involved in this alternative P cell nuclear migration pathway, a forward genetic screen was carried out for enhancers of the unc-84 nuclear migration defect.120 toca-1 was the first gene identified.120 Transducer of Cdc42-dependent actin assembly (TOCA-1) contains an F-BAR domain thought to bind curved membranes, a predicted Cdc42-interacting domain, and a domain proposed to interact with WASP/WAVE to nucleate actin filaments.120,121 TOCA-1 and its close ortholog TOCA-2 function redundantly to play an essential role in endocytosis,122 but only TOCA-1 functions to facilitate P-cell nuclear migration.120 It is likely that TOCA-1 and CDC-42 are regulating a branched actin network to help squeeze nuclei through constricted spaces. Branched actin networks are also thought to help dendritic cells move through small glass capillary tubes,118,123 suggesting a conserved mechanism. Elucidation of other players in the LINC-independent pathway will lead to a better understanding of how branched actin functions in P-cell nuclear migration.
Future directions
The network of LINC complexes, their effectors at the surface of nuclei, and the ways they interact with the cytoskeleton are better understood in C. elegans than in any other multi-cellular system. Most of what has been found in C. elegans has been conserved at a functional level in other animal systems, demonstrating its value as a model. Many questions must still be addressed in order to fully understand LINC networks. C. elegans continues to be well suited to address the following remaining gaps in the LINC field: (1) We do not fully understand how interactions between SUN and KASH proteins are regulated or how SUN proteins are prevented from interacting with KASH proteins until they are trafficked from the peripheral ER to the inner nuclear membrane. (2) The recent finding that anc-1 mutants have a much more severe nuclear anchorage phenotype than unc-84 mutants68Click or tap here to enter text. suggests that the giant cytoplasmic domains of ANC-1 function through unknown mechanisms. (3) It remains unknown how the KASH protein UNC-83 favors kinesin-1 or dynein in different nuclear migration events. (4) Our understanding of how LINC complexes mediate DNA damage repair and gene regulation is still in its infancy. (5) How LINC complexes work with other mechanisms, including branched actin networks and other means of softening the nucleoskeleton, to squeeze nuclei through constricted spaces warrants further investigation. Furthermore, how LINC complexes relate to human health and disease progression is poorly understood. Answering these open questions and more will require many more years of research on the mechanisms and regulation of LINC complexes.
Acknowledgments
I thank Ellen Gregory for help with drawing Figure 1 and Sean Burgess for discussions leading to the organization Figure 2. I thank past and present members of the Starr lab for many fruitful discussions and successful studies on nuclear positioning. I thank members of the field for discussions and continually pushing the field forward. I apologize to studies left out of this brief review.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Studies in the Starr lab are supported by the National Institutes of Health (grant number R01 GM073874).
References
- 1.Bone CR, Starr DA. Nuclear migration events throughout development. J Cell Sci 2016; 129:1951–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol 2005; 6:21–31 [DOI] [PubMed] [Google Scholar]
- 3.Ungricht R, Kutay U. Mechanisms and functions of nuclear envelope remodelling. Nat Rev Mol Cell Biol 2017; 18:229–45 [DOI] [PubMed] [Google Scholar]
- 4.Magistris P, Antonin W. The dynamic nature of the nuclear envelope. Curr Biol 2018; 28:R487–97 [DOI] [PubMed] [Google Scholar]
- 5.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 1983; 100:64–119 [DOI] [PubMed] [Google Scholar]
- 6.Sulston J, Horvitz H. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 1977; 56:110–56 [DOI] [PubMed] [Google Scholar]
- 7.White J. Obituary: John sulston (1942–2018). Development 2018; 145:dev166538. [DOI] [PubMed] [Google Scholar]
- 8.Kimble J. John sulston (1942–2018). Science 2018; 360:157. [DOI] [PubMed] [Google Scholar]
- 9.Horvitz H, Sulston J. Joy of the worm. Genetics 1990; 126:287–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sulston JE. Caenorhabditis elegans: the cell lineage and beyond (nobel lecture). Chembiochem 2003; 4:688–96 [DOI] [PubMed] [Google Scholar]
- 11.Waterston R, Sulston J. The genome of Caenorhabditis elegans. Proc Natl Acad Sci 1995; 92:10836–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sulston JE, Horvitz RH. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev Biol 1981; 82:41–55 [DOI] [PubMed] [Google Scholar]
- 13.Horvitz H, Sulston J. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 1980; 96:435–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.elegans Consortium TC. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 1998; 282:2012–8 [DOI] [PubMed] [Google Scholar]
- 15.Gerace L, Tapia O. Messages from the voices within: regulation of signaling by proteins of the nuclear lamina. Curr Opin Cell Biol 2018; 52:14–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Heras JI, Batrakou DG, Schirmer EC. Cancer biology and the nuclear envelope: a convoluted relationship. Semin Cancer Biol 2013; 23:125–37 [DOI] [PubMed] [Google Scholar]
- 17.Starr D, Hermann G, Malone C, Fixsen W, Priess J, Horvitz H, Han M. Unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development 2001; 128:5039–50 [DOI] [PubMed] [Google Scholar]
- 18.Mosley-Bishop KL, Qinghong L, Patterson K, Fischer JA. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the drosophila eye. Curr Biol 1999; 9:1211–20 [DOI] [PubMed] [Google Scholar]
- 19.Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol 1995; 129:1033–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malone C, Fixsen W, Horvitz H, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development 1999; 126:3171–81 [DOI] [PubMed] [Google Scholar]
- 21.Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science 2002; 298:406–9 [DOI] [PubMed] [Google Scholar]
- 22.Starr DA. A nuclear-envelope bridge positions nuclei and moves chromosomes. J Cell Sci 2009; 122:577–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 2010; 26:421–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luxton GG, Starr DA. KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr Opin Cell Biol 2014; 28:69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tapley EC, Starr DA. Connecting the nucleus to the cytoskeleton by SUN–KASH bridges across the nuclear envelope. Curr Opin Cell Biol 2013; 25:57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang W, Worman HJ, Gundersen GG. Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol 2015; 208:11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y, Burke B. LINC complexes and nuclear positioning. Semin Cell Dev Biol 2017;82:67–76 [DOI] [PubMed]
- 28.Gundersen GG, Worman HJ. Nuclear positioning. Cell 2013; 152:1376–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burke B, Roux KJ. Nuclei take a position: managing nuclear location. Dev Cell 2009; 17:587–97 [DOI] [PubMed] [Google Scholar]
- 30.Razafsky D, Hodzic D. Nuclear envelope: positioning nuclei and organizing synapses. Curr Opin Cell Biol 2015; 34:84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baum DA, Baum B. An inside-out origin for the eukaryotic cell. BMC Biol 2014; 12:1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burke B. LINC complexes as regulators of meiosis. Curr Opin Cell Biol 2018; 52:22–9 [DOI] [PubMed] [Google Scholar]
- 33.Hiraoka Y, Dernburg A. The SUN rises on meiotic chromosome dynamics. Dev Cell 2009; 17:598–605 [DOI] [PubMed] [Google Scholar]
- 34.Dawe H, Adams M, Wheway G, Szymanska K, Logan C, Noegel A, Gull K, Johnson C. Nesprin-2 interacts with meckelin and mediates ciliogenesis via remodelling of the actin cytoskeleton. J Cell Sci 2009; 122:2716–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potter C, Razafsky D, Wozniak D, Casey M, Penrose S, Ge X, Mahjoub M, Hodzic D. The KASH-containing isoform of Nesprin1 giant associates with ciliary rootlets of ependymal cells. Neurobiol Dis 2018; 115:82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gough L, Fan J, Chu S, Winnick S, Beck K. Golgi localization of syne-1. Mol Biol Cell 2003; 14:2410–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence KS, Tapley EC, Cruz VE, Li Q, Aung K, Hart KC, Schwartz TU, Starr DA, Engebrecht J. LINC complexes promote homologous recombination in part through inhibition of nonhomologous end joining. J Cell Biol 2016; 215:801–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lottersberger F, Karssemeijer R, Dimitrova N, de Lange T. 53BP1 and the LINC complex promote microtubule-dependent DSB mobility and DNA repair. Cell 2015; 163:880–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luxton GG, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 2010; 329:956–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang Y, Zhu F, Wang L, Ouyang Y, Dong M, Liu C, Zhao H, Cui X, Dongyuan M, Zhang Z, Yang X, Guo Y, Liu F, Yuan L, Gao F, Guo X, Sun Q, Cao Y, Wei L, Shang Y, Zhu F, Wang L, Ouyang Y, Dong M, Liu C, Zhao H, Cui X, Dongyuan M, Zhang Z, Yang X, Guo Y, Liu F, Yuan L, Gao F, Guo X, Sun Q, Cao Y, Wei L. Essential role for SUN5 in anchoring sperm head to the tail. Elife 2017; 6:e28199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elkhatib RA, Paci M, Longepied G, Saias-Magnan J, Courbière B, Guichaoua M-R, Lévy N, Metzler-Guillemain C, Mitchell MJ. Homozygous deletion of SUN5 in three men with decapitated spermatozoa. Hum Mol Genet 2017; 26:3167–71 [DOI] [PubMed] [Google Scholar]
- 42.Talamas JA, Hetzer MW. POM121 and Sun1 play a role in early steps of interphase NPC assembly. J Cell Biol 2011; 194:27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stroud MJ. Linker of nucleoskeleton and cytoskeleton complex proteins in cardiomyopathy. Biophys Rev 2018; 10:1033–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan K, Haelterman NA, Kwartler CS, Regalado ES, Lee P-T, Nagarkar-Jaiswal S, Guo D-C, Duraine L, Wangler MF, Bamshad MJ, Nickerson DA, Lin G, Milewicz DM, Bellen HJ. Ari-1 regulates myonuclear organization together with parkin and is associated with aortic aneurysms. Dev Cell 2018; 45:226–44.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, Ragnauth CD, Qijian Y, Mellad JA, Warren DT, Wheeler MA, Ellis JA, Skepper JN, Vorgerd M, Schlotter-Weigel B, Weissberg PL, Roberts RG, Wehnert M, Shanahan CM. Nesprin-1 and -2 are involved in the pathogenesis of emery dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet 2007; 16:2816–33 [DOI] [PubMed] [Google Scholar]
- 46.Puckelwartz MJ, Kessler E, Zhang Y, Hodzic D, Randles NK, Morris G, Earley JU, Hadhazy M, Holaska JM, Mewborn SK, Pytel P, McNally EM. Disruption of nesprin-1 produces an emery dreifuss muscular dystrophy-like phenotype in mice. Hum Mol Genet 2009; 18:607–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horn HF, Brownstein Z, Lenz DR, Shivatzki S, Dror AA, Dagan-Rosenfeld O, Friedman LM, Roux KJ, Kozlov S, Jeang K-T, Frydman M, Burke B, Stewart CL, Raham K. The LINC complex is essential for hearing. J Clin Invest 2013; 123:740–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu J, Lei K, Zhou M, Craft C, Xu G, Xu T, Zhuang Y, Xu R, Han M. KASH protein syne-2/nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum Mol Genet 2011; 20:1061–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Green E, Grozeva D, Forty L, Gordon-Smith K, Russell E, Farmer A, Hamshere M, Jones I, Jones L, McGuffin P, Moran J, Purcell S, Sklar P, Owen M, O’Donovan M, Craddock N. Association at SYNE1 in both bipolar disorder and recurrent major depression. Mol Psychiatry 2012; 18:614–7 [DOI] [PubMed] [Google Scholar]
- 50.Gros-Louis F, Dupré N, Dion P, Fox MA, Laurent S, Verreault S, Sanes JR, Bouchard J-P, Rouleau GA. Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nat Genet 2007; 39:80–5 [DOI] [PubMed] [Google Scholar]
- 51.Gonzalez B, Billion K, Rous S, Pavie B, Lange C, Goodchild R. Excess LINC complexes impair brain morphogenesis in a mouse model of recessive TOR1A disease. Hum Mol Genet 2018; 27:2154–70 [DOI] [PubMed] [Google Scholar]
- 52.Yu W, Chahrour H, Coulter E, Jiralerspong S, Okamura-Ikeda K, Ataman B, Schmitz-Abe K, Harmin A, Adli M, Malik N, Gama A, Lim T, Sanders J, Mochida H, Partlow N, Nu Christine Felie M, Rodriguez J, Nasir H, Ware J, Joseph M, Hill R, Kwan Y, Al-Saffar M, Mukaddes M, Hashmi A, Balkhy S, Gascon G, Hisama M, Leclair E, Poduri A, Oner O, Al-Saad S, Al-Awadi A, Bastaki L, Ben-Omran T, Teebi S, Al-Gazali L, Eapen V, Stevens R, Rappaport L, Gabriel B, Markianos K, State W, Greenberg E, Taniguchi H, Braverman E, Morrow M, Walsh A. Using whole-exome sequencing to identify inherited causes of autism. Neuron 2013; 77:259–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen C-Y, Chi Y-H, Mutalif R, Starost MF, Myers TG, Anderson SA, Stewart CL, Jeang K-T. Accumulation of the inner nuclear envelope protein sun1 is pathogenic in progeric and dystrophic laminopathies. Cell 2012; 149:565–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim PH, Luu J, Heizer P, Tu Y, Weston TA, Chen N, Lim C, Li RL, Lin P-Y, Dunn JC, Hodzic D, Young SG, Fong LG. Disrupting the LINC complex in smooth muscle cells reduces aortic disease in a mouse model of Hutchinson-Gilford progeria syndrome. Sci Transl Med 2018; 10:7163–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell 2007; 12:863–72 [DOI] [PubMed] [Google Scholar]
- 56.Doherty JA, Rossing MA, Cushing-Haugen KL, Chen C, Berg Djvd AW, Pike MC, Ness RB, Moysich K, Chenevix-Trench G, Beesley J, Webb PM, Chang-Claude J, Wang-Gohrke S, Goodman MT, Lurie G, Thompson PJ, Carney ME, Hogdall E, Kjaer SK, Hogdall C, Goode EL, Cunningham JM, Fridley BL, Vierkant RA, Berchuck A, Moorman PG, Schildkraut JM, Palmieri RT, Cramer DW, Terry KL, Yang HP, Garcia-Closas M, Chanock S, Lissowska J, Song H, Pharoah PD, Shah M, Perkins B, McGuire V, Whittemore AS, Cioccio RAD, Gentry-Maharaj A, Menon U, Gayther SA, Ramus SJ, Ziogas A, Brewster W, Anton-Culver H, Pearce CL. ESR1/SYNE1 polymorphism and invasive epithelial ovarian cancer risk: an Ovarian Cancer Association Consortium study. Cancer Epidem Biomar 2010; 19:245–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science 2006; 314:268–74 [DOI] [PubMed] [Google Scholar]
- 58.Huang W, Huang H, Wang L, Hu J, Song W. SUN1 silencing inhibits cell growth through G0/G1 phase arrest in lung adenocarcinoma. Oncotargets Ther 2017; 10:2825–33 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Matsumoto A, Hieda M, Yokoyama Y, Nishioka Y, Yoshidome K, Tsujimoto M, Matsuura N. Global loss of a nuclear lamina component, lamin a/C, and LINC complex components SUN1, SUN2, and nesprin-2 in breast cancer. Cancer Med 2015; 4:1547–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lv X, Liu L, Cheng C, Yu B, Xiong L, Hu K, Tang J, Zeng L, Sang Y. SUN2 exerts tumor suppressor functions by suppressing the Warburg effect in lung cancer. Sci Rep 2015; 5:17940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol 2009; 10:75–82 [DOI] [PubMed] [Google Scholar]
- 62.Crisp M, Liu Q, Roux K, Rattner J, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol 2006; 172:41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cain NE, Tapley EC, Nald KL, Cain BM, Starr DA. The SUN protein UNC-84 is required only in force-bearing cells to maintain nuclear envelope architecture. J Cell Biol 2014; 206:163–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cain NE, Starr DA. SUN proteins and nuclear envelope spacing. Nucleus 2015; 6:2–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 2012; 149:1035–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang W, Shi Z, Jiao S, Chen C, Wang H, Liu G, Wang Q, Zhao Y, Greene MI, Zhou Z. Structural insights into SUN-KASH complexes across the nuclear envelope. Cell Res 2012; 22:1440–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sosa BA, Kutay U, Schwartz TU. Structural insights into LINC complexes. Curr Opin Struct Biol 2013; 23:285–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cain NE, Jahed Z, Schoenhofen A, Valdez VA, Elkin B, Hao H, Harris NJ, Herrera LA, Woolums BM, Mofrad MR, Luxton GG, Starr DA. Conserved SUN-KASH interfaces mediate LINC complex-dependent nuclear movement and positioning. Curr Biol 2018; 28:3086–97.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 is a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell 2006; 17:1790–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hao H, Starr DA. SUN/KASH interactions facilitate force transmission across the nuclear envelope. Nucleus 2019; 10:73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jahed Z, Hao H, Thakkar V, Vu UT, Valdez VA, Rathish A, Tolentino C, Kim SC, Fadavi D, Starr DA, Mofrad MR. Role of KASH domain lengths in the regulation of LINC complexes. Mol Biol Cell 2019; 30:2076–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bone CR, Tapley EC, Gorjanacz M, Starr DA. The Caenorhabditis elegans SUN protein UNC-84 interacts with lamin to transfer forces from the cytoplasm to the nucleoskeleton during nuclear migration. Mol Biol Cell 2014; 25:2853–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.King MC, Drivas TG, Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell 2008; 134:427–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee KK, Starr D, Cohen M, Liu J, Han M, Wilson KL, Gruenbaum Y. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol Biol Cell 2002; 13:892–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meyerzon M, Gao Z, Liu J, Wu J-C, Malone CJ, Starr DA. Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Dev Biol 2009; 327:433–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tapley EC, Ly N, Starr DA. Multiple mechanisms actively target the SUN protein UNC-84 to the inner nuclear membrane. Mol Biol Cell 2011; 22:1739–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Anahan C, Shackleton S. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem 2010; 285:3487–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Borrego-Pinto J, Jegou T, Osorio DS, Aurade F, Gorjanacz M, Koch B, Mattaj IW, Gomes ER. Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci 2012; 125:1099–105 [DOI] [PubMed] [Google Scholar]
- 79.Silva N, Jantsch V. UNC-84: ‘LINC-ing’ chromosome movement and double strand break repair. J Cell Biol 2016; 215:753–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Janota CS, Calero-Cuenca FJ, Costa J, Gomes ER. SnapShot: Nucleo-cytoskeletal interactions. Cell 2017; 169:970–e1 [DOI] [PubMed] [Google Scholar]
- 81.Fridolfsson HN, Ly N, Meyerzon M, Starr DA. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev Biol 2010; 338:237–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meyerzon M, Fridolfsson HN, Ly N, McNally FJ, Starr DA. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development 2009; 136:2725–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fridolfsson HN, Starr DA. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol 2010; 191:115–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Starr DA. Watching nuclei move: Insights into how kinesin-1 and dynein function together. Bioarchitecture 2011; 1:9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and syne/nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron 2009; 64:173–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wilson MH, Holzbaur EL. Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development 2015; 142:218–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci 2009; 106:2194–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Horn HF, Kim D, Wright GD, Wong E, Stewart CL, Burke B, Roux KJ. A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J Cell Biol 2013; 202:1023–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Starr DA, Han M. A genetic approach to study the role of nuclear envelope components in nuclear positioning. Novartis Found Symp 2005; 264:208–19, discussion 219–30. [PubMed] [Google Scholar]
- 90.Bone CR, Chang Y-T, Cain NE, Murphy SP, Starr DA. Nuclei migrate through constricted spaces using microtubule motors and actin networks in C. elegans hypodermal cells. Development 2016; 143:4193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ho J, Valdez VA, Ma L, Starr DA. Characterizing dynein’s role in P-cell nuclear migration using an auxin-induced degradation system. MicroPubl Biol Epub ahead of print 2018. DOI: 10.17912/w2w96j [DOI] [PMC free article] [PubMed]
- 92.Hedgecock E, Thomson J. A gene required for nuclear and mitochondrial attachment in the nematode Caenorhabditis elegans. Cell 1982; 30:321–30 [DOI] [PubMed] [Google Scholar]
- 93.Fridolfsson HN, Herrera LA, Brandt JN, Cain NE, Hermann GJ, Starr DA. Genetic analysis of nuclear migration and anchorage to study LINC complexes during development of Caenorhabditis elegans. Methods Mol Biol 2018; 1840:163–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Elhanany-Tamir H, Yu YV, Shnayder M, Jain A, Welte M, Volk T. Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J Cell Biol 2012; 198:833–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reinsch S, Gonczy P. Mechanisms of nuclear positioning. J Cell Sci 1998; 111:2283–95 [DOI] [PubMed] [Google Scholar]
- 96.Malone CJ, Misner L, Bot N, Tsai M-C, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 2003; 115:825–36 [DOI] [PubMed] [Google Scholar]
- 97.Zhou K, Rolls MM, Hall DH, Malone CJ, Hanna-Rose W. A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J Cell Biol 2009; 186:229–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jaspersen SL, Martin AE, Glazko G, Giddings TH, Morgan G, Mushegian A, Winey M. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J Cell Biol 2006; 174:665–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilhelmsen K, Ketema M, Truong H, Sonnenberg A. KASH-domain proteins in nuclear migration, anchorage and other processes. J Cell Sci 2006; 119:5021–9 [DOI] [PubMed] [Google Scholar]
- 100.Minn I, Rolls MM, Hanna-Rose W, Malone CJ. SUN-1 and ZYG-12, mediators of centrosome-nucleus attachment, are a functional SUN/KASH pair in Caenorhabditis elegans. Mol Biol Cell 2009; 20:4586–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Priti A, Ong H, Toyama Y, Padmanabhan A, Dasgupta S, Krajnc M, Zaidel-Bar R. Syncytial germline architecture is actively maintained by contraction of an internal actomyosin corset. Nat Commun 2018; 9:4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wolke U, Jezuit EA, Priess JR. Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development 2007; 134:2227–36 [DOI] [PubMed] [Google Scholar]
- 103.Kozono T, Tadahira K, Okumura W, Itai N, Tamura-Nakano M, Dohi T, Tonozuka T, Nishikawa A. Jaw1/LRMP has a role in maintaining nuclear shape via interaction with SUN proteins. J Biochem 2018; 164:303–11 [DOI] [PubMed] [Google Scholar]
- 104.Koszul R, Kleckner N. Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections?. Trends Cell Biol 2009; 19:716–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 2006; 125:59–69 [DOI] [PubMed] [Google Scholar]
- 106.Meier I. LINCing the eukaryotic tree of life – towards a broad evolutionary comparison of nucleocytoplasmic bridging complexes. J Cell Sci 2016; 129:3523-31 [DOI] [PubMed] [Google Scholar]
- 107.Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 2009; 139:907–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Penkner A, Fridkin A, Gloggnitzer J, Baudrimont A, Machacek T, Woglar A, Csaszar E, Pasierbek P, Ammerer G, Gruenbaum Y, Jantsch V. Meiotic chromosome homology search involves modifications of the nuclear envelope protein matefin/SUN-1. Cell 2009; 139:920–33 [DOI] [PubMed] [Google Scholar]
- 109.Penkner A, Tang L, Novatchkova M, Ladurner M, Fridkin A, Gruenbaum Y, Schweizer D, Loidl J, Jantsch V. The nuclear envelope protein matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev Cell 2007; 12:873–85 [DOI] [PubMed] [Google Scholar]
- 110.Rog O, Dernburg AF. Direct visualization reveals kinetics of meiotic chromosome synapsis. Cell Rep 2015; 10:1639–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Baudrimont A, Penkner A, Woglar A, Machacek T, Wegrostek C, Gloggnitzer J, Fridkin A, Klein F, Gruenbaum Y, Pasierbek P, Jantsch V. Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PloS Genet 2010; 6:e1001219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Phillips C, Wong C, Bhalla N, Carlton P, Weiser P, Meneely P, Dernburg A. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 2005; 123:1051–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 2005; 123:1037–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Paouneskou D, Jantsch V. Meiotic chromosome movement: what’s lamin got to do with it? Nucleus. 2019; 10:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Link J, Paouneskou D, Velkova M, Daryabeigi A, Laos T, Labella S, Barroso C, Piñol SP, Montoya A, Kramer H, Woglar A, Baudrimont A, Markert SM, Stigloher C, Martinez-Perez E, Dammermann A, Alsheimer M, Zetka M, Jantsch V. Transient and partial nuclear lamina disruption promotes chromosome movement in early meiotic prophase. Dev Cell 2018; 45:212–25.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Woglar A, Daryabeigi A, Adamo A, Habacher C, Machacek T, Volpe A, Jantsch V. Matefin/SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. Plos Genet 2013; 9:e1003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kirby TJ, Lammerding J. Emerging views of the nucleus as a cellular mechanosensor. Nat Cell Biol 2018; 20:373–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Raab M, Gentili M, de Belly H, Thiam H, Vargas P, Jimenez A, Lautenschlaeger F, Voituriez R, Lennon-Dumenil A, Manel N, Piel M. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 2016; 352:7611 362 [DOI] [PubMed] [Google Scholar]
- 119.Nais C, Gilbert RM, Isermann P, McGregor AL, Te Lindert M, Weigelin B, Vidson P, Friedl P, Wolf K, Lammerding J. Nuclear envelope rupture and repair during cancer cell migration. Science 2016; 352:353–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chang Y-T, Dranow D, Kuhn J, Meyerzon M, Ngo M, Ratner D, Warltier K, Starr DA. Toca-1 is in a novel pathway that functions in parallel with a SUN-KASH nuclear envelope bridge to move nuclei in Caenorhabditis elegans. Genetics 2013; 193:187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ho H-Y, Rohatgi R, Lebensohn AM, Le M, Li J, Gygi SP, Kirschner MW. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell 2004; 118:203–16 [DOI] [PubMed] [Google Scholar]
- 122.Giuliani C, Troglio F, Bai Z, Patel FB, Zucconi A, Malabarba M, Disanza A, Stradal TB, Cassata G, Confalonieri S, Hardin JD, Soto MC, Grant BD, Scita G. Requirements for F-BAR proteins TOCA-1 and TOCA-2 in actin dynamics and membrane trafficking during Caenorhabditis elegans oocyte growth and embryonic epidermal morphogenesis. Plos Genet 2009; 5:e1000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Thiam H-R, Vargas P, Carpi N, Crespo C, Raab M, Terriac E, King MC, Jacobelli J, Alberts AS, Stradal T, Lennon-Dumenil A-M, Piel M. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat Commun 2016; 7:10997. [DOI] [PMC free article] [PubMed] [Google Scholar]