Domestic cats (Felis silvestris catus) were domesticated from wildcats approximately 9,000 years ago via close interaction between humans and cats. During cat evolution, various exogenous retroviruses infected different cat lineages and generated numerous ERVs in the host genome, some of which remain replication competent. Here, we detected several ERV-DC loci in Felis silvestris silvestris. Notably, a species-specific single nucleotide polymorphism in the ERV-DC14 env gene, which results in a replication-defective product, is highly prevalent in European wildcats, unlike the replication-competent ERV-DC14 that is commonly present in domestic cats. The presence of the same lethal mutation in the env genes of both FeLV and murine ERV provides a common mechanism shared by endogenous and exogenous retroviruses by which ERVs can be inactivated after endogenization. The antiviral role of Refrex-1 predates cat exposure to feline retroviruses. The existence of two ERV-DC14 phenotypes provides a unique model for understanding both ERV fate and cat domestication.
KEYWORDS: ERV-DC, endogenous retrovirus, Fv-4, FeLV, Felis, MuLV, domestic cat, domestication, evolution, wildcat
ABSTRACT
Endogenous retroviruses (ERVs) of domestic cats (ERV-DCs) are one of the youngest feline ERV groups in domestic cats (Felis silvestris catus); some members are replication competent (ERV-DC10, ERV-DC18, and ERV-DC14), produce the antiretroviral soluble factor Refrex-1 (ERV-DC7 and ERV-DC16), or can generate recombinant feline leukemia virus (FeLV). Here, we investigated ERV-DC in European wildcats (Felis silvestris silvestris) and detected four loci: ERV-DC6, ERV-DC7, ERV-DC14, and ERV-DC16. ERV-DC14 was detected at a high frequency in European wildcats; however, it was replication defective due to a single G → A nucleotide substitution, resulting in an E148K substitution in the ERV-DC14 envelope (Env). This mutation results in a cleavage-defective Env that is not incorporated into viral particles. Introduction of the same mutation into feline and murine infectious gammaretroviruses resulted in a similar Env dysfunction. Interestingly, the same mutation was found in an FeLV isolate from naturally occurring thymic lymphoma and a mouse ERV, suggesting a common mechanism of virus inactivation. Refrex-1 was present in European wildcats; however, ERV-DC16, but not ERV-DC7, was unfixed in European wildcats. Thus, Refrex-1 has had an antiviral role throughout the evolution of the genus Felis, predating cat exposure to feline retroviruses. ERV-DC sequence diversity was present across wild and domestic cats but was locus dependent. In conclusion, ERVs have evolved species-specific phenotypes through the interplay between ERVs and their hosts. The mechanism of viral inactivation may be similar irrespective of the evolutionary history of retroviruses. The tracking of ancestral retroviruses can shed light on their roles in pathogenesis and host-virus evolution.
IMPORTANCE Domestic cats (Felis silvestris catus) were domesticated from wildcats approximately 9,000 years ago via close interaction between humans and cats. During cat evolution, various exogenous retroviruses infected different cat lineages and generated numerous ERVs in the host genome, some of which remain replication competent. Here, we detected several ERV-DC loci in Felis silvestris silvestris. Notably, a species-specific single nucleotide polymorphism in the ERV-DC14 env gene, which results in a replication-defective product, is highly prevalent in European wildcats, unlike the replication-competent ERV-DC14 that is commonly present in domestic cats. The presence of the same lethal mutation in the env genes of both FeLV and murine ERV provides a common mechanism shared by endogenous and exogenous retroviruses by which ERVs can be inactivated after endogenization. The antiviral role of Refrex-1 predates cat exposure to feline retroviruses. The existence of two ERV-DC14 phenotypes provides a unique model for understanding both ERV fate and cat domestication.
INTRODUCTION
Endogenous retroviruses (ERVs) are integrated retroviral elements that make up 6% to 10% of cat, human, and mouse genome sequences (1–3). Most ERVs are disrupted subsequent to the original retroviral integration through the accumulation of mutations, deletions, or insertions in their genes during viral and host genome replication (4–8). However, some infectious ERVs have been identified in mice (5) and cats (9, 10). Feline endogenous retroviruses have been identified and grouped phylogenetically into different classes (11–13). For example, endogenous feline leukemia viruses (enFeLV) became integrated within the genomes of members of the Felis genus more than 2 million years ago (14–17). These ERVs are present at 6 to 12 copies per haploid genome in domestic cats (18–20), while fluorescent in situ hybridization detected 9 to 16 distinct autosomal enFeLV loci per domestic cat (21). enFeLVs can recombine with exogenous feline leukemia virus (FeLV) to yield recombinant FeLV subgroup B (22, 23). Additional feline endogenous ERVs have been characterized, including RD-114 (24), MAC-1 (25, 26), and feline endogenous retrovirus gamma4 (27).
One of the youngest feline ERV groups, called ERVs in domestic cats (ERV-DCs), is estimated to have integrated within the cat genome approximately 2.8 million years ago. ERV-DCs are classified as endogenous gammaretroviruses (10–12, 28, 29). We previously identified and cloned 13 ERV-DC loci and estimated that there were 7 to 17 ERV-DC copies present in each domestic cat. ERV-DCs have a simple retroviral structure, including gag, pol, and env genes enclosed between two noncoding long terminal repeats (LTRs) (10, 30). A unique feature of the ERV-DC family is that proviruses can be phylogenetically classified into three genotypes (Fig. 1A): genotype I (ERV-DC1, -DC2, -DC3, -DC4, -DC8, -DC14, -DC17, and -DC19), genotype II (ERV-DC7 and -DC16), and genotype III (ERV-DC6, -DC10, and -DC18). Among the ERV-DCs, ERV-DC10, -DC14, and -DC18 are infectious proviruses. ERV-DC18 may have been generated by retrotransposition during ERV-DC10 reintegration or reinfection in different members of one cat family (10). ERV-DC14 showed low promoter activity in its 5′ LTR due to a single A-to-T mutation. Reverting this mutation in ERV-DC14 (called ERV-DC14TA) enhanced its replication and enabled the ERV to persistently infect HEK293T cells (9). A survey of insertional polymorphisms within ERV-DCs in Japanese domestic cats indicated that a low proportion (2.5%) of cats tested carried ERV-DC14 (10). Notably, FeLV-positive cells were transduced with the env gene from a genotype I provirus, generating a novel interference subgroup called FeLV subgroup D (FeLV-D) (10). Genotype II proviruses were disrupted by mutations and deletions in the pol and env genes. However, the gene for the truncated Env protein of these proviruses (ERV-DC7 and -DC16) encoded an antiviral factor, called Refrex-1, that specifically inhibits ERV-DC genotype I and FeLV-D infections. Refrex-1 is efficiently secreted from feline cells as a soluble protein and may interfere with virus interaction with host cell receptors (31). ERV-DC6, -DC7, and -DC16 were apparently fixed in Japanese domestic cats, while the other ERV-DCs were polymorphic (10). Other examples of ERV env genes conferring resistance to viral infection have been demonstrated in the laboratory and in-house and wild mice; these include Fv-4, Rmcf, and Rmcf2, which inhibit infection by ecotropic, polytropic, and xenotropic murine leukemia virus (MLV), respectively (5, 32–35). The Mus musculus castaneus endogenous virus clone MmCN (MLV/MmCN; located in the qE1 region of chromosome 8) was amplified from the DNA of a mouse (M. musculus castaneus strain CAST/Ncr) trapped in Lake Casitas, CA (36). The sequence of this Cas subtype Env resembled that of Fv-4 (36), a defective endogenous MLV encoding a truncated Env that acts as a host restriction factor to block infection by ecotropic MLVs (33).
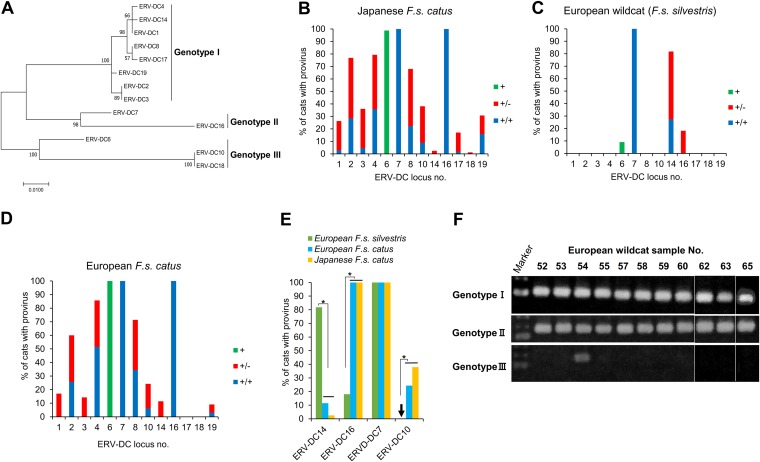
FIG 1.
Detection of ERV-DC proviruses in domestic cat and wildcat genomes. (A) A phylogenetic tree of the ERV-DC 3′ LTR was constructed using maximum likelihood methods. The percentages at the branch junctions indicate bootstrap values (1,000 replicates). Thirteen ERV-DC loci were classified into three genotypes: genotype I (ERV-DC1, -DC2, -DC3, -DC4, -DC8, -DC14, -DC17, and -DC19), genotype II (ERV-DC7 and -DC16), and genotype III (ERV-DC6, -DC10, and -DC18). (B to D) Insertional polymorphisms of 13 ERV-DCs in Japanese domestic cats (B), European wildcats (C), and European domestic cats (D). Green and +, provirus detected; red and +/−, heterozygous (the copy is present on one of two chromosomes); blue and +/+, homozygous (the copy is present on both chromosomes). (E) Comparison of genotype frequencies for three loci (ERV-DC14, -DC16, and -DC7) among different cat populations. (F) PCR detection of ERV-DC genotypes in European wildcats (F. s. silvestris). Statistical analyses were conducted using Student’s t tests and one-way ANOVAs. *, P < 0.0001.
We previously reconstructed the full-length env genes of ERV-DC7 and ERV-DC16 (called ERV-DC7fl and ERV-DC16fl, respectively) to assess the role of Refrex-1 in virus-host coevolution. ERV-DC7fl and ERV-DC16fl were unable to produce infectious viral particles due to defects in Env cleavage. Defects in ERV-DC7fl Env resulted from three determinant residues (R407, I421, and T429). Reverse genetics methods were used to successfully reconstruct an infectious ERV-DC7fl env bearing the ERV-DC14 Env consensus residues at these three positions (R407G, I427N, and T429A). Analyses of ERV-DC7 env sequence diversity in Japanese domestic cats indicated that the determinants of ERV-DC7fl dysfunction were not fixed in the population. Four variants with different combinations of residues at these positions were identified: 407G and 427N-429A (G-NA), 407R and 427N-429A (R-NA), 407G and 427I-429T (G-IT), and 407R and 427I-429T (R-IT). These variants are present because the integration of ERV-DC7 into the host genome and the sequence of ERV-DC7 env noncoding regions underwent purifying selection between the time of its integration and its truncation. The T nucleotide at position 801 encodes a latent stop codon in ERV-DC7fl Env and was predominant in most Japanese domestic cats examined. Two animals were exceptions (animals ON-C and ON-R) and bore a C nucleotide at this position, which did not affect the function of ERV-DC7fl Env (37). Thus, characterization of ERV-DC7 in the Felis genus could help clarify the diversity of ERV-DCs in cat lineages.
The wildcat (Felis silvestris) population is dispersed throughout the Old World, but there has been little description of its subspecies (38). The coexistence of three subspecies, including the European wildcat (F. s. silvestris), the African wildcat (F. s. lybica), and the domestic cat (F. s. catus), was reported in different regions across Europe (38–40). The earliest archaeological evidence indicates that the European wildcat appeared in Europe ∼230,000 years ago (41). The domestic cat (F. s. catus) was domesticated from F. s. lybica approximately 131,000 years ago in the Near East (41, 42). The results of a mitochondrial DNA analysis suggested that a common ancestor of both European wildcats and sand cats (F. s. margarita) was genetically distinct from the ancestor of domestic cats (F. s. catus) (41). ERV-DCs were demonstrated to be phylogenetically distinct from enFeLVs in both domestic cat (F. s. catus) and wildcats (10–12, 14). The domestic cat and its immediate progenitor, the wildcat (F. s. silvestris/F. s. lybica), showed a tremendous diversity of enFeLVs (14). To date, the relationships between European wildcats and domestic cats remain controversial. For example, the question of whether domestic cats and wildcats evolved from a common ancestor or whether domestic cats descended from wildcats remains unresolved. In addition, the existence of ERV-DCs and their functionality in wildcats remains unclear, as does the overall evolution of ERV-DC in members of the Felis genus. Therefore, we undertook a study of ERVs and of ERV-DCs in particular in the Felis genus to begin to elucidate these issues.
In this study, we assessed the presence of ERV-DCs and analyzed their insertional polymorphisms in European wildcats and domestic cats. Ours is the first report showing that a species-specific inactivation of infectious endogenous retroviruses also contributes to a common mechanism of viral inactivation employed by the host against both endogenous and exogenous retroviruses. Additional analyses of ERV-DCs in wildcats and domestic cats revealed unexpected retroviral diversity and clarified several other issues regarding the fate of ERV endogenization, retroviral pathogenesis, and host-virus interactions.
RESULTS
Insertional polymorphic distribution of ERV-DC in European wildcats and domestic cats.
The ERV-DC provirus insertions at 13 loci that were previously identified in domestic cats in Japan (Fig. 1A and B) (10) were investigated in 11 European wildcats in Spain. We detected ERV-DC proviral insertions of ERV-DC6, -DC7, -DC14, and -DC16 at frequencies of 10%, 100%, 82%, and 18%, respectively, in European wildcats (Fig. 1C). The ERV-DC provirus insertion was also investigated in 35 domestic cats in Spain. Eleven proviruses were detected at frequencies of 9.1 to 100%, and the insertional patterns were similar to those previously described for Japanese domestic cats (Fig. 1B and D). ERV-DC6 was detected in only one European wildcat (<10%), but it was fixed in domestic cats in Japan and Spain. ERV-DC17 and ERV-DC18 were detected only in Japanese domestic cats. ERV-DC14 was detected at a frequency of 82% (9 cats) in European wildcats; this rate was significantly higher than that in domestic cats in Japan (2.4%) and Spain (11.4%). ERV-DC10 was not detected in European wildcats; it was detected only in domestic cats in Japan and Spain at frequencies of 38% and 24.5%, respectively. ERV-DC7 was fixed (100% frequency) in all cat populations. ERV-DC16 was detected at a frequency of 18% in European wildcats, whereas it was fixed in domestic cats (Fig. 1E).
Next, we performed a PCR analysis to detect ERV-DC in a genotype-specific manner. As shown in Fig. 1F, ERV-DCs of genotypes I and II were detected in all European wildcats. However, ERV-DC6 (genotype III) was detected in only one wildcat (wildcat 54), which was also positive for ERV-DC14 (genotype I). These results demonstrated that ERV-DC was present in European wildcats and that the ERV-DC insertional polymorphic pattern was quite variable among European wildcats. ERV-DCs of genotypes I and II were fixed in all wildcats, but ERV-DC of genotype III had a limited spread.
Cloning of ERV-DC14 from European wildcats and analysis of viral replication.
Of the replication-competent proviruses (ERV-DC10, -DC14, and -DC18), ERV-DC14 was the only one detected at a high frequency in European wildcats. Thus, we attempted to isolate the ERV-DC14 provirus to compare the properties of ERV-DC14 between domestic cats and European wildcats. We successfully amplified two full-length ERV-DC14 proviruses from European wildcats (wildcats 54 and 63) via PCR and then cloned them (ERV-DC14/F. s. silvestris/wildcat54 and ERV-DC14/F. s. silvestris/wildcat63, respectively) and determined their direct sequences. Two single nucleotide polymorphism (SNPs) were found between these two clones, including G4367A and C4633T (data not shown). In this study, we use the terms ERV-DC14/F. s. silvestris and ERV-DC14 to refer to the clones amplified from European wildcat 63 and Japanese domestic cat SO38, respectively. Sequence analysis indicated that intact full-length open reading frames (ORFs) for all ERV-DC14/F. s. silvestris genes (gag, pol, and env) were present. We observed a difference of 7 nucleotides compared with the sequence of ERV-DC14 from domestic cats (Fig. 2A). Next, we determined if ERV-DC14/F. s. silvestris could replicate in cultured cells by infection of fresh HEK293T cells. As shown in Fig. 2B, all tested ERV-DC14 clones from Japanese domestic cats (cats SO38, GM21, IK19, and FO16) could infect HEK293T cells, and their viral titers were approximately 102 to 103 infectious units per ml. In contrast, ERV-DC14/F. s. silvestris clones (clones 54 and 63) could not infect HEK293T cells. Thus, unlike ERV-DC14 from domestic cats, ERV-DC14/F. s. silvestris from European wildcats was replication incompetent.
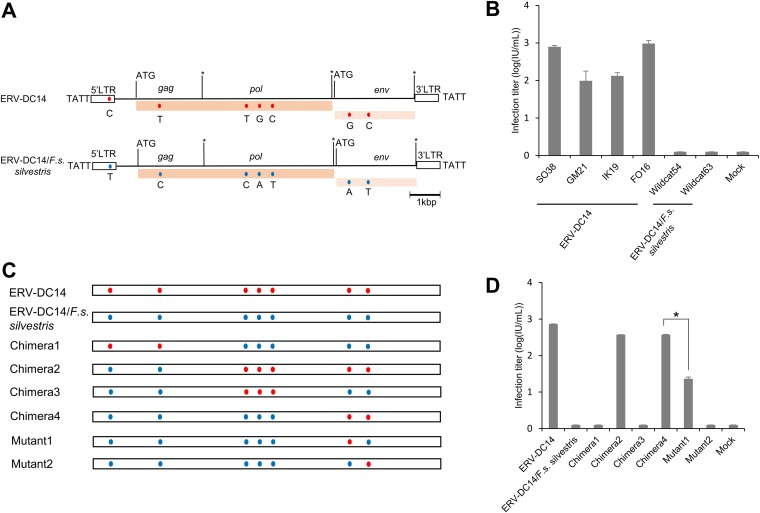
FIG 2.
Characterization and assessment of ERV-DC14/F. s. silvestris. (A) Schematic image of the full-length ERV-DC14/F. s. silvestris provirus. The ERV-DC14 clone SO38 strain was used as the ERV-DC14 reference genome. The gag, pol, and env genes are illustrated together with the 5ʹ and 3ʹ LTRs and the positions of the gag and env translational initiation codons (ATG). Asterisks, stop codons; dark pink box, open reading frame (ORF) of the Gag-Pol polyprotein; light pink box, Env protein; blue and red round circles, single nucleotide polymorphisms (SNPs) between the two proviruses ERV-DC14/SO38 and ERVDC14/F. s. silvestris. Nucleotide substitutions are shown. Flanking 4-bp target duplicate site (TSD) sequences are shown for each provirus. (B) Assessment of the replication-competent activity of ERV-DC14 in European wildcats (F. s. silvestris) and Japanese domestic cats. All tested proviral clones, including ERV-DC14 from different Japanese domestic cats (SO38, GM21, IK19, FO16), ERV-DC14/F. s. silvestris (wildcat 54, and wildcat 63), or the empty vector (mock transfection), were transfected into 293Lac cells, and then their infectivity was tested with fresh HEK293T cells. The viral titers are illustrated as the log number of infectious units (IU) per milliliter with standard deviations. (C) Schematic representation of the chimeric structures of the two proviruses. Chimera1 to Chimera4 were constructed via recombination between the two proviruses using restriction enzyme digestion, and the two mutants were constructed by site-directed mutagenesis. Blue and red round circles indicate single nucleotide polymorphisms (SNPs) between the two proviruses ERV-DC14/SO38 and ERVDC14/F. s. silvestris. (D) Assessment of the replication competence of chimeric ERV-DC14. 293Lac cells were transfected with plasmids containing different chimeric ERV-DC14s or mock transfected (with the empty vector), and the resulting supernatants were collected and used to infect fresh HEK293T cells. The viral titers are illustrated as the log number of infectious units (IU) per milliliter with standard deviations. *, P < 0.0001 (one-way ANOVA).
Identification of the mutation responsible for the replication incompetence of ERV-DC14/F. s. silvestris.
We next investigated potential reasons for the replication incompetence of the ERV-DC14/F. s. silvestris provirus. Sequence analyses indicated that several nucleotide differences existed in the 5ʹ LTR and the gag, pol, and env genes between ERV-DC14 from domestic cats and ERV-DC14/F. s. silvestris (Fig. 2A). We constructed four chimeric full-length proviruses (Chimera1, Chimera2, Chimera3, and Chimera4), consisting of ERV-DC14 and ERV-DC14/F. s. silvestris sequences (Fig. 2C). Next, we tested whether these four chimeric proviruses were infectious by infection of fresh HEK293T cells. As shown in Fig. 2D, Chimera2 and Chimera4 exhibited viral infectivity, whereas the other chimeras were not infectious. Thus, the env gene that contained two nucleotide differences (at nucleotide positions 6735 and 7110) could be responsible for the replication incompetence of ERV-DC14/F. s. silvestris.
Next, Mutant1 and Mutant2 were constructed (Fig. 2C and Fig. 3A), and their infectivity was tested in fresh HEK293T cells. Mutant1 showed viral infectivity, but Mutant2 did not (Fig. 2D). This result demonstrates that 148K in ERV-DC14/F. s. silvestris Env is critical for the infectivity of the proviruses. Notably, the viral titer of Mutant1 was significantly different from that of Chimera4 (P < 0.0001). To clarify the mechanism of Env dysfunction, we tested the infectivity of pseudotyped viruses produced from cells transfected with Env expression plasmids carrying the Env of ERV-DC14, ERV-DC14/F. s. silvestris (wild-type [WT] Env), Mutant1 (ERV-DC14/F. s. silvestris/K148E [K148E Env]), and Mutant2 (ERV-DC14/F. s. silvestris/S273P [S273P Env]) against fresh HEK293T cells. As shown in Fig. 3B, both the K148E Env-pseudotyped virus and the ERV-DC14 Env-pseudotyped virus from domestic cats could efficiently infect cells, and their titers (103.4 and 104, respectively) were significantly different (P < 0.0001). In contrast, neither the ERV-DC14/F. s. silvestris nor S273P Env-pseudotyped virus was able to infect HEK293T cells. These findings confirm that 148K in Env is responsible for the replication dysfunction of the ERV-DC14/F. s. silvestris provirus.
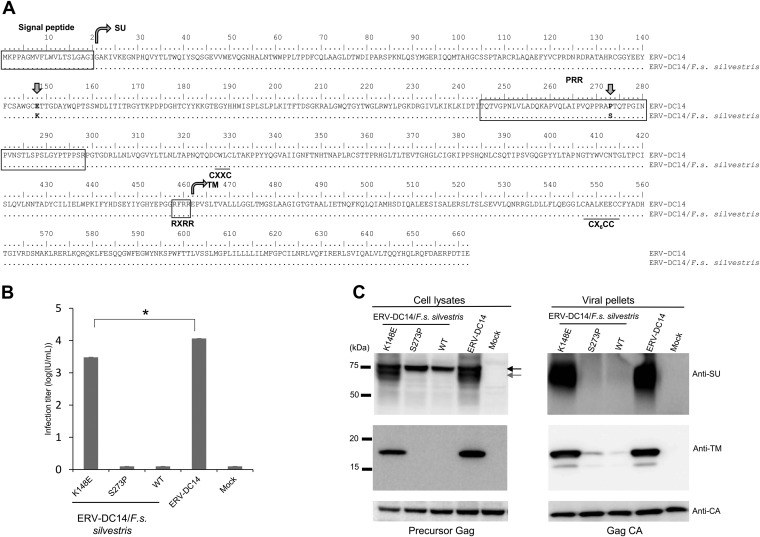
FIG 3.
Determination of the mutation responsible for ERV-DC14/F. s. silvestris Env dysfunction. (A) Amino acid sequence alignment of Env proteins of ERV-DC14 and ERV-DC4/F. s. silvestris. SU, surface subunit; PRR, proline-rich region; TM, transmembrane subunit. RXRR is the cleavage motif. CXXC and CX6CC are sites of covalent interaction. Arrows indicate the positions of amino acids 148 and 273, which differ between these two ERV-DC14 proviruses. (B) Assessment of Env-pseudotyped viruses based on the ERV-DC14/F. s. silvestris wild type (WT), Mutant1 (K148E), and Mutant2 (S273P) or on ERV-DC14. GPLac cells were transfected with the indicated Env expression plasmids. The corresponding Env-pseudotyped viruses were used to infect fresh HEK293T cells. The viral titers are illustrated as the log number of infectious units (IU) per milliliter with standard deviations. *, P < 0.0001 (one-way ANOVA). (C) Western blotting of GPLac cells expressing ERV-DC14/F. s. silvestris Env (K148E, S273P, or WT) or ERV-DC14 Env. The cell lysates and viral pellets from culture supernatants were analyzed. A goat polyclonal anti-FeLV SU (gp70) antibody was used to detect ERV-DC14 SU, and a mouse monoclonal anti-FeLV TM protein (p15E) antibody was used to detect the ERV-DC14 TM protein. The black arrow indicates immature SU; the gray arrow indicates mature SU. Precursor Gag (Pr65) and Gag CA (p30) were both detected with a goat anti-Raucher MLV CA antibody.
Mechanism of Env dysfunction of ERV-DC14/F. s. silvestris.
Using a goat polyclonal anti-FeLV surface glycoprotein (SU) antibody that detects ERV-DC Env, we conducted a Western blot analysis in GPLac cells that had been transfected with one of the Env expression vectors (ERV-DC14, ERV-DC14/F. s. silvestris, K148E, or S273P). As shown in Fig. 3C (top left), Env proteins were detected in cells transfected with either the ERV-DC14 or the K148E Env expression vector as multiple bands of approximately 75 kDa and 70 kDa (representing the precursor and mature SU protein, respectively). In contrast, cells transfected with the ERV-DC14/F. s. silvestris or S273P Env expression vector produced only a single 75-kDa band corresponding to the Env protein. Both the ERV-DC14/F. s. silvestris and S273P Env expression proviruses were highly expressed in cells.
These findings suggest that the ERV-DC/F. s. silvestris Env protein, which consists of the SU and transmembrane (TM) protein, had a cleavage dysfunction. Thus, we looked for antibodies that cross-reacted with the ERV-DC14 TM protein. Among seven monoclonal antibodies against the FeLV TM protein, we found two suitable monoclonal antibodies (PF6J-2A and EC6-6B1) which also cross-reacted with the FeLV TM protein (data not shown). Using the anti-FeLV TM protein antibody, a Western blot analysis was conducted, and a TM protein of approximately 17 kDa was detected as a single band in cells transfected with either the ERV-DC14 or the K148E Env expression vector. In contrast, the TM protein was not detected in cells expressing either ERV-DC14/F. s. silvestris or S273P Env (Fig. 3C, middle left). These results suggest that ERV-DC14/F. s. silvestris Env is not cleaved into the SU and TM proteins. Therefore, we again used two antibodies, anti-FeLV SU and anti-FeLV TM, to detect Env proteins in viral pellets prepared by ultracentrifugation of the supernatants (Fig. 3C, top and middle right). The Env SU protein (approximately 70 kDa) and the Env TM protein (approximately 17 kDa) were detected in the viral pellets of ERV-DC14 and K148E Env-pseudotyped viruses, whereas we failed to detect these proteins or even visible viral pellets of ERV-DC14/F. s. silvestris and S273P Env-pseudotyped viruses. Gag proteins were detected using a goat anti-Raucher MLV CA antibody and detected bands representing the Gag precursor (Pr65; approximately 65 kDa) in cell lysates or the Gag CA protein (p30; approximately 30 kDa) in cell supernatants (Fig. 3C, bottom). These Env proteins did not participate in the production of infectious viral particles and were not incorporated into virions, even though they were highly expressed in the cultured cells. These results indicate that the dysfunction of ERV-DC14/F. s. silvestris was caused by defects in the cleavage of the Env protein and that infectious viral particles were not produced from cells exposed to ERV-DC14/F. s. silvestris due to the nonfunctionality of Env.
ERV-DC14/F. s. silvestris Env localizes on the cell surface.
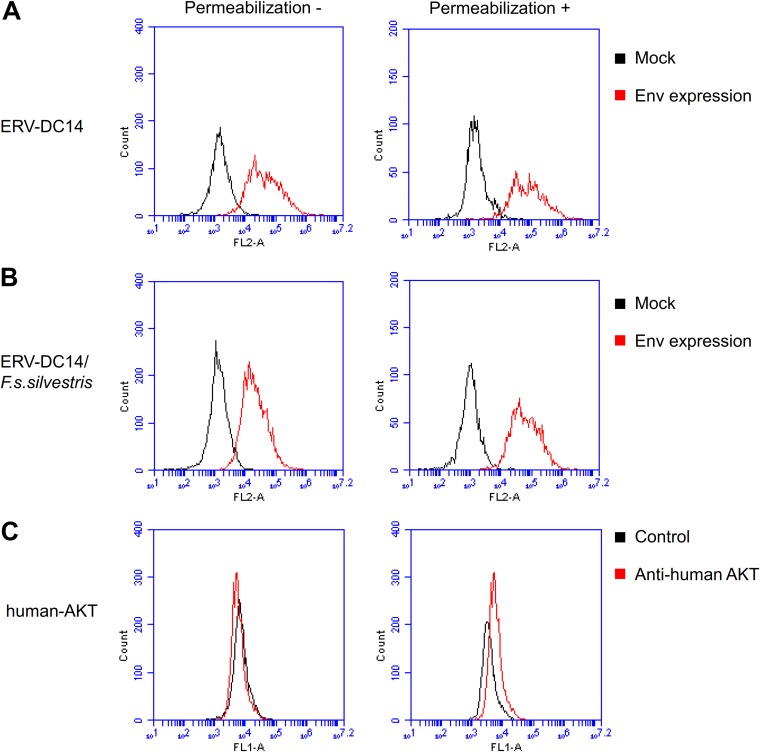
To better understand the mechanism of Env dysfunction, the subcellular localization of ERV-DC14/F. s. silvestris Env was investigated by flow cytometry. As shown in Fig. 4A and B, ERV-DC14- and ERV-DC14/F. s. silvestris Env-expressing cells had higher signals than the mock-transfected cells in both permeabilized (for detection of protein in the cytoplasm) and nonpermeabilized (for detection of protein on the cell surface) samples. These results suggest that ERV-DC14/F. s. silvestris Env is transported to and expressed on the cell surface. To confirm the results of this assay, we detected the serine/threonine-protein kinase (AKT) present in the cytoplasm in both permeabilized and nonpermeabilized samples from mock-transfected cells. The permeabilized samples had higher fluorescent signals than the control samples (without primary antibody) samples, whereas the nonpermeabilized samples had fluorescent signals similar to those of control (Fig. 4C).
FIG 4.
Flow cytometry analysis of ERV-DC14/F. s. silvestris cell surface expression. (A and B) Detection of Env on the interior and exterior of cells. HEK293T cells expressing ERV-DC14 Env (A) or ERV-DC14/F. s. silvestris Env (B) were permeabilized with 0.2% Triton X-100 in PBS (right) or were not permeabilized (left), and intracellular (right) and cell surface (left) Env proteins were stained with goat anti-FeLV SU (gp70) and phycoerythrin (PE)-conjugated anti-goat IgG antibody. Fluorescent signals were detected using the FL-2 channel of a flow cytometer. Histograms of Env-expressing cells (red) and mock-transfected cells (black) are overlaid in each graph. The x axis shows the signal intensity in FL-2; the y axis shows the cell counts. (C) Staining of human AKT in permeabilized (right) or nonpermeabilized (left) samples of mock-transfected cells using rabbit anti-human AKT and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG antibodies. Histograms of cells treated with anti-human-AKT antibody (red) and not treated with antibody (black) are overlaid in each graph. The x axis shows the signal intensity in FL-1; the y axis shows the cell counts.
Mutations in the ERV-DC14 env gene among European wildcats.
The results presented above indicated that ERV-DC14 is highly prevalent in European wildcats but not in domestic cats from Japan or Spain (Fig. 1). We next investigated whether or not the ERV-DC14 env gene mutations were evolutionally conserved in European wildcats and domestic cats. Sequence analyses showed that all domestic cats in Japan that were positive for ERV-DC14 (n = 6) displayed the 6735G (148E in Env) and 7110C (273P in Env) polymorphisms in the env gene. In contrast, all European wildcats that were positive for ERV-DC14 (n = 9) displayed the G6735A (E148K in Env) and C7110T (P273S in Env) polymorphisms in ERV-DC14 (Table 1). Three of the four ERV-DC14-positive domestic cats from Spain had the same sequences in the ERV-DC14 env gene as Japanese domestic cats, whereas the other ERV-DC14-positive domestic cat in Spain (cat 317) had the same ERV-DC14 env mutations as European wildcats. These results suggest that both ERV-DC14 phenotypes (i.e., an active ERV-DC14 encoding 148E and 273P in Env that is mainly present in the domestic cat population and an inactive ERV-DC14 encoding 148K and 273S in Env that is abundantly distributed in the European wildcat population) are present in cat populations.
TABLE 1.
Properties of the ERV-DC14 provirus in different cat populationsa
| Cat species | Sample size (no. of cats) | No. of cats whose provirus had the following characteristics: |
||
|---|---|---|---|---|
| ERV-DC14 (+) | ERV-DC14 env gene |
|||
| 6735A/148K and 7110T/273S | 6735G/148E and 7110C/273P | |||
| F. s. silvestris | 11 | 9 | 9 | 0 |
| European F. s. catus | 35 | 4 | 1 | 3 |
| Japanese F. s. catus | 247 | 6 | 0 | 6 |
(+), ERV-DC14 genotyping positive; 6735 and 7710, nucleotide positions of ERV-DC14; 148 and 273, amino acid positions of Env; A, G, T, and C are nucleotides, and K, E, S, and P are the corresponding amino acids.
Identification of specific mutations in FeLV and murine ERV corresponding to the ERV-DC14 Env 148K mutation.
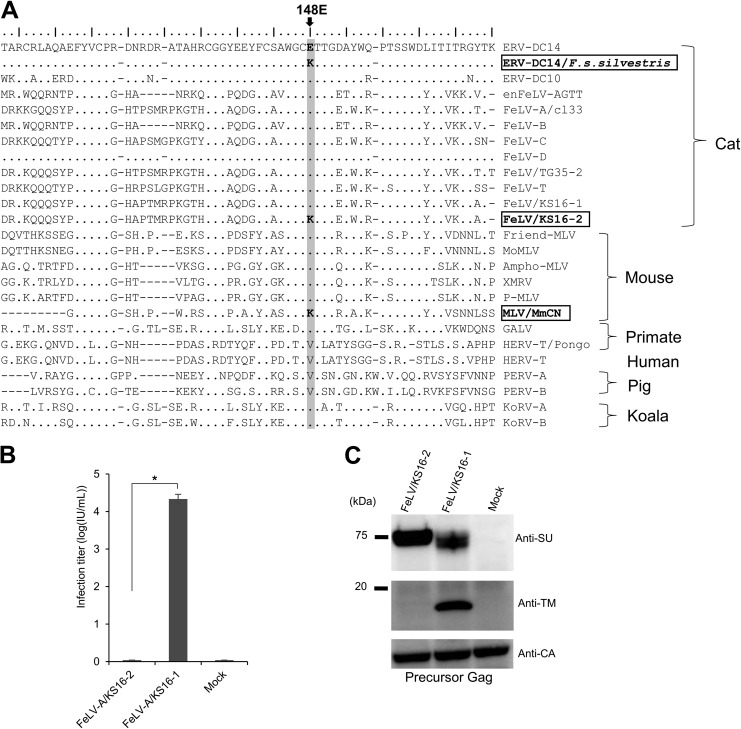
To identify specific mutations corresponding to the 148K mutation in the SU N-terminal domain of ERV-DC14 Env, we next analyzed the sequences of gammaretroviruses in different species, including our previous data on the major FeLV strains circulating in Japan (43). As shown in Fig. 5A, we identified two virus sequences bearing this specific mutation. One is the Mus musculus castaneus endogenous virus (MLV/MmCN). The other was the FeLV from cat KS16, a 5-year-old, neutered male with no history of FeLV vaccination; this animal presented with dyspnea and was diagnosed with thymic lymphoma. Two FeLV env variants isolated from peripheral blood mononuclear cell DNA of Japanese cat KS16 (43) showed single nucleotide changes at position 148, resulting in an E residue in FeLV/KS16-1 Env and a K residue in FeLV/KS16-2 Env (Fig. 5A).
FIG 5.
Sequence analysis of gammaretrovirus Env proteins and dysfunction of a FeLV variant induced by a single mutation in the SU N-terminal domain. (A) Sequence alignment of Env in gammaretroviruses, constructed by use of the mafft tool (70). The amino acid sequences of the gammaretrovirus SU N-terminal regions are also presented. The critical amino acid position 148E is shaded in light gray. The GenBank accession numbers of the reference sequences are as follows: BBL19108.1 for ERV-DC14/F. s. silvestris, BAM33599.1 for ERV-DC14, BAM33597.1 for ERV-DC10, AY364318.1 for enFeLV-AGTT, BAB63924.2 for FeLV-A clone 33 (cl33), AAA43052.1 for FeLV-B, AAA43049.1 for FeLV-C, BAM33588.1 for FeLV-D, BAU61794.1 for FeLV/TG35-2, AAA43050.1 for FeLV-T, BAK41670.2 for FeLV/KS16-1, BBL19109.1 for FeLV/KS16-2, AAA46480.1 for Friend MLV, NP_057935.1 for MoMLV, AAA46515.1 for Ampho-MLV, ADU55755.1 for XMRV, ARB03464.1 for P-MLV, AMK06448.1 for MLV/MmCN, AAA46811.1 for GALV, CAI15393.1 for HERV-T/Pongo, XP_011526770.1 for HERV-T, AAQ83899.1 for PERV-A, BAM67147.1 for KoRV-A, and AGO86848.1 for KoRV-B. (B) Infection assay using pseudotyped viruses of two FeLV variants (KS16-1 and KS16-2). GPLac cells were transfected with Env expression plasmids for FeLV/KS16-1 and FeLV/KS16-2. The filtered viral supernatants were used to infect fresh HEK293T cells. The viral titers are illustrated as the log number of infectious units (IU) per milliliter with standard deviations. *, P < 0.0001 (one-way ANOVA). (C) Immunoblotting analysis using cell lysates from GPLac cells transfected with the Env expression plasmids which are presented in panel B. Env proteins were detected by use of a mouse anti-FeLV SU (gp70) antibody and an anti-FeLV TM protein (p15E) antibody. Precursor Gag (Pr65) was detected with a goat anti-Raucher MLV CA antibody. The filter exposure time differed for each antibody.
To determine the infectivity of the FeLV/KS16-2 variant, we tested the infection of fresh HEK293T cells by pseudotyped viruses produced by transfection of cells with Env expression plasmids harboring the Env of the FeLV/KS16-1 variant (as a positive control), the FeLV/KS16-2 variant, and the empty vector (mock transfection). The FeLV subgroup A (FeLV-A)/KS16-2 variant was unable to infect cells, whereas the FeLV/KS16-1 variant successfully infected cells (Fig. 5B), consistent with a previous report (43). The transfected cells were analyzed by Western blotting with specific anti-FeLV SU and TM antibodies. Both the mature SU protein (approximately 70 kDa) and TM protein (approximately 17 kDa) were detected in the FeLV/KS16-1 variant. However, only the precursor SU protein was detected in the FeLV/KS16-2 variant (the TM protein was not detected) (Fig. 5C). The precursor Gag (Pr65) protein was detected in all samples. These results reveal that the specific mutation causing Env dysfunction and viral inactivation occurred not only in ERVs but also in exogenous retroviruses.
Mutational analysis of 148E within the SU N-terminal domain of Env conserved among gammaretroviruses.
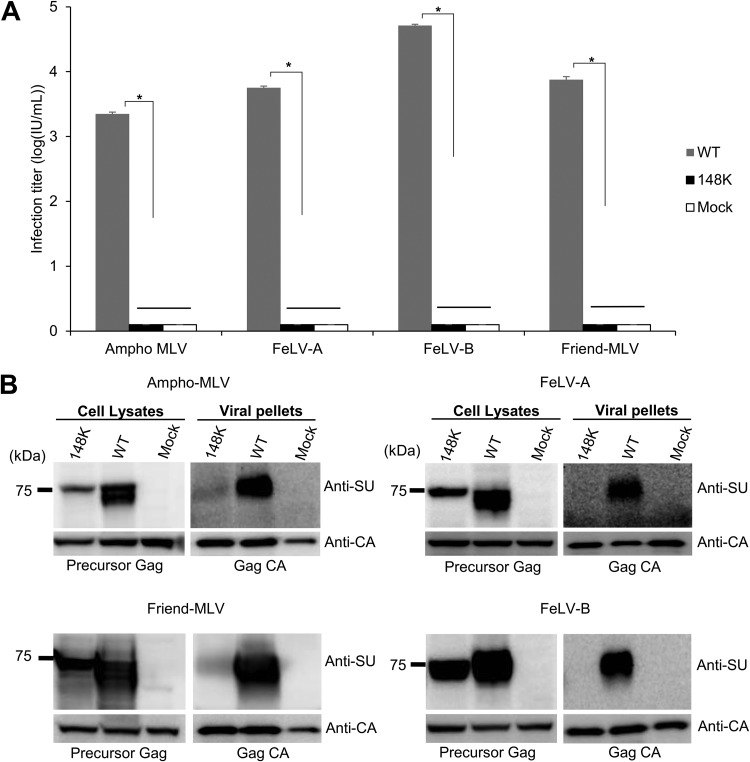
The 148E residue in ERV-DC14 Env is mainly conserved within gammaretroviruses (Fig. 5A). To determine if this mutation causes Env dysfunction in other gammaretroviruses, we constructed Env expression plasmids for amphotropic murine leukemia virus (Ampho-MLV) (4070A), Friend MLV (clone57), FeLV-A (FeLV clone 33), and FeLV-B (Gardner-Arnstein) bearing the E148K mutation and tested their infectivity. Ampho-MLV, FeLV-A, FeLV-B, or Friend MLV Env-pseudotyped viruses bearing the E148K mutation were unable to infect fresh HEK293T cells or Mus dunni tail fibroblast (MDTF) cells, whereas wild-type (WT) Ampho-MLV, FeLV-A, FeLV-B, or Friend MLV Env-pseudotyped viruses successfully infected HEK293T cells and MDTF cells (Fig. 6A). Western blotting with a specific anti-SU antibody detected two bands or a broad band of Env proteins from GPLac cells transfected with Ampho-MLV, Friend MLV, FeLV-A, or FeLV-B Env (WT) expression plasmids, whereas only a single band was detected in cells transfected with any of the Env-pseudotyped viruses bearing the E148K mutation (Fig. 6B), even though these Env-pseudotyped viruses were highly expressed in cells. The precursor Gag (Pr65) protein was detected using an anti-MuLV CA antibody in cell lysates as a control.
FIG 6.
Dysfunction caused by mutations within the SU N-terminal domain in gammaretroviruses. (A) Infection assay using pseudotyped viruses of WT or mutant Env (E148K) of FeLV-A clone 33, FeLV-B GA, Ampho-MLV 4070A, and Friend MLV clone 57. Expression plasmids were constructed for Env mutants and expressed in GPLac cells. After 72 h, the cell lysates and viral pellets were harvested from the culture supernatants. Fresh MDTF cells were inoculated with viral supernatants of Friend MLV, and fresh HEK293T cells were inoculated with viral supernatants of the other pseudotyped viruses. After 48 h, the X-Gal-positive cells were counted, and the viral titers are illustrated as the log number of infectious units (IU) per milliliter with standard deviations; *, P < 0.0001 (one-way ANOVA). (B) Immunoblotting analysis of cell lysates (left) and viral pellets (right) from GPLac cells expressing wild-type (WT) Env or Env mutants. FeLV-A Env and FeLV-B Env were detected with an anti-FeLV SU (gp70) antibody, Ampho-MLV Env was detected with an anti-Ampho-MLV SU (gp70) antibody, and Friend MLV Env was detected with an anti-MLV SU (gp70) antibody. Precursor Gag (Pr65) and Gag CA (p30) were both detected with a goat anti-Raucher MLV CA antibody. The filter exposure times differed between the cell lysates and viral pellets.
Virus pellets prepared by ultracentrifugation of the supernatants of transfected cells were analyzed by Western blotting with a specific anti-SU antibody. The Gag CA protein was detected, using goat anti-MLV CA antibody, in cell supernatants from all samples. Env SU proteins were detected in the viral pellets of Ampho-MLV, Friend MLV, FeLV-A, and FeLV-B Env-pseudotyped viruses, whereas these proteins were not detected in viral pellets of FeLV-A and FeLV-B Env-pseudotyped viruses bearing the E148K mutation or were detected as only a faint band in viral pellets of Ampho-MLV and Friend MLV Env-pseudotyped viruses bearing the E148K mutation (Fig. 6B). The mutant Env proteins did not participate in the production of infectious viral particles and were not incorporated into the virions, even though they were highly expressed in cultured cells. These results indicate that the E148K mutation caused the defect in Env cleavage in Ampho-MLV, Friend MLV, FeLV-A, and FeLV-B and that infectious viral particles were not produced from cells transfected with pseudotyped viruses bearing the E148K mutation. Additionally, these findings suggest that the critical amino acid substitution of E148K within the SU N-terminal domain caused in other gammaretroviruses the same dysfunctions observed in ERV-DC14/F. s. silvestris.
Analysis of Refrex-1 in European wildcats.
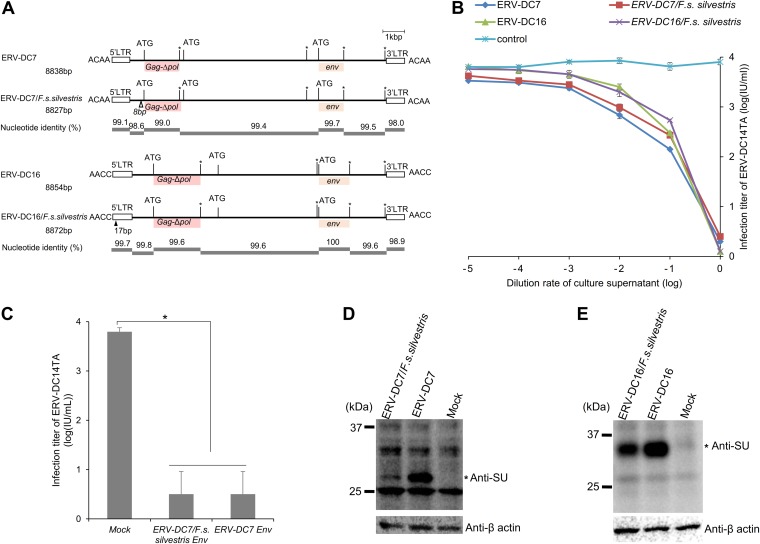
ERV-DC7 is fixed in both European wildcats and domestic cats, whereas ERV-DC16 is fixed only in domestic cats (Fig. 1E). To determine whether Refrex-1 was evolutionally conserved in European wildcats, we isolated the full-length ERV-DC7 and ERV-DC16 proviruses, termed ERV-DC7/F. s. silvestris and ERV-DC16/F. s. silvestris, respectively, from European wildcats. We found that these two proviruses had defective ORFs encoding gag, pol, and env, similar to ERV-DC7 and ERV-DC16 from domestic cats (31). However, SNPs, deletions, or insertions existed when the ERV-DC7 and ERV-DC16 sequences of European wildcats and domestic cats were compared (Fig. 7A). Next, the Refrex-1 activities of ERV-DC7/F. s. silvestris and ERV-DC16/F. s. silvestris were analyzed. The inhibition assay indicated that both the ERV-DC7/F. s. silvestris and the ERV-DC16/F. s. silvestris proviruses specifically inhibited the infection of ERV-DC14TA in a dose-dependent manner, showing Refrex-1 activity similar to that of ERV-DC from domestic cats (Fig. 7B).
FIG 7.
Analysis of Refrex-1 in European wildcats. (A) Schematic structure of ERV-DC7 and ERV-DC16 in domestic cat and European wildcats. The gag, deletion of pol (Δpol), and env genes are illustrated together with the 5ʹ and 3ʹ LTRs and the positions of the gag and env translational initiation codons (ATG). Asterisks, stop codons; dark pink boxes, predicted Gag; light pink boxes, truncated Env protein; black triangle, insertions; white triangle, deletions. Flanking 4-bp target duplicate site (TSD) sequences are shown for each provirus. (B) Dose-dependent inhibitory effect of Refrex-1 on viral infection. The supernatants of HEK293T cells transfected with each provirus (ERV-DC7, ERV-DC7/F. s. silvestris, ERV-DC16, ERV-DC16/F. s. silvestris, or the empty vector as a control [mock transfection]) were diluted and added to fresh HEK293T cells. After removing the supernatants, those cells were infected with the replication-competent ERV-DC14TA virus. X-Gal-positive cells were counted, and viral titers were calculated as the log number of infectious units (IU) per milliliter with standard deviations. (C) Inhibitory effect on viral infection of the Refrex-1 encoded by truncated env from ERV-DC7 and ERV-DC7/F. s. silvestris. The supernatants of HEK293T cells transfected with the indicated expression vectors encoding ERV-DC7 Env or ERV-DC7/F. s. silvestris Env or with the empty vector (mock transfection) were added to fresh HEK293T cells. After removing the supernatants, the cells were challenged with replication-competent ERV-DC14TA. X-Gal-positive cells were counted, and viral titers were calculated as the log number of infectious units per milliliter with standard deviations. (D and E) Detection of Refrex-1 expression in transfected cells. HEK293T cells were transfected with the provirus of ERV-DC7 or ERV-DC7/F. s. silvestris or with the empty vector (mock transfection). At 72 h posttransfection, the lysates were prepared for Western blotting with a polyclonal goat anti-FeLV SU (gp70) antibody. The Refrex-1 protein was detected as bands of ∼28 kDa for ERV-DC7 and of ∼32 kDa for ERV-DC16. The asterisk indicates the Refrex-1 protein. Human anti-β-actin antibody was used as an internal control.
To confirm if the truncated Env proteins conferred Refrex-1 activity, as reflected by the absence of differences in the Refrex-1-coding region between ERV-DC16 from the domestic cat and ERV-DC16/F. s. silvestris (Fig. 7A), we performed experiments with the Env expression vector ERV-DC7/F. s. silvestris Env. This expression vector encodes an Env bearing a difference of two amino acids in comparison with the sequence of ERV-DC7 from domestic cats. The supernatants of cells transfected with the Env expression plasmids of either ERV-DC7 or ERV-DC7/F. s. silvestris also specifically inhibited the infection of ERV-DC14TA (Fig. 7C). Western blotting of those cell lysates was conducted using anti-FeLV SU antibody. The Refrex-1 protein was detected as bands of ∼28 kDa in size for ERV-DC7 and of ∼32 kDa in size for ERV-DC16 from both domestic cat and wildcat sources. Interestingly, the amount of Refrex-1 from ERV-DC7/F. s. silvestris was slightly lower than that from ERV-DC7 (Fig. 7D). In addition, we also investigated whether Refrex-1 from the ERV-DC7/F. s. silvestris and ERV-DC16/F. s. silvestris proviruses could inhibit FeLV-D infection; the inhibition assay results showed that Refrex-1 expressed from European wildcats could also inhibit the pseudotyped FeLV-D/TY2.0 virus, and the viral titers decreased from 104.5 (mock-transfected cells) to 102 (data not shown). These results are consistent with those for Refrex-1 from domestic cats (31). Our data suggest that the genes for truncated Env from both ERV-DC7/F. s. silvestris and ERV-DC16/F. s. silvestris encode Refrex-1.
Sequence diversity of ERV-DC between domestic cats and wildcats.
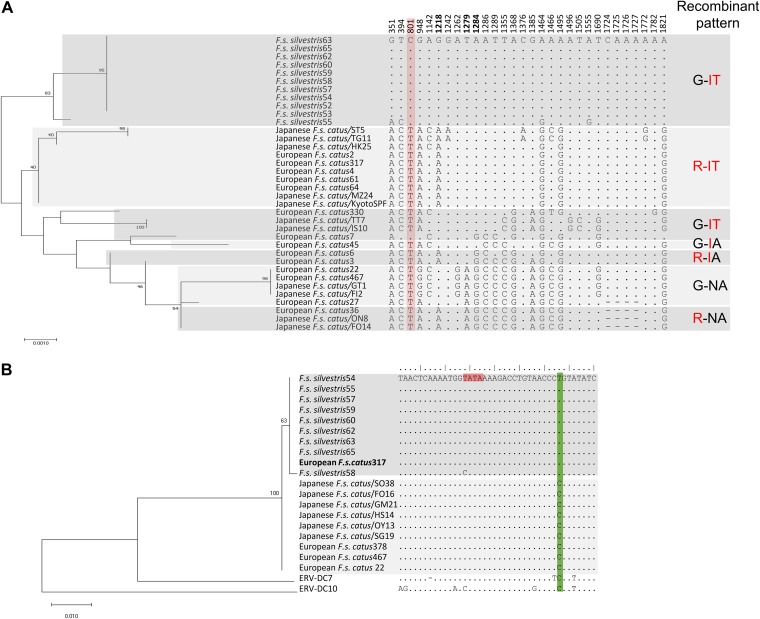
We previously ascertained the sequence diversity of ERV-DC7 env in Japanese domestic cats (37). In the present study, we investigated the sequence diversity of ERV-DC7 env in both European wildcats and domestic cats. The phylogenetic tree constructed from ERV-DC7 env sequences shows the evolutionary diversity of this provirus in each cat population (Fig. 8A). We identified a total of 16 alleles of ERV-DC7 env, including 9 alleles that were newly identified in this study. All of the alleles had the same stop codon mutation in the middle of env and encoded a truncated Env as Refrex-1. Among the 30 SNPs observed in European wildcats and Spanish domestic cats, two new SNPs were identified. These two SNPs (at nucleotide positions 1555 and 1782) caused nonsynonymous substitutions in ERV-DC7 env via deletion of a stop codon mutation. The nucleotide sequences of the ERV-DC7/F. s. silvestris env gene were mostly conserved among European wildcats, with the exception of one European wildcat (European wildcat 55), which showed four nucleotide differences from the sequences of the other European wildcats. The genetic diversity of the ERV-DC7 env sequences suggested that wildcats form a clade genetically different from that of domestic cats. Additionally, based on analyses of the amino acid sequence of defective ERV-DC7fl Env at positions 407, 427, and 429, we found that the combination 407G, 427I, and 429T was conserved in European wildcats. In contrast, the six combination variants (R-IT, G-IT, G-IA, R-IA, G-NA, and R-NA) existed only in domestic cats (Fig. 8A).
FIG 8.
Sequence diversity of ERV-DC in wildcats and domestic cats. (A) Sequence diversity of ERV-DC7 env in wildcats and domestic cats. The ERV-DC7 env sequences from 11 European wildcats were determined. A phylogenetic tree of the ERV-DC7 env sequences was constructed using maximum likelihood methods (left). The percentages at the branch junctions indicate their bootstrap values (1,000 replicates). Excerpts of polymorphic sites from the ERV-DC7 env sequence are shown in the middle. The numbers indicate nucleotide positions, and bold numbers indicate the nucleotide positions causing amino acid substitutions at positions 407, 427, and 429 (corresponding to R, I, and T, respectively) which cause defective Env cleavage in ERV-DC7fl. The nucleotide change generating a latent stop codon in domestic cats is shaded in pink. On the right, the sequence variations at positions 407 and 427 to 429 are shown. Amino acids suppressing Env cleavage are in red. (B) Phylogenetic and sequence analysis of ERV-DC14 based on the 5ʹ LTR. A phylogenetic tree of the ERV-DC14 5ʹ LTR in three different cat populations was constructed using maximum likelihood methods (left). The percentages at the branch junctions indicate their bootstrap values (1,000 replicates). The nucleotide sequences of the 5ʹ LTR are also presented (right). The position of the TATA box is shaded in pink, and the green shading indicates nucleotide determinants distinguishing wildcats and domestic cats.
Because LTR sequences may be involved in the integration and transcription of viral DNA, we were interested in whether ERV-DC14 LTRs had evolved in different cat populations. The 5ʹ LTRs of ERV-DC14 from different cat populations were amplified, and their sequences were determined. Figure 8B shows the phylogenetic tree constructed based on the ERV-DC14 5ʹ LTRs from different cat populations. These data showed that, based on the 5ʹ LTR sequences of ERV-DC14 proviruses, two cat populations exist: wildcats and domestic cats. Sequence analyses indicated that the 5ʹ LTR of ERV-DC14 is mainly conserved between these two cat populations. Only one nucleotide difference was identified between wildcats (T) and domestic cats (C) (Fig. 8B). Interestingly, one wild cat (cat 58) displayed a mutation (underlined) in the TATA box (TATA to CATA), and this mutation was also observed in the replication-competent ERV-DC10 and ERV-DC18 proviruses (Fig. 8B). In addition, we also analyzed the sequence diversity of the 3ʹ LTRs of ERV-DC14 in European wildcats (n = 2) and Japanese domestic cats (n = 4). The 3ʹ LTR of ERV-DC14 was conserved between these two cat populations (data not shown).
DISCUSSION
Here, we identified ERV-DC/F. s. silvestris in the European wildcat (F. s. silvestris), which is evolutionally close to the domestic cat. Our study detected four ERV-DC loci (ERV-DC14, -DC6, -DC7, and -DC16) in wildcats and 11 ERV-DC loci in domestic cats in Spain (Fig. 1). Several ERV-DC loci were less prevalent in European wildcats than in domestic cats (Fig. 1). Although two European wildcats (cats 52 and 53) were negative for ERV-DC14 (genotype I) and showed no evidence of any other known genotype I ERV-DC, genotype-specific PCR still amplified a fragment of genotype I ERV-DC. This finding may suggest the existence of an unknown genotype I ERV-DC locus in European wildcats (Fig. 1F). Furthermore, genotype-specific PCR also suggested that genotype I and genotype II ERV-DCs have invaded European wildcats, whereas genotype III ERV-DCs have not. As our previous results demonstrated, genotype I and genotype II ERV-DCs use the same receptor for viral entry, while genotype III ERV-DC uses a distinct receptor (37). Genotype I and genotype II ERV-DCs might have infected the ancestors of wild and domestic cats, whereas genotype III ERV-DC might have more recently infected domestic cats. The phenomenon of retrotransposition of ERV-DC18, which belongs to genotype III, was observed only in Japanese domestic cats and not in European wildcats or European domestic cats (Fig. 1C and D). This result suggests that this phenomenon may be specific to one cat family. ERV-DC14 was found to have significantly invaded European wildcats, where it was detected at a prevalence (80%) higher than that in domestic cats (2.4% and 11.4% in domestic cats from Japan and Spain, respectively) (Fig. 1). This result may be explained by the existence of two different ERV-DC14 phenotypes.
The identification and characterization of replication-defective ERV-DC14/F. s. silvestris indicate that a feline ancestor with an E148K mutation in ERV-DC14 Env probably gave rise to European wildcats, whereas a cat ancestor with a replication-competent version of ERV-DC14 probably gave rise to domestic cats. Figure 1 shows that the integration pattern of ERV-DC in wildcats was distinct from the patterns in European and Japanese domestic cats. One of four European domestic cats (cat 317) had a replication-defective ERV-DC14 (Table 1); the ERV-DC14 locus found in this animal could be derived from European wildcats due to the high potential for interbreeding (44–46). The 5ʹ LTR sequence of ERV-DC14 from this cat showed a single nucleotide polymorphism (C → T), which was observed in ERV-DC14/F. s. silvestris, further supporting this hypothesis (Fig. 8B). These results also suggest that the ERV-DC14 genotype is conserved among all cats, but its phenotype differs between wildcats and domestic cats.
The replication-defective ERV-DC14/F. s. silvestris provirus in European wildcats was found to result from a single G6735A mutation in env that resulted in an E148K substitution in ERV-DC14 Env. The other mutation (C7110T) in env, resulting in a P273S amino acid substitution (Table 1), also might cooperatively contribute to inactivation of the ERV-DC14/F. s. silvestris provirus (Fig. 2D). This lethal mutation may have been caused by apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3 proteins, which are mammal-specific cellular deaminases (47, 48) that play important roles as viral restriction factors (49). Interestingly, the same mutation observed in FeLV/KS16-2 and MLV/MmCN was also due to a G → A substitution, indicating that this phenomenon occurred in ERV-DC14/F. s. silvestris, MLV/MmCN, and FeLV/KS16-2, despite two of these retroviruses being endogenous and the other being exogenous. A reverse genetics technique revealed that a common mutation within the SU domain (E148K) resulted in defective Env cleavage and Env incorporation into virions. The 148E at Env was found to be relatively conserved among gammaretroviruses (Fig. 6), and the substitution of this residue (E148K) in FeLV-A, FeLV-B, Ampho-MLV, and Friend MLV caused the same dysfunctions of Env. This result suggests that the three-dimensional structure and/or folding pathway of the N terminus of SU is generally conserved among gammaretroviruses. Furthermore, our findings also indicate that any exogenous retrovirus, like FeLV-D, that is newly generated in wildcats would potentially be inactivated by this mechanism, although specific ERV-DC loci for FeLV-D generation have not yet been identified. In this study, the same mutation occurring in the SU region of MLV/MmCN resulted in a defective endogenous MLV encoding a truncated Env which functioned as a known restriction factor (Fv-4) in Mus musculus castaneus (36). The Fv-4 env-encoded protein is processed normally and can be incorporated into viral particles but is unable to produce infectious virus (50, 51). Conversely, Refrex-1 is a soluble antiretroviral factor and is not incorporated into viral particles (31).
The Env E148K substitution is close to the N terminus of SU and is distant from the SU-TM protein cleavage motif recognized by furin. One possible mechanism through which the E148K mutation causes Env dysfunction is by altering the protein’s structure in a way that prevents furin from accessing Env. Flow cytometry results indicated that the ERV-DC14/F. s. silvestris provirus affected by this conformational change was unable to be incorporated into viral particles, even though Env was transported to the cell surface (Fig. 4); this mechanism is similar to that of Env dysfunction in human endogenous retrovirus K (HERV-K) (52), ERV-DC7fl, and ERV-DC16fl (37).
The ERV-DC7 and ERV-DC16 loci are fixed in domestic cats, and the presence of Refrex-1 is evidence that ERVs are involved in the coevolution between host and virus (31). Our study found that only ERV-DC7/F. s. silvestris was fixed in all European wildcats, while ERV-DC16/F. s. silvestris was polymorphic in European wildcats (Fig. 1C). We demonstrated that ERV-DC7/F. s. silvestris and ERV-DC16/F. s. silvestris from European wildcats also encoded the antiretroviral factor Refrex-1 (Fig. 7B). The results of our work suggest that Refrex-1 has had an antiviral role throughout Felis evolution, predating the exposure of cats to feline retroviruses. European wildcats still maintain Refrex-1 in the population to avoid the threat of infection by genotype I ERV-DCs and FeLV subgroup D. One interesting observation was that the amount of Refrex-1 in ERV-DC7/F. s. silvestris was slightly lower than that in ERV-DC7 (Fig. 7D), which may be due to the genetic diversity of ERV-DC7s between domestic cats and wildcats.
Many intact ERVs are targeted for transcriptional silencing by modification or mutation of their LTRs, and species-specific ERV LTRs play important roles in regulatory effects during development by modulating the transcriptome (9, 53–56). Here, ERV-DC14/F. s. silvestris probably still had a low level of promoter activity due to a mutation (A → T) in the 5′ LTR, similar to that of ERV-DC14 in domestic cats (9). As shown in Fig. 8B, ERV-DC14/F. s. silvestris from cat 58 had a mutation in the TATA box (TATA → CATA), a sequence which was observed in the replication-competent ERV-DC10 and ERV-DC18 proviruses, suggesting that the LTR was probably not inactivated in this animal.
ERVs are known to have different fates in the host germ line following endogenization (57). ERVs are mainly inactive, acting much like fossil records in both vertebrate and invertebrate animals. Thus, there are very few infectious endogenous retroviruses in mammals (5, 58–62). Replication-competent ERV-DC14 in the domestic cat genome may be mobile and interact with other exogenous retroviruses to generate new recombinant viruses. Any replication-competent retrovirus in the host genome poses a potential risk to the host. Thus, the host will exert negative selection to eliminate deleterious endogenous elements during host-virus coevolution. Our data indicate that inactivated ERV-DC14/F. s. silvestris is potentially being selected during the evolutionary process of European wildcats, whereas this ERV-DC locus has not yet been inactivated in domestic cats. Although ERV-DC14/F. s. silvestris is inactivated, infectious ERV could be generated by recombination events, as reported in mice (63). Moreover, the promoter activity of ERV-DC14 in domestic cats is also low due to a mutation in the 5ʹ LTR (9). In contrast, the frequency of inactive ERV-DC14/F. s. silvestris in European wildcats was high (82%) (Fig. 1C), so the presence of ERV-DC14 in the wildcat genome may play an important role for the host.
Here, a broad sequence analysis of ERV-DC7 from European wildcat and domestic cat populations showed different sequence combinations, and the determinants of ERV-DC7fl env dysfunction were found to consist of six combination variants (G-IT, R-IT, G-IA, R-IA, G-NA, and R-NA), represented in Fig. 8A by different panel patterns. Interestingly, we found that ERV-DC7 env was highly conserved in the wildcat population, with only a single combination (G-IT) being observed in these animals. In contrast, six combination variants of ERV-DC7 were observed in domestic cats. This result suggests that ERVs, in general, and ERV-DC, in particular, genetically diverged during the process of domestication. The sequence diversity of ERV-DCs depends on the locus and the cat subspecies; in particular, ancestral ERV-DC7 may be conserved in European wildcat populations (Fig. 8). As previously described, a reconstructed ERV-DC7/F. s. silvestris full-length env did not contain a latent stop codon (37). Nucleotide position 801 was a conserved C in all European wildcats, whereas in domestic cats, a T nucleotide existed in all cats examined but one (European cat [F. s. catus] 45) (Fig. 8). This result concurred with that of our previous study (37). This latent stop codon suggests that ERV-DC7fl may evolve to a different prototype in domestic cat populations during endogenization.
In conclusion, this study revealed the existence of ERV-DC in the European wildcat population. Notably, this is the first report showing that an infectious endogenous retrovirus is inactivated in a feline species-specific manner. The strategy of tracking infectious ERVs that have invaded cat lineages can reveal the different fates of ERVs and uncover new properties of retroviruses. Our study may contribute to an understanding of the evolution and domestication of cat lineages. Furthermore, our findings provide insights into retroviral pathogenesis and virus-host interactions.
MATERIALS AND METHODS
Cell lines.
HEK293T (64) and MDTF cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 1× penicillin-streptomycin. GPLac cells (10), an env-negative packaging cell line containing a β-galactosidase (LacZ)-coding pMXs retroviral vector, and 293Lac cells (65), containing a LacZ-coding pMXs retroviral vector, were cultured in DMEM supplemented with 10% FCS and 1× penicillin-streptomycin.
Samples.
This study on European wildcats was performed with the permission and support of local authorities (Navarra, Spain, government) to update information on the health status of this population. All samples were collected from the carcasses of European wildcats between 2000 and 2007 and stored frozen at −18°C. The collection of this material did not require the approval of the Ethics Committee for Animal Experimentation because it was considered a routine veterinary practice without planned experimentation. Tissue samples were collected from domestic cats that had died and that were brought to the Department of Animal Pathology, Faculty of Veterinary Medicine, University of Zaragoza, for necropsy. Blood samples were provided by clinical veterinarians with the owners’ permission. Muscle tissues from European wildcats and blood from domestic cats in Spain were used for DNA extraction with a DNeasy blood and tissue kit (Qiagen, Osaka, Japan) or with phenol and chloroform extraction (66).
Cloning of ERV-DC proviruses from European wildcats.
ERV-DC7, ERV-DC14, and ERV-DC16 full-length proviral genomes were amplified from the splenic DNA of European wildcats using different primer pairs (Fe-58S/Fe-42R, Fe-66S/Fe-53R [10], and Fe-219S/Fe-44R [31], respectively). Each full-length DNA fragment was cloned into a pCR4 blunt-TOPO (Invitrogen) vector and sequenced.
PCR.
We used KOD FX Neo (Toyobo, Japan), KOD plus Neo (Toyobo, Japan), and GoTaq (Promega, Madison, WI, USA) polymerases for genotyping of insertional polymorphisms and various cloning procedures. The primers were designed based on unique sequences both outside and inside each ERV-DC provirus to determine ERV-DC haplotypes and are listed in a previous publication (10) and in Table 2.
TABLE 2.
Sequences of the primers used in this study
| Target | Primer orientation | Primer name | Sequence (5′–3′) or reference |
|---|---|---|---|
| ERV-DC14/F. s. silvestris provirus | Forward | Fe-603S | AGTTAAGGGACTGTGGACTT |
| Reverse | Fe-587R | GCTGGGCATTGTTCTCCTTT | |
| pFUΔss DC14 env | Forward | Fe-610S | GGATCCGGATCCCACCATGAAACCCCCAGCGGGAAT |
| Reverse | Fe-184R | GAATTCGAATTCTATTCGATTGTATCTGGCCTTT | |
| ERV-DC14/F. s. silvestris/K148E | Forward | Fe-619S | GCATCGCCAGTAGTCTCGCAGCCCCATGCCG |
| Reverse | Fe-597R | CGGCATGGGGCTGCGAGACTACTGGCGATGC | |
| ERV-DC14/F. s. silvestris/S273P | Forward | Fe-620S | GGTGTCTGAGTTGGGGCCCTTGGTGGC |
| Reverse | Fe-598R | GCCACCAAGGGCCCCAACTCAGACACC | |
| pFUΔss Ampho-MLV env mutant | Forward | Fe-636S | GGTAAATGGGGGTGTAAAACCACCGGACAGG |
| Reverse | Fe-614R | CCTGTCCGGTGGTTTTACACCCCCATTTACC | |
| pFUΔss Friend MLV env mutant | Forward | Fe-637S | CCTCTTGGGGCTGCAAGACAACCGGTAGA |
| Reverse | Fe-615R | TCTACCGGTTGTCTTGCAGCCCCAAGAGG | |
| pFUΔss FeLV-A env mutant | Forward | Fe-638S | GCTGCATGGGGTTGCAAAACTACGGGAGAAG |
| Reverse | Fe-616R | CTTCTCCCGTAGTTTTGCAACCCCATGCAGC | |
| pFUΔss FeLV-B env mutant | Forward | Fe-639S | CTGTATGGGGTTGCAAGACCACCGGGGAA |
| Reverse | Fe-617R | TTCCCCGGTGGTCTTGCAACCCCATACAG | |
| ERV-DC14/pol-3′fla | Forward | Fe-501S | AAGGAATTGCCAAAGGAGTTCTAA |
| Reverse | Fe-587R | GCTGGGCATTGTTCTCCTTT | |
| pFUΔss DC7 env | Forward | Fe-650S | GGATCCGGATCCCACCATGAAACCCCCAACAGGAAT |
| Reverse | Fe-626R | GCGCGAATTCGAATTCTCATTCGATTGTATCTGGCC | |
| ERV-DC14 5′ LTR | Forward | Fe-603S | AGTTAAGGGACTGTGGACTT |
| Reverse | Fe-140R | ACAAAACATAGAACACAATACC | |
| ERV-DC7/F. s. silvestris provirus | Forward | Fe-66S | CCGAAAAMTTCCTGACTGTTTAAGA |
| Reverse | Fe-53R | AGAGGAAATAAACCGGGTAGTGTGT | |
| ERV-DC16/F. s. silvestris provirus | Forward | Fe-219S | GCCACGGTCATGAAAATAAAAA |
| Reverse | Fe-44R | TGCAGACAGAACATACTGTGACAAA |
Construction of chimera proviruses.
We constructed chimera proviruses between ERV-DC14/SO38 (10) and ERV-DC14/F. s. silvestris using both restriction enzymes and site-directed mutagenesis. First, two restriction enzymes, SalI (TaKaRa) and NotI (TaKaRa), were used to make Chimera1 and Chimera2. Second, we used two restriction site pairs (SalI-XhoI and XhoI-NotI) to produce Chimera3 and Chimera4. Finally, Mutant1 (A → G at nucleotide position 6735) and Mutant2 (T → C at nucleotide position 7110), resulting in K148E and S273P changes in ERV-DC14/F. s. silvestris Env, respectively, were constructed using a QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA) following the manufacturer’s protocol. The primer pairs used for site-directed mutagenesis were 619S-597R and 620S-598R for Mutant1 and Mutant2, respectively. The chimeras and mutants were confirmed by direct sequencing.
Construction of expression vectors.
The expression vector pFUΔss (10) was used for constructing the expression plasmids pFUΔss DC7 Env/F. s. silvestris and DC14 Env/F. s. silvestris. env fragments were amplified from the ERV-DC7/F. s. silvestris and ERV-DC14/F. s. silvestris proviruses using the primer pairs Fe-650S/Fe-626R and Fe-610S/Fe-184R (31), respectively, and cloned into the pFUΔss vector between the BamHI and EcoRI restriction sites. The site-directed mutagenesis method described above was also used to construct expression plasmids for the Env point mutants pFUΔss ERV-DC14/F. s. silvestris/K148E, pFUΔss ERV-DC14/F. s. silvestris/S273P, pFUΔss Ampho-MLV/148K, pFUΔss Friend MLV/148K, pFUΔss FeLV-A/148K, and pFUΔss FeLV-B/148K. The primers used for site-directed mutagenesis are shown in Table 2. The resulting mutants and Env expression plasmids were confirmed by sequencing.
Transfection.
HEK293T, 293Lac, and GPLac cells were transfected with plasmids using the TransIT-293 transfection reagent (Mirus) or the Lipofectamine 3000 reagent (Invitrogen) in accordance with each manufacturer’s instructions.
Viruses.
To obtain infectious viruses, 293Lac cells containing a LacZ-coding pMXs retroviral vector were first seeded at a concentration of 1 × 106 cells in a six-well plate 1 day prior to transfection. The cells were then transfected with ERV-DC14TA (9), ERV-DC14/clone SO38 (10), ERV-DC14/clone GM21, ERV-DC14/clone IK19, ERV-DC14/clone FO16, ERV-DC14/F. s. silvestris clone wildcat54, ERV-DC14/F. s. silvestris clone wildcat63, Chimera1, Chimera2, Chimera3, Chimera4, Mutant1, and Mutant2. Three days later, the corresponding supernatants were collected, filtered through a 0.22-μm-pore-size filter (Merck Millipore, Burlington MA), and stored at −80°C. To obtain LacZ-carrying Env-pseudotyped viruses, GPLac cells were transfected with the corresponding Env expression plasmids of ERV-DC14, ERV-DC14/F. s. silvestris/WT, ERV-DC14/F. s. silvestris/K148E (K148E), ERV-DC14/F. s. silvestris/S273P (S273P), Ampho-MLV (4070A)/WT, Friend MLV (clone 57)/WT, FeLV-A (FeLV clone 33)/WT, FeLV-B (Gardner-Arnstein)/WT, Ampho-MLV (4070A)/148K, Friend MLV (clone57)/148K, FeLV-A (FeLV clone 33)/148K, FeLV-B (Gardner-Arnstein)/148K, FeLV/KS16-1, and FeLV/KS16-2. The culture supernatants were also collected after 72 h, filtered through a 0.22-μm-pore-size filter, and stored at −80°C.
Infection assay.
The target cells, HEK293T or MDTF cells, were seeded at a concentration of 3 × 104 cells in 24-well plates 1 day prior to infection. HEK293T or MDTF cells (the target for Friend MLV) were separately incubated with 250 μl of each virus listed above in the presence of Polybrene (8 μg/ml). After 48 h of incubation, the cells were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and single-cycle infectivity was assessed by counting blue-stained nuclei, as visualized under a microscope. The viral titers are illustrated as the log number of infectious units (IU) per milliliter with standard deviations.
Viral infection assay in the presence of Refrex-1.
HEK293T cells were transfected with the provirus clones (ERV-DC7, ERV-DC16, ERV-DC7/F. s. silvestris, and ERV-DC16/F. s. silvestris), Env expression vectors (ERV-DC7/F. s. silvestris or ERV-DC7), or the empty vector (mock transfection). The resulting culture supernatants were collected, filtered, and stored at −80°C. Fresh HEK293T cells were incubated with serial dilutions of the supernatants for 6 h at 37°C under a humidified atmosphere containing 5% CO2. After removing the supernatants, the cells were inoculated with a replication-competent virus, ERV-DC14TA (9), which produces a persistent infection in HEK293T cells. At 48 h postinfection, the infected cells were stained with X-Gal, and viral titers were calculated as the log number of infectious units (IU) per milliliter with standard deviations.
Virus purification.
Culture supernatants (4 ml) were collected at approximately 72 h posttransfection, filtered through 0.45-μm-pore-size filters, and ultracentrifuged for 90 min at 29,000 × g at 4°C in an Optima Max-XP ultracentrifuge (Beckman Coulter KK, Ariake, Tokyo, Japan). The resulting virions were resuspended in 20 μl of phosphate-buffered saline (PBS) and then used for Western blotting.
Immunoblotting.
Immunoblotting was performed as previously described (67). The primary antibodies used in these assays were goat polyclonal anti-FeLV SU (gp70; National Cancer Institute [NCI], Frederick, MD, USA), mouse monoclonal anti-FeLV SU (gp70; antibody C11D8; Custom Monoclonals International, CA, USA), goat anti-Rauscher MLV SU (gp70; NCI, Frederick, MD, USA), goat anti-Rauscher MLV CA (p30; NCI), rat monoclonal anti-Ampho-MLV SU (gp70; antibody 83A25; a gift of Leonard Evans from NIH, NIAID), mouse anti-FeLV TM protein (p15E; antibody PF6J2A; Custom Monoclonals International, CA, USA), mouse anti-FeLV TM protein (p15E; antibody (EC6-6B1; Custom Monoclonals International, CA, USA), and mouse monoclonal anti-human β-actin (Santa Cruz Biotechnology, Dallas. TX). The secondary antibodies used in these assays were horseradish peroxidase (HRP)-conjugated anti-mouse IgG (Cell Signaling Technology, Danvers, MA) or HRP-conjugated donkey anti-goat IgG (Santa Cruz Biotechnology).
Flow cytometry analysis.
HEK293T cells were seeded in six-well plates and transfected with the empty vector or Env expression plasmids for either ERV-DC14 env or ERV-DC14/F. s. silvestris env. After 48 h, the cells were harvested with PBS containing 5 mM EDTA and washed with PBS containing 0.1% bovine serum albumin (BSA). The cells were fixed for 30 min at room temperature with PBS containing 1% FCS and 10% formaldehyde. Some cells were permeabilized with 0.2% Trion X-100 at room temperature for 15 min. Permeabilization was used to detect the protein in the cytoplasm (intracellular), while nonpermeabilization was used to detect the protein expressed at the cell membrane (cell surface). All cells were blocked with 1% BSA in PBS for 30 min at 4°C. Goat anti-FeLV SU (gp70; NCI) or rabbit anti-human serine/threonine-protein kinase (AKT; Cell Signaling, Danvers, MA) was used as the primary antibody, and phycoerythrin (PE)-conjugated mouse anti-goat IgG (Santa Cruz Biotechnology) or fluorescein isothiocyanate (FITC)-conjugated goat F(ab′)2 anti-rabbit IgG Fc (Abcam, Cambridge, UK) was used as the secondary antibody. The cells were treated with each antibody for 30 min at 4°C. Finally, the cells were resuspended in 500 μl of wash buffer and analyzed using a BD Accuri C6 flow cytometer (Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
Phylogenetic and sequencing analysis.
We retrieved the 3′ LTR sequences of 13 loci of ERV-DC from the National Center for Biotechnology Information (NCBI). As a nucleotide substitution model, a Kimura 2-parameter model (68) with uniform rates was used because it had the lowest Bayesian information criterion (BIC) score. All positions containing gaps and missing data were eliminated from the analysis. Tree robustness was evaluated using the bootstrap method (1,000 replicates). All programs used for phylogenetic analyses were packaged in MEGA X software (69). The GenBank accession numbers are AB674439.1 for ERV-DC1, AB674449.1 for ERV-DC2, AB674440.1 for ERV-DC3, AB674441.1 for ERV-DC4, AB674450.1 for ERV-DC6, AB807599.1 for ERV-DC7, AB674443.1 for ERV-DC8, AB674444.1 for ERV-DC10, AB674445.1 for ERV-DC14, AB807600.1 for ERV-DC16, AB674446.1 for ERV-DC17, AB674447.1 for ERV-DC18, and AB674448.1 for ERV-DC19.
The Env amino acid sequences of ERV-DC (ERV-DC10 and -DC14), gibbon ape leukemia virus (GALV), Friend murine leukemia virus (Friend MLV), feline leukemia virus subgroup D (FeLV-D) strain TY26peL, feline leukemia virus TG35-2 (FeLV TG35-2), Moloney murine leukemia virus (MoMLV), koala retrovirus type A (KoRV-A) KoRV-B, endogenous FeLV AGTT locus (enFeLV/AGTT), feline leukemia virus subgroup B (FeLV-B) Gardner-Arnstein, amphotropic murine leukemia virus (Ampho-MLV), porcine endogenous retrovirus type A (PERV-A), feline leukemia virus subgroup A (FeLV-A) clone 33, feline leukemia virus subgroup C (FeLV-C) Sarma strain, feline leukemia virus subgroup T (FeLV-T), xenotropic murine leukemia virus-related virus (XMRV), polytropic murine leukemia virus AKR13 (P-MLV), FeLV/KS16-1, murine endogenous virus clone MmCN (MLV/MmCN), human endogenous retrovirus type T in Pongo pygmaeus (HERV-T/Pongo), and human endogenous retrovirus type T (HERV-T) were obtained from the NCBI database. Multiple alignments of the above-described amino acid sequences were generated using the mafft tool (70).
We next obtained the ERV-DC7 env sequences of 20 Japanese domestic cats (31, 37) from National Center for Biotechnology Information (NCBI) databases. We also used PCR to amplify ERV-DC7 env from the chromosomal DNA of 11 European wildcats and 14 European domestic cats with the primer pair Fe-615S (5ʹ-CCTCCAAGCCCTTTATCCTC-3ʹ) and Fe-53R, designed for the pol region and 3ʹ-flanking region, respectively, and directly determined their env region sequences. The accession numbers for the nucleotide sequences of ERV-DC7 env are included below. As a nucleotide substitution model, a Kimura 2-parameter model (68) with a discrete gamma-distributed rate variation and inferred proportion of invariable sites (+ G, + I; α = 0.0500, invar = 0.4934) was used. This model was selected because it had the lowest BIC score. All positions containing gaps and missing data were eliminated from the analysis. Tree robustness was evaluated using the bootstrap method (1,000 replicates). All programs used for phylogenetic analyses were packaged in MEGA X software (69).
We next used PCR to amplify the 5ʹ LTR of ERV-DC14 from the chromosomal DNA of nine European wildcats, four European domestic cats, and six Japanese domestic cats with the primer pair Fe-603S and Fe-140R. These primers were designed to anneal to the 5ʹ-flanking region and the gag region, respectively, and we directly determined the 5ʹ LTR region sequences. The accession numbers for the nucleotide sequences of the ERV-DC14 5ʹ LTR are included below. As a nucleotide substitution model, a Kimura 2-parameter model (68) with uniform rates was used because it had the lowest BIC score. All positions containing gaps and missing data were eliminated from the analysis. Tree robustness was evaluated using the bootstrap method (1,000 replicates). All programs used for phylogenetic analyses were packaged in MEGA X software (69).
We also used PCR with the primer pair Fe-609S (5′-ATATGCCCTCCCTAAGACTTCAAG-3′) and Fe-591R (5′-GATTCCATGGCCCTGAAGTAAGAA-3′) to amplify the gene encoding cytochrome b, including a partial control region in mitochondrial DNA, to confirm the results for the European wildcat samples. As a nucleotide substitution model, a Hasegawa-Kishino-Yano model (71) with uniform rates was used because it had the lowest BIC score. All positions containing gaps and missing data were eliminated from the analysis. Tree robustness was evaluated using the bootstrap method (1,000 replicates). All programs used for phylogenetic analyses were packaged in MEGA X software (69).
Statistical analysis.
The results of infection assays were considered statistically significant if P values were <0.05 by a Student's t test and one-way analysis of variance (ANOVA).
Ethical approval.
Animal studies were conducted following the guidelines for the care and use of laboratory animals of the Ministry of Education, Culture, Sports, Science and Technology, Japan. All experiments were approved by the Genetic Modification Safety Committee of Yamaguchi University, Yamaguchi, Japan.
Accession numbers.
The nucleotide sequences reported in this study were deposited in the DDBJ, EMBL, and GenBank databases under accession numbers LC485052 to LC485064 and LC485155.
ACKNOWLEDGMENTS
We thank Toshio Kitamura for providing the PLAT-GP cells. We also thank Leonard Evans (NIH, NIAID) for providing the monoclonal antibody against Ampho-MLV gp70. We also thank Fermín Urra and Veterinary Clinic Parque Bruil for providing European cat samples. We also thank Katie Oakley from the Edanz Group for editing a draft of the manuscript.
This study was funded by the Japan Society for the Promotion of Science KAKENHI to K.N. (grant number 15H04602).
REFERENCES
- 1.Pontius JU, Mullikin JC, Smith DR, Agencourt Sequencing Team, Lindblad-Toh K, Gnerre S, Clamp M, Chang J, Stephens R, Neelam B, Volfovsky N, Schäffer AA, Agarwala R, Narfström K, Murphy WJ, Giger U, Roca AL, Antunes A, Menotti-Raymond M, Yuhki N, Pecon-Slattery J, Johnson WE, Bourque G, Tesler G, NISC Comparative Sequencing Program, O’Brien SJ. 2007. Initial sequence and comparative analysis of the cat genome. Genome Res 17:1675–1689. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, et al. . 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, et al. . 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Nakaya Y, Miyazawa T. 2014. Dysfunction of bovine endogenous retrovirus K2 envelope glycoprotein is related to unsuccessful intracellular trafficking. J Virol 88:6896–6905. doi: 10.1128/JVI.00288-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozak CA. 2014. Origins of the endogenous and infectious laboratory mouse gammaretroviruses. Viruses 7:1–26. doi: 10.3390/v7010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliveira NM, Satija H, Kouwenhoven IA, Eiden MV. 2007. Changes in viral protein function that accompany retroviral endogenization. Proc Natl Acad Sci U S A 104:17506–17511. doi: 10.1073/pnas.0704313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee YN, Bieniasz PD. 2007. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog 3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh AK, Roy-Burman P. 1989. Characterization of enhancer elements and their mutations in the long terminal repeat of feline endogenous RD-114 proviruses. J Virol 63:4234–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuse K, Ito J, Miyake A, Kawasaki J, Watanabe S, Makundi I, Ngo MH, Otoi T, Nishigaki K. 2016. Existence of two distinct infectious endogenous retroviruses in domestic cats and their different strategies for adaptation to transcriptional regulation. J Virol 90:9029–9045. doi: 10.1128/JVI.00716-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anai Y, Ochi H, Watanabe S, Nakagawa S, Kawamura M, Gojobori T, Nishigaki K. 2012. Infectious endogenous retroviruses in cats and emergence of recombinant viruses. J Virol 86:8634–8644. doi: 10.1128/JVI.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song N, Jo H, Choi M, Kim JH, Seo HG, Cha SY, Seo K, Park C. 2013. Identification and classification of feline endogenous retroviruses in the cat genome using degenerate PCR and in silico data analysis. J Gen Virol 94:1587–1596. doi: 10.1099/vir.0.051862-0. [DOI] [PubMed] [Google Scholar]
- 12.Mata H, Gongora J, Eizirik E, Alves BM, Soares MA, Ravazzolo AP. 2015. Identification and characterization of diverse groups of endogenous retroviruses in felids. Retrovirology 12:26. doi: 10.1186/s12977-015-0152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiss RA. 2013. On the concept and elucidation of endogenous retroviruses. Philos Trans R Soc Lond B Biol Sci 368:20120494. doi: 10.1098/rstb.2012.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polani S, Roca AL, Rosensteel BB, Kolokotronis SO, Bar-Gal GK. 2010. Evolutionary dynamics of endogenous feline leukemia virus proliferation among species of the domestic cat lineage. Virology 405:397–407. doi: 10.1016/j.virol.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Weiss RA. 2006. The discovery of endogenous retroviruses. Retrovirology 3:67. doi: 10.1186/1742-4690-3-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roca AL, Pecon-Slattery J, O'Brien SJ. 2004. Genomically intact endogenous feline leukemia viruses of recent origin. J Virol 78:4370–4375. doi: 10.1128/JVI.78.8.4370-4375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coffin JM. 2004. Evolution of retroviruses: fossils in our DNA. Proc Am Philos Soc 148:264–280. [PubMed] [Google Scholar]
- 18.Niman HL, Akhavi M, Gardner MB, Stephenson JR, Roy-Burman P. 1980. Differential expression of two distinct endogenous retrovirus genomes in developing tissues of the domestic cat. J Natl Cancer Inst 64:587–594. [PubMed] [Google Scholar]
- 19.Koshy R, Gallo RC, Wong-Staal F. 1980. Characterization of the endogenous feline leukemia virus-related DNA sequences in cats and attempts to identify exogenous viral sequences in tissues of virus-negative leukemic animals. Virology 103:434–445. doi: 10.1016/0042-6822(80)90202-0. [DOI] [PubMed] [Google Scholar]
- 20.Benveniste RE, Sherr CJ, Todaro GJ. 1975. Evolution of type C viral genes: origin of feline leukemia virus. Science 190:886–888. doi: 10.1126/science.52892. [DOI] [PubMed] [Google Scholar]
- 21.Roca AL, Nash WG, Menninger JC, Murphy WJ, O'Brien SJ. 2005. Insertional polymorphisms of endogenous feline leukemia viruses. J Virol 79:3979–3986. doi: 10.1128/JVI.79.7.3979-3986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boomer S, Eiden M, Burns CC, Overbaugh J. 1997. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J Virol 71:8116–8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overbaugh J, Riedel N, Hoover EA, Mullins JI. 1988. Transduction of endogenous envelope genes by feline leukaemia virus in vitro. Nature 332:731–734. doi: 10.1038/332731a0. [DOI] [PubMed] [Google Scholar]
- 24.McAllister RM, Nicolson M, Gardner MB, Rongey RW, Rasheed S, Sarma PS, Huebner RJ, Hatanaka M, Oroszlan S, Gilden RV, Kabigting A, Vernon L. 1972. C-type virus released from cultured human rhabdomyosarcoma cells. Nat New Biol 235:3–6. doi: 10.1038/newbio235003a0. [DOI] [PubMed] [Google Scholar]
- 25.Bonner TI, Todaro GJ. 1979. Carnivores have sequences in their cellular DNA distantly related to the primate endogenous virus, MAC-1. Virology 94:224–227. doi: 10.1016/0042-6822(79)90454-9. [DOI] [PubMed] [Google Scholar]
- 26.Todaro GJ, Benveniste RE, Sherwin SA, Sherr CJ. 1978. MAC-1, a new genetically transmitted type C virus of primates: “low frequency” activation from stumptail monkey cell cultures. Cell 13:775–782. doi: 10.1016/0092-8674(78)90227-1. [DOI] [PubMed] [Google Scholar]
- 27.Kawasaki J, Kawamura M, Ohsato Y, Ito J, Nishigaki K. 2017. Presence of a shared 5′-leader sequence in ancestral human and mammalian retroviruses and its transduction into feline leukemia virus. J Virol 91:e00829-17. doi: 10.1128/JVI.00829-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Kuyl AC, Dekker JT, Goudsmit J. 1999. Discovery of a new endogenous type C retrovirus (FcEV) in cats: evidence for RD-114 being an FcEV(Gag-Pol)/baboon endogenous virus BaEV(Env) recombinant. J Virol 73:7994–8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beyer W, Mohring R, Drescher B, Nötzel U, Rosenthal S. 1987. Molecular cloning of an endogenous cat retroviral element (ECE1)—a recombinant between RD-114 and FeLV-related sequences. Arch Virol 96:297–301. doi: 10.1007/BF01320971. [DOI] [PubMed] [Google Scholar]
- 30.Kawasaki J, Nishigaki K. 2018. Tracking the continuous evolutionary processes of an endogenous retrovirus of the domestic cat: ERV-DC. Viruses 10:E179. doi: 10.3390/v10040179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito J, Watanabe S, Hiratsuka T, Kuse K, Odahara Y, Ochi H, Kawamura M, Nishigaki K. 2013. Refrex-1, a soluble restriction factor against feline endogenous and exogenous retroviruses. J Virol 87:12029–12040. doi: 10.1128/JVI.01267-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeda H, Sugimura H. 1989. Fv-4 resistance gene: a truncated endogenous murine leukemia virus with ecotropic interference properties. J Virol 63:5405–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikeda H, Laigret F, Martin MA, Repaske R. 1985. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J Virol 55:768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu T, Yan Y, Kozak CA. 2005. Rmcf2, a xenotropic provirus in the Asian mouse species Mus castaneus, blocks infection by polytropic mouse gammaretroviruses. J Virol 79:9677–9684. doi: 10.1128/JVI.79.15.9677-9684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung YT, Lyu MS, Buckler-White A, Kozak CA. 2002. Characterization of a polytropic murine leukemia virus proviral sequence associated with the virus resistance gene Rmcf of DBA/2 mice. J Virol 76:8218–8224. doi: 10.1128/jvi.76.16.8218-8224.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bamunusinghe D, Naghashfar Z, Buckler-White A, Plishka R, Baliji S, Liu Q, Kassner J, Oler AJ, Hartley J, Kozak CA. 2016. Sequence diversity, intersubgroup relationships, and origins of the mouse leukemia gammaretroviruses of laboratory and wild mice. J Virol 90:4186–4198. doi: 10.1128/JVI.03186-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito J, Baba T, Kawasaki J, Nishigaki K. 2016. Ancestral mutations acquired in Refrex-1, a restriction factor against feline retroviruses, during its cooption and domestication. J Virol 90:1470–1485. doi: 10.1128/JVI.01904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nowell K, Jackson P. 1996. European wildcat, Felis silvestris, silvestris group Schreber, 1775. Wild cats: status survey and conservation action plan, p 110–113. IUCN, Gland, Switzerland. [Google Scholar]
- 39.Hemmer H. 1978. The evolutionary systematics of living Felidae: present status and current problems. Carnivora 1:71–79. [Google Scholar]
- 40.Guggisberg CAW. 1975. Wild cats of the world. David & Charles, Newton Abbot, England. [Google Scholar]
- 41.Driscoll CA, Menotti-Raymond M, Roca AL, Hupe K, Johnson WE, Geffen E, Harley EH, Delibes M, Pontier D, Kitchener AC, Yamaguchi N, O'Brien SJ, Macdonald DW. 2007. The Near Eastern origin of cat domestication. Science 317:519–523. doi: 10.1126/science.1139518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Driscoll CA, Macdonald DW, O’Brien SJ. 2009. From wild animals to domestic pets, an evolutionary view of domestication. Proc Natl Acad Sci U S A 106:9971–9978. doi: 10.1073/pnas.0901586106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe S, Kawamura M, Odahara Y, Anai Y, Ochi H, Nakagawa S, Endo Y, Tsujimoto H, Nishigaki K. 2013. Phylogenetic and structural diversity in the feline leukemia virus env gene. PLoS One 8:e61009. doi: 10.1371/journal.pone.0061009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattucci F, Oliveira R, Bizzarri L, Vercillo F, Anile S, Ragni B, Lapini L, Sforzi A, Alves PC, Lyons LA, Randi E. 2013. Genetic structure of wildcat (Felis silvestris) populations in Italy. Ecol Evol 3:2443–2458. doi: 10.1002/ece3.569. [DOI] [Google Scholar]
- 45.Hertwig S, Schweizer M, Stepanow S, Jungnickel A, Böhle UR, Fischer M. 2009. Regionally high rates of hybridization and introgression in German wildcat populations (Felis silvestris, Carnivora, Felidae). J Zool Syst Evol Res 47:283–297. doi: 10.1111/j.1439-0469.2009.00536.x. [DOI] [Google Scholar]
- 46.Hubbard AL, McOris S, Jones TW, Boid R, Scott R, Easterbee N. 1992. Is survival of European wildcats Felis silvestris in Britain threatened by interbreeding with domestic cats? Biol Conserv 61:203–208. doi: 10.1016/0006-3207(92)91117-B. [DOI] [Google Scholar]
- 47.Koito A, Ikeda T. 2011. Intrinsic restriction activity by AID/APOBEC family of enzymes against the mobility of retroelements. Mob Genet Elements 1:197–202. doi: 10.4161/mge.1.3.17430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LaRue RS, Andresdottir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jonsson SR, Landau NR, Lochelt M, Malik HS, Malim MH, Munk C, O'Brien SJ, Pathak VK, Strebel K, Wain-Hobson S, Yu X-F, Yuhki N, Harris RS. 2009. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol 83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris RS, Liddament MT. 2004. Retroviral restriction by APOBEC proteins. Nat Rev Immunol 4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 50.Taylor GM, Gao Y, Sanders DA. 2001. Fv-4: identification of the defect in Env and the mechanism of resistance to ecotropic murine leukemia virus. J Virol 75:11244–11248. doi: 10.1128/JVI.75.22.11244-11248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeda A, Matano T. 2007. Inhibition of infectious murine leukemia virus production by Fv-4 env gene products exerting dominant negative effect on viral envelope glycoprotein. Microbes Infect 9:1590–1596. doi: 10.1016/j.micinf.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 52.Dewannieux M, Blaise S, Heidmann T. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J Virol 79:15573–15577. doi: 10.1128/JVI.79.24.15573-15577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robbez-Masson L, Rowe HM. 2015. Retrotransposons shape species-specific embryonic stem cell gene expression. Retrovirology 12:45. doi: 10.1186/s12977-015-0173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gifford WD, Pfaff SL, Macfarlan TS. 2013. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol 23:218–226. doi: 10.1016/j.tcb.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cowley M, Oakey RJ. 2013. Transposable elements re-wire and fine-tune the transcriptome. PLoS Genet 9:e1003234. doi: 10.1371/journal.pgen.1003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Isbel L, Whitelaw E. 2012. Endogenous retroviruses in mammals: an emerging picture of how ERVs modify expression of adjacent genes. Bioessays 34:734–738. doi: 10.1002/bies.201200056. [DOI] [PubMed] [Google Scholar]
- 57.Greenwood AD, Ishida Y, O'Brien SP, Roca AL, Eiden MV. 2017. Transmission, evolution, and endogenization: lessons learned from recent retroviral invasions. Microbiol Mol Biol Rev 82:e00044-17. doi: 10.1128/MMBR.00044-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fiebig U, Hartmann MG, Bannert N, Kurth R, Denner J. 2006. Transspecies transmission of the endogenous koala retrovirus. J Virol 80:5651–5654. doi: 10.1128/JVI.02597-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tarlinton R, Meers J, Hanger J, Young P. 2005. Real-time reverse transcriptase PCR for the endogenous koala retrovirus reveals an association between plasma viral load and neoplastic disease in koalas. J Gen Virol 86:783–787. doi: 10.1099/vir.0.80547-0. [DOI] [PubMed] [Google Scholar]
- 60.Tacke SJ, Kurth R, Denner J. 2000. Porcine endogenous retroviruses inhibit human immune cell function: risk for xenotransplantation? Virology 268:87–93. doi: 10.1006/viro.1999.0149. [DOI] [PubMed] [Google Scholar]
- 61.Hanger JJ, Bromham LD, McKee JJ, O'Brien TM, Robinson WF. 2000. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: a novel type C endogenous virus related to Gibbon ape leukemia virus. J Virol 74:4264–4272. doi: 10.1128/JVI.74.9.4264-4272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Armstrong JA, Porterfield JS, de Madrid AT. 1971. C-type virus particles in pig kidney cell lines. J Gen Virol 10:195–198. doi: 10.1099/0022-1317-10-2-195. [DOI] [PubMed] [Google Scholar]
- 63.Young GR, Eksmond U, Salcedo R, Alexopoulou L, Stoye JP, Kassiotis G. 2012. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature 491:774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Graham FL, Smiley J, Russell WC, Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 65.Miyake A, Watanabe S, Hiratsuka T, Ito J, Ngo MH, Makundi I, Kawasaki J, Endo Y, Tsujimoto H, Nishigaki K. 2016. Novel feline leukemia virus interference group based on the env gene. J Virol 90:4832–4837. doi: 10.1128/JVI.03229-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 67.Umehara D, Watanabe S, Ochi H, Anai Y, Ahmed N, Kannagi M, Hanson C, Ruscetti S, Nishigaki K. 2010. Role of Phosphatidylinositol 3-kinase in Friend spleen focus-forming virus-induced erythroid disease. J Virol 84:7675–7682. doi: 10.1128/JVI.00488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/bf01731581. [DOI] [PubMed] [Google Scholar]
- 69.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174. doi: 10.1007/bf02101694. [DOI] [PubMed] [Google Scholar]