APOBEC3 proteins are host factors that protect the integrity of the host genome by inhibiting retroelements as well as retroviruses, such as HIV-1. To do this, the APOBEC3H protein has evolved unique interactions with structured RNAs. Here, we studied the importance of these interactions in driving antiviral activity of APOBEC3H. Our results provide a clear picture of how RNA binding drives the ability of APOBEC3H to infiltrate new viruses and prevent synthesis of viral DNA. We also explore how RNA binding by APOBEC3H influences recognition and deamination of viral DNA and describe two possible routes by which APOBEC3H might hypermutate the HIV-1 genome. These results highlight how one protein can sense many nucleic acid species for a variety of antiviral activities.
KEYWORDS: APOBEC3H, RNA binding, deamination, heteroduplex, human immunodeficiency virus
ABSTRACT
APOBEC3 proteins APOBEC3F (A3F), APOBEC3G (A3G), and APOBEC3H (A3H) are host restriction factors that inhibit HIV-1 through DNA cytidine deaminase-dependent and -independent mechanisms and have either one (A3H) or two (A3F and A3G) zinc-binding domains. A3H antiviral activity encompasses multiple molecular functions, all of which depend on recognition of RNA or DNA. A3H crystal structures revealed an unusual interaction with RNA wherein an RNA duplex mediates dimerization of two A3H proteins. In this study, we sought to determine the importance of RNA-binding amino acids in the antiviral and biochemical properties of A3H. We show that the wild-type A3H-RNA interaction is essential for A3H antiviral activity and for two deaminase-independent processes: encapsidation into viral particles and inhibition of reverse transcription. Furthermore, an extensive mutagenesis campaign revealed distinct roles for two groups of amino acids at the RNA binding interface. C-terminal helix residues exclusively bind RNA, and loop 1 residues play a dual role in recognition of DNA substrates and in RNA binding. Weakening the interface between A3H and RNA allows DNA substrates to bind with greater affinity and enhances deamination rates, suggesting that RNA binding must be disrupted to accommodate DNA. Intriguingly, we demonstrate that A3H can deaminate overhanging DNA strands of RNA/DNA heteroduplexes, which are early intermediates during reverse transcription and may represent natural A3H substrates. Overall, we present a mechanistic model of A3H restriction and a step-by-step elucidation of the roles of RNA-binding residues in A3H activity, particle incorporation, inhibition of reverse transcriptase inhibition, and DNA cytidine deamination.
IMPORTANCE APOBEC3 proteins are host factors that protect the integrity of the host genome by inhibiting retroelements as well as retroviruses, such as HIV-1. To do this, the APOBEC3H protein has evolved unique interactions with structured RNAs. Here, we studied the importance of these interactions in driving antiviral activity of APOBEC3H. Our results provide a clear picture of how RNA binding drives the ability of APOBEC3H to infiltrate new viruses and prevent synthesis of viral DNA. We also explore how RNA binding by APOBEC3H influences recognition and deamination of viral DNA and describe two possible routes by which APOBEC3H might hypermutate the HIV-1 genome. These results highlight how one protein can sense many nucleic acid species for a variety of antiviral activities.
INTRODUCTION
APOBEC3 (A3) proteins are a family of restriction factors that protect vertebrates from retroviruses and retroelements (1). In primates, APOBEC3F (A3F), APOBEC3G (A3G), and APOBEC3H (A3H) potently restrict lentiviruses such as HIV-1 through a series of nucleic acid binding events (1, 2). First, RNA binding facilitates A3 encapsidation into new virions, which is a requirement for initiation of A3 restriction activities (3–7). Much of the antiviral potency of A3 proteins arises from their cytidine deaminase enzymatic function, which causes extensive hypermutation of the minus strand (−) DNA (single-stranded DNA [ssDNA]) product of HIV-1 reverse transcriptase (RT), leading to G-to-A transitions in the plus strand (+) DNA (1, 2, 8–10). While deaminase function is a critical aspect of A3 antiviral activity, catalytically inactive variants of A3F, A3G, and A3H retain partial antiviral activity, potentially through interactions with RNA or RT (11–17). Hence, two distinct mechanisms of restriction are described for antiviral APOBEC3 proteins, a deaminase-dependent mechanism and a deaminase-independent mechanism.
Deaminase-dependent restriction activity in HIV-1-infected cells has been characterized for A3F, A3G, and A3H (10, 18–20) in which a conserved zinc-binding domain deaminates a cytosine base (substrate) to uracil (product) in single-stranded DNA. A3 proteins have various dinucleotide requirements (21) for deamination, i.e., 5ʹ-CC for A3G (20) and 5ʹ-TC for A3F (21) and A3H (22) (the target C is bold and underlined); however, the determinants of dinucleotide specificity remain elusive. Aside from this distinction, DNA substrate preferences for each antiviral A3 have not been characterized extensively. Furthermore, because each antiviral A3 protein binds both RNA and DNA, RNA is thought to inhibit DNA binding and deaminase activity (23, 24), even though biochemical characterization of the interplay between RNA and DNA binding in the context of deaminase activity has not been thoroughly explored.
RNA binding plays many roles in A3 restriction. In addition to mediating virion incorporation of A3 proteins, interactions with RNA are important for deaminase-independent restriction (16, 25). Thus far, mechanisms of reverse transcription inhibition have been explored most thoroughly for A3G (12, 13, 16, 17, 25–29). Several groups have suggested that A3G inhibits reverse transcription through interactions with RT (13, 17, 26), while others suggested that A3G-RNA interactions drive inhibition of reverse transcription (25). These mechanisms have not been distinguished biochemically. In contrast to A3G, little is known about deaminase-independent mechanisms of restriction of A3H, although the ability of A3H to recognize structured RNAs has led to many hypotheses for an RNA-driven mechanism for A3H inhibition of reverse transcription (11, 30, 31).
The highly multifunctional nature of A3 proteins requires a variety of crucial nucleic acid interactions, which are poorly understood on a molecular level. Nucleic acid-binding functions are modular in A3 restriction factors that possess two Zn-binding domains (A3F and A3G), i.e., an N-terminal domain binds RNA, and a C-terminal domain catalyzes cytidine deamination (32–34). For the didomain antiviral A3 proteins (A3F and A3G), RNA binding and DNA binding (deaminase) functions are thought to be structurally independent, and although structures of individual domains have been reported, there are none of a full-length didomain alone or in complex with any nucleic acid. In contrast, A3H is functionally and evolutionarily distinct from A3F and A3G. As the only single-domain anti-HIV A3 protein, it must bind both RNA and ssDNA using one Zn-binding domain (35–37). The mechanism by which this domain either recognizes RNA and DNA simultaneously or toggles between RNA- and DNA-bound states remains a mystery.
Thus far, the structural repertoire of A3 proteins in complex with nucleic acids is limited. Recently reported structures of A3H-RNA complexes, the first of any A3 protein bound to RNA, revealed that A3H selectively binds to RNA duplexes (24, 30, 38). This unexpected binding mode lends itself to several A3H antiviral activities, including recognition of viral RNAs for incorporation into virions and inhibition of reverse transcription (24, 30, 38). In crystal structures of A3 catalytic domains in complex with DNA substrates, only 2 or 3 DNA nucleotides are visible, limiting the interpretation of these structures (39–41). In all cases, however, the nucleotide upstream of the target C, the −1 nucleotide, of the 5ʹ-XC dinucleotide motif binds near loop 7. In the A3H-RNA complex, this loop binds an unpaired RNA base (30).
Here, we present a compilation of antiviral and biochemical assays using the active human A3H haplotype II (huA3H II) as well as a pigtailed macaque (pgt) A3H variant (pgtA3Hα) to support a model in which a highly basic region of A3H, consisting of loop 1 and the C-terminal helix, promiscuously binds a variety of nucleic acid species, thereby driving both deaminase-dependent and deaminase-independent antiviral activities. Promiscuity in nucleic acid recognition may confer an evolutionary advantage for A3H in optimizing its ability to recognize packaged RNAs, structured regions of the viral genome, and a wide variety of substrates, potentially including heteroduplexes. This property of A3H is functionally and biochemically distinct from A3F and A3G, consistent with nonredundant functions for these related antiviral factors (36, 37).
RESULTS
A3H relies on RNA binding for antiviral activity.
Using structure-guided mutagenesis of A3H, we focused on amino acids that contact RNA (24, 30, 38). Sites for mutagenesis were selected based on the number of direct interactions of each amino acid with the RNA backbone or bases and were grouped based on their location in A3H. The first group is located on A3H extended loop 1 (R17, R18, Y23, and R26) (Fig. 1a, shown in pink). Loop 1 of A3H is longer than loop 1 of all other A3 proteins and harbors a greater number of positively charged residues (30). The second group is located on the A3H C-terminal helix (R175 and R176) (Fig. 1a, shown in blue), which is a highly basic surface that provides a favorable platform for binding phosphates in the RNA backbone. The distribution of positive charge on the surface of A3H is distinct from that on the surfaces of other antiviral A3 family members such as A3F and A3G (32). The effects of single and double amino acid substitutions on antiviral activity against HIV-1ΔVif (10) were initially tested in the context of the pig-tailed macaque (pgt) A3Hα, the variant with the highest levels of activity against HIV-1 (Fig. 1).
FIG 1.
Cellular restriction activities of A3H are mediated by RNA binding. (a) An RNA duplex bound to one A3H monomer (27) is shown; the second A3H monomer in the complex is omitted for visibility. Structure-guided mutagenesis of two basic patches on A3H that provide a platform for duplex RNA recognition, loop 1 in pink and the C-terminal helix in blue, is depiected. All data related to these mutants are color coded as such. (b) Antiviral activity of mutagenized pgtA3Hα at sites of RNA binding compared to wild-type pgtA3Hα. HIV-1ΔVif stocks were generated in 293T cells in the presence of increasing amounts of transiently transfected HA-tagged pgtA3Hα (0 [control], 80, 200, 500 ng expression plasmid for the wild type [wt] or 500 ng for each pgtA3Hα RNA-binding mutant) and titrated on indicator cell lines expressing nanoluciferase under the control of the HIV-1 promoter. Luciferase activity was measured and expressed as relative light units (RLU). Values are means plus standard deviations (error bars) from two independent experiments. (c) Antiviral activity of huA3H II mutants tested as in panel b. (d) Levels of all HA-tagged pgtA3Hα mutants in panel b were analyzed by immunoblotting in comparison to HIV-1 Gag and its proteolytic products (anti-HA [red] and anti-HIV-1 CA [green]). (e) Levels of HA-tagged huA3H II mutants in panel c tested as in panel d. (f) Virion incorporation of pgtA3Hα. 293T cells stably expressing wild-type HA-tagged A3H and mutants were infected with HIV-1ΔVif. Infected cells and sucrose-purified virion particles were analyzed by immunoblotting as in panel d. The specificity of virion incorporation was verified using supernatants from uninfected cells expressing selected A3H variants. Infected cells lacking A3H expression plasmid are the control. (g) Virion incorporation of huA3H tested as in panel f. Levels of A3H incorporation are shown under the gels, expressed as the ratio of signals (in arbitrary units) of A3H bands to the viral capsid (CA) bands.
Given the number of positively charged residues on the basic surface of A3H, it was surprising that certain substitutions of single amino acids abolished pgtA3Hα restriction activity in cells (R26E, R176E), as did nearly all double substitutions (R17E/R18E, Y23A/R26A, Y23A/R26E, R175A/R176A, and R175E/R176E) (Fig. 1b). Charge reversal substitutions at R17/R18 and R26 in loop 1 each resulted in near complete loss of antiviral activity. Less drastic alanine substitutions at these positions had either weaker effects (R26A) or no effect (R17A/R18A). Substitution of alanine for Y23, which stabilizes the 5ʹ end of the RNA (30), had no effect on antiviral activity. Y23A in combination with either R26A or R26E, marginally enhanced the effects of either of the R26 single substitutions (Fig. 1b). Two residues on the C-terminal helix, R175 and R176, each provide several contacts to the RNA backbone. R175A, R175E, and R176A were active; however, a single charge reversal at R176 (R176E) eliminated restriction activity. Double substitutions at these positions (R175A/R176A and R175E/R175E) resulted in complete loss of activity (Fig. 1b). In contrast, substitution of alanine for the catalytic residue E56 abolishes deaminase activity but only partially diminished antiviral activity (Fig. 1b), consistent with an earlier finding that A3H restriction is not solely dependent on deaminase activity (11).
Interestingly, the substitutions with the greatest effects on antiviral activity also resulted in different degrees of reduction in protein levels, most evident with R17E/R18E and R175E/R176E (Fig. 1d). However, the decrease in the levels of mutagenized pgtA3Hα in infected cells relative to wild-type levels did not account for the full extent of loss of antiviral activity. We also introduced selected substitutions in the huA3H haplotype II. huA3H II is less potent than pgtA3Hα in restricting HIV-1ΔVif (30), but as with pgtA3Hα, substitutions targeting key residues R26 and R175/R176 completely abolished antiviral activity, whereas the E56A substitution resulted in only partial loss of virus inhibition (Fig. 1c). The double substitutions at the C-terminal helix also reduced protein levels of both pgtA3Hα and huA3H II in infected cells (Fig. 1d and e). Together, these data suggest that, in addition to restriction activity, A3H binding to RNA affects protein stability in the cell, and that RNA may serve as an A3H chaperone, consistent with the build-up of similar RNA-binding mutants of chimpanzee A3H in cells upon treatment with MG132, a proteasome inhibitor (38).
RNA binding is critical for virion incorporation of A3H.
Loss of RNA binding would impair the ability of A3H to recognize viral RNA, thereby preventing A3H incorporation into nascent viral particles, which in turn would result in lower levels of antiviral activity. Here, transient expression of A3H in 293T cells resulted in significant accumulation of the protein in the culture supernatant even in the absence of infection. Therefore, to determine the effects of RNA binding on A3H incorporation into HIV-1 particles, we generated cell lines stably expressing each A3H mutant, infected them with HIV-1ΔVif (30), and measured the virion incorporation of each mutant. This approach eliminated A3H from supernatant samples in the absence of virus, detected with an antibody against the HIV-1 capsid protein (CA) (Fig. 1f and g).
Overall, the levels of antiviral activity of the RNA-binding A3H mutants for both pigtailed macaque and human proteins correlated with the levels of A3H incorporation into virion particles, in agreement with previous studies (6, 30). Specifically, pgtA3Hα mutants that exhibited complete loss of antiviral activity (Y23A/R26E, R26E, R17E/R18E, R176E, R175A/R176A, and R175E/R176E) were not incorporated into virions (Fig. 1f). Of note, trace amounts of the huA3H R26A mutant could be detected in virions in some experiments (Fig. 1g and data not shown). In contrast, the catalytically inactive E56A was efficiently incorporated into virions (Fig. 1f and g), confirming that virion incorporation requires RNA binding and is not dependent on cytidine deaminase activity.
A3H inhibits strong-stop DNA synthesis during reverse transcription.
Previous cross-linking immunoprecipitation sequencing (CLIP-seq) experiments demonstrated that A3H, but not A3F or A3G, preferentially binds elements in the structured 5ʹ leader of the viral genome (6, 30). Specific localization to the 5ʹ leader also suggests a mechanism for the observed deaminase-independent restriction through inhibition of reverse transcription (11). Accordingly, the catalytically inactive E56A A3H mutants maintained good levels of antiviral activity (Fig. 1b and c), in agreement with previous studies (11), and were efficiently packaged into virions (Fig. 1f and g).
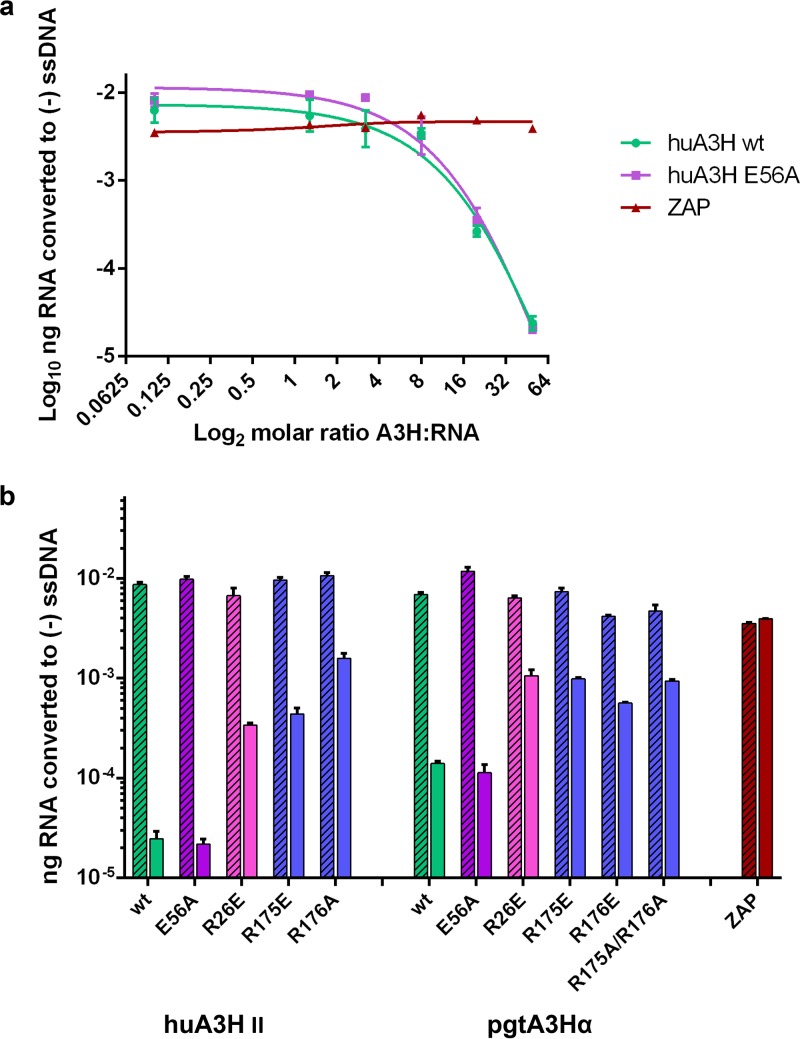
In light of these observations, we hypothesized that duplex RNA binding may drive inhibition of reverse transcription. Thus, we developed a modified quantitative PCR-based RT assay (42) to directly measure the impact of A3H on the formation of the 100- to 150-nucleotide (nt) early RT product known as “strong stop” DNA [(−) ssDNA] using purified, in vitro-synthesized HIV-1 NL4.3 5ʹ-leader RNA as the template and an 18-nt tRNA mimic as the primer for reverse transcription. The RNA template and primer were hybridized and preincubated with recombinant A3H at various molar ratios prior to the addition of RT (viral lysate). To avoid background from viral genomes, lysates were made using a virus construct lacking the 5ʹ leader. The presence of huA3H II in this reaction mixture resulted in a dose-dependent reduction in RNA converted to (−) ssDNA of up to 3 log units (Fig. 2a). The same trend occurred for the catalytically inactive huA3H E56A, demonstrating that RT inhibition is independent of deaminase activity (Fig. 2a). In contrast, even at great molar excess, the antiviral RNA-binding protein ZAP (zinc finger antiviral protein) did not inhibit (−) ssDNA formation (Fig. 2a).
FIG 2.
A3H inhibits the formation of strong-stop DNA during early stages of reverse transcription. (a) Dose effect of wild-type huA3H and E56A on production of (−) ssDNA in a viral lysate reconstitution of reverse transcription. A3H was preincubated with the 5ʹ-leader RNA prior to initiation of a 10-min reverse transcription reaction. The amount of RNA (in nanograms) converted to (−) ssDNA was calculated from a standard curve of known RNA concentrations. ZAP is an RNA-binding, antiviral protein that was used as a negative control. (b) A panel of RNA-binding mutants of huA3H and pgtA3Hα was tested in the RT-qPCR assay to determine the contribution of single amino acids to A3H inhibition of RT. Means and standard errors (error bars) are reported for three independent experiments. Slashed bars represent those with no A3H added, while solid bars represent the effects of 50:1 A3H-RNA.
Together, these results support a mechanism for deaminase-independent A3H inhibition of (−) ssDNA formation. We hypothesized that this inhibition was driven by the A3H propensity to bind to duplex RNA. To address this, we tested the RT inhibitory activity of mutagenized huA3H and pgtA3Hα at RNA-interacting residues. Because the largest inhibition of reverse transcription, within the detection limit of the assay, occurred at a 50:1 molar ratio of A3H to RNA, we used this ratio to assess whether disruption of RNA binding alleviated RT inhibition. The addition of purified, recombinant, mutagenized A3H resulted in significantly lower levels (1 to 2 log units) of RT inhibition (Fig. 2b). Consistent effects were observed in huA3H and pgtA3Hα but were substantially greater in huA3H.
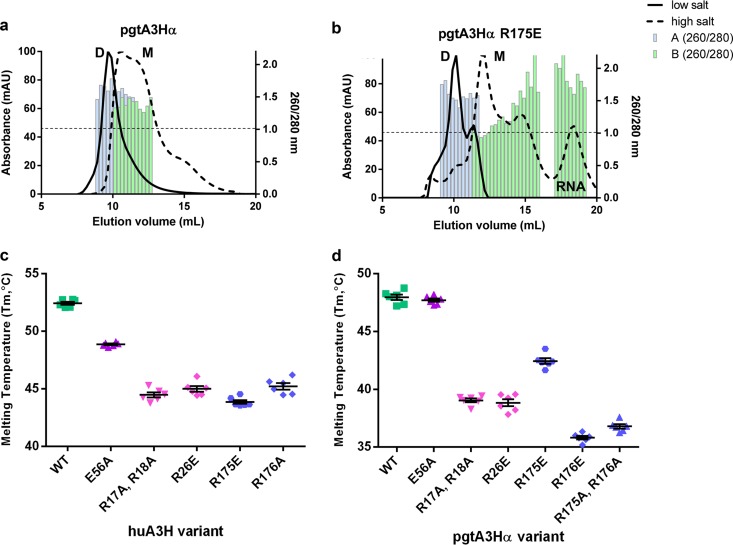
RT inhibition was not completely eliminated for any of the tested A3H mutants. We attribute this effect to a reduced, but not abolished, RNA affinity. All of the huA3H and pgtA3Hα RNA-binding mutants used in the RT assays purified as RNA-bound dimers with absorbance ratios (260 nm/280 nm) of 1.4 to 1.5 (compared to 0.5 to 0.6 for protein only), indicating that a single amino acid substitution does not abolish RNA binding (Fig. 3a and b). A3H mutants with multiple charge reversal substitutions at the RNA binding surface resulted in unstable proteins that could not be purified, suggesting that RNA is a stabilizing chaperone for A3H. The stable mutants with single substitutions of RNA-binding amino acids were more sensitive to dissociation by high salt (2 M NaCl), as detected by analytical gel filtration (Fig. 3a and b), and all single and double substitutions (R17A/R18A and R175A/R176A) exhibited substantially decreased thermostability relative to the wild type (Fig. 3c and d), illustrating the importance of RNA to A3H stability.
FIG 3.
RNA-binding mutants of A3H are destabilized. (a and b) Analytical gel filtration analysis of pgtA3Hα. pgtA3Hα (a) and pgtA3Hα R175E (b) were subjected to low salt (300 mM NaCl [solid traces]) or high salt (2 M NaCl [dashed traces]) before and during analytical gel filtration. Absorbance ratios (260 nm/280 nm) were measured for each fraction (blue for low salt and green for high salt). Wild-type pgtA3Hα was purely dimeric (D) under low-salt conditions and partially dissociated into RNA-associated monomeric (M) units (A260/A280 ratio of ≥1) under high-salt conditions. In contrast, under low-salt conditions, the R175E mutant was predominantly dimeric with a shoulder corresponding to a lower-molecular-weight species. Under high-salt conditions, the R175E mutant completely dissociated from RNA, resulting in a protein-only peak (A260/A280 ratio of < 1) and a distinct RNA peak (A260/A280 ratio of 1.5 to 2), which was not present for the wild-type protein under high-salt conditions. mAU, milli absorbance units. (c and d) Thermostability of A3H analyzed by differential scanning fluorimetry. Melting temperatures (Tm) were determined for each wild-type or mutant huA3H (c) and pgtA3Hα (d). Means and standard errors for six wells are reported.
A3H RNA-binding residues play a role in ssDNA substrate recognition.
While virion incorporation and inhibition of reverse transcription are likely driven by A3H recognition of RNA-RNA duplexes, A3H also binds single-stranded DNA, the substrate for cytidine deamination. Early results with partially purified A3H suggested that RNA binding is inhibitory to deaminase activity (11). However, highly purified A3H was bound to copurifying RNA and retained potent cytidine deaminase activity even though the bound RNA is near the Zn active site (Fig. 4a) (30). How the single domain of A3H senses and recognizes multiple nucleic acid species is unknown.
FIG 4.
RNA-binding residues play distinct roles in substrate recognition and deamination of single-stranded DNA. (a) A3H structure near two groups of amino acids at the protein-RNA interface (loop 1 side chains in pink, C-terminal helix side chains in blue). (b and d) Deaminase activity of pgtA3Hα (100 nM) and huA3H (50 nM) mutants with a 40-nt ssDNA (1 μM). Products were detected by denaturing gel electrophoresis. Data are shown as log2 fold changes compared to the wild type. Note the axis gap in panel b, highlighting the significant reduction in deaminase activity for the double Y23A/R26E mutant. (c and e) Affinities (Kd [in nanomolar]) of pgtA3Hα (c) and huA3H (e) variants for a 40-nt ssDNA substrate (10 nM) determined by fluorescence polarization. Means ± standard errors from three experiments are shown. Mutagenesis data in panels b to e are colored according to the amino acid coloring in panel a, with data for the wild type in green.
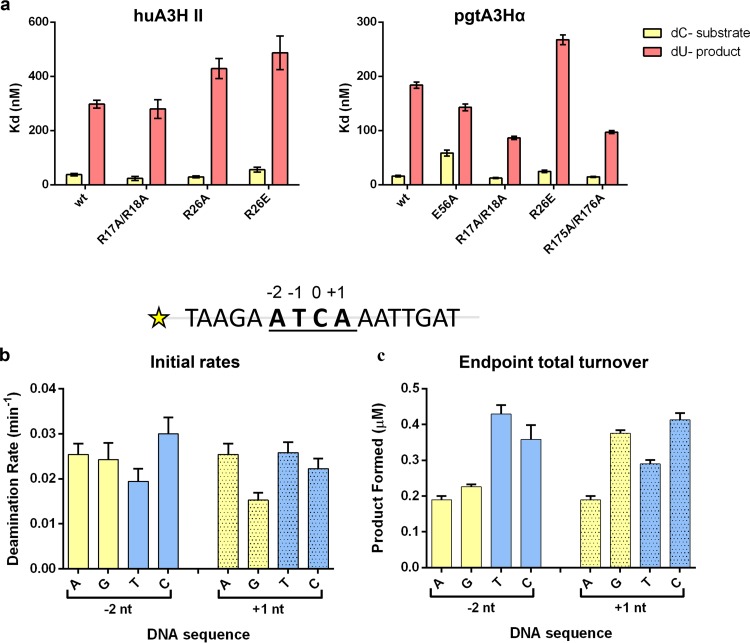
To address this question, we used the huA3H and pgtA3Hα RNA-binding mutants that were tested for antiviral activity (Fig. 1). Purified, recombinant A3H proteins and a 40-nucleotide single-stranded DNA substrate were used in assays of cytidine deaminase activity and DNA binding. Our data show that the two groups of RNA-binding amino acids that were defined by their location in A3H (Fig. 1a) also have distinct biochemical functions. First, loop 1 residues involved in RNA binding (R17, R18, Y23, and R26) (Fig. 4a, amino acids in pink) are also important for effective DNA binding and catalysis. The double R17A/R18A mutant had increased deaminase activity and DNA binding relative to wild-type A3H, while the double R17E/R18E mutant behaved similarly to wild type (Fig. 4b). A or E substitutions at R26 slightly reduced DNA affinity compared to wild-type A3H; however, R26A and R26E had markedly reduced deaminase activity, suggesting a role for R26 in catalysis (Fig. 4b to e). In contrast, the C-terminal helix exclusively supports RNA binding (Fig. 4a, amino acids in blue). Single A or E substitutions at R175 or R176 increased deaminase activity and DNA affinity relative to wild-type A3H, and the effect was only slightly enhanced in the double R175A/R176A mutant (Fig. 4b to e). Reduced A3H affinity for RNA with the C-terminal helix substitutions (Fig. 3) was correlated with increased DNA affinity and catalysis, suggesting that A3H either partially or fully releases RNA during DNA substrate recognition. Although the effects were greater for pgtA3Hα, they were consistent between huA3H and pgtA3Hα, implying a conserved mechanism of weakened RNA binding in the presence of a single-stranded DNA substrate.
Determinants of substrate selection by A3H.
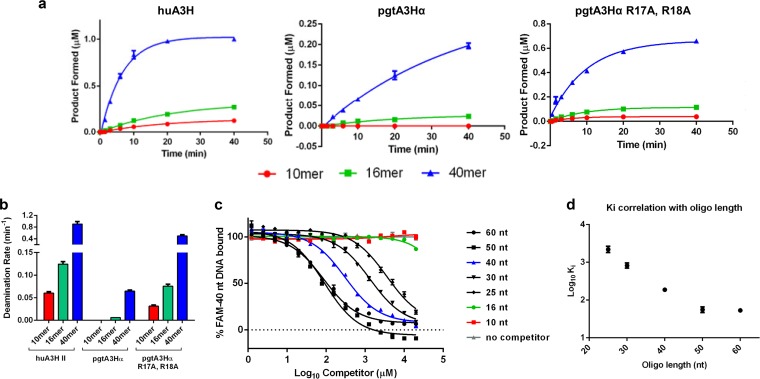
To probe A3H recognition of DNA in the presence of copurifying, bound RNA, we examined A3H preferences for deamination and binding using ssDNA substrates of different lengths (40, 16, and 10 nt), each containing a single 5ʹ-TC dinucleotide (Fig. 5a and b). Both huA3H and pgtA3Hα exhibited potent cytidine deaminase activity against the 40-nt substrate, as described previously (30), while initial deaminase rates for the 16-nt and 10-nt substrates were 8-fold and 16-fold lower, respectively. A similar length dependence of deamination rate was observed for the hyperactive deaminase pgtA3Hα R17A/R18A (Fig. 5a and b), suggesting that, although pgtA3Hα R17A/R18A had higher overall deaminase activity, it retained a preference for longer substrates. To determine whether the DNA length dependence was due to substrate affinity, competition binding experiments were performed with the catalytically inactive variant, pgtA3Hα E56A. First, we determined the apparent binding affinity (Kd) of 20 nM for pgtA3Hα E56A and the fluorescently tagged 40-nt ssDNA 5ʹ-TC-containing substrate using fluorescence polarization. For competition binding, a fixed concentration of 80 nM A3H (90 to 100% of the 40-nt substrate was bound) was preincubated with 10 nM labeled 40-nt ssDNA to ensure complete binding of the labeled DNA. Unlabeled competitor DNA oligomers of different lengths, all with a single 5ʹ-TC dinucleotide recognition motif located in the center of the sequence (10, 16, 25, 30, 40, 50, and 60 nt) were titrated into the labeled DNA-A3H complex and inhibitory constants (Ki) were determined (Fig. 5c). The ability to displace the labeled DNA (Ki) was strongly correlated with the length of the competing oligomer. The 60-, 50-, 40-, and 30-nt oligomers fully displaced the fluorescently labeled 40-nt ssDNA (Fig. 5c), with length-dependent behavior saturating at 50 nt (Fig. 5d). Interestingly, A3H displayed a preference for longer ssDNA substrates despite these substrates containing a single TC dinucleotide motif flanked by the same nucleotides as in shorter substrates. This suggests that A3H favors recognition of dinucleotide motifs that are flanked by longer sequences or simply greater DNA mass.
FIG 5.
A3H prefers longer single-stranded DNA substrates. (a) Time course of huA3H (50 nM), pgtA3Hα (100 nM), and pgtA3Hα R17A R18A (50 nM) cytidine deamination of single-stranded DNAs (1 μM) of different lengths (10, 16, and 40 nt) in an assay coupled with 40 nM uracil DNA glycosylase. Products were detected by denaturing gel electrophoresis. (b) Comparison of initial deamination rates for data in panel a. (c) Fluorescence polarization-based competition binding assay for pgtA3Hα E56A and DNA. Unlabeled DNA oligomers of different lengths were titrated into a mixture of 10 nM fluorescently labeled 40-nt DNA oligomer and 80 nM pgtA3Hα E56A (Kd = 16 nM). (d) Correlation of apparent Ki with competing DNA oligomer (oligo) length. Means ± standard errors from three experiments are reported.
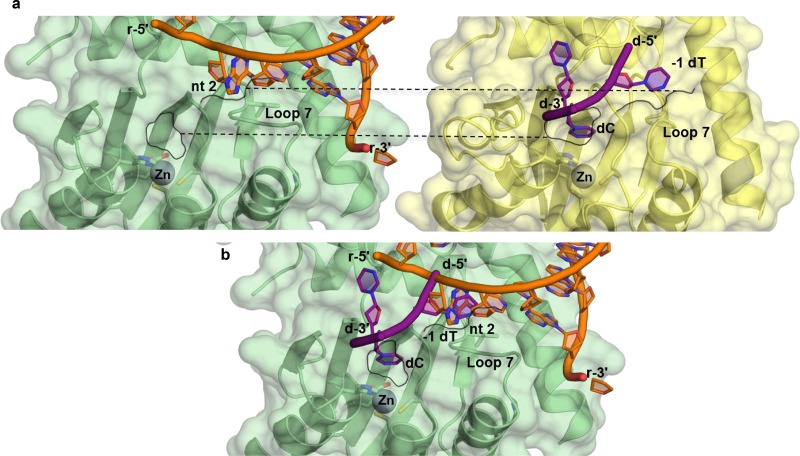
We sought a structural explanation for these results by comparing structures of A3-DNA substrate complexes and our A3H-RNA complex (30). In complexes of A3A (39, 41), A3B (41, 43), and A3G (40) with DNA substrates, the A3 enzymes form specific interactions with only the dinucleotide recognition motif, i.e., the target C and the −1 nucleotide. Although longer oligomers were used in all cases, only two or three nucleotides are resolved in published A3-DNA structures. Interestingly, in the A3 substrate complexes, the binding site for the DNA −1 nucleotide of the dinucleotide motif is equivalent to an aromatic cage occupied by an unpaired RNA base in the A3H-RNA complex (Fig. 6a and b). Thus, effective DNA substrate binding seems to require the displacement of the RNA base from loop 7. This could be facilitated by general disruption of RNA binding, which may be a simpler task for longer DNA substrates, as the primary RNA interaction is at the C-terminal helix, remote from loop 7, thus explaining the binding and catalytic preference for longer DNAs.
FIG 6.
Structural comparison of RNA-bound A3H with DNA-bound A3A. (a) Surface views of RNA-bound pgtA3H (PDB accession no. 5W3V [27]) (A3H [green]; RNA [magenta]) and DNA-bound A3A (PDB accession no. 5KEG [34]) (A3A [yellow]; DNA [blue]) shown in the same orientation. The Zn active site and loop 7 are shown in stick form and labeled. Black outlines highlight the binding pockets for dC and −1 dT (RNA nt 2). The −1 dT in A3A-DNA and nt 2 in A3H-RNA occupy equivalent clefts, cradled by loop 7. (b) Superposition of A3A-DNA and A3H-RNA structures, showing both nucleic acids and the surface of A3H.
While A3G has a unique CC dinucleotide signature, the identical TC signatures of A3F and A3H render their effects indistinguishable in hypermutation profiles of the HIV-1 genome in infected cells. Understanding additional requirements for substrate recognition by these proteins may help to parse hypermutation effects of A3F versus those of A3H. Indeed, we found that not only DNA length but also sequence affected A3H binding to ssDNA. Specifically, the affinity of huA3H and pgtA3Hα for a 40-nt dC-containing substrate was 10-fold greater than for a 40-nt dU-containing product of otherwise identical sequence (Fig. 7a). Furthermore, the sequence context of the TC dinucleotide, namely, the nucleotides at the −2 and +1 positions, had only modest effects on deaminase activity. The initial deamination rates (linear phase of the reaction time course) did not change significantly when the −2 or +1 nucleotides were varied among A, C, T, and G (Fig. 7b). However, the endpoint total turnover values revealed a twofold preference for pyrimidines (Fig. 7c, solid blue bars) over purines (Fig. 7c, solid yellow bars) at the −2 position and a slight preference at the +1 position for G/C over A/T (Fig. 7c, stippled bars).
FIG 7.
Sequence preferences for A3H substrate selection. (a) A3H affinities (Kd [in nanomolar]) for a 40-nt dC-substrate ssDNA and a 40-nt dU-product. huA3H II (left) and pgtA3Hα (right) were titrated into 10 nM DNA oligomer, and binding was detected by fluorescence polarization. (b and c) Sequence preference for deamination of single-stranded DNA substrates. 16-nt ssDNA oligomers with different −2 and +1 nucleotides (1 μM) were incubated with 100 nM huA3H and 40 nM uracil DNA glycosylase (UDG). Products were detected by denaturing gel electrophoresis over a 20-min time course. (b and c) Initial rates in the linear range of the reaction time course (0 to 5 min) are shown in panel b, and product formed (in micromolar) at the final 20-min time point is shown in panel c. Means plus standard errors from three experiments are reported.
Taken together, the DNA affinity and sequence selectivity results demonstrate that A3H deamination favors longer ssDNAs and is selective for a 5ʹ-TC dinucleotide-containing substrate compared to an analogous dU-containing product mimic. Subtle sequence preferences at the surrounding −2 or +1 positions are not critical in substrate selection. Several mechanistic possibilities are consistent with the data. The A3H-bound RNA duplex may be removed or remodeled in order for an ssDNA of preferred length and sequence to bind. Alternatively, A3H may also recognize and engage A-form RNA/DNA heteroduplexes that are generated during reverse transcription. Heteroduplex binding may position the A3H active site near newly synthesized minus-strand DNA following degradation of the RNA template strand by the RNase H domain of RT. These possibilities are not mutually exclusive, and A3H may act on both ssDNA and RNA/DNA heteroduplexes with ssDNA overhangs.
A3H recognizes and can catalyze deamination on select heteroduplex substrates.
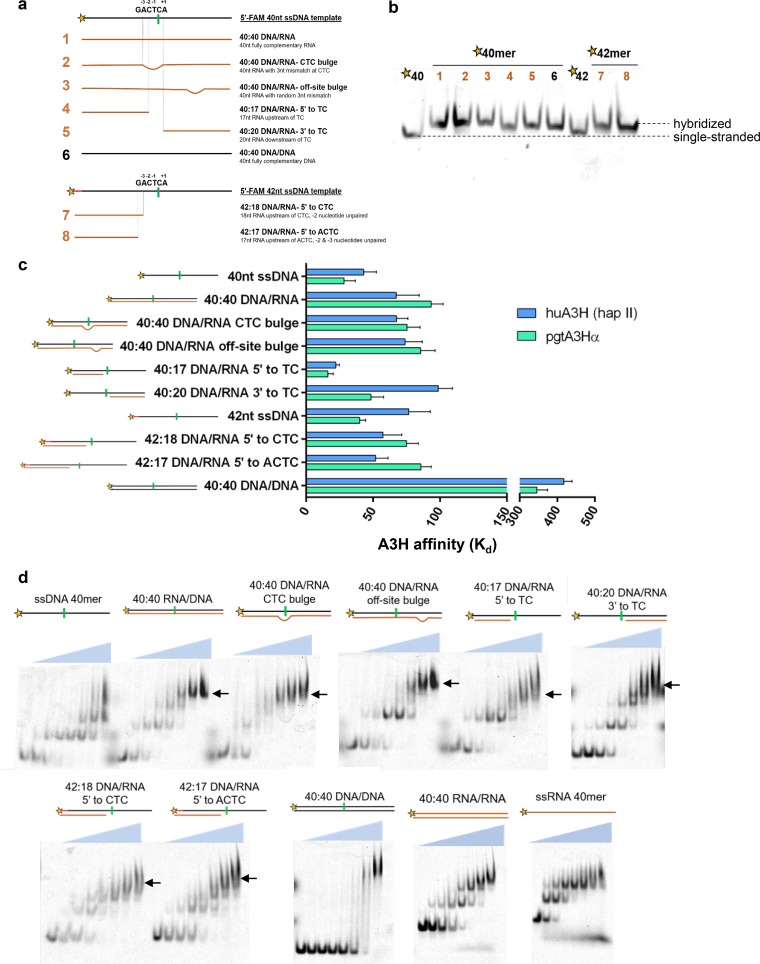
RNA/DNA heteroduplexes with ssDNA overhangs are formed by the combined polymerase and RNase H activities of RT during the minus-strand phase of reverse transcription (44). To test whether such heteroduplexes can serve as A3H substrates, we developed a heteroduplex library (Fig. 8a and b; also see Table 1). Either a 40- or 42-nucleotide fluorescently labeled ssDNA oligomer served as the template for hybridization of RNA oligonucleotides, labeled 1 to 5 for the 40-nt oligomers and 7 and 8 for the 42-nt oligomers (Fig. 8a). As a control, a complementary 40-nt DNA oligomer, labeled 6, was used (Fig. 8a). Hybrid formation was confirmed by native gel electrophoresis (Fig. 8b), and A3H affinities were determined by fluorescence polarization (Fig. 8c). Both human and pigtail macaque A3H proteins bound all heteroduplexes with Kd values comparable to the Kd of the 40-nt ssDNA oligomer, whereas double-stranded DNA (dsDNA) was bound very weakly (Fig. 8c). These fluorescence polarization results were recapitulated in the electrophoretic mobility shift data (Fig. 8d). The preference for DNA/RNA heteroduplex over dsDNA is consistent with the clear preference of A3H for A-form duplex, as in the A3H-RNA complex structure (30). In general, RNA/RNA and RNA/DNA duplexes favor the A-form, whereas DNA/DNA duplexes favor the B-form. In the electrophoretic mobility shift experiments, we confirmed that A3H binds RNA/RNA duplexes as well as single-stranded RNA (ssRNA), but not dsDNA (Fig. 8d).
FIG 8.
A3H binding to RNA/DNA heteroduplexes. (a) Cartoon representation of RNA/DNA hybrids tested for cytidine deaminase activity and binding by fluorescence polarization and electrophoretic mobility shift assays (DNA [black] and RNA [orange]). (b) Confirmation of hybrid formation by 15% TBE native gel electrophoresis. (c) A3H affinities (Kd [in nanomolar]) for all heteroduplexes. Binding was detected by fluorescence polarization by titrating huA3H (light blue) or pgtA3Hα (light green) into 10 nM nucleic acid. A3H bound to all heteroduplexes with high affinity, but it bound to the dsDNA control with 10-fold-weaker affinity. (d) Heteroduplex binding visualized by electrophoretic mobility shift assays. A fixed nucleic acid concentration of 10 nM was incubated with huA3H at various concentrations (0, 2.7, 8.2, 24.7, 74, 222, 667, and 2,000 nM). Additional experiments surveyed other nucleic acid species: DNA/DNA duplex, RNA/RNA duplex, and 40-nt ssRNA. Supershifted species in panel d are indicated with black arrows.
TABLE 1.
Calculated melting temperatures (Tm) for duplex substrates
| Fluorescent oligonucleotide |
Hybrid ID | Unlabeled oligonucleotide | Tm (°C) |
|---|---|---|---|
| 40-nt ssDNA (JBs01) | 1 | 40-nt fully complementary RNA (JBs27) | 47.4 |
| 2 | 40-nt RNA with 3-nt mismatch at –CTC (JBs28) | 43.2 | |
| 3 | 40-nt RNA with off-site 3-nt mismatch (JBs29) | 45.1 | |
| 4 | 17-nt RNA upstream of –TC (JBs30) | 22.7 | |
| 5 | 20-nt RNA downstream of –TC (JBs31) | 25.1 | |
| 6 | 40-nt fully complementary DNA (JBs32) | 66.6 | |
| 42-nt ssDNA (JBs33) | 7 | 18-nt RNA upstream of CTC (JBs34) | 23.8 |
| 8 | 17-nt RNA upstream of ACTC (JBs35) | 22.7 | |
| 40-nt ssDNA (JBs01) | 16-nt RNA upstream of CTCa | 18.3 | |
| 15-nt RNA upstream of ACTCa | 16.0 | ||
Not used due to predicted instability (low calculated Tm) of the hybrid.
The electrophoretic mobility shift assays also revealed the presence of multiple oligomer states of A3H bound to nucleic acid duplexes and single strands (Fig. 8d). This suggests that heteroduplex substrates can displace the copurifying RNA duplex from A3H. Moreover, because the footprint of A3H on the A-form duplex is minimally 7 bp, the 17- to 40-bp heteroduplex substrates in our panel are large enough to accommodate multiple A3H proteins, resulting in supershifted species in these assays (Fig. 8d). Similar observations were first noted during purifications of A3H from Escherichia coli and mammalian expression systems where, in the absence of RNase treatment, higher-order oligomeric species were detected by gel filtration (23, 30, 38). The ability of A3H to oligomerize when bound to both RNA/RNA and RNA/DNA duplexes may be a facet of its antiviral activity (45, 46).
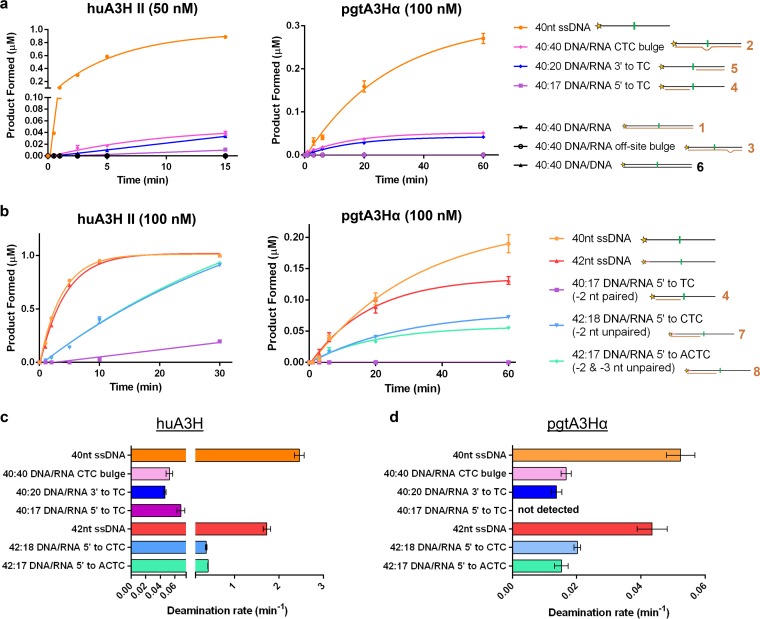
We next asked whether the heteroduplexes could serve as substrates for deamination (Fig. 9). The DNA strand of each hybrid contained the dinucleotide for cytidine deamination and thus, if the dinucleotide were exposed, we expected that A3H would deaminate these substrates. The heteroduplexes may represent A3H substrates encountered during reverse transcription, as the RT RNase H activity is slower than the polymerase activity, leaving behind short stretches of RNA/DNA hybrids during synthesis of minus-strand DNA (47, 48). A3H catalyzed deamination of some but not all heteroduplex substrates and did not deaminate the DNA/DNA duplex (Fig. 9a). Under conditions where the A3H concentration was limiting, A3H deaminated ssDNA more efficiently than heteroduplexes 2 and 5 but had no detectable activity with duplex substrates in which the TC dinucleotide was base paired (duplexes with DNA oligomers 1, 3, and 6 in Fig. 8a). Both huA3H and pgtA3Hα deaminated heteroduplex 2 with a mismatch/bulge in which the −2, −1, and target C nucleotides were unpaired as well as heteroduplex 5 where the hybridized 20-nt RNA was downstream of the target dinucleotide but the 5ʹ half of the DNA was single stranded, with unpaired −2, −1, and target C nucleotides (Fig. 9a). Interestingly, heteroduplex 4, where the hybridized 17-nt RNA is upstream of the target dinucleotide and sequesters the −2 nucleotide, was not a substrate for A3H deamination (Fig. 9a). This led to the hypothesis that an unpaired −2 nucleotide is important for substrate recognition and catalysis. Addressing this hypothesis required a longer DNA for stable hybridization upstream of the target TC site, so we generated a set of hybrids with a fluorophore-tagged 42-nt ssDNA (Fig. 8a and b) (see the melting temperatures of hybrids in Table 1). These hybrids contained annealed RNAs upstream of the dinucleotide motif, but the −2 nucleotide or both the −2 and −3 nucleotides were unpaired. Surprisingly, removing the base pair for the −2 nucleotide enabled A3H to deaminate these hybrids, but removing the base pair for both −2 and −3 nucleotides had no added effect (Fig. 9b). Initial deamination rates for huA3H and pgtA3Hα with ssDNA and heteroduplex substrates under multiple-turnover kinetic conditions are summarized in Fig. 9c and d, respectively.
FIG 9.
Heteroduplexes with accessible dinucleotides are substrates for A3H. (a) Comparison of heteroduplexes and 40-nt ssDNA as the substrates for A3H deamination. Duplexes 1 to 6 in panel a (1 μM) were incubated with 50 nM huA3H or 100 nM pgtA3Hα and 40 nM uracil DNA glycosylase (UDG). No activity was detected for substrates shown in black. (b) Comparison of A3H activities on substrates with DNA strands with a length of 40 and 42 nt. Duplexes 7 and 8 in panel a (1 μM) were incubated with 100 nM huA3H or pgtA3Hα and 40 nM UDG. Products were detected by denaturing gel electrophoresis. (c and d) Comparison of initial rates for data in panels a and b. Means ± standard errors (error bars) from three experiments are reported.
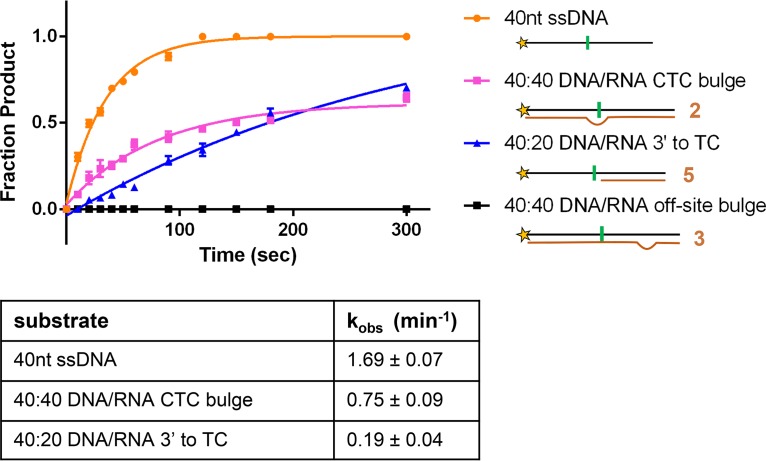
While A3H had lower activity on heteroduplexes than on the 40-nt or 42-nt ssDNA substrates, we tested the heteroduplexes under multiple-turnover kinetic conditions, i.e., the substrate was in large excess over the enzyme (Fig. 9). Under single-turnover conditions where enzyme is not limiting, huA3H catalyzed deamination on the heteroduplex substrates at rates similar to the rate with the ssDNA substrate (Fig. 10). Thus, if the local concentration of A3H is higher during reverse transcription, A3H would act equally well on ssDNA and heteroduplex substrates. In the context of an infection, A3H may recognize both single-stranded DNA and DNA/RNA heteroduplexes with single-stranded overhangs containing the TC dinucleotide recognition motif and an unpaired −2 nucleotide.
FIG 10.
Single-turnover kinetic analysis of A3H with heteroduplex substrates. 40-nt ssDNA and select heteroduplex substrates were tested for deaminase activity under single-turnover conditions (500 nM huA3H II, 50 nM substrate). 40 nM UDG was included in the reaction mixtures, and products were detected by denaturing gel electrophoresis. Data were fit to a single exponential to determine observed rate constants (kobs), summarized in the table below the graph. Means ± standard errors (error bars) from three experiments are reported.
DISCUSSION
A3H interactions with nucleic acid are critical for antiviral activities.
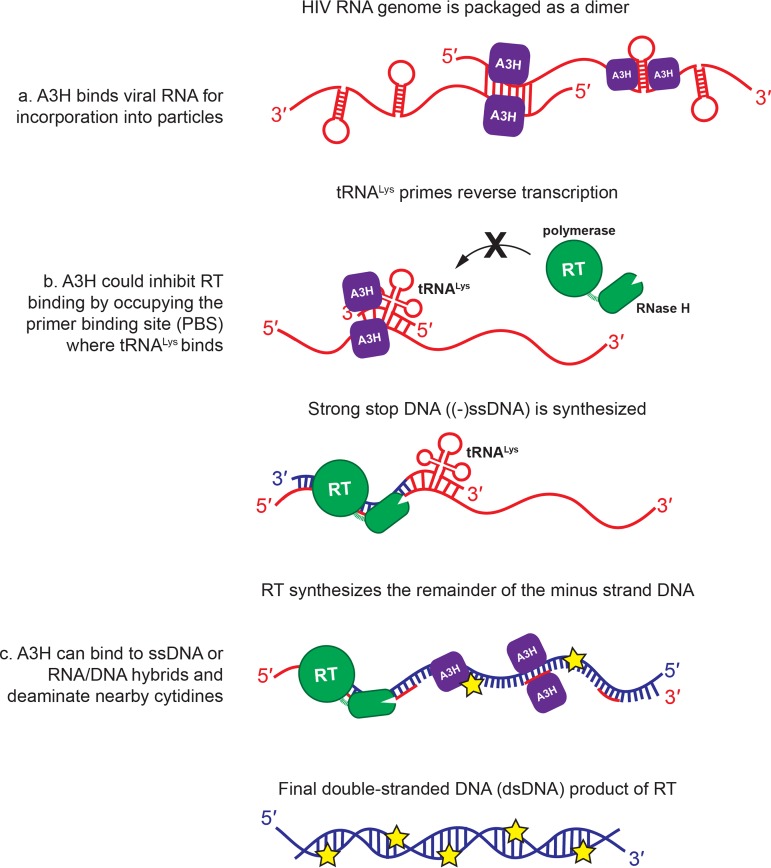
APOBEC3H is unique among A3 proteins that exhibit HIV-1 antiviral activity (A3F, A3G, and A3H) (49). A3H differs fundamentally from A3F and A3G in that it employs a single canonical A3 zinc-binding domain to bind RNA and to catalyze cytidine deamination of single-stranded DNA. In contrast, the didomain A3F and A3G proteins consist of an N-terminal RNA-binding domain (NTD) and a C-terminal catalytic domain (CTD) (8). While A3H and the RNA-binding A3F and A3G NTDs are highly basic, the A3F and A3G NTD sequences are more closely related to each other (62% identity) than either is to A3H (∼30% identity), and sequence similarity is even lower between A3H and the A3F/A3G catalytic CTDs (50, 51). Consistent with the low sequence identity, the charge distributions on the surfaces of the A3H and A3G NTD structures are strikingly different (30, 52), indicative of different sites for RNA binding. Furthermore, the binding profile of A3H on the viral genome differs significantly from those of A3G and A3F (6, 30). Together, these analyses suggest that A3H recognizes RNA in a manner that is distinct from that of A3F and A3G. Prior results from structural biology and cell-based cross-linking immunoprecipitation sequencing (CLIP-seq) experiments together with the antiviral assays and biochemical analyses presented here highlight these differences. For example, CLIP-seq data on A3F, A3G, and A3H binding to RNAs in cells and virions were initially interpreted as promiscuous binding of A3 proteins to RNA without a particular sequence recognition motif, likely targeting single-stranded regions of RNA (6, 7). The A3H-RNA complex structure (30) revealed that the lack of sequence specificity can be attributed to A3H recognition of the shape of a short A-form RNA duplex and not its sequence. Reanalysis of the CLIP-seq data provided biological relevance for the structural observations, as in HIV-1 virions, A3H bound most frequently to the highly structured 5′-leader region, which is replete with base-paired stem structures. RNA structure plays a central role in successful viral replication (53), a feature that A3H has evolved to exploit in a number of effective ways: (i) for selective virion encapsidation, (ii) for inhibition of early stages of reverse transcription, and (iii) for substrate recognition (Fig. 11).
FIG 11.
Mechanistic overview of the nucleic acid-dependent restriction activities of A3H. Restriction activity of A3H is driven by nucleic acid binding. (a) A3H binds to structured regions of viral RNA for incorporation into particles. (b) A3H inhibits reverse transcription, likely by binding to base-paired stems near the primer binding site (PBS) in the 5ʹ leader, sterically inhibiting RT binding and synthesis of proviral DNA. (c) During successful reverse transcription, A3H senses ssDNA for cytidine deamination either by binding directly to ssDNA or by recognizing RNA/DNA heteroduplexes and acting on proximal ssDNA overhangs. Cytidine deamination of TC dinucleotides in the minus strand DNA results in A3-driven hypermutation of the viral genome.
Following our determination of an A3H-RNA crystal structure (30), we sought to understand how the observed protein-RNA interactions modulate A3H antiviral activity. Using site-directed mutagenesis, we found that substitutions of RNA-binding amino acids abolished the A3H restriction activity, severely impaired virion incorporation, and destabilized the protein. Virion incorporation is essential for antiviral activity because genomic RNA binding is the first of a series of A3H-nucleic acid interactions in viral restriction. Additionally, A3H binds preferentially to sites in the 5ʹ leader of the HIV-1 genome in virions, specifically, at or near the primer binding site (PBS) of the 5ʹ leader (6). Thus, we hypothesized that this binding may interfere with reverse transcription and reduce minus-strand strong-stop DNA formation by RT (44). Indeed using a modified, in vitro reverse transcriptase quantitative PCR (RT-qPCR) assay, we show that A3H significantly impairs the conversion of a 5ʹ-leader RNA template to strong-stop DNA independently of its deaminase activity (Fig. 2). Our results are consistent with previously published data showing that both wild-type and catalytically inactive A3H proteins inhibit strong-stop DNA synthesis in virus particles (11).
A3H can toggle between tight and loose RNA-bound states.
We performed a detailed mechanistic analysis of A3H substrate selection and offer new insights into its substrate preferences. First, we showed that loop 1 residues, specifically R26, play a dual role in RNA binding and substrate recognition. Proteins with substitutions at A3H R26 were not only defective in virion incorporation but also had reduced deaminase activity compared to wild type. This suggests a role for R26 in positioning the DNA substrate in the A3H active site. In contrast, C-terminal helix residues seem to be involved in RNA binding by providing a positively charged platform for the RNA backbone, but not for DNA. Eliminating positively charged side chains on the C-terminal helix reduced RNA affinity but resulted in higher-affinity DNA binding and more-efficient cytidine deamination. This is consistent with a DNA-dependent toggle between A3H states with tightly bound or loosely bound RNA. These results also raise the possibility that DNA binding might liberate or destabilize one A3H from the RNA duplex-mediated dimer, and while one A3H remains bound to RNA, the other deaminates the target DNA. We also found that, when presented with substrates with a single TC flanked by identical nucleotides, A3H prefers longer DNA substrates (Fig. 5c). While the basis for this preference is unclear, it could be due to the ability of longer DNA substrates to disrupt RNA binding more effectively than shorter DNA substrates do. Our data also show that A3H lacks DNA sequence specificity beyond the well-established 5ʹ-TC dinucleotide motif for deamination (Fig. 7b) (22, 46, 54).
An attractive mechanism for substrate recognition involves interactions with the RNA/DNA heteroduplex product of the initial steps of reverse transcription. Importantly, the window of opportunity for cytoplasmic A3H to hypermutate the viral genome is limited to the lifetime of the minus-strand DNA, which includes both single-stranded regions and segments of heteroduplex (Fig. 11). We show that A3H binds duplex RNA, single-stranded DNA, single-stranded RNA, and intriguingly, RNA/DNA heteroduplexes. A3H also catalyzes cytidine deamination of heteroduplexes with ssDNA overhangs in which the −2 nucleotide, −1 nucleotide, and the target C are unpaired, without regard to the direction of the overhang (Fig. 9b). This is consistent with the crystal structure of A3H in complex with A-form duplex RNA (24, 30, 38). Given the finding that nucleic acid stabilizes A3H, a heteroduplex could both stabilize the protein and facilitate cytidine deamination of overhanging deoxy TC dinucleotides by providing a high local concentration of the enzyme relative to its substrate. Notably, we tested heteroduplex substrates where the TC dinucleotide was located on the single-stranded overhang directly adjacent to the heteroduplex, and thus, it is remarkable that activity was observed considering the potential steric hurdles for the target C to access the active site. We anticipate that A3H could more effectively catalyze deamination of substrates where the TC dinucleotide is further from the hybrid region, allowing the DNA to access the active site with more flexibility. The discovery that A3H can bind and deaminate heteroduplexes expands our understanding of the substrate scope of A3H.
This study highlights the versatility of nucleic acid recognition by A3H and the diversity of antiviral functions that are facilitated by these interactions. Future structural and biochemical studies of A3H in complex with a variety of nucleic acid substrates as well as of other full-length antiviral A3s will help to elucidate the assortment of A3-nucleic acid interactions and the functional consequences of these interactions during a viral infection.
MATERIALS AND METHODS
Plasmids and cells.
Human A3H haplotype II (huA3H II) and pigtailed macaque A3H variant (pgtA3Hα) were expressed using a modified LHCX (Clontech) vector, that expresses hygromycin, such that the A3H stop codon was removed and three copies of a hemagglutinin (HA) tag were fused to the C terminus of the protein (A3H-3xHA), as previously described for huA3H (6). Mutations at specific residues were introduced using overlapping PCR. The HIV-1NL4.3ΔVif plasmid is a full-length virus plasmid lacking Vif and has been previously described (6). 293T cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). Helios cells are a HeLa-based indicator cell line expressing CD4, CCR5, and also a nanoluciferase reporter gene under the control of an HIV-2 long terminal repeat (LTR) (55). They were maintained in DMEM supplemented with 10% FCS and G418 (100 ng/ml), hygromycin (100 ng/ml), and puromycin (0.6 ng/ml).
Antiviral activity assay for A3H RNA-binding mutants.
For transient-transfection experiments, 293T cells (2.3 × 105 cells/well in 24 wells) were transfected using PEI (polyethylenimine) (Polysciences, Inc.) with 300 ng of NL4.3Δvif and 500 ng of each A3H expression plasmid or control empty LHCX vector. For wild-type (wt) A3H, the amount of plasmid was varied between 500, 200, and 80 ng of A3H expression plasmid with empty vector making up the total concentration to 500 ng. Forty-eight hours after transfection, the supernatant was harvested and filtered through a 0.22-μm filter and titrated onto Helios cells. At 72 h after inoculation of the Helios cells, infectivity was quantified by measuring nanoluciferase expression using the Nano-Glo luciferase assay (Promega) and is expressed as relative light units (RLU).
Virion incorporation assay.
Murine leukemia virus (MLV)-based retroviral stocks of selected A3H mutants were generated by transfection of 293T cells (8 × 105 cells/well in six wells) using PEI, with 500 ng of each A3H-3xHA expression plasmid or the LHCX control plasmid together with 800 ng of an MLV GagPol expression plasmid and 100 ng of a vesicular stomatitis virus glycoprotein (VSV.G) expression plasmid. These stocks were used to generate 293T cells stably expressing each wt A3H or mutant, as previously described (6). Briefly, 293T cells were inoculated with each virus stock, and stable cells were selected using hygromycin.
For the generation of HIV-1NL4.3ΔVif virus stocks, 293T cells (8.5 × 106 cells/plate [10-cm plates]) were transfected using PEI with 10 μg of HIV-1NL4.3ΔVif and 0.5 μg of VSV.G. Forty-eight hours posttransfection, supernatant was harvested and filtered through a 0.22-μm filter, aliquoted, and frozen. Titers were determined on TZM-bl indicator cell lines.
293T cells stably expressing A3H proteins (7 × 105 cells/well in six wells) were infected with VSV.G-pseudotyped HIV-1NL4.3ΔVif at a multiplicity of infection (MOI) of ∼2. At 16 h postinfection, supernatants were collected, clarified by centrifugation at 2,500 rpm, and filtered through a 0.22-μm filter, and virions were purified via ultracentrifugation through a 25% sucrose cushion. Supernatants from uninfected A3H-expressing cells were used as a control for the specificity of A3H incorporation into viral particles. Purified virions and cell lysates were analyzed by immunoblotting using a rabbit anti-HA (catalog no. 600-401-384; Rockland) and mouse anti-HIV-1 p24 capsid (catalog no. 183-H12-5C; NIH AIDS Reagent Program) antibodies, followed by an anti-rabbit IgG conjugated to IRDye680 and an anti-mouse IgG conjugated to IRDye800 (Li-Cor).
Mutagenesis.
All A3H mutants listed in Table 2 were generated by site-directed mutagenesis PCR and confirmed by Sanger sequencing. Primers used to generate these mutants are listed in Table 2 and were designed with the QuikChange primer design tool website (Agilent).
TABLE 2.
Protein mutants and primers for plasmid construction
| Plasmid IDa | Mutant | Primer nameb | Primer sequence |
|---|---|---|---|
| ppgtA3H001 | pgtα wt | ||
| ppgtA3H030 | pgtα E56A | JBp30 pgtα E56A-F | AACAAGAAAGAGGACCATGCAGCAATTCGCTTTATTAACAAGATC |
| JBp31 pgtα E56A-R | GATCTTGTTAATAAAGCGAATTGCTGCATGGTCCTCTTTCTTGTT | ||
| ppgtA3H054 | pgtα R17A, R18A | JBp123 pgt R17,18A-F | CGGGTAGTAAGGCTTGTTGACTGCGGCCTTGTTGTTAAACTGTAAGCTG |
| JBp124 pgt R17,18A-R | CAGCTTACAGTTTAACAACAAGGCCGCAGTCAACAAGCCTTACTACCCG | ||
| ppgtA3H055 | pgtα R17E, R18E | JBp125 pgt R17,18E-F | CTTCCTCGGGTAGTAAGGCTTGTTGACCTCCTCCTTGTTGTTAAACTGTAAGCTGAATGT |
| JBp126 pgt R17,18E-R | ACATTCAGCTTACAGTTTAACAACAAGGAGGAGGTCAACAAGCCTTACTACCCGAGGAAG | ||
| ppgtA3H056 | pgtα Y23A | JBp172 pgt Y23A-F | GGCCTTCCTCGGGTAGGCAGGCTTGTTGACTCGG |
| JBp173 pgt Y23A-R | CCGAGTCAACAAGCCTGCCTACCCGAGGAAGGCC | ||
| ppgtA3H057 | pgtα Y23A, R26A | JBp174 pgt Y23A, R26A-F | ACACAAGAGGGCCTTCGCCGGGTAGGCAGGCTTGTTGACTCGG |
| JBp175 pgt Y23A, R26A-R | CCGAGTCAACAAGCCTGCCTACCCGGCGAAGGCCCTCTTGTGT | ||
| ppgtA3H058 | pgtα Y23A, R26E | JBp176 pgt Y23A, R26E-F | AACACAAGAGGGCCTTCTCCGGGTAGGCAGGCTTGTTGACTCGGC |
| JBp177 pgt Y23A, R26E-R | GCCGAGTCAACAAGCCTGCCTACCCGGAGAAGGCCCTCTTGTGTT | ||
| ppgtA3H059 | pgtα R26A | JBp127 pgt R26A-F | ACACAAGAGGGCCTTCGCCGGGTAGTAAGGCTTG |
| JBp128 pgt R26A-R | CAAGCCTTACTACCCGGCGAAGGCCCTCTTGTGT | ||
| ppgtA3H060 | pgtα R26E | JBp129 pgt R26E-F | GTAACACAAGAGGGCCTTCTCCGGGTAGTAAGGCTTGTTG |
| JBp130 pgt R26E-R | CAACAAGCCTTACTACCCGGAGAAGGCCCTCTTGTGTTAC | ||
| ppgtA3H061 | pgtα R175A | JBp131 pgt R175A-F | CTTTATCCTTTCAAGCCGTGCCTTTATGGCTTGGCTGTTTTTAT |
| JBp132 pgt R175A-R | ATAAAAACAGCCAAGCCATAAAGGCACGGCTTGAAAGGATAAAG | ||
| ppgtA3H062 | pgtα R175E | JBp133 pgt R175E-F | GGGACTTTATCCTTTCAAGCCGCTCCTTTATGGCTTGGCTGTTTTTAT |
| JBp134 pgt R175E-R | ATAAAAACAGCCAAGCCATAAAGGAGCGGCTTGAAAGGATAAAGTCCC | ||
| ppgtA3H063 | pgtα R176A | JBp135 pgt R176A-F | ACTTTATCCTTTCAAGCGCTCGCTTTATGGCTTGGCTGTT |
| JBp136 pgt R176A-R | AACAGCCAAGCCATAAAGCGAGCGCTTGAAAGGATAAAGT | ||
| ppgtA3H064 | pgtα R176E | JBp137 pgt R176E-F | GACTTTATCCTTTCAAGCTCTCGCTTTATGGCTTGGCTGTTT |
| JBp138 pgt R176E-R | AAACAGCCAAGCCATAAAGCGAGAGCTTGAAAGGATAAAGTC | ||
| ppgtA3H065 | pgtα R175A, R176A | JBp139 pgt R175,176A-F | CCGGGACTTTATCCTTTCAAGCGCTGCCTTTATGGCTTGGCTGTTTTTA |
| JBp140 pgt R175,176A-R | TAAAAACAGCCAAGCCATAAAGGCAGCGCTTGAAAGGATAAAGTCCCGG | ||
| ppgtA3H066 | pgtα R175E, R176E | JBp141 pgt R175,176E-F | CACTCCGGGACTTTATCCTTTCAAGCTCCTCCTTTATGGCTTGGCTGTTTTTATCTA |
| JBp142 pgt R175,176E-R | TAGATAAAAACAGCCAAGCCATAAAGGAGGAGCTTGAAAGGATAAAGTCCCGGAGTG | ||
| phA3H032 | hu wt | JBp189 del mocr huA3H-F | GGTACCGAGAACCTGTACTTCCAATCC |
| JBp188 del mocr huA3H-R | CAGATCTACACCAGAAGAATGATGATGATG | ||
| phA3H033 | hu E56A | JBp46 hu E56A-F | GAAAACAAGAAAAAGTGCCATGCAGCAATTTGCTTTATTAACGAGATCAAGTC |
| JBp47 hu E56A-R | GACTTGATCTCGTTAATAAAGCAAATTGCTGCATGGCACTTTTTCTTGTTTTC | ||
| phA3H034 | hu R17A, R18A | JBp103 hu R17,18A-F | GGTAGTAAGGCCTTCTGAGGGCGGCCTTGTTGTTAAACTGTAAGCG |
| JBp104 hu R17,18A-R | CGCTTACAGTTTAACAACAAGGCCGCCCTCAGAAGGCCTTACTACC | ||
| phA3H038 | hu R17E, R18E | JBp105 hu R17,18E-F | CCTCGGGTAGTAAGGCCTTCTGAGCTCCTCCTTGTTGTTAAACTGTAAGCGGAA |
| JBp106 hu R17,18E-R | TTCCGCTTACAGTTTAACAACAAGGAGGAGCTCAGAAGGCCTTACTACCCGAGG | ||
| phA3H039 | hu R26A | JBp107 hu R26A-F | CACAAGAGGGCCTTCGCCGGGTAGTAAGGCCT |
| JBp108 hu R26A-R | AGGCCTTACTACCCGGCGAAGGCCCTCTTGTG | ||
| phA3H040 | hu R26E | JBp109 hu R26E-F | AACACAAGAGGGCCTTCTCCGGGTAGTAAGGCCTTC |
| JBp110 hu R26E-R | GAAGGCCTTACTACCCGGAGAAGGCCCTCTTGTGTT | ||
| phA3H041 | hu R175A | JBp111 hu R175A-F | TTTATCCTGTCAAGCCGTGCCTTTATGGCTCGACTGTTTT |
| JBp112 hu R175A-R | AAAACAGTCGAGCCATAAAGGCACGGCTTGACAGGATAAA | ||
| phA3H042 | hu R175E | JBp113 hu R175E-F | CTGCTTTATCCTGTCAAGCCGCTCCTTTATGGCTCGACTGTTTTTA |
| JBp114 hu R175E-R | TAAAAACAGTCGAGCCATAAAGGAGCGGCTTGACAGGATAAAGCAG | ||
| phA3H043 | hu R176A | JBp115 hu R176A-F | CTTTATCCTGTCAAGCGCTCGCTTTATGGCTCGACTGTTT |
| JBp116 hu R176A-R | AAACAGTCGAGCCATAAAGCGAGCGCTTGACAGGATAAAG | ||
| phA3H044 | hu R176E | JBp117 hu R176E-F | CTTTATCCTGTCAAGCTCTCGCTTTATGGCTCGACTGTTT |
| JBp118 hu R176E-R | AAACAGTCGAGCCATAAAGCGAGAGCTTGACAGGATAAAG | ||
| phA3H045 | hu R175A, R176A | JBp119 hu R175,176A-F | GGACTGCTTTATCCTGTCAAGCGCTGCCTTTATGGCTCGACTGTTTTTA |
| JBp120 hu R175,176A-R | TAAAAACAGTCGAGCCATAAAGGCAGCGCTTGACAGGATAAAGCAGTCC |
ID, identifier.
At the end of the primer name, whether the primer is a forward (F) or reverse (R) primer is indicated after the hyphen.
Expression of A3H proteins.
Recombinant huA3H II, pgtA3Hα, and all related mutants were produced in Escherichia coli using codon-optimized synthetic DNAs (30). Ligation-independent cloning (LIC) was used with the expression vectors pMCSG7 (56) (N-terminal His6 tag and tobacco etch virus [TEV] protease cleavage site), including all pgtA3Hα and huA3H II mutants. All sequences were confirmed by Sanger sequencing. Plasmids in Table 2 were transformed into the E. coli strain BL21(DE3) carrying the pGro7 plasmid for coexpression of chaperones GroES-GroEL (TaKaRa) (57). Transformed cells were grown in terrific broth (TB) medium containing 4% glycerol, ampicillin (0.1 mg/ml), and chloramphenicol (0.035 mg/ml) at 37°C until the optical density at 600 nm (OD600) reached 0.6 to 0.8. Chaperone expression was induced with l-arabinose (1 g/liter) at 37°C for 1 h; then, the cultures were cooled to 20°C, induced with 200 μM isopropyl-β-d-thiogalactopyranoside (IPTG), and grown overnight (14 to 16 h). Cells were harvested via centrifugation and stored at –20°C.
Purification of A3H proteins.
Bacterial cell pellets were resuspended in buffer A (50 mM Tris [pH 8.5], 300 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol) with 2 mM MgCl2, 0.5 mg/ml DNase I, 0.5 mg/ml RNase A, and 0.1 mg/ml lysozyme (lysis conditions). Under these conditions, resuspended cells from a 1-liter culture were incubated on ice for at least 1 h, followed by sonication. Supernatants of centrifuged cell lysates were batch-bound to 5 ml nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) overnight at 4°C. The resin was washed with 5 column volumes (CV) of buffer A followed by a 5-CV wash using buffer A with 1 M NaCl and 20 mM imidazole. Protein was eluted with 6 CV buffer A with 200 mM imidazole. Purification tags were removed by overnight incubation with TEV protease (plus 5 mM dithiothreitol [DTT]) in dialysis against buffer A. Tag-free protein was separated from uncleaved protein and TEV protease with a 5-ml His Trap column (GE Healthcare), and the flowthrough was concentrated and incubated for 4 h with 25 μg/ml RNase A at 25°C. Concentrated protein was loaded onto a Superdex 16/60 S200 gel filtration column and eluted in 50 mM Tris (pH 8), 300 mM NaCl, 5% glycerol, and 1 mM tris(2-carboxyethyl)phosphine (TCEP). All APOBEC3H proteins eluted as monodisperse complexes with an apparent molecular weight (MW) of ∼60 kDa (calculated A3H MW = 25.2 kDa). Peak fractions were pooled, concentrated to 10 mg/ml, and flash frozen in liquid N2. All purified proteins, despite removal efforts, contained nucleic acid as shown by the absorbance ratio (A260/A280) of ∼1.4 to 1.5. This was true of wild-type A3H as well as RNA-binding mutants. Tag-free protein was used for all biochemical experiments. All protein concentrations were determined using a Bradford assay kit (Bio-Rad).
Quantitative PCR-based reverse transcriptase assay.
293T cells were grown at 37°C and 5% CO2 in DMEM containing 10% FBS and 50 μg/ml gentamicin. For production of virus stocks, 293T cells were cultured to 70% confluence in 10-cm2 dishes and transfected with 6 μg pCMV-ΔR8.2 (58) and 24 μg polyethylenimine (PEI). The medium was changed 24 h posttransfection, and virus-containing supernatant was harvested 48 h posttransfection and filtered through a 0.22-μm filter. Virions were purified and concentrated via ultracentrifugation through a 25% sucrose cushion. The concentration of harvested RT-containing virus was quantified using a real-time PCR-based reverse transcriptase (RT) assay where HIV-1-containing medium with known concentrations of capsid protein (CA) (p24) was used as a standard, and thus, the number of virions could be quantified (42, 59, 60). A virus concentration of ∼7.84 × 105 particles/μl, corresponding to ∼3.92 × 108 U of RT/μl for the RT assays described below, was used.
The intact NL4-3 5ʹ-leader RNA (nt 1 to 356) was in vitro transcribed and purified as described previously (59) and was a kind gift from Michael Summers (University of Maryland Baltimore County [UMBC]). The 5ʹ-leader RNA was the template for annealing of a synthetic 18-nt RNA primer (IDT) complementary to the primer binding site (PBS) region where tRNALys3 binds in vivo (61), and this annealed RNA hybrid served as the template for reverse transcription. We note that the first step in the RT reaction will synthesize the strong-stop DNA that will serve as the template in the subsequent PCR steps. The RNA template (8 ng) was preincubated with various molar ratios of A3H at room temperature in binding buffer (10 mM Tris [pH 7.5], 140 mM KCl, 50 mM NaCl) with added RNase inhibitor for 20 min prior to the addition of RT-containing viral lysate.
Quantitative PCR was conducted using PrimeTime Gene Expression Master Mix (IDT), primers (CCCACTGCTTAAGCCTCAATA [sense] and GTTACCAGAGTCACACAACAGA [antisense]) and a double-quenched probe that specifically recognized strong-stop DNA (TTATATGCAGCATCTGAGGGCTCGC). Cycling conditions were as follows: (i) 37°C for 10 min for RT synthesis of strong-stop DNA, (ii) 95°C for 3 min, (iii) 95°C for 15 s, (iv) 60°C for 60 s, and (v) 80°C for 5 s. Steps (ii) through (iv) were repeated for a total of 45 cycles. Negative-control reactions were performed in the absence of reverse transcriptase (viral lysate) as well as in the absence of added A3H. Cycle thresholds (Ct) were measured using the Roche LightCycler, and Ct values calculated with the vendor’s software were quantitated [nanograms of RNA template converted to (−) ssDNA] with a standard curve from a dilution series of the RNA template with no added A3H.
Cytidine deaminase assays.
Cytidine deaminase activity was evaluated in a coupled assay with uracil-DNA glycosylase (UDG) (22, 62). Recombinant UDG (63) was produced in E. coli from pUDG001 generated by ligation-independent cloning of a synthetic DNA into pMCSG7, and purified by Ni-affinity chromatography (30). All substrates contained a 5ʹ fluorescent tag (6-carboxyfluorescein [6-FAM]): 5ʹ-(6-FAM) tag and are summarized in Table 3.
TABLE 3.
Substrate library
| Assay and substrate name | Length (nt) | RNA or DNA | Fluorophore | Sequencea |
|---|---|---|---|---|
| Deaminase assays for substrate length | ||||
| JBs01b | 40 | DNA | 5'-6-FAM | AATGAAAGATATAAGACTCAAATTGAAATAGTTAAGATTA |
| JBs02 | 16 | DNA | 5'-6-FAM | TAAGAATCAAATTGAT |
| JBs03 | 10 | DNA | 5'-6-FAM | AGACTCAAAT |
| Competition binding fluorescence polarization experiments |
||||
| JBs13 | 60 | DNA | TATAATGAAAAATGAAAGATATAAGACTCAAATTGAAATAGTTAAGATTAAATAGATTAA | |
| JBs12 | 50 | DNA | TGAAAAATGAAAGATATAAGACTCAAATTGAAATAGTTAAGATTAAATAG | |
| JBs05 | 40 | DNA | AATGAAAGATATAAGACTCAAATTGAAATAGTTAAGATTA | |
| JBs06 | 30 | DNA | AAGATATAAGACTCAAATTGAAATAGTTAA | |
| JBs07 | 25 | DNA | ATATAAGACTCAAATTGAAATAGTT | |
| JBs08 | 16 | DNA | TAAGACTCAAATTGAA | |
| JBs09 | 10 | DNA | AGACTCAAAT | |
| Heteroduplex deaminase assays and fluorescence polarization |
||||
| JBs27 | 40 | RNA | UAAUCUUAACUAUUUCAAUUUGAGUCUUAUAUCUUUCAUU | |
| JBs28 | 40 | RNA | UAAUCUUAACUAUUUCAAUUUAUUUCUUAUAUCUUUCAUU | |
| JBs29 | 40 | RNA | UAAUCUUAUACAUUUCAAUUUGAGUCUUAUAUCUUUCAUU | |
| JBs30 | 17 | RNA | GUCUUAUAUCUUUCAUU | |
| JBs31 | 20 | RNA | UAAUCUUAACUAUUUCAAUU | |
| JBs32 | 40 | DNA | TAATCTTAACTATTTCAATTTGAGTCTTATATCTTTCATT | |
| JBs33 | 42 | DNA | 5'-6-FAM | TGAATGAAAGATATAAGACTCAAATTGAAATAGTTAAGATTA |
| JBs34 | 18 | RNA | UCUUAUAUCUUUCAUUCA | |
| JBs35 | 17 | RNA | CUUAUAUCUUUCAUUCA | |
| JBs36 | 40 | RNA | 5'-6-FAM | AAUGAAAGAUAUAAGACUCAAAUUGAAAUAGUUAAGAUUA |
| Deaminase assays for sequence specificity |
||||
| JBs37 | 16 | DNA | 5'-6-FAM | TAAGAATCTAATTGAT |
| JBs38 | 16 | DNA | 5'-6-FAM | TAAGAATCGAATTGAT |
| JBs39 | 16 | DNA | 5'-6-FAM | TAAGAATCCAATTGAT |
| JBs40 | 16 | DNA | 5'-6-FAM | TAAGATTCAAATTGAT |
| JBs41 | 16 | DNA | 5'-6-FAM | TAAGAGTCAAATTGAT |
| JBs42 | 16 | DNA | 5'-6-FAM | TAAGACTCAAATTGAT |
| JBs43 | 40 | DNA | 5'-6-FAM | AATGAAAGATATAAGACTUAAATTGAAATAGTTAAGATTA |
The TC dinucleotide motif and −2, −1, 0, or +1 nucleotides are shown in boldface type and underlined.
JBs01 is the main substrate used in deaminase assays and fluorescence polarization experiments with huA3H and pgtA3Hα mutants.
(i) Multiple-turnover kinetic assays. For multiple-turnover kinetic assays, reaction mixtures (total volume 40 μl) contained 1 μM substrate, 40 nM UDG, 50 mM Bis-Tris (pH 6.5), 40 mM NaCl, 5 mM MgCl2, 1 mM DTT, and either 50 or 100 nM A3H (protein concentrations are noted in figure legends). Reactions at 37°C were initiated by the addition of A3H. At each time point, 3 μl of the reaction mixture was quenched with 6 μl of 0.3 M NaOH. Next, samples were heated 15 min at 70°C to hydrolyze DNA at the deglycosylated nucleotide, augmented with 21 μl formamide loading solution, heated 10 min at 95°C, and resolved by 15% urea denaturing polyacrylamide gel electrophoresis.
(ii) Single-turnover kinetic assays. For single-turnover kinetic assays, reaction mixtures (total volume of 80 μl) contained 50 nM substrate, 40 nM UDG, 50 mM Bis-Tris (pH 6.5), 40 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 500 nM A3H. Reactions at 37°C were initiated by the addition of A3H. At each time point, 6 μl of the reaction mixture was quenched with 3 μl of 0.6 M NaOH. Samples were heated 15 min at 70°C to hydrolyze DNA at the deglycosylated nucleotide, augmented with 6 μl formamide loading solution, heated 10 min at 95°C, and resolved by 15% urea denaturing polyacrylamide gel electrophoresis. Gels were scanned using a Typhoon fluorescence imager, and bands corresponding to cleaved and uncleaved DNA were quantified using the ImageQuant software (GE Healthcare Life Sciences).
Analytical gel filtration.
Pure, dimeric, RNA-bound wt pgtA3Hα and pgtA3Hα R175E were each incubated in either low-salt buffer (50 mM Tris [pH 8], 300 mM NaCl, 5% glycerol) or high-salt buffer (50 mM Tris [pH 8], 2 M NaCl, 5% glycerol) for 16 h at 4°C. Protein (0.5 mg) was loaded onto a 24-ml Superdex 200 10/300 gel filtration column preequilibrated in either low- or high-salt buffer. Absorbance ratios (260 nm/280 nm) were measured for each 250-μl fraction. A260/A280 ratios of ≥1 indicated nucleic acid association, while ratios of ≤1 indicated relatively nucleic-acid free protein. Gel filtration chromatograms were overlaid with absorbance ratio measurements to assess the effect of high-salt incubation on A3H-RNA association.
Differential scanning fluorimetry.
Purified huA3H II and pgtA3Hα mutants were diluted to a concentration of 3 μM in reaction buffer (50 mM Bis-Tris [pH 6.5], 40 mM NaCl, 5 mM MgCl2, 1 mM DTT) containing 5× SYPRO Orange dye (10-μl final volume in a 384-well PCR plate). Reactions were run on the Applied Biosystems QuantStudio 7 Flex qPCR machine where the temperature ramped from 25 to 85°C at a rate of 0.01°C/s. Melting temperature (Tm) values were determined in GraphPad Prism.
Fluorescence polarization binding assay.
Serial 2× dilutions (0.3 nM to 5 μM) of A3H stocks were made in binding buffer (50 mM Bis-Tris [pH 6.5], 40 mM NaCl, 5 mM MgCl2, 1 mM DTT). A no-protein, buffer-only solution was a negative control. Triplicate samples were incubated with a 10 nM fixed concentration of 5ʹ-6-FAM-labeled substrate in a black 384-well plate (Corning). Binding reactions (50-μl total volume) were incubated for 10 min at room temperature, and fluorescence polarization (FP) was measured with excitation at 485 nm and emission at 525 nm. The data were fit to a one-site binding model (GraphPad Prism) to determine apparent affinities of A3H for a library of substrates (Table 3).
For competition binding experiments, an apparent binding affinity (Kd) of 16 nM, was first determined for the catalytically inactive variant pgtA3Hα E56A. A pgtA3Hα E56A concentration at which 90 to 100% of fluorescently labeled DNA was bound was selected for competition binding experiments (80 nM). A fixed concentration of 10 nM 5ʹ-6-FAM-labeled 40-nt DNA substrate was incubated with 80 nM pgtA3Hα E56A for 15 min prior to the addition of serial twofold dilutions of unlabeled DNA oligomers (1.2 nM to 20 μM). Mixtures with unlabeled oligomers were incubated for 15 min at room temperature prior to FP measurement. The data were fitted to a one-site Ki model (GraphPad Prism) to determine apparent inhibitory constants (Ki) for each unlabeled oligomer.
Hybridization of RNA and DNA oligomers.
5ʹ-6-FAM-labeled 40-nt ssDNA, 42-nt ssDNA, and 40-nt ssRNA served as the templates for annealing complementary oligomers (Table 3). Equimolar template and complementary oligomers were mixed together in annealing buffer (10 mM Tris [pH 7.5], 50 mM NaCl, 1 mM EDTA) to a final concentration of 10 μM. The mixture was heated 2 min at 95°C and then slowly cooled (0.1°C/s) to 25°C, incubated for 20 min, and stored at –20°C. Calculated melting temperatures (Northwest oligonucleotide calculator) for the hybrid substrates are listed in Table 1.
Electrophoretic mobility shift assays (EMSAs).
Serial threefold dilutions (2.7 nM to 2 μM) of huA3H II were made in binding buffer (50 mM Bis-Tris [pH 6.5], 40 mM NaCl, 5 mM MgCl2, 1 mM DTT, 20% glycerol) ranging. Additionally, a no-protein control was tested. Protein samples were incubated with a 10 nM fixed concentration of 5ʹ-6-FAM-labeled substrate. The binding reaction mixtures (20-μl total volume) were incubated for 1 h at 4°C. While binding reactions incubated, polyacrylamide gels were prerun at 50 V. Bound complexes were resolved by 5% TBE (Tris-borate-EDTA) native gel electrophoresis (50 V, 2.5 h, 4°C). Gels were scanned using a Typhoon fluorescence imager. For EMSAs involving RNA-based hybrids or single-stranded RNA, the protein dilutions were preincubated with RNase inhibitor for 20 min prior to mixing with substrate.
ACKNOWLEDGMENTS
The research was supported by NIH grant U54 AI150470 to A.T., T.H., and J.L.S., R01 AI078788 to T.H., NIH fellowship F31 AI129697 to J.A.B., and NIH T32 GM008353 to J.A.B.
We thank Jennifer Meagher at the University of Michigan Life Sciences Institute for kindly providing purified ZAP protein as well as Pengfei Ding and Michael Summers at University of Maryland Baltimore County (UMBC) for providing in vitro-transcribed 5ʹ-leader RNA for our RT assays.
REFERENCES
- 1.Harris RS, Dudley JP. 2015. APOBECs and virus restriction. Virology 479-480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malim MH, Bieniasz PD. 2012. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med 2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhen A, Du J, Zhou X, Xiong Y, Yu X-F. 2012. Reduced APOBEC3H variant anti-viral activities are associated with altered RNA binding activities. PLoS One 7:e38771. doi: 10.1371/journal.pone.0038771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng Y-H. 2011. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol 85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris RS, Liddament MT. 2004. Retroviral restriction by APOBEC proteins. Nat Rev Immunol 4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 6.York A, Kutluay SB, Errando M, Bieniasz PD. 2016. The RNA binding specificity of human APOBEC3 proteins resembles that of HIV-1 nucleocapsid. PLoS Pathog 12:e1005833. doi: 10.1371/journal.ppat.1005833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Apolonia L, Schulz R, Curk T, Rocha P, Swanson CM, Schaller T, Ule J, Malim MH. 2015. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog 11:e1004609. doi: 10.1371/journal.ppat.1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Refsland EW, Harris RS. 2013. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol 371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stavrou S, Ross SR. 2015. APOBEC3 proteins in viral immunity. J Immunol 195:4565–4570. doi: 10.4049/jimmunol.1501504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 11.Mitra M, Singer D, Mano Y, Hritz J, Nam G, Gorelick RJ, Byeon I-J, Gronenborn AM, Iwatani Y, Levin JG. 2015. Sequence and structural determinants of human APOBEC3H deaminase and anti-HIV-1 activities. Retrovirology 12:3. doi: 10.1186/s12977-014-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol 15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 13.Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res 35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bishop KN, Holmes RK, Malim MH. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J Virol 80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes RK, Koning FA, Bishop KN, Malim MH. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem 282:2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- 16.Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. 2008. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog 4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollpeter D, Parsons M, Sobala AE, Coxhead S, Lang RD, Bruns AM, Papaioannou S, McDonnell JM, Apolonia L, Chowdhury JA, Horvath CM, Malim MH. 2018. Deep sequencing of HIV-1 reverse transcripts reveals the multifaceted antiviral functions of APOBEC3G. Nat Microbiol 3:220–233. doi: 10.1038/s41564-017-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cullen BR. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J Virol 80:1067–1076. doi: 10.1128/JVI.80.3.1067-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang Y, Siew LM, Wang X, Han Y, Lampen R, Zheng YH. 2008. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J Biol Chem 283:11606–11614. doi: 10.1074/jbc.M707586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol 11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 21.Kohli RM, Abrams SR, Gajula KS, Maul RW, Gearhart PJ, Stivers JT. 2009. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J Biol Chem 284:22898–22904. doi: 10.1074/jbc.M109.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu J, Chen Q, Xiao X, Ito F, Wolfe A, Chen XS. 2016. Biochemical characterization of APOBEC3H variants: implications for their HIV-1 restriction activity and mC modification. J Mol Biol 428:4626–4638. doi: 10.1016/j.jmb.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDougall WM, Smith HC. 2011. Direct evidence that RNA inhibits APOBEC3G ssDNA cytidine deaminase activity. Biochem Biophys Res Commun 412:612–617. doi: 10.1016/j.bbrc.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaban NM, Shi K, Lauer KV, Carpenter MA, Richards CM, Salamango D, Wang J, Lopresti MW, Banerjee S, Levin-Klein R, Brown WL, Aihara H, Harris RS. 2018. The antiviral and cancer genomic DNA deaminase APOBEC3H is regulated by an RNA-mediated dimerization mechanism. Mol Cell 69:75–86.e9. doi: 10.1016/j.molcel.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belanger K, Savoie M, Rosales Gerpe MC, Couture JF, Langlois MA. 2013. Binding of RNA by APOBEC3G controls deamination-independent restriction of retroviruses. Nucleic Acids Res 41:7438–7452. doi: 10.1093/nar/gkt527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Ao Z, Chen L, Kobinger G, Peng J, Yao X. 2012. The cellular antiviral protein APOBEC3G interacts with HIV-1 reverse transcriptase and inhibits its function during viral replication. J Virol 86:3777–3786. doi: 10.1128/JVI.06594-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaurasiya KR, McCauley MJ, Wang W, Qualley DF, Wu T, Kitamura S, Geertsema H, Chan DS, Hertz A, Iwatani Y, Levin JG, Musier-Forsyth K, Rouzina I, Williams MC. 2014. Oligomerization transforms human APOBEC3G from an efficient enzyme to a slowly dissociating nucleic acid-binding protein. Nat Chem 6:28–33. doi: 10.1038/nchem.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morse M, Huo R, Feng Y, Rouzina I, Chelico L, Williams MC. 2017. Dimerization regulates both deaminase-dependent and deaminase-independent HIV-1 restriction by APOBEC3G. Nat Commun 8:597. doi: 10.1038/s41467-017-00501-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adolph MB, Ara A, Chelico L. 2019. APOBEC3 host restriction factors of HIV-1 can change the template switching frequency of reverse transcriptase. J Mol Biol 431:1339–1352. doi: 10.1016/j.jmb.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 30.Bohn JA, Thummar K, York A, Raymond A, Brown WC, Bieniasz PD, Hatziioannou T, Smith JL. 2017. APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA. Nat Commun 8:1021. doi: 10.1038/s41467-017-01309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng Y, Wong L, Morse M, Rouzina I, Williams MC, Chelico L. 2018. RNA-mediated dimerization of the human deoxycytidine deaminase APOBEC3H influences enzyme activity and interaction with nucleic acids. J Mol Biol 430:4891–4907. doi: 10.1016/j.jmb.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aydin H, Taylor MW, Lee JE. 2014. Structure-guided analysis of the human APOBEC3-HIV restrictome. Structure 22:668–684. doi: 10.1016/j.str.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 33.LaRue RS, Jonsson SR, Silverstein KA, Lajoie M, Bertrand D, El-Mabrouk N, Hotzel I, Andresdottir V, Smith TP, Harris RS. 2008. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol 9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 35.Feng Y, Baig TT, Love RP, Chelico L. 2014. Suppression of APOBEC3-mediated restriction of HIV-1 by Vif. Front Microbiol 5:450. doi: 10.3389/fmicb.2014.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol 22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 37.OhAinle M, Kerns JA, Malik HS, Emerman M. 2006. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol 80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuoka T, Nagae T, Ode H, Awazu H, Kurosawa T, Hamano A, Matsuoka K, Hachiya A, Imahashi M, Yokomaku Y, Watanabe N, Iwatani Y. 2018. Structural basis of chimpanzee APOBEC3H dimerization stabilized by double-stranded RNA. Nucleic Acids Res 46:10368–10379. doi: 10.1093/nar/gky676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kouno T, Silvas TV, Hilbert BJ, Shandilya SMD, Bohn MF, Kelch BA, Royer WE, Somasundaran M, Kurt Yilmaz N, Matsuo H, Schiffer CA. 2017. Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity. Nat Commun 8:15024. doi: 10.1038/ncomms15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maiti A, Myint W, Kanai T, Delviks-Frankenberry K, Sierra Rodriguez C, Pathak VK, Schiffer CA, Matsuo H. 2018. Crystal structure of the catalytic domain of HIV-1 restriction factor APOBEC3G in complex with ssDNA. Nat Commun 9:2460. doi: 10.1038/s41467-018-04872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi K, Carpenter MA, Banerjee S, Shaban NM, Kurahashi K, Salamango DJ, McCann JL, Starrett GJ, Duffy JV, Demir Ö, Amaro RE, Harki DA, Harris RS, Aihara H. 2017. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat Struct Mol Biol 24:131–139. doi: 10.1038/nsmb.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vermeire J, Naessens E, Vanderstraeten H, Landi A, Iannucci V, Van Nuffel A, Taghon T, Pizzato M, Verhasselt B. 2012. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PLoS One 7:e50859. doi: 10.1371/journal.pone.0050859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi K, Carpenter MA, Kurahashi K, Harris RS, Aihara H. 2015. Crystal structure of the DNA deaminase APOBEC3B catalytic domain. J Biol Chem 290:28120–28130. doi: 10.1074/jbc.M115.679951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Telesnitsky A, Goff SP. 1997. Reverse transcriptase and the generation of retroviral DNA, p 121–160 In Coffin JM, Hughes SH, Varmus HE (ed), Retroviruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 45.Li J, Chen Y, Li M, Carpenter MA, McDougle RM, Luengas EM, Macdonald PJ, Harris RS, Mueller JD. 2014. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. J Mol Biol 426:1296–1307. doi: 10.1016/j.jmb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito F, Yang H, Xiao X, Li SX, Wolfe A, Zirkle B, Arutiunian V, Chen XS. 2018. Understanding the structure, multimerization, subcellular localization and mC selectivity of a genomic mutator and anti-HIV factor APOBEC3H. Sci Rep 8:3763. doi: 10.1038/s41598-018-21955-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li A, Li J, Johnson KA. 2016. HIV-1 reverse transcriptase polymerase and RNase H (ribonuclease H) active sites work simultaneously and independently. J Biol Chem 291:26566–26585. doi: 10.1074/jbc.M116.753160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian L, Kim MS, Li H, Wang J, Yang W. 2018. Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid. Proc Natl Acad Sci U S A 115:507–512. doi: 10.1073/pnas.1719746115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. 2011. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol 85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sievers F, Higgins DG. 2018. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci 27:135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao X, Li S-X, Yang H, Chen XS. 2016. Crystal structures of APOBEC3G N-domain alone and its complex with DNA. Nat Commun 7:12193. doi: 10.1038/ncomms12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bieniasz P, Telesnitsky A. 2018. Multiple, switchable protein:RNA interactions regulate human immunodeficiency virus type 1 assembly. Annu Rev Virol 5:165–183. doi: 10.1146/annurev-virology-092917-043448. [DOI] [PubMed] [Google Scholar]
- 54.Ito F, Fu Y, Kao SA, Yang H, Chen XS. 2017. Family-wide comparative analysis of cytidine and methylcytidine deamination by eleven human APOBEC proteins. J Mol Biol 429:1787–1799. doi: 10.1016/j.jmb.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Del Prete GQ, Keele BF, Fode J, Thummar K, Swanstrom AE, Rodriguez A, Raymond A, Estes JD, LaBranche CC, Montefiori DC, KewalRamani VN, Lifson JD, Bieniasz PD, Hatziioannou T. 2017. A single gp120 residue can affect HIV-1 tropism in macaques. PLoS Pathog 13:e1006572. doi: 10.1371/journal.ppat.1006572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. 2002. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif 25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 57.Mogk A, Mayer MP, Deuerling E. 2002. Mechanisms of protein folding: molecular chaperones and their application in biotechnology. Chembiochem 3:807–814. doi:. [DOI] [PubMed] [Google Scholar]
- 58.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 59.Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, Barton S, Hosic A, Florwick A, Santos J, Bolden NC, McCowin S, Case DA, Johnson BA, Salemi M, Telesnitsky A, Summers MF. 2015. RNA structure. Structure of the HIV-1 RNA packaging signal. Science 348:917–921. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kharytonchyk S, Monti S, Smaldino PJ, Van V, Bolden NC, Brown JD, Russo E, Swanson C, Shuey A, Telesnitsky A, Summers MF. 2016. Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc Natl Acad Sci U S A 113:13378–13383. doi: 10.1073/pnas.1616627113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masuda T, Sato Y, Huang YL, Koi S, Takahata T, Hasegawa A, Kawai G, Kannagi M. 2015. Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci Rep 5:17680. doi: 10.1038/srep17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nair S, Rein A. 2014. In vitro assay for cytidine deaminase activity of APOBEC3 protein. Bio Protoc 4:e1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dmitrochenko AE, Turiianskaia OM, Gilep AA, Usanov SA, Iantsevich AV. 2014. An effective scheme to produce recombinant uracil-DNA glycosylase of Escherichia coli for PCR diagnostics. Prikl Biokhim Mikrobiol 50:398–407. (In Russian.) [PubMed] [Google Scholar]