Newcastle disease virus (NDV) substantially impacts the poultry industry worldwide and causes viral encephalitis and neurological disorders leading to brain damage, paralysis, and death. The mechanism of interaction between this neurotropic virus and the avian central nervous system (CNS) is largely unknown. Here, we report that host protein CARD11 presented brain-specific upregulated expression that inhibited NDV replication, which was not due to CARD11-Bcl10-MALT1 (CBM) complex-triggered activation of its downstream signaling pathways. The inhibitory mechanism of viral replication is through the CARD11 CC1 domain, and the viral large polymerase protein (L) competitively interacts with the X domain of the viral phosphoprotein (P), which hampers the P-L interaction, suppressing the viral polymerase activity and viral replication. An in vivo study indicated that CARD11 alleviated neuropathological lesions and reduced viral replication in chicken brains. These results provide insight into the interaction between NDV infection and the host defense in the CNS and a potential antiviral target for viral neural diseases.
KEYWORDS: avian neurons, brain-specific upregulation, CARD11, competitive binding, inhibition to viral replication, Newcastle disease virus, viral RNP, viral polymerase activity
ABSTRACT
Host factors play multiple essential roles in the replication and pathogenesis of mammalian neurotropic viruses. However, the cellular proteins of the central nervous system (CNS) involved in avian neurotropic virus infection have not been completely elucidated. Here, we employed a gene microarray to identify caspase recruitment domain-containing protein 11 (CARD11), a lymphoma-associated scaffold protein presenting brain-specific upregulated expression in a virulent neurotropic Newcastle disease virus (NDV)-infected natural host. Chicken primary neuronal cells infected with NDV appeared slightly syncytial and died quickly. CARD11 overexpression inhibited viral replication and delayed cytopathic effects; conversely, depletion of CARD11 enhanced viral replication and cytopathic effects in chicken primary neuronal cells. The inhibition of viral replication by CARD11 could not be blocked with CARD11-Bcl10-MALT1 (CBM) signalosome and NF-κB signaling inhibitors. CARD11 was found to interact directly with the viral phosphoprotein (P) through its CC1 domain and the X domain of P; this X domain also mediated the interaction between P and the viral large polymerase protein (L). The CARD11 CC1 domain and L competitively bound to P via the X domain that hindered the P-L interaction of the viral ribonucleoprotein (RNP) complex, resulting in a reduction of viral polymerase activity in a minigenome assay and inhibition of viral replication. Animal experiments further revealed that CARD11 contributed to viral replication inhibition and neuropathology in infected chicken brains. Taken together, our findings identify CARD11 as a brain-specific antiviral factor of NDV infection in avian species.
IMPORTANCE Newcastle disease virus (NDV) substantially impacts the poultry industry worldwide and causes viral encephalitis and neurological disorders leading to brain damage, paralysis, and death. The mechanism of interaction between this neurotropic virus and the avian central nervous system (CNS) is largely unknown. Here, we report that host protein CARD11 presented brain-specific upregulated expression that inhibited NDV replication, which was not due to CARD11-Bcl10-MALT1 (CBM) complex-triggered activation of its downstream signaling pathways. The inhibitory mechanism of viral replication is through the CARD11 CC1 domain, and the viral large polymerase protein (L) competitively interacts with the X domain of the viral phosphoprotein (P), which hampers the P-L interaction, suppressing the viral polymerase activity and viral replication. An in vivo study indicated that CARD11 alleviated neuropathological lesions and reduced viral replication in chicken brains. These results provide insight into the interaction between NDV infection and the host defense in the CNS and a potential antiviral target for viral neural diseases.
INTRODUCTION
Neurotropic viruses, such as Japanese encephalitis virus, Zika virus, rabies virus, pseudorabies virus, highly pathogenic avian influenza virus, and Newcastle disease virus (NDV), can irreversibly disrupt the central nervous system (CNS), negatively affecting human and animal health (1–3). The CNS plays a major role in organisms, as it is associated with the regulation of specific physiological functions that are essential for survival. Neurotropic viruses specifically invade the CNS in vertebrates, resulting in dramatic neural degeneration and damage (1). In humans and other mammals, cellular factors of the CNS play a vital role in the neuropathogenesis of neurotropic viruses. For instance, the neuronal N-methyl-d-aspartate receptor (NMDAR), E3 ubiquitin ligase Nedd4, and P21-activated kinase 4 (PAK4) signaling activities facilitate Japanese encephalitis virus-induced neuronal cell damage (4–6). The neural RNA-binding protein Musashi-1 (MSI1) and upregulation of AXL participate in the replication and cellular entry of Zika virus, respectively (7, 8), and the metabotropic glutamate receptor subtype 2 (mGluR2) mediates rabies virus entry and replication (9). However, in avian neurotropic viruses, such as NDV, the avian CNS factors involved in viral neuropathogenesis have not been elucidated, and identifying these factors will help us to understand the host response and viral neurovirulence in avian species.
NDV is an avian-specific highly virulent virus that poses a major threat to the poultry industry worldwide. NDV, also called avian avulavirus 1 or avian paramyxovirus serotype 1, is a member of the genus Avulavirus in the family Paramyxoviridae (10, 11). The NDV genome comprises nonsegmented, single-stranded negative-sense RNA that contains six genes encoding the nucleocapsid (NP), matrix protein (M), phosphoprotein (P), fusion protein (F), hemagglutinin-neuraminidase protein (HN), and large polymerase protein (L); the additional proteins V and W are generated by RNA editing during P mRNA synthesis (12). Viral RNA synthesis is driven by the ribonucleoprotein (RNP) complex that utilizes the viral NP, P, and L for viral transcription and replication (13). NDV strains have been classified into three pathotypes based on their pathogenicity in chickens: lentogenic (avirulent or low virulent), mesogenic (moderately virulent), and velogenic (highly virulent) (14). Velogenic strains are further classified into velogenic viscerotropic (VVNDV) and velogenic neurotropic (VNNDV) strains. VVNDV strains produce lethal hemorrhagic lesions in the viscera, causing breathing increases, weakness, clonospasm, and muscle tremor, whereas VNNDV strains can cause viral encephalitis and neurological disorders, such as paralysis of the legs or wings and retractable head, as well as respiratory and digestive system disorders, such as respiratory distress, cough, asthma, and diarrhea (15).
Caspase recruitment domain family, member 11 (CARD11), also known as caspase recruitment domain-containing C-terminal membrane-associated guanylate kinase (MAGUK) protein-1 (CARMA1), belongs to both the CARD family and the MAGUK family and serves as a scaffold protein (16, 17). Chicken CARD11 is a 1,170-amino-acid protein containing the N-terminal CARD domain, LATCH, a coiled-coil (CC) domain, and an autoinhibitory domain (ID), followed by a PSD-95/Dlg/ZO-1 (PDZ) domain, an SRC homology 3 (SH3) domain, and a guanylate kinase (GUK) domain that together constitute the MAGUK domain (16). Functionally, CARD11 plays a crucial role in signaling downstream from the T cell receptor (TCR) and B cell receptor (BCR). Most studies report that after TCR or BCR stimulation, CARD11 is activated and constitutively localized to lipid rafts at the plasma membrane and serves as a nucleation center for the CARD11/B cell lymphoma 10 (Bcl10)/mucosa-associated lymphoid tissue lymphoma-translocation gene 1 (MALT1) (CBM) complex, interacting with the partner proteins Bcl10 and MALT1, to trigger downstream pathways (18, 19). MALT1 in the CBM signalosome is believed to be essential for activation of the canonical nuclear factor κB (NF-κB), c-Jun N-terminal kinase (JNK), and mammalian target of rapamycin (mTOR) pathways in lymphocytes (20). While gain-of-function mutations of CARD11 are linked to lymphoproliferative disorders (18, 21, 22), loss-of-function mutations result in severe human immunodeficiency conditions (22). The normal structures of CARD11 and the CBM complex are believed to be essential for lymphocytes, diffuse large B cell lymphoma (DLBCL), primary gastric B cell lymphoma, primary lymphoma of the central nervous system (PCNSL), and adult T cell leukemia (23, 24). However, until now, little was known about the exact role of CARD11 in virus infection.
To identify the cellular proteins in the avian CNS that are involved in NDV infection, we used gene microarray screening to identify CARD11 as a brain-specific upregulated protein that influences the replication of NDV. We investigated the inhibition of CARD11 upregulation on NDV replication in chicken primary neuronal cells (chPNCs) and chicken brain. This antiviral effect of CARD11 in NDV was shown by the suppression of viral RNA polymerase activity via competitively interacting with the viral P to hamper the P-L interaction. This study is the first to show that CARD11 is involved in neuron-specific antiviral activity and provides novel insight into the neuropathogenesis of NDV in avian species.
RESULTS
A gene microarray analysis identifies CARD11 with brain-specific upregulation in NDV-infected chickens.
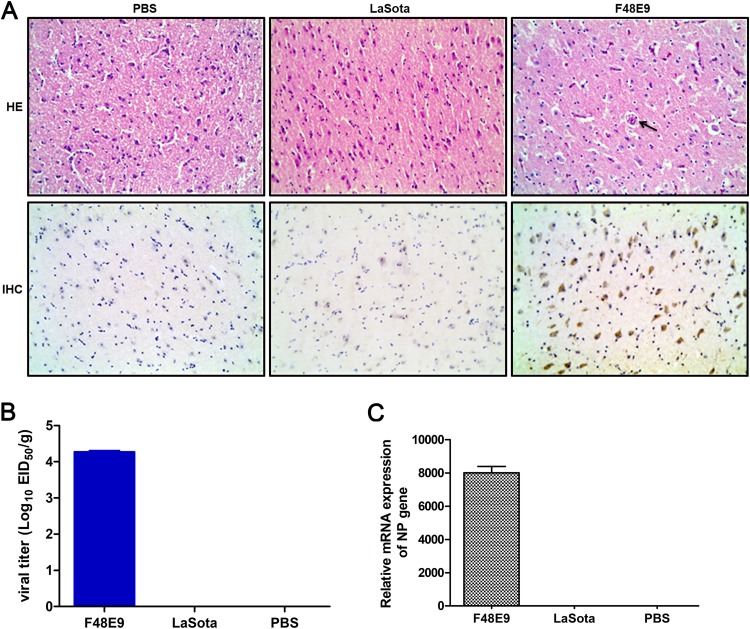
Our previous study reported that the neurotropic virulent NDV strain F48E9 caused encephalitis and neurological disorders in chickens, the natural host (25). In this study, specific-pathogen-free (SPF) chickens infected with virulent strain F48E9 via intraocular-nasal routes showed diarrhea, depression, cough, asthma, and respiratory distress at 3 days postinfection (dpi) and neurological symptoms (such as somnolence, retractable head, legs, or wings, paralysis, and limping) at 4 to 5 dpi. All animals died before 6 dpi. Conversely, the SPF chickens infected with the avirulent strain LaSota or the phosphate-buffered saline (PBS) control showed no apparent clinical signs. For the pathological changes in the infected chicken brains, perivascular cuffing formed by lymphocyte infiltration, congestion, encephalomalacia, and loose nerve fibers were observed in the cerebra of virulent F48E9-infected chicken brains via hematoxylin-eosin staining (HE) assay, but these changes were not observed in the LaSota and PBS groups (Fig. 1A, top). Similarly, the virus was only detected in the F48E9 strain-infected brains in an immunohistochemistry (IHC) assay (Fig. 1A, bottom). Moreover, the F48E9 virus was detected by viral titration and NP gene expression in the infected brains at 5 dpi, but LaSota was not detected (Fig. 1B and C), suggesting that the virulent virus, but not the avirulent virus, was capable of replication in the brain.
FIG 1.
Histopathology of the brains in NDV-infected chickens. (A) Four-week-old SPF chickens were infected with virulent F48E9, avirulent LaSota (105 PFU/100 μl/chicken), and PBS control (100 μl/chicken) via the intraocular-nasal routes. The brain samples were collected for HE and IHC analyses with anti-F48E9 mouse PAb (1:500) and anti-LaSota mouse PAb (1:500) at 5 dpi. The black arrows indicate vascular cuffing lesions in the cerebrum. Bars, 50 μm. (B) The viral titers were titrated in 9- to 11-day-old SPF embryonated chicken eggs. The 50% egg infective dose (EID50) values were calculated using the Reed-Muench method. (C) The relative mRNA expression of viral NP genes in F48E9- and LaSota-infected chicken brains, normalized to that of 28S rRNA, was determined by RT-qPCR. The results are presented as the means ± SDs from three independent experiments.
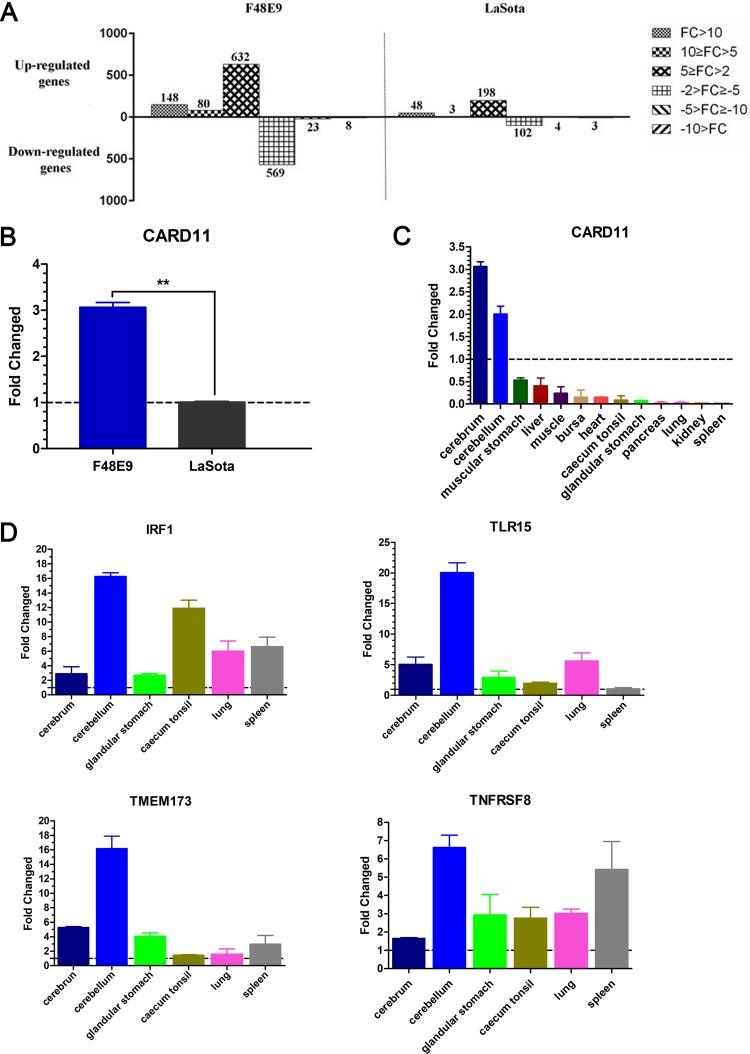
To analyze the host gene expression profiles in the infected chicken brains at 5 dpi, we used Agilent 44K chicken whole-genome chips. All gene microarray files were available and uploaded from the Gene Expression Omnibus database (accession number GSE121368). Genes were determined to be differentially expressed if they exhibited a fold change (FC) of >2 and a P value of <0.05. A total of 860 and 249 differentially expressed genes (DEGs) were found to be upregulated in the F48E9- and LaSota-infected brains, respectively, compared to those in the mock-infected brains. For the downregulated genes, 600 and 109 DEGs were identified in the F48E9 and LaSota groups, respectively (Fig. 2A). Fourteen of the DEGs involved in the immune response were significantly induced or repressed by the F48E9 virus (Table 1). Among these genes, one upregulated gene, CARD11, has not been presented in any published transcriptome databases of NDV-infected chicken cells or tissues, which include no neurons (26–28). We confirmed that CARD11 was upregulated in the brains of infected chickens and found that the relative mRNA expression of CARD11 was significantly increased by F48E9 infection but not by LaSota infection (Fig. 2B). Surprisingly, the upregulated expression of CARD11 induced by F48E9 infection was found only in the brain, cerebrum, and cerebellum and not in nonneural tissues (Fig. 2C), indicating that CARD11 is brain-specifically upregulated by the virulent virus. To further confirm this brain-specific upregulation, we found that IRF1, TLR15, TMEM173, and TNFRSF8 were upregulated in other nonneural tissues (Fig. 2D). These results suggested that CARD11 could play an important role in NDV infection in the brain.
FIG 2.
Identification of CARD11, a host factor with brain-specific upregulation in NDV-infected chickens. (A) The numbers of DEGs with different FCs (P < 0.05) in virus-infected brains are shown in the columns. Four-week-old SPF chickens were infected with virulent F48E9 and avirulent LaSota (105 PFU/100 μl) via the intraocular-nasal route. Brain samples were collected at 5 dpi. Total RNA was extracted from the brains for gene microarray analysis. The relative mRNA expression of CARD11 in NDV-infected chicken brains (B) and other tissues (C) normalized to that of 28S rRNA was determined by RT-qPCR. (D) The relative mRNA expression of IRF1, TLR15, TMEM173, and TNFRSF8 in NDV-infected chicken tissues was determined by RT-qPCR and normalized to that of 28S rRNA. Representative data shown as the means ± SDs (n = 3) were analyzed by two-tailed Student's t tests. **, P < 0.01.
TABLE 1.
Differentially expressed immunity-related genes
| No. | Gene symbol | Accession no. | Description | Function | Change factor (in F48E9 relative to that in LaSota) |
|---|---|---|---|---|---|
| 1 | B2M | NM_001001750 | β-2-Microglobulin | Immune response | 11.72 |
| 2 | BF2 | NM_001031338 | Major histocompatibility complex class I antigen BF2 | Immune response | 24.52 |
| 3 | CARD11 | NM_001006161 | Caspase recruitment domain family, member 11 | Positive regulation of NF-κB transcription factor activity | 6.12 |
| 4 | HSPD1 | NM_001012916 | Heat shock 60 kDa protein 1 (chaperonin) | MyD88-dependent Toll-like receptor signaling pathway; positive regulation of IFN-α production; positive regulation of macrophage activation | −2.03 |
| 5 | IL18 | NM_204608 | Interleukin 18 (IFN-γa-inducing factor) | Positive regulation of IFN-γ production; IFN-γ biosynthetic process; positive regulation of natural killer cell proliferation | 38.63 |
| 6 | IRF1 | NM_205415 | Interferon regulatory factor 1 | Induction of IFN; antivirus; anti-tumor | 27.46 |
| 7 | IRF7 | NM_205372 | Interferon regulatory factor 7 | Toll-like receptor signaling pathway; activation of innate immune response | 10.63 |
| 8 | IRF8 | NM_205416 | Interferon regulatory factor 8 | Immune response; positive regulation of IFN-γ production | 4.73 |
| 9 | JMJD6 | NM_001030703 | Jumonji domain containing 6 | Macrophage activation | −3.15 |
| 10 | MYD88 | NM_001030962 | Myeloid differentiation primary response gene (88) | MyD88-dependent Toll-like receptor signaling pathway | 2.91 |
| 11 | TLR15 | NM_001037835 | Toll-like receptor 15 | Critical component of the innate immune response | 59.64 |
| 12 | TMEM173 | XM_001232170 | Transmembrane protein 173 | Innate immune response; defense response to virus; IFN-β production | 18.03 |
| 13 | TNFRSF8 | NM_204444 | Tumor necrosis factor receptor superfamily, member 8 | Cytokine-mediated signaling pathway; positive regulation of tumor necrosis factor biosynthetic process; tumor necrosis factor-mediated signaling pathway | 17.47 |
| 14 | UBA52 | NM_205075 | Ubiquitin A-52 residue ribosomal protein fusion product 1 | Toll-like receptor signaling pathway; MyD88-dependent Toll-like receptor signaling pathway; activation of MAPK activity involved in innate immune response | −2.02 |
IFN-γ, gamma interferon.
Replication and infection of NDV in chPNCs.
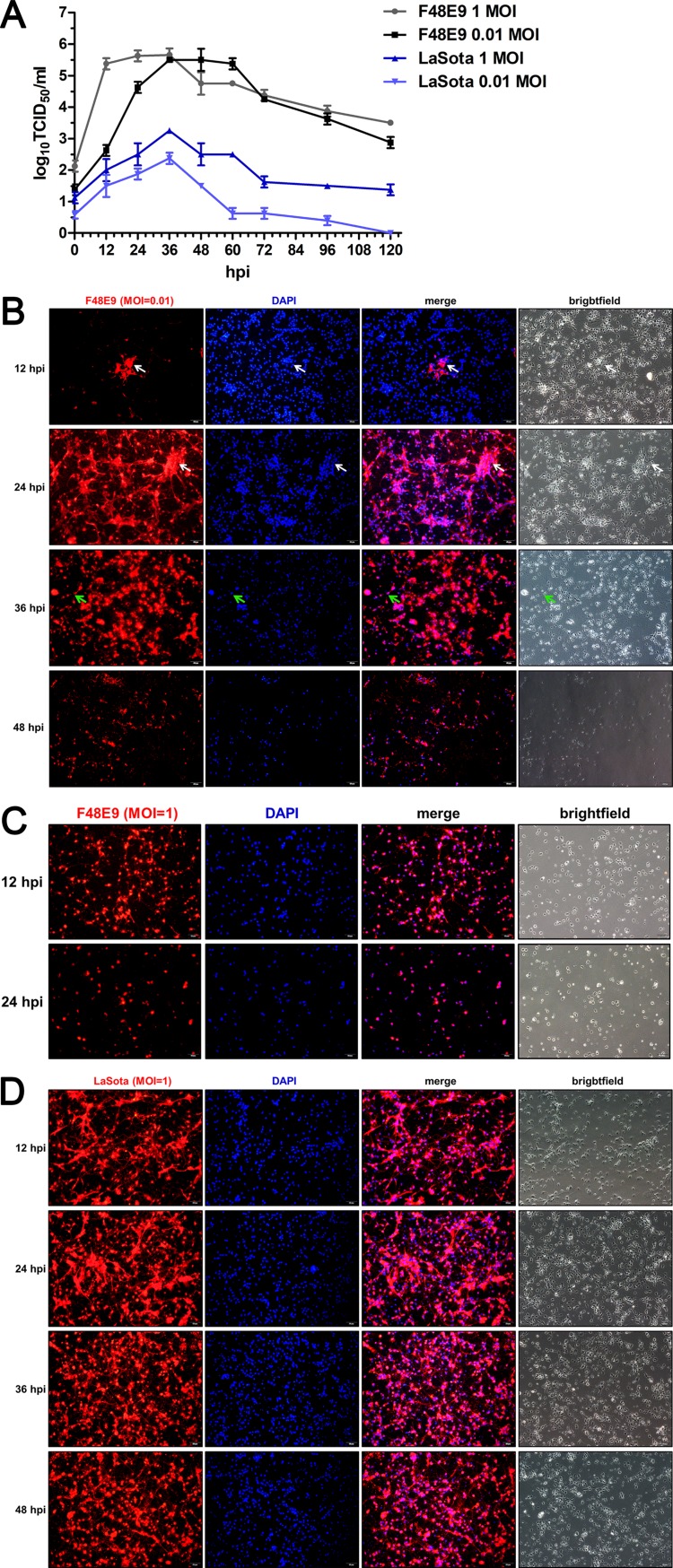
The chPNCs were infected with the virulent F48E9 strain and avirulent LaSota strain. The virus titers in the culture supernatants were measured according to the 50% tissue culture infective dose (TCID50) assay method. The growth of F48E9 with a high multiplicity of infection (MOI) of 1 in single-step replication was faster than that with a low multiplicity of infection (MOI) of 0.01 in multiple-step replication during early infection at 12 and 24 h postinfection (hpi). The growth titers of LaSota with an MOI of 1 were higher than those with an MOI of 0.01 throughout the infection period (Fig. 3A). Importantly, the growth kinetics of F48E9 were significantly higher than those of LaSota at either a high MOI or a low MOI. Meanwhile, F48E9 infection with an MOI of 0.01 caused slight syncytia among the cytopathic effects (CPEs) in chPNCs at 24 hpi. Axon disruption and cell body detachment appeared at 36 hpi, and most of the cells were lysed and dead at 48 hpi (Fig. 3B). Severe CPEs rapidly appeared at 12 hpi in F48E9-infected chPNCs with an MOI of 1, and the cells were dead at 24 hpi (Fig. 3C). However, LaSota-infected chPNCs exhibited no CPEs (Fig. 3D). These results suggested that the virulent virus is capable of replication and induces severe CPE in neuronal cells.
FIG 3.
Virulent NDV shows better replication than lentogenic NDV in chPNCs. (A) Replication of NDV in chPNCs. The cells were infected with F48E9 (MOI = 0.01 or 1) and LaSota (MOI = 0.01 or 1) for 1 h at 37°C. The viral replicates of the culture supernatants at different times after infection were titrated in DF-1 cells. The results are presented as the means ± SDs from three independent experiments. (B to D) The infection of NDV strains in the chPNCs. chPNCs were infected with F48E9 (MOI = 0.01) (B), F48E9 (MOI = 1) (C), and LaSota (MOI = 1) (D) at 12, 24, 36, and 48 hpi. The cells were examined by immunofluorescence assay (IFA) using an anti-NDV mouse PAb (1:200). The white arrows indicate syncytia, and the green arrows indicate axon disruption. Bars, 50 μm.
CARD11 is upregulated by NDV infection in chPNCs.
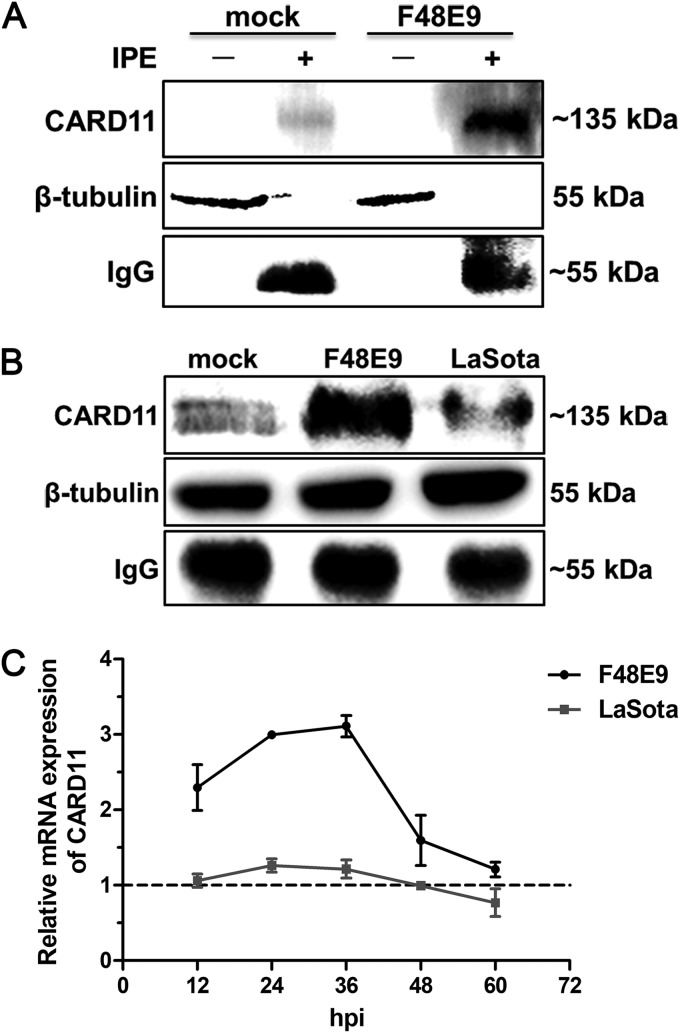
To examine whether the upregulated expression of CARD11 is induced by viral infection in chPNCs, we used an immunoprecipitation enrichment (IPE) method to detect CARD11 expression in chPNCs. CARD11 could not be detected by conventional Western blotting in either mock-infected or F48E9-infected chPNCs (Fig. 4A), suggesting an extremely low level of CARD11 expression in chPNCs. Using an IPE method, we detected CARD11 and showed it was dramatically upregulated by viral infection in chPNCs (Fig. 4A and B). F48E9 infection induced a higher level of CARD11 expression than LaSota infection (Fig. 4B). The mRNA expression of CARD11 (Fig. 4C) was consistent with these IPE data. These results indicated that CARD11 expression could be upregulated by NDV infection in neurons.
FIG 4.
CARD11 is upregulated in NDV-infected chPNCs. (A) The expression of cellular CARD11 was only detected by IPE. The chPNCs in 60-mm-diameter dishes (∼107 per dish) at day 3 infected with F48E9 (MOI = 0.01) and LaSota (MOI = 1) were collected at 24 hpi. For collection of the samples without IPE, the mock- and NDV-infected cells were scraped and centrifuged at 300 × g for 5 min at 4°C. The cells were lysed in 50 μl 1× SDS loading buffer. For the IPE assay samples, the mock- and NDV-infected cells were lysed in 500 μl of ice-cold RIPA buffer with PMSF (1:1,000) for 30 min and centrifuged at 13,000 × g for 15 min at 4°C. The supernatants were incubated with an anti-CARD11 mouse PAb (10 μl) at 4°C for 4 h and captured by the addition of 40 μl of a protein A/G-agarose (Abmart) slurry and gentle rotation overnight at 4°C. The agarose beads were further washed three times and resuspended in 50 μl of 1× SDS loading buffer and boiled for 10 min. All the nonimmunoprecipitated and immunoprecipitated CARD11 samples were analyzed by Western blotting with anti-CARD11 mouse PAb (1:1,000). The protein loading controls are listed: β-tubulin for samples without IPE and IgG for IPE proteins. (B) The upregulated expression of CARD11 protein in NDV-infected chPNCs with IPE assays. The immunoprecipitated CARD11 was analyzed by Western blotting with anti-CARD11 mouse PAb. The protein loading controls are listed: β-tubulin for input proteins, and IgG for IP proteins. (C) The upregulation of CARD11 mRNA expression in the infected chPNCs. The relative mRNA expression of CARD11 was analyzed by RT-qPCR in chPNCs infected with F48E9 (MOI = 0.01) and LaSota (MOI = 1) at 12, 24, 36, 48 and 60 hpi. The results are presented as the means ± SDs from three independent experiments.
CARD11 overexpression inhibits viral replication.
To test whether CARD11 upregulation affected viral replication in chPNCs, we used a recombinant adenovirus (rAdV) system overexpressing CARD11. CARD11 overexpression was examined both in HEK293A cells and chPNCs (Fig. 5A). The infection efficiency of rAdV-CARD11 was measured by flow cytometry in chPNCs (Fig. 5B). The rAdV-CARD11 infected 79.4% and 89.3% of the cells at MOIs of 100 and 200, respectively. Because the chPNCs infected with rAdV-CARD11 at an MOI of 200 were detached and died during infection (Fig. 5C), rAdV-CARD11 infection at an MOI of 100 was chosen for the subsequent experiments. The chPNCs were then infected with F48E9 (MOI = 0.01) or LaSota (MOI = 1) at 48 h after rAdV-CARD11 infection. However, the CPEs in F48E9-infected CARD11-overexpressing and control chPNCs were not obvious (Fig. 6A). The viral titers of NDV in CARD11-overexpressing chPNCs were significantly decreased at 36 to 60 hpi with F48E9 (Fig. 6B) and at 60 to 72 hpi with LaSota (Fig. 6C).Consistently, the levels of F48E9 and LaSota viral proteins, especially M and NP, in the infected cells were lower in the CARD11-overexpressing chPNCs than in the control cells (Fig. 6D). These results suggested that CARD11 overexpression could inhibit viral replication.
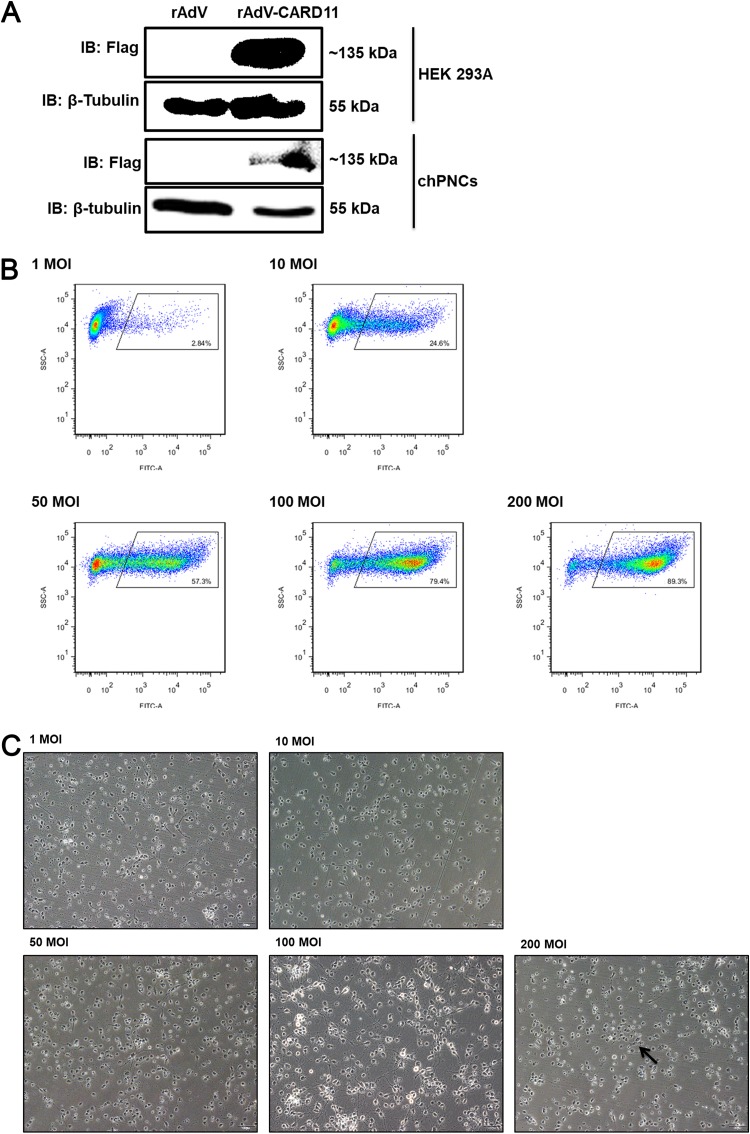
FIG 5.
The infection efficiency of rAdVs in chPNCs. (A) Detection of rAdV-CARD11 (MOI = 100) infected HEK293A cells and chPNCs. At 36 hpi in HEK293A cells and 48 hpi in chPNCs, the cell lysates were analyzed by Western blotting with an anti-Flag mouse MAb. β-Tubulin was used as a protein loading control. (B and C) Infection efficiency of rAdV-CARD11 in chPNCs. The chPNCs were infected with rAdV-CARD11 at different MOIs (1, 10, 50, 100, and 200) for 2 h at 37°C. The rAdV-CARD11-infected chPNCs at 48 hpi were collected by 0.25% trypsin digestion and incubated with an anti-Flag mouse MAb and goat anti-mouse IgG/FITC at 4°C with minimal exposure to light. (B) After the cells were washed, they were assessed by flow cytometry. (C) The cells were observed under a microscope. The black arrow indicates cell bodies with no axons. Bars, 50 μm.
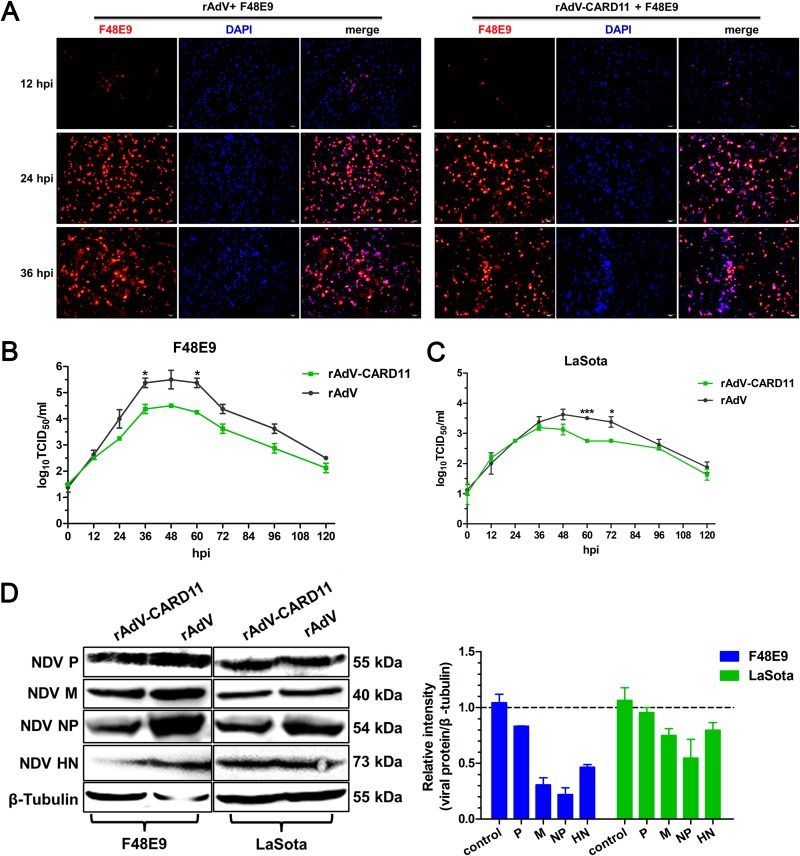
FIG 6.
CARD11 overexpression inhibits viral replication in chPNCs. (A) CPEs of NDV infection in the CARD11-overexpressing chPNCs. The cells were infected with rAdV and rAdV-CARD11 (MOI = 100). After 48 hpi, the cells were infected with F48E9 (MOI = 0.01) and observed at 12, 24, and 36 hpi by IFA with an anti-NDV mouse PAb. Bars, 50 μm. (B and C) NDV replication in CARD11-overexpressing chPNCs. The cells were infected with rAdV and rAdV-CARD11 (MOI = 100). At 48 hpi, the cells were infected with F48E9 (MOI = 0.01) (B) and LaSota (MOI = 1) (C). The viral titers in the supernatants of chPNCs were analyzed via the TCID50 method. Representative data, shown as the means ± SDs (n = 3), were analyzed with a two-tailed Student's t tests. *, P < 0.05; ***, P < 0.001. (D) The expression of viral proteins in CARD11-overexpressing chPNCs. The F48E9- (MOI = 0.01) and LaSota-infected (MOI = 1) CARD11-overexpressing cells at 36 hpi were analyzed by Western blot analysis with a guinea pig anti-LaSota P antibody, an anti-NP guinea pig PAb, an anti-HN guinea pig PAb, and an anti-M synthetic peptide (KLEKGHTLAKYNPFK) mouse PAb. The expression levels of viral proteins relative to that of β-tubulin were analyzed by densitometry. The results are presented as the means ± SDs from three independent experiments.
CARD11 depletion increases viral replication.
Next, an rAdV-mediated RNA interference (RNAi) system was used to knockdown CARD11 expression. Of the two rAdV-shRNA constructs, short hairpin RNA 1 (shRNA1) more effectively knocked down CARD11 expression than the negative control (NC) (Fig. 7A and B). The CPEs caused by F48E9 in the rAdV-shRNA1 cells appeared earlier than those in the rAdV-NC cells (Fig. 7C). NDV replication was significantly increased in the rAdV-shRNA1 cells with F48E9 infection (Fig. 7D) and LaSota infection at 36 hpi (Fig. 7E). Consistently, the expression of viral proteins, especially HN and NP, in the rAdV-shRNA1 cells was significantly increased compared to that in the rAdV-NC cells (Fig. 7F). These data revealed that CARD11 participated in the inhibition of viral replication in neuronal cells.
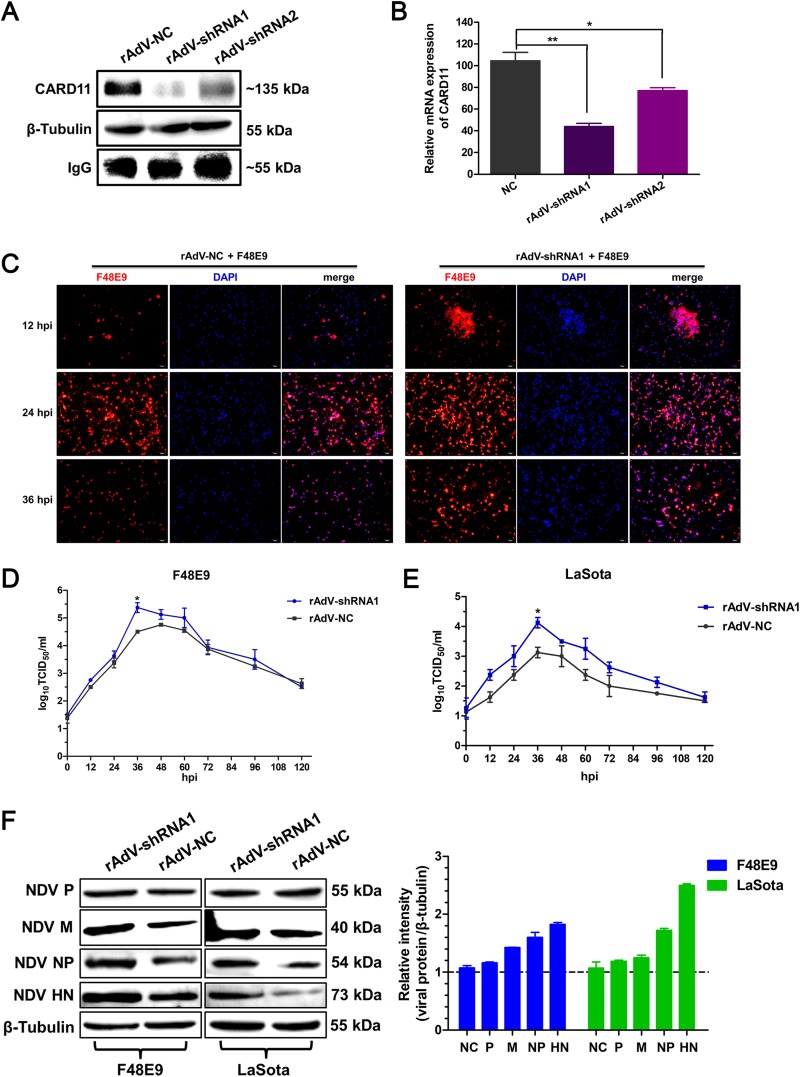
FIG 7.
CARD11 depletion increases viral replication in chPNCs. (A and B) The downregulation of CARD11 in rAdV-shRNA-infected chPNCs. The cells were infected with rAdV-NC, rAdV-shRNA1, and rAdV-shRNA2 (MOI = 100) in 100-mm dishes. (A) At 48 hpi, CARD11 was detected using the IPE assay. (B) The relative mRNA expression of CARD11 was determined by RT-qPCR. The data were normalized to 28S rRNA. Representative data, shown as the means ± SDs (n = 3), were analyzed by two-tailed Student's t tests. *, P < 0.05; **. P < 0.01. (C) A virulent virus causes CPEs in CARD11-knockdown chPNCs. The rAdV-NC and rAdV-shRNA1 cells were infected with F48E9 (MOI = 0.01) and observed by IFA with a mouse anti-LaSota PAb. Bars, 50 μm. (D and E) Growth kinetics of NDV in CARD11-knockdown chPNCs. The cells were transfected with rAdV-shRNA1 (MOI = 100). At 48 hpi, the cells were infected with virulent F48E9 (MOI = 0.01) (D) and avirulent LaSota (MOI = 1) (E). The viral titers in the supernatants were analyzed with the TCID50 method. Representative data, shown as the means ± SDs (n = 3), were analyzed by two-tailed Student's t tests. *, P < 0.05. (F) Expression of viral proteins in the infected CARD11-knockdown cells. The F48E9- (MOI = 0.01) and LaSota-infected (MOI = 1) CARD11-shRNA1 cells at 36 hpi were analyzed by Western blot analysis as described for Fig. 6D. The expression level of viral proteins relative to that of β-tubulin was analyzed by densitometry. The results are presented as the means ± SDs from three independent experiments.
The CBM signalosome has no effect on inhibiting viral replication.
CARD11 interacts with Bcl10 and MALT1 to form the CBM complex. Then, MALT1 triggers the activation of the NF-κB, JNK, and mTOR signaling pathways that are involved in inflammation, immune regulation, cell proliferation, and cell survival (29). We used three inhibitors, MI-2 for MALT1, BAY 11-7082 for NF-κB, and BMS-345541 for IκB kinase (IKK), to evaluate whether CBM signalosome-induced activation of the signaling pathways plays a role in inhibiting viral replication. The cytotoxicity of these three inhibitors was first assessed in the chPNCs. BAY 11-7082 at 1 μM, BMS-345541 at 10 μM, and MI-2 at 1 μM had no cytotoxicity or cell effects (Fig. 8A), and these doses of the three inhibitors were used for the following experiments. During NDV infection in the chPNCs, the viral replication of F48E9 and LaSota was not affected by the three inhibitors (Fig. 8B). MI-2, as an irreversible MALT1 inhibitor, prevented the formation of the CBM complex but did not affect the growth of F48E9 in either the CARD11 overexpression (Fig. 8C) or knockdown (Fig. 8D) chPNCs. Furthermore, the viral titers of F48E9 in the CARD11-overexpressing chPNCs were increased by treatment with the NF-κB signaling-specific inhibitors BAY 11-7082 and BMS-345541 but were significantly decreased without treatment (Fig. 8E). However, the viral titers of F48E9 showed no differences in CARD11 knockdown chPNCs treated with or without the two inhibitors (Fig. 8F). These data suggested that the inhibition of viral replication by NF-κB signaling was CBM independent. Moreover, a chicken NF-κB reporter assay was performed to confirm that BAY 11-7082 and BMS-345541 inhibited NF-κB promoter activity in chicken cells (Fig. 8G). These results revealed that the downstream CBM signalosome signaling pathways had no role in inhibiting NDV replication.
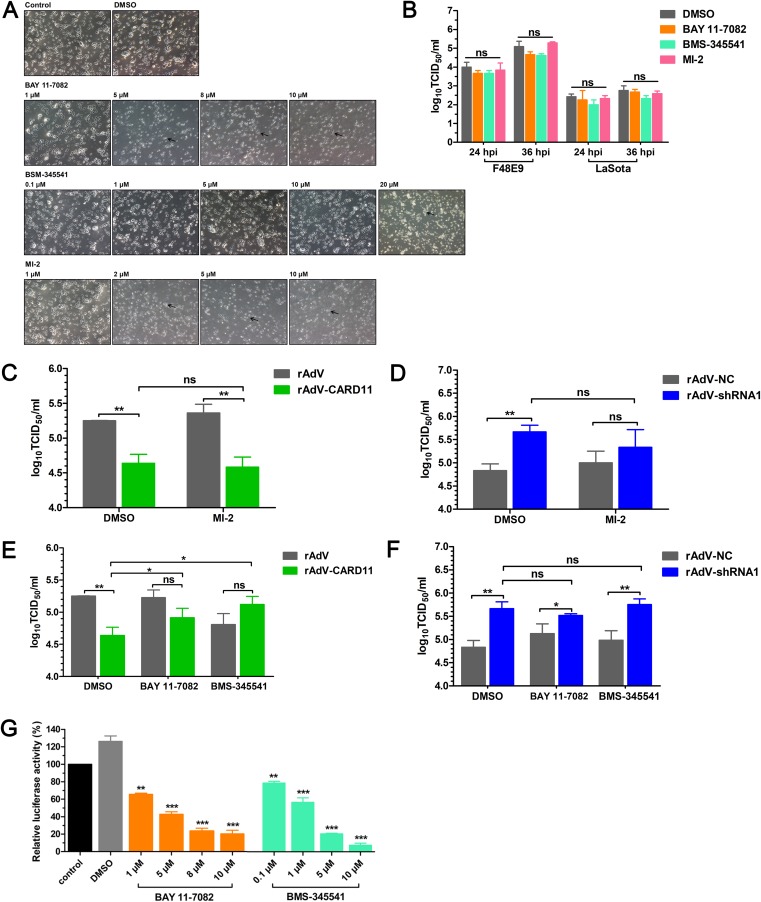
FIG 8.
The CBM signalosome plays no role in inhibiting NDV replication in chPNCs. (A) The CPEs of inhibitors in chPNCs. The chPNCs in 12-well plates at day 3 were incubated with different concentrations of inhibitors for 24 h. The treated chPNCs were observed after 24 h. The black arrows indicate axon disappearance and cell body disruption. Bars, 50 μm. (B) NDV replication in inhibitor-treated chPNCs. The cells at day 3 were treated with BAY 11-7082 (1 μM), BMS-345541 (10 μM), MI-2 (1 μM), and DMSO for 24 h, and then the treated chPNCs were infected with the F48E9 strain (MOI = 0.01). The culture supernatants were harvested at 24 and 36 hpi. (C to F) F48E9 replication in CARD11-overexpressing or knockdown chPNCs treated with inhibitors. The cells at day 3 were infected with rAdV, rAdV-CARD11, rAdV-NC, and rAdV-shRNA1 at the same MOI (MOI = 100). At 48 hpi, the cells were treated with MI-2 (1 μM), BAY 11-7082 (1 μM), BMS-345541 (10 μM), and DMSO for 24 h. Then, the treated chPNCs were infected with the F48E9 strain (MOI = 0.01). The culture supernatants were harvested at 36 hpi for titration of F48E9. The viral titers in the supernatants of chPNCs were analyzed via the TCID50 method in DF-1 cells. (G) The inhibitory role of BAY 11-7082 and BMS-345541 in DF-1 cells. The DF-1 cells in 12-well plates were cotransfected with pchNF-κB-TA-luc and pRL-SV40-N at a ratio of 10:1. Twenty-four hours later, the cells were incubated with 1, 5, 8, or 10 μM BAY 11-7082, 0.1, 1, 5, or 10 μM BMS-345541, or DMSO for 24 h and were lysed to quantify the luciferase activity. Renilla luciferase expressed by pRL-SV40-N was used as the normalizing standard. All the representative data, shown as the means ± SDs (n = 3), were analyzed by two-tailed Student's t tests. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
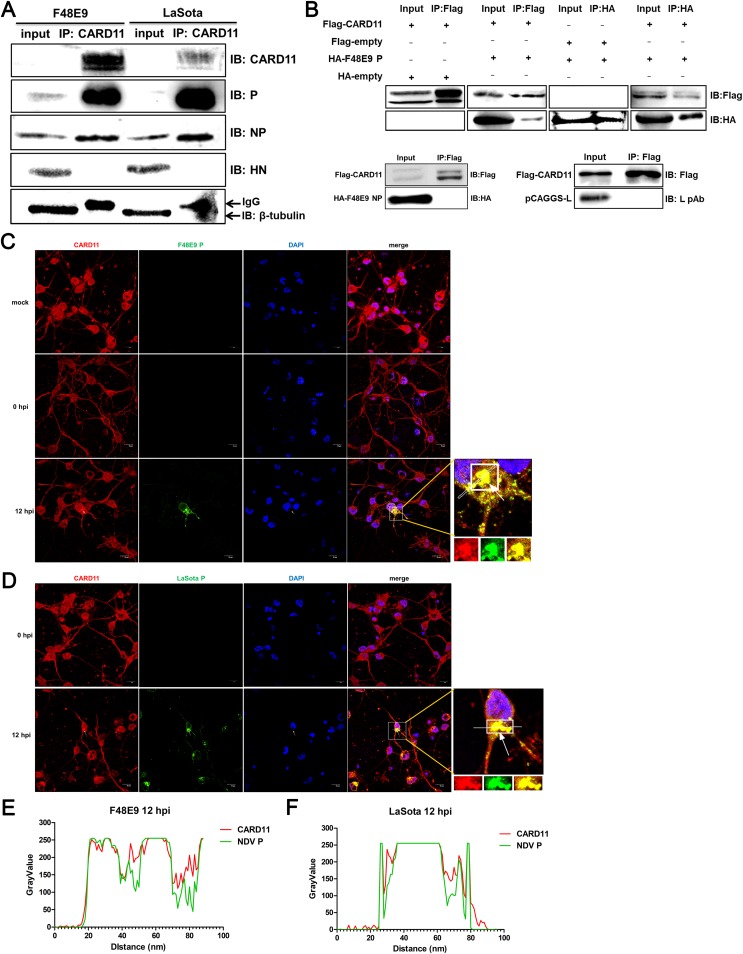
Endogenous CARD11 interacts with viral P in chPNCs.
We next investigated how CARD11 inhibits viral replication. We hypothesized that CARD11 may interact with viral proteins. To verify this hypothesis, we performed an immunoprecipitation experiment in NDV-infected chPNCs. Cellular CARD11 coimmunoprecipitated with P and NP in either F48E9- or LaSota-infected cells. However, the HN, a non-RNP complex protein used as a control, did not interact with the CARD11 protein (Fig. 9A). The P and NP protein of NDV interacted directly as previously described (30). To determine which viral protein interacted with CARD11, we performed coimmunoprecipitation (co-IP) experiments in transfected HEK293T cells with expression constructs of CARD11 with P or NP. We found that CARD11 coimmunoprecipitated with P but not with NP in the F48E9 strain (Fig. 9B). In addition, L, a subunit of the RNP complex, did not coimmunoprecipitate with CARD11 (Fig. 9B). Furthermore, the subcellular colocalization of endogenous CARD11 with the viral P aggregating around the nucleus was observed in both F48E9- (Fig. 9C) and LaSota-infected (Fig. 9D) chPNCs. The colocalization of CARD11 and the viral P at 12 hpi was qualitative in the F48E9- (Fig. 9E) and LaSota-infected (Fig. 9F) neuronal cells. These results revealed that endogenous CARD11 interacted with the viral P in the NDV-infected neurons.
FIG 9.
CARD11 interacts with the viral P in chPNCs. (A) Endogenous CARD11 coimmunoprecipitates with viral P and NP. The chPNCs infected with F48E9 (MOI = 0.01) and avirulent LaSota (MOI = 1) were lysed at 48 hpi, and the proteins were immunoprecipitated using an anti-CARD11 mouse PAb. Before incubation, 40 μl of supernatants as input sample was treated with 10 μl 5× SDS loading buffer. The immunoprecipitated proteins were analyzed by Western blotting using an anti-LaSota P guinea pig PAb, an anti-LaSota NP guinea pig PAb, and an anti-LaSota HN guinea pig PAb. β-Tubulin and IgG were used as protein loading controls for the input and immunoprecipitated proteins. (B) Validation of CARD11 interaction with the viral P by co-IP assays in HEK293T cells. HEK293T cells were cotransfected with pcDNA3-2×Flag-CARD11 and pcDNA3-KHA-P, pcDNA3-KHA-NP, or pCAGGS-L. At 36 h posttransfection, the cell lysates were coimmunoprecipitated using an anti-Flag mouse MAb or an anti-HA rabbit Ab. Before incubating with antibodies, 40 μl of the supernatants as input sample was treated with 10 μl 5× SDS loading buffer. (C and D) Subcellular localization of CARD11 and the viral P in chPNCs. The cells were infected with F48E9 (MOI = 0.01) (C) and LaSota (MOI = 1) (D) and were analyzed at 0 and 12 hpi by IFA with a mouse anti-CARD11 PAb and an anti-LaSota P guinea pig PAb. CARD11 (red), P (green), and the nucleus (blue) were observed by confocal microscopy. The white arrow depicts colocalization. Bars, 10 μm. (E and F) Colocalization analysis of the CARD11 and viral P in the infected chPNCs at 12 hpi using ImageJ.
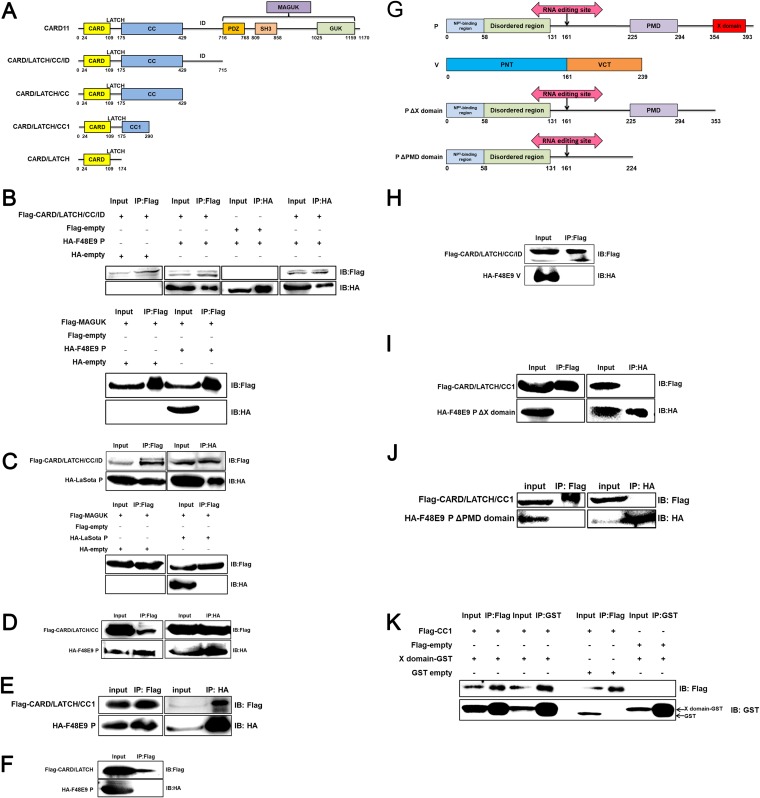
The CARD11 CC1 domain interacts with the viral P X domain.
To determine which domains of CARD11 and the viral P are involved in the interaction, we first truncated the CARD11 protein into four parts from the C terminus based on its functional domains (Fig. 10A). The CARD/LATCH/CC/ID was found to coimmunoprecipitate with the viral Ps of F48E9 (Fig. 10B) and LaSota (Fig. 10C), but not with MAGUK. The CARD/LATCH/CC and CARD/LATCH/CC1 were coimmunoprecipitated with P (Fig. 10D and E) but not the CARD/LATCH domain (Fig. 10F), indicating that only the CC1 domain of CARD11 interacted with the viral P. Correspondingly, P was divided into four regions (NP0-binding region, disordered region, PMD, and X domain) according to a previous report (31). To identify which domain of P interacts with CARD11, we constructed P truncations (Fig. 10G). In addition, V is produced from the viral P gene by the RNA editing machinery. V was unable to coimmunoprecipitate with CARD/LATCH/CC/ID (Fig. 10H), indicating that the N terminus of the viral P does not interact with the CC1 domain. The deletion of the X domain and PMD domain resulted in a failure to coimmunoprecipitate with CARD/LATCH/CC1 (Fig. 10I and J). The single X domain enabled the interaction with the single CC1 domain, which is direct evidence for the interaction of the two domains (Fig. 10K). These results demonstrated the mechanism of physical interaction between CARD11 and the viral P.
FIG 10.
The CARD11 CC1 domain interacts with the X domain of the viral P. (A) Schematic representation of chicken CARD11. The domains are shown as different colored boxes. The amino acid size is depicted below the boxes. The abbreviation of each truncation is shown to the left of each schematic representation. (B to F) The interaction between the CARD11 CC1 domain and the P of F48E9 and the LaSota strain was validated using the co-IP assays. HEK293T cells were transfected with pcDNA3-2×Flag-CARD/LATCH/CC/ID, MAGUK, and pcDNA3-HA-P (F48E9) (B) and pcDNA3-HA-P (LaSota) (C). HEK293T cells were transfected with CARD/LATCH/CC (D), CARD/LATCH/CC1 (E), CARD/LATCH (F), and pcDNA3-HA-P (F48E9). The empty plasmids were used as negative controls. (G) Schematic representation of viral P and V. The domains are shown as different colored boxes. The amino acid size is depicted below the boxes. The abbreviation of each truncation is shown to the left of each schematic representation. PMD, a coiled-coil P-P multimerization domain; PNT, P N terminus; VCT, V protein C terminus. (H) No interaction of CARD/LATCH/CC/ID with viral V as determined by the co-IP assays. P with the deleted X domain (I) and P with the deleted PMD domain (J) did not interact with the CARD/LATCH/CC1 domain of CARD11. (K) The interaction between the CARD11 CC1 domain and the X domain of the F48E9 P. HEK293T cells were transfected with pcDNA3-2×Flag-CC1 and pCAGGS-X-GST. The empty plasmids were used as negative controls. All cell lysates were prepared at 36 h posttransfection and proteins were immunoprecipitated using an anti-Flag mouse MAb, an anti-HA rabbit MAb, or an anti-GST mouse MAb. The immunoprecipitated proteins were analyzed by Western blotting.
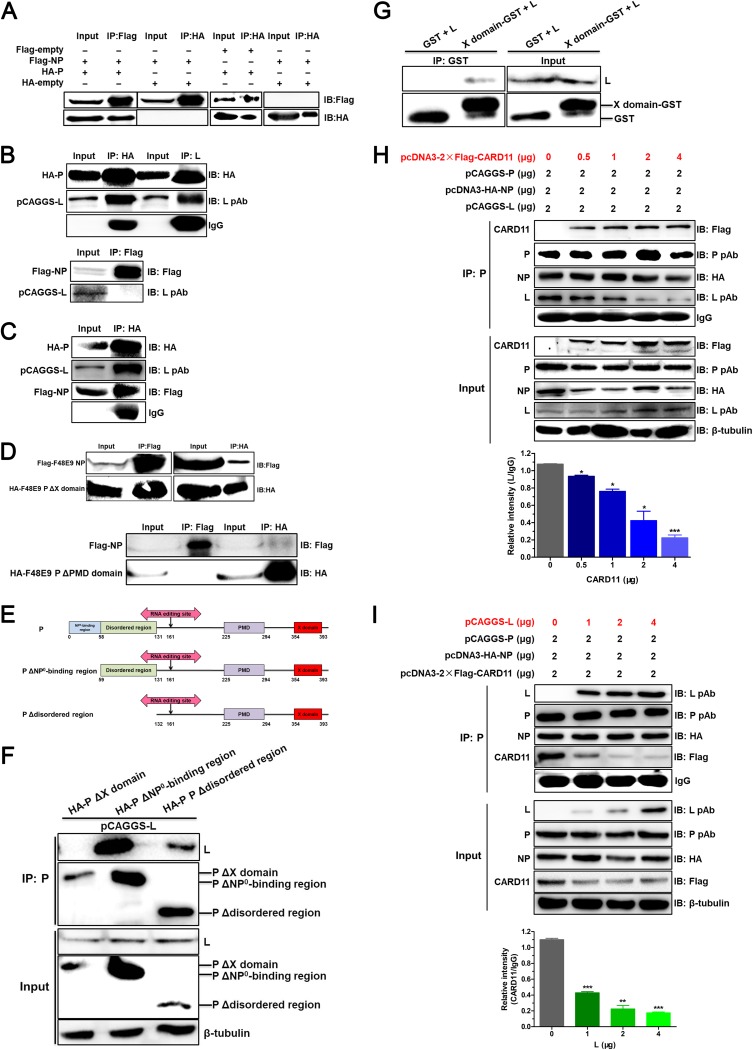
CARD11 competitively hampers the viral P-L interaction mediated by the X domain of P.
Viral RNA synthesis of NDV is driven by the RNP complex for viral transcription and replication. We hypothesized that CARD11 binding to the viral P may kinetically affect NP-P-L interactions of the viral RNP complex. We found that viral P did coimmunoprecipitate with viral NP (Fig. 11A) and L, but NP failed to coimmunoprecipitate with L (Fig. 11B). These interactions of NP and L with P were confirmed by co-IP under coexpression of NP, P, and L (Fig. 11C). These data indicated that viral P served as a bridge of the binding between viral NP and L. The PMD domain of viral P mediated the interaction between P and NP, but the X domain of viral P did not (Fig. 11D). To detect the region of P that interacted with L, P was then further truncated from the N terminus (Fig. 11E). We found that P with the deletion of the NP0-binding region or disordered region interacted with L, but P with the X domain deletion was unable to interact with L (Fig. 11F). Among them, the amount of the immunoprecipitated L in the P with the disordered region deletion was less than that with the NP0-binding region deletion, suggesting that the disordered region might affect the P-L interaction. Furthermore, the single X domain of enabled P to interact with L (Fig. 11G). Based on CARD11 CC1 domain binding to the X domain of P, CARD11 and L may competitively bind to P in the viral RNP complex. To examine this hypothesis, we determined the levels of the coimmunoprecipitated NP, P, and L with different expression levels of CARD11 in the cells. The level of L binding to P was remarkably reduced (Fig. 11H). Likewise, the levels of the coimmunoprecipitated NP, P, and CARD11 were analyzed with different expression levels of L, and the level of CARD11 binding to P was also remarkably reduced (Fig. 11I). These results indicated that the competitive binding of CARD11 to the viral P with L could hamper the P-L interaction of the RNP via the X domain of P in a dose-dependent manner.
FIG 11.
The viral P-L interaction is competitively hampered by CARD11. HEK293T cells were transfected with pcDNA3-2×Flag-NP (F48E9) (2 μg) and pcDNA3-HA-P (F48E9) (2 μg) (A), pcDNA3-HA-P (F48E9) (2 μg) and pCAGGS-L (2 μg), pcDNA3-2×Flag-NP (F48E9) (2 μg), and pCAGGS-L (2 μg) (B), pcDNA3-2×Flag-NP (F48E9) (2 μg), pcDNA3-HA-P (F48E9) (2 μg), and pCAGGS-L (2 μg) (C), and pcDNA3-2×Flag-NP (F48E9) (2 μg) and pcDNA3-HA-P ΔX domain (2 μg) or pcDNA3-HA-P ΔPMD domain (2 μg) (D). The cell lysates were prepared, and proteins were immunoprecipitated using DYKDDDK tag (3B9) mouse MAb or HA tag (C29F4) rabbit MAb. (E) Schematic representation of the truncations of N terminus in viral P. The domains are shown as different colored boxes. The amino acid size is depicted below the boxes. The abbreviation of each truncation is shown to the left of each schematic representation. (F) HEK293T cells were transfected with pCAGGS-L (2 μg) and pcDNA3-HA-P ΔX domain (2 μg), pcDNA3-HA-P ΔNP0-binding region (2 μg), or pcDNA3-HA-P Δdisordered region (2 μg). The cell lysates were prepared, and proteins were immunoprecipitated using a guinea pig anti-LaSota P PAb. (G) The interaction between the X domain of viral P and L. The cell lysates were prepared and immunoprecipitated using anti-GST mouse MAb. (H and I) HEK293T cells were transfected with different doses of pcDNA3-2×Flag-CARD11 or pCAGGS-L and pCAGGS-P and pcDNA3-HA-NP. The cells were lysed and immunoprecipitated using an anti-LaSota P guinea pig PAb. The protein loading controls are β-tubulin for input proteins and IgG for IP proteins. The expression levels of the L and CARD11 protein after immunoprecipitation relative to that of IgG were analyzed by densitometry. All the representative data, shown as the means ± SDs (n = 3), were analyzed by two-tailed Student's t tests. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
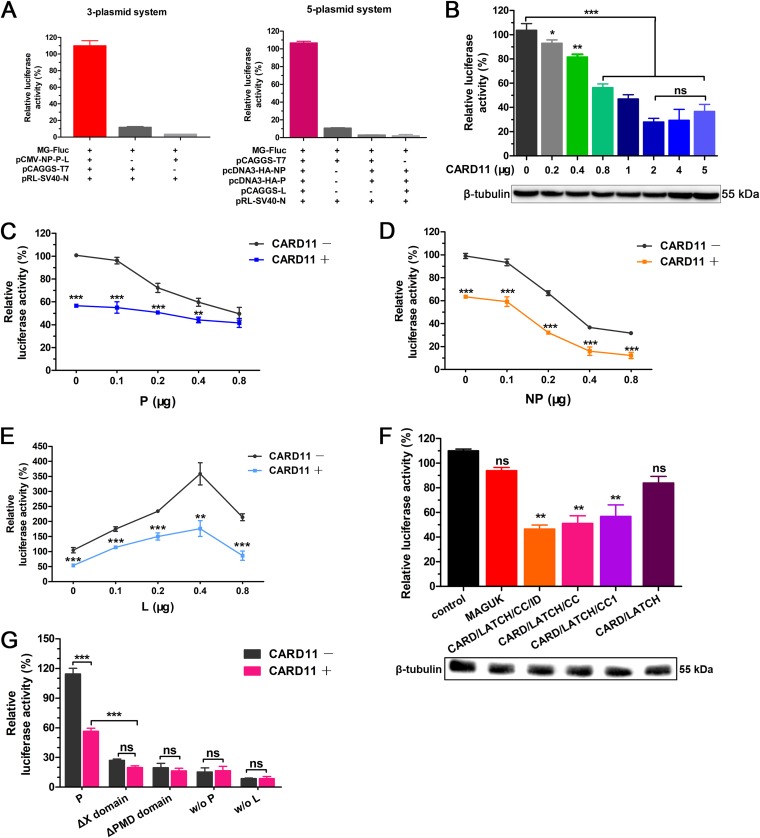
CARD11 inhibits viral polymerase activity.
The NP-P-L interaction of the RNP complex is essential for viral RNA-dependent RNA polymerase activity in viral transcription and replication. To test whether the interaction of CARD11 and the viral P affected viral polymerase activity, we established 5-plasmid and 3-plasmid minigenome (MG) systems with a firefly luciferase (Fluc) reporter for viral polymerase activity assays (Fig. 12A). The luciferase activity was apparently inhibited by CARD11 in a dose-dependent manner in the 3-plasmid MG system (Fig. 12B). The inhibition of the luciferase activity was not affected by increasing P expression with a constant amount of CARD11 in the 3-plasmid MG system but decreased without CARD11 (Fig. 12C), indicating that the inhibition of viral polymerase activity was mediated by the CARD11-P-specific interaction. However, the trends of the luciferase activity with inhibition by NP or an increase by L were not altered with or without CARD11 (Fig. 12D and E). The suppression of luciferase activity by viral P and NP as well as the increase in luciferase activity by L in a dose-dependent manner was consistent with a previous report of other negative-strand RNA viruses (32), but the mechanism has not been elucidated. Furthermore, the luciferase activity was suppressed by the CARD/LATCH/CC/ID, CARD/LATCH/CC, and CARD/LATCH/CC1 domains rather than by the CARD/LATCH and MAGUK domains (Fig. 12F), indicating that the CC1 domain of CARD11 played a key role in inhibiting viral polymerase activity. During the deletion of the X or PMD domain of P, the luciferase activity was inhibited with or without CARD11 (Fig. 12G), indicating that the X domain was critical for P function in viral RNA polymerase activity. These results demonstrated that the specific suppression of viral RNA polymerase activity by CARD11 could be through competitively hampering the P-L interaction in the viral RNP complex.
FIG 12.
CARD11 inhibits viral polymerase activity. (A) Establishment of NDV MG systems. The LaSota strain MG system was constructed under the T7 promoter with a firefly luciferase reporter (MG-Fluc). For the 3-plasmid system, the BHK-21 cells were cotransfected with pCAGGS-T7, pCMV-NP-P-L, and MG-Fluc at a ratio of 2:1:1. For the 5-plasmid system, the BHK-21 cells were cotransfected with pCAGGS-T7, MG-Fluc, pcDNA3-HA-NP, pcDNA3-HA-P, and pCAGGS-L at a ratio of 5:5:2:2:1 or without the three support plasmids, pCAGGS-L, and pCAGGS-T7. (B) CARD11 dose-dependently inhibits viral RNA polymerase activity using the 3-plasmid system. The BHK-21 cells were cotransfected with the 3-plasmid system and different doses of pcDNA3-2×Flag-CARD11. β-Tubulin was used as a protein loading control. (C to E) Viral RNA polymerase activity with different doses of P, NP, or L and CARD11 (1 μg) using the 3-plasmid system. The BHK-21 cells were cotransfected with the 3-plasmid system, pcDNA3-2×Flag (1 μg) or pcDNA3-2×Flag-CARD11 (1 μg), and additional P, NP, or L constructs (pcDNA3-HA-NP, pcDNA3-HA-P, or pCAGGS-L). (F) Inhibition of viral RNA polymerase activity by the CARD11 CC1 domain using a 3-plasmid system. The BHK-21 cells were cotransfected with the 3-plasmid system and truncated CARD11 constructs (1 μg) as well as with negative-control plasmids. β-Tubulin was used as a protein loading control. (G) The viral polymerase activity of P truncations and CARD11 (1 μg) using the 5-plasmid system. The BHK-21 cells were cotransfected with pCAGGS-T7, MG-Fluc, pcDNA3-HA-NP, pcDNA3-HA-P or pcDNA3-HA-P ΔX or pcDNA3-HA-P ΔPMD, and pCAGGS-L at a ratio of 5:5:2:2:1 or with pcDNA3-2×Flag-CARD11 (1 μg). In addition, the 5-plasmid system without the P was detected as previously described. All the cells above were cotransfected with pRL-SV40-N expressing Renilla luciferase as a normalizing standard and were lysed at 36 h posttransfection. All the representative data, shown as the means ± SDs (n = 3), were analyzed by two-tailed Student's t tests. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
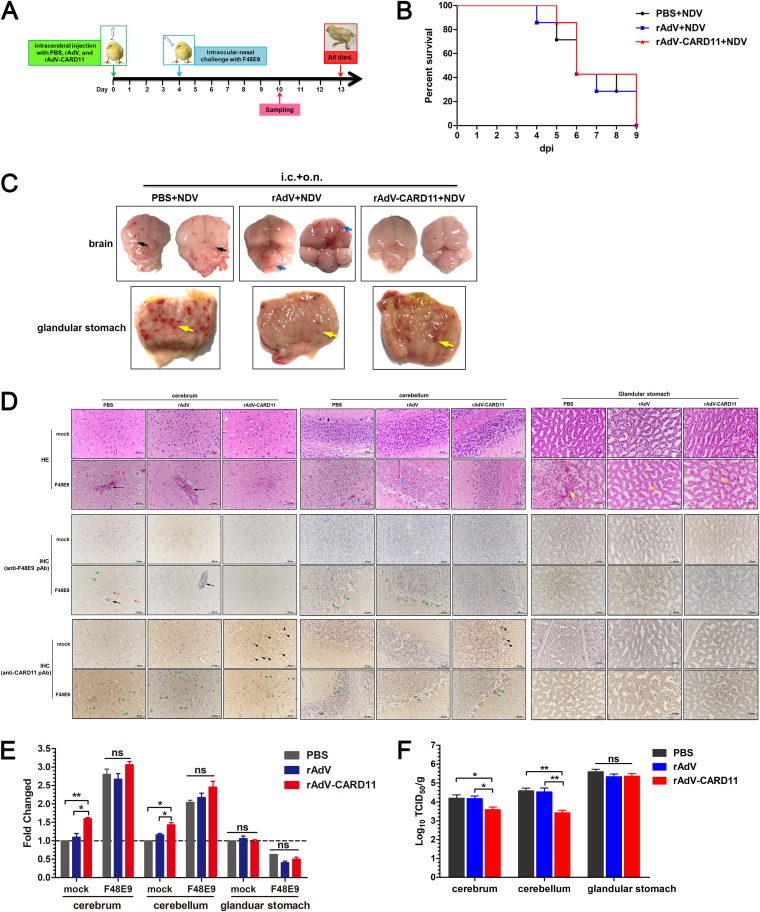
Intracerebrally overexpressed CARD11 alleviates neuropathological lesions and reduces viral replication in the brain.
For analysis of the antiviral effect of CARD11 in vivo, rAdV-CARD11, which overexpressed CARD11, was intracerebrally injected into 1-day-old chicks following VNNDV challenge on day 4 (Fig. 13A). The rAdV-CARD11-injected chicks showed no clinical signs or pathological changes, indicating that rAdV-CARD11 was not toxic or pathogenic to the birds. After challenge with the F48E9 virus via the intraocular-nasal route, chicks in the rAdV and rAdV-CARD11 groups exhibited neurological disorder symptoms at 4 days postchallenge (dpc). Birds in the PBS control group showed neurological disorder symptoms at 3 dpc. All birds in the challenge groups were dead at 9 dpc (Fig. 13B). In clinical anatomies (Fig. 13C), the F48E9-challenged brains in the PBS intracerebrally injected group showed severe liquefaction and an indistinct boundary between the cerebrum and cerebellum. Hyperemia was obvious in the brains of the rAdV group. However, these effects were mild and even absent in the rAdV-CARD11 group. In addition, the pathological symptoms in the glandular stomach, such as glandular papillary hemorrhage, were not significantly different among these three groups (Fig. 13C). These results indicated that CARD11 overexpression could alleviate the pathological damage of VNNDV infection in the chicken brain. The HE assays showed apparent perivascular cuffing and edema in the cerebra of the PBS and rAdV groups but not in the rAdV-CARD11 group (Fig. 13D, top). The stratum granulosum of the cerebellum became loose, hemorrhaging in the substantia alba was obvious, and the detachment of Purkinje cells from the stratum granulosum was obvious in the PBS and rAdV groups, but not in the rAdV-CARD11 group (Fig. 13D, top). The compound tubular glands of the glandular stomach in all F48E9-infected chicks exhibited the same histopathological changes, such as epithelium swelling, necrosis, and even exfoliation (Fig. 13D, top). IHC assays with an anti-F48E9 antibody showed that the virus distribution was consistent with the location of the lesion in the tissues (Fig. 13D, middle). CARD11 was successfully overexpressed via rAdV-CARD11 in the cerebrum and cerebellum, but not in the PBS and rAdV groups (Fig. 13D, bottom). After virus challenge, CARD11 was upregulated in the brains of all groups rather than in the glandular stomachs (Fig. 13D, bottom). These data were confirmed by real-time quantitative (RT-qPCR) for the relative mRNA expression of CARD11 (Fig. 13E). The viral titers of F48E9 in both the cerebra and cerebella of rAdV-CARD11-infected animals were significantly inhibited compared to those in the PBS and rAdV groups, unless a glandular stomach was present (Fig. 13F). Thus, although intracerebral CARD11 was not sufficient to protect the birds infected with VNNDV from dying due to of the lack of systemic overexpression, these animal experiments further demonstrated that CARD11 could help alleviate the neuropathological lesions caused by NDV and inhibit viral replication in the brain.
FIG 13.
Intracerebrally overexpressed CARD11 reduces neuropathology and viral replication in the brain. (A) Schematic representation of animal experiments. One-day-old chicks were intracerebrally injected with rAdV-CARD11, rAdV, and PBS. After 4 days, the birds were challenged with F48E9. Tissue samples were collected on day 10. (B) Survival rates of the F48E9-challenged chicks. (C) Lesions on the cerebrum, cerebellum, and glandular stomach at 6 dpc. The black arrows indicate the indistinct boundary between the cerebrum and cerebellum. The blue arrows indicate hyperemia in F48E9-infected brains. The yellow arrows indicate the glandular papillary hemorrhage. i.c., intracerebral injection; o.n., ocular-nasal route. (D) HE and IHC assays at 6 dpc. The black arrows indicate perivascular cuffing in the cerebrum. The red arrows indicate edema in the cerebrum. The blue arrows indicate hemorrhage in the substantia alba of the cerebellum. The yellow arrows indicate the epithelial hemorrhage, swelling, necrosis, and even exfoliation in the compound tubular glands of glandular stomach. The blue arrowhead indicates the detachment of Purkinje cells from the stratum granulosum in the cerebellum. The black arrowheads indicate the detected CARD11 in cells of mock chicks. The green arrowheads indicate the detected virus or CARD11 in the cells of F48E9-challenged chicks. (E) CARD11 expression in the cerebrum, cerebellum, and glandular stomach before and after the F48E9 challenge. (F) The viral replication of F48E9 in the cerebrum, cerebellum, and glandular stomach. The tissues were homogenized, and viral titers in the supernatants were measured in DF-1 cells using the TCID50 method. Representative data from three independent experiments, shown as the means ± SDs (n = 3), were analyzed by two-tailed Student's t tests. ns, not significant; *, P < 0.05; **, P < 0.01.
DISCUSSION
Avian neurotropic virus infection of the CNS is a severe problem affecting the health of animals, especially poultry. NDV, a predominantly fatal fetal virus, has evolved to effectively infect the CNS (33). This work aimed to investigate the avian neuronal response induced by NDV infection to illuminate the interactions between cellular molecules and viruses. This study was the first to investigate the transcriptomics of NDV-infected chicken brains by gene microarray. CARD11, a unique host protein, was only brain-specifically upregulated by VNNDV infection, which has not been reported in any published transcriptome databases, such as those of the spleen, primary chicken fibroblast cells, and embryonic visceral cells (26–28). In addition, the basal expression of CARD11 mRNA in each chicken tissue had no significant differences, as shown using an absolute quantification real-time PCR assay (data not shown). In mammals, CARD11 upregulation is only reported in clinical case monitoring of one human disease, T cell acute lymphoblastic leukemia (T-ALL) (34), which is not related to the virus. These findings suggest that chicken CARD11 likely plays a special role in the neurotropism of VNNDV. Further results found that CARD11 inhibits NDV replication and delays the occurrence of CPE in chPNCs, indicating a relationship between neural protein and NDV in the avian nervous system.
CARD11 as a subunit of the CBM complex is essential for MALT1-mediated activation of the NF-κB, JNK, and mTOR signaling pathways, which are vital to the cell immune response, inflammatory response, and proliferation (20). Whether the signaling pathways of the CBM signalosome play a crucial role in the inhibition of viral replication has not been reported. MI-2, a MALT1 inhibitor, did not affect the NDV replication in chPNCs. Prevention of CBM complex formation inhibited the MALT1-mediated activation of the NF-κB, JNK, and mTOR signaling pathways. However, blocking MALT1-mediated signaling pathways did not affect viral replication. These findings implied that the CBM signalosome did not play a role in the inhibition of NDV replication in avian neuronal cells, although CBM downstream signaling pathways, such as NF-κB, can induce high levels of interferon (IFN)-stimulating genes (ISGs), cytokines, and chemokines for antiviral effects (35).
The P of NDV acts as a noncatalytic subunit of the viral RNA polymerase, which tethers the catalytic subunit of L to the genomic template during RNA synthesis (36). For the NP-P interaction of paramyxoviruses, such as Sendai virus (SeV), parainfluenza virus (PIV), and measles virus (MeV), there are two independent N-binding sites in P: (i) the P N terminus (PNT) contains the site (residues 34 to 41) that binds newly synthesized soluble N during the nascent chain assembly step of genome replication (37); and (ii) the X domain of P binds the N of the N:RNA template (37–39). In the present study, we found that the PMD domain (residues 225 to 294) of P, rather than the X domain (Fig. 11E), interacted with NP. Moreover, the X domain of P binds to L in NDV (Fig. 11F and G). Our data are the first co-IP evidence to show the interactions of NP, P, and L in NDV. The X domain-mediated P-L interaction of paramyxovirus is consistently reported in MeV (40), human parainfluenza virus type 1 (HPIV1) (41), and respiratory syncytial virus (RSV) (42). In our study, the CARD11 CC1 domain was found to interact with the X domain of NDV P. The CARD11 and L can competitively bind to P through the X domain (Fig. 11H and I) that hampers the P-L interaction, resulting in the suppression of the viral RNA polymerase activity (Fig. 12). However, the precise regions of the X domain of P binding to CARD11 and L should be investigated in the future to understand this competitive interaction affecting the functions of the viral RNP complex.
Notably, in MG assays, increasing amounts of viral P and NP expression resulted in a reduction in the reporter luciferase activity (Fig. 12C and D). This result was consistent with those of other negative-strand RNA viruses, such as PIV5 (43), RSV (44), and a plant negative-strand RNA virus (32), indicating that the relative molar ratios of NP, P, and L for the viral RNA polymerase are vitally important. Transient expression of L (∼250 kDa) may be less efficient than that of the smaller NP (54 kDa) and P (55 kDa). Additionally, the redundant NP or P forms a homo-oligomer or hetero-oligomer, which may be toxic for cells and reduce reporter gene transcription.
In conclusion, this study first identified the interactions between CARD11 and virus infection, especially in neurons. CARD11 upregulation induced by VNNDV is brain-specific and serves as an antiviral suppressor to inhibit viral replication and CPEs in vitro as well as to reduce neuropathological lesions and virus propagation in vivo. The inhibitory roles could be mediated by the following mechanisms. The interaction of the CARD11 CC1 domain with the X domain of the viral P suppresses viral RNA polymerase activity via hindering the interaction of P-L. This study helps elucidate the biological functions of CARD11 by identifying the interactions between hosts and NDV and provides a potential host antiviral target against neurotropic viruses.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were approved by the Animal Care and Use Committee of Northwest A&F University, China, and were conducted in accordance with guidelines established by the Chinese Committee for Animal Experiments (approval numbers: 2016ZX09011008-003 for chickens, 2018ZX04007009-004 for mice and guinea pigs). Nine- to 11-day-old SPF embryonated chicken eggs were purchased from Jinan SAIS Poultry Co., Ltd. (China). Intracerebral injection in chicks was performed under anesthesia that was induced and maintained with isoflurane. Bleeding of mice and guinea pigs was under anesthesia induced and maintained with ketamine and xylazine. All efforts were made to minimize suffering. All animals were euthanized by using carbon dioxide.
Viruses and cells.
The NDV strains F48E9 and LaSota were obtained from the China Institute of Veterinary Drug Control. The F48E9 strain is a standard highly virulent virus that has been used in vaccine evaluations in China since 1948 (25), and as an avirulent virus, the LaSota strain is globally used for live poultry vaccines. The viruses were propagated in 9- to 11-day-old SPF embryonated chicken eggs. Human embryonic kidney cells (HEK293T and HEK293A; ATCC CRL-11268 and ATCC CRL-1573, respectively), chicken fibroblast cells (DF-1, ATCC CRL-12203), and BHK-21 cells (ATCC CCL-10) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P/S).
The chPNCs were prepared from 7- to 9-day-old SPF chicken embryos as described previously (25). Briefly, the brains were washed with DMEM/F12, cut into pieces, and then digested with 0.25% trypsin and 27.5 IU/ml DNase (Sigma-Aldrich) at 37°C for 12 min. The cells were suspended in DMEM/F12 supplemented with 10% FBS and 0.5% P/S and then filtered through 149-μm, 74-μm, and 47-μm cell strainers. The cells were cultured approximately at 2 × 106 cells per well in poly-d-lysine (Sigma)-coated 6-well plates. The culture medium was replaced with Neurobasal medium (Gibco) containing B27 supplement (1:50; Gibco), 1% l-glutamine, and 1% P/S after incubating at 37°C for 4 h, and then, the medium was changed every 2 days. All cells were maintained at 37°C in a 5% CO2 atmosphere.
Gene microarray analysis.
The initial microarray analysis was performed at LC-Bio (Hangzhou, China). An Agilent 44K chicken whole-genome chip (43,803 probes; Agilent Technologies, USA) was used for gene microarray analysis of virus-infected and mock-infected brains. The significance analysis was used to evaluate the differences in gene expression. P and fold change (FC) values are used to describe the alteration of gene expression between the experimental and control groups. The genes (FC > 2) were subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses to identify DEGs. The identified DEGs were then annotated for the analysis of relevant biological pathways.
Reverse transcription-PCR and RT-qPCR.
Total RNA was isolated using RNAiso Plus (TaKaRa, Japan). For determination of the relative expression of DEGs and the NDV NP gene, 5 μg of RNA from NDV-infected tissues or cells was reverse-transcribed using StarScript II First-strand cDNA Synthesis Mix (GenStar). PCR was then performed using the primers described in Table 2. Real-time qPCR was performed on the Applied Biosystems QuantStudio 6 Flex real-time PCR system (Applied Biosystems) using EvaGreen 2× qPCR MasterMix-ROX (Applied Biological Materials). The relative RNA levels of specific RNAs were normalized to those of 28S rRNA and calculated using the comparative threshold cycle (ΔΔCT) method. The FC of gene expression between the treatment and control groups was calculated as 2−ΔΔCT.
TABLE 2.
Primers used in this study
| Target | GenBank accession no. | Primera | 5′→3′ sequenceb | Size (bp) |
|---|---|---|---|---|
| RT-qPCR | ||||
| CARD11 | NM_001006161 | F | GCTTCTGACACGGCAGCATCT | 195 |
| R | CATTTCCTTTAGCCCAATCCTC | |||
| F48E9 NP | MG456905 | F | CAACAACAGGAGTGGAGTGTCTGA | 145 |
| R | TAGAGTATCAGTGATATCTTCT | |||
| IRF1 | NM_205415 | F | GAAGGGAATGGCAGAAGTCG | 256 |
| R | CAAGTGAGCAGCAACCAACAG | |||
| TLR15 | NM_001037835 | F | AACCTGGTGCATTTGAGAACCTGC | 270 |
| R | TTTCCTCTGTTCTTCTTTGTCTGAATC | |||
| TMEM173 | XM_001232170 | F | CGCAGCGTCTGCACCCAGCTGG | 350 |
| R | TCGCTCACGGCCTTGCCTGGTC | |||
| TNFRSF8 | NM_204444 | F | GCCAAGTTGAACAAAGTAGCACA | 322 |
| R | CCACAGGTAGCACCATCACAGA | |||
| 28S rRNA | BQ037787 | F | GGTATGGGCCCGACGCT | 160 |
| R | CCGATGCCGACGCTCAT | |||
| rAdV-CARD11 | NM_001006161 | F | GGATCCATGAGCACCCAAGGAGGAGAGCCA | 3,561 |
| R | GTCGACTTATTTATCGTCATCGTCTTTGTAGTCTTTATCGTCATCGTCTTTGTAGTCGAGCTGATCTTCATCTATCCAGATGG | |||
| pET-30a(+)-L2 | JF950510 | F | GAGCTCACTAATCGCCTCTTGATAGAGT | 1,704 |
| R | GCGGCCGCCCTAGTAAGTGCAATCTTAATTACG | |||
| pET-30a(+)-MAGUK | NM_001006161 | F | GGATCCCTTGCAGAAAAAGCAGGTCTTCG | 1,368 |
| R | GTCGACCGAGCTGATCTTCATCTATCCAGATG | |||
| pET-30a(+)-LaSota P | JF950510 | F | GAATTCATGGCCACCTTTACAGATGC | 1,188 |
| R | GAGCTCGCCATTTAGAGCAAGGCGCTTGATT | |||
| Co-IP | ||||
| CARD11 | NM_001006161 | F | GGATCCAGCACCCAAGGAGGAGAGCCA | 3,513 |
| R | GCGGCCGCTAGAGCTGATCTTCATCTATCCAG | |||
| CARD/LATCH/CC/ID | NM_001006161 | F | GGATCCAGCACCCAAGGAGGAGAGCCA | 2,145 |
| R | GAATTCTTATGATCCTGATACGACAGAGCTAACA | |||
| MAGUK | NM_001006161 | F | GGATCCCTTGCAGAAAAAGCAGGTCTTCGGG | 1,368 |
| R | GAATTCTTAGAGCTGATCTTCATCTATCC | |||
| CARD/LATCH/CC | NM_001006161 | F | GGATCCAGCACCCAAGGAGGAGAGCCA | 1,287 |
| R | GAATTCTCAGCAGGCTTCTTTCCGAACCATT | |||
| CARD/LATCH/CC1 | NM_001006161 | F | GGATCCAGCACCCAAGGAGGAGAGCCA | 870 |
| R | GAATTCTCACTGAATGATGGACTGTAACTCCT | |||
| CARD/LATCH | NM_001006161 | F | GGATCCAGCACCCAAGGAGGAGAGCCA | 336 |
| R | GAATTCCACTCCTTCCCTGTCACCAGTTT | |||
| CC1 | NM_001006161 | F | GGATCCATGCTGATGACCTTCCAGGAGAGATACAAC | 354 |
| R | GAATTCTTACTGAATGATGGACTGTAACTCCTGGAT | |||
| F48E9 P | MG456905 | F | GAATTCATGGCCACCTTTACAGACGCGGAG | 1,188 |
| R | GCGGCCGCTCAGCCATTCAGCGCAAGGCGCTTG | |||
| F48E9 P ΔX domain | MG456905 | F | GGATCCATGGCCACCTTTACAGACGCGGAG | 1,059 |
| R | GAATTCTCAGTCCCTCTCCACTCCTATAT | |||
| F48E9 P ΔPMD domain | MG456905 | F | GGATCCATGGCCACCTTTACAGACGCGGAG | 672 |
| R | GGATCCTCACACAAAGTCGACAGGTAGCT | |||
| F48E9 P X domain | MG456905 | F | GAATTCATGACTGTCCGTGCATTGATCACCTCACGT | 129 |
| R | TAACCTAGTATAGGGGACATGCCATTCAGCGCAAGGCGC | |||
| GST | U78872.1 | F | GCGCCTTGCGCTGAATGGCATGTCCCCTATACTAGGTTA | 735 |
| R | CTCGAGTCAGTCACGATGCGGCCGCTCGA | |||
| F48E9 V | MG456905 | F | GAATTCATGGCCACCTTTACAGACGCGGAG | 720 |
| R | GCGGCCGCTTACTTACCCTCTGTGATATCGCCT | |||
| F48E9 NP | MG456905 | F | GAATTCATGTCTTCCGTATTTGATGAGTA | 1,470 |
| R | GCGGCCGCTCAATACCCCCAGTCGGTGTCGTTA | |||
| LaSota P | JF950510 | F | GAATTCGCCACCTTTACAGATGCAGAG | 1,188 |
| R | GCGGCCGCCTTAGCCATTTAGAGCAAGGCGCT | |||
| LaSota L1 | JF950510 | F | CCATCGATATGGCGAGCTCCGGTCCTGAAAG | 2,931 |
| R | GACTAGCATGCGAATGCGGCCGCCCCAATCTCCATTCCCAGGC | |||
| LaSota L2 | JF950510 | F | TCCCCCGGGGACTACTGCTTTTGCAGAG | 3,686 |
| R | GAATGCGGCCGCTTAAGAGTCACAGTTACTGTAATATCC |
F, forward primer; R, reverse primer.
Flag tag sequences are underlined and restriction enzyme sites are in italic font.
For construction of plasmids overexpressing CARD11 and viral proteins, 5 μg of RNA from chPNCs or NDV was reverse-transcribed using the StarScript II First-strand cDNA Synthesis kit (GenStar). PCR was then performed using the primers described in Table 2.
Recombinant adenoviruses expressing shRNAs and CARD11.
The adenoviral vectors pDC315 and pBHGlox (delta) E1, 3Cre were used for adenovirus packaging. For RNAi assays, the chicken U6 promoter (chU6; GenBank number DQ531569) RNAi cassette fragment with XbaI and EcoRI was synthesized (Synbio Tech, China) first and cloned into pDC315 with XbaI and EcoRI to generate pDC315-chU6. Then, the pDC315-chU6 encoding CARD11 shRNAs (shRNA1 and shRNA2) and a nonsilencing negative control (NC) were cloned into the EcoRI and BamHI sites, respectively. The NC sequence was 5′-AATTCCGTGATCTTCACCGACAAGATTTCAAGAGAATCTTGTCGGTGAAGATCACGTTTTTTG-3′, and the shRNA1 and shRNA2 sequences were 5′-AATTCGCAGATGACTCCTCCACATCATTCAAGAGATGATGTGGAGGAGTCATCTGCTTTTTTG-3′ and 5′-AATTCGCTCCTTCAGTGCACTCTTCTTTCAAGAGAAGAAGAGTGCACTGAAGGAGCTTTTTTG-3′, respectively. For expression of CARD11, the target fragment with a 2×Flag tag was amplified from chPNCs and inserted into the BamHI and SalI sites of pDC315.
These constructed plasmids were cotransfected into the HEK293A cell line with pBHGlox (delta) E1, 3Cre at a ratio of 3:5 to package rAdV-NC, rAdV-shRNA1, rAdV-shRNA2, and rAdV-CARD11 using TurboFect reagent (Invitrogen) according to the manufacturer’s recommendations. Cells showing CPEs were collected, freeze-thawed, and propagated for 3 to 4 passages. The corresponding rAdVs were purified by CsCl gradient centrifugation and titrated in HEK293A cells using the TCID50 assay (45).
Virus infection in chPNCs.
The chPNCs at day 3 were infected with rAdVs at different MOIs (10 to 200) for 2 h at 37°C or with the F48E9 (MOI = 0.01) and LaSota (MOI = 1) strains for 1 h at 37°C, washed three times with Neurobasal medium (Gibco), and cultured in complete medium. The rAdV-infected cells at 48 hpi were infected with the F48E9 (MOI = 0.01) or LaSota (MOI = 1) strains. The culture medium was harvested at different times after infection. CPEs were visualized under an Olympus IX73 research inverted microscope. The titers of F48E9-infected cells were determined by CPEs in DF-1 cells, while those of LaSota-infected DF-1 cells were determined by immunofluorescence analysis in DF-1 cells. The TCID50 titer was calculated according to the Reed-Muench method (25).
Chicken NF-κB reporter system.
The four copies of the NF-κB promoter fragment (sequence GGGAATTTCC) in human pNF-κB-TA-luc (Beyotime Biotechnology, China) were replaced with four copies of a chicken NF-κB promoter fragment (chNF-κB; sequence GGGAATTCTC) as described previously (46); this construct was named pchNF-κB-TA-luc. In preliminary studies, we determined the inhibitory role of inhibitors of chNF-κB according to previous reports (47, 48). Briefly, DF-1 cells were seeded in 12-well plates and cotransfected with pchNF-κB-TA-luc and pRL-SV40-N at a ratio of 10:1. Twenty-four hours later, the cells were incubated with increasing concentrations (1, 5, 8, or 10 μM BAY 11-7082; 0.1, 1, 5, or 10 μM BMS-345541 or dimethyl sulfoxide [DMSO]) for 24 h and harvested to quantify the luciferase activity using the Spark multimode microplate reader (Tecan, Switzerland). The relative luciferase activity was calculated according to the manufacturer’s manual (Beyotime Biotechnology).
Inhibitor treatment.
The NF-κB inhibitors, BAY 11-7082 (MedChemExpress) for NF-κB and BMS-345541 (MedChemExpress) for IKK were dissolved in DMSO to provide 10 mM stock solutions and stored at −20°C until use. MI-2 (MedChemExpress) is an irreversible MALT1 inhibitor (29). The chPNCs were observed after incubating with increasing concentrations of the inhibitors (1, 5, 8, or 10 μM BAY 11-7082; 0.1, 1, 5, or 10 μM BMS-345541; 1, 2, 5, or 10 μM MI-2 or DMSO) for 24 h. For analysis of the effects of these inhibitors on NDV replication, the normal or rAdV-infected chPNCs in 12-well plates were incubated with different concentrations of inhibitors for 24 h. Then, the treated chPNCs were infected with the F48E9 (MOI = 0.01) or LaSota (MOI = 1) strain. After removal of the virus inoculum from infected cultures, fresh medium containing inhibitor was added and remained in contact with cells for the duration of the culture. The culture supernatants were harvested at 24 and 36 hpi for titration of NDV.
Construction of CARD11 and viral genes in eukaryotic expression vectors.
The fragments of the CARD11, CARD/LATCH/CC/ID, MAGUK, CARD/LATCH/CC, CARD/LATCH/CC1, CARD/LATCH, and CC1 genes were cloned into pcDNA3-2×Flag (Table 2). The NP, V, and P truncations of viral genes were cloned into pcDNA3-KHA or pcDNA3-2×Flag. The sequence encoding a fusion protein composed of the X domain of P and glutathione transferase (GST) was cloned into pCAGGS. The viral L gene was cloned into pCAGGS by overlapping four parts of the L gene (L1, L2, L3, and L4) (Table 2).
Preparation of polyclonal antibody.
For production of the anti-CARD11, anti-LaSota P polyclonal antibodies (PAbs), anti-LaSota L PAbs, the MAGUK domain of chicken CARD11, LaSota P, and the L2 gene were cloned into pET-30a(+) (Table 2). The prokaryotic expression was processed in Trans BL21(DE3) cells (Transgen Biotech, China). The inclusion bodies (P and MAGUK) in a 12% SDS-PAGE gel were stained with 0.25 M KCl for 10 min. The white protein band was crushed and lysed in gel extraction buffer (0.05 M Tris base, 0.1 mM EDTA, 1 mM dithiothreitol [DTT], 0.15 M NaCl, 1% SDS) and boiled for 10 min. The supernatants were collected by centrifugation at 13,000 × g for 15 min. The soluble LaSota L2 protein was purified with a His tag protein purification kit (Solarbio, China). All protein concentrations were calculated according to the Bradford method. Six-week-old SPF BALB/c mice were immunized with these proteins at 20 μg per animal. One-month-old SPF guinea pigs were immunized with P at 50 μg per animal. The immunization procedure for both mice and guinea pigs was as follows: the animals were subcutaneously injected with a mixture of the protein and complete Freund’s adjuvant (Sigma). Fourteen days later, the animals were subcutaneously injected with a mixture of the protein and incomplete Freund’s adjuvant (Sigma). After another fourteen days, the animals were intraperitoneally injected with a mixture of the protein and incomplete Freund’s adjuvant.
For preparation of an anti-LaSota mouse PAb, animals were subcutaneously injected with inactivated LaSota (YEBIO, China) at 200 μl per animal. After the mice and guinea pigs were anesthetized with ketamine and xylazine, their antisera were collected and stored at −80°C for Western blot and immunofluorescence analyses.
Western blot assay.
The infected or transfected cells were lysed in ice-cold RIPA buffer with the protease inhibitor phenylmethylsulfonyl fluoride (PMSF; 1:1,000; Solarbio, China) and then boiled for 10 min. After centrifugation, the supernatants were collected and subjected to 12% SDS-PAGE. The proteins were transferred to a polyvinylidene fluoride (PVDF) membrane. Immunoblotting was performed using the following primary antibodies: an anti-CARD11 mouse PAb, a guinea pig anti-LaSota P, an anti-NP guinea pig PAb (25), an anti-HN guinea pig PAb, an anti-M synthetic peptide (KLEKGHTLAKYNPFK) mouse PAb (49), an anti-L mouse PAb, a DYKDDDK tag (3B9) mouse monoclonal antibody (MAb) (Abmart), an anti-DDDDK tag rabbit MAb (Abcam), a hemagglutinin (HA) tag (C29F4) rabbit MAb (Cell Signaling Technology), a GST tag mouse (3B10) MAb (Sungene Biotech), or β-tubulin (3G7) mouse MAb (Sungene Biotech). The secondary antibodies included goat anti-mouse IgG conjugated to horseradish peroxidase (HRP) (Sungene Biotech) and goat anti-rabbit IgG (H&L) HRP antibody (ZETA). An enhanced chemiluminescence (ECL) peroxidase substrate (Millipore) was used for the detection of proteins using the Tanon 5200 Chemiluminescent Imaging System (Tanon, China).
IPE of cellular CARD11.
Because CARD11 endogenous protein expression cannot be detected in normal chPNCs by Western blotting, we constructed a detection method to enrich cellular CARD11, named IPE. Briefly, the chPNCs were seeded into 60-mm- or 100-mm-diameter poly-d-lysine-coated dishes at the same number of cells (∼107 per dish). The cells at day 3 were infected with F48E9 (MOI = 0.01) and LaSota (MOI = 1) as described above. For detection of the efficiency of RNA knockdown in CARD11 by rAdVs, the chPNCs seeded into poly-d-lysine-coated 100-mm-diameter dishes (∼5 × 107 per dish) at day 3 were infected with rAdV-shRNAs (MOI = 100). The mock- and NDV-infected cells at 24 hpi and the rAdV-shRNA-infected cells at 48 hpi were lysed in 500 μl of ice-cold RIPA buffer with PMSF (1:1,000) for 30 min and centrifuged at 13,000 × g for 15 min at 4°C. The supernatants were incubated with an anti-CARD11 mouse PAb (10 μl) at 4°C for 4 h. The immunoprecipitated protein was captured by the addition of 40 μl of a protein A/G-agarose (Abmart) slurry and gentle rotation overnight at 4°C. The agarose beads were further washed three times and resuspended in 50 μl of 1× SDS loading buffer and boiled for 10 min. After centrifugation, the supernatants were collected and subjected to SDS-PAGE for Western blot analysis with the anti-CARD11 PAb and β-tubulin (3G7) mouse MAb.
Immunofluorescence assay.
For analysis of the colocalization of CARD11 and viral P in chPNCs, the NDV-infected cells were fixed with 4% paraformaldehyde in PBS for 15 min, washed three times with PBS, and permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature. After blocking with PBS containing 1% bovine serum albumin (BSA) for 1 h at 37°C, the cells were incubated with the primary antibodies mouse anti-CARD11 PAb (1:100) and guinea pig anti-P PAb (1:100) in PBS at 4°C overnight. After being washed with PBS three times, the samples were incubated with the secondary antibodies goat anti-guinea pig IgG/fluorescein isothiocyanate (FITC) (Bioss, China) and goat anti-mouse IgG (H&L) (Alexa Fluor 647, preadsorbed, ab150115; Abcam) for 30 min at 37°C. Cell nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI, 1:1,000; Molecular Probes). The cells were observed and photographed under an Olympus FV3000 confocal microscope.
For titration of avirulent LaSota in chPNCs, the primary antibody mouse anti-LaSota PAb (1:200) and secondary antibody goat anti-mouse IgG (H&L) (Alexa Fluor 647, preadsorbed) were used. All cells were observed and photographed under an Olympus IX73 research inverted microscope.
Co-IP.
For analysis of the interaction between endogenous CARD11 and viral protein(s), CARD11 in NDV-infected chPNCs was immunoprecipitated using anti-CARD11 mouse PAb as described. The immunoprecipitated complexes of every sample were captured by the addition of protein A/G-agarose (Abmart) slurry and gentle rotation overnight at 4°C. The immunoprecipitated supernatants were analyzed by Western blotting with an anti-CARD11 mouse PAb, an anti-P mouse PAb, an anti-NP guinea pig PAb, and an anti-HN guinea pig PAb.
For the co-IP assay of CARD11 and viral proteins, HEK293T cells in 60-mm-diameter dishes were cotransfected with two plasmids at a dose of 2 μg. The cells were lysed in 500 μl ice-cold RIPA buffer with PMSF (1:1,000) for 30 min at 36 h posttransfection. After centrifugation at 13,000 × g for 15 min at 4°C, 40 μl of the supernatants as the input sample was treated with 10 μl 5× SDS loading buffer, and the remaining supernatants were incubated with DYKDDDK tag (3B9) mouse MAb (Abmart) or HA tag (C29F4) rabbit MAb (Cell Signaling Technology) at 4°C for 4 h. The immunoprecipitated complexes of every sample were captured by the addition of 50 μl protein A/G-agarose or glutathione beads (Smart-Life Sciences, China) to the slurry and gentle rotation overnight at 4°C.
For the co-IP assay of CARD11, P, NP, and L, HEK293T cells in 60-mm-diameter dishes were cotransfected with pcDNA3-2×Flag-CARD11 (0, 0.5, 1, 2, and 4 μg), pcDNA3-HA-P (F48E9) (2 μg), pcDNA3-2×Flag-NP (F48E9) (2 μg), and pCAGGS-L (2 μg) or pcDNA3-2×Flag-CARD11 (2 μg), pcDNA3-HA-P (F48E9) (2 μg), pcDNA3-2×Flag-NP (F48E9) (2 μg), and pCAGGS-L (0, 1, 2, 4 μg). The cells were lysed in 500 μl ice-cold RIPA buffer with PMSF (1:1,000) for 30 min at 36 h posttransfection. After centrifugation at 13,000 × g for 15 min at 4°C, 40 μl of the supernatants as the input sample was treated with 10 μl 5× SDS loading buffer; the remaining supernatants were incubated with a guinea pig anti-LaSota P PAb. The immunoprecipitated complexes of every sample were captured by the addition of 50 μl protein A/G-agarose slurry and gentle rotation overnight at 4°C.
The agarose beads were further washed three times on ice, resuspended in 50 μl 1× SDS loading buffer, and boiled for 10 min. After centrifugation, the supernatants were subjected to SDS-PAGE for Western blot analysis.
MG assay.
The LaSota strain MG system was constructed according to a previous report (50, 51). An MG cassette, T7 promoter-Trailer-MCS-Leader-HDV/Rbz-T7 terminator, was synthesized and cloned into the pUC19 vector (Synbio Tech, China). The luciferase fragment amplified from pNF-κB-TA-luc (Beyotime Biotechnology, China) was inserted into the multiple cloning sites (MCS) between the trailer and leader sequences via SphI and ClaI to generate the MG-firefly luciferase reporter plasmid (MG-Fluc). For analysis of the function of truncations of P with reverse transcription, the 5-plasmid system was used: pCAGGS-T7 (expressing T7 RNA polymerase, GenBank number FJ881694), MG-Fluc, pcDNA3-HA-NP, pcDNA3-HA-P or P truncations, and pCAGGS-L. Briefly, these plasmids were cotransfected into BHK-21 cells at a ratio of 5:5:2:2:1. For the dose-dependent assays of CARD11, NP, P, and L, we used a 3-plasmid system: pCAGGS-T7, pCMV-NP-P-L (expressing the LaSota strain’s NP, P, and L), and MG-Fluc, which were cotransfected into BHK-21 cells at a ratio of 2:1:1. The rest of the plasmids, pcDNA3-2×Flag-CARD11 or CARD11 truncations, were cotransfected into BHK-21 cells with the 3-plasmid system at different concentrations. The relative luciferase activity of all samples was normalized to that of Renilla luciferase by transfecting pRL-SV40-N (Beyotime Biotechnology, China). The cells were harvested at 36 h posttransfection, and the relative luciferase activity was measured by the Spark multimode microplate reader (Tecan, Switzerland) and calculated using the ratio of relative light units according to the manufacturer’s manual (Beyotime Biotechnology).
Flow cytometry.
For determination of the infectious efficiency of rAdVs in chPNCs, the infected cells at 48 hpi were digested with 0.25% trypsin and washed with sterile PBS at 300 × g for 5 min three times. The chPNCs were treated with DYKDDDK tag (3B9) mouse MAb and goat anti-mouse IgG/FITC at 4°C with minimal exposure to light. The percentage of rAdV-CARD11-infected chPNCs was analyzed by a FACS-Calibur instrument (BD FACSAriaTM III; BD Biosciences, USA).
Animal experiments.
All SPF white Leghorn chickens (Jinan Sais Poultry, China) were raised in biosafety isolators. For gene microarray analysis of NDV infection, fifteen 4-week-old chickens were equally divided into three groups; two of the groups were inoculated with F48E9 and LaSota viruses (105 PFU/100 μl/chicken) via an intraocular-nasal route, and the control group was inoculated with PBS. All chickens were observed for clinical signs daily throughout the experimental period. The F48E9-infected chickens were euthanized when they displayed severe neurological symptoms or became moribund. The brains from each group were collected at 5 dpi for gene microarray analysis, pathological analysis, and virus titration (25).
For CARD11 intracerebral overexpression virus challenge experiments, intracerebral injection in all chicks was performed under anesthesia that was induced and maintained with isoflurane. Nine 1-day-old SPF chicks were equally divided into three groups and served as rAdV controls without NDV challenge. The three groups were inoculated via an intracerebral route with rAdV (2 × 1010 PFU/20 μl/chick), rAdV-CARD11 (2 × 1010 PFU/20 μl/chick), and PBS. Another 21 1-day-old SPF chicks were equally divided into three groups and intracerebrally inoculated with equal doses of rAdV, rAdV-CARD11, and PBS. After 4 days, these three groups were challenged with the F48E9 virus (103 PFU/100 μl/chick) via intraocular-nasal routes and observed for clinical signs and survivals daily. All chicks were euthanized on day 10. The cerebrum (near the cerebellar junction), cerebellum (close to the cerebral junction), and glandular stomach were collected for RT-qPCR, virus titration, and pathological analysis.
Histopathology and IHC assays.
The HE and IHC assays were performed by Shaanxi Yike Biotechnology Service Co., Ltd. All harvested tissues were fixed in phosphate-buffered formalin (10%), embedded in paraffin, and sectioned. The tissues were deparaffinized, rehydrated, and subsequently stained with HE. The IHC assay was performed to detect virus and CARD11 using anti-F48E9 mouse PAb (1:500), anti-LaSota mouse PAb (1:500), and anti-CARD11 mouse PAb (1:500). All samples were observed and photographed under a Nikon Ni-U microscope.
Statistical analysis.
Two-tailed Student's t tests were used to estimate the statistical significance between two columns in GraphPad Prism 5. Data from three independent experiments are presented as the means ± standard deviations (SDs) from triplicate samples (n = 3); P < 0.05 was considered statistically significant.
Data availability.
All gene microarray files are available from the Gene Expression Omnibus database (accession number GSE121368). All relevant data are within the paper.
ACKNOWLEDGMENTS
We thank Yanqing Jia for excellent technical assistance and Yi Zhang and Ying Wang for preparing the anti-NDV viral protein PAbs.
This work was supported by the National Natural Science Foundation of China (31572533 to S.X., 31572538 to Z.Y.) and the Faculty Support Fund of Northwest A&F University (Z111021401 to S.X.). The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Ludlow M, Kortekaas J, Herden C, Hoffmann B, Tappe D, Trebst C, Griffin DE, Brindle HE, Solomon T, Brown AS, van Riel D, Wolthers KC, Pajkrt D, Wohlsein P, Martina BEE, Baumgartner W, Verjans GM, Osterhaus A. 2016. Neurotropic virus infections as the cause of immediate and delayed neuropathology. Acta Neuropathol 131:159–184. doi: 10.1007/s00401-015-1511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dahm T, Rudolph H, Schwerk C, Schroten H, Tenenbaum T. 2016. Neuroinvasion and inflammation in viral central nervous system infections. Mediators Inflamm 2016:8562805. doi: 10.1155/2016/8562805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soung A, Klein RS. 2018. Viral encephalitis and neurologic diseases: focus on astrocytes. Trends Mol Med 24:950–962. doi: 10.1016/j.molmed.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, Wang X, Ashraf U, Zheng B, Ye J, Zhou D, Zhang H, Song Y, Chen H, Zhao S, Cao S. 2018. Activation of neuronal N-methyl-d-aspartate receptor plays a pivotal role in Japanese encephalitis virus-induced neuronal cell damage. J Neuroinflammation 15:238. doi: 10.1186/s12974-018-1280-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Q, Zhu N, Chen S, Zhao P, Ren H, Zhu S, Tang H, Zhu Y, Qi Z. 2017. E3 ubiquitin ligase Nedd4 promotes Japanese encephalitis virus replication by suppressing autophagy in human neuroblastoma cells. Sci Rep 7:45375. doi: 10.1038/srep45375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He W, Zhao Z, Anees A, Li Y, Ashraf U, Chen Z, Song Y, Chen H, Cao S, Ye J. 2017. p21-activated kinase 4 signaling promotes Japanese encephalitis virus-mediated inflammation in astrocytes. Front Cell Infect Microbiol 7:271. doi: 10.3389/fcimb.2017.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavali PL, Stojic L, Meredith LW, Joseph N, Nahorski MS, Sanford TJ, Sweeney TR, Krishna BA, Hosmillo M, Firth AE, Bayliss R, Marcelis CL, Lindsay S, Goodfellow I, Woods CG, Gergely F. 2017. Neurodevelopmental protein Musashi-1 interacts with the Zika genome and promotes viral replication. Science 357:83–88. doi: 10.1126/science.aam9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowakowski TJ, Pollen AA, Di Lullo E, Sandoval-Espinosa C, Bershteyn M, Kriegstein AR. 2016. Expression analysis highlights AXL as a candidate Zika virus entry receptor in neural stem cells. Cell Stem Cell 18:591–596. doi: 10.1016/j.stem.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Wang Z, Liu R, Shuai L, Wang X, Luo J, Wang C, Chen W, Wang X, Ge J, He X, Wen Z, Bu Z. 2018. Metabotropic glutamate receptor subtype 2 is a cellular receptor for rabies virus. PLoS Pathog 14:e1007189. doi: 10.1371/journal.ppat.1007189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Yu W, Huo N, Wang W, Guo Y, Wei Q, Wang X, Zhang S, Yang Z, Xiao S. 2017. Comprehensive analysis of amino acid sequence diversity at the F protein cleavage site of Newcastle disease virus in fusogenic activity. PLoS One 12:e0183923. doi: 10.1371/journal.pone.0183923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng C, Qiu X, Yu S, Li C, Sun Y, Chen Z, Liu K, Zhang X, Tan L, Song C, Liu G, Ding C. 2016. Evolution of Newcastle disease virus quasispecies diversity and enhanced virulence after passage through chicken air sacs. J Virol 90:2052–2063. doi: 10.1128/JVI.01801-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aldous EW, Alexander DJ. 2008. Newcastle disease in pheasants (Phasianus colchicus): a review. Vet J 175:181–185. doi: 10.1016/j.tvjl.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu X, Cheng J, He Z, Li C, Song Y, Xue J, Yang H, Zhang R, Zhang G. 2017. The glutamic residue at position 402 in the C-terminus of Newcastle disease virus nucleoprotein is critical for the virus. Sci Rep 7:17471. doi: 10.1038/s41598-017-17803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexander DJ. 2000. Newcastle disease and other avian paramyxoviruses. Rev Sci Tech 19:443–462. doi: 10.20506/rst.19.2.1231. [DOI] [PubMed] [Google Scholar]
- 15.Dortmans JC, Koch G, Rottier PJ, Peeters BP. 2011. Virulence of Newcastle disease virus: what is known so far? Vet Res 42:122. doi: 10.1186/1297-9716-42-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beziat V, Jouanguy E, Puel A. 2019. Dominant negative CARD11 mutations: beyond atopy. J Allergy Clin Immunol 143:1345–1347. doi: 10.1016/j.jaci.2018.12.1006. [DOI] [PubMed] [Google Scholar]
- 17.Knittel G, Liedgens P, Korovkina D, Pallasch CP, Reinhardt HC. 2016. Rewired NFkappaB signaling as a potentially actionable feature of activated B-cell-like diffuse large B-cell lymphoma. Eur J Haematol 97:499–510. doi: 10.1111/ejh.12792. [DOI] [PubMed] [Google Scholar]
- 18.Roche MI, Ramadas RA, Medoff BD. 2013. The role of CARMA1 in T cells. Crit Rev Immunol 33:219–243. doi: 10.1615/CritRevImmunol.2013007056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juilland M, Thome M. 2016. Role of the CARMA1/Bcl10/MALT1 complex in lymphoid malignancies. Curr Opin Hematol 23:402–409. doi: 10.1097/MOH.0000000000000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bedsaul JR, Carter NM, Deibel KE, Hutcherson SM, Jones TA, Wang Z, Yang C, Yang YK, Pomerantz JL. 2018. Mechanisms of regulated and dysregulated CARD11 signaling in adaptive immunity and disease. Front Immunol 9:2105. doi: 10.3389/fimmu.2018.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno-Garcia ME, Sommer K, Shinohara H, Bandaranayake AD, Kurosaki T, Rawlings DJ. 2010. MAGUK-controlled ubiquitination of CARMA1 modulates lymphocyte NF-kappaB activity. Mol Cell Biol 30:922–934. doi: 10.1128/MCB.01129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brohl AS, Stinson JR, Su HC, Badgett T, Jennings CD, Sukumar G, Sindiri S, Wang W, Kardava L, Moir S, Dalgard CL, Moscow JA, Khan J, Snow AL. 2015. Germline CARD11 mutation in a patient with severe congenital B cell lymphocytosis. J Clin Immunol 35:32–46. doi: 10.1007/s10875-014-0106-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scudiero I, Vito P, Stilo R. 2014. The three CARMA sisters: so different, so similar: a portrait of the three CARMA proteins and their involvement in human disorders. J Cell Physiol 229:990–997. doi: 10.1002/jcp.24543. [DOI] [PubMed] [Google Scholar]
- 24.Deckert M, Montesinos-Rongen M, Brunn A, Siebert R. 2014. Systems biology of primary CNS lymphoma: from genetic aberrations to modeling in mice. Acta Neuropathol 127:175–188. doi: 10.1007/s00401-013-1202-x. [DOI] [PubMed] [Google Scholar]
- 25.Guo Y, Zhao J, Chang X, Yao W, Wang H, Wang W, Wang X, Zhang S, Yang Z, Xiao S. 2018. Alpha2,3- and alpha2,6-linked sialic acids are important for cell binding and replication of Newcastle disease virus in chicken primary neuronal cells. Acta Virol 62:235–245. doi: 10.4149/av_2018_217. [DOI] [PubMed] [Google Scholar]
- 26.Hu Z, Hu J, Hu S, Song Q, Ding P, Zhu J, Liu X, Wang X, Liu X. 2015. High levels of virus replication and an intense inflammatory response contribute to the severe pathology in lymphoid tissues caused by Newcastle disease virus genotype VIId. Arch Virol 160:639–648. doi: 10.1007/s00705-014-2301-2. [DOI] [PubMed] [Google Scholar]
- 27.Rue CA, Susta L, Cornax I, Brown CC, Kapczynski DR, Suarez DL, King DJ, Miller PJ, Afonso CL. 2011. Virulent Newcastle disease virus elicits a strong innate immune response in chickens. J Gen Virol 92:931–939. doi: 10.1099/vir.0.025486-0. [DOI] [PubMed] [Google Scholar]
- 28.Munir S, Sharma JM, Kapur V. 2005. Transcriptional response of avian cells to infection with Newcastle disease virus. Virus Res 107:103–108. doi: 10.1016/j.virusres.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Fontan L, Yang C, Kabaleeswaran V, Volpon L, Osborne MJ, Beltran E, Garcia M, Cerchietti L, Shaknovich R, Yang SN, Fang F, Gascoyne RD, Martinez-Climent JA, Glickman JF, Borden K, Wu H, Melnick A. 2012. MALT1 small molecule inhibitors specifically suppress ABC-DLBCL in vitro and in vivo. Cancer Cell 22:812–824. doi: 10.1016/j.ccr.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jahanshiri F, Eshaghi M, Yusoff K. 2005. Identification of phosphoprotein:phosphoprotein and phosphoprotein:nucleocapsid protein interaction domains of the Newcastle disease virus. Arch Virol 150:611–618. doi: 10.1007/s00705-004-0439-z. [DOI] [PubMed] [Google Scholar]
- 31.Qiu X, Zhan Y, Meng C, Wang J, Dong L, Sun Y, Tan L, Song C, Yu S, Ding C. 2016. Identification and functional analysis of phosphorylation in Newcastle disease virus phosphoprotein. Arch Virol 161:2103–2116. doi: 10.1007/s00705-016-2884-x. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Ma X, Qian S, Zhou X, Sun K, Chen X, Zhou X, Jackson AO, Li Z. 2015. Rescue of a plant negative-strand RNA virus from cloned cDNA: insights into enveloped plant virus movement and morphogenesis. PLoS Pathog 11:e1005223. doi: 10.1371/journal.ppat.1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ecco R, Susta L, Afonso CL, Miller PJ, Brown C. 2011. Neurological lesions in chickens experimentally infected with virulent Newcastle disease virus isolates. Avian Pathol 40:145–152. doi: 10.1080/03079457.2010.544289. [DOI] [PubMed] [Google Scholar]
- 34.Ma Y, Liao Z, Xu Y, Zhong Z, Wang X, Zhang F, Chen S, Yang L, Luo G, Huang X, Huang S, Wu X, Li Y. 2014. Characteristics of CARMA1-Bcl10-MALT1-A20-NF-kappaB expression in T cell-acute lymphocytic leukemia. Eur J Med Res 19:62. doi: 10.1186/s40001-014-0062-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulmann D, Bortmann S, Grimm F, Berk I, Kraemer L, Vallbracht A, Dotzauer A. 2014. NF-kappaB activation induced by hepatitis A virus and Newcastle disease virus occurs by different pathways depending on the structural pattern of viral nucleic acids. Arch Virol 159:1723–1733. doi: 10.1007/s00705-014-1993-7. [DOI] [PubMed] [Google Scholar]
- 36.Fearns R, Plemper RK. 2017. Polymerases of paramyxoviruses and pneumoviruses. Virus Res 234:87–102. doi: 10.1016/j.virusres.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowman MC, Smallwood S, Moyer SA. 1999. Dissection of individual functions of the Sendai virus phosphoprotein in transcription. J Virol 73:6474–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolakofsky D, Le Mercier P, Iseni F, Garcin D. 2004. Viral DNA polymerase scanning and the gymnastics of Sendai virus RNA synthesis. Virology 318:463–473. doi: 10.1016/j.virol.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 39.Brunel J, Chopy D, Dosnon M, Bloyet LM, Devaux P, Urzua E, Cattaneo R, Longhi S, Gerlier D. 2014. Sequence of events in measles virus replication: role of phosphoprotein-nucleocapsid interactions. J Virol 88:10851–10863. doi: 10.1128/JVI.00664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bloyet LM, Schramm A, Lazert C, Raynal B, Hologne M, Walker O, Longhi S, Gerlier D. 2019. Regulation of measles virus gene expression by P protein coiled-coil properties. Sci Adv 5:eaaw3702. doi: 10.1126/sciadv.aaw3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bousse T, Takimoto T, Matrosovich T, Portner A. 2001. Two regions of the P protein are required to be active with the L protein for human parainfluenza virus type 1 RNA polymerase activity. Virology 283:306–314. doi: 10.1006/viro.2001.0881. [DOI] [PubMed] [Google Scholar]
- 42.Sourimant J, Rameix-Welti M-A, Gaillard A-L, Chevret D, Galloux M, Gault E, Eléouët J-F. 2015. Fine mapping and characterization of the L-polymerase-binding domain of the respiratory syncytial virus phosphoprotein. J Virol 89:4421–4433. doi: 10.1128/JVI.03619-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, Zengel J, Sun M, Sleeman K, Timani KA, Aligo J, Rota P, Wu J, He B. 2015. Regulation of viral RNA synthesis by the V protein of parainfluenza virus 5. J Virol 89:11845–11857. doi: 10.1128/JVI.01832-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fearns R, Peeples ME, Collins PL. 1997. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology 236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 45.Luo J, Deng ZL, Luo X, Tang N, Song WX, Chen J, Sharff KA, Luu HH, Haydon RC, Kinzler KW, Vogelstein B, He TC. 2007. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc 2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Wang X, Jia Y, Wang C, Han Q, Lu ZH, Yang Z. 2017. Adenoviral-expressed recombinant granulocyte monocyte colony-stimulating factor (GM-CSF) enhances protective immunity induced by inactivated Newcastle disease virus (NDV) vaccine. Antiviral Res 144:322–329. doi: 10.1016/j.antiviral.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 47.Winkler C, Ferdous F, Dimmick M, Scott T. 2017. Lipopolysaccharide induced interleukin-6 production is mediated through activation of ERK 1/2, p38 MAPK, MEK, and NFkappaB in chicken thrombocytes. Dev Comp Immunol 73:124–130. doi: 10.1016/j.dci.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 48.Khatri M, Sharma JM. 2006. Infectious bursal disease virus infection induces macrophage activation via p38 MAPK and NF-kappaB pathways. Virus Res 118:70–77. doi: 10.1016/j.virusres.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 49.Bi Y, Jin Z, Wang Y, Mou S, Wang W, Wei Q, Huo N, Liu S, Wang X, Yang Z, Chen H, Xiao S. 2019. Identification of two distinct linear B cell epitopes of the matrix protein of the Newcastle disease virus vaccine strain LaSota. Viral Immunol 32:221. doi: 10.1089/vim.2019.0007. [DOI] [PubMed] [Google Scholar]
- 50.Liu H, de Almeida RS, Gil P, Albina E. 2017. Comparison of the efficiency of different Newcastle disease virus reverse genetics systems. J Virol Methods 249:111–116. doi: 10.1016/j.jviromet.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 51.Liu H, Albina E, Gil P, Minet C, de Almeida RS. 2017. Two-plasmid system to increase the rescue efficiency of paramyxoviruses by reverse genetics: the example of rescuing Newcastle disease virus. Virology 509:42–51. doi: 10.1016/j.virol.2017.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All gene microarray files are available from the Gene Expression Omnibus database (accession number GSE121368). All relevant data are within the paper.