The MT-4 human T-cell line expresses HTLV-1 Tax and is permissive for replication of an HIV-1 gp41 mutant lacking the cytoplasmic tail. MT-4 cells (lot 150048), distributed by the NIH AIDS Reagent Program (NIH-ARP), were found to be Tax deficient and unable to host replication of the gp41-truncated HIV-1 mutant. These findings, together with short tandem repeat profiling, established that lot 150048 are not bona fide MT-4 cells.
KEYWORDS: MT-4, STR profiling, authentication, short tandem repeat
ABSTRACT
The MT-4 human T-cell line expresses HTLV-1 Tax and is permissive for replication of an HIV-1 gp41 mutant lacking the cytoplasmic tail. MT-4 cells (lot 150048), distributed by the NIH AIDS Reagent Program (NIH-ARP), were found to be Tax deficient and unable to host replication of the gp41-truncated HIV-1 mutant. These findings, together with short tandem repeat profiling, established that lot 150048 are not bona fide MT-4 cells.
TEXT
MT-4 cells were originally established by cocultivating cells from a 50-year-old, human T-cell lymphotropic virus type 1 (HTLV-1)-seropositive Japanese male with adult T-cell leukemia (ATL) with human cord blood lymphocytes (1). As a consequence of their derivation from an HTLV-1-infected individual, MT-4 cells express the HTLV-1 Tax protein (2). MT-4 cells were previously reported to express low levels of APOBEC3G (A3G) (3) and are known to support the rapid replication of HIV-1, with wild-type (WT) virus levels peaking at approximately 2 to 4 days postinfection (4). Furthermore, MT-4 cells are uniquely capable of supporting the replication of an HIV-1 mutant, CTdel-144, in which the cytoplasmic tail (CT) of the transmembrane glycoprotein gp41 has been deleted (5–7).
MT-4 cells are an important tissue culture tool for the study of HIV-1 and hepatitis C virus infection. The cells are widely used for cytotoxicity inhibition assays with antiviral compounds (8–10), HIV-1 growth competition assays (11), syncytium formation assays (12), and experiments in which defective HIV-1 mutants are propagated for the selection of viral revertants (7, 13–15).
In 1997, the NIH-ARP issued a recall for a lot of MT-4 cells that were found to be Tax-deficient. These cells were traced to lot 3/9/92. Recently, the National Institutes of Health AIDS Reagent Program (NIH-ARP) released a new lot of MT-4 cells (lot 150048) that we observed to be morphologically and phenotypically different from authentic MT-4 cells. In the course of investigating this issue, we found that lot 150048 cells were derived from the same Tax-deficient MT-4 cells recalled by the NIH-ARP over 2 decades ago (2) and confirmed that these cells are a T-cell line other than MT-4.
BONA FIDE MT-4 CELLS EXPRESS A3G AND HTLV-1 Tax
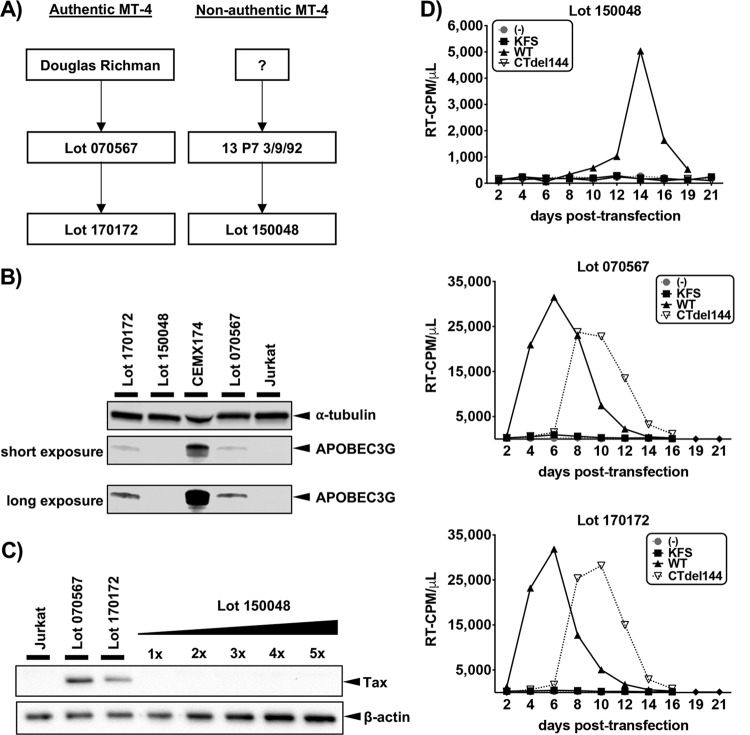
We realized that a recent lot of MT-4 cells (lot 150048) distributed by the NIH-ARP were morphologically distinct from bona fide MT-4 cells and, unlike bona fide MT-4 cells, did not express A3G. The NIH-ARP provided a new lot of MT-4 cells (170172) to replace lot 150048 and traced lot 150048 to a previous lot, 13 P7 3/9/92 (Fig. 1A), which in 1997 was reported to be Tax deficient (2). Western blot analysis of cell lysates obtained from three different MT-4 lots provided by the NIH-ARP were analyzed (Fig. 1B). CEMX174 (A3G high-expressing) and Jurkat (A3G-deficient) cells were included as controls. Lot 150048 cells did not express A3G at detectable levels, while lots 070567 and 170172 did express low levels of A3G.
FIG 1.
Analysis of cell lines by Western blotting. (A) Flow chart indicating the source of four different MT-4 lots. (B) Western blot of A3G expression. (C) Western blot of Tax expression with increasing volumes of lot 150048 cell lysate relative to the volume loaded on the gel for Jurkat, lot 070567, and lot 170172. All Western blots are representative of three independent experiments. (D and E) Spreading infection kinetics for the various MT-4 lots transfected with WT, KFS [Env(–)], or CTdel144 molecular clones. The data are representative of three independent experiments.
MT-4 cells are HTLV-1 transformed and thus express HTLV-1 Tax (2). Both lots that expressed low levels of A3G also expressed Tax (Fig. 1C), as expected for MT-4 cells. Increasing amounts of lot 150048 lysate were analyzed by Western blot in an attempt to detect low levels of Tax. No Tax protein could be detected in lot 150048 samples. Altogether, these findings suggest that lot 150048 are not MT-4 cells. The NIH-ARP was notified, and lot 150048 was pulled from distribution. Tax expression in cells from the currently distributed lot, 170172, is similar to the previously distributed lot 070567 from which they are derived (Fig. 1C).
LOT 150048 CELLS SUPPORT REPLICATION OF WT BUT NOT CTdel144 HIV-1
Whereas HIV-1 replication in most T-cell lines is abrogated by truncation of the long gp41 CT, the gp41 CT truncation mutant, CTdel144, can replicate in the MT-4 T-cell line (5–7). To determine whether lot 150048 is permissive for the CTdel144 mutant, these cells were transfected with the WT pNL4-3 molecular clone or the Env(–) (KFS) (16) or CTdel144 (5) derivatives (Fig. 1D). Cells from lots 070567 and 170172 supported rapid HIV-1 replication and were permissive for the CTdel144 mutant (Fig. 1E). In contrast, replication in cells from lot 150048 was markedly slower, and these cells did not support replication of CTdel144 (Fig. 1D). These results are consistent with lot 150048 being a T-cell line other than MT-4.
LOT 150048 CELLS ARE SMALLER THAN BONA FIDE MT-4 CELLS
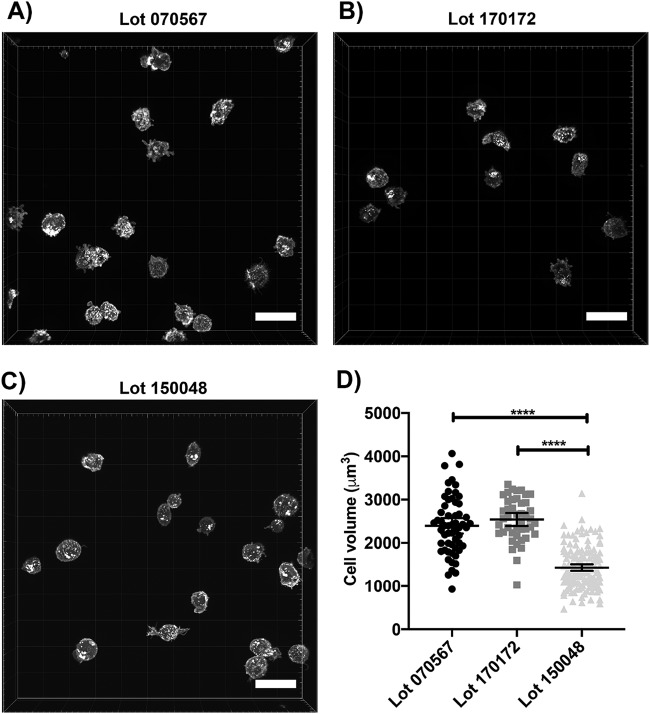
Another indication that lot 150048 cells are not bona fide MT-4 cells is their non-MT-4-like morphology. Tax upregulates expression of cell adhesion molecules, resulting in cell clustering (17). While lot 150048 did grow in suspension, the cells did not form cell clusters/clumps characteristic of authentic MT-4 (data not shown). Rather, they maintained their status as single cells during cellular expansion. To analyze the morphological characteristics and volumes of cells from lot 150048, fluorescence microscopy, followed by cell segmentation, was conducted (Fig. 2). Lots 070567 and 170172 show a more spherical shape and uniform cell volume (Fig. 2A and B) than lot 150048 (Fig. 2C). After segmentation of cells from all three lots, it was determined that the volume of cells from lot 150048 is significantly smaller than that of cells from lots 070567 and 170172 (Fig. 2D). In addition, relative to cells from lots 070567 and 170172, cells from lot 150048 are morphologically more diverse with a subset of cells displaying more irregular and larger protrusions on the plasma membrane (Fig. 2C), consistent with the conclusion that these cells are not MT-4.
FIG 2.
Lot 150048 cells are smaller in volume than authentic MT-4 cells. (A to C) Representative micrographs of cells from lot 070567 (A), lot 170172 (B), and lot 150048 (C). (D) Graph of cell volumes (****, P < 0.0001). Segmentation for cell volume was performed on 44 cells from lot 170172, 57 cells from lot 070567, and 147 cells from lot 150048. One-way ANOVA, followed by a Tukey-Kramer test, was used to analyze the statistics. Error bars depict the 95% confidence intervals. Scale bars, 30 μm.
STR PROFILING ESTABLISHES THAT LOT 150048 CELLS ARE NOT MT-4
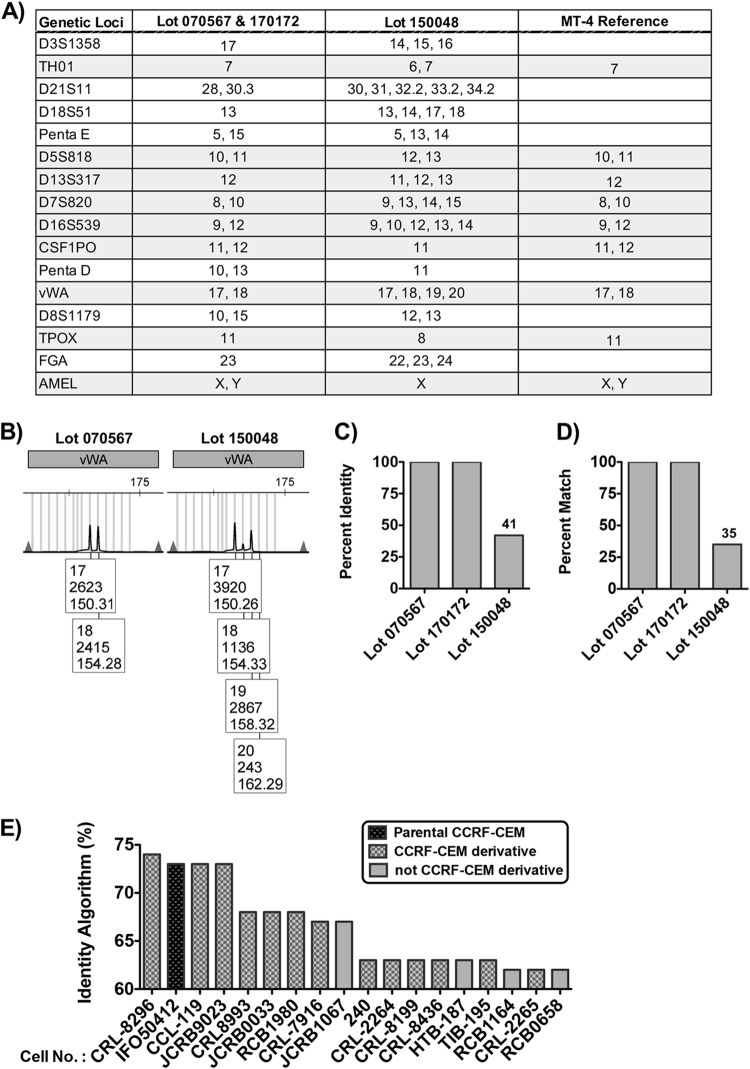
The human genome contains many repetitive DNA elements of various lengths, including DNA regions with short repeating units (2 to 6 bp/unit), known as short tandem repeats (STRs). An individual inherits one copy of an STR for a gene locus from each parent, resulting in two STR values (one for each allele) of similar or different sizes. Furthermore, the number of STRs can be highly variable between individuals in a population, making STR profiling a highly effective cell line identification tool. When applied to cell line authentication and identification, determination of the percent identity (18) and percent match (19) can facilitate determining whether cell samples are pure (100%), related but divergent due to genomic instability (≥80%), contaminated with a second cell line (55 to 79%), or misidentified (< 55%). Genomic instability is also exemplified by one or more loci on the electropherogram with more than two alleles present at positions +1 or −1 off the main peak with peaks of variable height.
To confirm that lots 170172 and 070567 are authentic MT-4 cells and lot 150048 is not, STR profiling was performed (Fig. 3). The STR profile for all lots was compared against the Cellosaurus (20) reference STR profile for MT-4 (Fig. 3A). Lots 070567 and 170172 were 100% matches to the MT-4 reference. Consistent with MT-4 cells being derived from a male ATL patient, STR profiling of these cells confirmed the presence of a Y chromosome. Lot 1500048 did not match the MT-4 reference STR profile. Furthermore, lot 150048 displayed markers of genetic instability on the electropherogram, indicated by multiple peaks of variable height at +1 off the main peaks (e.g., gene locus vWA [Fig. 3B]). The percent identity (see equation 1 below) (Fig. 3C) and percent match (see equation 2 below) (Fig. 3D) for lot 150048 with MT-4 were found to be below the threshold (55%) for the value that would suggest possible cell line contamination as an explanation for the mismatch. We therefore conclude that lot 150048 is a T-cell line other than MT-4.
FIG 3.
STR profiling confirms that lot 150048 are not MT-4 cells. (A) Table listing the STR identities at various genetic loci for query lots of MT-4 and the MT-4 reference provided by Cellosaurus. (B) Electropherogram for the STR profile at the vWA gene locus for lots 070567 and 150048. Numbers in the box below each peak represent the following: top, allele call/STR peak; middle, peak height in relative fluorescent units; bottom, size of STR fragment in base pairs (distance traveled in the capillary). (C and D) Percent identity (C) and percent match (D) of query MT-4 lots to the MT-4 reference STR profile. (E) Percent identity of lot 150048 to best-match cell lines. Parental (black diamond grid) and derivative CCRF-CEM (gray diamond grid) are distinguished by bars with different patterns, while cell lines not related to CCRF-CEM are differentiated by bars with no pattern.
To define the origins of lot 150048, the STR profile was analyzed on the Leibniz-Institut Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) STR Profile Search (21). The percent identity algorithm was used to determine the best match to reference STR profiles in the database (Fig. 3E). Lot 150048 most closely matched the T-cell line CCRF-CEM, but the percent identity value for the best-matching CCRF-CEM derivative, 74%, is below the threshold necessary for lot 150048 to be identified as CCRF-CEM that has diverged due to genomic instability. Therefore, the true identity of this T-cell line remains unknown.
SUMMARY OF FINDINGS
Two decades ago, the NIH-ARP recalled a nonauthentic lot of MT-4 cells derived from lot 13 P7 3/9/92 (Fig. 1A) after it was discovered that this lot did not express HTLV-I Tax and did not contain HTLV-I DNA (2). Recently, the NIH-ARP distributed MT-4 lot 150048, which contained cells that were found to be phenotypically and genetically different from authentic MT-4 cells. Our communications with the NIH-ARP suggested that lot 150048 was derived from the same source of Tax-deficient cells that were distributed in 1997. Lot 150048 is a suspension T-cell line that is able to host HIV-1 replication but with delayed kinetics relative to bona fide MT-4 cells. In addition, unlike bona fide MT-4 cells, lot 150048 is unable to support replication of HIV-1 bearing a gp41 CT truncation. Lot 150048 also lacks expression of A3G and HTLV-I Tax, two proteins expressed by bona fide MT-4 cells. Finally, STR profiling verified that lot 150048 cells are not MT-4 but was unable to conclusively identify this cell line.
Laboratories that received lot 150048 were recently contacted by the NIH-ARP to inform researchers that this lot of cells are not MT-4. It is unknown how many researchers might have received lot 150048 indirectly from colleagues. To prevent lots 150048 and 13 P7 3/9/92 from being distributed again, the NIH-ARP has destroyed these cells. Furthermore, the NIH-ARP has contacted all researchers who received lot 150048 to inform them that they are not MT-4 cells.
We hope that our evaluation of these cells highlights various cost-effective methods for determining whether the MT-4 cells investigators have currently in their labs are bona fide MT-4 cells. We have communicated with the NIH-ARP to trace various lots of MT-4 and identified their source. Furthermore, we confirmed that the current lot of MT-4 in distribution (lot 170172) are bona fide MT-4.
CELL CULTURE AND CELL LINES
All cells were obtained from the NIH-ARP. Bona fide MT-4 cells (catalog no. 120) were contributed by Douglas Richman. CEMX174 (catalog no. 272) (22) were contributed by Peter Cresswell. The original contributor and source of the nonauthentic MT-4 cell line are unknown. Cells were grown in Roswell Park Memorial Institute (RPMI) 1640 medium (CellGro) supplemented with 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin (penicillin [50 U/ml] and streptomycin [50 μg/ml], final concentrations; Lonza, Walkersville, MD). All cells were maintained in humidified 37°C incubators with 5% CO2.
WESTERN BLOTS
For detection of A3G, cell lysates from 106 cells were prepared using CelLytic M (Sigma) solution (125 μl) containing Protease Inhibitor Cocktail (Roche), followed by a 10-min, 10,000 × g spin to remove cellular debris. The cell lysates were mixed with NuPAGE LDS sample buffer (Invitrogen) containing β-mercaptoethanol and heated for 5 min at 95°C. Samples were analyzed on 4 to 20% Tris-glycine gels (Invitrogen) using standard Western blotting techniques. Proteins were detected with primary antibodies against A3G (1:5,000 dilution, from Klaus Strebel [NIH-ARP, catalog no. 10082]) (23, 24) and α-tubulin (1:10,000 dilution; Sigma, catalog no. T9026). An IRDye 800CW-labeled goat anti-rabbit secondary antibody (LI-COR, catalog no. 926-32211) was used at a 1:10,000 dilution to detect rabbit primary antibodies, and an IRDye 680-labeled goat anti-mouse secondary antibody (LI-COR, catalog no. 926-68070) was used at a 1:10,000 dilution to detect mouse primary antibodies. Protein bands were visualized using an Odyssey infrared imaging system (LI-COR).
For detection of Tax, cell lysates from 106 cells were prepared using 100 μl of Triton X-100 lysis buffer containing 300 mM NaCl, 50 mM Tris-HCl (pH 7.5), 0.5% Triton X-100, 10 mM iodoacetamide, and 10 tablets/0.5 liter of complete protease inhibitor (Roche, catalog no. 11836145001). Cell lysates were mixed with 30 μl of a 6× denaturing buffer (25) and boiled for 10 min at 95°C. Proteins were separated on hand-cast 12% Tris-glycine gels and analyzed using standard Western blotting techniques. Proteins were detected with primary antibodies as follows: α-Tax (1:5,000 dilution; Abcam, catalog no. ab26997), and α-β-actin conjugated directly to horseradish peroxidase (HRP; Abcam, catalog no. ab49900). HRP-conjugated secondary antibodies and goat anti-mouse (Invitrogen, catalog no. 32230) antisera were used at a 1:10,000 dilution to detect Tax. Protein bands were visualized using chemiluminescence with a Bio-Rad Universal Hood II Chemidoc and then analyzed with ImageLab v5.1 software.
HIV-1 REPLICATION ASSAYS
Virus replication kinetics were determined in Jurkat and MT-4 cell lines as previously described (26). Briefly, T cells were transfected with proviral clones (1 μg DNA/106 cells) in the presence of 700 μg/ml DEAE-dextran. Lot 150048 was split 1:3, and authentic MT-4 cells were split 1:2 every 2 days with fresh media. Virus levels were quantified by measuring reverse transcriptase (RT) activity in collected supernatants over time. RT activity values were plotted using GraphPad Prism.
MICROSCOPY STAINING AND IMAGE ANALYSIS
Cells (106) were fixed with 4% paraformaldehyde (Boston BioProducts, BM-155) for 20 min at room temperature and washed three times with a glycine quench solution (0.112 g of glycine in 50 ml of 1× phosphate-buffered saline). To identify the cell plasma membrane and nucleus, the cells were stained with 5 μg/ml wheat germ agglutinin (WGA; Invitrogen, catalog no. W11261) at room temperature for 15 min. After labeling, the cells were washed to remove excess labeling solution, and 300 nM DAPI (4′,6′-diamidino-2-phenylindole; Thermo Fisher, catalog no. D1306) was used to counterstain cells for 10 min to detect nuclei. Excess counterstain was then washed off. Suspension cells were placed in 35-mm glass-bottom dishes (MatTek, P35GCol-1.5-14-C). Suspension cells were imaged with a 63× oil objective using a Leica DMI-8 equipped with a Yokagawa-CSU-W1 spinning disk confocal with a Zyla sCMOS camera (Andor). Images were captured for WGA conjugated to AF488 (488 nm) and DAPI (405 nm) at 2,048 × 2,048 pixels with a z-step of 200 nm and a 75-ms exposure time. At least ten fields of view from each cell type were imaged in three dimensions. Cells unattached to other cells were segmented using IMARIS software version 9.3.0 (Bitplane) using the surfaces function. Cell surfaces were identified using the channel for WGA, which was smoothed to detect structures of 300 nm and was set to a manual threshold value of 190 (parameters set in the IMARIS surface wizard). Surfaces with an area below 200 μm2 or above 3,000 μm2 were excluded. Also, all cells within 250 nm of the XYZ border were excluded to ensure that the entire cell was being segmented. Segmentation resulted in cell volume measurements. One-way analysis of variance (ANOVA), followed by a Tukey-Kramer test, was used to assess statistical significance between the samples using GraphPad Prism (v8.1.1).
STR PROFILING AND CELL LINE IDENTIFICATION
Genomic DNA from all MT-4 lots (5 × 106 cells per sample) provided by the NIH-ARP was extracted using the QIAamp DNA blood minikit (Qiagen, catalog no. 51106). STR profiling of genomic DNA was performed by Genetica (LabCorp). Electropherograms were analyzed to obtain the STR profile at the indicated gene loci and compared to an MT-4 reference STR profile from Cellosaurus (CVCL 2632). The percent identity algorithm (equation 1) (18) and percent match algorithm (equation 2) (19) were used to calculate the percent match and the percent identity to the reference profile using the following equations:percent identity =
| (1) |
and
| (2) |
To attempt identification of lot 150048, the STR profile was entered into the DSMZ Short Tandem Repeat Profile Search. Best-match returned hits were restricted to a percent identity (called EV on the search engine) score of ≥60%. The cell number was plotted against the percent identity score using GraphPad Prism 5.
ACKNOWLEDGMENTS
We thank Schuyler van Engelenburg (University of Denver) for bringing to our attention the morphological differences between bona fide and nonauthentic MT-4. We also thank Bruce K. Brown and Jennifer Greisler of the NIH-ARP for tracing the various MT-4 lots and coordinating the destruction of lot 150048 and lot 13 P7 3/9/92 located in the NIH-ARP inventory.
The NIH’s AIDS Reagent Program was managed by Fisher Clinical Services and Thermo Fisher Scientific.
This study was supported by the Intramural AIDS Targeted Antiviral Program and the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH.
The views expressed in this article do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Miyoshi I, Taguchi H, Kubonishi I. 1982. Type C virus-producing cell lines derived from adult T cell leukemia. Gan Mongr 28:219–228. doi: 10.1007/978-1-4615-8336-3_17. [DOI] [Google Scholar]
- 2.Jeang KT, Derse D, Matocha M, Sharma O. 1997. Expression status of Tax protein in human T-cell leukemia virus type 1-transformed MT4 cells: recall of MT4 cells distributed by the NIH AIDS Research and Reference Reagent Program. J Virol 71:6277–6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mulder LC, Ooms M, Majdak S, Smedresman J, Linscheid C, Harari A, Kunz A, Simon V. 2010. Moderate influence of human APOBEC3F on HIV-1 replication in primary lymphocytes. J Virol 84:9613–9617. doi: 10.1128/JVI.02630-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harada S, Koyanagi Y, Yamamoto N. 1985. Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science 229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 5.Murakami T, Freed EO. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc Natl Acad Sci U S A 97:343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tedbury PR, Novikova M, Ablan SD, Freed EO. 2016. Biochemical evidence of a role for matrix trimerization in HIV-1 envelope glycoprotein incorporation. Proc Natl Acad Sci U S A 113:E182–E190. doi: 10.1073/pnas.1516618113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joshi A, Ablan SD, Soheilian F, Nagashima K, Freed EO. 2009. Evidence that productive human immunodeficiency virus type 1 assembly can occur in an intracellular compartment. J Virol 83:5375–5387. doi: 10.1128/JVI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szucs G, Melnick JL, Hollinger FB. 1988. A simple assay based on HIV infection preventing the reclustering of MT-4 cells. Bull World Health Organ 66:729–737. [PMC free article] [PubMed] [Google Scholar]
- 9.Van Hout A, D’huys T, Oeyen M, Schols D, Van Loy T. 2017. Comparison of cell-based assays for the identification and evaluation of competitive CXCR4 inhibitors. PLoS One 12:e0176057. doi: 10.1371/journal.pone.0176057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonckers TH, Lin TI, Buyck C, Lachau-Durand S, Vandyck K, Van Hoof S, Vandekerckhove LA, Hu L, Berke JM, Vijgen L, Dillen LL, Cummings MD, de Kock H, Nilsson M, Sund C, Rydegard C, Samuelsson B, Rosenquist A, Fanning G, Van Emelen K, Simmen K, Raboisson P. 2010. 2010. 2′-Deoxy-2′-spirocyclopropylcytidine revisited: a new and selective inhibitor of the hepatitis C virus NS5B polymerase. J Med Chem 53:8150–8160. doi: 10.1021/jm101050a. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez-Merino V, Farrow MA, Brewster F, Somasundaran M, Luzuriaga K. 2008. Identification and characterization of HIV-1 CD8+ T cell escape variants with impaired fitness. J Infect Dis 197:300–308. doi: 10.1086/524845. [DOI] [PubMed] [Google Scholar]
- 12.Nishino Y, Nakaya T, Fujinaga K, Kishi M, Azuma I, Ikuta K. 1994. Persistent infection of MT-4 cells by human immunodeficiency virus type 1 becomes increasingly likely with in vitro serial passage of wild-type but not nef mutant virus. J Gen Virol 75:2241–2251. doi: 10.1099/0022-1317-75-9-2241. [DOI] [PubMed] [Google Scholar]
- 13.Lopez CS, Tsagli SM, Sloan R, Eccles J, Barklis E. 2013. Second site reversion of a mutation near the amino terminus of the HIV-1 capsid protein. Virology 447:95–103. doi: 10.1016/j.virol.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noviello CM, Lopez CS, Kukull B, McNett H, Still A, Eccles J, Sloan R, Barklis E. 2011. Second-site compensatory mutations of HIV-1 capsid mutations. J Virol 85:4730–4738. doi: 10.1128/JVI.00099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tedbury PR, Ablan SD, Freed EO. 2013. Global rescue of defects in HIV-1 envelope glycoprotein incorporation: implications for matrix structure. PLoS Pathog 9:e1003739. doi: 10.1371/journal.ppat.1003739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freed EO, Delwart EL, Buchschacher GL Jr, Panganiban AT. 1992. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci U S A 89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi T, Higuchi M, Fukushi M, Oie M, Ito M, Fujii M. 2002. Homotypic cell–cell adhesion induced by human T cell leukemia virus type 1 Tax protein in T cell lines. Virology 302:132–143. doi: 10.1006/viro.2002.1629. [DOI] [PubMed] [Google Scholar]
- 18.Tanabe H, Takada Y, Minegishi D, Kurematsu M, Masui T, Mizusawa H. 1999. Cell line individualization by STR multiplex system in the cell bank found cross-contamination between ECV304 AND EJ-1/T24. Tissue Culture Res Commun 18:329–338. [Google Scholar]
- 19.American Type Culture Collection Standards Development Organization Workgroup ASN-0002, Alston-Roberts C, Bauer SR, Butler J, Capes-Davis A, Dirks WG, Elmore E, Furtado M, Kerrigan L, Kline MC, Kohara A, Los GV, MacLeod RAF, Masters JR, Nardone M, Nims RW, Price PJ, Reid YA, Shewale J, Steuer AF, Storts DR, Sykes G, Taraporewala Z, Thomson J. 2011. Authentication of human cell lines: standardization of STR profiling: ANSI/ATCC ASN-0002–2011. http://webstore.ansi.org/RecordDetail.aspx?sku=ANSI%2fATCC%2bASN-0002-2011.
- 20.Bairoch A. 2018. The Cellosaurus, a cell-line knowledge resource. J Biomol Tech 29:25–38. doi: 10.7171/jbt.18-2902-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dirks WG, MacLeod RA, Nakamura Y, Kohara A, Reid Y, Milch H, Drexler HG, Mizusawa H. 2010. Cell line cross-contamination initiative: an interactive reference database of STR profiles covering common cancer cell lines. Int J Cancer 126:303–304. doi: 10.1002/ijc.24999. [DOI] [PubMed] [Google Scholar]
- 22.Salter RD, Howell DN, Cresswell P. 1985. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics 21:235–246. doi: 10.1007/bf00375376. [DOI] [PubMed] [Google Scholar]
- 23.Kao S, Miyagi E, Khan MA, Takeuchi H, Opi S, Goila-Gaur R, Strebel K. 2004. Production of infectious human immunodeficiency virus type 1 does not require depletion of APOBEC3G from virus-producing cells. Retrovirology 1:27. doi: 10.1186/1742-4690-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan MA, Kao S, Miyagi E, Takeuchi H, Goila-Gaur R, Opi S, Gipson CL, Parslow TG, Ly H, Strebel K. 2005. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J Virol 79:5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallagher SR. 2003. Current protocols in molecular biology, p 10.2A.28 In Ausubel MBR, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (ed), Analysis of proteins. John Wiley & Sons, Inc, Media, PA. [Google Scholar]
- 26.Freed EO, Martin MA. 1994. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J Virol 68:2503–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]