Infections with bat-origin coronaviruses (CoVs) (severe acute respiratory syndrome CoV [SARS-CoV] and Middle East respiratory syndrome CoV [MERS-CoV]) have caused severe illness in humans after “host jump” events. Recently, a novel bat-HKU2-like CoV named swine acute diarrhea syndrome CoV (SADS-CoV) has emerged in southern China, causing lethal diarrhea in newborn piglets. It is important to assess the species barriers of SADS-CoV infection since the animal hosts (other than pigs and bats) and zoonotic potential are still unknown. An in vitro susceptibility study revealed a broad species tropism of SADS-CoV, including various rodent and human cell lines. We established a mouse model of SADS-CoV infection, identifying its active replication in splenic dendritic cells, which suggests that SADS-CoV has the potential to infect rodents. These findings highlight the potential cross-species transmissibility of SADS-CoV, although further surveillance in other animal populations is needed to fully understand the ecology of this bat-HKU2-origin CoV.
KEYWORDS: interspecies transmission, Coronavirus, SADS-CoV, mouse infection model
ABSTRACT
Outbreaks of severe diarrhea in neonatal piglets in Guangdong, China, in 2017 resulted in the isolation and discovery of a novel swine enteric alphacoronavirus (SeACoV) derived from the species Rhinolophus bat coronavirus HKU2 (Y. Pan, X. Tian, P. Qin, B. Wang, et al., Vet Microbiol 211:15–21, 2017). SeACoV was later referred to as swine acute diarrhea syndrome CoV (SADS-CoV) by another group (P. Zhou, H. Fan, T. Lan, X.-L. Yang, et al., Nature 556:255–258, 2018). The present study was set up to investigate the potential species barriers of SADS-CoV in vitro and in vivo. We first demonstrated that SADS-CoV possesses a broad species tropism and is able to infect cell lines from diverse species, including bats, mice, rats, gerbils, hamsters, pigs, chickens, nonhuman primates, and humans. Trypsin contributes to but is not essential for SADS-CoV propagation in vitro. Furthermore, C57BL/6J mice were inoculated with the virus via oral or intraperitoneal routes. Although the mice exhibited only subclinical infection, they supported viral replication and prolonged infection in the spleen. SADS-CoV nonstructural proteins and double-stranded RNA were detected in splenocytes of the marginal zone on the edge of lymphatic follicles, indicating active replication of SADS-CoV in the mouse model. We identified that splenic dendritic cells (DCs) are the major targets of virus infection by immunofluorescence and flow cytometry approaches. Finally, we demonstrated that SADS-CoV does not utilize known CoV receptors for cellular entry. The ability of SADS-CoV to replicate in various cells lines from a broad range of species and the unexpected tropism for murine DCs provide important insights into the biology of this bat-origin CoV, highlighting its possible ability to cross interspecies barriers.
IMPORTANCE Infections with bat-origin coronaviruses (CoVs) (severe acute respiratory syndrome CoV [SARS-CoV] and Middle East respiratory syndrome CoV [MERS-CoV]) have caused severe illness in humans after “host jump” events. Recently, a novel bat-HKU2-like CoV named swine acute diarrhea syndrome CoV (SADS-CoV) has emerged in southern China, causing lethal diarrhea in newborn piglets. It is important to assess the species barriers of SADS-CoV infection since the animal hosts (other than pigs and bats) and zoonotic potential are still unknown. An in vitro susceptibility study revealed a broad species tropism of SADS-CoV, including various rodent and human cell lines. We established a mouse model of SADS-CoV infection, identifying its active replication in splenic dendritic cells, which suggests that SADS-CoV has the potential to infect rodents. These findings highlight the potential cross-species transmissibility of SADS-CoV, although further surveillance in other animal populations is needed to fully understand the ecology of this bat-HKU2-origin CoV.
INTRODUCTION
The spread of zoonotic pathogens remains among the leading threats to global public health. Coronaviruses (CoVs) can infect a wide variety of animals and humans, resulting in several diseases with respiratory, enteric, and neurological pathologies of various severity (1–4). Because of the various routes of infection and extensive phagocytosis in tissues, close contact between humans and animals provides potential scenarios for adaptive mutation and interspecies transmission (5).
The source of the severe acute respiratory syndrome CoV (SARS-CoV) was traced to civets in animal markets and, ultimately, to bats, leading to more than 8,000 human infections and 774 deaths after its emergence in 2002 (5–7). The emergence of Middle East respiratory syndrome CoV (MERS-CoV) in 2012 (2) resulted in more than 1,000 clinical cases with a mortality rate of 35%, making it the second marked threatening CoV of the 21st century (8, 9). Although camels can be infected with MERS-CoV, bats are also thought to be the original host of MERS-CoV (5). Both SARS-CoV and MERS-CoV originated in bats, illustrating the damage caused by CoVs during interspecies transmission events and highlighting the need for increased global vigilance of CoV-associated disease (1, 5, 10).
In February 2017, outbreaks of severe diarrhea of suckling piglets occurred in swine herds in Guangdong Province, China (11). Clinical signs consisted of acute vomiting and watery diarrhea, but porcine viruses commonly associated with diarrhea, including porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV), and porcine deltacoronavirus (PDCoV), were not detected in any of the clinical samples (11). The new enteric pathogen of commercial pigs that was isolated was finally identified as a new porcine CoV belonging to the species Rhinolophus bat coronavirus HKU2 (11–13). Our research group tentatively designated this newly emerged virus as swine enteric alphacoronavirus (SeACoV) (11), and it was later named swine acute diarrhea syndrome CoV (SADS-CoV) by Zhou et al. (14). It is also known by other names, such as porcine enteric alphacoronavirus (PEAV) (13). For purposes of unity, SADS-CoV is the name used to refer to this new virus in the current study. The expanded host range of bat-origin HKU2 to pigs indicates that bats play an important role in the ecology and evolution of SADS-CoV, although the mechanism of bat-to-swine transmission remains unclear. In view of the damage caused by SARS and MERS for both animal and public health, careful attention must be paid to the prevalence of CoV-associated disease among humans and domestic animals (15).
Therefore, there is an urgent need for more information on the details of SADS-CoV infection. It is critically important to assess potential species barriers of SADS-CoV transmission since the animal hosts (other than pigs and bats) and zoonotic potential are still unknown. In the present study, we demonstrated that SADS-CoV possesses a very broad species tropism in vitro and is able to infect cell lines from diverse species, including rodents and humans. Furthermore, in vivo evidence from experimental infection of mice with SADS-CoV identified splenic dendritic cells (DCs) as the major site of SADS-CoV replication in mice. Finally, we demonstrated that SADS-CoV does not utilize known CoV protein receptors for cellular entry. These results present the possibility that rodents are among the susceptible hosts of SADS-CoV, highlighting the potential cross-species transmissibility of SADS-CoV.
RESULTS
SADS-CoV can infect cell lines originating from various species.
Previously, we reported that SADS-CoV was isolated in Vero cells supplemented with trypsin (11). Since exogenous trypsin is essential for propagation of PEDV isolates in vitro (16), likely by mediating activation of membrane fusion by S glycoprotein proteolysis (17), we were interested to know whether it is also required for SADS-CoV growth in cell culture. A total of 24 cell lines originating in various tissues of humans and different animal species were tested for susceptibility to SADS-CoV treated with or without trypsin (Table 1). As a brief summary of the results, 21 of the 24 cell lines showed significant susceptibility to SADS-CoV infection, defined by efficient viral replication, antigen expression, and the appearance of cytopathic effect (CPE). The three cell lines that were not infected by SADS-CoV were MDCK, BFK, and RAW 264.7.
TABLE 1.
Summary of human and animal cell lines and their susceptibility to SADS-CoV infection as determined by CPE and IFA
| Cell line information |
Results of in vivo infection with SADS-CoVa |
|||||
|---|---|---|---|---|---|---|
| Without trypsin |
With trypsin |
|||||
| Species and/or tissue origin | Name | ATCC no. | IFA | CPE | IFA | CPE |
| Human | ||||||
| Hepatocellular carcinoma | Huh-7 | N/Ab | ++ | − | ++ | − |
| Hepatocellular carcinoma | HepG2/C3A | HB-8065 | + | − | ++ | − |
| Embryonic kidney | 293T | CRL-11268 | + | − | N/Dc | N/D |
| Lung carcinoma | A549 | CCL-185EMT | + | − | ++ | − |
| Cervix adenocarcinoma | HeLa | CCL-2 | + | − | + | − |
| Monkey | ||||||
| African green monkey kidney | Marc-145 | N/A | + | − | ++ | + |
| Cos-7 | CRL-1651 | ++ | − | +++ | ++ | |
| BSC-40 | CRL-2761 | ++ | − | +++ | ++ | |
| Vero | CRL-1586 | ++ | + | +++ | +++ | |
| Swine | ||||||
| Testis | ST | CRL-1746 | + | + | ++ | ++ |
| Kidney | PK15 | CCL-33 | + | − | ++ | + |
| LLC-PK1 | CL-101 | + | − | ++ | + | |
| Small intestinal epithelium | IPEC-J2 | N/A | + | − | + | − |
| Bat | ||||||
| Myotis petax, fetal kidney | BFK | N/A | − | − | − | − |
| Tadarida brasiliensis, lung | Tb-1 | CCL-88 | + | − | ++ | − |
| Canine | ||||||
| Kidney | MDCK | CCL-34 | − | − | − | − |
| Mouse | ||||||
| Embryo fibroblasts | NIH/3T3 | CRL-1658 | + | − | N/D | N/D |
| Monocyte/macrophage | RAW 264.7 | TIB-71 | − | − | − | − |
| Hamster | ||||||
| Syrian golden hamster, kidney | BHK-21 | CCL-10 | + | − | +++ | + |
| Chinese hamster, ovary | CHO | CCL-61 | + | − | N/D | N/D |
| Rat | ||||||
| Liver | BRL 3A | CRL-1442 | ++ | + | N/D | N/D |
| Kidney | NRK-52E | CRL-1571 | + | − | N/D | N/D |
| Gerbil | ||||||
| Primary kidney cells | N/A | ++ | − | N/D | N/D | |
| Chicken | ||||||
| Embryo fibroblasts | DF-1 | CRL-12203 | + | − | N/D | N/D |
Degree of infection as determined by IFA or CPE (−, no infection or obvious lesion [≤1%]; +, ≤25%; ++, ≤50%; +++, ≤75%; ++++, ≤100%).
N/A, Not available.
N/D, Not detected due to cell sensitivity to trypsin.
First, CPE was examined by inverted light microscopy at 48 hours postinfection (hpi), and scores are shown in Table 1. As the 293T, NIH/3T3, CHO, BRL-3A, and NRK-52E cell lines were sensitive to trypsin, they could not be tested for SADS-CoV infection in MMT cells. Apart from that, CPE was visible in Vero, ST, and BRL-3A cell lines without trypsin, and prominent CPE appeared in or was enhanced with trypsin in Marc-145, Cos-7, BSC-40, Vero, ST, PK15, LLC-PK1, and BHK-21 cell lines (Table 1).
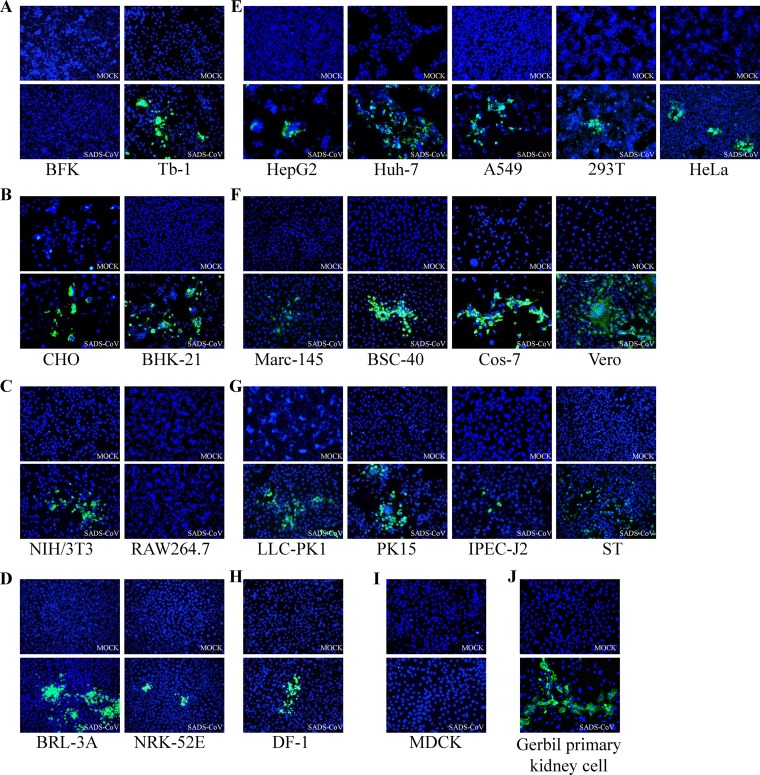
As some cells did not display CPE after SADS-CoV infection, all cell lines were subsequently tested for viral M protein expression by immunofluorescence assay (IFA) (Fig. 1), revealing the same range as seen by CPE in the different cell lines (data not shown). Syncytium formation was prominent in Huh-7, Vero, and BHK-21 cells, whereas in MDCK, BFK and RAW 264.7 cells, the antigen expression was much less prominent than in the other cell lines (Fig. 1A, C, and I). Most cell lines tested showed evidence of productive infection, as indicated by expression of the M protein, while the inefficient antigen expression in Marc-145, LLC-PK1, and IPEC-J2 cells suggested only a limited infection.
FIG 1.
Immunofluorescence assay showing susceptibility of different cell lines to SADS-CoV infection. Immunofluorescence assay of cells infected with SADS-CoV at an MOI of 0.01 was performed using rabbit anti-SADS-CoV-M polyclonal Ab (200× magnification) and Alexa Fluor 488-conjugated anti-rabbit IgG as secondary antibody, with DAPI for visualization of cell nuclei. Mock-infected cells were treated with the same procedures as appropriate. Cells were tested from different species of origin, including bats (BFK and Tb-1) (A), hamsters (CHO and BHK-21) (B), mice (NIH/3T3 and RAW264.7) (C), rats (BRL-3A and NRK-52E) (D), humans (Huh-7, HepG2, 293T, A549, and HeLa) (E), monkeys (Marc-145, Cos-7, BSC-40, and Vero) (F), pigs (ST, PK15, LLC-PK1, and IPEC-J2) (G), chickens (DF-1) (H), dogs (MDCK) (I), and gerbil primary kidney cells (J).
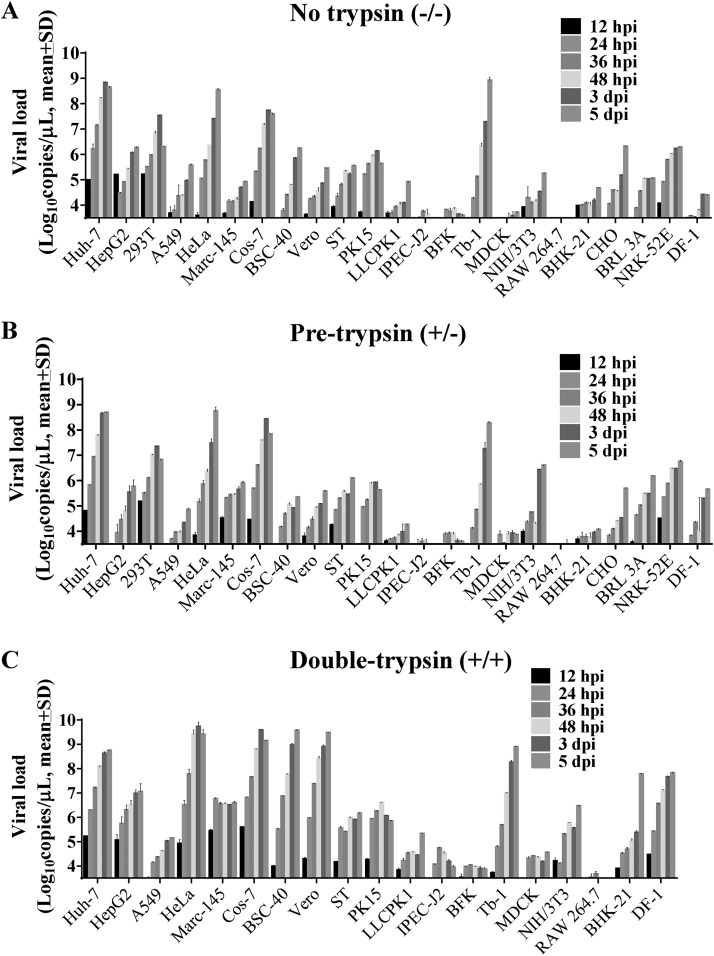
Next, viral load in the culture supernatants was detected over 5 days postinfection (dpi) by reverse transcriptase quantitative PCR (qRT-PCR) (Fig. 2A to C). A higher mean viral load was detected by qRT-PCR after trypsin treatment in HepG2, HeLa, Marc-145, Cos-7, BSC-40, Vero, LLC-PK1, IPEC-J2, BHK-21, and DF-1 cells. Therefore, trypsin contributes to but is not essential for SADS-CoV propagation in these cell lines. There was no difference after trypsin treatment in the other cell lines, although Huh-7 and Tb-1 cells had high levels of SADS-CoV RNA regardless of trypsin treatment.
FIG 2.
Growth of SADS-CoV in different cell lines through 5 days postinfection. To determine the effect of trypsin on SADS-CoV infection, each cell line was infected in the following three conditions: “no trypsin” treatment, inoculated with SADS-CoV diluted in maintenance medium (MM) for 2 h and subsequently replaced with MM (A); “pretrypsin” treatment, inoculated with SADS-CoV diluted in MM containing 5 μg/ml trypsin (MMT) for 2 h and subsequently replaced with MM (B); and “double-trypsin” treatment, inoculated with SADS-CoV in MMT and subsequently replaced with MMT (C). Infection supernatants were collected at 12, 24, 36, 48, 72, and 120 hpi for viral load detection by a qRT-PCR assay targeting the viral N gene. Data are expressed as the mean viral load (log10 copies/μl) ± standard deviation (SD), and all experiments were performed in triplicate. The 293T, CHO, BRL-3A, and NRK-52E cell lines did not survive in the presence of trypsin. (D) Infectious titers (TCID50/ml) of SADS-CoV secreted from HeLa, Vero, Tb-1, BHK-21 PK-15, and MDCK cells were determined on Vero cells.
The progressive release of infectious SADS-CoV into the culture medium of six representative cell lines infected with SADS-CoV was determined by titration of supernatants in Vero cells (Fig. 2D). Unlike in MDCK cells, SADS-CoV infection of HeLa, Vero, Tb-1, BHK-21, and PK-15 cells was productive, with HeLa cells showing the greatest susceptibility (Fig. 2D).
Wild-type C57BL/6J mice can be infected by SADS-CoV via oral and intraperitoneal routes.
With the observation that SADS-CoV could infect diverse rodent cell lines (from mice, rats, and hamsters as well as gerbil primary kidney cells), we hypothesized that mice may be susceptible to SADS-CoV. To test this, we inoculated 6- to 8-week-old wild-type B6 mice with 5 × 105 50% tissue culture infective dose (TCID50) of SADS-CoV by the per oral (p.o.) or intraperitoneal (i.p.) route and monitored them for 28 days for clinical symptoms. The mice did not succumb to the infection nor did they develop diarrhea or experience weight loss during the incubation period (data not shown).
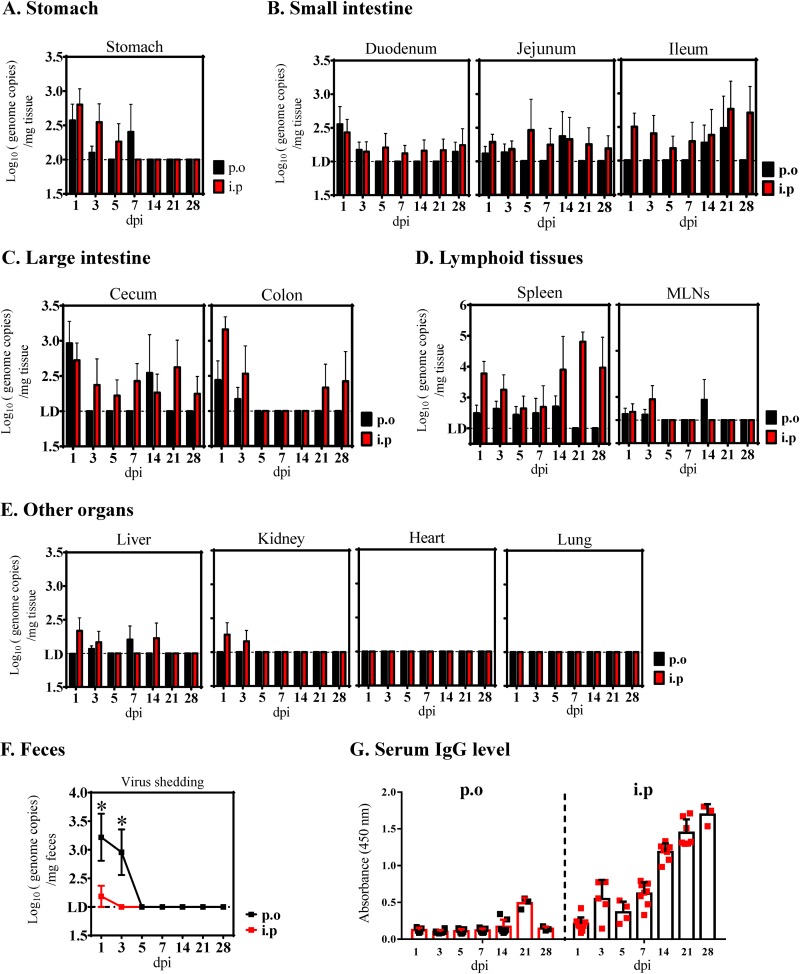
To determine whether SADS-CoV infected the animals asymptomatically, tissue and fecal samples from inoculated mice were collected at 1, 3, 5, 7, 14, 21, and 28 dpi to determine viral growth kinetics and shedding. Analysis of tissue samples by qRT-PCR suggested that SADS-CoV replicated modestly in the stomach early after i.p. or p.o. infection, declining and reaching undetectable levels at 7 or 14 dpi and thereafter (Fig. 3A). A very limited viral replication was observed in each region of the small intestine, with the ileum via i.p. infection showing continuous and decent detectable viral RNA (Fig. 3B). In the large intestine, i.p. infection also resulted in viral RNA loads slightly above the limit of detection at each time point in the ceca, whereas it led to higher viral RNA levels at 1 to 3 dpi, and much lower viral RNA at 21 to 28 dpi in the colon than that of the p.o. route (Fig. 3C). However, this replication in the large intestine did not translate into higher shedding, as hardly any viral genomes were detected, even at 1 dpi in the fecal samples collected from i.p.-infected mice (Fig. 3F). On the contrary, significantly more virus was detected in the feces of p.o.-infected mice at 1 and 3 dpi, indicating that i.p. inoculation does not lead to higher virus shedding.
FIG 3.
SADS-CoV infection of mice. C57BL/6J WT mice were infected per orally (p.o.; black) or intraperitoneally (i.p.; red) with 5 × 105 TCID50 of purified SADS-CoV. Viral loads in different tissue samples, including stomach (A), small intestinal segments (B), large intestinal segments (C), lymphoid tissues (D), the other organs (liver, kidney, heart, and lung) (E), and feces (F) collected at 1, 3, 5, 7, 14, 21, and 28 days postinfection (dpi) were determined by qRT-PCR. MLN, mesenteric lymph nodes. Data are from three independent experiments, and each symbol represents titers from an individual sample (*, P < 0.05). The limit of detection was 1 × 102 genome copies/mg. (G) SADS-CoV IgG antibodies were detected in serum samples collected at euthanasia using an ELISA based on purified SADS-CoV virus particles.
Finally, SADS-CoV replicated more efficiently in the spleen following the i.p. route, with significantly higher viral RNA loads at 21 dpi (Fig. 3D). More importantly, the virus was not cleared from this tissue by 28 dpi in the i.p.-infected group and by 14 dpi in the p.o.-infected group, suggestive of a SADS-CoV prolonged infection in the spleen independent of inoculation route. In contrast to the spleen, only very low levels of viral RNA were detected in the local lymphoid tissue of mesenteric lymph nodes (MLNs) at 1 to 3 dpi, and no virus was detectable at later time points (Fig. 3D). We also looked for virus in other extraintestinal sites, including the heart, lungs, liver, kidneys, and blood, but they were all negative or had extremely low levels (Fig. 3E). IgG antibody levels after 7 days detected by SADS-CoV virion-based enzyme-linked immunosorbent assay (ELISA) showed that the i.p. route could effectively elicit host immune responses (Fig. 3G).
Splenocytes support SADS-CoV replication.
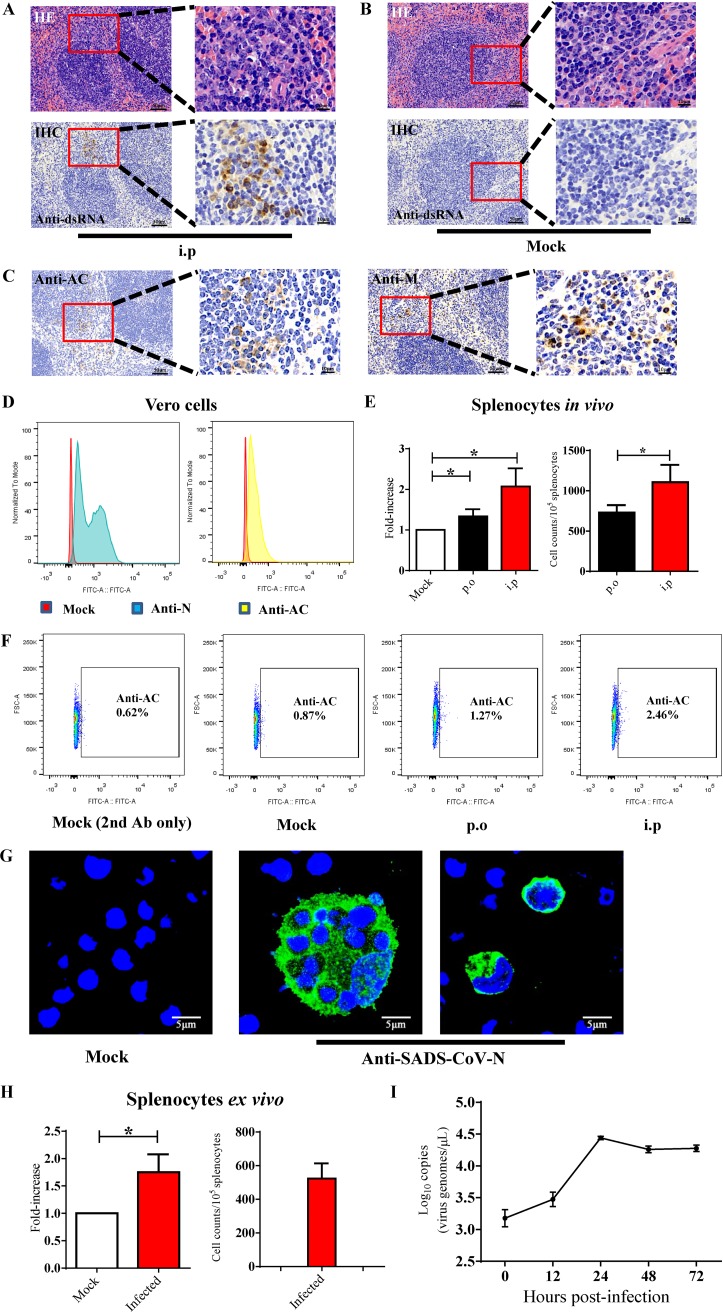
With the mouse infection model described above, our next step was to determine the cell tropism of SADS-CoV in vivo. Thus, we performed immunohistochemistry (IHC) on sections of small and large intestine and spleen from mice infected i.p. with 5 × 105 TCID50 of SADS-CoV at 3 dpi. A monoclonal antibody against double-stranded RNA (dsRNA) was used to identify cells that supported active virus replication, as dsRNA is an intermediate that only exists during intracellular viral replication. SADS-CoV dsRNA signals were observed in the splenic white pulp in the marginal zone on the edge of lymphatic follicles and in the margins of the periarteriolar lymphocyte sheath (Fig. 4A). Staining of tissue sections from mock-infected mice were used as a control (Fig. 4B). In addition to dsRNA, we also used rabbit polyclonal antibodies (pAbs) to detect the expression of the viral structural protein (M) or nonstructural protein (Nsp3-AC). At 3 dpi, anti-M or anti-AC staining was observed in the white pulp around the lymphatic nodules (Fig. 4C), similar to the localization of dsRNA staining (Fig. 4A). Tissue sections from SADS-CoV or mock-infected mice probed with preimmune sera were negative, indicating the specificity of the SADS-CoV antibody. Unfortunately, neither viral proteins (structural or nonstructural) nor dsRNA were detected in the intestine of infected mice, consistent with the detection of only very low levels of viral RNA in these tissues by qRT-PCR (Fig. 3).
FIG 4.
SADS-CoV replication in mouse splenocytes. Hematoxylin and eosin staining (HE) and immunohistochemistry (IHC) were performed on sections of spleen from intraperitoneally infected mice (A) and Mock-infected mice at 3 dpi (B) using anti-dsRNA antibodies to identify splenic cells that support active virus replication. (C) SADS-CoV infection could also be detected by IHC using SADS-CoV nonstructural protein antibody (anti-AC) and structural protein antibody (anti-M). Scale bars = 50 μm, except for magnified fields shown on the right side, with scale bars = 10 μm. (D) SADS-CoV antibody validation in Vero cells for developing the flow cytometry assay. Flow cytometry plots of Vero cells infected with SADS-CoV (MOI, 0.1) at 24 hpi; staining with anti-N or anti-AC. (E) Flow cytometry detection of Nsp3-AC antigens of SADS-CoV in splenocytes from infected mice using anti-AC antibody at 3 dpi. The data are presented as the fold increase in staining splenocytes from infected mice relative to the mock-infected group for statistical purposes (left); *, P < 0.05. (F) Representative FACS plots of (E). The solid-line frame-gated anti-AC positive splenocytes from p.o. or i.p. inoculated mice. The plot of mock-infected cells stained only with secondary antibody was also shown. (G) Isolated mouse splenocytes were infected with SADS-CoV at an MOI of 1, and SADS-CoV N protein expression was detected by IFA with anti-N antibody. (H) Flow cytometry detection of nonstructural protein antigens of SADS-CoV in infected splenocytes at 48 hpi using anti-AC antibody. The data are presented as the fold increase in staining cells relative to the mock-infected cells for statistical purposes (left); *, P < 0.05. (I) Growth of SADS-CoV in ex vivo splenocytes was monitored over 72 hpi by qRT-PCR targeting the SADS-CoV N gene.
Next, SADS-CoV infection was quantified in the spleen using flow cytometry. We inoculated B6 wild-type mice with 5 × 105 TCID50 of virus either i.p. or p.o. and extracted the bulk immune cells from the spleen of infected animals at 3 dpi. The flow cytometry method was first validated in Vero cells infected with SADS-CoV at a multiplicity of infection (MOI) of 0.01, followed by staining with a pAb against the N or AC protein at 24 hpi (Fig. 4D). As the anti-AC pAb exhibited optimal intracellular staining for viral signals (Fig. 4D), it was used to determine the percentage of infected splenocytes. There were approximately 1.5- and 2.5-fold increases of total splenocytes positive for virus replication after p.o. and i.p. inoculation, respectively (Fig. 4E, left; Fig. 4F), with a significant increase in the total number of AC-positive splenocytes in i.p.-infected mice compared with that of p.o. (Fig. 4E, right). These data are consistent with the significantly lower viral loads in the spleen at 1 and 3 dpi in p.o.-inoculated mice (Fig. 3D), suggesting better virus dissemination and replication and escape from mucosal immune clearance.
We then evaluated the growth characteristics of SADS-CoV in splenocytes by assessing antigen production and replication kinetics ex vitro. Splenocytes were first extracted from naive mice, plated in 100-mm dishes, and infected with 1 × 105 TCID50 of SADS-CoV. We observed clusters of infected cells that appeared to have been engulfed by phagocytes (Fig. 4G, middle), and the structural N protein was shown in the cytoplasm of infected cells by confocal microscopy (Fig. 4G, middle and right). The percentage of infected cells was quantified by flow cytometry using anti-AC pAb, revealing a nearly 2-fold increase in the splenocytes positive for viral signals (Fig. 4H), very similar to the percentage of infection observed in vivo. To further characterize the growth kinetic of SADS-CoV in primary splenocytes, cells were infected with 1 × 105 TCID50 of SADS-CoV, and culture supernatants were harvested at 0, 12, 24, 48, and 72 hpi. Active viral replication was confirmed, with a 1.5-log time-dependent increase in genomic RNA equivalents, plateauing from 24 to 72 hpi (Fig. 4I). These data suggest that although only ∼2% of splenocytes were infected, and these cells supported a decent level of viral replication. Together, these results indicate that SADS-CoV productively infects mouse splenocytes.
Splenic DCs support SADS-CoV replication.
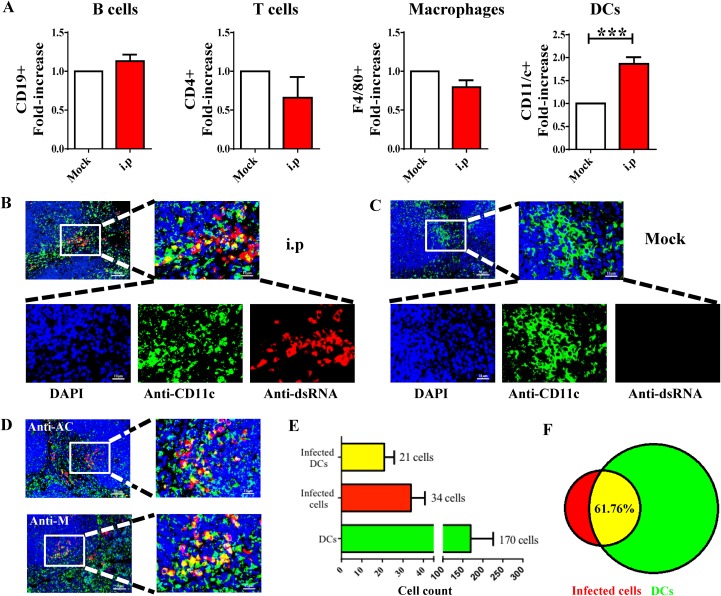
Splenocytes were harvested from i.p.-infected mice at 3 dpi, and the extracted cells were costained with antibodies against SADS-CoV-AC and each of four cell surface markers (anti-CD19 for B cells, anti-CD4 for T cells, anti-CD11/c+ for DCs, and anti-F4/80+ for macrophages) using flow cytometry (Fig. 5A). The percentage of infected CD11/c+ cells was significantly higher than the other cell subgroups, indicating that DCs are the major targets of SADS-CoV infection in the spleen.
FIG 5.
SADS-CoV infection of dendritic cells in the spleen of mice inoculated via i.p. route. (A) Splenocytes were extracted from infected mice at 3 dpi, and flow cytometry was used to detect nonstructural antigen AC of SADS-CoV with immune cell markers on splenocytes, including B cells (CD19+), T cells (CD4+), macrophages (F4/80+), and dendritic cells (DCs; CD11c+). The data are presented as the fold increase in positive staining cells from infected mice relative to the mock-infected cells for statistical purposes; ***, P < 0.001. (B) Immunofluorescence assay of SADS-CoV dsRNA and DC marker CD11c in sections of spleen from intraperitoneally (i.p.) infected mice (B) and mock-infected mice (C) at 3 dpi. (D) SADS-CoV infection could also be detected by IFA using anti-AC and anti-M antibodies. The numbers of SADS-CoV-positive DCs in i.p.-infected mice were counted and averaged from 10 to 15 different visual fields (E), and the proportion of DCs in infected cells was presented with a Venn diagram (F). Scale bars = 50 μm, except for magnified fields shown on the right side, with scale bars = 10 μ m.
The phenotype was further confirmed by double-staining IFA with anti-dsRNA, anti-M, or anti-AC antibody plus anti-CD11/c+ in splenic sections. As expected, dsRNA staining overlapped with the CD11/c surface marker on the edges of lymphatic follicles (Fig. 5B), whereas no viral signals were seen in the mock-infected control (Fig. 5C). Similar patterns of costaining were detected by M and AC antibodies (Fig. 5D). To gain insight into the relative quantity of DCs compared with other undefined target cells, cells positive for dsRNA and CD11/c were counted in 10 to 15 different microscope fields of spleens from 3 infected mice (Fig. 5E), showing that 61.76% of SADS-CoV-infected cells were DCs (Fig. 5F).
SADS-CoV does not utilize known CoV protein receptors for cellular entry.
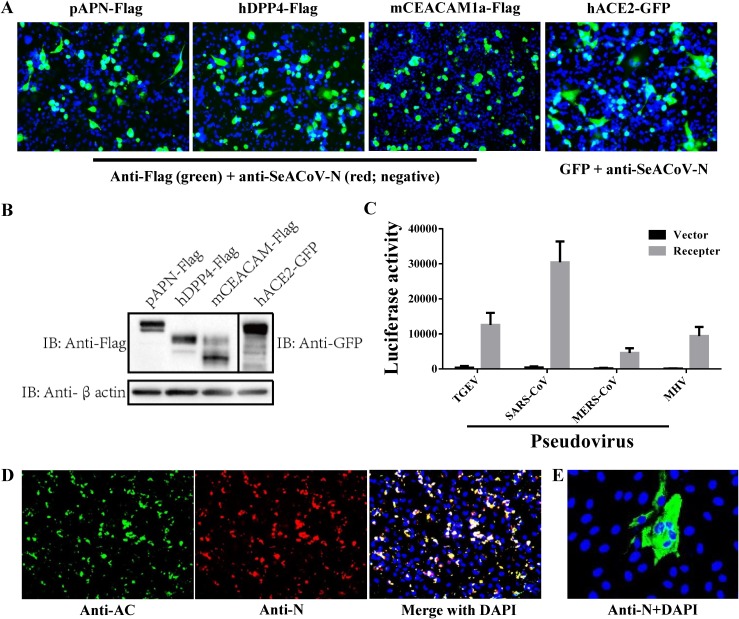
To our knowledge, these results reveal the most extensive cell tropism among known CoVs, suggesting the functional receptor(s) for SADS-CoV is likely to be a very common molecule. In order to test this hypothesis, it was first necessary to find a cell line that was refractory to infection only at the internalization step. MDCK cells, which showed undetectable virus production in early infection tests (Fig. 1 and 2), were chosen as a potential candidate. There are four known types of functional CoV protein receptors, including angiotensin converting enzyme 2 (ACE2) for SARS-CoV (18), dipeptidyl peptidase 4 (DPP4) for MERS-CoV (19), aminopeptidase N (APN) for TGEV (20) and PDCoV (21, 22), and mouse carcinoembryonic antigen-related cell adhesion molecule 1a (mCEACAM1a) for mouse hepatitis virus (MHV) (23). To test whether one of these molecules serves as the SADS-CoV receptor, we attempted to inoculate nonsusceptible MDCK cells overexpressing porcine APN, human DPP4, mouse CEACAM1a, or human ACE2 with SADS-CoV, but none of them allowed infection, as staining with anti-SADS-CoV-N pAb was negative (Fig. 6A). Meanwhile, the expression of each receptor in MDCK cells was confirmed by IFA (Fig. 6A) and Western blot analysis (Fig. 6B) using antibodies against the tags fused to the receptors. We confirmed that, as positive controls, lentiviruses pseudotyped with TGEV, SARS-CoV, MERS-CoV, or MHV spike (i.e., pseudoviruses) efficiently entered MDCK cells exogenously expressing the respective receptors (Fig. 6C).
FIG 6.
SADS-CoV utilizes an unknown receptor for cellular entry. (A) MDCK cells overexpressing each of the four known CoV receptors fused with detectable tags (pAPN-Flag, hDPP4-Flag, mCEACAM1a-Flag, or hACE2-GFP) did not confer SADS-CoV infection at 24 h posttransfection of the expression plasmids. At 48 h, SADS-CoV-inoculated cells transfected with pAPN-Flag, hDPP4-Flag, or mCEACAM1a-Flag were costained with a mouse anti-FLAG MAb and a rabbit anti-SADS-CoV-N pAb. Alexa Fluor 488-conjugated anti-mouse IgG and Alexa Fluor 594-conjugated anti-rabbit IgG were costained for secondary antibody detection, followed by DAPI incubation. For challenged cells transfected with hACE2-GFP, anti-SADS-CoV-N pAb and Alexa Fluor 594-conjugated anti-rabbit IgG were used; magnification = 200×. (B) Western blot analysis also confirmed the expression of CoV receptors in transfected MDCK cells. (C) TGEV-, SARS-CoV-, MERS-CoV-, or MHV-spike-mediated pseudovirus entry into MDCK cells overexpressing the corresponding receptor. The pseudovirus entry efficiency was characterized as luciferase activity accompanying the entry. Cells transfected with the empty backbone vector were used as controls. (D) Rescue of SADS-CoV in MDCK cells transfected with a SeACoV infectious cDNA clone. Detection of expression of Nsp3-AC and N proteins of SADS-CoV was conducted at 72 h posttransfection by costaining with a rabbit anti-AC pAb and a mouse anti-N pAb (magnification = 200×). (E) Infection of fresh Vero cells with progeny SADS-CoV rescued in MDCK cells. The expression of SADS-CoV N protein was detected by staining with anti-N pAb at 36 hpi.
Next, we demonstrated that MDCK cells can confer SADS-CoV replication competency by transfection of a SADS-CoV/SeACoV infectious cDNA clone established recently (24), as simultaneous expression of Nsp3-AC and N proteins were clearly detected by IFA (Fig. 6D). Moreover, passaging of supernatants from pSEA-transfected MDCK cells onto fresh Vero cells resulted in progeny SADS-CoV infection, as evidenced by expression of the N protein (Fig. 6E), indicating that MDCK cells can also support infectious SADS-CoV production without cell-to-cell spread. Therefore, SADS-CoV apparently does not utilize any of the known CoV receptors for cellular entry. The same conclusion was reached using HeLa cells overexpressing each of the four classical CoV receptors followed by SADS-CoV inoculation by Zhou et al. (14); however, the HeLa cell line itself was most susceptible to SADS-CoV infection in the present study (Fig. 2D).
DISCUSSION
In order to assess the potential species barriers of SADS-CoV infection, a cell line susceptibility study was first conducted using 24 different cell lines. As SADS-CoV probably originated from a bat SADSr-CoV (14) derived from HKU2-CoV identified in Rhinolophus sinicus (Chinese horseshoe bats) (12), we commenced testing viral susceptibility in two available bat cell lines, namely, BFK from Myotis daubentonii (25) and Tb-1 from Tadarida brasiliensis. Although BFK cells did not support SADS-CoV replication, it replicated efficiently in Tb-1 cells (Fig. 1A and Fig. 2), suggesting that other bat species in addition to horseshoe bats are likely susceptible to SADS-CoV infection.
Interestingly, SADS-CoV protein expression was detected in almost all of the rodent cells (hamster, gerbil, mouse, and rat) including BHK-21, which is not susceptible to other known human CoVs, such as SARS-CoV and MERS-CoV (26, 27), as well as three swine enteric CoVs, namely, PEDV, PDCoV, and TGEV (21). Given the fact that SADS-CoV infects both primary and passaged or primary cell lines originating from rodents, we hypothesized that rodents may be susceptible to SADS-CoV infection. To explore this possibility, we challenged wild-type B6 mice with SADS-CoV by two different inoculation routes.
The challenged animals neither succumbed to infection nor manifested any signs of gastroenteritis. In fact, experimental infection of neonatal piglets with a higher dose of purified SADS-CoV in our laboratory resulted only in mild diarrheal signs or subclinical infection (11). Also, there was a lack of robust viral replication in the intestines during infection, and no tissue damage was detected throughout the intestines (Fig. 3B and C), reflecting the suboptimal infection by SADS-CoV in immunocompetent wild-type mice. On the contrary, the virus had more efficient replication within the spleen, which was reflected by a continuous detection of viral genomic RNA in the immune cells at all time points over a 28-day period (Fig. 3D). The phenotype was also consistent with the replication kinetics in extracted splenocytes in vitro, in which viral genomic RNA peaked and plateaued at 72 hpi (Fig. 4G and I). These data collectively led to speculation that SADS-CoV favors splenic cells over other tissues. The most logical explanation for these tissue-specific discrepancies in virus replication is (i) target cells are more concentrated in the spleen and more sporadic in the intestine or (ii) splenic immune cells have enhanced expression of the unknown receptor(s) over intestinal cells. The animals were more susceptible to i.p. infection, resulting in higher virus replication in the distal section of the small intestine, large intestine, and spleen, and perhaps a delayed clearance of viral infection in the cecum (Fig. 3B to E), suggesting the important role of mucosal immunity for controlling early infection in SADS-CoV in mice. It should be noted that mice (C57BL/6J mice in this study) may not be the optimal rodent species for SADS-CoV infection, as wild rats are more commonly seen in Chinese pig farms. In addition, other transmission routes may be considered. Recently, PDCoV has been shown to possibly spread via the respiratory route in addition to fecal-oral transmission (28). Therefore, it will be interesting to try intranasal route for the inoculation in rats or the other rodent species to mimic SADS-CoV natural transmission in future studies.
More interestingly, we identified DCs to be the precise cell population that supported SADS-CoV replication (Fig. 5). There have been a few reports of immune cell tropism for CoVs. Macrophages are susceptible to MHV infection, representing the largest group of innate immune cells that infiltrate the central nervous system after infection with neurotropic MHV strains (29). In addition, based on the fact that SARS-CoV spike-pseudotyped HIV-based vectors can efficiently transduce human DCs, Kobinger et al. hypothesized that SARS-CoV infection in immature DCs contributes to viral pathogenesis (30). Yang et al. demonstrated that SARS-CoV can infect myeloid DCs via S glycoprotein-associated cell entry and DC infection-mediated viral transmission to other cells in vivo (31). These previous evidences support our present results, showing that SADS-CoV can efficiently replicate in DCs.
Furthermore, this study gives us a novel inspiration that rodents may potentially serve as susceptible hosts for SADS-CoV in addition to bats and pigs. Of note, the species Rhinolophus bat α-CoV HKU2, including SADS-CoV, possesses unique S genes closely related to the betacoronavirus (β-CoV), in a manner similar to some globally distributed rodent α-CoVs (11, 32, 33), implying an unknown evolutionary connection between the bat α-CoV HKU2 and rodent α-CoVs. Under the field conditions of China, direct contact between pigs and flying bats is a low probability; however, rodents (especially rats) are frequently visible in the swine industry, causing great nuisance due to feed loss. It is possible that as bats prey on insects near pig facilities, they leave feces containing HKU2-like CoVs that contaminate pig feed, which is then eaten by pig and rodents that subsequently become carriers of SADS-CoV. Rats and mice are increasingly implicated as external vectors for a wide range of different pig pathogens, such as Lawsonia intracellularis (34). Rodents not only spread pathogens but also harm the practitioners of the swine industry, as they are thought to be the major source of leptospirosis in pigs and piggery workers (35). Future studies on identifying SADS-CoV-positive samples in rodents near pig farms are warranted to test this hypothesis.
In addition to rodents, we also measured the SADS-CoV susceptibility of cell lines from humans, monkeys, chickens, and dogs, revealing a remarkably broad spectrum of tropism (Table 1 and Fig. 1). As for the ability of SADS-CoV to grow efficiently in human cell lines, we should not underestimate the risk that this bat-origin CoV may “jump” from pigs to humans. It is noteworthy that camel workers with high rates of exposure to camel nasal and oral secretions had evidence of MERS-CoV infection (36). Considering that SARS-CoV and MERS-CoV originated from bats and spread from one species to another through intermediate hosts (civets and camels, respectively), SADS-CoV may pose a similar risk to human health through transmission from pigs or other susceptible hosts.
The cell susceptibility study and testing of the overexpression of four known CoV receptors in nonsusceptible MDCK cells (Fig. 6) demonstrated that SADS-CoV might use a new receptor molecule that is conserved in bats, pigs, rodents, chickens, monkeys, and humans, indicating a low barrier to cross-species transmission. This is in line with the unusual feature of broad species tropism of SADS-CoV.
In summary, these results provide important insights into the ecology of this bat-origin CoV, highlighting the possibility of its ability to jump interspecies barriers and the potential role of rodents as susceptible hosts in the field. Identification of the unknown SADS-CoV cellular receptor and further surveillance of other animal populations are needed to fully understand the biology of SADS-CoV.
MATERIALS AND METHODS
Virus stocks and viral antibodies.
The SADS-CoV isolate CH/GD-01/2017 at passage 10 was used in all experiments and cultured in Vero cells (24). The virus was passaged serially using the culture supernatant to infect fresh Vero cells at a multiplicity of infection (MOI) of 0.1, and viral titers were determined in Vero cells by endpoint dilution as the 50% tissue culture infective dose (TCID50). Rabbit polyclonal antibodies (pAb) against the membrane (M), nucleocapsid (N), and the nonstructural protein 3 (Nsp3) acidic domain (AC) of SADS-CoV were generated in-house and validated in SADS-CoV-infected Vero cells (24). A mouse anti-SADS-CoV-N pAb was also produced to allow double staining when mixed with the rabbit pAb. A monoclonal antibody (MAb) against dsRNA (anti-dsRNA MAb J2; catalog number J2-1702, Scicons, Hungary) was used to specifically detect viral replication of SADS-CoV.
Cell lines and cell culture.
Twenty-four cell lines derived from tissues of different species were used (Table 1), including human (Huh-7, HepG2/C3A, 293T, A549, and HeLa), monkey (Marc-145, Cos-7, BSC-40, and Vero), swine (ST, PK15, LLC-PK1, and IPEC-J2 [37]), bat (BFK [25] and Tb-1), canine (MDCK), mouse (NIH/3T3 and RAW 264.7), hamster (BHK-21 and CHO), rat (BRL-3A and NRK-52E), and chicken (DF-1) cell lines and a primary kidney cell line from Mongolian gerbils (prepared in-house). The BFK cell line was a generous gift from Changchun Tu at the Institute of Military Veterinary Medicine, Changchun, China. Each cell line was cultured in Dulbecco’s modified Eagle’s medium (DMEM; HyClone) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Biological Industries), 100 U/ml penicillin, and 100 U/ml streptomycin under 37°C, 5% CO2, and water-saturated humidity conditions.
To determine viral susceptibility, each cell line was cultured at 70% confluence in 12-well plates with maintenance medium (MM) containing DMEM, 0.3% tryptose phosphate broth (TPB), and 1% penicillin-streptomycin or MM with addition of 5 μg/ml trypsin (MMT) (catalog number T7186-50TAB; Sigma, St. Louis, MO, USA). After cells were washed with phosphate-buffered saline (PBS), they were inoculated with SADS-CoV diluted in MM or MMT at an MOI of 0.01 for 2 h. Nonattached viruses were removed by washing the cells three times with DMEM, and cell monolayers were subsequently incubated in MM or MMT at 37°C for 5 days. To determine the effect of trypsin on viral entry, cell monolayers were infected by SADS-CoV in the following three conditions: (i) no trypsin treatment, infected with SADS-CoV diluted in MM, subsequently incubated in MM; (ii) pretrypsin treatment, inoculated with SADS-CoV diluted in MMT, subsequently incubated in MM; and (iii) double-trypsin treatment, inoculated with SADS-CoV in MMT, subsequently incubated in MMT. Supernatants from cells were collected at 12, 24, 36, 48, 72, and 120 hours postinfection (hpi) for one-step quantitative RT-PCR analysis. Cell cultures were examined for cytopathic effects (CPEs) and immunofluorescence assay at 48 to 72 hpi.
IFA for cell line susceptibility.
Different cells infected with SADS-CoV in 12-well plates were washed twice with PBS and fixed in 4% paraformaldehyde in PBS and then permeabilized with 0.1% Triton X-100 in PBS. Cells were then incubated with the rabbit anti-SADS-CoV-M pAb at 1:5,000 dilution for 1 h at 37°C, washed with PBS, and stained with the Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (Thermo Fisher Scientific, USA) at 1:1,000 dilution. After incubation for 1 h at 37°C, the cells were washed with PBS, stained with 4ʹ,6-diamidino-2-phenylindole (DAPI) at 1:1,000 dilution and visualized on a fluorescence microscope.
One-step quantitative RT-PCR analysis targeting the N gene.
The full-length SADS-CoV N gene was inserted into an appropriately digested pET-28a vector using two unique restriction sites, namely, NdeI and XhoI, and then linearized with XhoI. The N gene was in vitro transcribed using the T7 high-efficiency transcription kit (TransGen Biotech Co., Ltd., Beijing China). Standard curves were generated using dilutions of a known quantity of the N gene RNA to allow absolute quantitation of SADS-CoV RNA copy numbers in samples.
Total RNA was extracted from culture supernatants or tissue homogenates using Trizol (ThermoFisher Scientific, USA) following the manufacturer’s instructions. SADS-CoV RNA titer was determined by one-step qRT-PCR (Toyobo Co., Ltd.) targeting the N gene with the primers 5ʹ-CTAAAACTAGCCCCACAGGTC-3′ and 5ʹ-TGATTGCGAGAACGAGACTG-3′ and the probe 6-carboxyfluorescein (FAM)-GAAACCCAAACTGAGGTGTAGCAGG-6-carboxytetramethylrhodamine (TAMRA). Samples with a cycle threshold value of <35 were considered positive based upon validation data using the RNA standards.
Mouse infections, tissue harvest, and viral load determination.
Wild-type C57BL/6J mice (catalog number 000664; Jackson Laboratory) were purchased from the Model Animal Research Center of Nanjing University and housed in animal facilities at the Zhejiang University under specific-pathogen-free conditions. For SADS-CoV infections, 6- to 8-week-old female and male mice were inoculated with 5 × 105 TCID50 (equal to 6 × 108 genome copies) of SADS-CoV, either per oral (p.o.) infection with 25-μl inoculum (2 × 107 TCID50/ml) or intraperitoneal (i.p.) infection with 200-μl inoculum (2.5 × 106 TCID50/ml). For viral load determination in specific tissues, mice were euthanized at 1, 3, 5, 7, 14, 21, and 28 days postinfection (dpi), and tissues were harvested, including stomach, duodenum, jejunum, ileum, cecum, colon, mesenteric lymph nodes, spleen, kidney, liver, heart, lung, blood, and feces. Tissues were weighed and homogenized in medium (DMEM contained 2% FBS) by bead beating using sterile zirconium oxide beads (catalog number ZrOB20; MidSci). Total RNA was extracted from tissue homogenates and tested by quantitative RT-PCR analysis targeting the SADS-CoV N gene, as described above. Blood samples were collected from the heart, and serum was separated for virus-specific antibody detection.
Enzyme-linked immunosorbent assay.
SADS-CoV virus particles were purified from infected cell culture supernatants by sucrose density gradient centrifugation, and protein concentration was determined by the bicinchoninic acid (BCA) protein assay kit (Beyotime Biotechnology, Shanghai, China). The optimal dilution of antigen was determined by square titration. The IgG antibodies contained in serum at a 1:100 dilution were detected in wells coated with purified SADS-CoV virus particles (6.25 ng/well) as antigen.
Histopathology, immunohistochemistry, and immunofluorescence assay for murine spleen.
Mice were infected i.p. with SADS-CoV, and at 3 dpi, spleens were harvested and fixed in 4% paraformaldehyde for 24 h and embedded in paraffin. Tissue sections were then deparaffinized and rehydrated in three changes of xylene, 15 min each, dehydrated in two changes of pure ethanol for 5 min, followed by rehydration in an ethanol gradient of 85% and 75% ethanol. After tissues were washed in distilled water, they were subjected to hematoxylin and eosin staining for histopathological examinations.
For antigen retrieval, deparaffinized and rehydrated sections were immersed in sodium citrate antigen retrieval solution (pH 6.0) and maintained at a subboiling temperature for 8 min, were left at 98°C for 8 min, and then incubated again at subboiling temperature for 7 min. After the sections were allowed to cool to room temperature (RT) and were washed three times with PBS (pH 7.4), endogenous peroxidase was blocked by immersion in 3% hydrogen peroxide at RT for 30 min, and sections were again washed with PBS. Tissue sections were blocked in 3% bovine serum albumin (BSA) at RT for 30 min and then incubated with a 1:500 dilution of each primary antibody (anti-dsRNA MAb, anti-SADS-CoV-M pAb, or anti-SADS-CoV-AC pAb) overnight at 4°C. After slides were washed three times with PBS (pH 7.4), they were stained with appropriate secondary antibodies labeled with horseradish peroxidase at RT for 50 min. Freshly prepared diaminobenzidine chromogenic reagent was added and counterstained with hematoxylin and then dehydrated and visualized on a light microscope.
Spontaneous fluorescence quenching reagent (Wuhan Servicebio Technology Co., Ltd., Wuhan, China) was added to the tissue sections and incubated for 5 min after antigen retrieval. The sections were then washed in running water, followed with blocking and antibody staining as described above. In addition, the primary antibody was supplement with a CD11c antibody (Wuhan Servicebio Technology) at a 1:200 dilution and then stained with appropriate secondary antibodies. Finally, DAPI was added, and sections were visualized on a fluorescence microscope; nuclei labeled with DAPI appear blue, and positive cells are green by labeling with CD11/c or red by labeling with a virus-specific antibody.
Preparation of murine splenocytes and flow cytometry.
Mice infected with SADS-CoV were euthanized at 3 dpi, and spleens were removed and placed in 5 ml complete DMEM. After the excised spleen was ground through a 100-μm cell strainer using the plunger end of a 5-ml syringe, cells were washed with an excess of DMEM and centrifuged at 200 × g for 5 min. After cells were resuspended in 3 ml of red blood cell lysis buffer (Solarbio Life Sciences, Beijing, China) and incubated at RT for 10 min, 5 ml of DMEM was added, and cells were passed through another strainer to remove clumps. After centrifugation at 200 × g for 5 min, the supernatant was discarded, and cells were resuspended in 10 ml fresh DMEM for cell counting and viability checks using trypan blue and a hemocytometer.
For flow cytometry, cultured cells were resuspended in Fc Block buffer (containing anti-mouse CD16 Fc Block antibody at 1:500 dilution) and incubated on ice. Cells in Fc Block buffer were added to 96-well plates at 1 × 106 cells/well. After a 30-min incubation, cells were centrifuged at 200 × g for 10 min, the supernatant was discarded, and pellets were resuspended in a 100-μl Cytofix/Cytoperm solution (Cytofix/Cytoperm soln kit; BD Biosciences, San Jose, CA, USA) to fix cells. After incubation on ice for 20 min protected from light, cells were centrifuged at 800 × g for 5 min at 4°C, supernatant was removed without disturbing cell pellets, and cells were washed twice in 150 μl of 1× Perm/Wash buffer. After the addition of 50-μl virus-specific primary antibody (anti-dsRNA MAb, anti-SADS-CoV-N pAb, or anti-SADS-CoV-AC pAb) diluted in 1× Perm/Wash buffer with 3% BSA and incubation at 4°C for 30 min, cells were centrifuged at 200 × g for 10 min. Cells were washed twice in 150 μl of 1× Perm/Wash buffer followed by staining with appropriate secondary antibodies conjugated to Alexa Fluor 488 (Thermo Fisher Scientific, USA) at 4°C for 30 min. After pellets were centrifuged at 800 × g for 5 min at 4°C and washed with 1× Perm/Wash buffer, they were resuspended in 0.2 ml fluorescence-activated cell sorter (FACS) buffer and analyzed by flow cytometry.
Infection of splenocytes in vitro.
To detect replication of SADS-CoV in mouse splenic cells in vitro, splenocytes were extracted from naive mice, plated in 100- or 35-mm dishes, and infected with SADS-CoV at an MOI of 0.1. At 48 hpi, splenocytes were harvested and placed in a 15-ml tube, centrifuged at 200 × g for 10 min at 4°C, and analyzed by flow cytometry as described above. Infected mouse splenic cells in 35-mm dishes were detected by immunofluorescence assay with anti-SADS-CoV-N antibodies, and infection supernatants were collected at 0, 12, 24, 36, 48, and 72 hpi for one-step quantitative RT-PCR analysis.
FACS analysis of splenocytes with cell marker staining.
Mice infected with SADS-CoV were euthanized at 3 dpi, and splenocytes were prepared for flow cytometry by staining with the following appropriate antibodies: anti-SADS-CoV-AC following secondary antibodies conjugated to Alexa Fluor 647 (Thermo Fisher Scientific, USA), anti-CD19-FITC (catalog number 4318813; eBioscience) for B cells, anti-CD4-PE (catalog number 4329629; eBioscience) for T cells, anti-CD11/c-PE-Cy7 (catalog number 561022; BD Bioscience) for DCs, and anti-F4/80-PE/Cy5 (catalog number 123111; Biolegend) for macrophages. Stained cells were resuspended in 0.2 ml FACS buffer and analyzed by flow cytometry.
Production and transduction of S protein-pseudotyped lentiviruses.
Pseudovirions with various CoV spike proteins were produced as described previously (38). Briefly, each of the plasmids encoding TGEV, SARS-CoV, MERS-CoV, and mouse hepatitis virus (MHV) S proteins were cotransfected into 293T cells with pLenti-Luc-green fluorescent protein (GFP) and psPAX2 plasmids (kindly provided by Zhaohui Qian, Chinese Academy of Medical Sciences & Peking Union Medical College) at a molar ratio of 1:1:1 by using polyethylenimine (PEI). The cells were fed with fresh medium in the next 24 h, and the supernatant media containing pseudovirions were then collected and centrifuged at 800 × g for 5 min to remove debris. To quantify S protein-mediated entry of pseudovirions, MDCK cells were seeded at about 25% to 30% confluence in 24-well plates and transfected with either pAPN-Flag, hDPP4-Flag, mCEACAM1a-Flag, hACE2-GFP (kindly provided by Zhaohui Qian) (38), or the control backbone vector by using Lipofectamine 3000 (Thermo Fisher). The MDCK cells overexpressing each receptor were inoculated with 500 μl of 1:1 diluted corresponding pseudovirions at 24 h posttransfection. At 40 hpi, cells were lysed at room temperature with 110 μl of medium with an equal volume of Steady-Glo (Promega, Madison, WI). The cell lysates were also used to confirm the expression of each receptor by using Western blotting. Transduction efficiency was monitored by quantitation of luciferase activity using a Modulus II microplate reader (Turner Biosystems, Sunnyvale, CA). On the other hand, the MDCK cells overexpressing each receptor were inoculated with SADS-CoV (MOI, 1) at 24 h posttransfection. IFA was performed to test for SADS-CoV susceptibility using anti-N pAb. The replication competency of SADS-CoV in MDCK cells was further determined by a reverse genetics system. Development of a DNA-launched SADS-CoV (SeACoV) infectious cDNA clone (named pSEA) and rescue of SADS-CoV by transfection of cultured cells with pSEA followed by passaging on Vero cells have been described recently by our lab (24).
Ethics statement.
All animal experiments were performed in strict accordance with the Experimental Animal Ethics Committee of Zhejiang University (approval number ZJU20170026).
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2016YFD0500102), the National Natural Science Foundation of China (31872488), and the Fundamental Research Funds for the Central Universities of China (2019FZA6014).
The professional editing service NB Revisions was used for technical preparation of the text prior to submission.
REFERENCES
- 1.Graham RL, Baric RS. 2010. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol 84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3:e00473-12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soma T, Saito N, Kawaguchi M, Sasai K. 2018. Feline coronavirus antibody titer in cerebrospinal fluid from cats with neurological signs. J Vet Med Sci 80:59–62. doi: 10.1292/jvms.17-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang YW, Dickerman AW, Pineyro P, Li L, Fang L, Kiehne R, Opriessnig T, Meng XJ. 2013. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 4:e00737-13. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu G, Wang Q, Gao GF. 2015. Bat-to-human: spike features determining “host jump” of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol 23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group . 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 7.Drexler JF, Corman VM, Drosten C. 2014. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res 101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan JFW, Lau SKP, To KKW, Cheng VCC, Woo PCY, Yuen K-Y. 2015. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev 28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin Y, Wunderink RG. 2018. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology 23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hulswit RJG, de Haan CAM, Bosch BJ. 2016. Coronavirus spike protein and tropism changes. Adv Virus Res 96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan Y, Tian X, Qin P, Wang B, Zhao P, Yang YL, Wang L, Wang D, Song Y, Zhang X, Huang YW. 2017. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet Microbiol 211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau SKP, Woo PCY, Li KSM, Huang Y, Wang M, Lam CSF, Xu H, Guo R, Chan K-H, Zheng B-J, Yuen K-Y. 2007. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology 367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong L, Li J, Zhou Q, Xu Z, Chen L, Zhang Y, Xue C, Wen Z, Cao Y. 2017. A new bat-HKU2-like coronavirus in swine, China, 2017. Emerg Infect Dis 23:1607–1609. doi: 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou P, Fan H, Lan T, Yang X-L, Shi W-F, Zhang W, Zhu Y, Zhang Y-W, Xie Q-M, Mani S, Zheng X-S, Li B, Li J-M, Guo H, Pei G-Q, An X-P, Chen J-W, Zhou L, Mai K-J, Wu Z-X, Li D, Anderson DE, Zhang L-B, Li S-Y, Mi ZQ, He T-T, Cong F, Guo P-J, Huang R, Luo Y, Liu X-L, Chen J, Huang Y, Sun Q, Zhang X-L-L, Wang Y-Y, Xing S-Z, Chen Y-S, Sun Y, Li J, Daszak P, Wang L-F, Shi Z-L, Tong Y-G, Ma J-Y. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Su S, Bi Y, Wong G, Gao GF. 2018. Bat-origin coronaviruses expand their host range to pigs. Trends Microbiol 26:466–470. doi: 10.1016/j.tim.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann M, Wyler R. 1988. Propagation of the virus of porcine epidemic diarrhea in cell culture. J Clin Microbiol 26:2235–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wicht O, Li W, Willems L, Meuleman TJ, Wubbolts RW, van Kuppeveld FJ, Rottier PJM, Bosch BJ. 2014. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J Virol 88:7952–7961. doi: 10.1128/JVI.00297-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delmas B, Gelfi J, L'Haridon R, Vogel LK, Sjöström H, Norén O, Laude H. 1992. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B, Liu Y, Ji C-M, Yang Y-L, Liang Q-Z, Zhao P, Xu L-D, Lei X-M, Luo W-T, Qin P, Zhou J, Huang Y-W. 2018. Porcine deltacoronavirus engages the transmissible gastroenteritis virus functional receptor porcine aminopeptidase N for infectious cellular entry. J Virol 92:e00318-18. doi: 10.1128/JVI.00318-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Hulswit RJG, Kenney SP, Widjaja I, Jung K, Alhamo MA, van Dieren B, van Kuppeveld FJM, Saif LJ, Bosch BJ. 2018. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci U S A 115:E5135–E5143. doi: 10.1073/pnas.1802879115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams RK, Jiang GS, Holmes KV. 1991. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc Natl Acad Sci U S A 88:5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y-L, Liang Q-Z, Xu S-Y, Mazing E, Xu G-H, Peng L, Qin P, Wang B, Huang Y-W. 2019. Characterization of a novel bat-HKU2-like swine enteric alphacoronavirus (SeACoV) infection in cultured cells and development of a SeACoV infectious clone. Virology 536:110–118. doi: 10.1016/j.virol.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He B, Yang F, Yang W, Zhang Y, Feng Y, Zhou J, Xie J, Feng Y, Bao X, Guo H, Li Y, Xia L, Li N, Matthijnssens J, Zhang H, Tu C. 2013. Characterization of a novel G3P[3] rotavirus isolated from a lesser horseshoe bat: a distant relative of feline/canine rotaviruses. J Virol 87:12357–12366. doi: 10.1128/JVI.02013-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan JF-W, Chan K-H, Choi GK, To KK-W, Tse H, Cai J-P, Yeung ML, Cheng VC-C, Chen H, Che X-Y, Lau SK-P, Woo PC-Y, Yuen K-Y. 2013. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J Infect Dis 207:1743–1752. doi: 10.1093/infdis/jit123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller MA, Raj VS, Muth D, Meyer B, Kallies S, Smits SL, Wollny R, Bestebroer TM, Specht S, Suliman T, Zimmermann K, Binger T, Eckerle I, Tschapka M, Zaki AM, Osterhaus ADME, Fouchier RAM, Haagmans BL, Drosten C. 2012. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. mBio 3:e00515-12. doi: 10.1128/mBio.00515-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woo PC, Lau SK, Tsang CC, Lau CC, Wong PC, Chow FW, Fong JY, Yuen KY. 2017. Coronavirus HKU15 in respiratory tract of pigs and first discovery of coronavirus quasispecies in 5′-untranslated region. Emerg Microbes Infect 6:e53. doi: 10.1038/emi.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazaleuskaya L, Veltrop R, Ikpeze N, Martin-Garcia J, Navas-Martin S. 2012. Protective role of Toll-like receptor 3-induced type I interferon in murine coronavirus infection of macrophages. Viruses 4:901–923. doi: 10.3390/v4050901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobinger GP, Limberis MP, Somanathan S, Schumer G, Bell P, Wilson JM. 2007. Human immunodeficiency viral vector pseudotyped with the spike envelope of severe acute respiratory syndrome coronavirus transduces human airway epithelial cells and dendritic cells. Hum Gene Ther 18:413–422. doi: 10.1089/hum.2006.194. [DOI] [PubMed] [Google Scholar]
- 31.Yang Z-Y, Huang Y, Ganesh L, Leung K, Kong W-P, Schwartz O, Subbarao K, Nabel GJ. 2004. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol 78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsoleridis T, Chappell JG, Onianwa O, Marston DA, Fooks AR, Monchatre-Leroy E, Umhang G, Muller MA, Drexler JF, Drosten C, Tarlinton RE, McClure CP, Holmes EC, Ball JK. 2019. Shared common ancestry of rodent alphacoronaviruses sampled globally. Viruses 11:125. doi: 10.3390/v11020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W, Lin XD, Guo WP, Zhou RH, Wang MR, Wang CQ, Ge S, Mei SH, Li MH, Shi M, Holmes EC, Zhang YZ. 2015. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology 474:19–27. doi: 10.1016/j.virol.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins AM, Fell S, Pearson H, Toribio JA. 2011. Colonisation and shedding of Lawsonia intracellularis in experimentally inoculated rodents and in wild rodents on pig farms. Vet Microbiol 150:384–388. doi: 10.1016/j.vetmic.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 35.Everard CO, Ferdinand GA, Butcher LV, Everard JD. 1989. Leptospirosis in piggery workers on Trinidad. J Trop Med Hyg 92:253–258. [PubMed] [Google Scholar]
- 36.Alshukairi AN, Zheng J, Zhao J, Nehdi A, Baharoon SA, Layqah L, Bokhari A, Al Johani SM, Samman N, Boudjelal M, Ten Eyck P, Al-Mozaini MA, Zhao J, Perlman S, Alagaili AN. 2018. High prevalence of MERS-CoV infection in camel workers in Saudi Arabia. mBio 9:e01985-18. doi: 10.1128/mBio.01985-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji C-M, Wang B, Zhou J, Huang Y-W. 2018. Aminopeptidase-N-independent entry of porcine epidemic diarrhea virus into Vero or porcine small intestine epithelial cells. Virology 517:16–23. doi: 10.1016/j.virol.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ou XY, Zheng WL, Shan YW, Mu ZX, Dominguez SR, Holmes KV, Qian ZH. 2016. Identification of the fusion peptide-containing region in betacoronavirus spike glycoproteins. J Virol 90:5586–5600. doi: 10.1128/JVI.00015-16. [DOI] [PMC free article] [PubMed] [Google Scholar]