Abstract
The pre-mRNA splicing factor SF3b1 exhibits recurrent mutations among hematologic malignancies and cancers, and consequently is a major therapeutic target of clinically-advanced spliceosome inhibitors. In this review, we highlight and rigorously analyze emerging views of SF3b1 conformational transitions, including the human SF3b particle either in isolation or bound to spliceosome inhibitors, and human or yeast spliceosome assemblies. Among spliceosome states characterized to date, an SF3b1 α-helical superhelix significantly closes to surround a U2 small nuclear RNA duplex with the pre-mRNA branch point sequence. The SF3b1 torus is locally unwound at an active site adenosine, whereas protein cofactors appear to stabilize overall closure in the spliceosome. Network analyses demonstrates that the natural SF3b1 dynamics mimic its conformational change in the spliceosome, raising the possibility of conformational selection underpinning spliceosome assembly. These dynamic SF3b1 conformations have consequences for gatekeeping of spliceosome assembly and therapeutic targeting of its cancer-associated dysfunction.
Keywords: pre-mRNA splicing, cancer; myelodysplastic syndrome; ribonucleoprotein structure; Hsh155; HEAT repeat
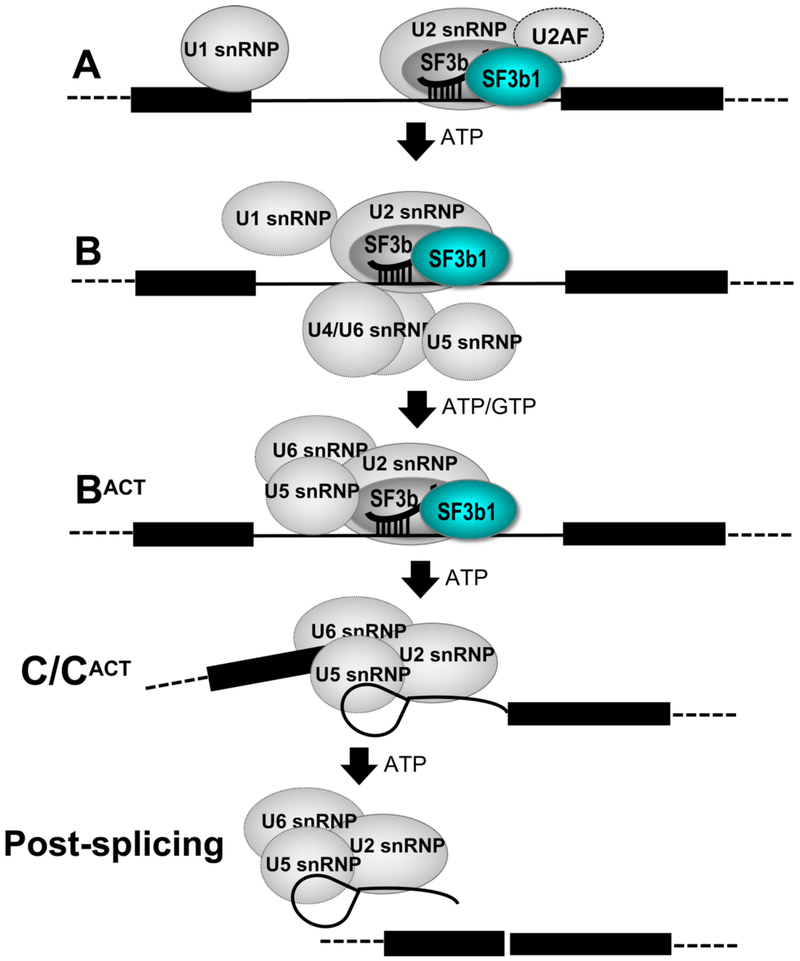
Defects in pre-mRNA splicing and acquired mutations of pre-mRNA splicing factors are an emerging hallmark of cancers and hematologic malignancies [1, 2]. SF3b1 is an outstanding pre-mRNA splicing factor due to its frequent cancer-associated mutations and targeting for drug therapy [3, 4]. In the early stages of spliceosome assembly, SF3b1 orchestrates an ATP-dependent series of structural and compositional rearrangements among small nuclear ribonucleoprotein particles (snRNP) at the pre-mRNA splice sites that ultimately accomplishes the act of pre-mRNA splicing. The SF3b1 subunit is a component of a multi-protein SF3b particle in the U2 snRNP of the major spliceosome [5] and also is found in the U11/U12 snRNP of the minor spliceosome [6]. In the major spliceosome, SF3b participates in the initial A, pre-B, B and BACT stages of pre-mRNA splicing, then dissociates at the C stage, during formation of branched intron lariat (Fig. 1). A key function of SF3b1 in this process is to stabilize a duplex between the spliceosomal U2 snRNA and a consensus branch point sequence (BPS), which subsequently offers the branch point adenosine as a nucleophile for splicing catalysis. This transient role of SF3b1 is driven by ATP-dependent RNA helicases, including DDX46/Prp5 (human/yeast homologues) for entry [7] and Prp2 for exit [8-10] of the SF3b particle from spliceosome assemblies. Several excellent reviews have summarized an emerging structural understanding of spliceosome architecture (e.g. [11-13]). Here, we focus significant conformational changes of the cancer-relevant SF3b1 subunit during integration with the assembling spliceosome.
Figure 1.
Diagram of SF3b1-containing complexes during spliceosome assembly.
1. Structural revelations of the SF3b1 subunit
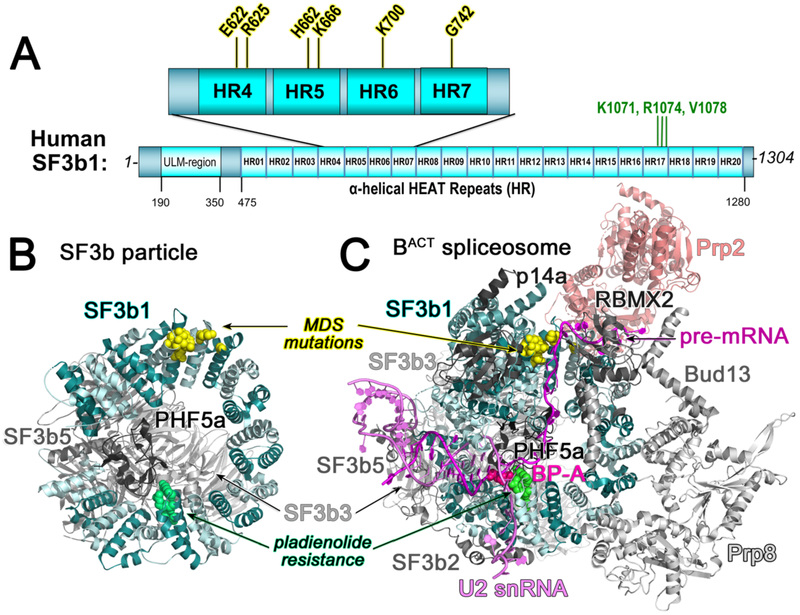
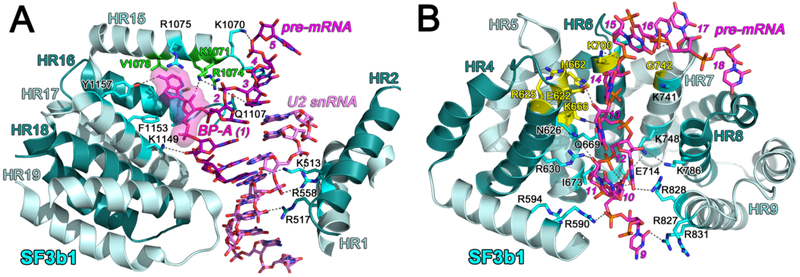
A crystal structure of the human SF3b core first showed that the SF3b1 α-helical repeat (HEAT) domain (Fig. 2A) serves as a C-shaped scaffold for three core SF3b subunits (SF3b3, SF3b5, and PHF5a) (Fig. 2B) [14]. Cryo-electron microscopy structures, initially of the Saccharomyces cerevisiae BACT spliceosome [15, 16] and soon followed by A, pre-B, and BACT human and yeast spliceosomes (Table 1), reveal SF3b1 and its yeast homologue, Hsh155 in the context of the spliceosome (e.g. Fig. 2C). In both the isolated particle and spliceosomes, the center of the SF3b1/Hsh155 torus is capped by the PHF5a subunit and bridged by the SF3b3 β-propeller domains as a heterodimer with SF3b5. As analyzed in detail below, the SF3b1 HEAT domain undergoes large conformational changes following integration in the spliceosome. Yet, among the stages of spliceosome assembly, the SF3b1/Hsh155 HEAT domain conformations and contacts remain similar (e.g. RMSD 0.85 Å between 775 matching Cα atoms of human B-complex, PDB ID 6AHD and BACT-complex, PDB ID 6FF4, Table 1). In the spliceosome, the SF3b1/Hsh155 N- and C-terminal α-helical repeats clamp an RNA duplex comprising the branchsite recognition region (BSRR) of the U2 snRNA and BPS of the pre-mRNA (Fig. 3A). The extrahelical branchsite adenosine packs between the PHF5a subunit and the central SF3b1/Hsh155 repeats, including hydrophobic/aromatic stacking and phosphodiester salt bridges. In contrast with the well-defined SF3b1-bound BPS, structures of isolated U2 – BPS-containing RNA duplexes are dynamic in the absence of protein chaperones [17-19], suggesting that SF3b1 functions to promote the bulged branchsite conformation. Several spliceosome structures (human PDB ID’s 6QX9, 6AH0, 6AHD, 5Z56, 5Z57, 5Z58; yeast PDB ID’s 5ZWM, 5ZWO, 5LQW, 5GM6) report intron sequences downstream of the BPS traversing the mutation-prone HEAT repeats of SF3b1 (or corresponding sites of Hsh155) (Fig. 3B). Structure-guided alanine-scanning mutagenesis has shown that the SF3b1/Hsh155 contacts with this intronic region are important for yeast growth, whereas most BPS – BSRR contacts are dispensable with the exception of a K1149/K818 side chain interaction near the branch point [20].
Figure 2.
(A) Domain organization of SF3b1. Recurrent cancer-associated mutations are expanded above (yellow highlight). Amino acids for which substitutions confer pladienolide-resistance are named (green font). HR, dihelical HEAT repeat; ULM, U2AF Ligand Motif. (B) Structure of the SF3b particle (PDB ID 5IFE). (C) Representative structure of the BACT spliceosome (PDB ID 5Z58 shown to illustrate intron position). SF3b1, cyan; U2 snRNA, violet; pre-mRNA, magenta. Mutational hotspots are yellow spheres; pladienolide-resistance sites are green spheres; branch point adenosine (BP-A) is shown as hot pink spheres.
Table 1.
SF3b1-containing structures
| Stage | Method | Resolution Limit (Å) 1 |
PDB ID |
SF3b1 Chain |
Reference | RMSD (Å) vs. PDB ID |
|---|---|---|---|---|---|---|
| HUMAN | ||||||
| Isolated SF3b | X-ray | 3.1 | 5IFE | C | [14] | 6.0 vs. 6FF4 (BACT) |
| Pre-B | cryoEM | 5.7 | 6AH0 | 1 | [24] | 0.9 vs. 6FF4 |
| B | cryoEM | 3.8 | 6AHD | 1 | [24] | 0.9 vs. 6FF4 |
| B | cryoEM | 4.5 | 5O9Z | V | [64] | 1.9 vs. 6FF4 |
| BACT, early | cryoEM | 4.9 | 5Z58 | 1 | [21] | 0.8 vs. 6FF4 |
| BACT, mature | cryoEM | 5.1 | 5Z56 | 1 | [21] | 0.8 vs. 6FF4 |
| BACT, late | cryoEM | 6.5 | 5Z57 | 1 | [21] | 0.8 vs. 6FF4 |
| BACT | cryoEM | 3.4 | 6FF4 | U | [22] | Baseline |
| Pladienolide B-bound SF3b | X-ray | 3.1 | 6EN4 | C | [57] | 0.2 vs. 5IFE (apo) |
| E7107-bound SF3b | cryoEM | 4.0 | 5ZYA | C | [58] | 1.4 vs. 6EN4 |
| YEAST | ||||||
| A-complex | cryoEM | 4.0 | 6G90 | O | [65] | 1.2 vs. 5GM6 |
| B-complex | cryoEM | 7.2 | 5NRL | O | [66] | 1.2 vs. 5GM6 |
| Pre-B complex | cryoEM | 4.6 | 5ZWM | 1 | [67] | 0.7 vs. 5GM6 |
| B-complex | cryoEM | 3.9 | 5ZWO | 1 | [67] | 0.4 vs. 5GM6 |
| BACT | cryoEM | 5.8 | 5LQW | 1 | [16] | 1.7 vs. 5GM6 |
| BACT | cryoEM | 3.5 | 5GM6 | G | [15] | 1.1 vs. 4FFE (human) |
As reported in citation.
Figure 3.
(A) SF3b1 interactions with the BPS – U2 snRNA duplex. The PHF5a subunit is omitted for clarity and would overlay the branch point adenosine (BP-A, magenta surface) facing the viewer. Sites of pladienolide-resistant mutations are green. (B) SF3b1 interactions with the pre-mRNA intron. PDB ID 5Z58. Mutational hotspots are colored yellow. For reference between panels, the pre-mRNA nucleotides are numbered relative to the BP-A. HR, HEAT repeat.
Additional protein subunits assemble on the SF3b1/Hsh155 scaffold in the context of the spliceosome (Fig. 2C). A few α-helices of the SF3b2/Cus1 subunit consistently bind the exterior of the SF3b1 torus and appear to stabilize SF3a and SF3b4-mediated contacts with the U2 snRNA. In the activated BACT spliceosome [15, 21, 22], massive rearrangements shift a major spliceosomal protein, Prp8 close to SF3b1/Hsh155, where it appears to contact α-helical repeats preceding the branchsite pocket. A Bud13 subunit of the pre-mRNA retention and splicing (RES) bridges Prp8 and the RBMX2/Snu17 RES subunit, which in turn sandwiches the intron RNA against the cancer-affected SF3b1 repeats. In human spliceosomes [21-24], a globular SF3b6 subunit (also called p14a) binds N-terminal to the SF3b1 α-helical repeat domain, further interactions among SF3b1, the RES complex and Prp8 following spliceosome rearrangements to the BACT stage [21]. Transient, early-stage splicing factors such as U2AF and its paralogues are thought to bind “U2AF Ligand Motifs” (ULM) of SF3b1 immediately adjacent its SF3b6-binding site [25], expanding a role for SF3b1 as a transient hub for multiple splicing factor interactions.
RNA helicases mediate an SF3b-dependent checkpoint for spliceosome activation, including DDX46/Prp5 for SF3b integration in the A-complex [7], snRNP200/Brr2 for dissociation of the U4 snRNP and snRNP-specific proteins at the B- to-BACT transition [26, 27], and DHX16/Prp2 for release SF3b at the time of the first transesterification reaction in the B* complex [8-10]. Although DDX46/Prp5 has yet to be resolved in spliceosome structures, in vitro interaction and yeast genetic experiments indicate that this helicase binds SF3b1/Hsh155 and thereby displaces a Tat-SF1/Cus2-mediated checkpoint for spliceosome assembly [28-30]. In the BACT complex, snRNP200/Brr2 packs against the SF3b3 subunit near SF3b1, and there bridges an additional interface between the SF3b particle and the activated Prp8 conformation [15, 21]. DHX16/Prp2 also becomes evident in the BACT complex, in which it binds adjacent to cancer-affected hotspots on the convex face of SF3b1. The DDX46/Prp5 and DHX16/Prp2 helicases have been proposed to modulate the curvature of the SF3b1 superhelix and hence compatibility with the BPS – U2 snRNA duplex [20, 28, 29, 31], thereby regulating SF3b association and release during spliceosome assembly.
2. SF3b1 conformational changes following spliceosome integration
2.1. Overall SF3b1 conformation in the isolated particle compared to the spliceosome
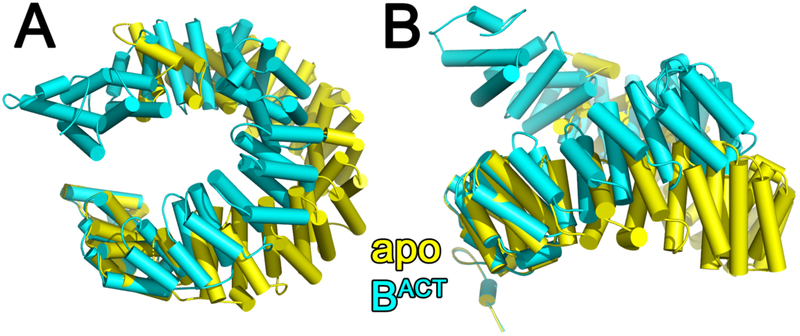
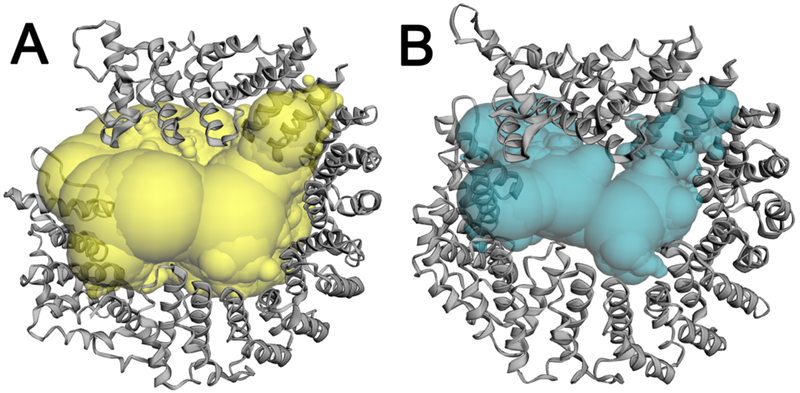
In the isolated SF3b particle, SF3b1 is in an “open” conformation with the N- and C-terminal α-helical repeats separated at a distance that is incompatible with simultaneous binding to both U2 snRNA and BPS strands [14]. In the spliceosome, “overwinding” by ~23° closes the SF3b1 superhelix to surround the BPS – U2 snRNA duplex (e.g. Fig. 4, Supplementary Movie S1, RMSD is 6.0Å among 735 matching Cα atoms of PDB ID’s 5IFE and 6FF4). Following closure in the spliceosome, the internal cavity within the SF3b1 torus decreases significantly (by nearly 25,000 Å3 calculated using CASTp [32], Fig. 5). This large conformational change is similar regardless of the homolog (Saccharomyces cerevisiae compared to human) or the stage of spliceosome assembly (RMSD’s in Table 1), with the caveat that locally lower resolutions of SF3b1-containing regions could contribute to the apparent similarity for a subset of structures. The conformational changes also appear to be independent of the structure determination method, since X-ray crystallographic and cryoEM structures of the SF3b particle are nearly identical (PDB ID 6EN4 vs. 5ZYA, Table 1).
Figure 4.
(A) Comparison of human SF3b1 conformation in the isolated SF3b particle (yellow, PDB ID 5IFE) with the BACT spliceosome (cyan, PBD ID: 6FF4, a representative structure of the human BACT at relatively high resolution, Table 1) following alignment of the C-terminal repeat (residues 1244-1285). (B) View rotated 90° about the x-axis relative to (A). Results are similar following comparison of SF3b1 of the SF3b particle with other spliceosome structures. A movie portraying the conformational transition is given as supplementary information (Supplementary Movie S1).
Figure 5.
Internal pocket of the SF3b1 torus in the (A) SF3b particle (yellow, PDB ID 5IFE) or (B) BACT spliceosome (cyan, PDB ID 6FF4) calculated using the Castp server (http://sts.bioe.uic.edu/castp/) with a 1.8 Å probe radius.
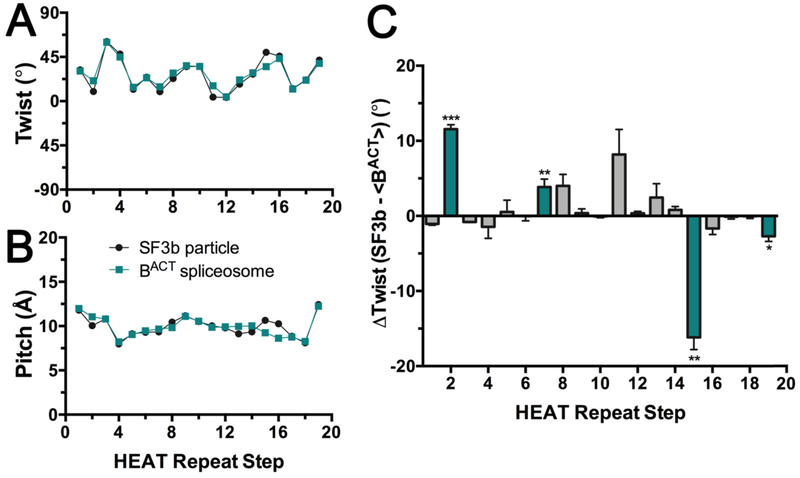
2.2. Local changes in twist and pitch are focused in a subset of consecutive SF3b1 repeats
Two mechanisms of action could account for the different SF3b1 curvature in the context of the spliceosome compared to the SF3b particle: Small, evenly-distributed changes among the HEAT repeats, or larger, locally-concentrated changes. To investigate these possibilities, we analyzed the twist and pitch relating consecutive SF3b1 repeats (Fig. 6, Fig. S1). The Lightweight Object-Oriented Structure (LOOS) library [33] was used to calculate the angle between the first principal axis and distance between the centroids of consecutive α-helical repeats (representative scripts in Supplementary Material). We maintained a consistent algorithm across a disordered SF3b1 segment in the SF3b particle (residues 1093 −1106) by focusing on the C-terminal α-helices of each HEAT repeat (Table S1), which produced similar results as analyzing dihelical units (Fig. S1). Results also were similar following comparison of the human BACT with the corresponding structure of the yeast homolog [15] (Fig. S1).
Figure 6.
Relationship between consecutive HEAT repeats of the SF3b1 superhelix in the context of the isolated particle (black, PDB ID 5IFE) or spliceosome (teal, PDB ID 6FF4). The plots analyze the intramolecular step from one HEAT repeat to the next in the indicated superhelix. The steps are numbered on the x-axis for clarity: Step 1 corresponds to the relationship of HEAT repeat 1 (HR1) to HR2, Step 2 corresponds to the relationship of HR2 to HR3, and so forth. (A) The rotational angle (twist) between the principal axes of α-helices from consecutive HEAT repeats. (B) Bar graph of the differences in the rotational angle of consecutive HEAT repeats between SF3b1 in the two contexts. The average values and standard deviation for the two relatively high resolution human BACT structures (PDB ID’s 6FF4 and 5Z58) compared to the SF3b particle are given. Unpaired, two-tailed t-tests with Welch’s correction analyze the rotational difference of each repeat relative to the average difference over all repeats: ***, p <0.0005; **, p <0.005; *, p <0.05; not significant, p >0.05. Repeats with significant differences are colored teal. (C) Translational shift (pitch) between centroids of α-helices from consecutive HEAT repeats. No significant differences in pitch of the superhelix are observed among structures. Results from comparison of the C-terminal α-helix of each di-helical HEAT repeat are shown (residue ranges in Table S1), which avoids a disordered region of SF3b1 in the isolated particle (residues 1093 – 1106). Similar results are obtained from comparison of the intact dihelical units as well as human or yeast homologues (Fig. S1). Representative scripts are given as Supplementary Materials.
We saw a small average difference in the twist of the SF3b1 superhelix between the two states (27 ± 17° in the SF3b particle and 28 ± 14° in the BACT spliceosome) (Fig. 6A), which in principle could account for most of the structural change through cumulative addition over the 20 HEAT repeats. However, our analysis instead shows that the different curvature of the two structures arises from significant local changes at a subset of repeats (Fig. 6B). Overwinding by ~10° each is observed at HR2/HR3 and HR11/HR12, which respectively interact with the pre-mRNA intron immediately downstream of the BPS, and the endonuclease/RNaseH-like domains of the core spliceosome protein Prp8. The central HR7/HR8 and HR8/HR9 steps, which contact the RBMX2 and Bud13 subunits of the retention and splicing (RES) complex, also show spliceosome-induced overwinding of ~5° each. Remarkably, the insertion of the branch point adenosine in the SF3b1 binding pocket (HR15/HR16) induces unwinding of the superhelix by −15°, in the opposite direction as other repeats in a manner reminiscent of DNA duplex unwinding by aromatic intercalators [34]. Indeed, the disordered region is near the branch point binding pocket (N-terminal α-helix of HR16, residues 1093 −1106) and folds on binding to the BPS – U2 snRNA duplex. Altogether, the overwinding and unwinding of key SF3b1 repeats sums to ~20°, consistent with the observed overall ~23° change in SF3b1 curvature following integration in the spliceosome.
2.3. Clamp-like action of the SF3b1 superhelix
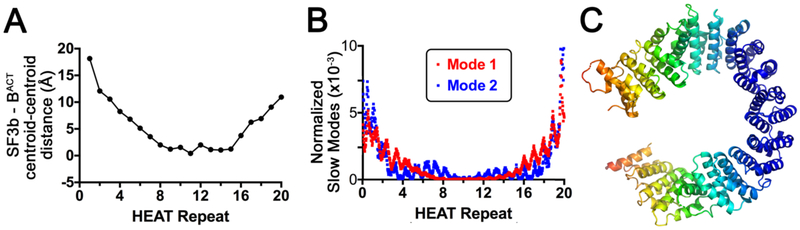
Comparison of the structures reveal that the SF3b1 conformational changes mimic a “clamp” surrounding the BPS – U2 snRNA duplex (Fig. 7A). The intermolecular distances between the centroids of the dihelix repeats between the aligned structures of human SF3b1 in the SF3b particle (PDB ID: 5IFE) compared to BACT spliceosome (PDB ID: 6FF4) show a hyperbolic increase at the termini. We explored the dynamics of the SF3b1 protein using an anisotropic network model [35] (Fig. 7B - C, Supplementary Movie S2 - S3). The major normal modes close and twist the SF3b1 superhelix analogous to the motion observed for binding to the BPS – U2 snRNA duplex and integration in the spliceosome. This natural flexibility of the SF3b1 protein raises the possibility that the RNA-compatible conformation is selected by the spliceosome from a pre-existing ensemble for the SF3b particle alone, thereby adding to an emerging theme of conformational selection among regulatory proteins [36]. Regardless, the network analysis supports the suitability of the SF3b1 structure to adopt dynamic conformations during spliceosome assembly and disassembly.
Figure 7.
(A) The distances between the centroids of the HEAT repeats following alignment of the human SF3b1 structures in the SF3b particle (PDB ID: 5IFE) compared to BACT spliceosome (PDB ID: 6FF4). (B-C), Anisotropic network model analysis (ANM 2.0, http://anm.csb.pitt.edu/) shows natural modes of SF3b1 motion similar to its conformational changes for spliceosome integration. The core HEAT domain (residues 463-1274) is analyzed. (B), Plot of the major normalized modes (fluctuations, a measure of flexibility) per SF3b1 residue. The directions of mode 1 (red) and mode 2 (blue) respectively close and twist the SF3b1 torus. (C), Heat map of normalized fluctuations in mode 1, in a color gradient from blue for no movement to orange for maximum movement. Mode 2 appears similar, although the direction of movement differs.
3. Cancer-associated SF3b1 hotspots clustered within the α-helical torus suggest hypotheses for altered splice site choice
Cancer-associated hotspots for acquired SF3b1 mutations cluster in consecutive repeats near the N-terminus of the HEAT domain (Fig. 2A, Fig. 3B). A K700E substitution accounts for the majority of SF3B1 mutations among hematologic malignancies (e.g. [37-40]), including acute myeloid leukemias, chronic myelomonocytic leukemias, chronic lymphocytic leukemias, myelodysplastic syndromes, and refractory anemias with ring sideroblasts, and also recurs in certain pancreatic and breast cancers (e.g. [41-44]). Among solid tumors, SF3B1 mutations are most common in uveal melanomas (occurring in 15-20% of patient samples tested) [45-47], and the amino acid changes are dominated by R625H/C or K666 substitutions rather than K700E. The SF3b1 K700, R625, or K666 substitutions promote use of cryptic 3′ splice sites separated by shorter branch point-spliced junction spacings compared to the normal counterparts [48, 49].
Considering the involvement of the affected SF3b1 residues in intramolecular hydrogen bonds, it was originally proposed that cancer-associated mutations alter splice site choice by changing the specific curvature of the superhelix [14], which in turn would influence interactions with spliceosome subunits or RNAs and lead to cryptic splice site choice. Subsequently, several spliceosome structures (human PDB ID’s: 6QX9, 6AH0, 6AHD, 5Z56, 5Z57, 5Z58; yeast PDB ID’s: 5ZWM, 5ZWO, 5LQW, 5GM6) reported interactions between the polypyrimidine splice site signal and the cancer-associated SF3b1 hotspots. Based on these structures, we suggested a possibly interleaved explanation of electrostatic repulsion between the phosphodiester backbone and the prevalent K700E substitution, which could destabilize pre-mRNA contacts, thereby shortening the preferred BPS – splice site spacing of mutant SF3b1 [2]. Yet, network analyses of K700, R625, or K666-mutated SF3b1 models (available on request), and binding of K700E SF3b1 to a strong splice site [14], lack detectable differences from wild-type, suggesting that additional factors are at play in the biological setting.
4. Spliceosome inhibitors target the branchsite-binding pocket of SF3b1
4.1. SF3b1 is a major target for spliceosome inhibition
Cancer-associated dysregulation highlights the spliceosome as an “Achilles heel” for selective targeting by anti-cancer therapeutics. Nature has taken advantage of this eukaryotic vulnerability and produced small molecule inhibitors of the pre-mRNA splicing pathway (reviewed in [3, 4]). Remarkably, SF3b1 is the target of the major known class of naturally-occurring spliceosome inhibitors comprising a central conjugated diene separating two variable moieties, including spliceostatin-A precursor from Pseudomonas (sp. 2663) [50], pladienolides from Streptomyces species [51, 52], and isoginkgetin isolated from the Metasequoia glyptostroboides dawn redwood tree [53]. Although high-throughput screens have identified other inhibitors of distinct aspects of the pre-mRNA splicing process (reviewed in [54]), the SF3b1-inhibitors remain the most clinically advanced to date. Phase I clinical trials first initiated in 2007 for treatment of solid tumors with a pladienolide-D derivative, E7107 (ClinicalTrials.gov ID: ) were suspended due to vision impairment of two participants [55]. More recently, an iteratively optimized derivative starting from a pladienolide scaffold [56] entered Phase I clinical trials for treatment of hematologic malignancies (ClinicalTrials.gov ID: ). Knowledge of SF3b1 structures and future characterizations of cancer-associated SF3b1 mutants are likely to aid future optimizations of these promising compounds.
4.2. Inhibitors bind the SF3b1 conformation corresponding to the isolated SF3b particle
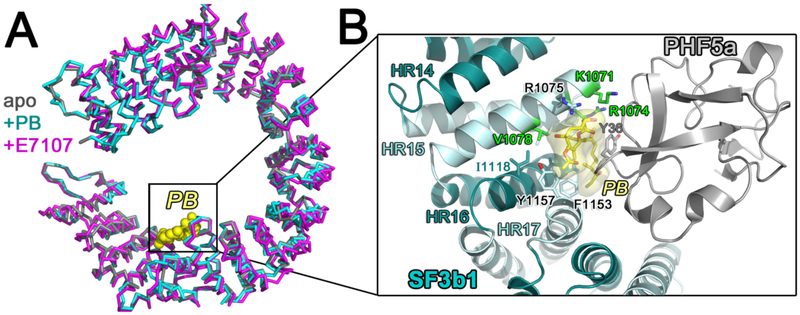
Currently, two structures are available of inhibitor-bound SF3b1 in the context of the core human SF3b particle: a co-crystal structure bound to pladienolide-B [57] and a cryoEM structure bound to E7107 [58]. The compounds bind the SF3b1 subunit without significant conformational rearrangement compared to the apo-SF3b particle (respective RMSD 0.2, 1.4, or 1.4 Å between 709, 787, or 795 matching SF3b1 Ca atoms between pairwise alignments of PDB ID’s: 5IFE (X-ray, apo) – 6EN4 (X-ray, pladienolide B-bound; 5IFE – 5ZYA, cryoEM, E7107-bound; 6EN4 – 5ZYA, Fig. 8A). The compound binding site is available only in the “open” conformation of the isolated SF3b particle. The relatively rigid diene group inserts as a molecular wedge between adjacent α-helices of SF3b1 near the BPS-binding site and is enclosed by the PHF5a subunit. Thereby, the bound compound stabilizes an open SF3b1 conformation, spreading the α-helical repeats at a distance that is incompatible with contacting the RNA duplex. An “hourglass” shape of the inhibitor-binding pocket further explains the ability of this site to accommodate various SF3b1 modulators. The rigid diene is bound within a constricted tunnel between PHF5a and residues near the BPS-binding site of the SF3b1 structure, whereas surrounding openings accommodate the flanking macrocyclic group and aliphatic chain. The locations of residues that confer resistance or sensitivity to pladienolide-based inhibitors [58-60], and the structure-activity relationships among different compounds [58, 61, 62], are structurally consistent. Notably, SF3b1-R1074 and PHF5a-Y36, for which R1074H and Y36C mutations confer resistance to pladienolide derivatives [58, 59], engage in respective cation-π and π-π interactions with the conjugated diene group. Several residues involved in pladienolide-resistance interact with the branchsite adenosine also form the pladienolide-binding pocket. Altogether, this SF3b-targeted family of spliceosome inhibitors appears to interfere with SF3b1 annealing of the BPS – U2 snRNA strands through a combination of mutually-exclusive interactions and stabilization of the “open” SF3b1 conformation.
Figure 8.
(A) Comparison of the SF3b1 conformation in the SF3b particle (gray, PDB ID 5IFE) compared to SF3b bound to inhibitors pladienolide B (PB) (cyan, PDB ID 6EN4) or E7107 (magenta, PDB ID 5ZYA). PB is shown as yellow spheres for reference. (B) Close view of SF3b1 interactions with PB (yellow surface) (PDB ID 6EN4). SF3b1 residues for which mutations confer pladienolide-resistance are green. PHF5a is shown in gray. The interactions of SF3b1 with E7107 (PDB ID 5ZYA) are nearly identical with the exception of an additional cycloheptyl-piperazine moiety.
5. Structural implications for spliceosome assembly and therapeutic targeting
Conformational changes in SF3b1 have been proposed to regulate spliceosome assembly and contribute to its cancer-associated defects. An emerging plethora of SF3b1-containing structures open the opportunity to rigorously analyze SF3b1 conformational changes for integration in the spliceosome. The SF3b1 superhelix undergoes significant closure following transition from the isolated SF3b particle to the spliceosome context, enabling the α-helical repeats to surround the BPS – U2 snRNA duplex and shield the branchsite adenosine (Fig. 4 - 5, Supplementary Movie S1). We find that the dramatic clamp-like action of the SF3b1 torus in the spliceosome arises from large local differences at key regulatory sites. The binding site of the branch point adenosine is locally unwound (opened) analogous to the structural effect of DNA intercalation [34] (Fig. 6), which rules out prior proposals that the a branch site acts as a wedge driving apart the terminal SF3b1 repeats [14, 57]. Instead, local overwinding at SF3b1 interfaces with the downstream intron and protein subunits more than compensates for the local unwinding at the branch site and accounts for the overall SF3b1 domain closure in the spliceosome. Consistent with the typical malleability of the HEAT repeat fold [63], network analysis reveals the inherent flexibility of the SF3b1 superhelix and raise the possibility of conformational selection [36] of the spliceosome-associated, “closed” SF3b1 conformation (Fig. 7).
Remarkably, the human SF3b1 and yeast Hsh155 structures appear similar among the different spliceosome stages resolved to date. This result is consistent with a major function of SF3b1, stabilization of the BPS – U2 snRNA duplex until release of this RNA duplex for catalytic activation. As such, the similarity of the SF3b1 superhelix conformations among the intermediate pre-B, B, and BACT spliceosome stages are consistent with an ongoing requirement for SF3b1 to chaperone the BPS – U2 snRNA duplex until the time of catalytic activation. The underlying drivers of the conformational changes during association and release of SF3b1 remains an outstanding topic of investigation. Network analysis suggests that SF3b1 has inherent flexibility (Fig. 7, Supplemental Movies S2 - S3) and the recombinant SF3b core particle is capable of binding pre-mRNA in vitro [14]. Yet, RNA helicases regulate the entry and exit of SF3b1 from the spliceosome. Several groups have correlated SF3b1/Hsh155 entry in the assembling spliceosome with the RNA helicase Prp5 [28, 29, 31], although the position of Prp5 has yet to be resolved among spliceosome structures. In later stages, Prp2 has been proposed to release SF3b1 from the B* spliceosome by inducing a change in SF3b1 architecture to a state that is incompatible with the surrounding spliceosomal proteins and RNAs [16, 20, 28, 31]. Accordingly, Prp2 can be visualized bound to the exterior of the SF3b1 torus in most BACT structures (PDB ID’s: 5Z58, 5LQW, 5GM6). However, whether the Prp2 (and Prp5) RNA helicases would directly modulate the SF3b1 protein conformation, as opposed to indirectly influencing access to its RNA binding site, awaits future investigations.
The conformational transitions of the SF3b1 superhelix are relevant to understanding the consequences of cancer-associated mutations for therapeutic targeting. One hypothesis is that cancer-associated mutations modify the choice of branch site by favoring an “open” SF3b1 conformation, which in turn influences activation by Prp5 or Prp2 [28, 31]. We have suggested an alternative hypothesis that the mutations disrupt local RNA interactions, thereby shortening the preferred BPS-to-3′ splice site spacing [2]. Although these possibilities remain to be experimentally tested, they are likely co-exist and even synergize. For example, an impaired RNA interface with the mutated SF3b1 site would circumvent the ATP-dependent Prp2 checkpoint for certain “weak” splice sites, consistent with the conditional impact of cancer-associated SF3B1 mutations on a subset of spliced junctions [48, 49]. The dynamic SF3b1 conformations provide an important target for drug discovery. Already, the clinically-developed class of pladienolide derivatives exploit the SF3b1 superhelix, rigidifying the “open” SF3b1 conformation and thereby interfering with stable BPS – U2 snRNA association [57, 58]. Future investigations of the structural consequences of SF3b1-targeted mutations and therapies, including contributions of induced fit versus conformational selection, and the detailed interplay of SF3b1 conformations with RNA helicases, will enhance a growing framework for understanding and targeting SF3b1 functions and cancer-associated dysregulation in pre-mRNA splicing.
Supplementary Material
Highlights:
An α-helical SF3b1 superhelix closes following integration in pre-B, B, or BACT spliceosomes.
The SF3b1 superhelix is locally unwound by insertion of the branch site adenosine.
Protein cofactors appear to stabilize closure of the SF3b1 torus in the spliceosome.
SF3b1 dynamics parallel its conformational change in the spliceosome.
Dynamic SF3b1 conformations are a target for drug discovery.
Acknowledgments
We apologize to those whose work was not cited in this paper due to space constraints. NIH R01 GM070503 and R01 GM117005 supported this project.
References
- [1].Dvinge H, Kim E, Abdel-Wahab O, Bradley RK, RNA splicing factors as oncoproteins and tumour suppressors, Nat Rev Cancer, 16 (2016) 413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jenkins JL, Kielkopf CL, Splicing factor mutations in myelodysplasias: Insights from spliceosome structures, Trends Genet, 33 (2017) 336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Agrawal AA, Yu L, Smith PG, Buonamici S, Targeting splicing abnormalities in cancer, Curr Opin Genet Dev, 48 (2018) 67–74. [DOI] [PubMed] [Google Scholar]
- [4].Effenberger KA, Urabe VK, Jurica MS, Modulating splicing with small molecular inhibitors of the spliceosome, Wiley Interdiscip Rev RNA, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Behrens SE, Tyc K, Kastner B, Reichelt J, Luhrmann R, Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions, Mol Cell Biol, 13 (1993) 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Will CL, Schneider C, Reed R, Luhrmann R, Identification of both shared and distinct proteins in the major and minor spliceosomes, Science, 284 (1999) 2003–2005. [DOI] [PubMed] [Google Scholar]
- [7].Ruby SW, Chang TH, Abelson J, Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA, Genes Dev, 7 (1993) 1909–1925. [DOI] [PubMed] [Google Scholar]
- [8].Warkocki Z, Odenwalder P, Schmitzova J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Luhrmann R, Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components, Nat Struct Mol Biol, 16 (2009) 1237–1243. [DOI] [PubMed] [Google Scholar]
- [9].Lardelli RM, Thompson JX, Yates JR 3rd, Stevens SW, Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing, RNA, 16 (2010) 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kim SH, Lin RJ, Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing, Mol Cell Biol, 16 (1996) 6810–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kastner B, Will CL, Stark H, Luhrmann R, Structural Insights into Nuclear pre-mRNA Splicing in Higher Eukaryotes, Cold Spring Harbor perspectives in biology, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang L, Vielle A, Espinosa S, Zhao R, RNAs in the spliceosome: Insight from cryoEM structures, Wiley Interdiscip Rev RNA, 10 (2019) e1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yan C, Wan R, Shi Y, Molecular Mechanisms of pre-mRNA Splicing through Structural Biology of the Spliceosome, Cold Spring Harbor perspectives in biology, 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cretu C, Schmitzova J, Ponce-Salvatierra A, Dybkov O, De Laurentiis EI, Sharma K, Will CL, Urlaub H, Luhrmann R, Pena V, Molecular architecture of SF3b and structural consequences of Its cancer-related mutations, Mol Cell, 64 (2016) 307–319. [DOI] [PubMed] [Google Scholar]
- [15].Yan C, Wan R, Bai R, Huang G, Shi Y, Structure of a yeast activated spliceosome at 3.5 A resolution, Science, 353 (2016) 904–911. [DOI] [PubMed] [Google Scholar]
- [16].Rauhut R, Fabrizio P, Dybkov O, Hartmuth K, Pena V, Chari A, Kumar V, Lee CT, Urlaub H, Kastner B, Stark H, Luhrmann R, Molecular architecture of the Saccharomyces cerevisiae activated spliceosome, Science, 353 (2016) 1399–1405. [DOI] [PubMed] [Google Scholar]
- [17].Lin Y, Kielkopf CL, X-ray structures of U2 snRNA-branchpoint duplexes containing conserved pseudouridines. , Biochemistry, 47 (2008) 5503–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kennedy SD, Bauer WJ, Wang W, Kielkopf CL, Dynamic stacking of an expected branch point adenosine in duplexes containing pseudouridine-modified or unmodified U2 snRNA sites, Biochem Biophys Res Commun, 511 (2019) 416–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Newby MI, Greenbaum NL, Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine, Nat Struct Biol, 9 (2002) 958–965. [DOI] [PubMed] [Google Scholar]
- [20].Carrocci TJ, Paulson JC, Hoskins AA, Functional analysis of Hsh155/SF3b1 interactions with the U2 snRNA/branch site duplex, RNA, 24 (2018) 1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang X, Yan C, Zhan X, Li L, Lei J, Shi Y, Structure of the human activated spliceosome in three conformational states, Cell Res, 28 (2018) 307–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Haselbach D, Komarov I, Agafonov DE, Hartmuth K, Graf B, Dybkov O, Urlaub H, Kastner B, Luhrmann R, Stark H, Structure and Conformational Dynamics of the Human Spliceosomal B(act) Complex, Cell, 172 (2018) 454–464 e411. [DOI] [PubMed] [Google Scholar]
- [23].Charenton C, Wilkinson ME, Nagai K, Mechanism of 5' splice site transfer for human spliceosome activation, Science, 364 (2019) 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhan X, Yan C, Zhang X, Lei J, Shi Y, Structures of the human pre-catalytic spliceosome and its precursor spliceosome, Cell Res, 28 (2018) 1129–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Loerch S, Kielkopf CL, Unmasking the U2AF homology motif family: a bona fide protein-protein interaction motif in disguise, RNA, 22 (2016) 1795–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Raghunathan PL, Guthrie C, RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2, Curr Biol, 8 (1998) 847–855. [DOI] [PubMed] [Google Scholar]
- [27].Laggerbauer B, Achsel T, Luhrmann R, The human U5–200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro, Proc Natl Acad Sci U S A, 95 (1998) 4188–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tang Q, Rodriguez-Santiago S, Wang J, Pu J, Yuste A, Gupta V, Moldon A, Xu YZ, Query CC, SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing, Genes Dev, 30 (2016) 2710–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Talkish J, Igel H, Hunter O, Horner SW, Jeffery NN, Leach JR, Jenkins JL, Kielkopf CL, Ares M Jr., Cus2 enforces the first ATP-dependent step of splicing by binding to yeast SF3b1 through a UHM-ULM interaction, RNA, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Perriman R, Barta I, Voeltz GK, Abelson J, Ares M Jr., ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA, Proc Natl Acad Sci U S A, 100 (2003) 13857–13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Carrocci TJ, Zoerner DM, Paulson JC, Hoskins AA, SF3b1 mutations associated with myelodysplastic syndromes alter the fidelity of branchsite selection in yeast, Nucleic Acids Res, 45 (2017) 4837–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tian W, Chen C, Lei X, Zhao J, Liang J, CASTp 3.0: computed atlas of surface topography of proteins, Nucleic Acids Res, 46 (2018) W363–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Romo TD, Leioatts N, Grossfield A, Lightweight object oriented structure analysis: tools for building tools to analyze molecular dynamics simulations, J Comput Chem, 35 (2014) 2305–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Werner MH, Gronenborn AM, Clore GM, Intercalation, DNA kinking, and the control of transcription, Science, 271 (1996) 778–784. [DOI] [PubMed] [Google Scholar]
- [35].Eyal E, Lum G, Bahar I, The anisotropic network model web server at 2015 (ANM 2.0), Bioinformatics, 31 (2015) 1487–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Boehr DD, Nussinov R, Wright PE, The role of dynamic conformational ensembles in biomolecular recognition, Nature Chem Biol, 5 (2009) 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Ishiyama K, Mori H, Nolte F, Hofmann WK, Miyawaki S, Sugano S, Haferlach C, Koeffler HP, Shih LY, Haferlach T, Chiba S, Nakauchi H, Miyano S, Ogawa S, Frequent pathway mutations of splicing machinery in myelodysplasia, Nature, 478 (2011) 64–69. [DOI] [PubMed] [Google Scholar]
- [38].Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, Godfrey AL, Rapado I, Cvejic A, Rance R, McGee C, Ellis P, Mudie LJ, Stephens PJ, McLaren S, Massie CE, Tarpey PS, Varela I, Nik-Zainal S, Davies HR, Shlien A, Jones D, Raine K, Hinton J, Butler AP, Teague JW, Baxter EJ, Score J, Galli A, Della Porta MG, Travaglino E, Groves M, Tauro S, Munshi NC, Anderson KC, El-Naggar A, Fischer A, Mustonen V, Warren AJ, Cross NC, Green AR, Futreal PA, Stratton MR, Campbell PJ, C. Chronic Myeloid Disorders Working Group of the International Cancer Genome, Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts, N Engl J Med, 365 (2011) 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, Zhang W, Vartanov AR, Fernandes SM, Goldstein NR, Folco EG, Cibulskis K, Tesar B, Sievers QL, Shefler E, Gabriel S, Hacohen N, Reed R, Meyerson M, Golub TR, Lander ES, Neuberg D, Brown JR, Getz G, Wu CJ, SF3B1 and other novel cancer genes in chronic lymphocytic leukemia, N Engl J Med, 365 (2011) 2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, Ramsay AJ, Bea S, Pinyol M, Martinez-Trillos A, Lopez-Guerra M, Colomer D, Navarro A, Baumann T, Aymerich M, Rozman M, Delgado J, Gine E, Hernandez JM, Gonzalez-Diaz M, Puente DA, Velasco G, Freije JM, Tubio JM, Royo R, Gelpi JL, Orozco M, Pisano DG, Zamora J, Vazquez M, Valencia A, Himmelbauer H, Bayes M, Heath S, Gut M, Gut I, Estivill X, Lopez-Guillermo A, Puente XS, Campo E, Lopez-Otin C, Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia, Nat Genet, 44 (2012) 47–52. [DOI] [PubMed] [Google Scholar]
- [41].Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, Ng S, Lin L, Crowder R, Snider J, Ballman K, Weber J, Chen K, Koboldt DC, Kandoth C, Schierding WS, McMichael JF, Miller CA, Lu C, Harris CC, McLellan MD, Wendl MC, DeSchryver K, Allred DC, Esserman L, Unzeitig G, Margenthaler J, Babiera GV, Marcom PK, Guenther JM, Leitch M, Hunt K, Olson J, Tao Y, Maher CA, Fulton LL, Fulton RS, Harrison M, Oberkfell B, Du F, Demeter R, Vickery TL, Elhammali A, Piwnica-Worms H, McDonald S, Watson M, Dooling DJ, Ota D, Chang LW, Bose R, Ley TJ, Piwnica-Worms D, Stuart JM, Wilson RK, Mardis ER, Whole-genome analysis informs breast cancer response to aromatase inhibition, Nature, 486 (2012) 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerod A, Oslo Breast Cancer C, Lee MT, Shen CY, Tee BT, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin SF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van 't Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SA, Salomon AV, Borresen-Dale AL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR, The landscape of cancer genes and mutational processes in breast cancer, Nature, 486 (2012) 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, Chang DK, Cowley MJ, Gardiner BB, Song S, Harliwong I, Idrisoglu S, Nourse C, Nourbakhsh E, Manning S, Wani S, Gongora M, Pajic M, Scarlett CJ, Gill AJ, Pinho AV, Rooman I, Anderson M, Holmes O, Leonard C, Taylor D, Wood S, Xu Q, Nones K, Fink JL, Christ A, Bruxner T, Cloonan N, Kolle G, Newell F, Pinese M, Mead RS, Humphris JL, Kaplan W, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chou A, Chin VT, Chantrill LA, Mawson A, Samra JS, Kench JG, Lovell JA, Daly RJ, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, I. Australian Pancreatic Cancer Genome, Kakkar N, Zhao F, Wu YQ, Wang M, Muzny DM, Fisher WE, Brunicardi FC, Hodges SE, Reid JG, Drummond J, Chang K, Han Y, Lewis LR, Dinh H, Buhay CJ, Beck T, Timms L, Sam M, Begley K, Brown A, Pai D, Panchal A, Buchner N, De Borja R, Denroche RE, Yung CK, Serra S, Onetto N, Mukhopadhyay D, Tsao MS, Shaw PA, Petersen GM, Gallinger S, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Schulick RD, Wolfgang CL, Morgan RA, Lawlor RT, Capelli P, Corbo V, Scardoni M, Tortora G, Tempero MA, Mann KM, Jenkins NA, Perez-Mancera PA, Adams DJ, Largaespada DA, Wessels LF, Rust AG, Stein LD, Tuveson DA, Copeland NG, Musgrove EA, Scarpa A, Eshleman JR, Hudson TJ, Sutherland RL, Wheeler DA, Pearson JV, McPherson JD, Gibbs RA, Grimmond SM, Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes, Nature, 491 (2012) 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grutzmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM, I. Australian Pancreatic Cancer Genome, Munzy DM, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey UM, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SM, Genomic analyses identify molecular subtypes of pancreatic cancer, Nature, 531 (2016) 47–52. [DOI] [PubMed] [Google Scholar]
- [45].Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM, Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma, Nat Genet, 45 (2013) 133–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Martin M, Masshofer L, Temming P, Rahmann S, Metz C, Bornfeld N, van de Nes J, Klein-Hitpass L, Hinnebusch AG, Horsthemke B, Lohmann DR, Zeschnigk M, Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3, Nat Genet, 45 (2013) 933–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, Turajlic S, Piperno-Neumann S, de la Grange P, Roman-Roman S, Stern MH, Marais R, SF3B1 mutations are associated with alternative splicing in uveal melanoma, Cancer Discov, 3 (2013) 1122–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Alsafadi S, Houy A, Battistella A, Popova T, Wassef M, Henry E, Tirode F, Constantinou A, Piperno-Neumann S, Roman-Roman S, Dutertre M, Stern MH, Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage, Nat Commun, 7 (2016) 10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, Bailey SL, Bhavsar EB, Chan B, Colla S, Corson L, Feala J, Fekkes P, Ichikawa K, Keaney GF, Lee L, Kumar P, Kunii K, MacKenzie C, Matijevic M, Mizui Y, Myint K, Park ES, Puyang X, Selvaraj A, Thomas MP, Tsai J, Wang JY, Warmuth M, Yang H, Zhu P, Garcia-Manero G, Furman RR, Yu L, Smith PG, Buonamici S, Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3' Splice Site Selection through Use of a Different Branch Point, Cell Rep, 13 (2015) 1033–1045. [DOI] [PubMed] [Google Scholar]
- [50].Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, Tani T, Horinouchi S, Yoshida M, Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA, Nat Chem Biol, 3 (2007) 576–583. [DOI] [PubMed] [Google Scholar]
- [51].Arai K, Buonamici S, Chan B, Corson L, Endo A, Gerard B, Hao MH, Karr C, Kira K, Lee L, Liu X, Lowe JT, Luo T, Marcaurelle LA, Mizui Y, Nevalainen M, O'Shea MW, Park ES, Perino SA, Prajapati S, Shan M, Smith PG, Tivitmahaisoon P, Wang JY, Warmuth M, Wu KM, Yu L, Zhang H, Zheng GZ, Keaney GF, Total synthesis of 6-deoxypladienolide D and Assessment of Splicing Inhibitory Activity in a Mutant SF3B1 cancer cell line, Org Lett, 16 (2014) 5560–5563. [DOI] [PubMed] [Google Scholar]
- [52].Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y, Splicing factor SF3b as a target of the antitumor natural product pladienolide, Nat Chem Biol, 3 (2007) 570–575. [DOI] [PubMed] [Google Scholar]
- [53].O'Brien K, Matlin AJ, Lowell AM, Moore MJ, The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing, J Biol Chem, 283 (2008) 33147–33154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Salton M, Misteli T, Small Molecule Modulators of Pre-mRNA Splicing in Cancer Therapy, Trends Mol Med, 22 (2016) 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hong DS, Kurzrock R, Naing A, Wheler JJ, Falchook GS, Schiffman JS, Faulkner N, Pilat MJ, O'Brien J, LoRusso P, A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors, Invest New Drugs, 32 (2014) 436–444. [DOI] [PubMed] [Google Scholar]
- [56].Seiler M, Yoshimi A, Darman R, Chan B, Keaney G, Thomas M, Agrawal AA, Caleb B, Csibi A, Sean E, Fekkes P, Karr C, Klimek V, Lai G, Lee L, Kumar P, Lee SC, Liu X, Mackenzie C, Meeske C, Mizui Y, Padron E, Park E, Pazolli E, Peng S, Prajapati S, Taylor J, Teng T, Wang J, Warmuth M, Yao H, Yu L, Zhu P, Abdel-Wahab O, Smith PG, Buonamici S, H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers, Nat Med, 24 (2018) 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Cretu C, Agrawal AA, Cook A, Will CL, Fekkes P, Smith PG, Luhrmann R, Larsen N, Buonamici S, Pena V, Structural Basis of Splicing Modulation by Antitumor Macrolide Compounds, Mol Cell, 70 (2018) 265–273 e268. [DOI] [PubMed] [Google Scholar]
- [58].Finci LI, Zhang X, Huang X, Zhou Q, Tsai J, Teng T, Agrawal A, Chan B, Irwin S, Karr C, Cook A, Zhu P, Reynolds D, Smith PG, Fekkes P, Buonamici S, Larsen NA, The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action, Genes Dev, 32 (2018) 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Teng T, Tsai JH, Puyang X, Seiler M, Peng S, Prajapati S, Aird D, Buonamici S, Caleb B, Chan B, Corson L, Feala J, Fekkes P, Gerard B, Karr C, Korpal M, Liu X, Mizui Y, T.L. J., Palacino J, Park E, Smith PG, Subramanian V, Wu ZJ, Zou J, Yu L, Chicas A, Warmuth M, Larsen N, Zhu P, Splicing modulators act at the branch point adenosine binding pocket defined by the PHF5A-SF3b complex, Nat Commun, 8 (2017) 15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yokoi A, Kotake Y, Takahashi K, Kadowaki T, Matsumoto Y, Minoshima Y, Sugi NH, Sagane K, Hamaguchi M, Iwata M, Mizui Y, Biological validation that SF3b is a target of the antitumor macrolide pladienolide, FEBS J, 278 (2011) 4870–4880. [DOI] [PubMed] [Google Scholar]
- [61].Effenberger KA, Urabe VK, Prichard BE, Ghosh AK, Jurica MS, Interchangeable SF3B1 inhibitors interfere with pre-mRNA splicing at multiple stages, RNA, 22 (2016) 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Effenberger KA, Anderson DD, Bray WM, Prichard BE, Ma N, Adams MS, Ghosh AK, Jurica MS, Coherence between cellular responses and in vitro splicing inhibition for the anti-tumor drug pladienolide B and its analogs, J Biol Chem, 289 (2014) 1938–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yoshimura SH, Hirano T, HEAT repeats - versatile arrays of amphiphilic helices working in crowded environments?, J Cell Sci, 129 (2016) 3963–3970. [DOI] [PubMed] [Google Scholar]
- [64].Bertram K, Agafonov DE, Dybkov O, Haselbach D, Leelaram MN, Will CL, Urlaub H, Kastner B, Luhrmann R, Stark H, Cryo-EM Structure of a Pre-catalytic Human Spliceosome Primed for Activation, Cell, 170 (2017) 701–713 e711. [DOI] [PubMed] [Google Scholar]
- [65].Plaschka C, Lin PC, Charenton C, Nagai K, Prespliceosome structure provides insights into spliceosome assembly and regulation, Nature, 559 (2018) 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Plaschka C, Lin PC, Nagai K, Structure of a pre-catalytic spliceosome, Nature, 546 (2017) 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Bai R, Wan R, Yan C, Lei J, Shi Y, Structures of the fully assembled Saccharomyces cerevisiae spliceosome before activation, Science, (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.