Abstract
Background:
Microbial contamination of cosmetics products is of incredible significance since it will not only cause significant health hazardous but also act as a potential source of infections. Contamination will cause spoilage of the item and when pathogenic they become a genuine threat for its users.
Aim:
To evaluate the bacterial contamination in regularly used lipsticks.
Objectives:
To identify the pathogenic organism present in the lipsticks. To understand the potential complications of the organisms identified. Comparing the microbial count in used and new lipsticks.
Keywords: Lipsticks, Mueller-Hinton agar, Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus saprophyticus
INTRODUCTION
The word “cosmetic” is derived from the antiquated Greek word “kosmein,” which implies adornment.[1] In modern era, the use of cosmetics is as important as the fundamental needs of life. Cosmetics have been characterized as any substance or preparation intended to be placed in contact with the various external parts of the human body or with the teeth and mucous membrane of oral cavity with a view exclusively or mainly to cleaning them, perfuming them, changing their appearance/correcting body odors or protecting them or keeping them in good conditions.[2]
Cosmetics need not be sterile due to the ubiquitous nature of microorganisms; hence, the cosmetics are incorporated with various preservatives to keep it sterile and also to avoid economic loss as well.[3] As the vast majority of the beautifying agents were utilized in delicate regions, the additives included ought to be nondangerous and ought not make any damage to the consumers.[1]
Unfortunately, some of the contents in preservatives may support the growth of organisms particularly water and supplements such as cocoa margarine, olive oil, and so on.[4,5] These contaminants will begin increasing and cause injurious impact to both the functional and utilitarian qualities of cosmetic products.[4]
Microorganisms which are present in the cosmetic products can lead to its deterioration as these organisms act by altering the physical and chemical properties of these cosmetics which inturn can be harmful for their users. Hence, it is ought to be vital to guarantee that the additive utilized has the ability to prevent the growth of microorganisms.[6]
Lipstick was being a standout among the most widely recognized cosmetic item regularly utilized by women.[7] It was seen as a wellspring of contamination because of the increased development of smaller scale microbial life forms.[8] Even though lipsticks contain preservatives, it harbors microorganisms, which will grow and multiply as it has moistened by breath after every usage.[9]
Different methods were used to analyze the microbial contamination in cosmetics and their raw materials based on traditional plate counts.[5]
In this context, the present study was undertaken to evaluate the microbial contamination in lipsticks which was regularly used by women.
MATERIALS AND METHODS
A total of 80 used lipsticks were incorporated in this investigation, With 10 unused ones which were kept as control.
Samples of used lipsticks were gathered randomly, and the ones which were out of expiry date were barred from the study.
A sterile cotton swab was used to wipe on surface of each lipsticks, and it was streaked onto the surfaces of three primary culture media [Figures 1 and 2]. Media used for primary isolation include nutrient agar, MacConkey agar, and blood agar. After inoculation, culture plates were incubated for 24 h at 37°C. After incubation, bacterial growth was observed in colonies in most of the culture media [Figure 3], and these bacterial colonies were transferred to nutrient agar slants to obtain pure culture. All pure cultures were subjected to characterization using different tests. Bacterial culturing and inoculation was done by following the standard protocol.[10]
Figure 1.

Sterile cotton swab wiped over the surface of lipstick
Figure 2.
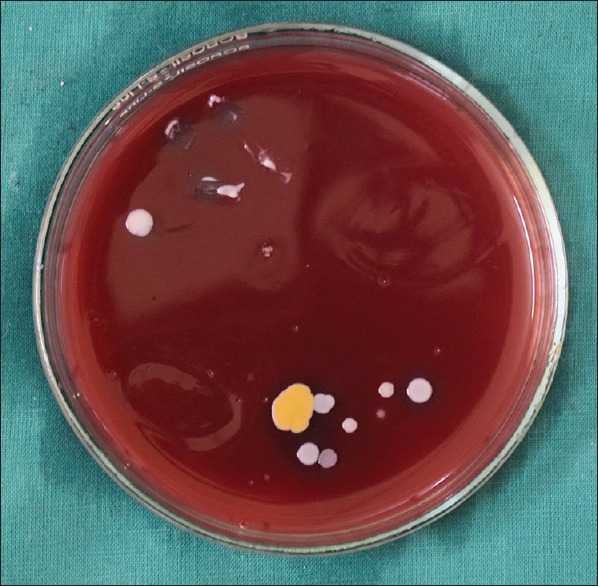
Bacterial colonies observed in MacConkey's media
Figure 3.

Bacterial colonies observed in nutrient media
Gram staining was done to differentiate between Gram-positive and Gram-negative organisms, and then, catalase test was done to identify whether the organisms were staphylococci or streptococci species, and to determine specific strains, coagulase test and antibiotic sensitivity test using novobiocin were done.
RESULTS
Eighty samples of lipsticks were selected randomly including a control group which comprised ten samples. Of these study samples, 93% yielded the growth of Gram-positive organisms [Graph 1], which was confirmed by Gram staining [Table 1].
Graph 1.

Bacterial growth observed in cultures
Table 1.
Bacterial growth observed in colonies
| n (%) | |
|---|---|
| Total number of lipsticks | 70 (100) |
| No growth | 5 (7) |
| Growth | 65 (93) |
These samples were further subjected to catalase test to differentiate between staphylococci and streptococci species [Table 2].
Table 2.
Bacterial species present in the culture
| Organisms | n (%) |
|---|---|
| Staphylococcus aureus | 21 (32) |
| Staphylococcus epidermidis | 26 (40) |
| Staphylococcus saprophyticus | 16 (25) |
| Streptococci | 2 (3) |
Only 3% of the organism showed negativity in catalase test and it was confirmed as streptococci species, and the remaining organism were of staphylococci species [Figure 4]. Coagulase test was performed and 32% showed positive result and confirmed as Staphylococcus aureus and novobiocin sensitivity test [Figure 5] confirmed 40% of the organism as Staphylococcus epidermidis and 25% as Staphylococcus saprophyticus [Graph 2].
Figure 4.

Gram-positive cocci observed in cultures (Grams stain, ×10)
Figure 5.

Antibiotic sensitivity test using novobiocin
Graph 2.
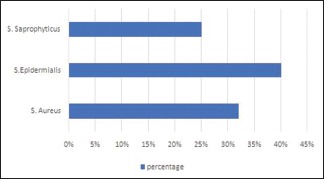
Bacterial strains being identified in cultures
DISCUSSION
Microbial contamination of cosmetics may result in its disintegration as well as economic loss for the consumers. Hence, it is important to ensure that cosmetic products are free from microorganisms, and the preservatives used in cosmetics should have the capability to eliminate these potential microbes.
The presence of different natural segments and the nearness of the vast amounts of water in cosmetics give a phenomenal domain to the development of microorganisms. In 1975 Wilson et al. concluded that microbial growth in cosmetic products was favored by atmospheric changes like temperature variation and moisture provided by humidity.[11]
The findings of Smart and Spooner in 1972 had additionally added that the cosmetic items are likewise discovered helpless to changes in physiochemical conditions because of microbial decontamination.[4]
In comparative investigations on various cosmetic products such as eyecare products, face care products, and child shampoos, it was found that Staphylococcus species and Escherichia coli were the ones that had been regularly detached.[2]
In 1989, Akin and Altanlar had carried out a study to investigate microbial quality and control of lipsticks, and they found that of 81 samples, 42% yield aerobic plate count and 23% were found to be consisting of mold and yeast.[1]
The pathogens observed in the lipsticks are opportunistic pathogens, and their effects varied among persons.
In the present study, the presence of streptococci was considered to be minimal (3%), but they are recognized as a causative factor for many infections.
Budzik and Schneewind in 2006 reported that the presence of streptococci was found to be associated with infective endocarditis.[12] According to Arciola et al., this organism was considered as a causative agent for orthopedic infections, and they published a report on it in 2007.[13] Routsi et al. in the year 2000 identified this organism as a factor for blood stream infections.[14]
Other potentially pathogenic organism identified in this study was staphylococci species. The predominant species present was S. epidermidis (40%). The presence of this organism was considered to be fatal in the oral cavity.
Michael Otto et al. in 2009 reported that the presence of S. epidermidis leads to the formation of biofilm in the prosthetic devices and implants, which may further lead to failure of these devices.[15] The exact mechanism related to the biofilm formation of this organism was not clearly understood, and further genomic studies are needed for the same.
The second predominant staphylococci strain observed in this study was S. aureus (32%). According to Pour et al. in 2007, this organism was considered as the normal flora of skin and mucous membrane. However, their high incidence has clinical significance and is considered as a well-recognized pathogen.[16] Everitt et al. published a report in 2006 which revealed the role of S. aureus in bacteremia which was complicated in patients with endocarditis and metastatic infections.[10]
Another strain of staphylococci which was observed to be minimal was S. saprophyticus (25%). The clinical significance of this organism in urinary tract infections has been reported by Elmanama et al. in 2006.[17]
Infections caused by cosmetics are significant, and it is important to identify and prevent it. From the present study, it was evident that the quality of cosmetic product may also have an important role in the process of contamination. The incorporation of poor-quality preservatives may be the reason for contamination.
Further investigations are needed to evaluate the pathogenesis of organism, to ensure the quality of the cosmetic products.
CONCLUSION
In spite of the fact that contamination of cosmetic items will be uncommon, it presents extraordinary importance for consumers as well as the industry. As lipsticks were used in the lips, the potential life forms present in this product venture into oral cavity and cause genuine well-being dangers. Hence, it is an incredible worry to consider the nature of additives utilized in the beauty care products to remain safe from the perilous impact.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Akin A, Altanlar N. Microbiological quality control of lipsticks which are on the market in our country. Mikrobiyol Bul. 1989;23:369–78. [PubMed] [Google Scholar]
- 2.Onurdag FK, Ozgen S, Abhasoglu D. Microbiological investigation of used cosmetic samples. Hacettepe Univ J Fac Pharm. 2010;30:1–16. [Google Scholar]
- 3.Saeed S, Asif K. Bacteriological analysis of lipsticks. RADS J Biol Res Appl Sci. 2011;2:21–6. [Google Scholar]
- 4.Smart R, Spooner DF. Microbiological spoilage in pharmaceuticals and cosmetics. J Soc Cosmet Chem. 1972;23:721–37. [Google Scholar]
- 5.Orús P, Leranoz S. Current trends in cosmetic microbiology. Int Microbiol. 2005;8:77–9. [PubMed] [Google Scholar]
- 6.Olson SW. The application of microbiology to cosmetic testing. J Soc Cosmet Chem. 1967;18:191–8. [Google Scholar]
- 7.Brannan DK, Dille JC. Type of closure prevents microbial contamination of cosmetics during consumer use. Appl Environ Microbiol. 1990;56:1476–9. doi: 10.1128/aem.56.5.1476-1479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ergun H, Tuncer I, Sengil AZ, Kirca NK. Microbiological analysis of some cosmetics on the market. Mikrobiyol Bul. 1987;21:301–7. [PubMed] [Google Scholar]
- 9.Parker MT. The clinical significance of the presence of microorganisms in pharmaceutical and cosmetic preparations. J Soc Cosmet Chem. 1972;23:415–26. [Google Scholar]
- 10.Everitt HA, Little PS, Smith PW. A randomised controlled trial of management strategies for acute infective conjunctivitis in general practice. BMJ. 2006;333:321. doi: 10.1136/bmj.38891.551088.7C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mateos-Rivera A, Yde JC, Wilson B, Finster KW, Reigstad LJ, Ovreas L. The effect of temperature change on the microbial diversity and community structure along the chronosequence of the sub-arctic glacier forefield of Styggedalsbreen (Norway) FEMS Microbiol Ecol. 2016;92 doi: 10.1093/femsec/fiw038. [DOI] [PubMed] [Google Scholar]
- 12.Budzik JM, Schneewind O. Pili prove pertinent to enterococcal endocarditis. J Clin Invest. 2006;116:2582–84. doi: 10.1172/JCI30088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arciola CR, Campoccia D, Baldassarri L, Pirini V, Huebner J, Montanaro L. The role of Enterococcus faecalis in orthopaedic periimplant infections demonstrated by automated ribotyping and cluster analysis. Biomaterials. 2007;28:3987–95. doi: 10.1016/j.biomaterials.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 14.Routsi C, Platsouka E, Paniara O, Dimitriadou E, Saroglou G, Roussos C, et al. Enterococcal infections in a Greek intensive care unit: A 5-y study.Scand. J Infect Dis. 2000;32:275–80. doi: 10.1080/00365540050165910. [DOI] [PubMed] [Google Scholar]
- 15.Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–28. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pour FZ, S Miri R, Ghaseni R. Farid and J Ghenaat Skin colonization with Staphylococcus aureus in patients with atopin dermatitis. Int J Dermatol. 2007;5:23–8. [Google Scholar]
- 17.Elmanama AA, Elaiwa NM, El-Ottol AE, Ahu-Elamreen FH. Antibiotic resistance of uropathogens isolated from Al-Shifa hospital in Gaza strip in 2002. J Chemother. 2006;18:298–302. doi: 10.1179/joc.2006.18.3.298. [DOI] [PubMed] [Google Scholar]


